Abstract
Pancreatic ductal adenocarcinoma (PDAC) is among the deadliest cancers. Dissecting the tumor cell proteome from that of the non-tumor cells in the PDAC tumor bulk is critical for tumorigenesis studies, biomarker discovery, and development of therapeutics. However, investigating the tumor cell proteome has proven evasive due to the tumor’s extremely complex cellular composition. To circumvent this technical barrier, we have combined bioorthogonal noncanonical amino acid tagging (BONCAT) and data-independent acquisition mass spectrometry (DIA-MS) in an orthotopic PDAC model to specifically identify the tumor cell proteome in vivo. Utilizing the tumor cell-specific expression of a mutant tRNA synthetase transgene, this approach provides tumor cells with the exclusive ability to incorporate an azide-bearing methionine analogue into newly synthesized proteins. The azide-tagged tumor cell proteome is subsequently enriched and purified via a bioorthogonal reaction and then identified and quantified using DIA-MS. Applying this workflow to the orthotopic PDAC model, we have identified thousands of proteins expressed by the tumor cells. Furthermore, by comparing the tumor cell and tumor bulk proteomes, we showed that the approach can distinctly differentiate proteins produced by tumor cells from those of non-tumor cells within the tumor microenvironment. Our study, for the first time, reveals the tumor cell proteome of PDAC under physiological conditions, providing broad applications for tumorigenesis, therapeutics, and biomarker studies in various human cancers.
Keywords: pancreatic ductal adenocarcinoma (PDAC), bioorthogonal noncanonical amino acid tagging (BONCAT), data-independent acquisition mass spectrometry (DIA-MS), methionyl-tRNA synthetase (MetRS), azidonorleucine (ANL), patient-derived xenografts (PDX)
Graphical Abstract
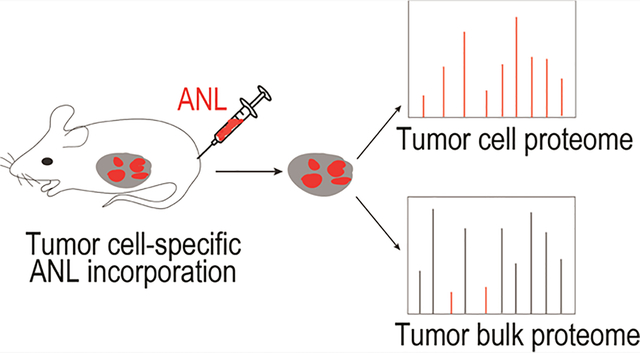
INTRODUCTION
The tumor bulk is not only a mass of proliferating tumor cells but also consists of a variety of non-tumor cells, secreted factors, and the extracellular matrix, which are collectively known as the tumor microenvironment (TME). The interaction between tumor cells and the surrounding TME has a profound impact on all stages of tumor development. Human pancreatic ductal adenocarcinoma (PDAC), in particular, has a highly complex TME, imparted by a dense desmoplastic stroma and a host of stromal fibroblasts and endothelial, inflammatory, and immune cells. The stromal components of human PDAC may account for up to 80% of the total tumor volume, with tumor cells constituting a minor population.1–3 Differentiating proteins produced by the PDAC tumor cells from those of non-tumor cells in the TME is critical for tumorigenesis and therapeutic studies. However, the heterogeneous and complex cellular composition of the PDAC tumor mass has, thus far, precluded precise isolation and identification of the PDAC tumor cell proteome in vivo.
Direct investigation of the PDAC tumor cell proteome requires selective purification of proteins from the tumor cells and not the non-tumor cells within the tumor bulk. Several recent studies have focused on cell-selective metabolic labeling of the proteomes.4 These approaches include cell-type-specific labeling using amino acid precursors (CTAP),5 bioorthogonal noncanonical amino acid tagging (BONCAT),6 and stochastic orthogonal recording of translation (SORT).7 BONCAT has been shown to label cell-selective proteomes in the fruit fly8 as well as mouse brain and muscle.9–11 BONCAT works through bioorthogonal chemical reactions that do not exist in nature and, thus, will not cross-react with any physiological processes in the cells.12,13 This technique relies on bioorthogonal incorporation of azide-bearing methionine analogues, such as azidonorleucine (ANL) and azidohomoalanine (AHA), into newly synthesized polypeptides. Due to the small size of the azide moiety, ANL or AHA incorporation has no apparent effect on protein function.9–11 During protein translation, ANL is preferentially recognized and charged onto tRNAMet by a mutant methionyl-tRNA synthetase (MetRSL274G) and is subsequently incorporated in the elongating polypeptide chains (Figure 1A,B).14 ANL-tagged proteins can be selectively conjugated and enriched through azide–alkyne cycloaddition.13 Further identification of the proteome is achieved through mass spectrometric (MS) analysis of the ANL-tagged proteins. ANL incorporation is unbiased, non-toxic, and biocompatible and does not affect protein stability.6 We therefore adopted the BONCAT approach for in vivo application to identify the tumor cell proteome of PDAC (Figure 1C). The ectopic expression of MetRSL274G transgene in PDAC tumor cells, but not the non-tumor cells in the tumor bulk, enables the exclusive tagging of the tumor cell proteome with ANL. Following ANL labeling, the tumor cell proteome is enriched and purified for MS analysis. Thus, applying BONCAT to the animal model of PDAC facilitates the identification of tumor cell proteome in a physiological context.
Figure 1.
