Abstract
Background
Cancer cachexia is a multifactorial metabolic syndrome in which bile acid (BA) metabolism might be involved. The aim of the present study was to clarify the contribution of liver and gut microbiota to BA metabolism disturbance in cancer cachexia and to check the possibility of targeting BA metabolism using agents such as tauroursodeoxycholic acid (TUDCA) for cancer cachexia therapy.
Methods
The BA profiles in liver, intestine, and serum of mice with cancer cachexia induced by inoculation of colon C26 tumour cells were analysed using metabolomics methods and compared with that of control mice. Proteomic analysis of liver protein expression profile and 16S rRNA gene sequencing analysis of gut microbiota composition in cancer cachexia mice were conducted. Expression levels of genes related to farnesoid X receptor (FXR) signalling pathway in the intestine and liver tissues were analysed using RT–PCR analysis. The BA profiles in serum of clinical colon cancer patients with or without cachexia were also analysed and compared with that of healthy volunteers. The effects of TUDCA in treating cancer cachexia mice were observed.
Results
In the liver of cancer cachexia mice, expression of BA synthesis enzymes was inhibited while the amount of total BAs increased (P < 0.05). The ratios of conjugated BAs/un‐conjugated BAs significantly increased in cancer cachexia mice liver (P < 0.01). Gut microbiota dysbiosis such as decrease in Lachnospiraceae and increase in Enterobacteriaceae was observed in the intestine of cancer cachexia mice, and microbial metabolism of BAs was reduced. Increase in expression of FGF15 in intestine (P < 0.01) suggested the activation of FXR signalling pathway which might contribute to the regulation of BA synthesis enzymes, transporters, and metabolic enzymes. Increase in the BA conjugation was observed in the serum of cancer cachexia mice. Results of clinical patients showed changes in BA metabolism, especially the increase in BA conjugation, and also suggested compensatory mechanism in BA metabolism regulation. Oral administration of 50 mg/kg TUDCA could significantly ameliorate the decrease in body weight (P < 0.001), muscle loss (P < 0.001), and atrophy of heart and liver (P < 0.05) in cancer cachexia mice without influence on tumour growth.
Conclusions
Bile acid metabolism dysregulation such as decrease in BA synthesis, increase in BA conjugation, and decrease in BA microbial metabolism was involved in development of cancer cachexia in mice. Targeting BA metabolism using agents such as TUDCA might be helpful for cancer cachexia therapy.
Keywords: Cancer cachexia, Bile acids, Liver, Gut microbiota, TUDCA
Introduction
Cancer cachexia is a multifactorial metabolic syndrome found in approximately 50–80% of advanced cancer patients and is directly attributable to more than 30% of cancer‐related deaths. 1 Muscle and adipose wasting along with progressive anorexia and multiple organ functional impairment in the later stages are characteristics of cancer cachexia. 2 Cancer cachexia is thought to be the consequence of a net of effects by a combination of different mechanisms, including alterations in physiological energy balance; hormonal and metabolic disturbances in both the periphery and the central nervous system; and a pro‐inflammatory environment accompanied by high levels of cytokines. 3 Bile acids (BAs) are endocrine molecules that regulate various metabolic processes and facilitate the absorption of fat‐soluble nutrients. Whole body BA homeostasis is maintained by efficient enterohepatic cycling of BAs between the liver and small intestine, where the majority (approximately 95%) of BAs released from the liver into the proximal duodenum are efficiently reabsorbed in the terminal ileum. 4 The remaining (about 5%) of BAs reach the colon, where they are deconjugated, dehydrogenated, and dehydroxylated by intestinal bacteria to form secondary BAs before reabsorption or excretion. 5 The involvement of dysregulation of BA metabolism in cancer, especially gastrointestinal cancer, has been reported. 6 However, some conflicting results have been reported in clarifying the crosstalk between BA regulation and cancer. Similarly, although several studies have attempted to determine the BA composition in cancer cachexia 7 , 8 or to study the possibility of targeting BA metabolism using ursodeoxycholic acid (UDCA) to treat cancer cachexia, 9 the exact roles and mechanisms of BA metabolism dysregulation in the development of cancer cachexia are still unclear and largely underexplored.
Liver dysfunction plays an important role in the development of cancer cachexia, 10 and gut microbiota dysbiosis has been observed in various preclinical models of cancer cachexia. 11 , 12 In the present study, we first profiled the BA composition in the liver, intestine, and serum of cancer cachexia mice bearing C26 mouse colon adenocarcinoma cells and compared them with those of healthy control mice. The C26 cancer cachexia model is a type of cancer cachexia model that has been well accepted and widely used in both the study of aetiopathogenesis of cancer cachexia 13 and screening drug candidates for cancer cachexia. 14 Two types of metabolite ratios of BAs were evaluated in the present study to elucidate the mechanisms leading to altered BA profiles in cancer cachexia. The ratios of conjugated/unconjugated BAs would indicate the extent of BA conjugation in the liver, while the ratios of secondary/primary BAs would indicate the extent of microbial metabolism of BAs in the intestine. The protein expression profile in the liver tissues and gut microbiota composition in the caecum of C26 cancer cachexia mice were further analysed using proteomic analysis and 16S rRNA gene sequencing, respectively. Expression levels of genes related to the farnesoid X receptor (FXR) signalling pathway, an important mediator in the gut‐liver axis, 15 , 16 were analysed in both the intestine and liver tissues of mice. FXR is a member of the steroid/thyroid hormone receptor family of ligand‐activated transcription factors expressed in the mammalian liver and intestines and can be activated or inhibited by BAs. The FXR signalling pathway is an important mediator of the gut–liver axis, and the activation of FXR in the intestine induces the expression of fibroblast growth factor 15 (FGF15). FGF15, secreted in the portal circulation and pass through the portal vein to the liver, could activate the liver FXR through fibroblast growth factor receptor 4 (FGFR4) thus regulate the expression of genes involved in BA synthesis and transport such as CYP7A1, the rate‐limiting enzyme in liver BA synthesis. 17 Subsequently, to confirm whether the findings in the animal studies could also be observed in clinical patients, the BA profiles in the serum of clinical colon cancer patients with or without cachexia were analysed and compared with those of healthy volunteers. Furthermore, the effects of TUDCA in treating cancer cachexia mice were assessed to determine the possibility of targeting BA metabolism in the treatment of cancer cachexia therapy.
Materials and methods
Animals and drugs
All animal care and experimental protocols for this study complied with the Chinese regulations and the Guidelines for the Care and Use of Laboratory Animals by the National Institutes of Health (United States) and were approved by the Institutional Animal Care and Use Committee of the East China Normal University. Male BALB/c mice (6–8 weeks old) were purchased from Shanghai SLAC Laboratory Animal Co. Ltd. In the first animal study, mice with the same initial body weight were randomly divided into two groups (eight mice per group): control group (healthy control mice without tumour inoculation) and, cachexia group (mice inoculated with C26 tumour cells to induce cancer cachexia). In the second animal study, mice with the same initial body weight were randomly divided into three groups (10 mice per group): control group (healthy control mice without tumour inoculation), cachexia group (mice inoculated with C26 tumour cells to induce cancer cachexia), and cachexia + TUDCA group (C26 tumour‐bearing mice treated with 50 mg/kg TUDCA, i.g.). Mice were maintained on a 12:12 light–dark cycle in a temperature‐controlled (21–23°C) and specific pathogen‐free (SPF) conditional room, and were provided standard rodent chow and water ad libitum. All animals were acclimatized for 1 week before beginning the study. TUDCA (purity > 98%) was purchased from MedChemExpress (MCE, USA).
Induction of cancer cachexia in mice
Cancer cachexia in mice was induced using C26 colon tumour cells according to a previously established protocol. 18 The complete procedure is described in Supporting Information, Data S1.
Patient cohorts
A total of 49 serum samples were collected from healthy volunteers (22 cases), non‐cachexia (13 cases), and cachexia patients (14 cases) with colon cancer at Shanghai Tenth People's Hospital, Tongji University School of Medicine between January 2014 and January 2019. A colon CSS was used to evaluate cachexia in patients with cancer. The cancer cachexia staging score consists of three components (as shown in Table S1): weight loss within 6 months (score range: 0–3); Eastern Cooperative Oncology Group performance status (score range: 0–2); and abnormal biochemistry (score range: 0–3). Patients with colon tumours were classified into two groups: non‐cachexia (score range: 0–4) and cachexia (score range: 5–8). Detailed information about the clinical characteristics of the patients with cancer is shown in Table S2 . The study protocol was approved by the Ethics Committee of the Shanghai Tenth People's Hospital, Tongji University School of Medicine (Approval No: SHSY‐IEC‐pap‐16‐24). The written informed consent was obtained from the patients and healthy volunteers. This study is registered with Clinical Trials.gov (NCT02917707). The detailed information is provided in Data S1 and Tables S1 and S2.
Bile acid analysis
The BA concentrations in the samples were quantified using ultra‐high‐performance liquid chromatography coupled with a QTRAP® 6500+ LC–MS/MS System from SCIEX (Framingham, MA, United States) according to a previously reported protocol. 19 In the animal study, eight samples from healthy control mice and eight samples from cancer cachexia mice were analysed. In clinical sample study, 22 samples from healthy volunteers, 13 samples from non‐cachexia cancer patients, and 14 samples from cancer cachexia patients were analysed. Detailed information about the BA analysis is provided in Data S1 and Table S3.
