Abstract
Arp2/3 complex, a crucial actin-filament nucleator, undergoes structural rearrangements during activation by nucleation promoting factors (NPFs). However, the conformational pathway leading to the nucleation-competent state is unclear due to lack of high-resolution structures of the activated state. Here we report a ~3.9 Å resolution cryo-EM structure of activated Schizosaccharomyces pombe Arp2/3 complex bound to the S. pombe NPF Dip1 and attached to the end of the nucleated actin-filament. The structure reveals global and local conformational changes that allow the two actin-related proteins in Arp2/3 complex to mimic a filamentous actin dimer and template nucleation. Activation occurs through a clamp twisting mechanism, in which Dip1 forces two core subunits in Arp2/3 complex to pivot around one another, shifting half of the complex into a new activated position. By showing how Dip1 stimulates activation, the structure reveals how NPFs can activate Arp2/3 complex in diverse cellular processes.
Regulated actin filament nucleation allows cells to assemble actin networks with the proper timing and localization to drive processes like endocytosis and cellular motility1,2. Arp2/3 complex is an essential actin filament nucleator that nucleates both linear and branched actin filaments and is conserved across eukaryotes1,3. The complex exists in an inactive state until bound by nucleation promoting factors (NPFs) that stimulate activating structural changes4–11. Two subunits in Arp2/3 complex, Arp2 and Arp3, are termed actin-related proteins (Arps) because of their homology to actin. Previous experiments suggest that upon activation, Arp2 and Arp3 move into or close to the side-by-side arrangement of consecutive actin subunits along the short pitch helical axis of an actin filament, hereafter called the short pitch conformation7–12. In this position, Arp2 and Arp3 are thought to mimic a filamentous actin dimer, thereby creating a template for new filament assembly. In the inactive complex, however, Arp2 and Arp3 do not mimic a filamentous actin dimer13,14. Instead, two core subunits of Arp2/3 complex, ARPC2 and ARPC4, form a clamp that holds the Arps in an end-to-end (splayed) arrangement distinct from the short pitch conformation. Furthermore, in the inactive complex, Arp2 and Arp3 adopt conformations that resemble unpolymerized actin monomers instead of filamentous actin subunits15,16. The structural differences between filamentous actin dimers and the actin-related proteins in inactive Arp2/3 complex suggest that both intra- and inter-subunit changes within the complex are required to create a nucleation-competent state. However, the lack of a high-resolution structure of activated Arp2/3 complex has prevented us from understanding how NPFs trigger these changes.
High resolution structures of activated Arp2/3 complex have been unobtainable because the activated complex is recalcitrant to crystallization and tends to adopt limited orientations in vitrified ice during cryo-EM studies. Recently it was discovered that WISH/DIP/SPIN90 (WDS) family NPFs activate Arp2/3 complex to nucleate linear actin filaments3, which are more amenable for single particle cryo-EM studies than the branched actin filaments created during activation by other NPFs. Furthermore, unlike WASP family NPFs, WDS proteins remain associated with Arp2/3 complex at the pointed end of the actin filament after nucleation17, making it possible to capture the activated, NPF bound state. Taking advantage of these biochemical properties we determined by cryo-EM the near-atomic resolution structure of activated Schizosaccharomyces pombe Arp2/3 complex bound to its nucleated actin-filament and the S. pombe WDS protein, Dip1. The structure provides a snapshot of the activated conformation of Arp2/3 complex and reveals the conformational pathway by which NPFs drive activation.
Results
Structure solution
To create short actin filaments capped with Dip1-bound Arp2/3 complex for single particle reconstruction, we mixed Dip1 with Arp2/3 complex and monomeric actin to nucleate new filaments and added capping protein to block barbed end elongation. Optimized reactions produced many short linear filaments with knobs of density corresponding to Arp2/3 complex at one end (Extended Data Fig. 1a,b). From these samples we obtained a ~3.9 Å resolution reconstruction of Dip1 bound to activated Arp2/3 complex, with the complex anchored to the pointed end of the filament it nucleated (Fig. 1a,b, Extended Data Fig. 1b-d, 2a and 3a,b,d-l, Table 1). The structure contains the first four actin subunits (n, n+1, n+2, n+3) in the nucleated actin filament. The first two actins (n, n+1) are attached to the barbed end of the Arps (Fig. 1a and 2a).
Fig. 1|. Overview of structures of inactive Arp2/3 complex and activated Arp2/3 complex bound to actin filament pointed end and Dip1.
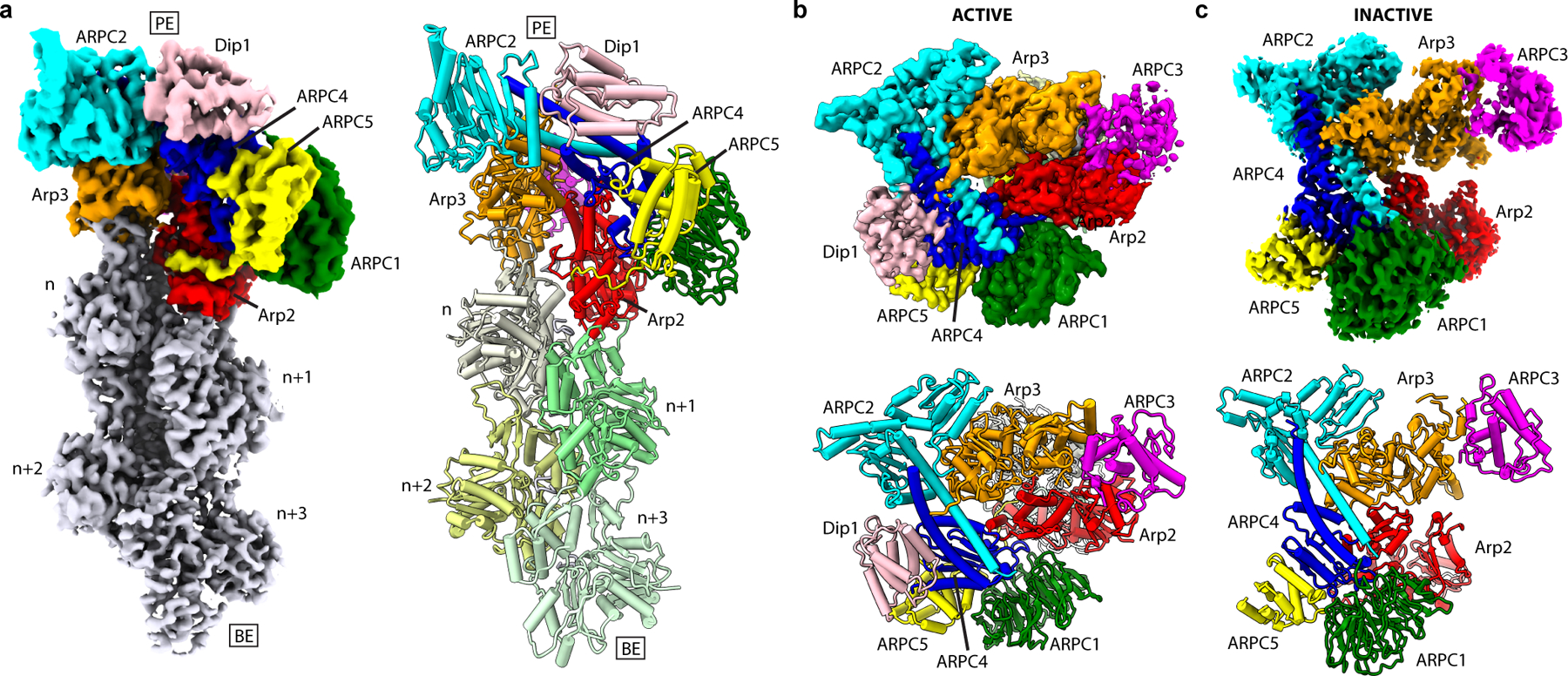
a, Reconstructed map (left) and atomic model (right) of actin filament pointed end with bound Arp2/3 complex and Dip1. PE: pointed end, BE: barbed end. b, Map (top) and atomic model (bottom) of activated Arp2/3 complex structure viewed from the pointed end looking down the helical axis of the nucleated actin filament. c, Map (top) and molecular model (bottom) of inactive Arp2/3 complex.
Table 1:
Cryo-EM data collection, refinement and validation statistics
| Dip1–Arp2/3–actin (EMD-21502, PDB 6W17) |
Inactive Arp2/3 complex (EMD-21503, PDB 6W18) | |
|---|---|---|
| Data collection and processing | ||
| Magnification | x92,000 | x120,000 |
| Voltage (kV) | 200 | 200 |
| Electron exposure (e–/Å2) | 36.35 | 44.34 |
| Defocus range (μm) | −1.00 – −1.75 | −0.70 – −1.30 |
| Pixel size (Å) | 1.12 | 0.8757 |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 3,500,000 | 3,336,981 |
| Final particle images (no.) | 110,433 | 112,170 |
| Map resolution (Å) | 3.9 | 4.2 |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range (Å) | 3.5 – 5.5 | 3.9 – 6.0 |
| Refinement | ||
| Initial models used | PDB 3DWL, PDB 3J8I | PDB 3DWL |
| Model resolution (Å) | 3.92 | 4.45 |
| FSC threshold | 0.5 | 0.5 |
| Model resolution range (Å) | 2.3 – 4.0 | 3.7 – 4.5 |
| Map sharpening B factor (Å2) | −45.12 | −116.22 |
| Model composition | ||
| Non-hydrogen atoms | 27,399 | 12,727 |
| Protein residues | 3,483 | 1,854 |
| Ligands | 6 Mg2+, 2 ATP, 4 ADP, 5 Phalloidin | Mg2+, 2 ATP |
| B factors (Å2) | ||
| Protein | 102.53 | 51.05 |
| Ligand | 92.30 | 82.27 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.004 | 0.004 |
| Bond angles (°) | 0.967 | 0.983 |
| Validation | ||
| MolProbity score | 1.65 | 1.42 |
| Clashscore | 5.63 | 4.67 |
| EMRinger score | 1.39 | 1.29 |
| Poor rotamers (%) | 0.00 | 0.00 |
| Ramachandran plot | ||
| Favored (%) | 95.00 | 96.96 |
| Allowed (%) | 5.00 | 3.04 |
| Disallowed (%) | 0.00 | 0.00 |
| CaBLAM outliers (%) | 4.2 | 3.8 |
Fig. 2|. Arp2 and Arp3 flatten and adopt the short pitch helical conformation upon activation.
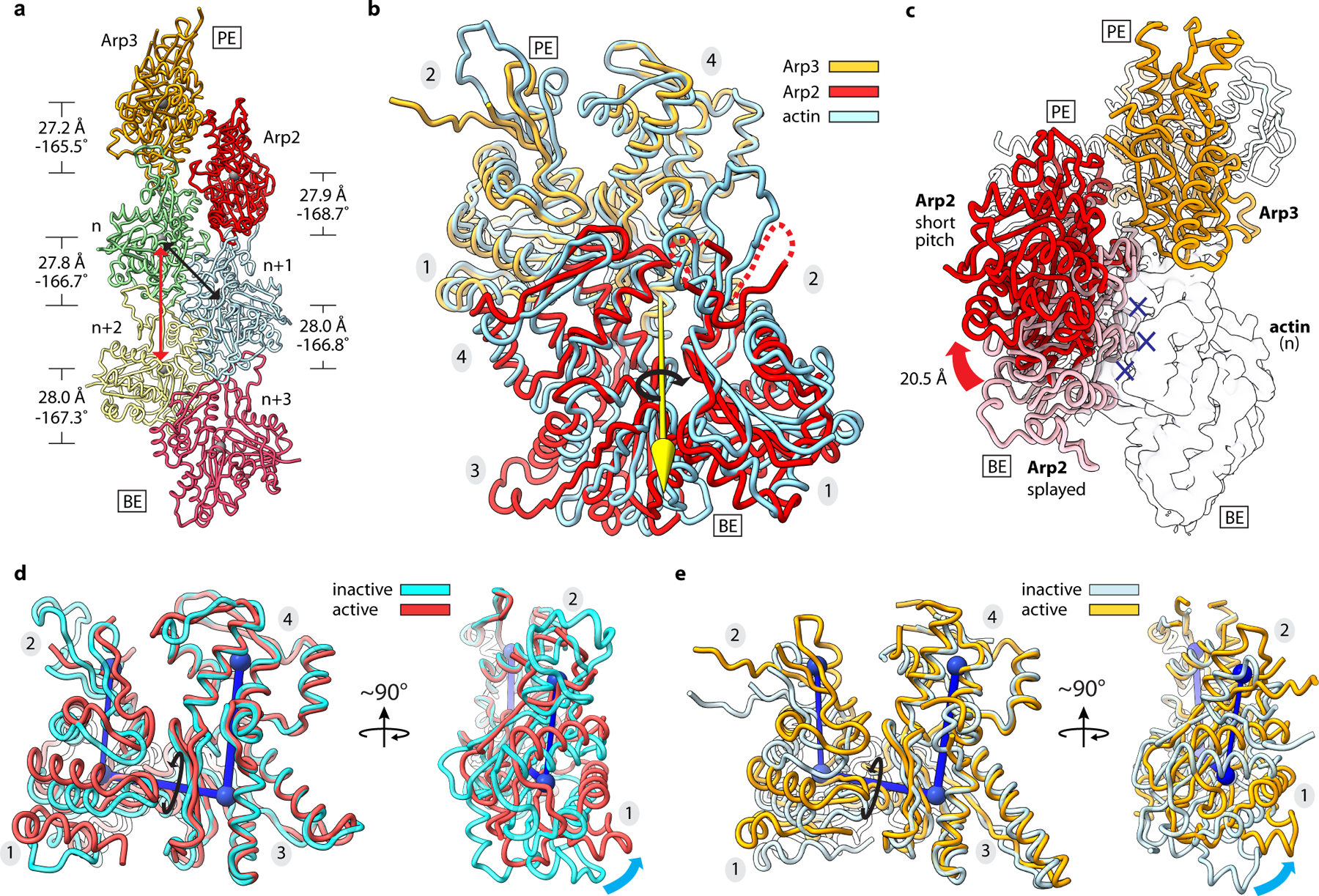
a, Ribbon diagram showing the two actin-related subunits at the pointed end of the nucleated actin-filament. The rise and twist of each subunit is indicated. Black and red arrows show short and long pitch helical interactions within the filament, respectively. b, Superposition of Arp2 and Arp3 from the active structure with a short pitch actin dimer from an actin filament (PDB 6DJO)16. Axis of rotation (yellow arrow) of Arp2 (9°) compared to its equivalent actin subunit in the dimer is shown. Subdomain numbers are shown in grey ovals throughout. c, Movement of Arp2 from the splayed (inactive) to short pitch conformation. Blue X’s show potential steric clash between Arp2 in the splayed conformation and the first actin subunit in the nucleated filament (transparent surface). d, Superposition of Arp2 in the active structure with subdomains 3 and 4 from the inactive structure. Cyan arrows show flattening motion. e, Superposition of Arp3 in the active structure with subdomains 3 and 4 from the inactive structure. PE: pointed end, BE: barbed end.
To understand the structural changes that occur during activation, we also determined a ~4.2 Å structure (Fig. 1c) of the inactive S. pombe Arp2/3 complex using a tilted cryo-EM data collection methodology18 (Extended Data Fig. 1e-h, 2b and 3c, Table 1). This structure is nearly identical to structures of inactive Arp2/3 complex from Bos taurus13,19 (Extended Data Fig. 4).
Arp2 and Arp3 adopt the short pitch conformation upon activation
Prior low-resolution EM reconstructions and biochemical/biophysical assays suggested Arp2 and Arp3 move into or near the short pitch conformation during activation8–12, but the structural rearrangements that allow these subunits to trigger nucleation have remained unclear.