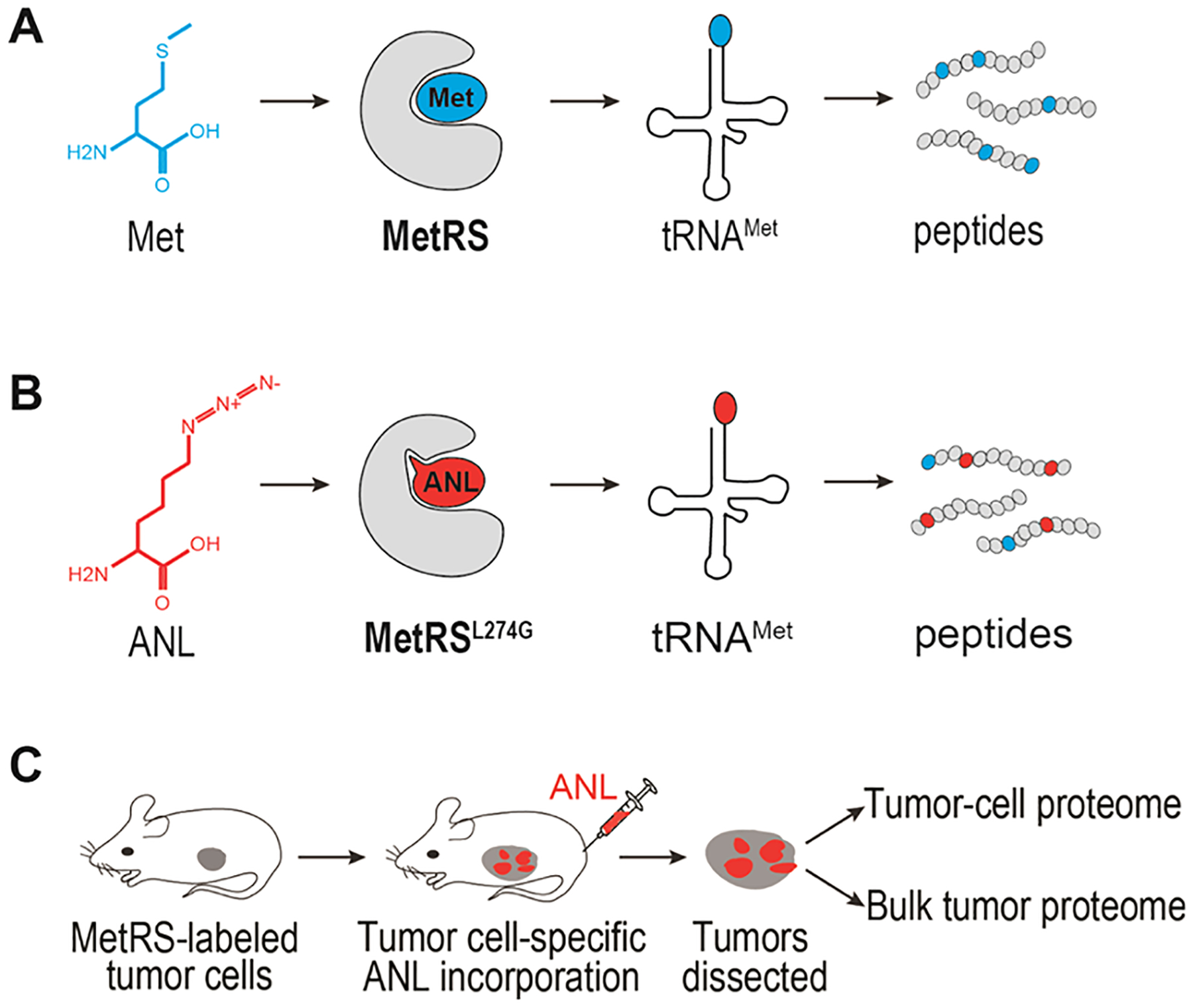
MetRSL274G-assisted peptide labeling and BONCAT reaction. (A, B) Diagram showing cell-selective labeling of proteome by BONCAT. (C) Overview of in vivo BONCAT for the identification of tumor cell proteome.
Data-independent acquisition mass spectrometry (DIA-MS) is a highly reproducible, state-of-the-art approach for quantitative proteomic analysis.15–19 Traditionally, data-dependent acquisition mass spectrometry (DDA-MS) has been used in a variety of label-free and label-based methods to measure quantitative changes in global protein levels in biological samples. However, the stochastic nature of DDA bears a bias toward higher abundance peptides. Under-sampling of medium- and low-abundance peptides causes inconsistencies in detection of peptides and hampers reproducibility among replicates. In the DIA-MS approach, all precursors are fragmented to yield tandem-MS data, providing sequence information from virtually all peptides in a sample with minimal loss of information. Due to its high accuracy and reproducibility, DIA-MS is a powerful method for comprehensive proteomic studies of complex samples, including tumor specimens.20–22
Here, we have combined BONCAT bioorthogonal chemistry and DIA-MS proteomics to specifically investigate the tumor cell proteome in an orthotopic transplantation model of PDAC. We have identified over 3000 proteins expressed in PDAC tumor cells, many of which are predominantly, if not exclusively, expressed in the tumor cells. Thus, we have established a robust technical platform for in vivo identification of the proteome of the tumor cells embedded within the bulk tumor, with broad applications in the studies of tumorigenesis, cancer therapeutics, and cancer detection.
MATERIALS AND METHODS
Cell Lines, Constructs, and Chemical Reagents
The 4292 murine PDAC cell line was a generous gift from Dr. Marina Pasca di Magliano.23 KRAS expression in this cell line is controlled by the Tet-ON system. The MetRSL274G coding sequence was PCR amplified using the pMarsL274G construct (Addgene 63177) as a template and inserted into BamHI and MluI sites within the pLV-EF1a-IRES-puro vector (Addgene 85132) to produce the Lentiviral-MetRSL274G vector. The FLAG M2 antibody used for the detection of the MetRSL274G protein was purchased from Sigma. ANL, H-L-Lys(N3)-OH*HCL (HAA1625), was obtained from Peptide Solutions (Tucson, AZ). DBCO-agarose beads (1034) and DBCO-TAMRA (A131) were purchased from Click Chemistry Tools and tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine from Fisher Scientific.
Cell Culture and ANL Labeling In Vitro
In total, 4292 murine cells and 4292 cells harboring the MetRSL274G construct were cultured to 80–90% confluency in RPMI 1640 supplemented with 10% FBS, 1% penicillin/streptomycin, and 1 μg/mL doxycycline. Growth media was then removed and replaced with the media supplemented with 4 mM ANL or Met. Cells were allowed to grow for an additional 5 h and subsequently harvested and resuspended in PBS (pH 7.4), 1% SDS, 100 mM chloroacetamide, and protease inhibitors. The homogenate was left at room temperature for 20–30 min to allow protein solubilization. Lysates were boiled for 10 min and centrifuged at room temperature at 16 000g for 10 min to remove cell debris.
Click Chemistry Reactions
TAMRA Reaction.
To examine labeling efficiency, 20 μg of protein lysate was incubated with 30 μM DBCO-TAMRA/PBS (pH 7.4) for 1 h at room temperature. Samples were boiled in Laemmli buffer and run on SDS-PAGE gels. Electrophoresed samples were visualized using the ChemiDoc imaging system (Bio-Rad) with Pro-Q Diamond filters (absorbance/emission of 548/562 nm). The gel was subsequently stained with Imperial stain (Coomassie dye R-250, Thermo Fisher 24615) according to the manufacturer’s recommendation and visualized on the ChemiDoc imaging system. Densitometric quantification of Coomassie blue and TARMA-alkyne stained SDS-PAGE gels was performed with Image Lab (version 6.0. Bio-Rad).
CuAAC.
A copper-assisted click reaction was performed on paraffin-embedded slides. Slides were deparaffinized in two changes of xylene, 5 min per change, and rehydrated sequentially in two changes, 5 min each of 100% ethanol and 95% ethanol, and 5 min of 70% ethanol, and changed into water. To quench endogenous peroxidase, slides were immersed in 3% H2O2 for 15 min at room temperature. Slides were washed three times with PBS/0.1% triton X-100. Copper-assisted reaction was essentially performed as described.24 Briefly, the orthogonal tagging reaction was assembled in the dark; 5 μL of 200 mM TBTA, 5 μL of 500 mM TCEP, 5 μL of 2 mM biotin-alkyne-tag (Click Chemistry Tools, 1266), and 5 μL of 200 mM CuSO4 were added in the specified order to 5 mL of PBS (pH 7.8), and the mixture was vortexed for 10 s after each addition. The slides were reacted with the mixture overnight at room temperature. Slides were subsequently washed three times, 20 min each, in PBS (pH 7.8), 0.5 mM EDTA, and 1% Tween 20, followed by two washes, 10 min each, of PBS (pH 7.8) and 0.1% Tween 20. Slides were finally washed twice with the PBS (pH 7.4). For signal amplification and HRP conjugation, samples were incubated with VECTASTAIN Elite ABC reagent for 30 min, washed for 15 min in PBS (pH 7.4), and changed into water. Signals were detected using the ImmPACT DAB Peroxidase (HRP) Substrate (Vector laboratories, SK-4105).