Protein extraction, enzymatic digest and tandem mass tags based proteomics
The analysis was performed using a shotgun proteomics approach based on liquid chromatography‐high resolution tandem mass spectrometry. After reduction and alkylation, liver proteins were digested with trypsin (Promega, Madison, Wisconsin, USA) overnight at 37°C, and then, the peptides were collected. Peptide mixtures from each sample were labelled using 16‐plex tandem mass tags (TMT) pro reagents (Thermo Scientific, MA, USA) to quantify up to 16 samples simultaneously in the same analysis. Eight isobaric compounds were used to label different samples of the control group (reporter ions at 126, 127N, 127C, 128N, 128C, 129N, 129C, and 130N), while other tags were used to label the cachexia group sample (reporter ion at 130C, 131N, 131C, 132N, 132C, 133N, 133C, and 134), according to the manufacturer's instructions. A high pH reversed‐phase peptide fractionation was used to fractionate TMT‐labelled peptides by increasing acetonitrile step‐gradient elution. Mass spectrometric data were collected on an Orbitrap QE HF‐X mass spectrometer coupled to an EASY‐nLC™ 1200 UHPLC system (Thermo Fisher Scientific, USA). Raw data were processed using Proteome Discoverer (v2.4) and searched using the Mascot (Matrix Science, London, UK; version 2.2) engine. Detailed MS procedure is provided in Data S1.
Parallel reaction monitoring analysis of CYP7A1 protein expression level
For proteins that were not detected in TMT‐based proteomic analysis, parallel reaction monitoring (PRM)‐based proteomic analysis was used to specifically search and analyse the proteins. Briefly, the proteins were extracted and enzymatically hydrolysed similar to the above method. Same amount of trypsin treated‐peptide was taken of each sample, and labelled peptide GAGSSEPVTGLDAK as an internal standard of each sample and the peptides were dissolved in solvent A (0.1% formic acid in 2% acetonitrile)and solvent B (0.1% formic acid in 80% acetonitrile). PRM mass spectrometric analysis was performed using an EASY‐nLC™ 1200 UHPLC system (Thermo Fisher Scientific, USA). The full scan mass spectrum resolution was set to 60 000 (200 m/z), the maximum C‐trap and IT were 3 × 106 and 50 ms, respectively. The PRM resolution was set to 30 000 (200 m/z), the maximum C‐trap and IT were 1 × 106 and 100 ms, respectively. Normalized collision energy is 28. The data were analysed using the Skyline software (MacCoss Lab, Univ. of Washington), and the peak area was corrected using the internal standard peptide.
LC–MS/MS quantification of 7α‐hydroxy‐4‐cholesten‐3‐one (C4) in mouse serum
The HPLC system consisted of ExionLC™ Series UHPLC and an ACQUITY UPLC BEH C18 (2.1 × 100 mm, 1.7 μm, Waters). Mobile phase A was 0.4% formic acid and 0.75 mM ammonium acetate in water, and mobile phase B was 0.4% formic acid and 0.75 mM Ammonium acetate in methanol. The gradient elution ramped linearly from 60% to 90% B in 2 min, followed ramped linearly from 90% to 100% B in 4 min, held at 100% for 2 min, and then returned to 60% B in 0.5 min and maintained at 60% B until the end of the run at 10 min. The HPLC flow rate was 0.4 mL/min, the injection volume was 1 μL. The oven temperature set for 40°C. The MSD system was interfaced with an QTRAP 6500+ mass spectrometer operated in positive electrospray mode. The ion source parameters in Turbo Ionspray mode were curtain gas 35 psi, GAS1 50 psi, GAS2 50 psi, Ionspray voltage 5500 V, and source temperature 550°C, respectively. The entrance potential was 10 V, decluster potential was 100 V, and exit potential was 15 V for all the ions. The multiple reaction monitoring (MRM) transitions for C4 were 401.3 → 177.1 and 401.3 → 97.0 for collision energy (CE) was 34 V. Data acquisition and processing were performed using Analyst software (Applied Biosystems, Foster City, CA, USA). Calibration curves were constructed by plotting the peak area of C4.
RT‐PCR assay of the expression levels of FXR‐related genes in mice liver and intestine samples
Total RNA was extracted using TRIzol (TaKaRa, Otsu, Japan) and stored at −80°C. The cDNA Synthesis Kit (TaKaRa) was used to reverse transcribe mRNA according to the manufacturer's instructions. β‐Actin was used as an endogenous control for mRNA expression. qRT‐PCR primers were designed using Primer Premier 5 and are listed in Table S4 . Reaction volume for qRT‐PCR was 20 μL, including cDNA 2 μL, F/R primers 0.2 μL, double‐distilled water 7.6 μL, and 10 μL FastStart Essential DNA Green Master PCR mix (Roche, USA).
Faecal DNA extraction and 16S rRNA sequencing
Faecal microbial DNA was extracted from the collected faeces using a QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to manufacturer's protocols. DNA integrity and size were verified by 1% agarose gel electrophoresis, and DNA concentrations were determined using NanoDrop spectrophotometry (NanoDrop, Germany). The V3–V4 regions of the 16S rRNA genes were amplified using the primers 338F (5′‐ACTCCTACGGGAGGCAGC‐3′) and 806R (5′‐GGACTACVVGGGTATCTAATC‐3′). Amplicons were purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, U.S.) and quantified using Qubit® 2.0 (Invitrogen, USA). The tags were sequenced by Shanghai Majorbio Bio‐pharm Technology Co., Ltd. (Shanghai, China) on the Illumina MiSeq platform (Illumina, Inc., CA, USA).
Quantification and statistical analysis
All data are expressed as mean and standard error of the mean. Unpaired t‐test or one‐way analysis of variance followed by Tukey's multiple comparison test was conducted by using GraphPad Prism Version 6.0 software (Graphpad Software Inc., La Jolla, CA, USA) with Bonferroni's post hoc test. The number of animals or replicates (n) for each group is indicated in the figure legends. P values < 0.05 were considered significant.
Data availability
16S rRNA sequencing data have been deposited in the NCBI SRA database 20 under accession number SRP301790 and SRP301551. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository 21 with the data set identifier PXD023497. Proteomic data were searched against a uniprot database containing Mus musculus proteins (https://www.uniprot.org/proteomes/); Metabolomics data have been deposited to the EMBL‐EBI MetaboLights database 22 with the identifier MTBLS2389. The complete data set can be accessed here https://www.ebi.ac.uk/metabolights/MTBLS2389.
Results
C26 tumour‐bearing mice exhibited cancer cachexia symptoms
To confirm the success in establishing cancer cachexia mice model, the physiological characteristics, such as body weight, food intake, and several other variables as shown in Figure 1, of cancer cachexia mice (C26 tumour‐bearing mice) were compared with those of healthy control mice. As shown in Figure 1A, during the experimental period, the body weight of healthy control mice gradually increased while the body weight of cancer cachexia mice did not increase and even decreased strongly from Day 11; thus, the experiment had to be terminated at Day 14. The results of the tumour‐free body weight also showed a similar tendency (Figure 1B). The cumulative food intake of the cancer cachexia model group was lower than that of the healthy control group (Figure 1C). At the end of the experiment, the body weight of cancer cachexia mice was significantly lower than that of healthy control mice. The tumour growth curve of the C26 tumour‐bearing mice is shown in Figure 1D. Furthermore, cachexia model mice exhibited a significant decrease in gastrocnemius (GA) muscle weight (Figure 1E) and epididymal white fat (eWAT) weight (Figure 1F). In the cancer cachexia mice, atrophy of the heart (Figure 1G) and liver (Figure 1H) were also observed. The significant decrease in body weight and the considerable loss in weight of muscle tissue, fat tissue, heart, and liver suggested the successful establishment of the cancer cachexia model in the present study.
Figure 1.
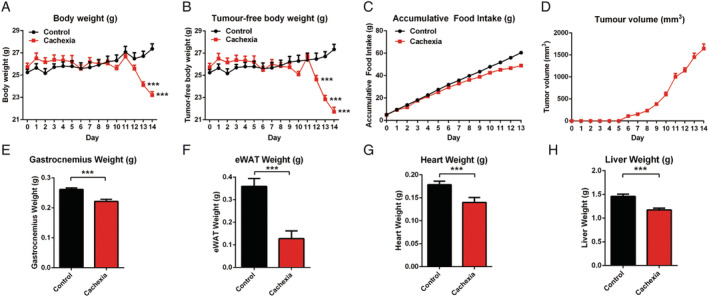
Cachexia symptoms of cancer cachexia model mice (C26 tumour‐bearing mice). (A) Body weight of cancer cachexia mice and healthy control mice. (B) Tumour‐free body weight of cancer cachexia mice and healthy control mice. (C) Accumulative food intake of cancer cachexia mice and healthy control mice. (D) Tumour growth curve of C26 tumour‐bearing mice. (E) GA muscle tissue weight of cancer cachexia mice and healthy control mice. (F) eWAT fat tissue weight of cancer cachexia mice and healthy control mice. (G) Heart weight of cancer cachexia mice and healthy control mice. (H) Liver weight of cancer cachexia mice and healthy control mice. Data presented are the mean ± SEM of results of 8 mice in each group. T‐test analysis was performed to assess the significance of the difference. *P < 0.05; **P < 0.01, ***P < 0.001 between the indicated two groups.