In the activated structure we present here, we find that the Arps form a short pitch heterodimer that closely mimics the filamentous short pitch actin dimer (Fig. 2a,b, Extended Data Fig. 5, Supplementary Video 1). Arp2 is rotated ~9° about an axis orthogonal to the filament axis (Fig. 2b), but remains in close contact with Arp3 (~450 Å2 buried). This contrasts with the low-resolution EM reconstruction of the branch junction, in which Arp3 and Arp2 are separated by a wide gap12. The 9° rotation of Arp2 does not cause long range distortions in the nucleated filament, as the actin subunits in the Dip1–Arp2/3–filament structure show helical parameters similar to those reported in high resolution cryo-EM structures of actin filaments (Fig. 2a)15,16. Besides creating a filament-like Arp2–Arp3 heterodimer, movement of Arp2 into the short pitch conformation releases Arp2 from its splayed (inactive) position at the barbed end of Arp3 (Fig. 2c). This rearrangement frees the barbed end of Arp3 to interact with the first actin subunit (n) in the nucleated actin filament and is one of multiple structural changes that trigger activation.
Flattening of the Arps gates interactions with the first actin subunits
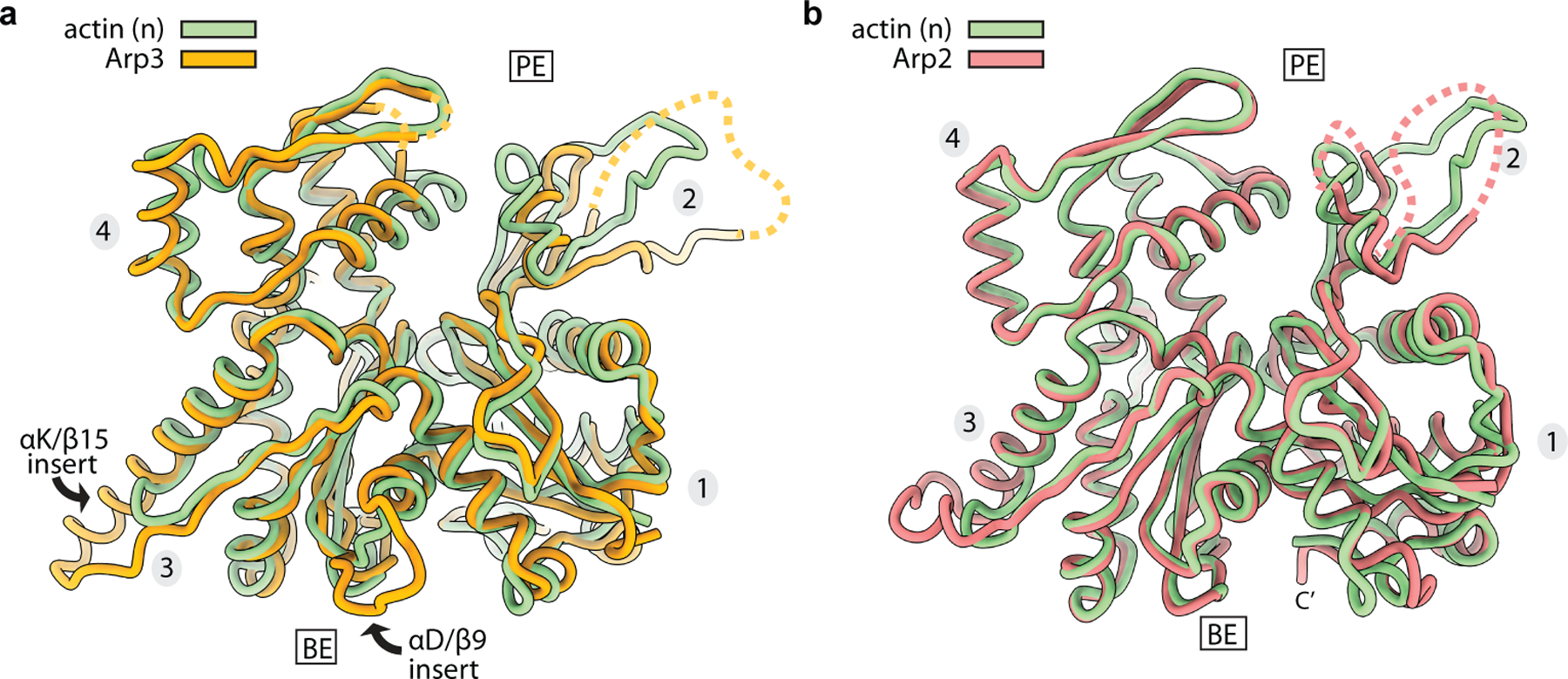
While inter-subunit structural changes in Arp2/3 complex juxtapose the Arps like two consecutive subunits along the short pitch helical axis of a filament, intra-subunit changes within the Arps are also important to create the nucleation-competent state. Upon activation both Arps undergo a scissor-like rotation of their small domain (subdomains 1 and 2) relative to their large domain (subdomains 3 and 4) that moves them from a twisted toward a flattened conformation (Fig. 2d,e). This conformational change also occurs in actin upon incorporation into filaments, and is important for facilitating both long- and short-pitch interactions between filamentous actin subunits15,16,20. In the activated structure, Arp2 flattens to the same extent as polymerizing actin subunits (Fig. 2d, Extended Data Fig. 6a). In contrast, Arp3 only partially flattens, adopting a conformation between the structures of monomeric versus filamentous actin subunits (Fig. 2e, Extended Data Fig. 6a). However, partial flattening allows Arp3 to bury ~200 Å2 more surface area and make increased interactions with Arp2 that mimic the actin-actin short pitch interface (Extended Data Fig. 6b-d).
At the Arp–actin long-pitch interfaces, three structural features from the pointed end of each actin subunit insert into three corresponding creases in the barbed ends of the Arps, making interactions analogous to long-pitch interactions between actin subunits in a filament15,16 (Fig. 3). Conformational changes at the barbed ends of the Arps that occur concurrently with flattening are important to permit a subset of these interactions. Specifically, flattening promotes insertion of the D-loop of actins n and n+1 into the crease between subdomains 1 and 3 in the Arps called the barbed end groove (BEG) (Extended Data Fig. 7a,b). Flattening of the Arps is accompanied by uncurling of the W-loop in subdomain 3, which opens a hydrophobic pocket in the BEG that accommodates Met44 of the actin D-loop (Extended Data Fig. 7a-d, Supplementary Fig. 1). Flattening also rotates a trio of residues that line the subdomain 1 side of the BEG into position to make a hydrophobic pocket that binds Val45 in the actin D-loop (Extended Data Fig. 8a-b, Supplementary Fig. 1). Finally, flattening pulls the C-terminus of both Arps away from the front of the BEG (Extended Data Fig. 8a-c). The Arp3 C-terminus is five residues longer than in actin and includes hydrophobic residues that pin it to the BEG in some inactive structures7. In this position, the Arp3 C-terminus would block the D-loop of actin monomer n from interacting with the Arp3 barbed end during activation (Extended Data Fig. 8b). By pulling the C-terminal extension out of the groove, flattening may allow the D-loop from monomer n to bind, thereby permitting nucleation.
Fig. 3|. Arp2 and Arp3 make filament-like long pitch interactions with actin.
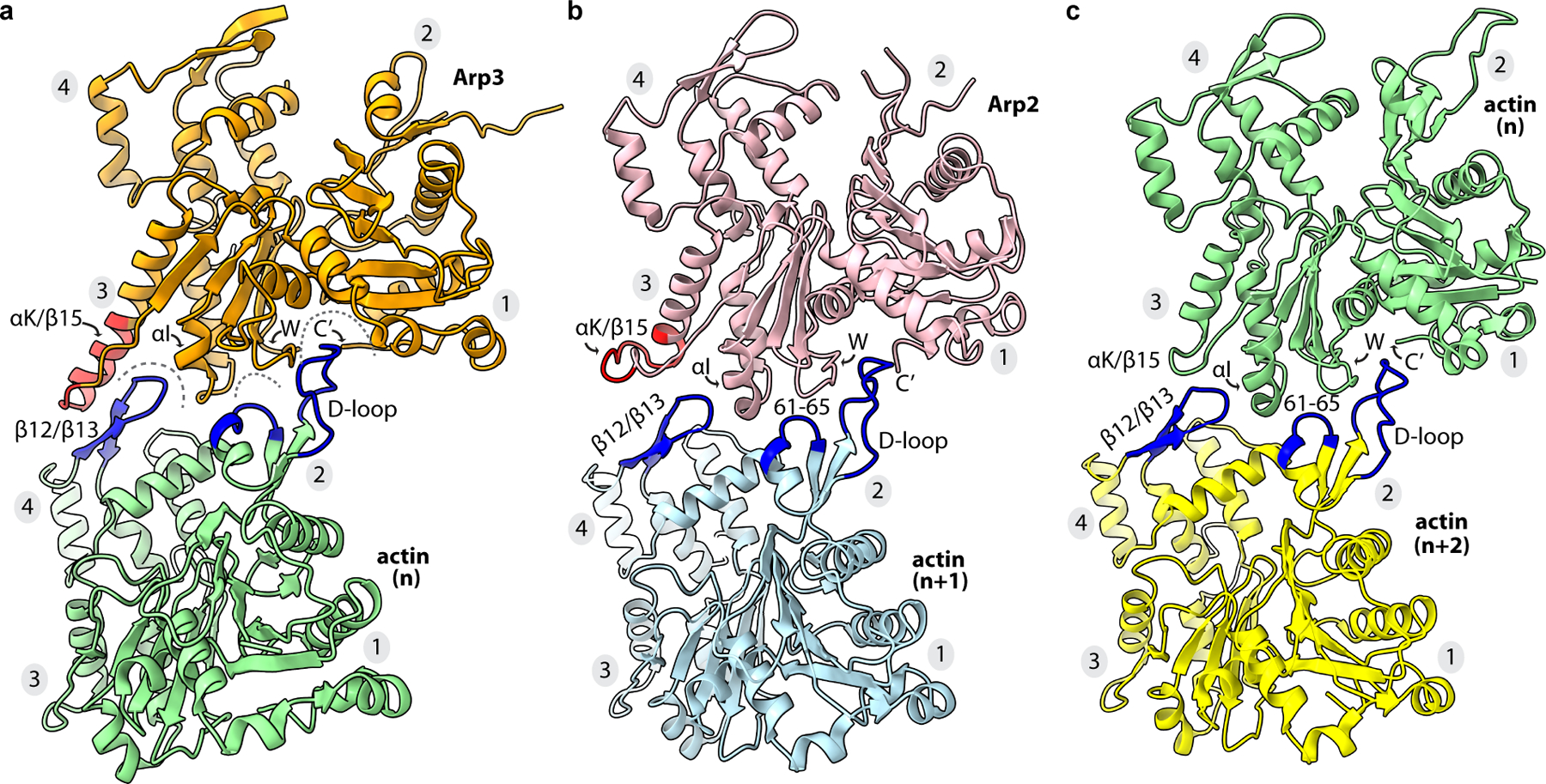
a, Long pitch interactions between Arp3 and actin (n). The αK/β15 insert is colored red. Structural features on the pointed end of actin that interact with the barbed end of Arp3 are colored blue. b, Long pitch interactions between Arp2 and actin (n+1) and c, actin (n) and actin (n+2) are shown with highlighted features colored as described for panel a. These interactions are described in detail in Extended Data Fig. 7,8.
Nucleotide binding state of the Arps in activated Arp2/3 complex
In the activated structure, both the Arp3 and Arp2 nucleotide binding clefts show density consistent with ATP (Fig. 4). In Arp3, the nucleotide density is weaker than in Arp2, but density that could be attributed to the gamma-phosphate was present in both Arps. In contrast, all four actin subunits in the reconstruction appear to have hydrolyzed their bound ATP and released the gamma-phosphate (Fig. 4). The apparent density for the gamma-phosphate of ATP in the Arp nucleotide clefts is consistent with previous biochemical experiments showing that ATP hydrolysis is not required for activation21,22.
Fig. 4|. Nucleotide binding states of the Arps and actin.
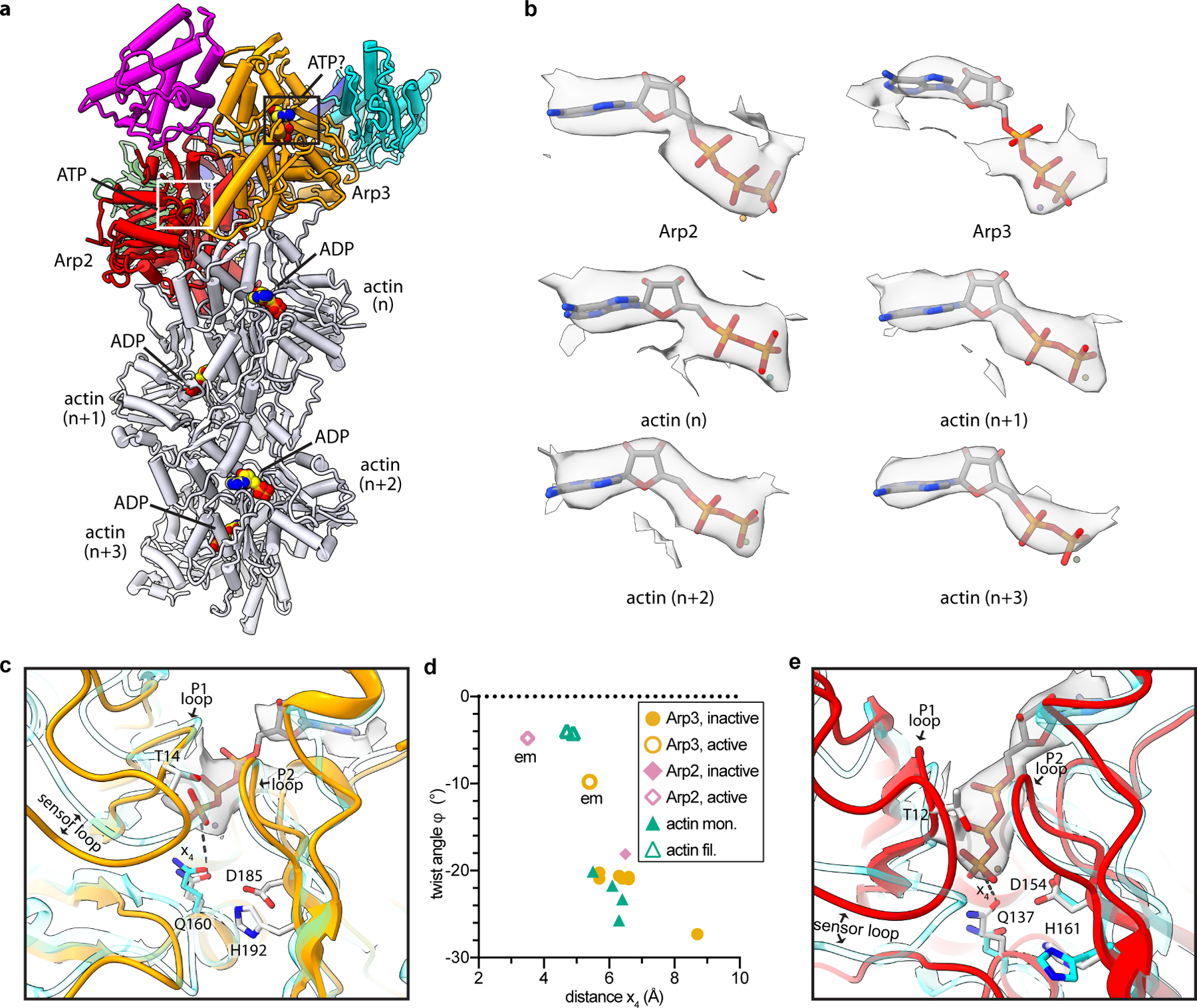
a, Structure of activated Arp2/3 complex with nucleated actin filament in which the bound nucleotides modeled into the Arps and actin subunits are shown as spheres with yellow carbon atoms. b, Individual maps of bound nucleotides (shown in gray) with the corresponding nucleotide structures inside the map. c, Close up of boxed region in panel a showing activated Arp3 (orange) superposed with an AMP-PNP-bound actin subunit from an actin filament PDB 6DJM16 (semi-transparent cyan) using subdomains 3 and 4. Distance x4 between the catalytic glutamine (Q160) and the gamma phosphate is shown as a dotted line. d, Plot of distance x4 for inactive or active Arp2 or Arp3 or monomeric or filamentous structures of actin. All structures had bound ATP or AMP-PNP. Data points labeled “em” are from the activated structure presented here. (See Online Methods for more details) e, Close up of white boxed region in panel a showing activated Arp2 (red) superposed onto inactive Arp2 from the structure PDB 4JD219 (cyan) using subdomains 3 and 4. Flattening rotates Q137Arp2 into or near its presumed catalytically active position.
