Abstract
Background
Sarcopenia is defined as muscle wasting, characterized by a progressive loss of muscle mass and function due to ageing. Diagnosis of sarcopenia typically involves both muscle imaging and the physical performance of people exhibiting signs of muscle weakness. Despite its worldwide prevalence, a molecular method for accurately diagnosing sarcopenia has not been established.
Methods
We develop an artificial intelligence (AI) diagnosis model of sarcopenia using a published transcriptome dataset comprising patients from multiple ethnicities. For the AI model for sarcopenia diagnosis, we use a transcriptome database comprising 17 339 genes from 118 subjects. Among the 17 339 genes, we select 27 features as the model inputs. For feature selection, we use a random forest, extreme gradient boosting and adaptive boosting. Using the top 27 features, we propose a four‐layer deep neural network, named DSnet‐v1, for sarcopenia diagnosis.
Results
Among isolated testing datasets, DSnet‐v1 provides high sensitivity (100%), specificity (94.12%), accuracy (95.83%), balanced accuracy (97.06%) and area under receiver operating characteristics (0.99). To extend the number of patient data, we develop a web application (http://sarcopeniaAI.ml/), where the model can be accessed unrestrictedly to diagnose sarcopenia if the transcriptome is available. A focused analysis of the top 27 genes for their differential or co‐expression with other genes implied the potential existence of race‐specific factors for sarcopenia, suggesting the possibility of identifying causal factors of sarcopenia when a more extended dataset is provided.
Conclusions
Our new AI model, DSnet‐v1, accurately diagnoses sarcopenia and is currently available publicly to assist healthcare providers in diagnosing and treating sarcopenia.
Keywords: Sarcopenia, Muscle wasting, Artificial intelligence, Transcriptome, Diagnosis
Introduction
Ageing involves progressive changes in an individual's physiology. Sarcopenia, defined as muscle loss, is a leading cause of frailty in the elderly 1 , 2 and hence affects human health by potentiating disease occurrence leading to type 2 diabetes, 3 heart and respiratory diseases and insufficiency in mechanical support. 3 , 4 , 5 , 6
The pathogenesis of sarcopenia is associated with ageing, which may impede either anabolic signalling to build components of the muscle, regenerative activity to repair damaged tissues, intricate circuits to deliver nervous signals in and out of the muscle, or cellular surveillance to maintain energy flexibility of the body. 7 , 8 , 9 , 10 In addition, malnutrition and immobility affect the onset and degree of sarcopenia. 11 , 12 , 13 Such phenomena may occur individually or simultaneously to induce muscle atrophy when imbalanced or out of control, thereby resulting in causal complexity and difficulty in treating sarcopenia. 2 Currently, no efficient drug for treating sarcopenia is available, 2 , 7 although efforts have been expended to identify biomarkers for early diagnosis to develop preventive medicines or alleviate sarcopenia. A recent study using multi‐ethnic aged muscle biopsies exemplifies the nuclear‐encoded mitochondrial genes as a typical gene set that can be applied across ethnicities. 14 In addition, serum biomarkers that can reliably predict sarcopenia has been reported. 15 Candidate biomarkers were selected from the literature, followed by comparative biochemical analyses of sera between sarcopenia and normal elderly groups. Four combined biomarkers indicated higher diagnostic accuracy for sarcopenia than individual biomarkers, thereby re‐emphasizing the causal complexity contributing to sarcopenia. Therefore, the identification of additional biomarkers that reliably reflect sarcopenia may warrant early diagnosis to facilitate the deployment of preventative medicine and early intervention therapy. 16
In this study, we aimed to develop an artificial intelligence (AI) model for sarcopenia diagnosis using a previously published transcriptome dataset that contain differentially expressed genes in muscle biopsies from patients with sarcopenia and age‐matched healthy individuals across three ethnic groups. The AI model developed in this study yielded a diagnostic accuracy of >94%, which is an unprecedented level. To the best of our knowledge, our current study is the first attempt to develop an AI model to diagnose sarcopenia based on a transcriptome dataset only.
Materials and methods
Datasets
We used a published transcriptomic dataset deposited in the Gene Expression Omnibus of the National Center for Biotechnology (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE111017. 14 (Table S1) summarizes the subject information from the dataset. A total of 118 subjects (mean age, 73.34 ± 5.43) participated in this study, of whom 86 were healthy and 32 were sarcopenic. Subjects of three different races from different studies participated in the study: 40 subjects from the Hertfordshire sarcopenia study (HSS; 36 healthy vs. 4 sarcopenic), 39 from the Jamaica sarcopenia study (JSS; 30 healthy vs. 9 sarcopenic) and 39 from the Singapore sarcopenia study (SSS; 20 healthy vs. 19 sarcopenic). Transcriptome analysis includes 17 339 genes, which can be potential biomarkers for the diagnosis of sarcopenia (Data S1).
For hold‐out validation, we segmented the data into training (80%) and testing (20%) datasets in a stratified manner based on both the race (HSS, JSS, and SSS) and outcome (normal and sarcopenic). Accordingly, we used 94 subjects as the training dataset (69 normal and 25 sarcopenic) and 24 data as the testing dataset (17 normal and 7 sarcopenic). The testing dataset was isolated and used only to evaluate the performance of the proposed AI model. For box or scatter plots, data are shown as median ± interquartile. *P < 0.05, **P < 0.01, ***P < 0.001; P values calculated using either two‐tailed Wilcoxon rank sum test or two‐way ANOVA followed by Tukey's multiple comparisons test as described. This study was approved by Institutional Review Board Wonkwang University (WKIRB‐202108‐SB‐060).
Preprocessing
In the dataset, each of the 17 339 genes can be the feature for the AI model. Some features (gene information) were missing from the training and testing datasets (Supporting information, Figure S1). To manage the missing features, we calculated the mean value from the training dataset for each feature and replaced the missing feature with the mean value in both the training and testing datasets. Subsequently, we standardized the dataset, which is a typical requirement for machine learning algorithms. The standardization changes the data distribution of each feature with a mean of zero and a standard deviation of one.
| (1) |
where mean(train) and SD(train) are the mean and standard deviation values, respectively, for each feature from the training dataset. Standardization was applied to both the training and testing datasets.