The liver bile acid profile was altered in cancer cachexia mice
The amounts of total liver BAs, total conjugated liver BAs, and total unconjugated liver BAs, which were normalized to the liver weight, are shown in Figure 2A. As shown in Figure 2A, the amount of total unconjugated BAs in the liver of cancer cachexia mice was significantly lower than that of control mice. The amounts of total liver BAs and total conjugated liver BAs in cancer cachexia mice were significantly higher than those in control mice. The profiles of BA pool composition in the liver of cancer cachexia mice and control mice are shown in Figure 2B. An increase in the proportion of tauro‐conjugated muricholic acid, including T‐α‐MCA and T‐β‐MCA, in the total BAs was observed in the liver of cancer cachexia mice (Figure 2B). The levels of individual conjugated or unconjugated BA are shown in Figure 2C and 2D, respectively. The ratio values of each conjugated vs. unconjugated BA are shown in Figure 2E. The levels of conjugated BAs such as TCA, T‐α‐MCA, and T‐β‐MCA significantly increased, while the levels of unconjugated BAs such as CA, α‐MCA, and β‐MCA significantly decreased in the liver of cancer cachexia mice. The ratios of T‐α‐MCA/α‐MCA, TCDCA/CDCA, TUDCA/UDCA, and THDCA/HDCA in cancer cachexia mice liver samples were significantly higher than those in healthy controls. Consistently, the ratio of total conjugated BAs/total unconjugated BAs also increased significantly in cancer cachexia mice liver (Figure 2E). These results indicate that the conjugation of BAs was enhanced in cancer cachexia mice liver.
Figure 2.
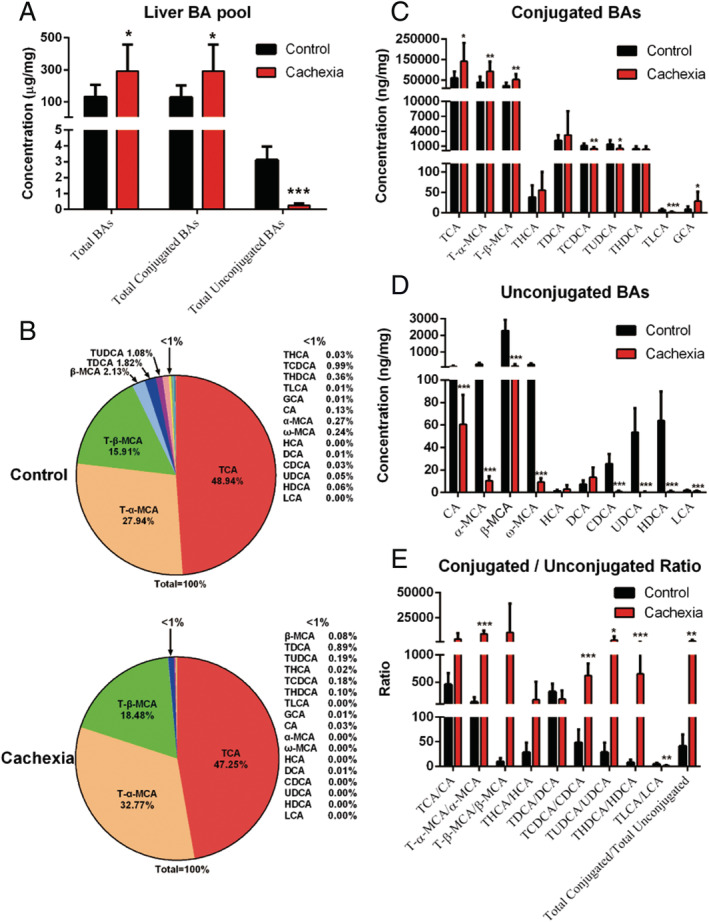
Liver BA profiles in cancer cachexia model mice and healthy control mice. (A) The amount of total BAs, total conjugated BAs and total unconjugated BAs in the liver of mice. (B) Pie graphs are the mean percentage of the individual bile acids in total BAs. (C) Bar graphs are the concentrations of the conjugated bile acids in liver. (D) Bar graphs are the concentrations of the unconjugated bile acids in liver. (E) Bar graphs are the ratios of the conjugated/unconjugated bile acids in liver. Data presented are the mean ± SEM of results of eight mice in each group. T‐test analysis was performed to assess the significance of the difference. *P < 0.05; **P < 0.01, ***P < 0.001. BA, bile acid.
Expression of proteins related to lipid metabolism, including BA synthesis enzymes, transporters, and metabolic enzymes, was altered in cancer cachexia mice liver
To clarify the contribution of the liver to BA metabolism dysregulation in cancer cachexia, a TMT‐based proteomic approach was used to compare the liver protein expression profiles of cancer cachexia mice and control mice. In total, 6209 proteins were identified, of which 6123 were quantified. The fold‐change cut‐off was set when proteins with quantitative ratios above 1.5, or below 0.67, were deemed to be significant. Among the quantified proteins, 123 proteins were up‐regulated and 222 proteins were down‐regulated. The differentially expressed proteins between the cachexia and control groups are shown by volcano plots (Figure 3A). Results of Kyoto Encyclopedia of Genes and Genomes pathway analysis showed enriched pathways of the down‐regulated proteins in cancer cachexia mice liver compared with healthy control mice liver (Figure 3B). As shown in Figure 3B, many lipid metabolism‐related pathways, such as fatty acid degradation metabolism, steroid hormone biosynthesis, and primary BA biosynthesis, were involved in the enriched pathways. These results suggest the inhibition of lipid metabolism‐related pathways in cancer cachexia.
Figure 3.
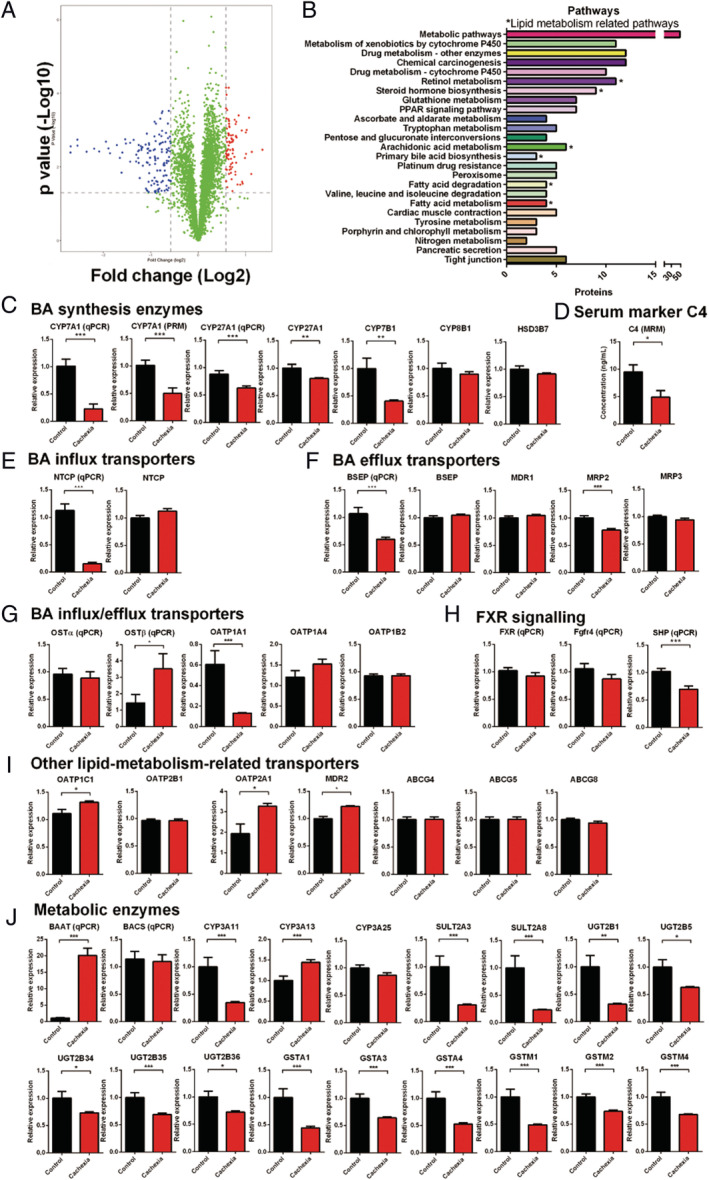
Results of liver proteomic analysis. (A) Volcano graph of the results of comparing protein expression profiles of cancer cachexia mice liver and healthy control mice liver. (B) Results of Kyoto Encyclopedia of Genes and Genomes enrichment analysis based on the down‐regulated proteins in the cancer cachexia group. *Lipid metabolism‐related pathways. (C) The expression levels of key enzymes involved in BA synthesis. (D) The expression level of C4 which is BA synthesis marker in serum. (E) The expression levels of BA influx transporters. (F) The expression levels of BA efflux transporters. (G) The expression levels of BA influx/efflux transporters. (H) The expression levels of genes involved in FXR signalling. (I) The expression levels of other key transporters involved in lipid metabolism. (J) The expression levels of BA metabolic enzymes. Data presented are the mean ± SEM of results of eight mice in each group. T‐test analysis was performed to assess the significance of the difference. *P < 0.05; **P < 0.01, ***P < 0.001. BA, bile acid.