IHC.
Staining was performed with the VECTASTAIN ABC Elite kit (Vector Labs) according to the accompanying protocol. Antibodies for α-SMA (19245), EZH2 (5246), RAS (67648), ZMIZ1 (89500), and ARID1B (65747) were purchased from Cell Signaling Technology. Ki67 antibody (ab 16667) was purchased from Abcam. Briefly, slides were deparaffinized in three changes of xylene, 15 min each, and rehydrated in changes of 100% ethanol, 95% ethanol, and 75% ethanol for 5 min each. Sections were then washed in water twice, 5 min per wash. Antigens were unmasked by boiling the slides for 15–20 min in antigen unmasking citrate buffer (Cell Signaling Technology, 14746) and cooled down to room temperature. Endogenous peroxidase activity was quenched by incubating the slides in 3% hydrogen peroxide/H2O. Slides were washed in H2O and PBS and subsequently blocked with normal blocking serum for 30 min at room temperature. Primary antibody incubation was performed overnight at 4 °C. Following three washes in PBS, tissues were incubated in secondary biotinylated antibody for 1 h at room temperature. Tissues were treated with the avidin/biotin complex reaction according to the manufacturer’s protocol. Slides were finally washed with PBS, changed into water, and developed using the ImmPACT DAB HRP substrate kit (Vector Labs SK-4105).
Animal Models, Orthotopic Transplantation, and ANL Labeling
Animal studies and experimental protocols were approved by the Institutional Animal Care and Use Committee at Houston Methodist Research Institute. All experimental methods were performed in accordance with the relevant national and institutional guidelines and regulations. Six- to eight-week-old NOD-scid IL2Rγnull (NSG) mice underwent surgical orthotopic injection of the pancreatic cancer cells in their pancreata. Carprofen medicated gel (5 mg/kg/day) was used for analgesia prior to the surgery, and within 3 days following the surgical procedure. Mice were anesthetized with isoflurane. The abdominal skin directly above the spleen was incised and the pancreas was retracted laterally and positioned outside the body. Direct injection of 1 × 106 cells was performed using a 28.5-G needle. The needle was inserted through the knot into the pancreas tail and passed into the pancreas head to deliver the cells. Following cell injection, the spleen and pancreas were returned to the peritoneal cavity, and the abdominal muscle and the skin layers were sequentially sutured. One day following surgery, doxycycline was administered through drinking water at a concentration of 0.2 g/L in a solution of 5% sucrose and replaced every 3–4 days. On day 4 post-surgery, experimental and control mice were IP injected with 0.1 mg/g per day of the amino acid analogue or normal saline, respectively, for 10 days.25
BONCAT Enrichment
Tumor nodules were harvested and snap-frozen in liquid nitrogen until further use. Frozen tumor samples were homogenized for 20–60 s in PBS (pH 7.4), 1% SDS, 100 mM chloroacetamide, and protease inhibitors. The homogenate was left at room temperature for 20–30 min to allow protein solubilization. Lysates were boiled for 10 min and centrifuged at room temperature at 16 000g for 10 min. The supernatant was separated and aliquoted. Protein concentration was determined using BCA protein assay. Supernatants were used for the identification of tumor bulk (Bulk-ANL and Bulk-Met) and tumor cell proteomes following BONCAT enrichment (BONCAT-ANL and BONCAT-Met). The conditions for BONCAT enrichment were based on a previously published protocol.26 Approximately 1.5 mg of protein was diluted 2× with 8 M urea/0.15 M NaCl/PBS (pH 7.4) to a total volume of 1 mL. Fifty microliters of DBCO-agarose bead 2× slurry was washed three times with 0.8% SDS in PBS (pH 7.4). The diluted protein sample was added to the washed resin and shaken at 1200 rpm at room temperature for more than 12 h. Unreacted DBCO was quenched by the addition of 2 mM ANL for 30 min. Resins were washed with 1 mL of water and reduced with 1 mM DTT (in 0.8% SDS in PBS) for 15 min at 70 °C with shaking at 1200 rpm. Free thiols were subsequently blocked with 40 mM iodoacetamide (in 0.8% SDS in PBS) for 30 min in the dark with shaking at 1200 rpm. Resins were then subjected to the following washes: 40 mL of 0.8% SDS in PBS, 40 mL of 8 M urea, and 40 mL of 20% acetonitrile. Beads were then washed with 10% acetonitrile in 50 mM ammonium bicarbonate. The beads were spun at 2000g to remove the liquid and resuspended in 100 μL of 10% acetonitrile in 50 mM ammonium bicarbonate and 100 ng of trypsin (Thermo Scientific Pierce, 90057). Beads were digested at 37 °C on a shaking platform overnight, washed three times with 20% acetonitrile, and subsequently removed using centrifuge columns (Thermo Scientific, 89868). Digested peptides were dried at 30 °C with a vacuum concentrator (Vacufuge Plus, Eppendorf) and subsequently subjected to DIA-MS analysis (see below).
Lysis and Digestion of Tumor Bulk
Tumor bulk cells were lysed in a buffer containing 5% SDS/50 mM triethylammonium bicarbonate (TEAB) in the presence of protease and phosphatase inhibitors (Halt; Thermo Scientific) and nuclease (Pierce Universal Nuclease for Cell Lysis; Thermo Scientific). Aliquots corresponding to 100 μg of protein (EZQ Protein Quantitation Kit; Thermo Scientific) were reduced with tris(2-carboxyethyl)phosphine hydrochloride (TCEP), alkylated in the dark with iodoacetamide, and applied to S-Traps (mini; Protifi) for tryptic digestion (sequencing grade; Promega) in 50 mM TEAB. Peptides were eluted from the S-Traps with 0.2% formic acid in 50% aqueous acetonitrile, quantified using Pierce Quantitative Fluorometric Peptide Assay (Thermo Scientific), and diluted as needed to achieve a concentration of 0.4 μg/μL.