The presumed stability of ATP in the Arp3 cleft could be caused by the intermediate conformational state of Arp3. Distance measurements in the active site suggest that partial flattening of Arp3 fails to fully rotate Gln160Arp3 into position near the gamma-phosphate of ATP, where it is thought to position the hydrolytic water molecule23 (Fig. 4c,d). It is important to note that the limited resolution of the map increases the uncertainty of these measurements (Extended Data Fig. 9a-b). Arp2, on the other hand, flattens as much as filamentous actin subunits, and its equivalent glutamine, Gln137Arp2, rotates into its proposed activated position within the nucleotide-binding cleft (Fig. 4d,e). However, in Arp2, the ATP is pushed deeper in the nucleotide-binding pocket, into a position that likely prevents the gamma-phosphate from properly aligning Gln137Arp2, His161Arp2 and the catalytic water molecule for hydrolysis (Extended Data Fig. 9c).
Previous data showed that WASP-mediated branching nucleation stimulates ATP hydrolysis on Arp2, and likely also on Arp3, to trigger branch destabilization and network disassembly21,22,24,25. Given that Arp2 hydrolyzes ATP seconds after branching nucleation24, we expected ATP to be hydrolyzed on Arp2, and possibly also Arp3, in our structure of Dip1-activated Arp2/3 complex, because we aged filaments for tens of minutes to hours before plunge freezing to image. The apparent presence of ATP in Arp2 and Arp3 of Dip1-activated Arp2/3 complex suggests that Dip1 may not trigger rapid ATP hydrolysis upon activation. Given the role of ATP hydrolysis in destabilizing branches, this biochemical feature could contribute to the stability of Dip1 bound to activated Arp2/3 complex on the pointed end of nucleated actin filaments17.
Twisting of the clamp rotates a block of subunits into the short pitch conformation
The structures we present here not only reveal the key features of nucleation-competent state, but they also show how Arp2/3 complex switches to this state. To adopt the short pitch conformation, the complex undergoes a dramatic conformational rearrangement in which the center of mass of Arp2 moves 20.5 Å relative to its position in inactive structures (Fig. 2c). A previously proposed “clamp release model” hypothesized that to reach this state, Arp2 releases from the clamp subunits (ARPC2 and ARPC4)—which remain rigid—and rebinds in a new position that approximates the short pitch conformation12. This model predicts that Arp2 will make a distinct set of contacts with the clamp in the short pitch state compared to the splayed state. In contrast, a second model, based on computational studies, suggested that Arp2 maintains its contacts with the clamp during activation, but the clamp twists to move Arp2 into the short pitch conformation26. The Dip1-Arp2/3-filament structure is incompatible with the clamp release model, since Arp2 makes identical contacts to the clamp in the inactive and active conformations (Extended Data Fig. 10a). Furthermore, we find that the clamp itself is not rigid during the activation process, a prerequisite of the clamp release model. Instead, during activation the clamp twists, rotating its bottom half along with a large block of subunits that includes Arp2, the globular portion of ARPC4, ARPC1, and ARPC5 to bring Arp2 into the short pitch position (Fig. 5a,b, Extended Data Fig. 10b, Supplementary Video S2). The twist axis runs parallel to the long helices in the clamp and rotates the block of subunits ~19° (Fig. 5b).
Fig. 5|. Dip1 bends the ARPC4 long helix to twist the clamp and rotate a block of subunits into the short pitch conformation.
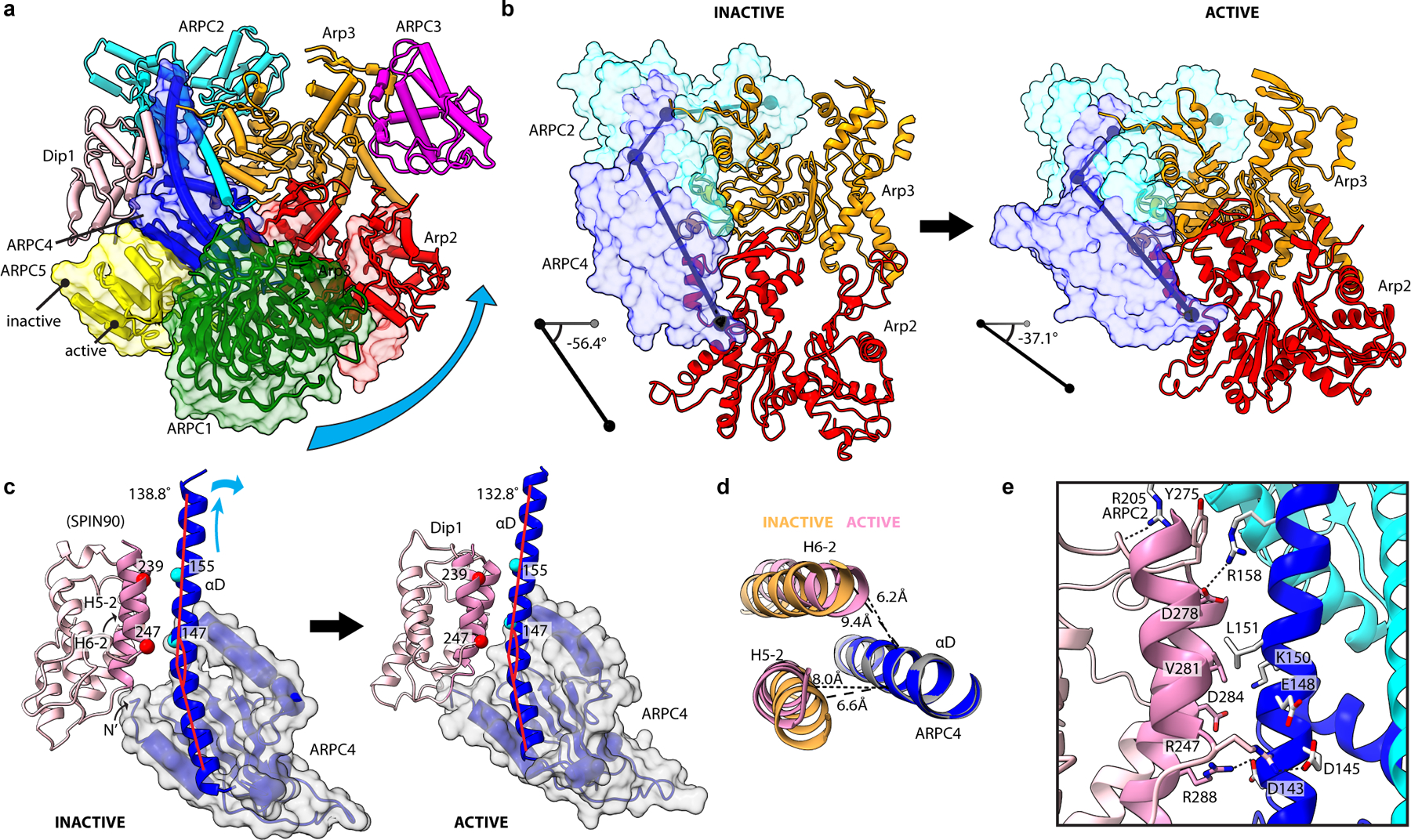
a, Comparison of the activate and inactive Arp2/3 complex structures. Subunits Arp2, ARPC1, ARPC4 and ARPC5 of the inactive structure are shown in semi-transparent surface representation. Cyan curved arrow shows direction of rotation during activation of a block consisting of Arp2, ARPC1, ARPC4 and ARPC5. b, The clamp (subunits ARPC2 and ARPC4, transparent surface) rotates to move the Arps into the short pitch conformation. c, Comparison of contacts between SPIN90/Dip1 in the inactive (PDB 6DEC) versus activate states of Arp2/3 complex. Cyan arrows show direction of motion of ARPC4 αD helix upon activation. Red lines show bend angle of ARPC4. d, End on view of helices H5–2 and H6–2 from Spin90/Dip1 and helix αD from ARPC4 in the activated and inactive (PDB 6DEC) states. e, Interactions between Dip1 (pink), ARPC4 (blue) and ARPC2 (cyan) in the activated structure.
To trigger clamp twisting, Dip1 binds the center of the clamp along a long helix that extends from the globular portion of the ARPC4 subunit (Fig. 5c). Its binding mode is similar to the human WDS protein, SPIN90, bound to inactive Arp2/3 complex observed in a recent crystal structure27. In the active structure we present here, Dip1 remains braced against the globular domain of ARPC4, but changes its register with the extended helix in ARPC4 to pack more tightly against it (Fig. 5c-e). This allows Dip1 to serve as a wedge that bends the helix and pushes it away from the globular domain of ARPC4 (Fig. 4c-e, Extended Data Fig. 3m and 10b). Bending of the extended ARPC4 helix triggers the ~19° clamp twist that rotates Arp2 into the short pitch conformation. Bending of this helix also appears to indirectly stimulate the intra-subunit domain rotation that partially flattens Arp3 because—once moved into the short pitch conformation—flattening of Arp3 increases its interactions with Arp2 (Extended Data Fig. 6).
Discussion
While WASP family NPFs bind to different sites on Arp2/3 complex than Dip16,11,27–30, biochemical and low-resolution structural data suggest that like Dip1, WASP stimulates movement of Arp2 into the short pitch conformation3,8–11. Based on the flexibility of the clamp we observed here and the requirement to move the Arps into the short pitch conformation during activation31, we expect clamp twisting is a general feature of activation triggered by WASP and other NPFs. Given that diverse NPFs regulate Arp2/3 complex across a broad range of biological functions32, we believe this conformational change to be a key step for controlling actin assembly in many different cellular processes.
Besides clamp twisting and adoption of the short pitch conformation, the structure of activated Arp2/3 complex reveals the other conformational changes required for activation, and along with previously published data allows us to propose a structural pathway for actin filament nucleation (Fig. 6). In the inactive complex, the Arp2 and Arp3 subunits adopt a splayed arrangement. This conformation is stabilized by inter-subunit interactions between the Arps and by their interactions with the clamp7 (Extended Data Fig. 10a). In the inactive conformation, the barbed ends of the Arps are not configured to make strong long-pitch interactions with actin monomers and Arp2 sterically blocks the barbed end of Arp3, thus preventing nucleation. The clamp twisting conformational change induced by Dip1 binding during activation disrupts the inactive arrangement of the Arps and moves Arp2 into the short-pitch conformation, thereby permitting association of actin monomers with the nascent filament.
Fig. 6|. Proposed mechanism of activation of Arp2/3 complex by Dip1.
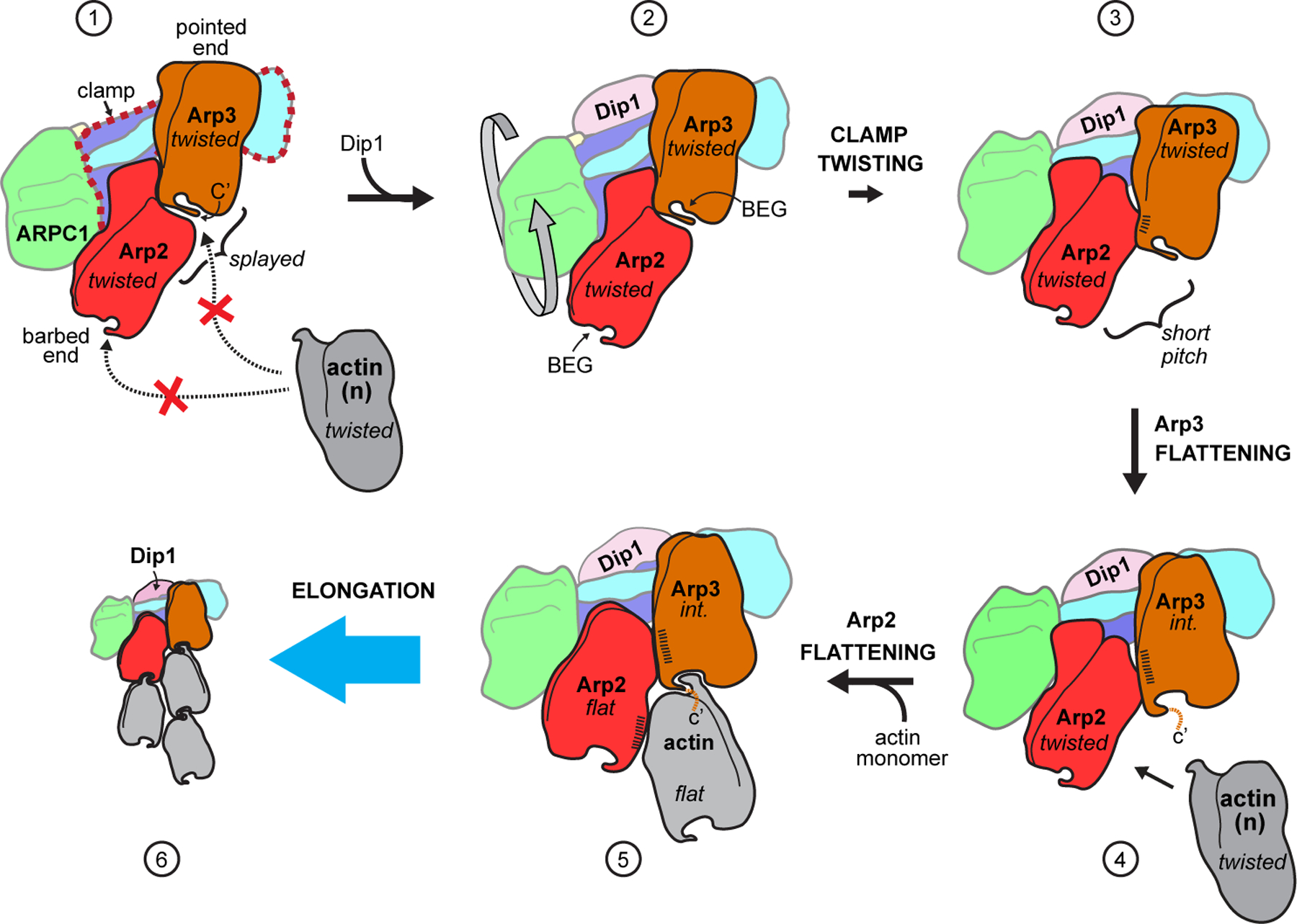
In state 1, Arp2/3 complex is in an inactive state with the barbed end of Arp3 blocked by Arp2 and the barbed end grooves (labeled BEG in state 2) are not properly arranged for interaction with actin. Dip1 binds to the clamp (red dashed outline in state 1) to stimulate clamp twisting and movement of Arp2 into the short pitch conformation (states 2 to 3). Arp2 in the short pitch conformation triggers partial flattening of Arp3, which opens up the Arp3 barbed end groove for optimal long pitch interactions with actin n (states 3 to 4). Steps 4 and 5 show actin monomer n flattens and stimulates flattening of Arp2 upon binding to the complex, however, it is possible that formation of the nucleus requires binding of additional actin monomers to the nascent filament. Dip1 binds tightly to activated Arp2/3 complex on the pointed end of the actin filament17 (state 6), unlike WASP, which must be released for nucleation to proceed. Conformational states are indicated in italicized text; int.: intermediate (partially flattened) state of Arp3. Hatching indicates relative surface area buried at short pitch interface. For clarity, ARPC3 is not depicted in this cartoon.