The expression levels of BA synthesis enzymes, BA influx transporters, BA efflux transporters, BA influx/efflux transporters, other lipid‐metabolism‐related transporters, and metabolic enzymes are shown in Figure 3C, 3E, 3F, 3G, 3I, and 3J, respectively. The data showed mostly the protein expression levels of genes detected in proteomic analysis while the mRNA expression levels of several important genes analysed using RT‐PCR are also shown. Both the mRNA and protein levels of CYP7A1 and CYP27A1 in the liver of cancer cachexia mice were significantly lower than those in the liver of control mice. The protein level of CYP7B1 was also significantly decreased in the liver of cancer cachexia mice compared with the control. The serum level of C4, a biomarker indicating the rate of BA synthesis, was also significantly decreased in cancer cachexia mice compared with control (Figure 3D). These results suggest the inhibition of the expression of BA synthesis enzymes and decrease in BA synthesis in cancer cachexia liver. There were also changes in the levels of BA influx transporters, efflux transporters, BA influx/efflux transporters, and other transporters related to lipid metabolism. The mRNA expression levels of sodium/BA cotransporter (NTCP) and bile salt export pump (BSEP) were significantly down‐regulated in cancer cachexia mice, although there was no difference in their protein levels between cancer cachexia mice and control mice. The protein level of MRP2 (ATP‐binding cassette subfamily G member 2) was also decreased in cancer cachexia mice. In the BA influx/efflux transporters, the level of organic solute transporter subunit beta (OSTβ) significantly increased while the level of solute carrier organic anion transporter family member 1A1 significantly decreased in cancer cachexia mice compared with the control (Figure 3G). Figure 3H shows the expression levels of FXR, FGFR4, and small heterodimer partner (SHP). The levels of FXR and FGFR4 in cancer cachexia mice liver were similar to those of control mice, while the level of SHP significantly decreased in the liver of cancer cachexia mice. As for other lipid metabolism‐related transporters, the levels of solute carrier organic anion transporter family member 1, solute carrier organic anion transporter family member 2A1, and phosphatidylcholine translocator ABCB4 (MDR2) were significantly increased in cancer cachexia mice. The levels of BA‐CoA amino acid N‐acetyltransferase (BAAT) and cytochrome P450 3A13 significantly increased in cancer cachexia mice. The levels of cytochrome P450 3A11, sulfotransferases (SULT2A3 and SULT2A8), UDP‐glucuronosyltransferases (UGT2B1, UGT2B5, UGT2B34, UGT2B35, and UGT2B36), glutathione S‐transferases (GSTA1, GSTA3, and GSTA4), and glutathione S‐transferase Mu (GSTM1, GSTM2, and GSTM4), were all significantly decreased in cancer cachexia mice. These results indicate that the lipid metabolism‐related proteins were significantly altered in cachexia model mice, in which BA synthesis, influx/efflux and modification were all affected.
The microbial metabolism of BAs by intestine microbiota was decreased in cancer cachexia mice
As shown in Figure 4A, there was no significant difference between the amount of total caecal BAs in the cancer cachexia mice and control mice. The amount of total secondary BAs in cancer cachexia mice intestine was lower than that of the control, although the decrease was not significant (P = 0.06). The profiles of BA composition in the caecal BA pool of cancer cachexia mice and control mice are shown in Figure 4B. The levels of individual BAs (primary or secondary) were shown in Figure 4C and 4D, respectively. The levels of CA and β‐MCA significantly increased, while the levels of TCA, LCA, UDCA, and TUDCA significantly decreased in cancer cachexia mice compared with the control. The ratio values of each secondary/primary BAs and conjugated/unconjugated BAs are shown in Figure 4E and 4F, respectively. The ratios of secondary/primary BAs such as DCA/CA, LCA/CDCA, and MDCA/β‐MCA were significantly lower in cancer cachexia mice than in control mice. These results suggest that microbial BA metabolism was inhibited in cancer cachexia, and there might be considerable changes in the gut microbiota composition in cancer cachexia mice. With regard to the ratios of conjugated/unconjugated BAs, there was no considerable difference between cancer cachexia mice and control mice, except for a decrease in the ratio of TCA/CA in cancer cachexia mice caecum (Figure 4F). To check the involvement of the FXR signalling pathway in cancer cachexia, the expression levels of FXR, FGF15, and other FXR‐targeted genes were analysed by RT‐PCR, and the results are shown in Figure 4G. The significant increase in FXR and FGF15 suggest the activation of the FXR signalling pathway in cancer cachexia mice caecum (Figure 4G). The levels of apical sodium‐dependent bile salt transporter and organic solute transporter subunit alpha (OSTα) were significantly decreased in the caecum of cancer cachexia mice (Figure 4G).
Figure 4.
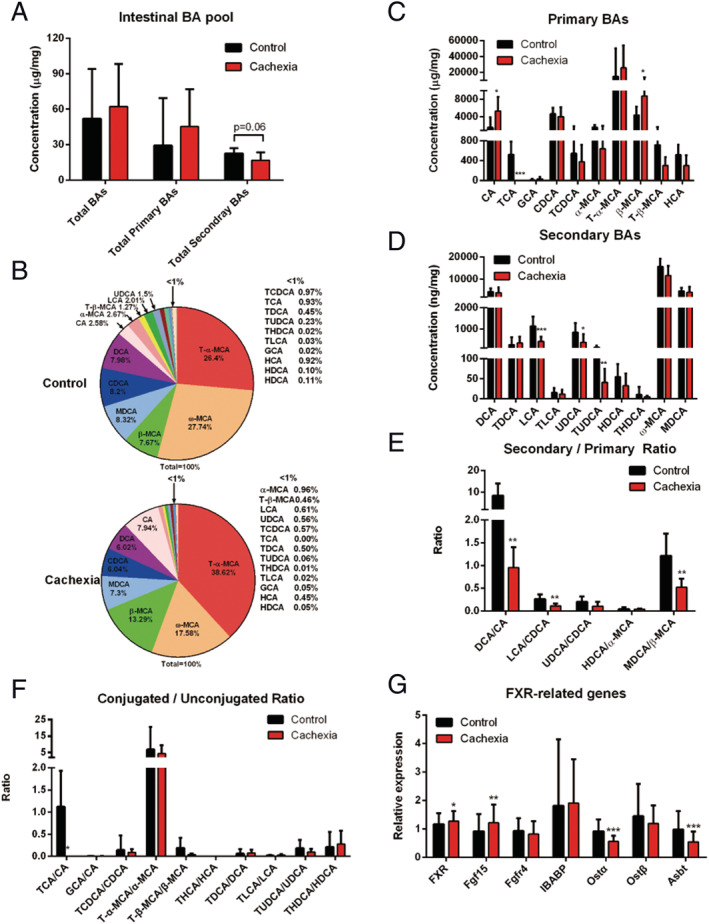
Intestinal BA profiles in cancer cachexia model mice and healthy control mice. (A) The amount of total BAs, total primary BAs and total secondary BAs in the caecum of cancer cachexia model mice and healthy control mice. (B) Pie graphs are the mean percentage of the individual bile acids in total BAs. (C) Bar graphs are the concentrations of the primary bile acids in caecum. (D) Bar graphs are the concentrations of the secondary bile acids in caecum. (E) Bar graphs are the ratios of the primary/secondary bile acids in caecum. (F) Bar graphs are the ratios of the conjugated/unconjugated bile acids in caecum. (G) The expression levels of FXR‐related genes in caecum. Data presented are the mean ± SEM of results of eight mice in each group. T‐test analysis was performed to assess the significance of the difference. *P < 0.05; **P < 0.01, ***P < 0.001. BA, bile acid; FXR, farnesoid X receptor.
Cachexia mice harboured an altered composition of the gut microbiota
Results of analysing the composition of the gut microbial community in the faeces of cancer cachexia mice collected every other day during the experimental period are shown in Figure 5A. Using OTUs to track the dynamics of the abundance of different bacterial phyla in cancer cachexia mice, time‐dependent changes in the gut microbiota profile in cancer cachexia mice were observed. At Day 7 after inoculation of C26 tumour cells, the proportion of Firmicutes increased considerably (from approximately 36.6% at Day 1 to approximately 49.5% at Day 7), while the abundance of Bacteroidetes markedly decreased (Figure 5A). Day 7 was also nearly the time point when the tumour‐bearing mice began to exhibit symptoms of cancer cachexia, such as a decrease in food intake and body weight. The high proportion of Firmicutes gradually decreased after Day 7; thus, the proportion of Firmicutes was only 46.7% on Day 13 after inoculation (Figure 5A). These results suggest that there might be a compensatory regulatory mechanism in maintaining the balance of the composition of the gut microbiota. To further understand the change in the composition of intestinal flora, the composition of the gut microbial community in the caecal contents collected on Day 14 (the end of the experiment) were thoroughly analysed. In total, 818 042 sequences and 345 683 952 bases from 16 samples (two groups) were obtained. The changes in gut microbiota profiles at phylum and family levels were shown in Figure 5B and 5C, respectively. At Day 14 in cancer cachexia mice, the abundance of Firmicutes was higher and the abundance of Bacteroidetes was lower than that of healthy control mice although the difference was not significant. At the family level, significant changes such as a decrease in Lachnospiraceae and an increase in Eubacteriaceae were observed in cancer cachexia mice. Importantly, the results of analysing the correlation between the intestinal microbial abundance and the intestinal levels of BAs showed a significantly high correlation (Figure 5D). For example, the change in Enterobacteriaceae was significantly positively correlated with the change in intestinal levels of BAs, including 3‐DHCA, GCA, CA, and TCDCA. (Figure 5D). Consistent with the results at the family level, changes in several species of Lachnospiraceae were highly correlated with the changes in the intestinal levels of BAs, especially changes in secondary BAs such as LCA and HCA (Figure 5E). Among them, a species of Lachnospiraceae was found to be significantly decreased in cancer cachexia mice (Figure 5F) although no more information was available for this unclassified species.