DIA-MS Proteomic Analyses
Experimental samples were randomized for sample preparation and analysis. DIA-MS analyses were conducted on an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific). On-line HPLC separation was accomplished with an RSLC NANO HPLC system (Thermo Scientific/Dionex): column, PicoFrit (New Objective; 75 μm i.d.) packed to 15 cm with C18 adsorbent (Vydac; 218MS 5 μm, 300 Å); mobile phase A, 0.5% acetic acid (HAc)/0.005% trifluoroacetic acid (TFA) in water; mobile phase B, 90% acetonitrile/0.5% HAc/0.005% TFA/9.5% water; gradient 3–42% B in 120 min; and flow rate, 0.4 μL/min. Separate pools were made of all of the samples in each experiment (equal volumes from the BONCAT-ANL/BONCAT-Met digests; equal quantities for the tumor bulk lysate digests). For the tumor bulk lysates, injections of 2 μg of peptides of the pooled samples were used for chromatogram library generation.27 For the BONCAT-ANL and BONCAT-Met samples, aliquots of the pool of equal volumes of the digests were injected. To create the DIA chromatogram library for each sample type, the indicated peptide quantities were analyzed using gas-phase fractionation and 4-m/z windows (staggered; 30-k resolution for precursor and product ion scans, all in the orbitrap) and the MS files processed in Scaffold DIA (v2.1.0; Proteome Software) and searched against a predicted spectral library generated from the UniProt_mouse (2019_01) protein database by Prosit.28 Injections of 2 μg of peptides were employed for DIA-MS analysis of the individual bulk tumor lysate digests, while injections corresponding to equal volumes were used for the BONCAT-ANL and BONCAT-Met samples. MS data for all individual digests were acquired in the orbitrap using 12-m/z windows (staggered; 30-k resolution for precursor and product ion scans) and searched against the chromatogram library. Scaffold DIA (v2.1.0; Proteome Software) was used for processing the DIA data from the experimental samples. Only peptides that were exclusively assigned to a protein were used for relative quantification, with two minimum peptides required for each protein and a protein-level FDR of 1%.
Correlations among different BONCAT-enriched samples and total lysate samples were analyzed by Pearson correlation. Protein intensity values (Supplementary Tables S1 and S2) were log10-transformed. Differentially abundant proteins were analyzed by a moderated Student’s t-test via the limma package.29 A paired design was used for the BONCAT-ANL vs Bulk-ANL analysis to account for samples being derived from the same tumor. For log2 fold change calculations, first, missing values were imputed via the “Quantile Regression for Imputation of Left-Censored data” (QRLIC) method implemented in Scaffold DIA software. Then, the difference in medians of the log10-transformed protein intensity values between each group was computed and converted to base 2. A twofold change cutoff and a false discovery rate (FDR) <0.05 threshold were used to identify differentially abundant proteins. Gene ontology term enrichment analysis was performed using enrichR.30
RESULTS
Construction and Validation of PDAC-BONCAT Cells
To label the proteome of PDAC tumor cells, the MetRSL274G mutant transgene was cloned into a lentiviral vector and delivered to a murine pancreatic cancer cell line (4292)23 via lentiviral infection. Single-cell clones were derived and the expression of FLAG-tagged MetRSL274G was confirmed by western blot analysis (Figure 2A, Supplementary Figure S1). ANL incorporation by MetRSL274G in the tumor cell proteome was visualized using the azide-reactive red-fluorescent tetramethylrhodamine dibenzocyclooctyne (TAMRA-DBCO) alkyne probe. First, metabolic labeling was achieved by growing 4292 or 4292-MetRSL274G cells in media containing ANL or control media for 5 h. Cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Next, to perform in-gel fluorescence, TAMRA was covalently reacted onto the ANL azide moiety of the newly synthesized proteins via a copper-free click reaction. ANL-labeled proteins were detected through direct in-gel fluorescence. The same SDS-PAGE gel was stained with Coomassie blue to visualize the total protein load and size distribution. In-gel fluorescence detected strong signals in 4292-MetRSL274G cells labeled with ANL, but not methionine (Met), while no fluorescence was detected in the parental 4292 cells labeled with either ANL or Met (Figure 2B, Supplementary Figure S2). Notably, ANL incorporation was evenly distributed across the proteome as judged by the similarity of the band patterns between TAMRA and Coomassie blue staining of the ANL-labeled samples (Figure 2B). Thus, utilizing the cell-specific expression of the MetRSL274G transgene, the PDAC-BONCAT system allows for effective and unbiased incorporation of ANL into the tumor cell proteome, facilitating subsequent enrichment and identification via mass spectrometry.
Figure 2.
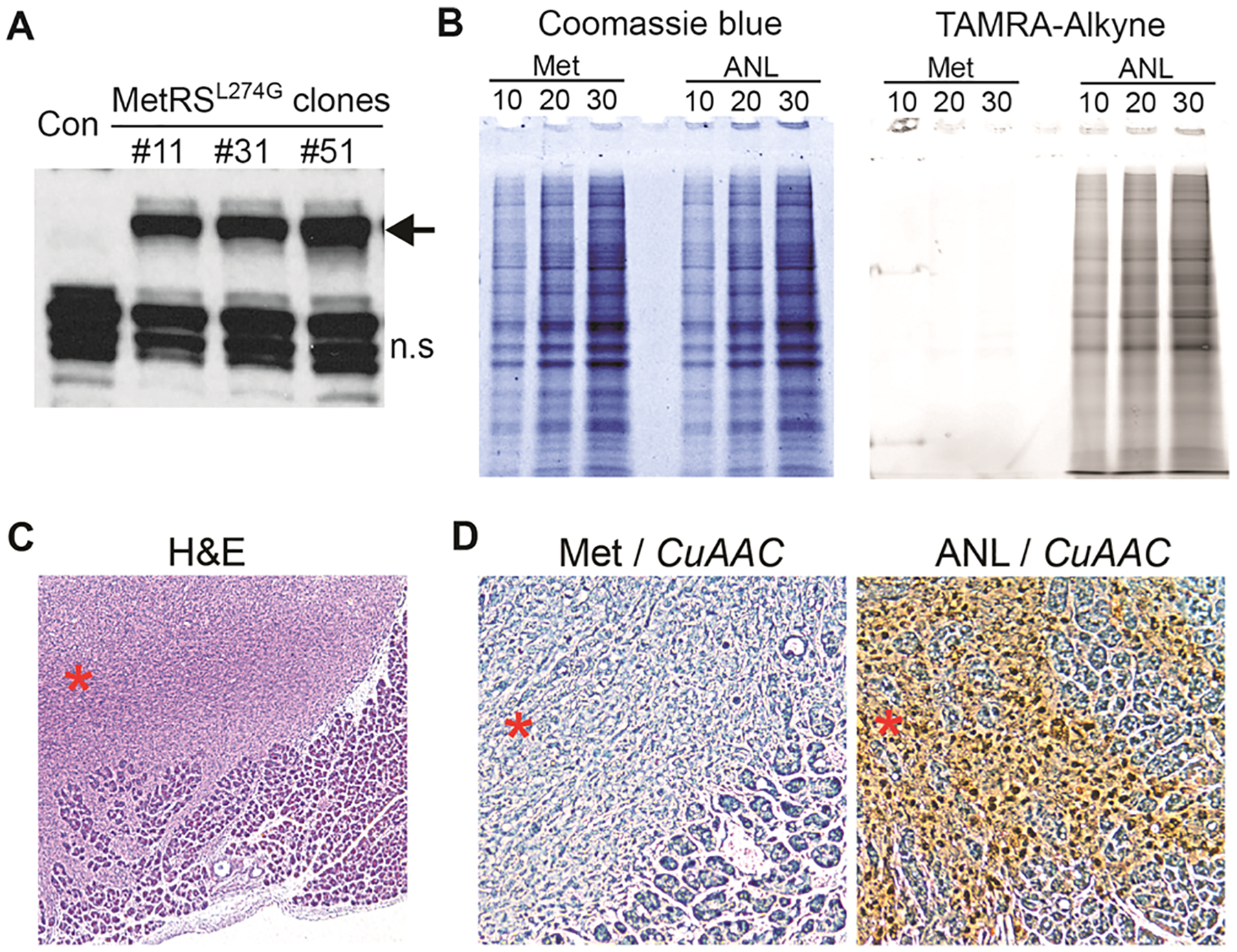
In vitro and in vivo validation of the PDAC-BONCAT system. (A) Western blot analysis of single-cell clones ectopically expressing the MetRSL274G transgene. The MetRSL274G protein is indicated by an arrow; n.s. indicates nonspecific bands recognized by the FLAG antibody. (B) Detection of ANL-labeled proteins by TAMRA-DBCO and SDS-PAGE in vitro; 10, 20, and 30 indicate micrograms of protein lysate loaded per lane. (C) H&E staining showing tumor infiltration in the adjacent normal acinar tissues. Tumor nodule is indicated by a red asterisk. (D) IHC analysis of ANL incorporation by CuAAC. Met samples serve as the negative control. Tumor nodules are indicated by red asterisks.