Intra-subunit changes are also important for activation, and the activated structure shows that flattening causes structural rearrangements at the barbed ends of both Arps that allow them to make increased long pitch interactions with actin (Fig. 6, states 3,4, Extended Data Fig. 7,8). Some of these changes—including W-loop uncurling and realignment of the two sides of the barbed end groove—occur in both the activated Arps and in polymerized actin subunits15,16, therefore constituting an evolutionarily conserved conformational switch that controls assembly. As in Arp3, residues in the C-terminal tail of both actin and Arp2 are pulled out of the barbed end groove and toward the filament helical axis during flattening (Extended Data Fig. 8). However, it is currently unclear whether, like Arp3, movement of the relatively short C-terminal tails in Arp2 or actin during flattening gates their long pitch interactions with actin subunits. Disorder in the inactive structures makes it unclear whether the C-terminus is long enough and positioned correctly to block interactions of the D-loop of an incoming actin monomer with the barbed end groove in these proteins16,19.
How Dip1 and other WDS proteins activate Arp2/3 complex without a pre-existing actin filament while WASP proteins cannot has been an important unresolved question, as this biochemical property allows WDS proteins to initiate the assembly of branched actin networks3,33,34. We postulate that a key difference between these two NPFs is rooted in the mechanism by which they stimulate the short pitch conformation. While WDS proteins activate through their interactions with the clamp, WASP binds to two sites far from the clamp6,8,11,28–30,35 (Supplementary Fig. 2). At these sites, the Arp2/3-interacting segment in WASP, WASP-CA, engages the complex to stimulate the short pitch conformation while the WASP V region recruits actin monomers to the complex6,8,11,28–30,35,36. The activated structure presented here, along with structural and biochemical data on WASP6,8,11,28–30,35, indicates that the WASP-C segment blocks the D-loops of the first two actin subunits (n, n+1) from inserting into the barbed end grooves of the Arps (Supplementary Fig. 1). These observations suggest that bound WASP blocks proper association of its recruited actin subunits with the barbed ends of the Arps. Therefore, while bound WASP, like Dip1, stimulates the short pitch conformation3,8–10, it likely blocks subsequent activating steps. This hypothesis is supported by recent observations that showed WASP must be released before nucleation37, and also by the observation that when Arp2/3 complex is crosslinked into the short-pitch conformation it is inhibited by WASP-CA8. We speculate that by stimulating the short pitch conformational change without blocking filament-like interactions with the first actin monomers, WDS proteins allow activation to proceed without Arp2/3 complex binding to actin filaments. During WASP-mediated branching nucleation, actin filaments may stimulate WASP release from the nascent branch junction to allow nucleation, but the precise role of actin filaments during WASP-mediated activation is still an important unresolved question.
In conclusion, the structure of activated Arp2/3 complex bound to its nucleated actin filament provides a new structural context for interpreting over two decades of biophysical and biochemical studies, while also providing the foundation for future experiments aimed towards understanding how the highly conserved Arp2/3 complex nucleates actin filament in response to different cellular signals.
Methods
Protein Purification
Rabbit skeletal muscle actin was extracted from acetone powder (Pel Freeze, Cat No. 41995) by stirring in G-buffer (20 mL/g, 2 mM Tris pH 8.0, 0.2 mM ATP, 0.5 mM DTT, 0.1 mM CaCl2) on ice for 30 min before spinning in a Beckman JA-20 rotor at 16,000 rpm for 30 min at 4 ˚C. Supernatant containing actin monomers was filtered through glass wool. The pellet was resuspended in the original volume of G-buffer and the extraction, centrifugation and filtration steps were repeated once more. Actin was polymerized from the combined supernatant by adding KCl to 50 mM and MgCl2 to 2 mM final concentrations and stirred on ice for 1 hr. Additional KCl was then added to bring the final concentration of KCl to 0.8 M. After 30 mins, the solution was centrifuged for 2 hrs in a Ti70 rotor at 31,700 rpm to pellet the polymerized actin filaments. The pellet was gently washed with G-buffer and resuspended in G-buffer in a proportion of 3 mL per gram of the starting stock of acetone powder. Actin was then diluted to 5 mg/mL and dialyzed for two days with three changes of G-buffer to depolymerize the actin filaments. The solution was then clarified by centrifugation in a Ti90 rotor at 36,000 rpm for 2 hrs and actin was purified by size exclusion chromatography over a Sephacryl S-300 column equilibrated with G-buffer containing 1 mM sodium azide. Fractions were stored on ice and used for experiments within one week. The concentration of actin was measured by absorbance using an extinction coefficient of 26,000 M−1cm−1 at 290 nm.
Dip1 was purified as described previously3. Briefly, Dip1 was overexpressed as a GST-fusion in E. coli using the pGV67-Dip1 plasmid. Cells were homogenized and the lysate was clarified by centrifugation. The clarified lysate was run over a glutathione Sepharose affinity column and eluted with reduced glutathione. The protein was then dialyzed overnight at 4 ˚C with TEV protease then loaded onto a resource Q column in 20 mM Tris pH 8.0, 50 mM NaCl and 1 mM dithiothreitol before eluting with a 50 – 500 mM NaCl gradient. The protein was then concentrated and loaded onto a Superdex 200 gel filtration column. The concentration of Dip1 was measured by absorbance using an extinction coefficient at 280 nm of 36,330 M−1cm−1.
S. pombe Arp2/3 complex was purified as previously described34. Briefly, S. pombe cells were lysed in a Microfluidics Model M-110EH-30 Microfluidizer and the lysate was clarified before adding 0.243 g/mL ammonium sulfate to the supernatant over 30 min with stirring. After 30 min, the pellet was resuspended in PKME (25 mM PIPES pH 6.8, 50 mM KCl, 1 mM EGTA, 3 mM MgCl2, 1 mM DTT and 0.1 mM ATP) then dialyzed overnight against the same buffer before adding to a glutathione sepharose column charged with GST-N-WASP-VCA. Arp2/3 complex was eluted using a gradient of PKME with a total of 150–1000 mM KCl. Pooled fractions were then run on a monoQ column equilibrated in 10 mM PIPES pH 6.8, 25 mM NaCl, 0.25 mM EGTA, 0.25 mM MgCl2 and eluted with a gradient of 25–500 mM NaCl. The dialysate was concentrated and loaded on a Superdex 200 gel filtration column equilibrated with Tris pH 8.0, 50 mM NaCl and 1 mM DTT. The final concentration was determined by measuring the absorbance at 290 nm (E290 = 139,030 M−1cm−1) and then the protein was aliquoted and flash frozen.
His-tagged mouse capping protein (alpha-1, beta-2) in a pRSF Duet vector (a gift from David Kovar) was overexpressed in E. coli. Cells were lysed in 50 mM NaH2PO4, pH 8.0, 500 mM NaCl, 10 % glycerol, 10 mM imidazole and 10mM β-mercaptoethanol with protease inhibitor tables. Lysate was run over a Talon Metal Affinity resin (Clontech 635501) and eluted with lysis buffer containing 250 mM imidazole. Peak fractions were dialyzed overnight against 20mM Tris pH 8.5, 50 mM NaCl, 5 % glycerol, 0.01 % NaN3 and 1mM DTT, then loaded onto a resource Q column, washed, and eluted with a salt gradient of 50–500 mM NaCl. Pure protein was dialyzed into storage buffer (20mM HEPES, 1mM EDTA, pH8, 200mM KCl, 0.01% NaN3, 10% glycerol, 1mM DTT) before flash freezing.
GST-Wsp1-VCA was overexpressed in E. coli, the cells lysed, and the lysate run over a glutathione sepharose column in 20 mM Tris, pH 8.0, 140 mM NaCl, 2 mM EDTA, and 2 mM DTT. The GST tag was cleaved on the column by adding TEV protease and incubating overnight at 4˚C with gentle inversion. The NaCl concentration was reduced to 100 mM by dilution and loaded on a MonoQ column, washed, and eluted with a gradient of 100–500 mM NaCl. Pooled fractions were concentrated and loaded on a Superdex 75 gel filtration column. The concentration was measured using the absorbance at 280 nm and an extinction coefficient of 5690 M−1cm−1 before flash freezing.
Sample preparation for cryo-EM
To generate filaments capped on their pointed-ends by Dip1-bound Arp2/3 complex, we mixed 1.0 μM S. pombe Arp2/3 complex, 5.0 μM Dip1, 1.0 μM Wsp1-VCA, 9.0 μM rabbit skeletal muscle actin monomers, and 0.6 μM mouse capping protein in a buffer containing 50 mM KCl, 1 mM EGTA, 1 mM MgCl2, 0.2 mM ATP, 20 mM HEPES pH 7.5 and 1 mM Dithiothreitol (DTT) at room temperature. To keep the filaments short, we added all components except actin monomers and capping protein first, then spiked in one third of the desired final capping protein and actin monomers to 1.4 μM. After polymerizing for 20 min, we added additional actin monomers to bring the total actin concentration to 2.3 μM. This process was repeated eight additional times to keep the concentration of unpolymerized actin monomers in the reaction relatively low throughout the reaction. The remaining capping protein was added (a third at a time) just before the third and seventh actin monomer addition steps. After the last addition of actin, the reaction was incubated for 20 min before adding phalloidin to a final concentration of 10 μM. For the inactive Arp2/3 complex sample, we attempted to form co-assemblies of Arp2/3 complex and Dip1 by mixing on ice 1.0 μM S. pombe Arp2/3 complex and 3.0 μM Dip1 in a buffer containing 50 mM NaCl, 1 mM MgCl2, 1 mM ATP, 10 mM Tris pH 8.0 and 1 mM dithiothreitol (DTT). Only Arp2/3 complex alone and no Dip1-Arp2/3 complex assemblies were resolved by both 2D and 3D classifications during cryo-EM image processing.
The same grid vitrification procedure was used for both samples. 4 μl of sample was applied to freshly glow discharged 300 mesh UltrAuFoil R1.2/1.3 holey Gold grid (Quantifoil). Excess sample was manually blotted off with a dry Whatman No.1 filter paper for 5–7 s. Immediately after the blotting step the sample containing grid was rapidly vitrified by plunge freezing in liquid-ethane at −179°C. The entire cryo-grid preparation process was carried out in a 4°C cold room and at 98% relative humidity to minimize excessive evaporation of sample from grid surface.
Cryo-EM data acquisition
Cryo-electron microscopy (Cryo-EM) data were acquired on a 200kV Talos Arctica transmission electron microscope, equipped with a Falcon 3EC direct electron detector (Thermo Fisher). Automated data acquisition software EPU (Thermo Fisher) was used for data collection. For the Dip1-activated Arp2/3-actin complex, each micrograph was collected as dose-fractionated movies, consisting of 45 fractions per movie and at a nominal magnification of 92,000X. This corresponded to a physical pixel size of 1.12 Å/pixel for each micrograph. The total exposure time per micrograph was 60 s and the total dose was 36.35 e−/Å2 (0.81 e−/fraction). Data was collected under-focused with a nominal defocus range between −1.00 and −1.75 µm (Extended Data Fig. 1a).
Electron micrographs of inactive Arp2/3 complex were collected by using a previously described tilted data acquisition method18. Micrographs were acquired by tilting the grids to 20˚, 30˚ and 40˚ angles. Additionally, data was also collected without tilting of the grids. All data were collected at a nominal magnification of 120,000X, corresponding to a pixel size of 0.8757 Å/pixel, with nominal defocus ranging from −0.7 and −1.3 µm (Extended Data Fig. 1e). Each micrograph was collected as dose-fractionated movie, consisting of 62 fractions over 40 s exposure and total dose of 44.34 e−/Å2 (0.72 e−/fraction).
Cryo-EM data processing
A total of 5,109 micrographs were collected for the Dip1-activated Arp2/3-actin complex (Extended Data Fig. 1a). Beam induced motion-correction and per-fraction radiation damage compensation over spatial-frequencies (dose-weighting) were performed using the motion-correction algorithm implemented in the RELION v3.0.6 (henceforth referred as RELION) suite38. Contrast Transfer Function (CTF) estimation for the micrographs from aligned and summed fractions were estimated using the program Gctf39. Based on the results for CTF estimation, we discarded 661 micrographs which had poor confidence of fit or were severely astigmatic. From 4,448 remaining micrographs particles were picked using the automated Laplacian Gaussian picking program implemented in RELION. This resulted in 3.5 million picked particles, which were extracted from micrographs using square box size of 304 pixels and further binned to box size of 76 pixels (voxel size of 4.48 Å/pixel). These particles were subjected to multiple rounds of 2D classification. 2,346,578 particles belonging to 2D classes of the Arp2/3 complex capped pointed-ends (Extended Data Fig. 1b) were selected for further processing. Using these particles, an ab intio 3D reference volume was created using cryoSPARC v240. This volume was used as initial reference for 3D processing. Multiple iterations of 3D refinements followed by 3D classifications without alignment (clustering) were performed to isolate the best resolving 3D volume. Following this, 125,359 particles from 3D classes representing the Arp2/3 capped pointed-ends of actin filament were then re-centered and re-extracted as unbinned particles (304 pixels box, 1.12 Å/pixel). Initial 3D refinement with this unbinned stack of particles resulted in a 4.3 Å resolution map. 3D binary masks for data processing were created by using the 3D reference volume of the complex as template and expanding it by 8 pixels along with Gaussian smoothing across 10 pixels. For more accurate per-particle CTF estimation, CTF refinement within RELION was performed. Particles were also subjected to per-particle motion correction by Bayesian polishing41. These polished particles were subjected to 3D refinement using mask, resulting in reconstruction at 4.1 Å resolution. The 3D refined particle stack was subjected to three class, clustering. 110,433 particles belonging to the best 3D class were selected and subjected to further 3D refinement. The final refined map was at a resolution of 3.9 Å (at 0.143 FSC) (Extended Data Fig. 3a). As the cap and the filament regions were flexible relative to each other, we decided to improve the quality of maps for these sub-regions by performing signal-subtraction based focused refinement42 (Extended Data Fig. 2a,3a). Both the sub-regions had the Arp2 and Arp3 subunits in common. In order to rebuild the composite map, focused maps for the two regions were first fitted to non-focused map of the full complex, with the Arp2 and Arp3 subunits as the overlapping subunits between the two maps. Then the composite map was generated by retaining the maximum valued voxel at each location from the two maps by using the “vop maximum” function within UCSF Chimera43 (Extended Data Fig. 2a, 1c,d). Local resolution of the map was estimated using RELION’s local resolution estimation program (Extended Data Fig. 1c). The final map was sharpened by applying a B-factor of −45.12 Å2 during the postprocessing step in RELION.