Figure 5.
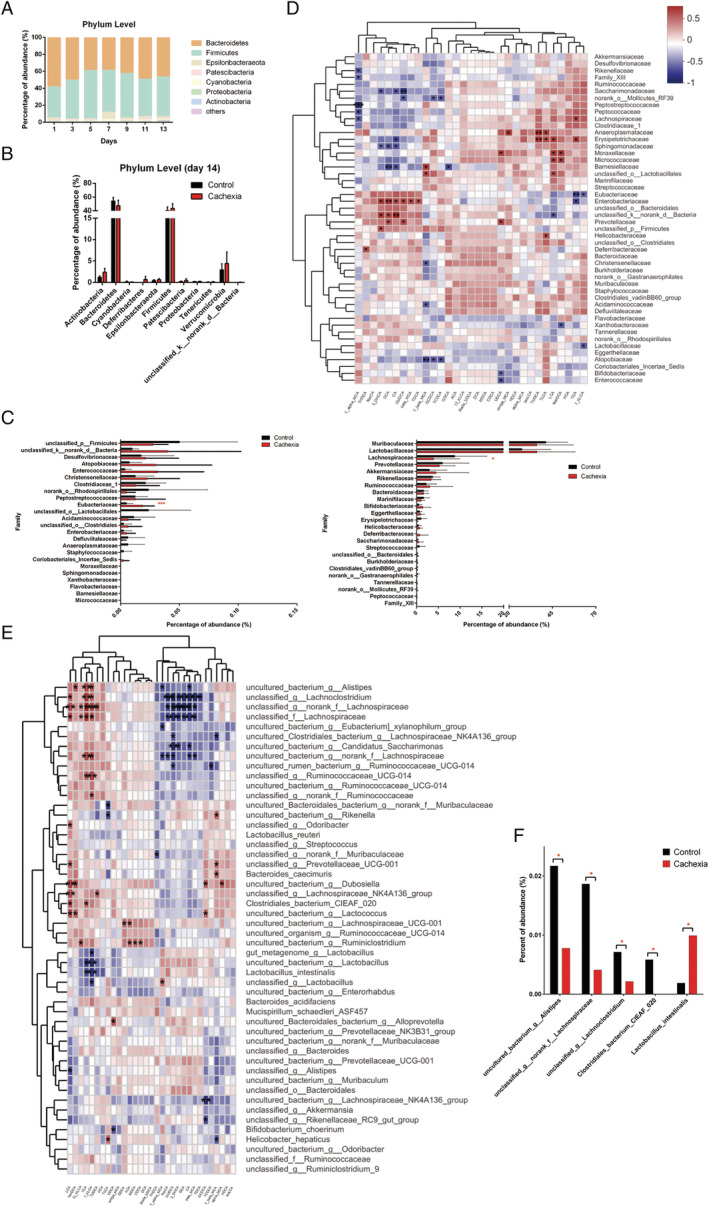
Gut microbiota profiles at the different levels. (A) Time‐related change in the intestinal microbiota profile at phylum level in cachexia mice. (B) Relative abundances of the 11 phylum of intestinal microbiota at Day 14 after tumour inoculation in cachexia mice compared with that of healthy control mice. (C) Relative abundances of the major family of intestinal microbiota in cachexia mice compared with that of healthy control mice. (D) Heatmap of correlation between the intestinal microbial abundance (at family level) and the intestinal levels of BAs. (E) Heatmap of correlation between the intestinal microbial abundance (at species level) and the intestinal levels of BAs. (F) Relative abundances of the major species of cachexia intestinal microbiota were listed. Data presented are the mean ± SEM of results of eight mice in each group. T‐test analysis was performed to assess the significance of the difference. *P < 0.05; **P < 0.01, ***P < 0.001. BA, bile acid.
The serum BA profile of cachexia model mice exhibited an increase in the proportion of conjugated BAs
As shown in Figure 6A, there was a slight increase in the serum level of total BAs in cancer cachexia mice compared with control mice, but the increase was not significant. The total conjugated BAs and total unconjugated BAs were also not significantly changed (Figure 6A). As shown in Figure 6B, in the serum BA pool of cancer cachexia mice, the proportions of TCA and T‐β‐MCA in the total BAs increased. The serum levels of TCA and T‐β‐MCA also increased in the serum of cancer cachexia mice compared with the control, although the increase was not significant (Figure 6C). The ratio of T‐β‐MCA/β‐MCA also increased in cancer cachexia mice, although the increase was not significant (Figure 6E). The ratio of THDCA/HDCA significantly increased, while the ratios of TDCA/DCA and GCA/CA significantly decreased in cancer cachexia mice serum compared with that of the control (Figure 6E).
Figure 6.
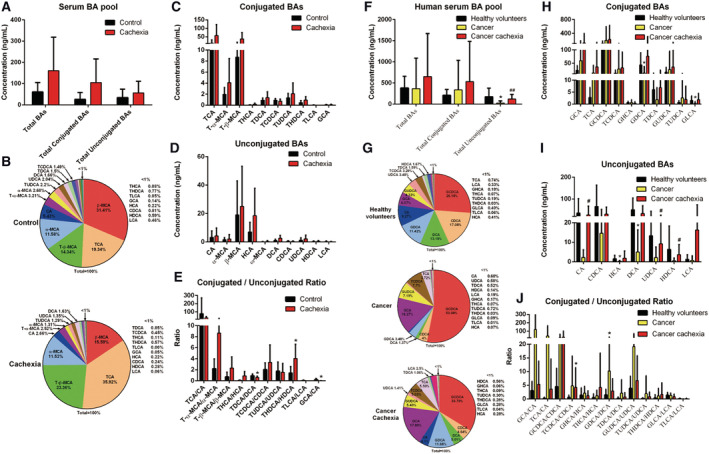
Serum BA profiles analysis of cancer cachexia mice and clinical cancer cachexia patients. (A) The amount of total BAs, total conjugated BAs and total unconjugated BAs in the serum of mouse with cachexia and healthy control mice. (B) Pie graphs are the mean percentage of the individual bile acids in total mice serum BAs. (C) Bar graphs are the concentrations of the conjugated bile acids in mice serum. (D) Bar graphs are the concentrations of the unconjugated bile acids in mice serum. (E) Bar graphs are the ratios of the conjugated/unconjugated bile acids in mice serum. Data presented are the mean ± SEM of results of eight mice in each group. T‐test was performed to assess the significance of the difference. *P < 0.05; **P < 0.01, ***P < 0.001. (F) The amount of total BAs, total conjugated BAs, and total unconjugated BAs in the serum of healthy volunteers, cancer patients without cachexia, and cancer patients with cachexia. (G) Pie graphs are the mean percentage of the individual bile acids in total clinical serum BAs. (H) Bar graphs are the concentrations of the conjugated bile acids in clinical serum. (I) Bar graphs are the concentrations of the unconjugated bile acids in clinical serum. (J) Bar graphs are the ratios of the conjugated/unconjugated bile acids in clinical serum. Data presented are the mean ± SEM of results of 13–22 clinical samples in each group. Ordinary one‐way analysis of variance for multiple comparisons was used to assess the significance of the difference. *P < 0.05; **P < 0.01, ***P < 0.001. BA, bile acid.
The serum BA profile of clinical cancer patients without cachexia was different from that of healthy volunteers as well as that of cancer cachexia patients
The results of the analysis of serum BA profiles of healthy volunteers, cancer patients without cachexia and cancer cachexia patients are shown in Figure 6F–6J. As shown in Figure 6F, there was no significant difference in the amounts of total BAs or total conjugated BAs among the three groups. Interestingly, the serum level of total unconjugated BAs in cancer patients without cachexia was significantly lower than that in healthy control volunteers while the serum level of total unconjugated BAs in cancer cachexia patients was significantly higher than that in cancer patients without cachexia (Figure 6F). The results indicate a possible compensatory response in the development of cachexia. The proportion of various individual BAs in total BAs (Figure 6G) and the serum level of each BA (Figure 6H and 6I) also exhibited a similar tendency. There was a difference between cancer patients without cachexia and healthy volunteers, while the difference was ameliorated between cancer cachexia patients and healthy volunteers. Importantly, compared with healthy volunteers, the only significant change that could be observed in both cancer patients without cachexia and cancer cachexia patients was the increase in BA conjugation, although the increase might also be ameliorated in cancer cachexia patients. For example, the proportion of GCDCA in total BAs was 26.18% in healthy volunteers, 53.08% in cancer patients, and 33.79% in cancer cachexia patients (Figure 6G). The proportion of GCA in total BAs was 6.07% in healthy volunteers, 16.27% in cancer patients, and 17.89% in cancer cachexia patients (Figure 6G). The ratios of conjugated/unconjugated BA also showed similar tendency (Figure 6J). An increase in the ratios of GCA/CA, TCA/CA, GCDCA/CDCA, TCDCA/CDCA, GHCA/HCA, and GDCA/DCA were observed both in comparison between cancer patients without cachexia and healthy controls, as well as in comparison between cancer patients with cachexia and healthy volunteers (Figure 6J). These results suggest that the conjugation of BAs is enhanced in cancer patients with or without cachexia.