In Vivo Validation of the PDAC-BONCAT System
To examine whether the PDAC-BONCAT system allows for in vivo tumor cell-specific proteome labeling, we set up an orthotropic transplantation model. The 4292 murine PDAC cells expressing the MetRSL274G transgene were surgically implanted in the pancreata of immunodeficient NOD-scid IL2Rγnull (NSG) mice. The tumor-bearing mice were then randomized into ANL and Met groups. Mice in the ANL group were metabolically labeled via daily intraperitoneal (IP) injection of ANL for 10 days, while those in the Met group were injected with normal saline. Throughout the experiment, animals were provided with a regular diet with no methionine depletion. At the end of the 10-day injection regimen, tumor samples were collected and processed for hematoxylin and eosin (H&E) and α-smooth muscle actin (α-SMA) immunohistochemistry (IHC) staining. Additionally, in situ detection of ANL-incorporated proteins was performed using copper-catalyzed azide–alkyne cycloaddition (CuAAC).
H&E staining of the tumor sections revealed a highly heterogeneous population of cancer cells invading the adjacent acinar tissues (Figure 2C). IHC analysis with α-SMA antibody identified abundant stromal fibroblasts in the tumor bulk (Supplementary Figure S3). Collectively, these features confirm the establishment of a murine PDAC model, capable of recapitulating the heterogeneous cellular composition and histological features of human pancreatic cancer. Further in situ detection of ANL incorporation via the CuAAC click reaction showed signals specifically localized in tumor cells but not in the adjacent normal cells and tissues (Figure 2D), confirming that the tumor cells but not the non-tumor cells in the tumor bulk can incorporate ANL into their proteome. Contrary to the ANL-labeled tissues, no signal was detected in tumor tissues isolated from animals in the Met group where ANL labeling had not taken place (Figure 2D). These in vivo observations demonstrate that BONCAT effectively tags the PDAC tumor cell proteome within its physiological milieu and that the proteome labeling is highly specific to the tumor cells, distinguishing them from the various non-tumor cells in the TME.
Defining the In Vivo Tumor Proteome through Coupling BONCAT and DIA-MS
For in vivo identification of tumor cell-specific proteins, surgical implantation of the PDAC-MetRSL274G cells was performed in a large cohort of NSG mice. Following the same 10-day injection regimen for metabolic labeling, tumors from both ANL and Met groups were collected, lysed, and subjected to BONCAT purification and DIA-MS proteomic analysis.
TAMRA-alkyne cycloaddition reaction detected ANL incorporation in the tumor bulk lysates collected from the ANL but not from the Met group (Figure 3A,B). Of note, the tumor bulk proteomes, represented by the relative intensities and overall pattern of the bands in lanes 5–11 of Coomassie blue staining showed poor correlation with the corresponding tumor cell proteomes visualized by TAMRA staining in the same lanes (Supplementary Figure S4). This observation suggests that the tumor bulk proteome, typically identified in preclinical and/or clinical analysis of tumor samples, does not adequately reflect the tumor cell proteome.
Figure 3.
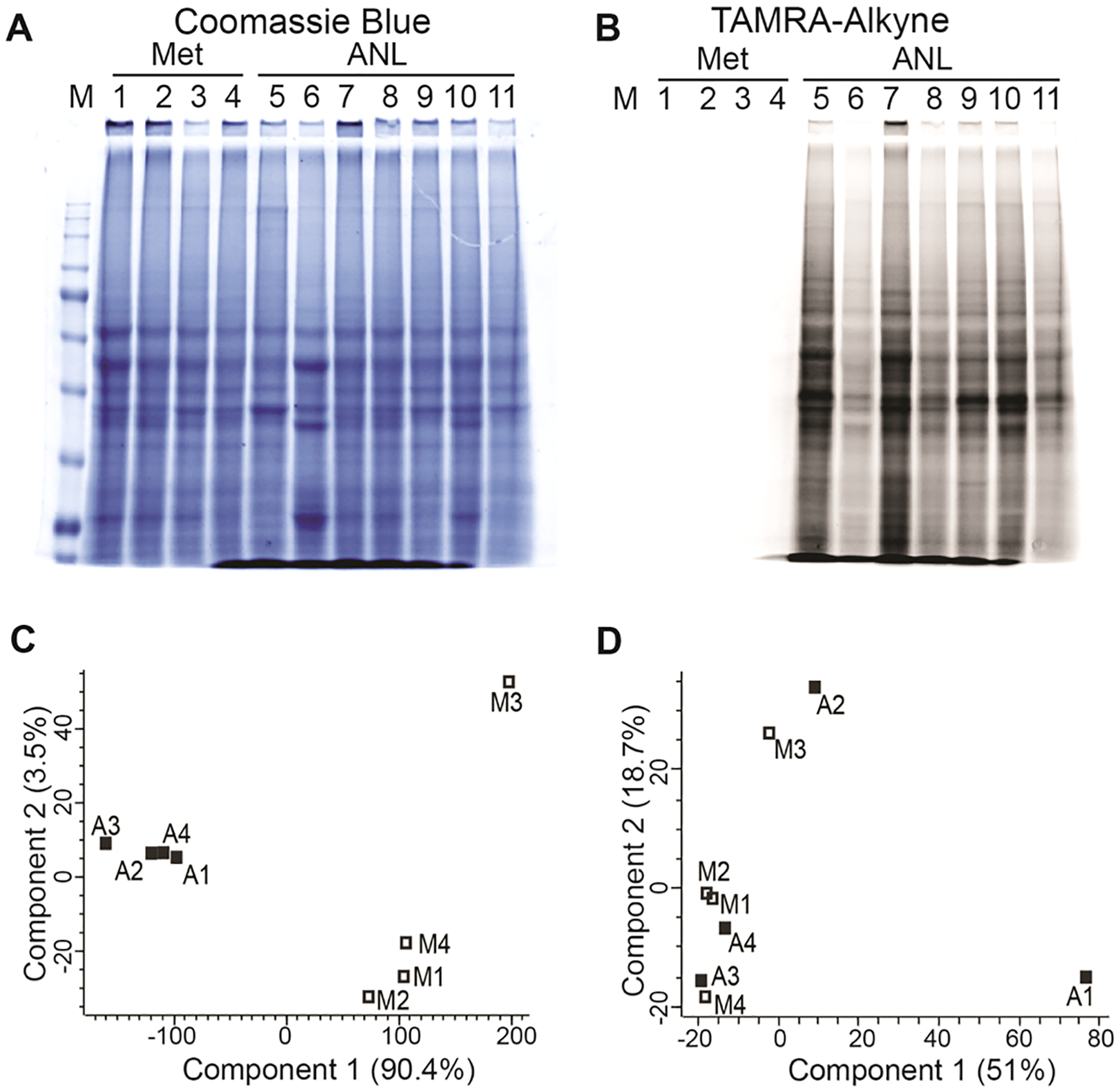
Quality assessment of BONCAT-enriched samples analyzed by DIA-MS. (A, B) TAMRA-alkyne and SDS-PAGE analysis of 11 in vivo tumor samples labeled with Met (lanes 1–4) or ANL (lanes 5–11). (C) PCA of DIA-MS data for BONCAT-ANL and BONCAT-Met samples. Four tumors are included in the Met (M1, 31C; M2, 30C; M3, 32C; and M4, 33C) and ANL groups (A1, 40A; A2, ANL1; A3, 38A; and A4, 29A). (D) PCA of DIA-MS data for Bulk-ANL and Bulk-Met samples.