Feature selection
To select important features that affect clinical severity, we investigated the contribution of each of the 17 339 input variables on sarcopenia diagnosis via feature importance analysis using a random forest (RF), 17 extreme gradient boosting (XGBoost), 18 and adaptive boosting (AdaBoost) 19 , 20 algorithms.
By repeating the five‐fold cross‐validation 10 times, we obtained the best hyperparameters. For AdaBoost, we set the hyperparameters as follows: number of tree estimators, 200; learning rate, 0.2. For the RF, we set the number of tree estimators to 100, maximum depth to four and maximum features to five. For XGBoost, we set the maximum depth to four, learning rate to 0.1, number of tree estimators to 100, regularization parameter α to 1.0, fraction of observations to 0.9 and fraction of columns to 0.9.
Based on the resultant 50 sets of feature importance values for each classifier (AdaBoost, RF and XGBoost), we averaged the values and normalized them such that the importance values from each classifier ranged from zero to one. Next, we averaged the importance values for the final ranked feature importance values. Finally, we determined the optimal number of top features to be incorporated into the AI diagnosis model based on the cross‐validation results.
Development of artificial intelligence model based on deep neural network
A deep neural network (DNN) was used to develop the final AI model for sarcopenia diagnosis. In the DNN approach, we investigated up to five hidden layers, and each layer depth (node) up to the previous layer depth (node). For the input layer, we first ranked the features based on their importance and increased the number of top features used in the input layer. Accordingly, we used the top 27 features as input layers. For the fully connected (FC) layers as hidden layers, we applied dropouts by changing the dropout rate from 0 to 0.5, at increments of 0.1. The last FC layer was fed into a sigmoid layer, which is an output layer that provides the probabilities of patient severity. We trained the models using the ADAM optimizer and binary cross‐entropy cost function with a learning rate of 0.0001 and a batch size of 64.
For each set of top features, we obtained the best cross‐validation accuracy using two metrics, that is, the area under the curve and the balanced accuracy (Equation 2).
| (2) |
Based on the cross‐validation accuracy analysis, we modelled a four‐layer DNN using the top 27 features, as shown in (Figure 1). The four‐layer DNN comprised an input layer, two FC layers as hidden layers, and an output layer. The input layer was fed into a series of two FC layers comprising 27 and 8 nodes, respectively. In the two FC layers, we used a dropout rate of 0.5. Subsequently, the last FC layer was fed into the sigmoid layer. Our proposed DNN model was named DSnet‐v1, which represents the DNN for sarcopenia diagnosis version 1.
Figure 1.
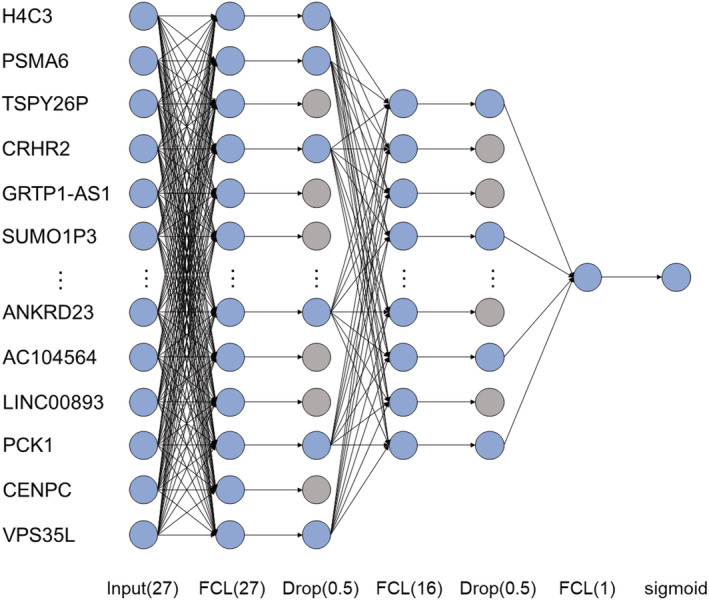
Proposed DSnet‐v1 with four‐layer deep neural network (DNN) for the diagnosis of sarcopenia.
Implementation
We implemented and trained the DNN using TensorFlow (version: tensorflow‐gpu 2.0), whereas we used NumPy (version: 1.16.4), Pandas (version: 0.25.3), Matplotlib (version 3.1.2) and Scikit‐learn (version 0.22.1) to build the model and analyse the results. We trained the models using the ADAM optimizer and the binary cross‐entropy cost function shown in Equation 3 by adjusting the learning rate to 0.0005 and 0.0001 and using a batch size of 64 on an NVIDIA GeForce GTX 1080 Ti GPU.
| (3) |
where y i is the label (1 for sarcopenia and 0 for normal) and p(y i ) is the classified probability of each subject being sarcopenic for a batch size comprising N patients.
Performance evaluation of AI models
To evaluate the performance of DSnet‐v1 for the diagnosis of sarcopenia, we used accuracy metrics of sensitivity, specificity, accuracy, balanced accuracy and area under the receiver operating characteristics (AUROC).
In the training dataset, our DSnet‐v1 was evaluated based on 5‐fold stratified cross‐validation. Subsequently, the diagnostic performance was evaluated independently using an isolated testing dataset. To compare the performance of DSnet‐v1 with those of other external AI models, we separately trained the models of RF, XGBoost and AdaBoost, each of which was evaluated with hyperparameter search.