The inactive Arp2/3 complex data was processed using CryoSPARC v2.12.440 (Extended Data Fig. 2b). A total of 5,309 micrographs, consisting of movies, were collected with and without tilting and subjected to full-frame motion correction followed by patch-motion correction (Extended Data Fig. 1e). CTF parameters were estimated locally using the patch CTF-estimation program. An automated Gaussian based blob picker was used to pick all the particles from the micrographs, and 3,336,981 picked particles were then extracted from the micrographs using a square box of 368 pixels (0.8757 Å/pixel). These particles were subjected to multiple rounds of 2D classification. After excluding particles belonging to classes having non-particle features and aggregates, 450,747 particles representing different views of Arp2/3 complex (Extended Data Fig. 1f) were selected for 3D processing. These selected particles were then subjected to another round of local-motion correction to more accurately correct per-particle drifts. These particles were subjected to three class ab initio 3D reconstructions. 185,772 particles belonging to the intact inactive Arp2/3 complex class was subjected to homogeneous refinement. To further separate heterogeneity, these particles were further subjected to 3D classification by heterogenous refinement. 112,170 particles belonging to well resolved, inactive Arp2/3 complex 3D class was finally refined using a static binary 3D mask, which resulted in a final reconstruction of 4.2 Å (at 0.143 FSC) (Extended Data Fig. 1g,h, 2b, 3b). Local resolution estimation and sharpening of the map was performed in RELION using the two unfiltered half maps (Extended Data Fig. 1g). The final map was sharpened by applying a B-factor of −116.219 Å2.
Model building and analysis
The final maps generated from 3D reconstructions mentioned above were used for atomic model building of each of the complexes. Structures of S. pombe inactive Arp2/3 complex, lacking the Arp2 subunit (PDB 3DWL), and F-actin subunits from rabbit (Oryctolagus cuniculus) ADP-actin filament (PDB 3J8I), were used as initial templates for atomic model building. Ab initio computational models for the S. pombe Arp2 and Dip1 subunits were generated using RaptorX44 (http://raptorx.uchicago.edu/StructPredV2/predict/) and I-TASSER45 (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) protein structure prediction servers. These models were initially rigid body fitted into the maps using “fit map” function in UCSF Chimera43. Flexible fitting of the models into their respective maps were performed using the Namdinator web server (https://namdinator.au.dk)36. Amino acid registers, backbone and side chain geometries for the models were manually fixed using Coot46. Density corresponding to the ARPC3 subunit was poor due to structural flexibility. The model for this region was truncated to Cß and rigid body fitted into the map. In our activated structure of the Arp2/3 complex, we were able to build residues 235–366 of full length Dip1, as the rest of the Dip1 molecule was missing from the density map. Repeated rounds of model fixing using Coot and real space refinement in Phenix47 were performed to fix all model geometries. Regions lacking side chain densities were truncated to Cß stubs, and for those regions where the density was missing or extremely broken, the polypeptide chain was deleted in the model. Accurate pixel size for the map was determined using the EMBuilder program48. The final corrected pixel size for the Dip1-bound Arp2/3 complex-actin filament map was 1.11 Å and the pixel size for the inactive Arp2/3 complex remained unchanged. Model validations were performed using the Molprobity server (http://molprobity.biochem.duke.edu/)49 and also the PDB Validation Service server (www.wwpdb.org)50. All figure and movie making were performed using UCSF Chimera33 and UCSF ChimeraX43.
The degree of flattening of Arp2, Arp3 or actin was measured using the angle φ. This angle was determined by treating the center of mass of each of the four subdomains as a point in a dihedral definition. The centers of mass of each subdomain were calculated using backbone atoms within the following subdomain definitions: S. pombe Arp3; 6–32, 94–169, 384–417 (Subdomain 1), 33–37, 74–93 (Subdomain 2), 170–212, 306–355, 371–382 (Subdomain 3), 213–293 (Subdomain 4), Bos taurus Arp3; 6–32, 78–153, 375–408 (Subdomain 1), 33–37, 60–77 (Subdomain 2), 154–196, 295–344, 362–374 (Subdomain 3), 197–282 (Subdomain 4), S. pombe Arp2; 4–30, 71–146, 348–385 (Subdomain 1), 31–35, 52–70 (Subdomain 2), 147–181, 273–322, 335–347 (Subdomain 3), 182–261 (Subdomain 4), Bos taurus Arp2; 7–33, 74–150, 352–387 (Subdomain 1), 34–38, 55–73 (Subdomain 2), 151–185, 277–326, 339–351 (Subdomain 3), 186–265 (Subdomain 4), Oryctolagus cuniculus actin; 6–32, 71–146, 337–373 (Subdomain 1), 33–37, 52–70 (Subdomain 2), 147–181, 273–336 (Subdomain 3), 182–261 (Subdomain 4).
Distance x1, which measures intra-subunit flattening (Extended Fig. 6a), was defined by the following pairs of atoms: S. pombe Arp3; Y92 Cα to K235 Cα, Bos taurus Arp3; Y76 Cα to E222 Cα, S. pombe Arp2; Y69 Cα to E207 Cα, B. taurus Arp2; Y72 Cα to E211 Cα, O. cuniculus actin; Y69 Cα to E207 Cα. Distance x2, which measures opening/uncurling of the W-loop (Extended Figure 7d), was defined by the following pairs of atoms: S. pombe Arp3; V159 Cα to Y200 Cα, Bos taurus Arp3; V143 Cα to Y184 Cα, S. pombe Arp2; I136 Cα to V169 Cα, Bos taurus Arp2; I140 Cα to F173 Cα, O. cuniculus actin; I136 Cα to Y169 Cα. Distance x3, which measures the position of the C-terminus relative to the barbed end groove (Extended Data Figure 8), was defined by the following pairs of atoms: S. pombe Arp3; L179 Cα to R418 Cα, Bos taurus Arp3; L163 Cα to R409 Cα, S. pombe Arp2; S148 Cα to D383 Cα, Bos taurus Arp2; L152 Cα to E387 Cα, O. cuniculus actin; T148 Cα to H371 Cα. Distance x4 measures the distance between the catalytic glutamine and the gamma phosphate (Figure 4d). This distance was measured between the gamma phosphate and the following residues: S. pombe Arp3; Q160, Bos taurus Arp3, Q144; S. pombe Arp2; Q137, Bos taurus Arp2; Q141, O. cuniculus actin; Q137. For distances x1, x2, x3 the following structures were used: 1) Arp2 and Arp3 from all available structures of Arp2/3 complex (PDB 1K8K, 1TYQ, 1U2V, 2P9I, 2P9K, 2P9L, 2P9N, 2P9P, 2P9S, 2P9U, 3DXK, 3DXM, 3DWL, 3RSE, 3UKU, 3UKR, 3ULE, 4JD2, 6DEC14,19,28,35,51–53, and the inactive and activated structures presented here). Subdomains 1 and 2 of Arp2 is disordered in most of these structures so fewer data points could be generated for the Arp2 subunit. 2) Two representative actin monomer x-ray crystal structures in either the ADP or the ATP bound state (PDB 1NWK, PDB 2J6Z)54,55. 3) The highest resolution cryo-EM structures of actin filaments with subunits in the ADP, ADP-Pi, or AMP-PNP bound states15,16. In Fig. 4d, in addition to the activated structure presented here, distance x4 was measured in the following structures: PDB 4JD2, 2P9K, 1TYQ, 3ULE, 3DWL, 2P9S, 2P9U, 1ATN, 1NWK, 2A40, 2A42, 2HBT, 6DJM, and 5OOE.
For analysis of the surface area buried at interfaces in Extended Data Fig. 6c, coordinate files were processed using the Dockprep module in Chimera to add missing side chains and then submitted to the PISA protein interaction server (https://www.ebi.ac.uk/msd-srv/prot_int/cgi-bin/piserver)43,56. The clamp twist angle for Fig. 5b was defined as the dihedral between Arg32 Cα (ARPC4), Ser 147 Cα (ARPC4), Ile262 Cα (ARPC2), and Arg18 Cα (ARPC2) in S. pombe Arp2/3 complex or the homologous residues in BtArp2/3 complex. The ARPC4 αD helix bend angle in Fig. 5c was defined using Lys130 Cα, Asp143 Cα and Ser166 Cα in the SpArp2/3 complex or the homologous residues/atoms in BtArp2/3 complex. In Fig. 2c the inactive Arp2/3 complex structure was overlaid on subdomains 1 and 2 of Arp3 in the activated structure to show movement of Arp2. In Fig. 5a the activated and inactive Arp2/3 complex were superposed using subdomains 1 and 2 of Arp3 and the globular portion of ARPC2. In Fig. 5d, the αD helix of ARPC4 from each structure was superposed.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
EM maps and atomic models were deposited in the Electron Microscopy Data Bank and wwPDB, respectively with accession entries EMD-21502 and PDB 6W17 (Dip1–Arp2/3–actin ) and EMD-21503 and PDB 6W18 (inactive Arp2/3 complex), respectively. Corresponding atomic models were deposited in the Protein Data Bank under accession IDs 6W17 and 6W18.
Extended Data
Extended Data Fig. 1|. Cryo-Electron Microscopy of Arp2/3 complex in active and inactive states.
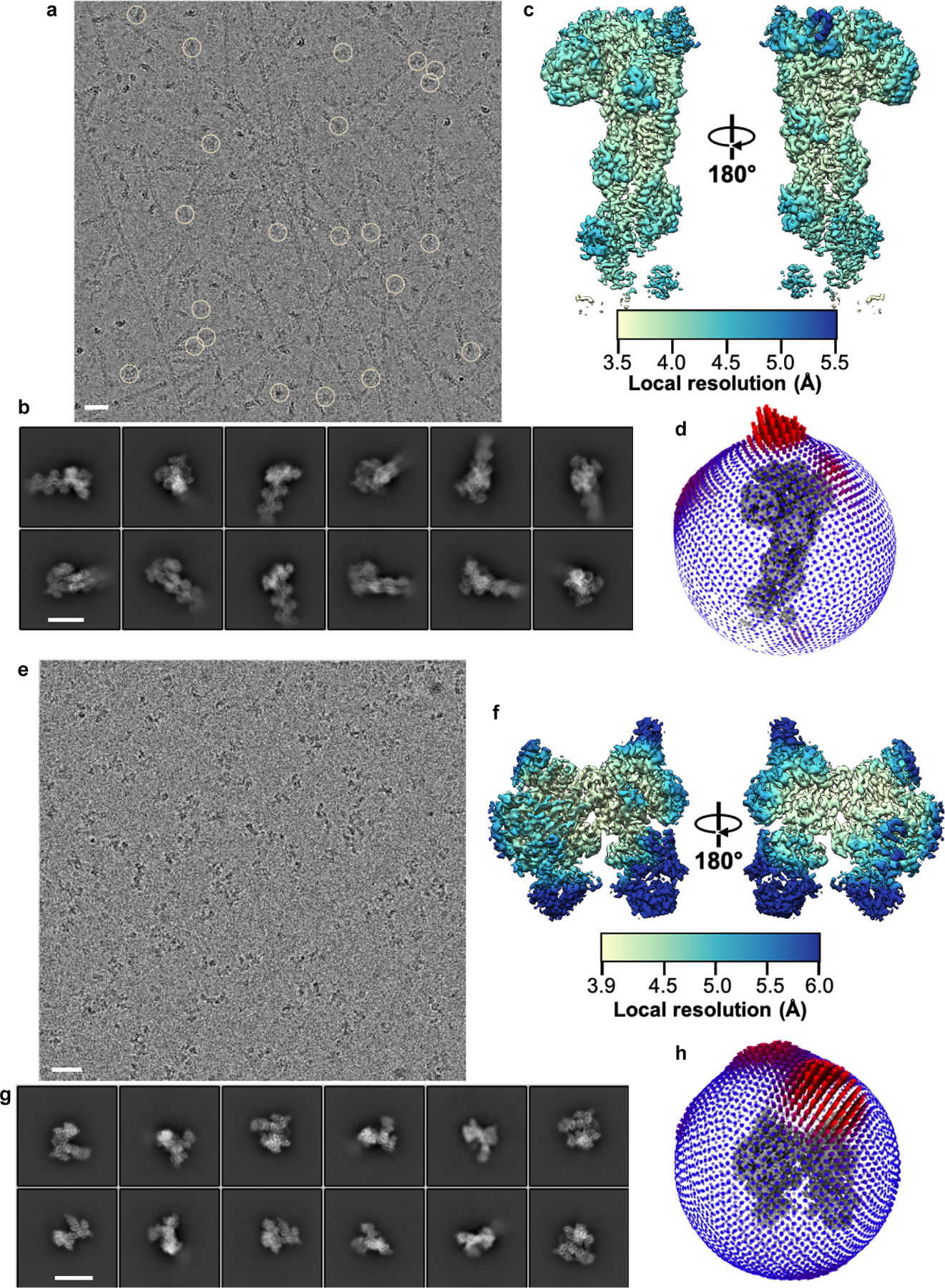
a, A representative micrograph containing actin filaments polymerized from Dip1-activated Arp2/3 complex. Circles mark the knob of density corresponding to Dip1-bound S. pombe Arp2/3 complex at the pointed ends of filaments. Dip1 and Arp2/3 complex bind weakly to the ends of spontaneously nucleated actin filaments but strongly to Dip1-Arp2/3-nucleated filaments34, so we concluded that most or all filaments with density for Arp2/3 complex were nucleated by Dip1-activated Arp2/3 complex. b, Reference-free 2D class averages obtained from particles corresponding to the knob shaped densities highlighted by circles in a. These class averages show Dip1-Arp2/3-actin-filament complex adopts different orientations in vitrified ice. c, Two different views of the reconstructed 3D map of active Arp2/3 complex bound to Dip1 and pointed end of nucleated actin filament, colored based on local resolution values of each voxel. d, Euler angle distribution plot for particles contributing to the reconstructed map in c (map colored in gray at the center). e, A micrograph of S. pombe Arp2/3 complex sample collected by tilting the stage to 40˚ showing uniform distribution of particles in ice. f, Representative reference free 2D class averages of Arp2/3 complex showing different orientations of the complex obtained from particles extracted from tilted and non-tilted micrographs. g, Two different rotated views of the 3D reconstruction of Arp2/3 complex in an inactive state colored based on per-voxel local resolution values. h, Plot showing the Euler angle distribution of particles that went into the final reconstructed inactive Arp2/3 complex map (colored in gray). Scale bars (white line) for the micrographs in a and e represent distance of 25 nm and for the 2D class averages b and f represent 12.5 nm distance.
Extended Data Fig. 2|. Single-particle cryo-EM data processing workflow for 3D reconstructions of activated and inactive Arp2/3 complex.