Four tumors from each group were randomly chosen for BONCAT enrichment and downstream DIA-MS proteomic analysis. Purification and enrichment of ANL-incorporated proteins in tumor lysates were achieved using the DBCO click chemistry reaction. Enriched proteins were next subjected to DIA-MS analysis. In addition to the BONCAT-enriched samples (BONCAT-ANL and BONCAT-Met), DIA-MS analysis was performed on the tumor bulk input lysates that were not subjected to BONCAT enrichment (Bulk-ANL and Bulk-Met). The proteomes of the four BONCAT-ANL samples were highly correlated among each other (Pearson’s correlation coefficient, r = 0.94–0.97) (Supplementary Figure S5A). Principal component analysis (PCA) categorized the samples into two distinct groups; BONCAT-ANL and BONCAT-Met samples, pointing to the specificity and efficacy of ANL labeling and BONCAT enrichment (Figure 3C). Interestingly, the proteomes of all bulk tumor samples (four Bulk-ANL and four Bulk-Met) were highly correlated with each other (Pearson’s correlation coefficient, r = 0.98–0.99) (Supplementary Figure S5B), and PCA was not able to differentiate Bulk-ANL from Bulk-Met samples (Figure 3D), suggesting that the ANL labeling does not affect the bulk tumor proteome.
Comparison of DIA-MS results from the BONCAT-ANL and BONCAT-Met samples confirmed that the majority of the proteins detected belong to the BONCAT-ANL samples, with some nonspecific background present in the BONCAT-Met samples (Figure 4A). Among the highly enriched candidates, many proteins critical for pancreatic tumorigenesis, including KRAS, YAP1, HMGB1, HMGB2, and LEG3 (galectin-3), were identified (Figure 4A). There were 4405 proteins identified in the BONCAT-ANL samples (Supplementary Table S1). Only proteins enriched by at least twofold in BONCAT-ANL compared to BONCAT-Met samples at a false discovery rate (FDR) <0.05 were considered as true proteins expressed in tumor cells. Subtracting the nonspecific background identified in the BONCAT-Met samples resulted in a total of 3727 BONCAT-ANL-specific proteins (Supplementary Figure S6). These proteins together represent the tumor cell proteome of the murine pancreatic cancer.
Figure 4.
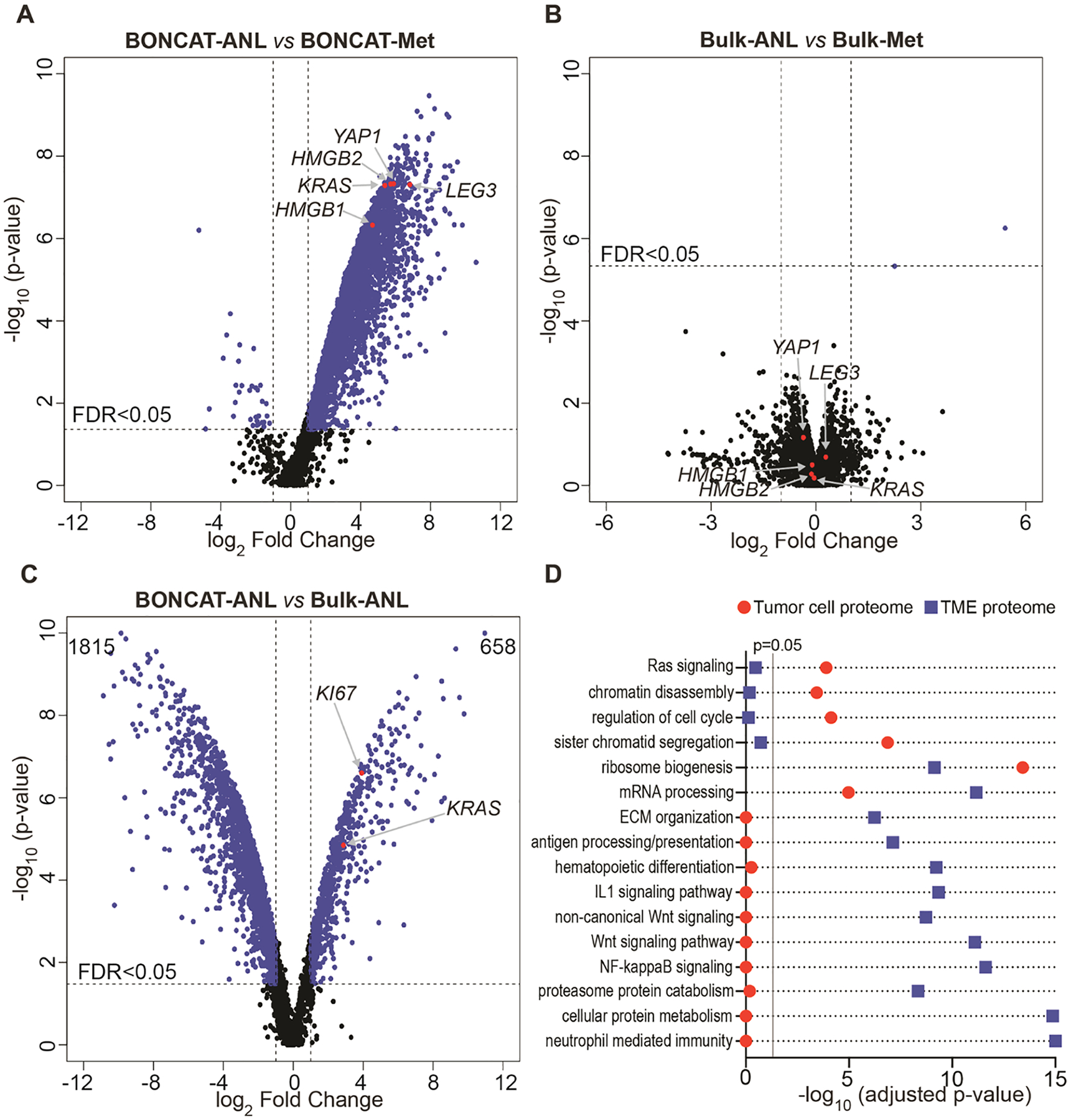
Tumor cell and tumor bulk proteomes. (A) Comparison of the BONCAT-ANL and BONCAT-Met samples. Selected proteins critical for pancreatic tumorigenesis are labeled. (B) Comparison of the bulk proteomes from ANL and Met total tumor lysates. (C) Comparison of BONCAT-ANL proteome and the corresponding Bulk-ANL proteome differentiates tumor cell-specific proteins from those of non-tumor cells within the TME. Tumor-specific marker proteins, including KRAS and KI67, are highlighted in the plot. Numbers on the top right and left corners (658 and 1815) indicate the sum of proteins identified as tumor cell- and TME-specific, respectively. (D) Selected GO terms enriched in the tumor cell or TME proteomes.