Box plots, correlation and network analysis
The evaluation and visualization of gene expression (boxplots), Spearman's correlation (correlogram matrices), and gene network for 27 AI‐featured genes and their associated genes were conducted using R packages ggpubr, ggplot2, igraph, ggraph, egg, corrr, corrplot, dplyr, tidyverse, and reshape (https://www.r‐project.org). The network visualization of human phenotype ontology was assessed using Enrichr. 21
Public website deployment
We deployed DSNet‐v1 on a public web server (http://sarcopeniaAI.site/) through Amazon Web Services (AWS), which provides secure, durable and scalable service. After accessing the website, a user enters the 27 genes, which are encoded to the website server and can immediately obtain the diagnosis result of sarcopenia. There is no need to enter any private information other than gene information, and the entered information is immediately deleted when the diagnosis result is derived, so there is no risk of information exposure.
Results
Feature selection and cross‐validation
(Table 1) summarizes the results of the ranked feature importance from the RF, XGBoost, and AdaBoost, as well as their combination: We ranked the top 27 features based on the combination among the 17,339 features. The RF results indicated that GRTP1‐AS1 possessed the highest importance value, followed by SUMO1P3, TEX261, SMIM26, and H4C3. The XGBoost results indicated that H4C3 possessed the highest importance value, followed by PSMA6, AC002070.1, PCK1, and CENPC. The AdaBoost results indicated that TSPY26P possessed the highest importance value, followed by STAG3L3, CRHR2, PEF1, and FKBP1C. By averaging the values obtained from the three models, H4C3 exhibited the highest importance value, followed by PSMA6, TSPY26P, CRHR2, and GRTP1‐AS1. The full list of ranked feature importance values is summarized in (Data S2).
Table 1.
Feature importance
| Rank | Feature name | Gene name/ensembl gene ID | Random forest | XGBoost | AdaBoost | Mean |
|---|---|---|---|---|---|---|
| 1 | H4C3 | H4 clustered histone 3 | 0.5715 | 1.0000 | 0.2857 | 0.6191 |
| 2 | PSMA6 | Proteasome subunit alpha 6 | 0.2263 | 0.8000 | 0.4286 | 0.4850 |
| 3 | TSPY26P | Testis specific protein, Y‐linked 26, pseudogene | 0.0000 | 0.0667 | 1.0000 | 0.3556 |
| 4 | CRHR2 | Corticotropin releasing hormone receptor 2 | 0.0943 | 0.2000 | 0.7143 | 0.3362 |
| 5 | GRTP1‐AS1 | Growth hormone regulated TBC protein 1‐antisense | 1.0000 | 0.0000 | 0.0000 | 0.3333 |
| 6 | SUMO1P3 | SUMO1 Pseudogene 3 | 0.9620 | 0.0000 | 0.0000 | 0.3207 |
| 7 | STAG3L3 | Stromal antigen 3‐like 3, transcribed_unprocessed_pseudogene | 0.0000 | 0.0000 | 0.8571 | 0.2857 |
| 8 | KAT2A | Lysine acetyltransferase 2A | 0.4174 | 0.0000 | 0.4286 | 0.2820 |
| 9 | PEF1 | Penta‐EF‐hand domain containing 1 | 0.0000 | 0.0667 | 0.7143 | 0.2603 |
| 10 | SMIM26 | Small integral membrane protein 26 | 0.5975 | 0.1333 | 0.0000 | 0.2436 |
| 11 | FKBP1C | FKBP prolyl isomerase family member 1C | 0.0000 | 0.0000 | 0.7143 | 0.2381 |
| 12 | TEX261 | Testis expressed 261 | 0.7084 | 0.0000 | 0.0000 | 0.2361 |
| 13 | PFKFB4 | 6‐Phosphofructo‐2‐kinase/fructose‐2,6‐biphosphatase 4 | 0.2055 | 0.3333 | 0.1429 | 0.2272 |
| 14 | AC116913.1 | No NCBI gene ID yet, novel noncoding transcript, antisense to MAP 2 K1 and SNAPC5/ENSG00000261351 | 0.2651 | 0.4000 | 0.0000 | 0.2217 |
| 15 | TBC1D8 | TBC1 domain family member 8 | 0.0690 | 0.0000 | 0.5714 | 0.2135 |
| 16 | MYF5 | Myogenic factor 5 | 0.0000 | 0.3333 | 0.2857 | 0.2063 |
| 17 | TPSAB1 | Tryptase alpha/beta 1 | 0.1825 | 0.0000 | 0.4286 | 0.2037 |
| 18 | AC002070.1 | LOC105370027/ENSG00000248636 | 0.0000 | 0.4667 | 0.1429 | 0.2032 |
| 19 | RASSF1 | RAS association domain family member 1 | 0.3554 | 0.2000 | 0.0000 | 0.1851 |
| 20 | AC006971.1 | No NCBI gene ID yet, novel noncoding transcript, ARHGAP5 pseudogene/ENSG00000218586 | 0.0000 | 0.2667 | 0.2857 | 0.1841 |
| 21 | SNX12 | Sorting nexin 12 | 0.4790 | 0.0000 | 0.0000 | 0.1597 |
| 22 | ANKRD23 | Ankyrin repeat domain 23 | 0.0000 | 0.3333 | 0.1429 | 0.1587 |
| 23 | AC104564.5 | No NCBI gene ID yet, novel noncoding transcript/ENSG00000265625 | 0.0430 | 0.0000 | 0.4286 | 0.1572 |
| 24 | LINC00893 | Long intergenic non‐protein coding RNA 893 | 0.0430 | 0.0000 | 0.4286 | 0.1572 |
| 25 | PCK1 | Phosphoenolpyruvate carboxykinase 1 | 0.0000 | 0.4667 | 0.0000 | 0.1556 |
| 26 | CENPC | Centromere protein C processed_pseudogene | 0.0000 | 0.4667 | 0.0000 | 0.1556 |
| 27 | VPS35L | VPS35 endosomal protein sorting factor like | 0.1680 | 0.0000 | 0.2857 | 0.1512 |
We investigated the cross‐validation performance using metrics of sensitivity, specificity, accuracy, balanced accuracy, and AUROC. (Figure 2) shows the values of accuracy, balanced accuracy, and AUROC based on each number of selected top features. Detailed results are summarized in (Table S2). For the accuracy metrics, we identified the optimal DNN, including the hyperparameters for each selected feature. The results show that all the accuracy metrics, that is, accuracy, balanced accuracy and AUROC, increased as the number of selected features increased until 27. As the number of selected features exceeded 27, all accuracy metrics decreased as the number of selected features increased. For the top 27 features, we obtained a sensitivity of 0.88, specificity of 0.97, accuracy of 0.95, balanced accuracy of 0.93, and AUROC of 0.97.