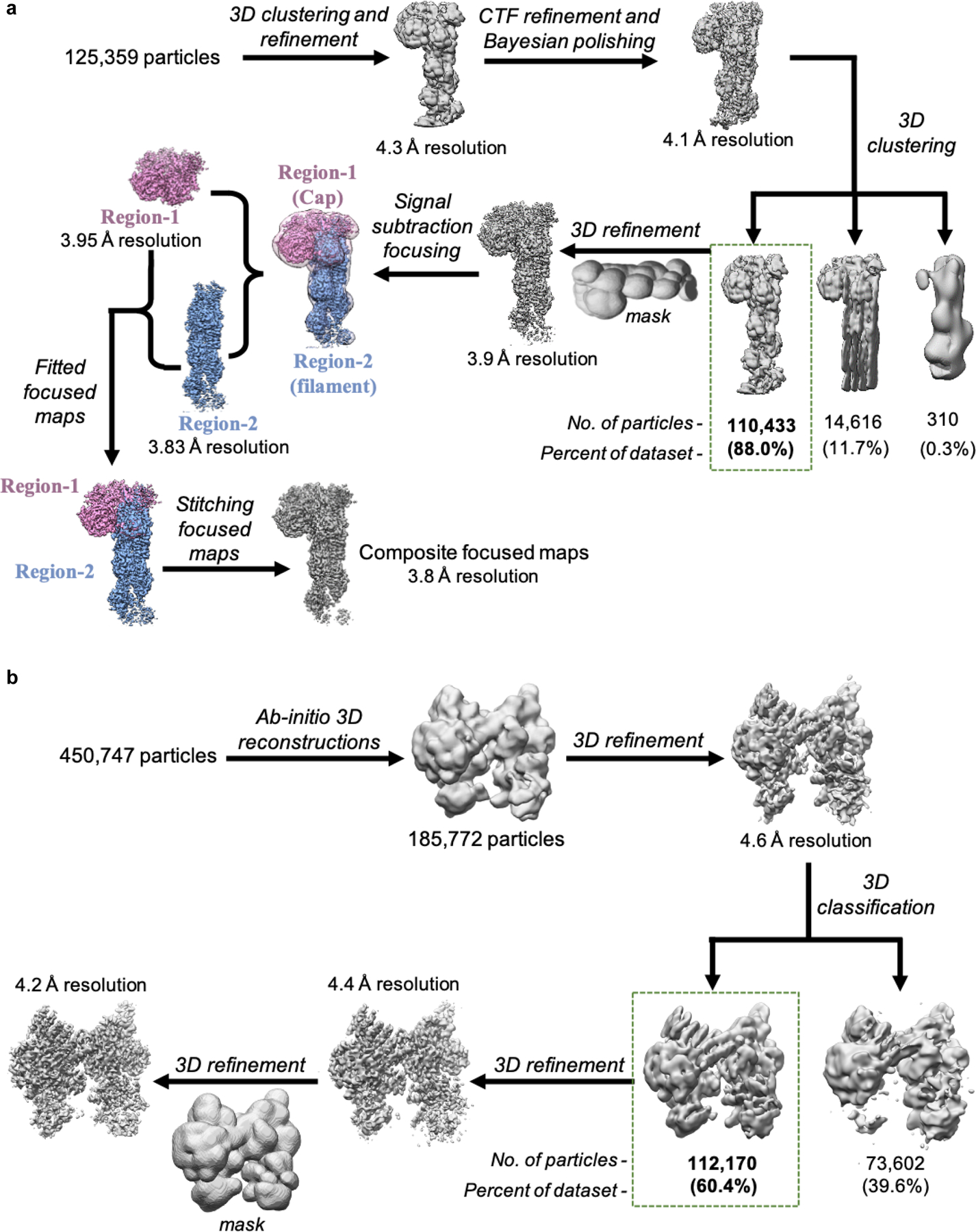
a, Data processing schematic for Dip1-Arp2/3-actin-filament complex. Particle stack was initially binned by factor of four and subjected to multiple rounds of 2D and 3D classification to discard particles that did not correspond to the complex. The cleaned particle stack was re-extracted without binning and further subjected to downstream 3D processing. All 3D classifications were performed without image alignment (referred to as clustering). Final composite map was generated by combining focused maps corresponding to the Dip1-Arp2/3 cap (Region-1, colored purple) and the Arp-actin filament (Region-2, colored blue). b, Schematic for processing of Arp2/3 complex (inactive state) cryo-EM data. Particles extracted from all micrographs (tilted and non-tilted) were cleaned by multiple rounds of 2D classification and then subjected to multiple ab initio 3D reconstructions. Particles corresponding to intact Arp2/3 complex were further subjected to downstream data processing that lead to a 4.2 Å 3D reconstruction of Arp2/3 complex in inactive state.
Extended Data Fig. 3|. Reconstructed maps and atomic models of subunits.
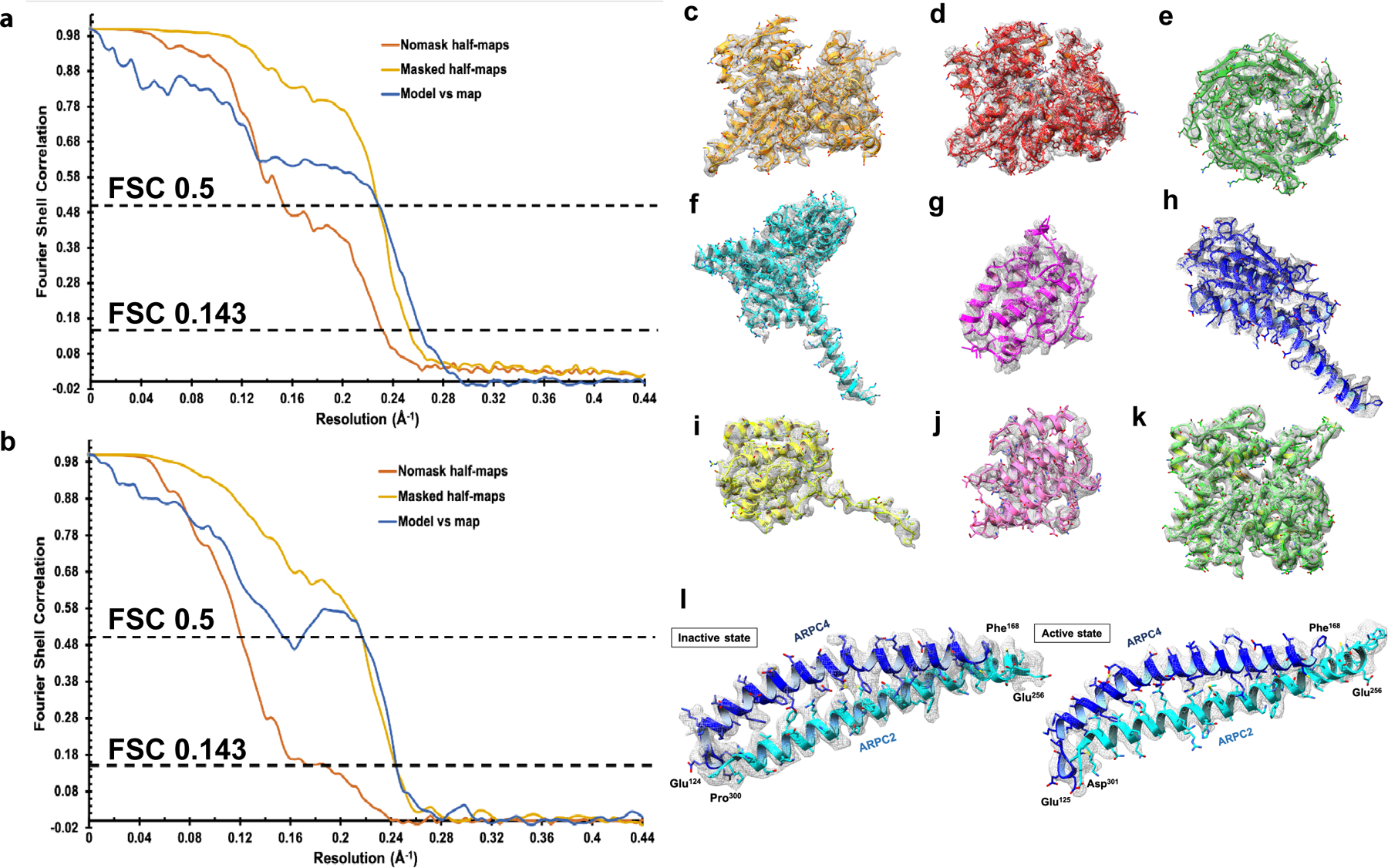
Fourier shell correlation (FSC) plots between masked (yellow) and unmasked (orange) half maps, and final map and model (blue) for global resolution estimates for a, Dip1–Arp2/3–actin-filament complex, and b, inactive Arp2/3 complex. EM density for each subunit is shown as gray mesh. c, Arp3, d, Arp2, e, ARPC1, f, ARPC2, g, ARPC3, h, ARPC4, i, ARPC5, j, Dip1, and k, actin maps with built models. l, Maps and models corresponding to the long helices from the “clamp” subunits, ARPC2 (cyan) and ARPC4 (blue) in the inactive state (left) and active state (right).
Extended Data Fig. 4|. The structure of inactive S. pombe Arp2/3 complex is nearly identical to other inactive Arp2/3 complex structures.
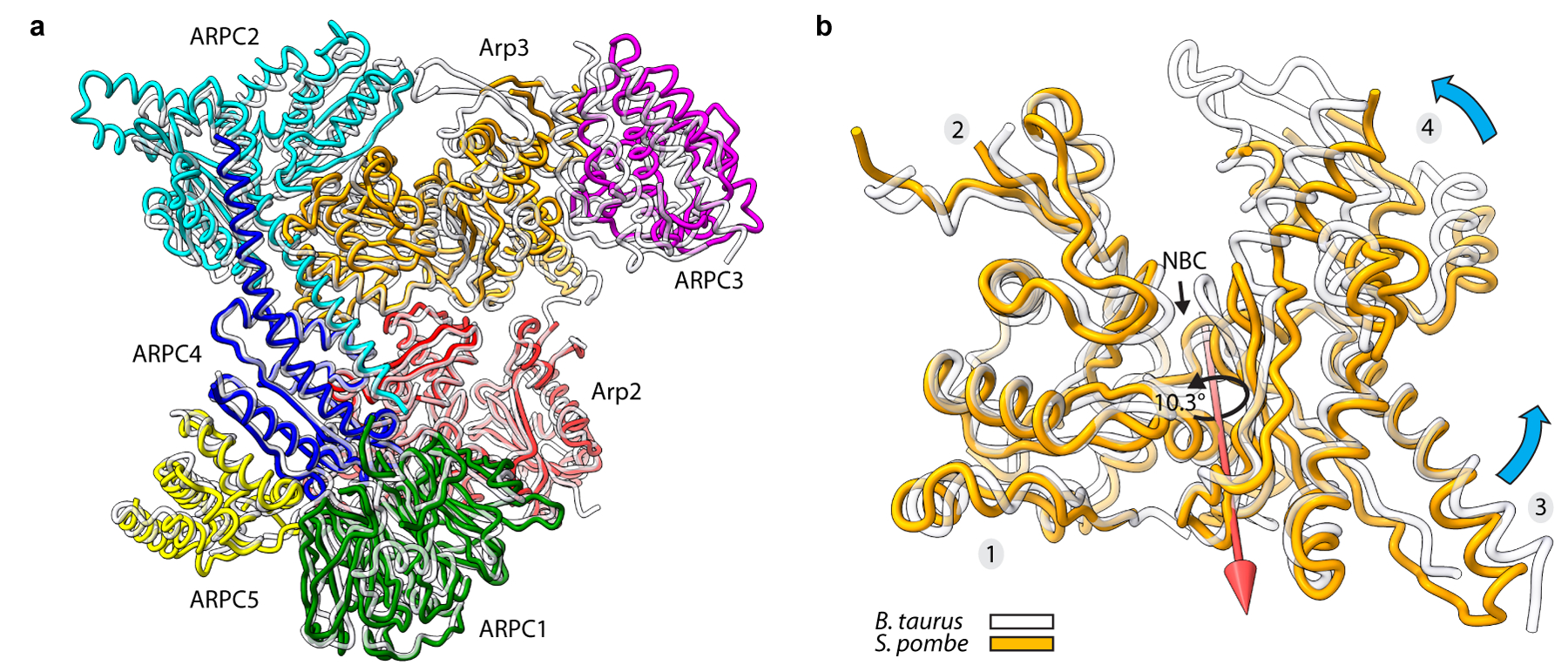
a, Structural superposition of inactive S. pombe Arp2/3 complex (colored subunits) from the cryo-EM reconstruction presented here with inactive Bos taurus Arp2/3 complex (semi-transparent gray subunits, PDB:4jd218). Structures were superposed using Arp3’s subdomains 1 and 2, Arp2, ARPC1, ARPC2 and ARPC4. b, Structural superposition showing that Arp3 from the S. pombe inactive Arp2/3 complex structure shows a more open nucleotide binding cleft than other ATP-bound structures. Subdomains 1 and 2 of inactive S. pombe Arp3 (orange) were superposed on inactive BtArp3 (transparent gray, 4jd2). Cyan arrow shows the direction of rotation of subdomains 3 and 4 toward subdomains 1 and 2 during cleft closure. Red arrow shows the axis of rotation of cleft closure. A rotation of 10.3º about this axis is required to close the S. pombe nucleotide cleft to the same extent as the BtArp2/3 structure (PDB 4jd2). The open cleft in the inactive S. pombe Arp2/3 complex is the major structural difference between the two structures.
Extended Data Fig. 5|. Comparison of the Arps in the activated complex to actin in the nucleated filament.
a, Superposition of Arp3 Cα atoms from the active structure to actin subunit n in the nucleated filament. The RMSD between 256 pruned atom pairs is 1.31 Å (across all 354 pairs: 3.45). b, Superposition of Arp2 Cα atoms from the active structure to actin subunit n in the nucleated filament. The RMSD between 314 pruned atom pairs is 0.92 Å (across all 352 pairs: 2.74).
Extended Data Fig. 6|. Partial flattening allows Arp3 to make short pitch interactions that mimic those of a short pitch actin dimer.