TUDCA ameliorated the decrease in body weight and atrophy of the muscle, heart, and liver in cachexia mice
Based on our finding that dysregulation BA metabolism was involved in the development of cancer cachexia and the levels of UDCA and TUDCA were both decreased in cancer cachexia mice intestine, we speculated that oral administration of TUDCA, which could increase the secretion of BA, 23 might be helpful in the treatment of cancer cachexia. The results of the animal experiment assessing the effects of TUDCA in treating cancer cachexia are shown in Figure 7. TUDCA administration successfully ameliorated the decrease in body weight and food intake of cachexia mice without affecting tumour growth (Figure 7A–7C). Figure 7D shows that TUDCA did not affect tumour growth in mice inoculated with C26 tumour cells. TUDCA reversed the muscle atrophy in cancer cachexia and significantly ameliorated the decrease in gastrocnemius muscle weight of cancer cachexia mice (Figure 7E). TUDCA also improved the decrease in adipose tissue weight, although the change was not statistically significant (Figure 7F). TUDCA significantly improved the decrease in heart and liver weights in the cancer cachexia model mice (Figure 7G and 7H). These results suggest the possible usefulness of providing TUDCA in the treatment of cancer cachexia.
Figure 7.
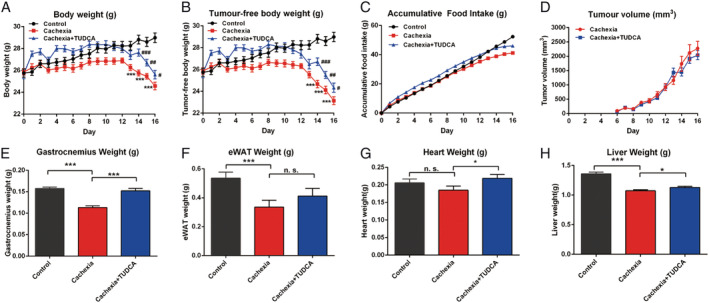
Tauroursodeoxycholic acid (TUDCA) ameliorated the cancer cachexia symptoms of the cancer cachexia mice with inoculation of C26 tumour cells. (A) Body weight of different groups of mice. (B) Tumour‐free body weight of different groups of mice. (C) Accumulative food intake of different groups of mice. (D) Tumour growth curve of C26 tumour‐bearing mice with or without TUDCA treatment. (E) GA muscle tissue weight of different groups of mice. (F) eWAT fat tissue weight of different groups of mice. (G) Heart weight of different groups of mice. (H) Liver weight of different groups of mice. Data presented are the mean ± SEM of results of 10 mice in each group. TUDCA (50 mg/kg) was i.g. administrated daily. Ordinary one‐way analysis of variance for multiple comparisons was used to assess the significance of the difference. *P < 0.05; **P < 0.01, ***P < 0.001 indicate the significance between the indicated two groups (*vs. control group mice; #vs. cachexia group mice.).
Discussion
The first finding in the present study was the decrease in BA synthesis in the liver of cancer cachexia mice. Both the protein and mRNA expression levels of BA synthesis enzymes such as CYP7A1 and CYP27A1 were significantly down‐regulated in the liver of cancer cachexia mice compared with the control. The significant decrease in C4 levels in the serum of cancer cachexia mice also suggested a decrease in BA synthesis. The inhibition of the expression of liver BA synthesis enzymes such as CYP7A1 might result from the activation of the intestinal FXR signalling pathway, as shown by the increase in the expression level of FGF15 in the intestine of cancer cachexia mice. Interestingly, in the liver of cancer cachexia mice, the level of SHP significantly decreased, while the levels of FXR and FGFR4 did not change. It is possible that the liver FXR signalling pathway was not activated or even inhibited (as shown by the decreased SHP), although the elevated FGF15 in the intestine could bind to liver FGFR4 and lead to SHP‐independent inhibition of the expression of BA synthesis enzymes.
Although BA synthesis was decreased, the amount of total liver BAs in the liver of cancer cachexia mice was significantly higher than that in control mice. Less than 10% of the liver BAs are newly synthesized, and most BAs have previously undergone efficient enterohepatic cycling. Although the newly synthesized BAs were decreased, the change in BA transportation, as shown by the changes in the expression of BA transporters, might cause accumulation of BAs in the liver of cancer cachexia mice. In hepatocytes, BAs are taken up across the sinusoidal membrane by sodium‐dependent and independent mechanisms involving NTCP and OATPs. 24 Secretion of monovalent BAs across the canalicular membrane occurs by BSEP, whereas divalent anionic substrates are transported by MRP2. 25 The OSTα‐OSTβ is a unique heteromeric transporter localized to the basolateral membrane of epithelial cells involved in sterol transport, and it prevents the accumulation of toxic compounds along with other basolateral membrane proteins. 26 MDR2 is also an energy‐dependent phospholipid efflux translocator that acts as a positive regulator of biliary lipid secretion. The mRNA levels of NTCP and BSEP were significantly decreased in the liver of cancer cachexia mice compared with those of controls. The levels of MRP2 and organic anion transporter family member 1A1 significantly decreased, while the levels of OSTβ, organic anion transporter family member 1, solute carrier organic anion transporter family member 2A1, and MDR2 significantly increased in the liver of cancer cachexia mice. The expression of transporters such as NTCP, OSTα‐OSTβ, and MDR2 could be regulated by the FXR signalling pathway. 17 The cooperation of the BA synthesis enzymes, transporters, and modification enzymes under concerted control by FXR and other BA‐related nuclear receptors might be the basis of regulation of BA metabolism, even in the presence condition of cancer cachexia.
The second finding in the present study is the increase in BA conjugation in the liver as well as in the serum of cancer cachexia mice. One of the reasons for the increase in BA conjugation in cancer cachexia mice might be the up‐regulation of BA conjugation enzymes such as BAAT. BAAT is the terminal enzyme in the synthesis of bile salts from cholesterol and is the sole enzyme responsible for the conjugation of primary and secondary BAs to taurine and glycine. 27 The expression level of BAAT in the liver of cancer cachexia mice was significantly higher than that in control mice. The increase in BAAT level in cancer cachexia mice liver might result from the activation of liver FXR by BAs and contribute to the increase in BA conjugation in cancer cachexia. Another possible reason for the increase in BA conjugation in cancer cachexia might be the decrease in the deconjugation of BAs by bile salt hydrolase (BSH) in the intestine. BSH, a constitutive intracellular enzyme responsible for the hydrolysis of an amide bond between glycine or taurine and the steroid nucleus of BAs, has been shown to be present in microorganisms of several bacterial genera. 28 Although it is possible that the change in the microbiota profile in the intestine of cancer cachexia mice such as the decrease in Lachnospiraceae might contribute to the possible decrease in BSH‐mediated deconjugation of BAs in cancer cachexia, no conclusion could be reached presently because of the lack of sufficient information about these microorganisms.
The third finding in the present study was the decrease in the microbial metabolism of BAs in cancer cachexia. The levels of secondary BAs and the ratios of secondary BAs to primary BAs were significantly decreased in cancer cachexia mice caecum. Most BAs such as CA are agonists of FXR while several BA species, including T‐β‐MCA and UDCA, were identified as FXR antagonists. 29 The significant increase in the level of CA and the decrease in T‐β‐MCA and UDCA in cancer cachexia mice intestine might contribute to the activation of intestinal FXR signalling pathway. Further 16S rRNA sequencing analysis confirmed microbiota dysbiosis in cancer cachexia mice. There was an increase in Firmicutes in cancer cachexia mice, and the increase reached its peak at Day 7 after inoculation of tumour cells. An altered gut microbiota, characterized by elevated levels of Firmicutes and depleted levels of Bacteroidetes, was reported to be associated with obesity. 30 Our results suggest a relationship between microbiota dysbiosis and weight change in cancer cachexia. At the family level, two significant changes, including a decrease in Lachnospiraceae and an increase in Eubacteriaceae, were observed in the caecum of cancer cachexia mice. Lachnospiraceae comprise of 58 genera and several unclassified strains and, importantly, belongs to the core of gut microbiota colonizing the intestinal lumen from birth and increasing during the host's life. 31 All members of Lachnospiraceae are anaerobic, fermentative, chemoorganotrophic, and some display strong hydrolysing activities. 31 Previous reports have shown that intestinal tumorigenesis is related to low levels of Lachnospiraceae; thus, reagents that could increase Lachnospiraceae are protective against intestinal tumorigenesis. 32 , 33 In the present study, the observed significant decrease in Lachnospiraceae might contribute to the decreased microbial metabolism of BAs in cancer cachexia. In the analysis of the correlation between the change in gut bacteria and the change in intestinal BA profile, Enterobacteriaceae was found to be the most important bacteria whose change was significantly associated with changes in BAs. Our results are in accordance with previous reports which showed that the phenotype of cancer cachexia is associated with increased levels of Enterobacteriaceae. 12 At the same time, the significant increase in Eubacteriaceae observed in cancer cachexia mice might also be interesting. An increase in Eubacteriaceae has been found in patients with depression 34 ; thus, the role of Eubacteriaceae in anorexia of cancer cachexia might deserve further study.