DIA-MS proteomic analysis of the bulk tumor, comprising tumor cells, non-tumor cell types, and the extracellular matrix components, identified >5800 proteins from ANL and Met groups (Bulk-ANL and Bulk-Met), with negligible differences between the two (Figure 4B, Supplementary Table S2), confirming that the ANL labeling process does not interfere with the general protein synthesis machinery in PDAC tumors. Notably, the protein levels of the candidates critical for pancreatic tumorigenesis, such as KRAS, YAP1, HMGB1, HMGB2, and LEG3 (galectin-3), were not different between Bulk-ANL and Bulk-Met samples, further suggesting that the tumors from ANL and Met groups are biologically identical (Figure 4B). Thus, coupling BONCAT and DIA-MS allows for the exclusive in vivo dissection of the PDAC tumor cell proteome in a physiological context.
Delineating Proteins Expressed in Tumor Cells from Non-tumor Cells within the PDAC Tumor Microenvironment
BONCAT-enriched proteins represent the tumor cell proteome, while the tumor bulk proteome (Bulk-ANL and Bulk-Met) encompasses the entire tumor and non-tumor proteins within the TME. Notably, many proteins expressed by the tumor cells, such as housekeeping proteins, may also be abundantly produced by other cell types within the tumor bulk. Identifying the proteins preferentially expressed in either tumor cells or non-tumor cells within the TME is critical for the study of tumor cell intrinsic carcinogenesis, dynamic interaction between tumor cells and their environment, and the discovery of novel therapeutic targets and biomarkers.
To identify tumor cell-specific and TME-specific proteins, we compared the BONCAT-enriched tumor cell proteome to the pre-enrichment tumor bulk proteome within the ANL-labeled group (BONCAT-ANL vs Bulk-ANL). For each protein, the ratio of abundance in BONCAT-enriched to tumor bulk indicates the preferential distribution in tumor or non-tumor cells. A high BONCAT-ANL/Bulk-ANL ratio points to tumor cell-specific expression, while a low ratio implies preferential expression in various non-tumor cells within the TME (Supplementary Table S3). Notably, the PDAC driver oncogene KRAS (RASK) was detected as a tumor cell-specific candidate with a fold change ratio of 7.3. Another protein marker highly expressed in tumor cells, KI67, was also enriched with a ratio of 15.1 (Figure 4C). IHC analysis of selective candidates with high ratios, including KI67, RAS, EZH2, ZMIZ1, and ARID1B, confirmed their preferential presence in the tumor cells but not the adjacent connective tissues (Supplementary Figure S7). These data provide evidence that our analysis indeed identifies tumor cell-specific protein expression.
To further validate our analysis, we examined signature proteins expressed exclusively in non-tumor cells within the TME. The PDAC TME of the NSG mice contains various cell types, such as stromal fibroblasts, monocytes/macrophages, dendritic cells, neutrophils, and endothelial cells. We, therefore, searched for the signature proteins of these non-tumor cell types in our BONCAT-ANL and Bulk-ANL data sets.31,32 Among 34 signature proteins present in the bulk tumor lysates, the majority were either not detected or detected at very low levels in the BONCAT-ANL tumor cell proteome. In contrast, housekeeping proteins, such as GAPDH and β-actin, did not differ between the BONCAT-enriched tumor cell proteome and the bulk proteome (Supplementary Table S4). Gene ontology (GO) term analysis of tumor cell-enriched proteins revealed general and pancreatic cancer-specific processes, including RNA metabolism, cell cycle/mitosis, apoptosis, chromatin remodeling, and RAS/MAPK signaling among the top terms (Supplementary Table S5). In contrast, GO analysis of the non-tumor cell proteins confirmed the overrepresentation of biological processes related to TME, such as immunity, extracellular matrix organization, WNT signaling, and antigen processing and presentation (Supplementary Table S6). Parallel comparison of the GO enrichments demonstrates the distinct separation of the tumor cell and TME proteomes (Figure 4D). These observations further support that our BONCAT-DIA-MS approach delineates proteins expressed in the tumor cells from non-tumor cells in the TME, compartmentalizing tumor and non-tumor proteins within the tumor bulk.
DISCUSSION
Coupling BONCAT bioorthogonal chemistry with DIA-MS proteomics analysis in an orthotopic pancreatic cancer model, we have developed an innovative technical framework that can specifically label, enrich, and identify the tumor cell proteome in vivo. The sensitivity and efficiency of this approach were validated through the identification of thousands of proteins expressed in pancreatic tumor cells within the tumor bulk. Comparative analysis of the BONCAT-enriched tumor cell proteome and the tumor bulk proteome facilitated the differentiation of proteins preferentially expressed in tumor cells from those of non-tumor cells within the TME.
Our approach has broad application in studies of tumorigenesis, cancer therapeutics, and biomarker discovery. Our platform may be applied to primary tumors isolated from human patients to systematically define their tumor cell proteomes. Patient-derived xenograft (PDX) models are increasingly utilized to investigate novel therapeutics and guide clinical cancer treatment.33–35 Following well-established protocols, primary PDX tumors can express the MetRSL274G enzyme via lentiviral infection for tumor cell-specific proteomic labeling and characterization.36,37 Our approach, therefore, enables in vivo tumor cell-specific proteomic characterization in PDX models, providing an unprecedented ability for systemic interrogation of therapeutic responses at the level of individual proteins. Additionally, this technical framework may be implemented to reveal the tumor cell-specific secretome.38 ANL labeling of the tumor cell proteome in PDX models allows for selective purification and enrichment via BONCAT of various proteins secreted by tumor cells into the systemic circulation. Subsequent identification of the tumor cell-specific secretome using mass spectrometry will open new opportunities for the development of novel biomarkers for early cancer detection, a particularly persistent challenge in pancreatic cancers.