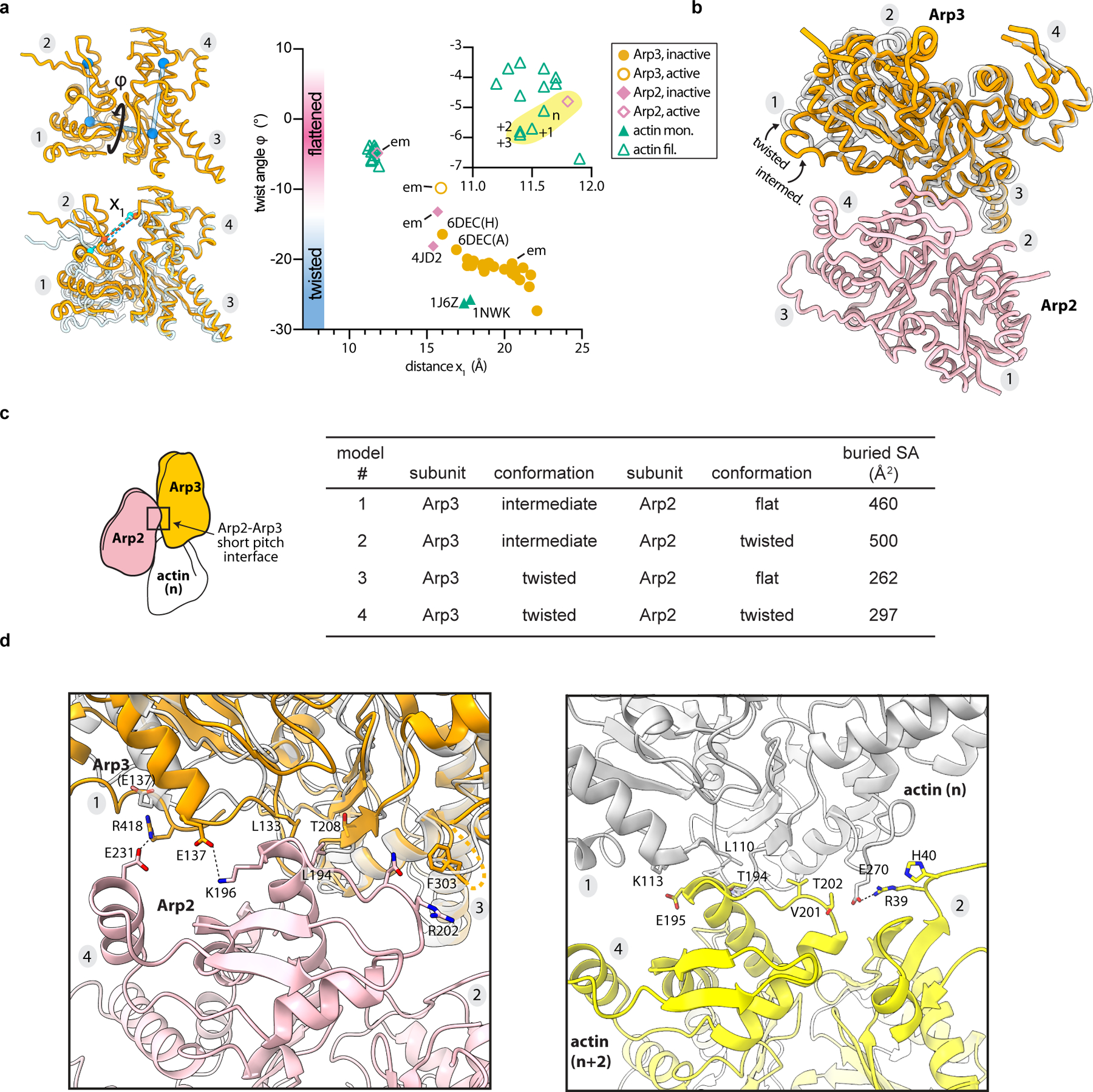
a, Left ,Arp3 from the active (orange) and inactive (semi-transparent gray) Arp2/3 complex structures, with definition of measurements in plot. Top, the twist angle (φ) is the dihedral angle between the centers of mass (blue spheres) of each of the four subdomains (see Online Methods). Bottom, distance x1 between Y92 Cα and K235 Cα in S. pombe Arp3. Right, plot of the twist (φ) versus distance x1 in inactive or active structures of actin, Arp2 or Arp3.. Data points highlighted in yellow are from the activated Arp2/3 structure. A subset of structures are labeled: em (from cryo-EM reconstructions here); 6dec (Spin90 bound to Bos taurus (Bt)Arp2/3 complex, chain ID in parentheses21); 4jd2 (BtArp2/3 complex bound to GMF18); 1nwk and 1j6z (representative actin monomer structures bound to ATP or ADP, respectively53,54). b, Superposition of inactive (twisted) Arp3 from the inactive S. pombe Arp2/3 complex cryo-EM structure and Arp3 from the activated structure overlaid using subdomains 3 and 4. Arp2 in the short pitch conformation from the activated structure is shown in pink. Partial flattening of Arp3 upon activation allows subdomains 1 and 2 to make closer contacts with Arp2 in the short pitch conformation. c, Surface area buried at the short pitch interface between Arp2 and Arp3 in the active structure (model 1) or theoretical models in which the Arps are modeled in the short pitch conformation and either Arp2 (model 2), Arp3 (model 3), or both (model 4) adopt the twisted conformation. Models 2, 3 and 4 were constructed by superposing either Arp2, Arp3, or both subunits from the inactive structure onto their corresponding subunit in the activated structure using Cα atoms from subdomains 3 and 4. d, Comparison of the short pitch interface between Arp3 and Arp2 (left panel) or actin (n) and actin (n+2) right panel. The 9° rotation of Arp2 brings the helix harboring E231 from Arp2 closer to subdomain 1 of Arp3, allowing E231 to interact with R418.
Extended Data Fig. 7|. Uncurling of the W-loop facilitates long-pitch interactions with actin.
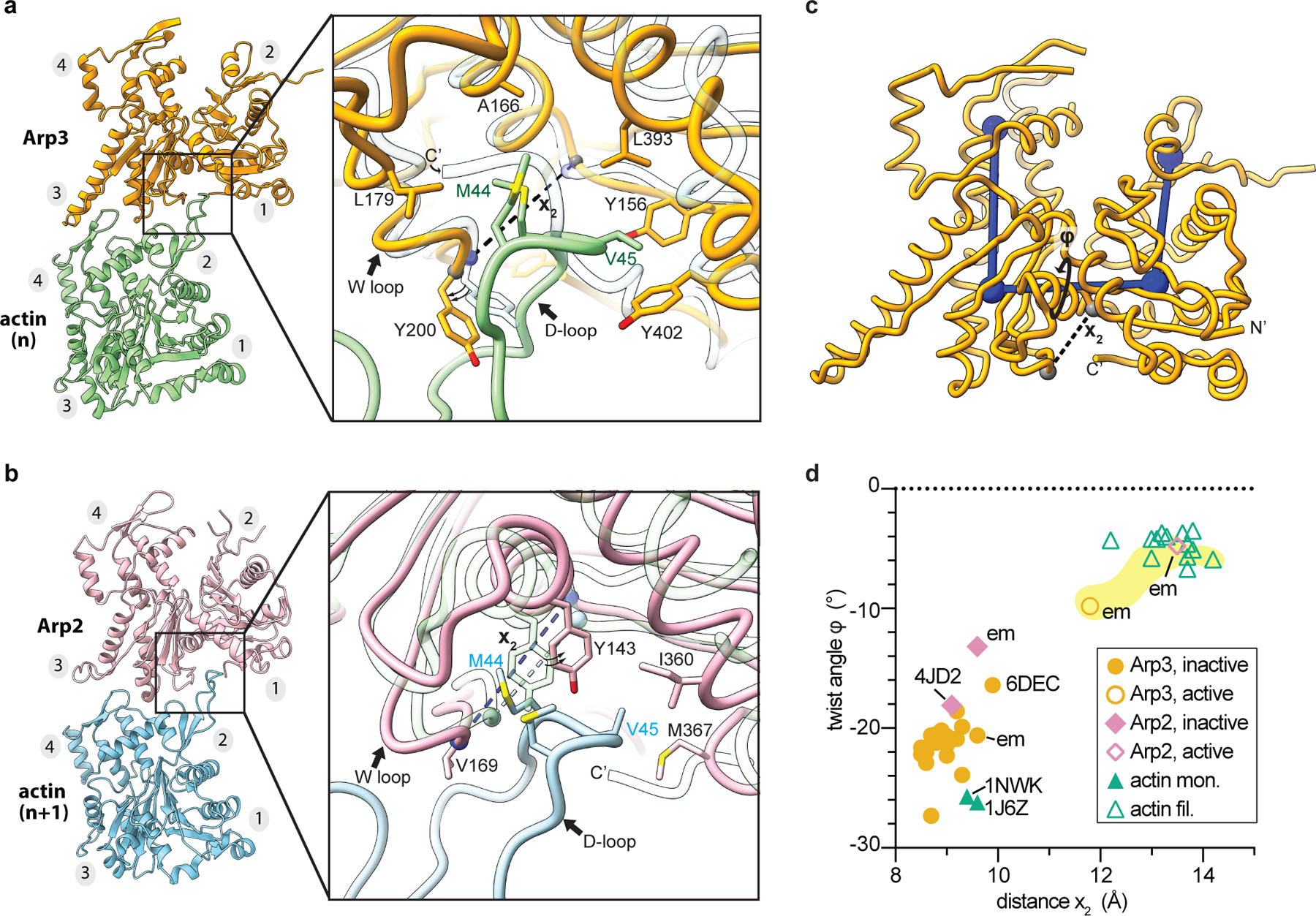
a, Close up of long pitch interactions between the Arp3 barbed end groove (BEG) and the actin D-loop. Activated Arp3 (orange) is overlaid on an inactive (transparent) structure using subdomains 3 and 4. A structure of Bos taurus Arp2/3 complex (PDB 4JD2) was used as the inactive structure in this and all analyses in this figure as some sidechains at the barbed end of Arp3 are missing in the inactive S. pombe Arp2/3 complex structure presented here. b, Close up of long pitch interactions between the Arp2 BEG in the activated (pink) or inactive (transparent) structure (PDB 4JD2). Arp2 from each structure was superposed using subdomains 3 and 4. c, Structure of Arp3 from activated Arp2/3 complex showing the definitions of the measurements made in panel d. d, Plot of the twist angle (φ) versus distance x2—which measures uncurling of the W loop—for inactive or active structures of actin, Arp2 or Arp3.: structures from the cryo-EM reconstructions presented here; 6dec: structure of Spin90 bound to Bos taurus (Bt)Arp2/3 complex26; PDB 4JD2: BtArp2/3 complex bound to GMF18; PDB 1NWK and PDB 1J6Z: representative actin monomer structures bound to ATP or ADP, respectively53,54.
Extended Data Fig. 8|. Flattening causes changes in the barbed end groove that facilitate long pitch interactions with actin subunits.
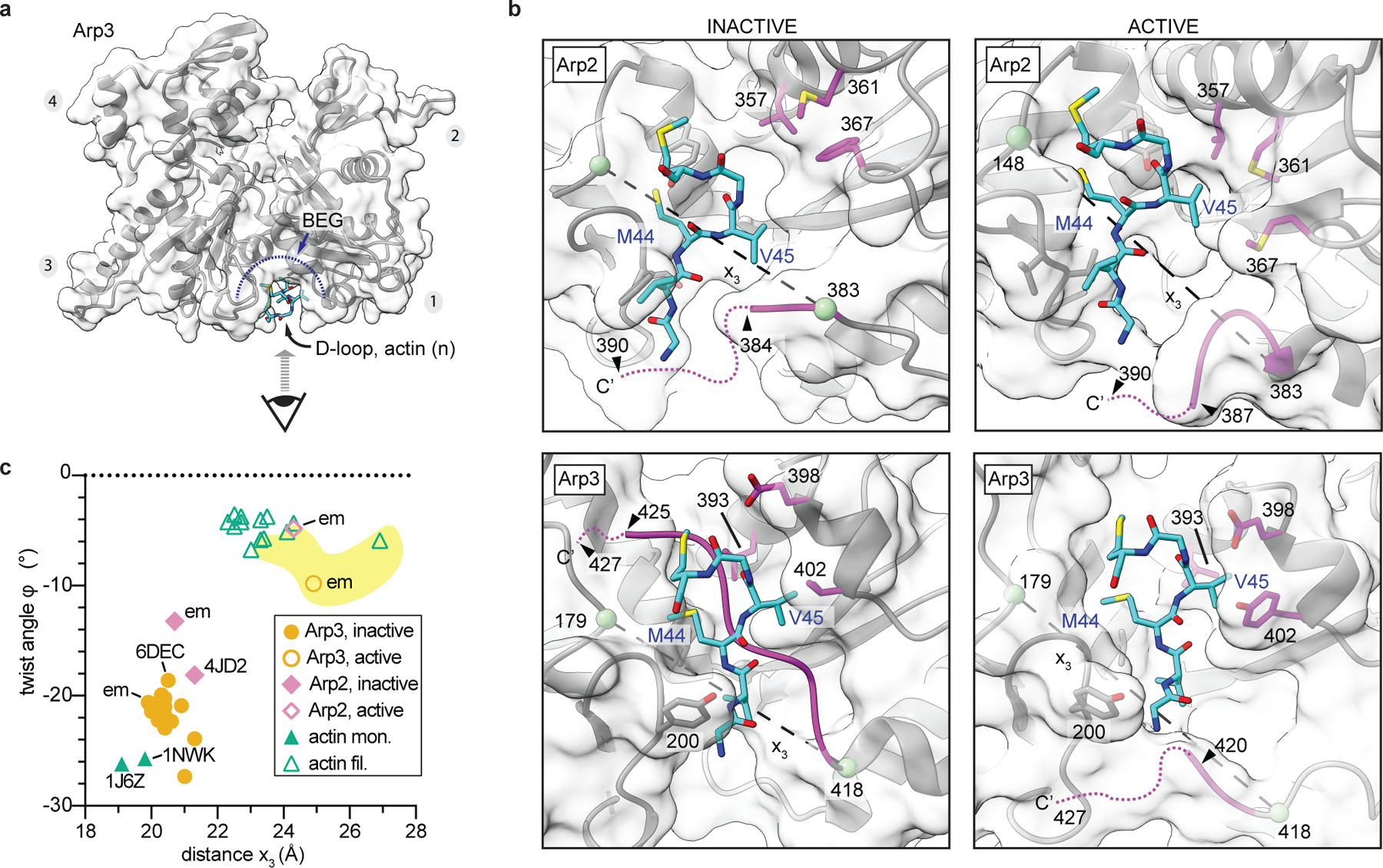
a, Molecular surface representation of active Arp3 showing perspective of close up views of the barbed end grooves (BEGs) depicted in b. b, BEG of inactive (PDB 4JD2) or active Arp2 or Arp3 with engaged D-loop from actin subunit n+1 or n. Residue numbers in left panels are for the S. pombe Arp2/3 complex. Green spheres and dashed line show distance x3, which measures the distance between the C-terminus and the front of the BEG. c, Plot of the twist angle versus distance x3 for inactive or active structures of actin, Arp2 or Arp3. Individually labeled data points are as described in Extended Data Fig. 7d.
Extended Data Fig. 9|. Close up of nucleotide clefts of Arp2 and Arp3.
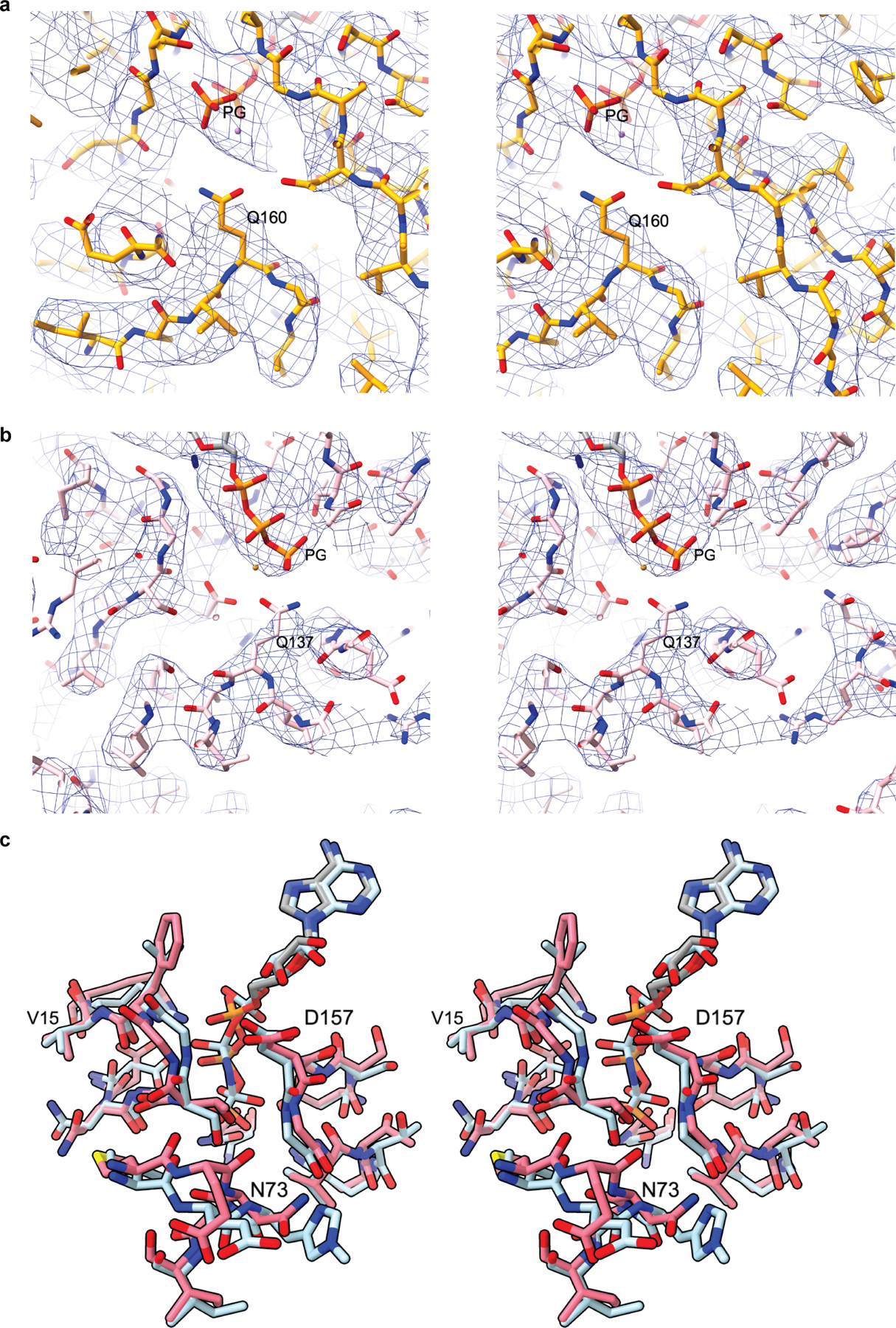
a, Stereo image of the nucleotide binding cleft of Arp3 showing the electron density of the modeled ATP phosphates and conserved catalytic residue Q160. b, Stereo image of the nucleotide binding cleft of Arp2 showing the electron density of the modeled ATP phosphates and conserved catalytic residue Q137. c, Stereo image of Arp2 from the activated structure (pink) superposed with actin (cyan) from the cryo-EM structure of AMP-PNP bound actin filaments15 showing the nucleotide and the nucleotide binding cleft.