Compared with the two groups (healthy control and cancer cachexia) in the animal study, the results of the three groups of clinical samples (healthy volunteers, cancer without cachexia, and cancer cachexia) were more complicated but could provide more information about both the influence of tumour load on BA profiles and the difference between the cancer and cancer cachexia groups. In summary, the changes in BA metabolism of cancer cachexia found in animal studies (such as decrease in unconjugated BA, increase in BA conjugation, and decrease in secondary BAs) could also be observed in comparison between cancer patients without cachexia and healthy volunteers. Because there might be compensatory self‐regulation mechanisms in BA metabolism regulation, the difference between the serum BA profile of cancer cachexia patients and healthy volunteers was mostly not significant. Interestingly, previous results have also shown stage‐related changes in liver protein metabolism and lipid metabolism during the development of cancer cachexia. In rats inoculated with hepatoma cells (Yoshida AH‐130), there was an initial enlargement and subsequent atrophy of the liver, firstly a decrease and then an augmented protein degradation, as well as elevated protein synthesis in the liver regression phase. 35 Compared with healthy volunteers, the only significant change that could be observed both in cancer patients without cachexia and cancer cachexia patients was the increase in BA conjugation. The increase in BA conjugation might be considered a serum marker of BA metabolism dysregulation in the development of cancer cachexia.
The results of the present study also suggest the possibility of targeting BA synthesis and metabolism in cancer cachexia therapy. UDCA and TUDCA, which have been popularly used in the clinical treatment of liver and biliary diseases, could increase the secretion of BA and show both protective effects on the liver and regulative effects on gut microbiota. 23 , 36 , 37 , 38 UDCA exhibited promising results in the treatment of cancer cachexia mice. 9 Our study examining the influence of TUDCA administration on C26 cancer cachexia mice (as shown in Figure 7) showed that TUDCA successfully ameliorated weight loss, muscle and fat loss, heart atrophy, and liver atrophy in cancer cachexia mice. The efficacy of UDCA and TUDCA in the treatment of cancer cachexia warrants further study.
Based on the findings of the present study, an illustration of the possible changes in BA metabolism in the development of cancer cachexia and the contribution of the liver and intestinal microbiota are shown in Figure 8. Although considerable changes such as increased BA conjugation were observed, there are many limitations in the present study. The mechanisms by which the amounts of total BAs in the liver, intestine, and serum were maintained at almost normal or even increased levels in cancer cachexia mice under the inhibition of liver BA synthesis are unclear. The lack of sufficient information about the mice gut microbiota, especially those of unclassified species, made it impossible to draw clear conclusions about the contribution of gut microbiota dysbiosis in cancer cachexia development. The change in BA metabolism (synthesis and modification) and the development of cancer cachexia might be a vicious cycle. The present study provides evidence about the involvement of dysregulation of BA metabolism in the development of cancer cachexia, both in cancer cachexia animals and in cancer cachexia patients. Comparison of the BA profiles in healthy volunteers, cancer patients without cachexia, and cancer cachexia patients indicated the adaptive compensatory mechanisms in the development of cancer cachexia. Furthermore, the results of the present study suggest the possibility of targeting BA metabolism using agents such as TUDCA for the treatment of cancer cachexia.
Figure 8.
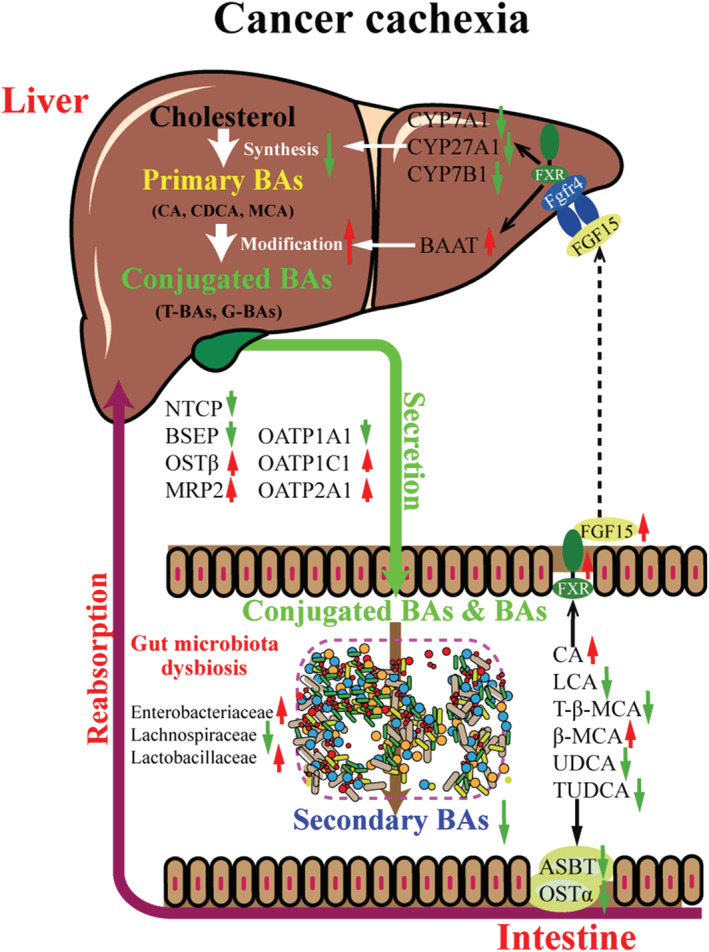
Illustration of the contribution of liver and gut microbiome to the BA metabolism dysregulation associates with cancer cachexia. Liver health, BA profile, and gut microbiota composition are closely connected. On the one hand, in liver, BA synthesis decreases while the conjugation of BAs is enhanced in the development of cancer cachexia. On the other hand, in intestine, there is microbiota dysbiosis which resulted in decrease in the microbial metabolism of BAs. Change in intestine BA profile induced activation of intestine‐specific FXR signalling pathway thus produces FGF15 to inhibit expression of BA synthesis enzymes and modification enzymes in liver. Expression of BA transporters in intestine and liver was also changed by FXR signalling pathway or other BA metabolism‐related signalling pathways. Importantly, there are feedback mechanisms in BA metabolism regulation and possible compensatory mechanisms in the development of cancer cachexia; thus; the BA metabolism might be dynamically changed in the different stages of cancer cachexia. BA, bile acid.
Funding
This work was supported by National Natural Science Foundation of China (81872496, 81873056, and 81772905) and the Science and Technology Commission of Shanghai Municipality (20S11902200 and 16DZ2280100).
Author contributions
Xiongwen Zhang and Xuan Liu conceived and designed the study. Lixing Feng, Wanli Zhang, Qiang Shen, Chunxiao Miao, Xiaofan Gu, Yiwei Li, Meng Fan, and Lijuan Chen generated the laboratory data. Hui Wang and Yushui Ma designed and performed the primary clinical study. Xuan Liu and Lixing Feng interpreted the data and wrote the manuscript. Xiongwen Zhang revised the manuscript. All authors critically reviewed the paper and approved of the final version of the paper for submission.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Table S1. The colon cancer cachexia stating score system for classifying cachexia stages.
Table S2. Clinical characteristics of colon cancer patients with Cachexia or Non‐cachexia.
Table S3. The targeted bile acid panel used to measure BAs.
Table S4. Primers used in RT‐PCR analysis.
Data S1. Supporting information.
Acknowledgements
The authors thank Shanghai Majorbio Bio‐pharm Technology Co., Ltd for Proteomics and Metabolomics and 16S rRNA sequencing services; all the multi‐omics date analysis was performed using the free online platform of Majorbio Cloud Platform (www.majorbio.com). The authors certify that they comply with the ethical guidelines for authorship and publishing of the Journal of Cachexia, Sarcopenia and Muscle. 39
Feng L., Zhang W., Shen Q., Miao C., Chen L., Li Y., Gu X., Fan M., Ma Y., Wang H., Liu X., and Zhang X. (2021) Bile acid metabolism dysregulation associates with cancer cachexia: roles of liver and gut microbiome, Journal of Cachexia, Sarcopenia and Muscle, 12, 1553–1569, 10.1002/jcsm.12798
Lixing Feng, Wanli Zhang, and Qiang Shen contributed equally to this work and are co‐first authors.
Contributor Information
Xuan Liu, Email: xuanliu@shutcm.edu.cn.
Xiongwen Zhang, Email: xwzhang@sat.ecnu.edu.cn.