Figure 2.
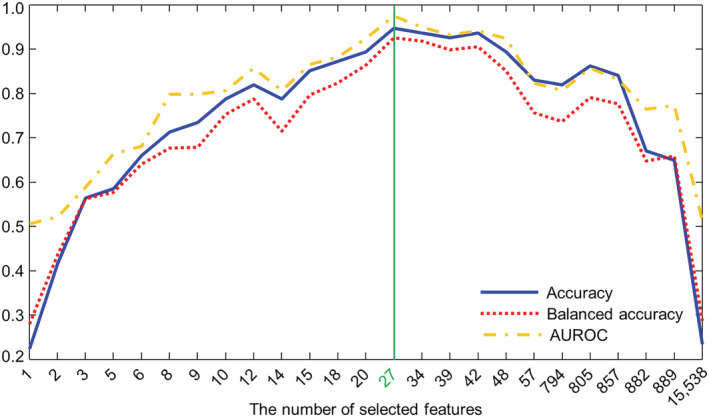
Accuracy, balanced accuracy and area under the receiver operating characteristics (AUROC) according each different number of selected top features.
(Table 2) summarizes the comparison of the cross‐validation accuracy. The results show that DSnet‐v1 provided the highest values of all accuracy metrics.
Table 2.
Comparison of cross‐validation evaluation metrics (mean ± standard deviation)
| Model | Cross‐validation results | |||
|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | Balanced accuracy (%) | |
| RF | 0.7448 ± 0.15832 | 0.8790 ± 0.1441 | 0.8503 ± 0.0.096 | 0.8119 ± 0.0813 |
| XGBoost | 0.7162 ± 0.1866 | 0.86485 ± 0.1415 | 0.8292 ± 0.1153 | 0.7905 ± 0.1014 |
| AdaBoost | 0.7848 ± 0.1705 | 0.8971 ± 0.0674 | 0.8719 ± 0.0487 | 0.8418 ± 0.0593 |
| DNN | 0.8772 ± 0.1072 | 0.9825 ± 0.0317 | 0.9583 ± 0.0439 | 0.9286 ± 0.0596 |
DNN, deep neural network; RF, random forest.
Performance of proposed DNN
Using the isolated testing dataset (n = 4), DSnet‐v1 showed a sensitivity of 1.00, specificity of 0.94, accuracy of 0.96, balanced accuracy of 0.97, and AUROC of 0.99. (Table 3) shows the diagnostic performances of various AI models; as shown, our proposed four‐layer DNN provided a higher accuracy, more balanced accuracy, and higher AUROC values than the other external AI models: the RF, XGBoost, and AdaBoost. (Figure S2 ) shows the ROC curves for model comparison.
Table 3.
Comparison of prediction performances among prediction models in test dataset
| Model | TN | FP | FN | TP | Sensitivity | Specificity | Accuracy | Balanced accuracy | AUROC |
|---|---|---|---|---|---|---|---|---|---|
| RF | 13 | 4 | 1 | 6 | 0.8571 | 0.7647 | 0.7917 | 0.8109 | 0.7479 |
| XGBoost | 12 | 5 | 1 | 6 | 0.8571 | 0.7059 | 0.7500 | 0.7815 | 0.7563 |
| AdaBoost | 14 | 3 | 1 | 6 | 0.8571 | 0.8235 | 0.8333 | 0.8403 | 0.8319 |
| DNN | 16 | 1 | 0 | 7 | 1.0000 | 0.9412 | 0.9583 | 0.9706 | 0.9916 |
AUROC, area under the receiver operating characteristics; DNN, deep neural network; RF, random forest.
The web application provides the probability of sarcopenia, as shown in (Figure 3). A user inputs his or her quantized gene information (as shown in Figure 3A), and the diagnosis results are presented (as shown in Figure 3B).
Figure 3.
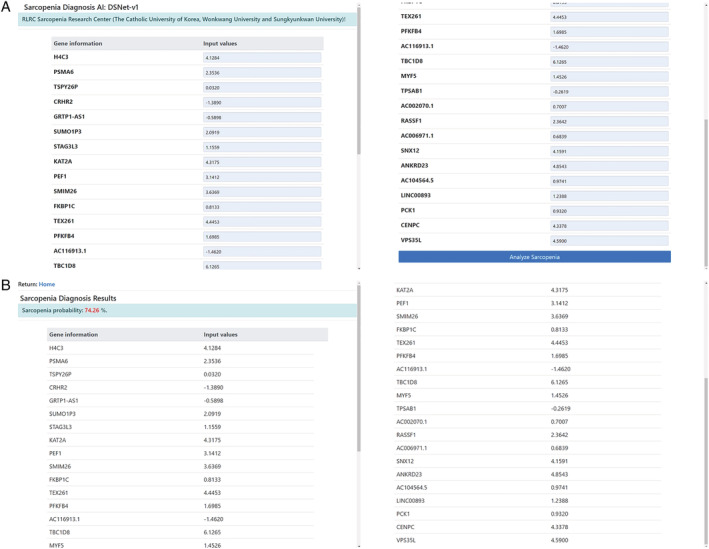
Developed AI model (DSNet‐v1) was successfully deployed on a public website (http://sarcopeniaAI.ml/)
Biological relevance of 27 AI‐featured genes
We evaluated the gene expression of 27 selected features (genes) in patients with sarcopenia (green box) and in age‐matched healthy individuals (white box) (Figure 4A). The gene expression of each individual for the three races (HSS, JSS and SSS) is indicated by red, blue and green dots, respectively. As shown in (Figure 4), the expression of seven genes (PCK1, AC116913.1, CENPC, KAT2A, RASSF1, PSMA6 and PEF1) was elevated in the sarcopenic muscle, whereas 10 genes (SNX12, TEX261, H4C3, PFKFB4, SUMO1P3, SMIM26, MYF5, AC002070.1, GRTP1‐AS1 and CRHR2) showed reduced expression. The remaining 10 genes did not show statistical differences between patients with sarcopenia and age‐matched healthy individuals. Twenty‐seven AI‐featured genes were positively (blue) or negatively (red) correlated with each other (the depth of colour for Spearman's rho between 0.0 and 0.5) (Figure 4B).