Extended Data Fig. 10|. The clamp subunits twist during activation of Arp2/3 complex.
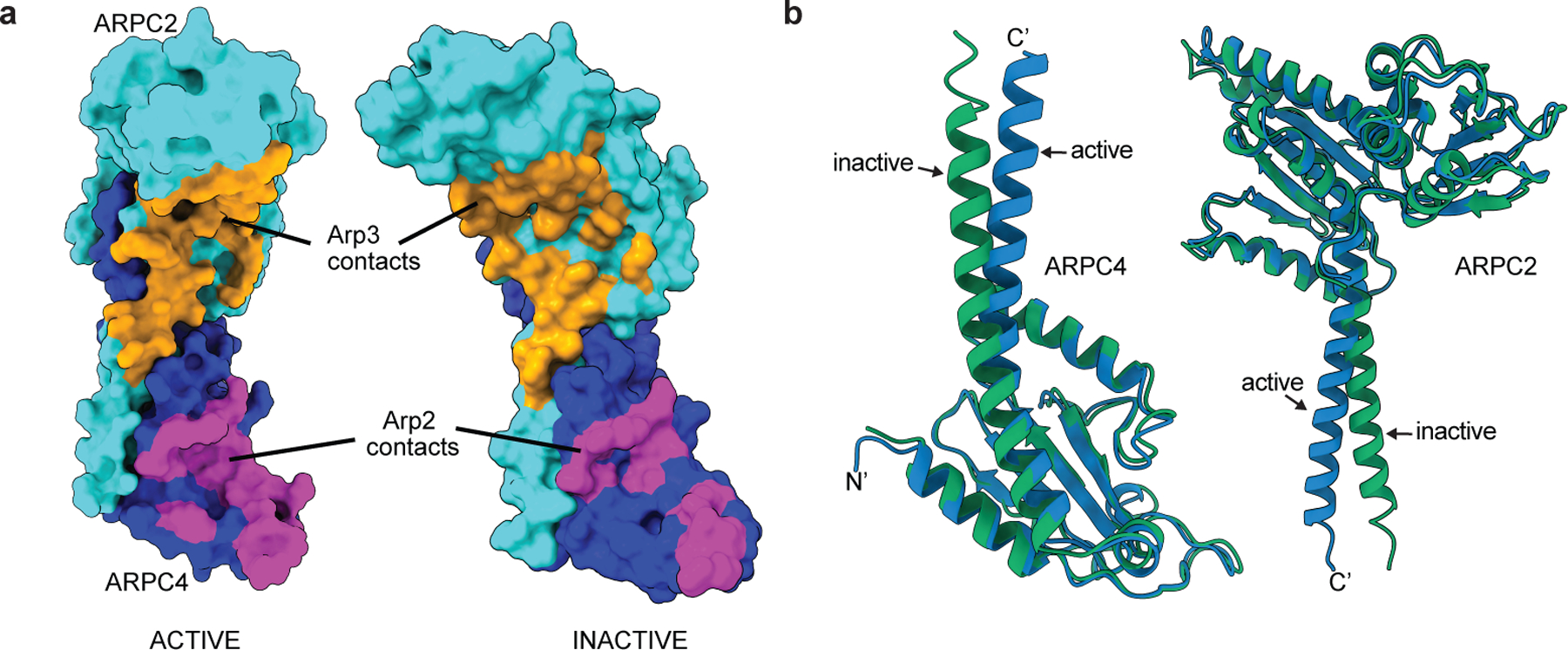
a, Model-based surface representation of ARPC2 and ARPC4 in active or inactive structure (PDB 4JD2) with residues contacting Arp2 or Arp3 (within 3.7 Å) colored magenta or orange, respectively. The Bos taurus inactive structure (PDB 4JD2) was used for the inactive structure in this analysis because the inactive S. pombe Arp2/3 complex structure presented here is missing some sidechains at the interface with the clamp. b, Overlay of ARPC2 and ARPC4 from the active structure on ARPC2 and ARPC4 from the inactive S. pombe Arp2/3 complex structure, respectively. Residues 1–271 of ARPC2 and 4–140 of ARPC4 were used to overlay the structures.
Extended Data Table 1.
| Sample name | Dip1-Arp2/3-Actin | Inactive Arp2/3 |
|---|---|---|
| EMDB ID | EMD-21502 | EMD-21503 |
| PDB ID | 6W17 | 6W18 |
| Microscope | Talos Arctica | Talos Arctica |
| Detector (Mode) | Falcon 3EC, counting mode | Falcon 3EC, counting mode |
| Voltage (kV) | 200 | 200 |
| Magnification (nominal) | 92,000X | 120,000X |
| Total electron fluence (e−/Å2) | 36.35 | 44.34 |
| Electron flux (e−/pixel/sec) | 0.76 | 0.85 |
| Defocus range (µm) | −1 – −1.75 | −0.7 – −1.3 |
| Pixel size (Å) | 1.12 | 0.8757 |
| Total exposure time (sec) | 60 | 40 |
| Total fractions/micrograph | 45 | 62 |
| Exposure per fraction (e−/Å2/fraction) | 0.81 | 0.72 |
| Micrographs collected (no.) | 5,109 | 5,309 |
| Total extracted particles (no.) | 3,500,000 | 3,336,981 |
| Refined particles (no.) | 2,346,578 | 450,747 |
| Final particles (no.) | 110,433 | 112,170 |
| Symmetry imposed | C1 | C1 |
| Global Resolution (Å) | ||
| FSC 0.5 (masked/unmasked) | 4.4/6.5 | 4.6/8.3 |
| FSC 0.143 (masked/unmasked) | 3.9/4.3 | 4.2/5.2 |
| Local resolution range (Å) | 3.5 – 5.5 | 3.9 – 6.0 |
| Map Sharpening B factors (Å2) | −45.12 | −116.219 |
| Model composition | ||
| Non-hydrogen atoms | 27,399 | 12,727 |
| Protein residues | 3,483 | 1,854 |
| Ligands | 6 Mg2+, 2 ATP, 4 ADP, 5 Phalloidin | Mg2+, 2 ATP |
| Refinement | ||
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.004 | 0.004 |
| Bond angles (°) | 0.967 | 0.983 |
|
| ||
| Validation | ||
| EMRinger score | 1.39 | 1.29 |
| MolProbity score | 1.65 | 1.42 |
| Clashscore | 5.67 | 4.66 |
| Poor rotamers (%) | 0.00 | 0.00 |
| Cβ deviations (%) | 0.00 | 0.00 |
| Ramachandran plot | ||
| Favored (%) | 95.00 | 96.96 |
| Allowed (%) | 5.00 | 3.04 |
| Disallowed (%) | 0.00 | 0.00 |
| CaBLAM outliers (%) | 4.2 | 3.8 |
Supplementary Material
Acknowledgments
Cryo-EM data were collected at the Stony Brook University (SBU) Cryo-EM center, and we thank G. Hu for providing support at this center. Computational infrastructures were provided by the cryo-EM and High-Performance Computing facilities at SBU. We thank N. Samaroo and B. Ding for help with experiments and acknowledge K. Prehoda and members of the Nolen and Chowdhury lab for comments on the manuscript. We are also grateful to G.C. Lander for providing access to the Scripps Research cryo-EM facility for determining feasibility of the project. We thank D. Kovar for the capping protein expression vector. This work was supported by SBU start-up funds to S.C and NIH grants R01GM092917 and R35GM136319 to B.J.N. The SBU cryo-EM center is supported by NIH grant S10 OD012272. M.S was supported by Fulbright Association.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Goley ED & Welch MD The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol 7, 713–726 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Higgs HN & Pollard TD Regulation of Actin Polymerization by Arp2/3 Complex and WASp/Scar Proteins. J. Biol. Chem 274, 32531–32534 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Wagner AR, Luan Q, Liu S-L & Nolen BJ Dip1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr. Biol 23, 1990–1998 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goley ED, Rodenbusch SE, Martin AC & Welch MD Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol. Cell 16, 269–279 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Rodal AA et al. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat. Struct. Mol. Biol 12, 26–31 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Boczkowska M, Rebowski G, Kast DJ & Dominguez R Structural analysis of the transitional state of Arp2/3 complex activation by two actin-bound WCAs. Nat Commun 5, 3308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodnick-Smith M, Liu S-L, Balzer CJ, Luan Q & Nolen BJ Identification of an ATP-controlled allosteric switch that controls actin filament nucleation by Arp2/3 complex. Nat Commun 7, 12226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodnick-Smith M, Luan Q, Liu S-L & Nolen BJ Role and structural mechanism of WASP-triggered conformational changes in branched actin filament nucleation by Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A 113, E3834–3843 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X-P et al. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J 31, 236–247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez Espinoza, Sofia Metskas, Ann Lauren, Chou Steven Z., Rhoades Elizabeth & Pollard Thomas D. Conformational changes in Arp2/3 complex induced by ATP, WASp-VCA and actin filaments. PNAS (2018) 10.1073/pnas.1717594115. [DOI] [PMC free article] [PubMed]
- 11.Zimmet A et al. Cryo-EM structure of NPF-bound human Arp2/3 complex and activation mechanism. Sci Adv 6, eaaz7651 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rouiller I et al. The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol 180, 887–895 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nolen BJ, Littlefield RS & Pollard TD Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc. Natl. Acad. Sci. U.S.A 101, 15627–15632 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson RC et al. Crystal structure of Arp2/3 complex. Science 294, 1679–1684 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Merino F et al. Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM. Nat. Struct. Mol. Biol 25, 528–537 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Chou SZ & Pollard TD Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc. Natl. Acad. Sci. U.S.A 116, 4265–4274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balzer CJ, Wagner AR, Helgeson LA & Nolen BJ Single-Turnover Activation of Arp2/3 Complex by Dip1 May Balance Nucleation of Linear versus Branched Actin Filaments. Curr. Biol 29, 3331–3338.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan YZ et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luan Q & Nolen BJ Structural basis for regulation of Arp2/3 complex by GMF. Nat. Struct. Mol. Biol 20, 1062–1068 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oda T, Iwasa M, Aihara T, Maéda Y & Narita A The nature of the globular- to fibrous-actin transition. Nature 457, 441–445 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Ingerman E, Hsiao JY & Mullins RD Arp2/3 complex ATP hydrolysis promotes lamellipodial actin network disassembly but is dispensable for assembly. J. Cell Biol 200, 619–633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin AC, Welch MD & Drubin DG Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat. Cell Biol 8, 826–833 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Vorobiev S et al. The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism. Proc. Natl. Acad. Sci. U.S.A 100, 5760–5765 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dayel MJ & Mullins RD Activation of Arp2/3 complex: addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol 2, E91 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Clainche C, Pantaloni D & Carlier M-F ATP hydrolysis on actin-related protein 2/3 complex causes debranching of dendritic actin arrays. Proc. Natl. Acad. Sci. U.S.A 100, 6337–6342 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalhaimer P & Pollard TD Molecular dynamics simulations of Arp2/3 complex activation. Biophys. J 99, 2568–2576 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luan Q, Liu S-L, Helgeson LA & Nolen BJ Structure of the nucleation-promoting factor SPIN90 bound to the actin filament nucleator Arp2/3 complex. EMBO J 37, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ti S-C, Jurgenson CT, Nolen BJ & Pollard TD Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A 108, E463–471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luan Q, Zelter A, MacCoss MJ, Davis TN & Nolen BJ Identification of Wiskott-Aldrich syndrome protein (WASP) binding sites on the branched actin filament nucleator Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A 115, E1409–E1418 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padrick SB, Doolittle LK, Brautigam CA, King DS & Rosen MK Arp2/3 complex is bound and activated by two WASP proteins. Proc. Natl. Acad. Sci. U.S.A 108, E472–479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hetrick B, Han MS, Helgeson LA & Nolen BJ Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem. Biol 20, 701–712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alekhina O, Burstein E & Billadeau DD Cellular functions of WASP family proteins at a glance. J. Cell. Sci 130, 2235–2241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basu R & Chang F Characterization of dip1p reveals a switch in Arp2/3-dependent actin assembly for fission yeast endocytosis. Curr. Biol 21, 905–916 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balzer CJ, Wagner AR, Helgeson LA & Nolen BJ Dip1 Co-opts Features of Branching Nucleation to Create Linear Actin Filaments that Activate WASP-Bound Arp2/3 Complex. Curr. Biol 28, 3886–3891.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jurgenson CT & Pollard TD Crystals of the Arp2/3 complex in two new space groups with structural information about actin-related protein 2 and potential WASP binding sites. Acta Crystallogr F Struct Biol Commun 71, 1161–1168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchand JB, Kaiser DA, Pollard TD & Higgs HN Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol 3, 76–82 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Smith BA et al. Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. Elife 2, e01008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheres SHW RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Zivanov J, Nakane T & Scheres SHW A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai X, Rajendra E, Yang G, Shi Y & Scheres SHW Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Källberg M, Margaryan G, Wang S, Ma J & Xu J RaptorX server: a resource for template-based protein structure modeling. Methods Mol. Biol 1137, 17–27 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Roy A, Kucukural A & Zhang Y I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afonine PV et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Cryst D 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou N, Wang H & Wang J EMBuilder: A Template Matching-based Automatic Model-building Program for High-resolution Cryo-Electron Microscopy Maps. Sci Rep 7, 2664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berman H, Henrick K, Nakamura H & Markley JL The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res 35, D301–303 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nolen BJ & Pollard TD Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol. Cell 26, 449–457 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nolen BJ & Pollard TD Structure and biochemical properties of fission yeast Arp2/3 complex lacking the Arp2 subunit. J. Biol. Chem 283, 26490–26498 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nolen BJ et al. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460, 1031–1034 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otterbein LR, Graceffa P & Dominguez R The crystal structure of uncomplexed actin in the ADP state. Science 293, 708–711 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Graceffa P & Dominguez R Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics. J. Biol. Chem 278, 34172–34180 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Krissinel E & Henrick K Inference of macromolecular assemblies from crystalline state. J. Mol. Biol 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
EM maps and atomic models were deposited in the Electron Microscopy Data Bank and wwPDB, respectively with accession entries EMD-21502 and PDB 6W17 (Dip1–Arp2/3–actin ) and EMD-21503 and PDB 6W18 (inactive Arp2/3 complex), respectively. Corresponding atomic models were deposited in the Protein Data Bank under accession IDs 6W17 and 6W18.