References
- 1. Brown J, Rosa‐Caldwell ME, Lee DE, Blackwell TA, Brown LA, Perry RA, et al. Mitochondrial degeneration precedes the development of muscle atrophy in progression of cancer cachexia in tumour‐bearing mice. J Cachexia Sarcopenia Muscle 2017;8:926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu R, Liu Z, Jiao R, Zhang C, Yu Q, Han S, et al. Updates on the pathogenesis of advanced lung cancer‐induced cachexia. Thorac Cancer 2019;10:8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baracos VE, Martin L, Korc M, Guttridge DC, Fearon KC. Cancer‐associated cachexia. Nat Rev Dis Primers 2018;4:17105. [DOI] [PubMed] [Google Scholar]
- 4. Kazgan N, Metukuri MR, Purushotham A, Lu J, Rao A, Lee S, et al. Intestine‐specific deletion of SIRT1 in mice impairs DCoH2‐HNF‐1α‐FXR signaling and alters systemic bile acid homeostasis. Gastroenterology 2014;146:1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Y, Gao X, Zhang X, Xiao Y, Huang J, Yu D, et al. Gut microbiota dysbiosis is associated with altered bile acid metabolism in infantile cholestasis. mSystems 2019;4:e00463–e00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kühn T, Stepien M, López‐Nogueroles M, Damms‐Machado A, Sookthai D, Johnson T, et al. Prediagnostic plasma bile acid levels and colon cancer risk: a prospective study. J Natl Cancer Inst 2020;112:516–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castellani C, Singer G, Kaiser M, Kaiser T, Huang J, Sperl D, et al. Neuroblastoma causes alterations of the intestinal microbiome, gut hormones, inflammatory cytokines, and bile acid composition. Pediatr Blood Cancer 2017;64. [DOI] [PubMed] [Google Scholar]
- 8. Yu B, Peng XH, Wang LY, Wang AB, Su YY, Chen JH, et al. Abnormality of intestinal cholesterol absorption in Apc (Min/+) mice with colon cancer cachexia. Int J Clin Exp Pathol 2019;12:759–767. [PMC free article] [PubMed] [Google Scholar]
- 9. Tschirner A, von Haehling S, Palus S, Doehner W, Anker SD, Springer J. Ursodeoxycholic acid treatment in a rat model of cancer cachexia. J Cachexia Sarcopenia Muscle 2012;3:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rosa‐Caldwell M, Brown JL, Lee DE, Wiggs MP, Perry RA Jr, Haynie WS, et al. Hepatic alterations during the development and progression of cancer cachexia. Appl Physiol Nutr Metab 2020;45:500–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Puppa MJ, White JP, Sato S, Cairns M, Baynes JW, Carson JA. Gut barrier dysfunction in the ApcMin/+ mouse model of colon cancer cachexia. Biochim Biophys Acta Mol Basis Dis 2011;1812:1601–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bindels LB, Neyrinck AM, Claus SP, Le Roy CI, Grangette C, Pot B, et al. Synbiotic approach restores intestinal homeostasis and prolongs survival in leukaemic mice with cachexia. ISME J 2016;10:1456–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Molinari F, Pin F, Gorini S, Chiandotto S, Pontecorvo L, Penna F, et al. The mitochondrial metabolic reprogramming agent trimetazidine as an ‘exercise mimetic’ in cachectic C26‐bearing mice. J Cachexia Sarcopenia Muscle 2017;8:954–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Villars FO, Pietra C, Giuliano C, Lutz TA, Riediger T. Oral treatment with the ghrelin receptor agonist HM01 attenuates cachexia in mice bearing colon‐26 (C26) tumors. Int J Mol Sci 2017;18:986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gadaleta R, Cariello M, Sabbà C, Moschetta A. Tissue‐specific actions of FXR in metabolism and cancer. Biochim Biophys Acta Mol Cell Biol Lipids 2015;1851:30–39. [DOI] [PubMed] [Google Scholar]
- 16. Modica S, Gadaleta R, Moschetta A. Deciphering the nuclear bile acid receptor FXR paradigm. Nucl Recept Signal 2010;8:e005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Modica S, Petruzzelli M, Bellafante E, Murzilli S, Salvatore L, Celli N, et al. Selective activation of nuclear bile acid receptor FXR in the intestine protects mice against cholestasis. 2012;142:355–65.e1‐4. [DOI] [PubMed] [Google Scholar]
- 18. Miao C, Lv Y, Zhang W, Chai X, Feng L, Fang Y, et al. Pyrrolidine dithiocarbamate (PDTC) attenuates cancer cachexia by affecting muscle atrophy and fat lipolysis. Front Pharmacol 2017;8:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shen H, Chen W, Drexler DM, Mandlekar S, Holenarsipur VK, Shields EE, et al. Comparative evaluation of plasma bile acids, dehydroepiandrosterone sulfate, hexadecanedioate, and tetradecanedioate with coproporphyrins I and III as markers of OATP inhibition in healthy subjects. Drug Metab Dispos 2017;45:908–919. [DOI] [PubMed] [Google Scholar]
- 20. Kodama Y, Shumway M, Leinonen R. The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res 2012;40:D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma J, Chen T, Wu S, Yang C, Bai M, Shu K, et al. iProX: an integrated proteome resource. Nucleic Acids Res 2019;47:D1211–D1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haug K, Cochrane K, Nainala VC, Williams M, Chang J, Jayaseelan KV, et al. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res 2020;48:D440–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baiocchi L, Tisone G, Russo MA, Longhi C, Palmieri G, Volpe A, et al. TUDCA prevents cholestasis and canalicular damage induced by ischemia‐reperfusion injury in the rat, modulating PKCalpha‐ezrin pathway. Transpl Int 2008;21:792–800. [DOI] [PubMed] [Google Scholar]
- 24. Li J, Dawson PA. Animal models to study bile acid metabolism. Biochim Biophys Acta Mol Basis Dis 2019;1865:895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Geier A, Kim SK, Gerloff T, Dietrich CG, Lammert F, Karpen SJ, et al. Hepatobiliary organic anion transporters are differentially regulated in acute toxic liver injury induced by carbon tetrachloride. J Hepatol 2002;37:198–205. [DOI] [PubMed] [Google Scholar]
- 26. Soroka C, Ballatori N, Boyer JL. Organic solute transporter, OSTalpha‐OSTbeta: its role in bile acid transport and cholestasis. Semin Liver Dis 2010;30:178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pellicoro A, van den Heuvel FA, Geuken M, Moshage H, Jansen PL, Faber KN. Human and rat bile acid‐CoA:amino acid N‐acyltransferase are liver‐specific peroxisomal enzymes: implications for intracellular bile salt transport. Hepatology 2007;45:340–348. [DOI] [PubMed] [Google Scholar]
- 28. Horáčková Š, Plocková M, Demnerová K. Importance of microbial defence systems to bile salts and mechanisms of serum cholesterol reduction. Biotechnol Adv 2018;36:682–690. [DOI] [PubMed] [Google Scholar]
- 29. Cao L, Che Y, Meng T, Deng S, Zhang J, Zhao M, et al. Repression of intestinal transporters and FXR‐FGF15 signaling explains bile acids dysregulation in experimental colitis‐associated colon cancer. Oncotarget 2017;8:63665–63679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Riva A, Borgo F, Lassandro C, Verduci E, Morace G, Borghi E, et al. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ Microbiol 2017;19:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vacca M, Celano G, Calabrese FM, Portincasa P, Gobbetti M, De Angelis M. The controversial role of human gut Lachnospiraceae. Microorganisms 2020;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang F, Liu M, Wang H, Shi G, Chen B, Chen T, et al. Wu Mei Wan attenuates CAC by regulating gut microbiota and the NF‐kB/IL6‐STAT3 signaling pathway. 2020;125:109982. [DOI] [PubMed] [Google Scholar]
- 33. Mai V, Colbert LH, Perkins SN, Schatzkin A, Hursting SD. Intestinal microbiota: a potential diet‐responsive prevention target in ApcMin mice. Mol Carcinog 2007;46:42–48. [DOI] [PubMed] [Google Scholar]
- 34. Barandouzi ZA, Starkweather AR, Henderson WA, Gyamfi A, Cong XS. Altered composition of gut microbiota in depression: a systematic review. Front Psych 2020;11:541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tessitore L, Bonelli G, Baccino F. Early development of protein metabolic perturbations in the liver and skeletal muscle of tumour‐bearing rats. A model system for cancer cachexia. Biochem J 1987;241:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cabrera D, Arab JP, Arrese M. UDCA, NorUDCA, and TUDCA in liver diseases: a review of their mechanisms of action and clinical applications. Handb Exp Pharmacol 2019;256:237–264. [DOI] [PubMed] [Google Scholar]
- 37. Lu Q, Jiang Z, Wang Q, Hu H, Zhao G. The effect of tauroursodeoxycholic acid (TUDCA) and gut microbiota on murine gallbladder stone formation. Ann Hepatol 2020;23:100289. [DOI] [PubMed] [Google Scholar]
- 38. Ma H, Zeng M, Han Y, Yan H, Tang H, Sheng J, et al. A multicenter, randomized, double‐blind trial comparing the efficacy and safety of TUDCA and UDCA in Chinese patients with primary biliary cholangitis. Medicine (Baltimore) 2016;95:e5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. von Haehling S, Morley JE, Coats AJ, Anker SD. Ethical guidelines for publishing in the journal of cachexia, sarcopenia and muscle: update 2019. J Cachexia Sarcopenia Muscle 2019;10:1143–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The colon cancer cachexia stating score system for classifying cachexia stages.
Table S2. Clinical characteristics of colon cancer patients with Cachexia or Non‐cachexia.
Table S3. The targeted bile acid panel used to measure BAs.
Table S4. Primers used in RT‐PCR analysis.
Data S1. Supporting information.
Data Availability Statement
16S rRNA sequencing data have been deposited in the NCBI SRA database 20 under accession number SRP301790 and SRP301551. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository 21 with the data set identifier PXD023497. Proteomic data were searched against a uniprot database containing Mus musculus proteins (https://www.uniprot.org/proteomes/); Metabolomics data have been deposited to the EMBL‐EBI MetaboLights database 22 with the identifier MTBLS2389. The complete data set can be accessed here https://www.ebi.ac.uk/metabolights/MTBLS2389.