Figure 4.
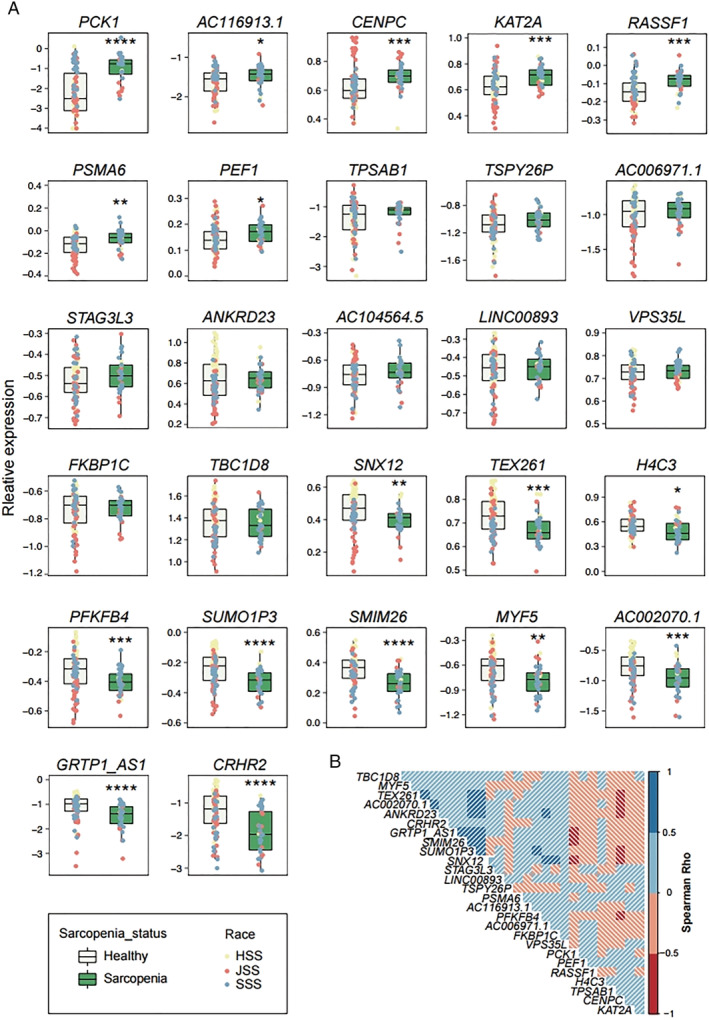
The visualization of expression and correlation of 27 AI‐featured gene. (A) Boxplots showing the relative expression of each gene in the healthy (white box) and sarcopenic elderly (green box). Yellow (HSS), pink (JSS) or blue (SSS) dots represent the gene expression of each individual subject in three races. Data are median ± interquartile. *P < 0.05, **P < 0.01, ***P < 0.001; P values calculated using two‐tailed Wilcoxon rank sum test. (B) Correlogram matrices display Spearman's rho between the genes facing each side of the square. The depth of the shading at the correlation matrices indicates the magnitude of the correlation as shown in the scale.
For a better understanding, we performed a transcriptome‐wide co‐expression analysis. We selected the top five correlative transcripts from each of the 27 featured genes and visualized them using a correlogram matrix (Figure 5A) and a gene network (Figure 5B). Three distinguishable groups (α, β and γ) were generated from Spearman's rank correlation coefficient (rho). Groups α and β generated geologically confined regions from other genes with a high positive correlation in each group (Spearman's rho between 0.5–1.0). Although group γ failed to form a highly associated cluster, unlike groups α and β, it was negatively associated (red edges) in both groups. Furthermore, three groups (Figure 5A,B) were generated via network analysis. Finally, the human phenotype ontology (HPO) (https://hpo.jax.org/) assay revealed the physiological and pathological relevance of the three groups (Figure 5C). The genes assigned to group α were linked to the HPO of exercise‐induced myalgia (HP:0003738), exercise intolerance (HP:0003546), exercise‐induced muscle cramps (HP:0003710), amyloidosis (HP:0011034), lactic acidosis (HP:0003128), axonal loss (HP:0003447), respiratory failure (HP:0002878) and mitochondrial respiratory chain (HP:0008972). Group β was associated with the HPO of rhabdomyosarcoma (HP:0002859), neoplasm of striated muscle (HP:0009728) and autosomal recessive inheritance (HP:0000007). Group γ comprised genes associated with the HPO of acanthosis nigricans (HP:0000956), oesophageal neoplasm (HP:0100751), gastrointestinal stromal tumour (HP:0100723), abnormality of eosinophils (HP:0001879), hypertriglyceridemia (HP:0002155), neoplasm of the small intestine (HP:0100833), large hands (HP:0001176), neoplasm of the small intestine (HP:0100833), neoplasm of the head and neck (HP:0012288) and neoplasm of the peripheral nervous system (HP:0100007). The seemingly separated group γ from groups α and β might be due to either a variable clinical history of sarcopenia patients or an as‐yet unknown biological interaction affecting sarcopenia pathogenesis. Further studies based on an extended dataset will be necessary to understand the co‐expression gene features comprehensively.
Figure 5.
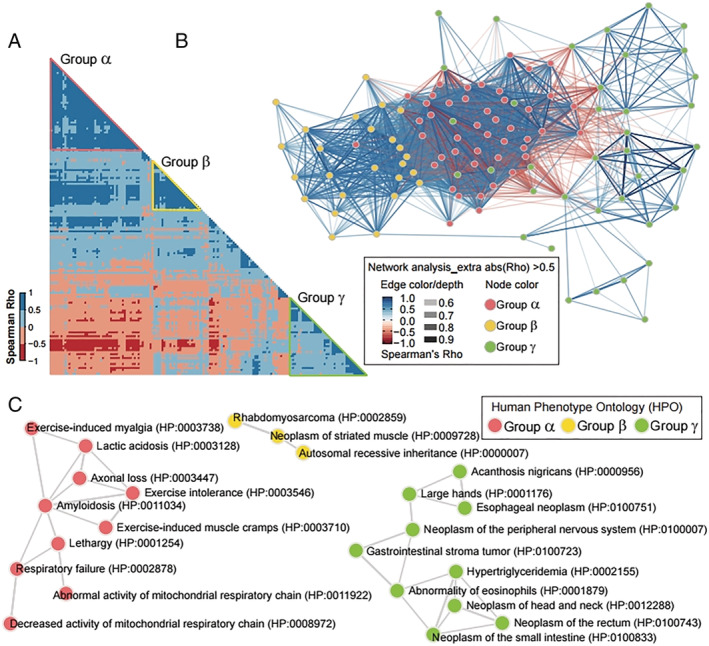
Co‐expression and human phenotype ontology assay categorizing 27 artificial intelligence (AI)‐featured genes into three groups related with skeletal muscle function, metabolism and diseases. (A) Correlogram matrices display Spearman's rho of two genes facing each side of the square. The shading intensity of the correlation matrices displays Spearman's Rho as presented in the scale (left‐hand side of the correlogram). The red, yellow and green triangles on the correlogram tie group α, β and γ, respectively. (B) Gene network showing co‐expression of group α, β and γ. The Spearman's rho of two node (gene) generates the colour and depth of each edge. The colour (pink, yellow and green) of node indicates each group (group α, β and γ). The gene symbols are indicated in Figure S2. (C) Three networks generated by Enrichr showing Human Phenotype Ontology that associated with each group. The colour of nodes (HPO terms) indicates each group and the edge means sharing common genes.
Discussion
Our proposed AI model, DSNet‐v1, successfully diagnosed sarcopenia accurately (100% sensitivity, 94.12% specificity, 95.83% accuracy, 97.06% balanced accuracy, and 0.99 AUROC). This model presents several unique characteristics. First, our model was developed based on subjects across three different continents: Europe, Africa and Asia. Second, our AI diagnosis model was developed using a vast amount of gene information (i.e. 17 339 genes), and feature importance analysis was performed to identify genes associated with sarcopenia. Finally, we created a web application (http://sarcopeniaAI.ml/) to access the model. We believe that allowing the public to access the AI model will facilitate the validation and improvement of the model.
We observed that the expression patterns of several genes varied among the three races (Figure 4A). For instance, the CENPC expression was elevated in the sarcopenia of SSS and HSS, but not in that of JSS. The expression of TEX261 reduced significantly in the sarcopenia of HSS and JSS, but not in that of SSS. This implies that regardless of race, certain factors might contribute to or be related to sarcopenia. Additionally, some other factors may be race‐specific sarcopenic factors. Indeed, we observed a race‐specific alteration of 27 genes in sarcopenic muscle compared to the healthy (Figure S3). For instance, AC104564.5, TPSAB1, PFKFB4 and GTTP1‐AS1 were altered only in the HSS. AC116913.1 (a.k.a. Lnc‐SNAPC5‐1), RASSF1, PSMA6 and CRHR2 were changed only in the JSS, whereas CENPC, KAT2A, PEF1, VPS35L, H4C3 and SUMO1P3 were altered only in the SSS. Although it might imply that there is a race‐specific pathology in sarcopenia, the power (n) might be not enough to lead to a clear conclusion. In the case of PCK1, it failed to generate a statistical significance in the HSS. However, the mean expression was escalated. Because PCK1 is known to be expressed in only gluconeogenic or glyceroneogenic tissues such as the liver, kidney, intestine and fat cells (https://www.gtexportal.org/), the altered expression of PCK1 might imply the anatomical loss of muscle and gain of adipocytes. Besides, Migliavacca et al. clearly demonstrated that gene sets associated with mitochondrial function and NAD+ biosynthesis pathway were tightly associated with the development/progression of sarcopenia. 14 Although our AI‐featured 27 genes did not include a gene directly involved mitochondrial and NAD+ biosynthesis metabolism, several genes could be associated indirectly with two pathways (i.e. mitochondria and NAD+ biosynthesis). For instance, it is well demonstrated that NAD+ biosynthesis and mitochondria function are tightly associated with sirtuin, a NAD+‐dependent protein deacetylase 22 , 23 and the protein acetyltransferase could be a count partner. 24 KAT2A is one of the well‐defined count partners of Sirtuins 25 and a very recent study proposed that KAT2A regulates muscle integrity. 26 In addition, the gene‐regulating glycolysis displayed altered gene expression. An additional intensive study of these 27 genes featured by our AI algorithm will expand our understanding of muscle ageing and sarcopenia.
Using Spearman's correlation assay, we identified three functional groups that were highly associated with 27 AI‐featured genes (Figure 5). Interestingly, groups α and β were associated with HPO terms of skeletal muscle function (e.g., exercise, lactate metabolism and abnormal mitochondrial function) and diseases with muscular symptoms (e.g., amyloidosis, axonal loss and rhabdomyosarcoma). Sarcopenia is often associated with physical frailty, 23 reduced muscle function, 27 and mitochondrial dysfunction. 14 In addition, recent studies revealed that amyloidosis in skeletal muscle is associated with mitochondrial dysfunction 28 , 29 and muscle diseases including inclusion body myositis, 30 , 31 indicating that both mitochondrial dysfunction and amyloidosis may trigger sarcopenia. In group γ, which generated a negative cluster against both groups α and β, most of the HPO terms were related to neoplasms. One HPO term was hypertriglyceridemia (HPO:0002155), which is known to be associated with sarcopenic obesity. 32
Our study has several limitations. First, to find the 27 genes, we used the three machine learning algorithms such as RF, XGBoost and AdaBoost, which result in opposing feature importance values for some genes. To improve generalization performance, we used an ensemble approach, which combines machine learning techniques into one stable model by reducing variance and bias. To investigate the effectiveness, we compared the performance when the selected features are obtained from RF, XGBoost, AdaBoost and their ensemble in Table S3. The results show that the ensemble approach provided the highest accuracy metrics. It implies that the ensemble may help to improve generalization performance. However, for the combination, we equally weighted the feature importance values. In the future work, we will further investigate the ways to combine the values more efficiently and accurately. Second, our proposed AI diagnosis model was validated using an isolated test dataset (n = 24), which was a dataset segregated from an entire dataset. It may be necessary to validate our AI model using external datasets, such as prospectively collected data. In addition, we plan to further develop DSnet‐v1 and change its name to DSnet‐v2 and DSnet‐v3. To update the model, we will use our developed web application to acquire additional data and validate the model. Currently, the application does not store any information entered by users. However, we plan to store information entered by users upon agreement to improve the AI model via a real‐time learning process. Third, our data included only three subjects of different races. In future studies, we will train and apply our AI model to more datasets comprising more diverse subjects.
Conclusion
Our AI model with 27 selected genes diagnosed sarcopenia accurately. We believe that it can facilitate healthcare providers in treating patients with early‐stage sarcopenia.
Conflict of interest
Heewon Chung, Yunju Jo, Dongryeol Ryu, Changwon Jeong, Seong‐Kyu Choe and Jinseok Lee declare that they have no conflict of interest.
Supporting information
Figure S1. Histogram for the number of available gene information. 39 subjects include approximately 16,100 gene information out of 17,339. 40 subjects include approximately 16,600 gene information. 39 subjects include approximately 16,700 gene information.
Figure S2. Receiver operating characteristic curves (ROCs); four different models of RF, XGBoost, AdaBoost, and DSnet‐v1based on testing data.
Figure S3. Scatter plots showing the relative expression of each gene in the healthy and sarcopenic elderly in the three different races.
Figure S4. Gene network showing co‐expression of group α, β, and γ. This figure is a replicate of Figure 6b and includes the name of each node (gene). The Spearman's Rho of two node generates the color and depth of each edge. The color (pink, yellow, and green) of node indicates each group (group α, β, and γ).
Table S1. Summary of training and testing datasets.
Table S2. The values of sensitivity, specificity, accuracy and balanced accuracy according each different number of selected features.
Table S3. Performance comparison based on the selected features from RF, XGBoost, AdaBoost and their ensemble.
Data S1. Gene information.
Data S2. Ranked Feature importance.
Acknowledgements
The authors of this manuscript certify that they comply with the ethical guidelines for authorship and publishing in the Journal of Cachexia, Sarcopenia and Muscle. 33 This study was supported by the grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare (HI18C1216), by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2021R1A5A8029876), and by the Korea Medical Device Development Fund grant funded by the Korea government (the Ministry of Science and ICT; Ministry of Trade, Industry and Energy; Ministry of Health and Welfare; and Ministry of Food and Drug Safety) (KMDF_PR_20200901_0095).
Chung H., Jo Y., Ryu D., Jeong C., Choe S.‐K., and Lee J. (2021) Artificial‐intelligence‐driven discovery of prognostic biomarker for sarcopenia, Journal of Cachexia, Sarcopenia and Muscle, 12, 2220–2230, 10.1002/jcsm.12840
Heewon Chung and Yunju Jo contributed equally to this work.
Contributor Information
Heewon Chung, Email: heewon1001@gmail.com.
Changwon Jeong, Email: mediblue@wku.ac.kr.
Seong‐Kyu Choe, Email: seongkyu642@wku.ac.kr.
Jinseok Lee, Email: gonasago@khu.ac.kr.
References
- 1. Rosenberg IH. Sarcopenia: origins and clinical relevance. J Nutr 1997;127:990S–991S. [DOI] [PubMed] [Google Scholar]
- 2. Hardee JP, Lynch GS. Current pharmacotherapies for sarcopenia. Expert Opin Pharmacother 2019;20:1645–1657. [DOI] [PubMed] [Google Scholar]
- 3. Mesinovic J, Zengin A, De Courten B, Ebeling PR, Scott D. Sarcopenia and type 2 diabetes mellitus: a bidirectional relationship. Diabetes Metab Syndr Obes 2019;12:1057–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pacifico J, Geerlings MAJ, Reijnierse EM, Phassouliotis C, Lim WK, Maier AB. Prevalence of sarcopenia as a comorbid disease: a systematic review and meta‐analysis. Exp Gerontol 2020;131:110801. [DOI] [PubMed] [Google Scholar]
- 5. Fletcher B, Gulanick M, Lamendola C. Risk factors for type 2 diabetes mellitus. J Cardiovasc Nurs 2002;16:17–23. [DOI] [PubMed] [Google Scholar]
- 6. Postma DS, Bush A, van den Berge M. Risk factors and early origins of chronic obstructive pulmonary disease. Lancet 2015;385:899–909. [DOI] [PubMed] [Google Scholar]
- 7. Dhillon RJ, Hasni S. Pathogenesis and management of sarcopenia. Clin Geriatr Med 2017;33:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rolland Y, Czerwinski S, Abellan Van Kan G, Morley JE, Cesari M, Onder G, et al. Sarcopenia: its assessment, etiology, pathogenesis, consequences and future perspectives. J Nutr Health Aging 2008;12:433–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim TN, Choi KM. Sarcopenia: definition, epidemiology, and pathophysiology. J Bone Metab 2013;20:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park SS, Seo YK, Kwon KS. Sarcopenia targeting with autophagy mechanism by exercise. BMB Rep 2019;52:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malafarina V, Uriz‐Otano F, Iniesta R, Gil‐Guerrero L. Sarcopenia in the elderly: diagnosis, physiopathology and treatment. Maturitas 2012;71:109–114. [DOI] [PubMed] [Google Scholar]
- 12. Cerri AP, Bellelli G, Mazzone A, Pittella F, Landi F, Zambon A, et al. Sarcopenia and malnutrition in acutely ill hospitalized elderly: prevalence and outcomes. Clin Nutr 2015;34:745–751. [DOI] [PubMed] [Google Scholar]
- 13. Pierik VD, Meskers CGM, Van Ancum JM, Numans ST, Verlaan S, Scheerman K, et al. High risk of malnutrition is associated with low muscle mass in older hospitalized patients—a prospective cohort study. BMC Geriatr 2017;17:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Migliavacca E, Tay SKH, Patel HP, Sonntag T, Civiletto G, McFarlane C, et al. Mitochondrial oxidative capacity and NAD+ biosynthesis are reduced in human sarcopenia across ethnicities. Nat Commun 2019;10:5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kwak JY, Hwang H, Kim SK, Choi JY, Lee SM, Bang H, et al. Prediction of sarcopenia using a combination of multiple serum biomarkers. Sci Rep 2018;8:8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scharf G, Heineke J. Finding good biomarkers for sarcopenia. J Cachexia Sarcopenia Muscle 2012;3:145–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Breiman L. Random forests. Mach Learn 2001;45:5–32. [Google Scholar]
- 18. Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. San Francisco, California, USA: Association for Computing Machinery; 2016. p 785–794. [Google Scholar]
- 19. Freund Y, Schapire RE. Game Theory, On‐line Prediction and Boosting. Desenzano del Garda, Italy: Association for Computing Machinery; 1996. p 325–332. [Google Scholar]
- 20. Ratsch G, Onoda T, Muller KR. Soft margins for AdaBoost. Mach Learn 2001;42:287–320. [Google Scholar]
- 21. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kang BE, Choi JY, Stein S, Ryu D. Implications of NAD+ boosters in translational medicine. Eur J Clin Invest 2020;50:e13334. [DOI] [PubMed] [Google Scholar]
- 23. Dao T, Green AE, Kim YA, Bae SJ, Ha KT, Gariani K, et al. Sarcopenia and muscle aging: a brief overview. Endocrinol Metab (Seoul) 2020;35:716–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mutlu B, Puigserver P. GCN5 acetyltransferase in cellular energetic and metabolic processes. Biochimica et Biophysica Acta (BBA)‐Gene Regulatory Mechanisms 2021;1864:194626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 2012;13:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Addicks GC, Zhang H, Ryu D, Vasam G, Marshall PL, Green AE, et al. GCN5 maintains muscle integrity by acetylating YY1 to promote dystrophin expression. bioRxiv. 2021;436986. 10.1101/2021.03.29.436986 [DOI] [PMC free article] [PubMed]
- 27. Cruz‐Jentoft AJ, Bahat G, Bauer J, Boirie Y, Bruyere O, Cederholm T, et al. Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing 2019;48:16–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sorrentino V, Romani M, Mouchiroud L, Beck JS, Zhang H, D'Amico D, et al. Enhancing mitochondrial proteostasis reduces amyloid‐beta proteotoxicity. Nature 2017;552:187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Romani M, Sorrentino V, Oh CM, Li H, de Lima TI, Zhang H, et al. NAD+ boosting reduces age‐associated amyloidosis and restores mitochondrial homeostasis in muscle. Cell Rep 2021;34:108660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buzkova J, Nikkanen J, Ahola S, Hakonen AH, Sevastianova K, Hovinen T, et al. Metabolomes of mitochondrial diseases and inclusion body myositis patients: treatment targets and biomarkers. EMBO Mol Med 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oldfors A, Moslemi AR, Jonasson L, Ohlsson M, Kollberg G, Lindberg C. Mitochondrial abnormalities in inclusion‐body myositis. Neurology 2006;66:S49–S55. [DOI] [PubMed] [Google Scholar]
- 32. Batsis JA, Villareal DT. Sarcopenic obesity in older adults: aetiology, epidemiology and treatment strategies. Nat Rev Endocrinol 2018;14:513–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. von Haehling S, Morley JE, Coats AJS, Anker SD. Ethical guidelines for publishing in the Journal of Cachexia, Sarcopenia and Muscle: update 2019. J Cachexia Sarcopenia Muscle 2019;10:1143–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Histogram for the number of available gene information. 39 subjects include approximately 16,100 gene information out of 17,339. 40 subjects include approximately 16,600 gene information. 39 subjects include approximately 16,700 gene information.
Figure S2. Receiver operating characteristic curves (ROCs); four different models of RF, XGBoost, AdaBoost, and DSnet‐v1based on testing data.
Figure S3. Scatter plots showing the relative expression of each gene in the healthy and sarcopenic elderly in the three different races.
Figure S4. Gene network showing co‐expression of group α, β, and γ. This figure is a replicate of Figure 6b and includes the name of each node (gene). The Spearman's Rho of two node generates the color and depth of each edge. The color (pink, yellow, and green) of node indicates each group (group α, β, and γ).
Table S1. Summary of training and testing datasets.
Table S2. The values of sensitivity, specificity, accuracy and balanced accuracy according each different number of selected features.
Table S3. Performance comparison based on the selected features from RF, XGBoost, AdaBoost and their ensemble.
Data S1. Gene information.
Data S2. Ranked Feature importance.