Supplementary Material
ACKNOWLEDGMENTS
We greatly appreciate Dr. Marina Pasca di Magliano for generously providing the murine pancreatic cancer cell line (4292). We would also like to thank members of David Tirrell and Erin Schuman labs for sharing protocols on BONCAT procedures. Mass spectrometry analyses were conducted in the Mass Spectrometry Laboratory at the University of Texas Health Science Center at San Antonio. This work was supported in part by NIH K22CA207598 (Y.L.), CPRIT RP200472 (Y.L.), and NIH GM008042 (D.K.S.; UCLA-Caltech Medical Scientist Training Program). Support from the University of Texas System Proteomics Core Network for the purchase of the Lumos mass spectrometer is gratefully acknowledged.
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jproteome.0c00666
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00666.
Full size image of the western blot presented in Figure 2A; molecular weight of the protein ladder is labeled on the left side (Figure S1); detection of ANL-labeled proteins by TAMRA-DBCO and SDS-PAGE in cell lines with or without MetRSL274G transgene (Figure S2); α-SMA antibody identifies stromal fibroblasts in the tumor bulk (Figure S3); densitometric analysis of Coomassie blue and TARMA-alkyne stained SDS-PAGE gel for tumor sample lanes 5–11 (Figure S4); Pearson correlation of DIA-MS data from the BONCAT (A) and the Bulk samples (B); the correlation coefficients are labeled above the dot plots; the dashed triangle indicates the correlation among BONCAT-ANL samples (Figure S5); Venn diagram of the number of proteins identified by BONCAT enrichment in the ANL-labeled samples compared to the Met-labeled samples (Figure S6); IHC analysis of protein candidates preferentially present in pancreatic tumor cells; the tumor cells with dark brown staining are indicated with “T” (Figure S7) (PDF) List of proteins identified in BONCAT-enriched samples by DIA-MS (Table S1); list of proteins identified in the tumor bulk lysates by DIA-MS (Table S2); ratio of protein abundance in BONCAT-ANL versus Bulk-ANL samples (Table S3); levels of marker proteins from tumor and non-tumor cells within the tumor microenvironment (Table S4); GO term analysis of proteins enriched in tumor cells (Table S5); GO term analysis of proteins enriched in the tumor microenvironment (Table S6) (XLSX)
The authors declare no competing financial interest.
The raw DIA-MS proteomic data have been uploaded to the MassIVE repository with the dataset identifier MSV000086021 and the ProteomeXchange accession number PXD021151. All code for data analysis is available at https://github.com/Yenaled/JPROBS_MetRS.
Contributor Information
Nancy G. Azizian, Center for Immunotherapy Research, Houston Methodist Research Institute, Houston, Texas 77030, United States; Department of Medicine, Weill Cornell Medical College, New York, New York 10065, United States
Delaney K. Sullivan, UCLA-Caltech Medical Scientist Training Program, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, California 90095, United States;.
Litong Nie, Department of Experimental Radiation Oncology, Division of Radiation Oncology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, United States.
Sammy Pardo, Department of Biochemistry and Structural Biology, The University of Texas Health Science Center at San Antonio, San Antonio, Texas 78229, United States.
Dana Molleur, Department of Biochemistry and Structural Biology, The University of Texas Health Science Center at San Antonio, San Antonio, Texas 78229, United States.
Junjie Chen, Department of Experimental Radiation Oncology, Division of Radiation Oncology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, United States;.
Susan T. Weintraub, Department of Biochemistry and Structural Biology, The University of Texas Health Science Center at San Antonio, San Antonio, Texas 78229, United States;.
Yulin Li, Center for Immunotherapy Research, Houston Methodist Research Institute, Houston, Texas 77030, United States;; Department of Medicine, Weill Cornell Medical College, New York, New York 10065, United States;
REFERENCES
- (1).Öhlund D; et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med 2017, 214, 579–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lafaro KJ; Melstrom LG The Paradoxical Web of Pancreatic Cancer Tumor Microenvironment. Am. J. Pathol 2019, 189, 44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Pothula SP; et al. Key role of pancreatic stellate cells in pancreatic cancer. Cancer Lett 2016, 381, 194–200. [DOI] [PubMed] [Google Scholar]
- (4).Stone SE; Glenn WS; Hamblin GD; Tirrell DA Cell-selective proteomics for biological discovery. Curr. Opin. Chem. Biol 2017, 36, 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Gauthier NP; et al. Cell-selective labeling using amino acid precursors for proteomic studies of multicellular environments. Nat. Methods 2013, 10, 768–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Dieterich DC; Link AJ; Graumann J; Tirrell DA; Schuman EM Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. U.S.A 2006, 103, 9482–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Elliott TS; Bianco A; Chin JW Genetic code expansion and bioorthogonal labelling enables cell specific proteomics in an animal. Curr. Opin. Chem. Biol 2014, 21, 154–160. [DOI] [PubMed] [Google Scholar]
- (8).Erdmann I; et al. Cell-selective labelling of proteomes in Drosophila melanogaster. Nat. Commun 2015, 6, No. 7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Liu Y; et al. Application of bio-orthogonal proteome labeling to cell transplantation and heterochronic parabiosis. Nat. Commun 2017, 8, No. 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Schanzenbächer CT; Langer JD; Schuman EM Time- and polarity-dependent proteomic changes associated with homeostatic scaling at central synapses. eLife 2018, 7, No. e33322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Alvarez-Castelao B; et al. Cell-type-specific metabolic labeling of nascent proteomes in vivo. Nat. Biotechnol 2017, 35, 1196–1201. [DOI] [PubMed] [Google Scholar]
- (12).Prescher JA; Dube DH; Bertozzi CR Chemical remodelling of cell surfaces in living animals. Nature 2004, 430, 873–877. [DOI] [PubMed] [Google Scholar]
- (13).Baskin JM; et al. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. U.S.A 2007, 104, 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Mahdavi A; et al. Identification of secreted bacterial proteins by noncanonical amino acid tagging. Proc. Natl. Acad. Sci. U.S.A 2014, 111, 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Venable JD; Dong MQ; Wohlschlegel J; Dillin A; Yates JR Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 2004, 1, 39–45. [DOI] [PubMed] [Google Scholar]
- (16).Bruderer R; et al. Optimization of Experimental Parameters in Data-Independent Mass Spectrometry Significantly Increases Depth and Reproducibility of Results. Mol. Cell. Proteomics 2017, 16, 2296–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Gillet LC; et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 2012, 11, No. O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Michalski A; Cox J; Mann M More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J. Proteome Res 2011, 10, 1785–1793. [DOI] [PubMed] [Google Scholar]
- (19).Wilson RS; et al. Development of Targeted Mass Spectrometry-Based Approaches for Quantitation of Proteins Enriched in the Postsynaptic Density (PSD). Proteomes 2019, 7, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Nguyen EV; et al. Identification of Novel Response and Predictive Biomarkers to Hsp90 Inhibitors Through Proteomic Profiling of Patient-derived Prostate Tumor Explants. Mol. Cell. Proteomics 2018, 17, 1470–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Keam SP; et al. Exploring the oncoproteomic response of human prostate cancer to therapeutic radiation using data-independent acquisition (DIA) mass spectrometry. Prostate 2018, 78, 563–575. [DOI] [PubMed] [Google Scholar]
- (22).Kim YJ; et al. Data-Independent Acquisition Mass Spectrometry To Quantify Protein Levels in FFPE Tumor Biopsies for Molecular Diagnostics. J. Proteome Res 2019, 18, 426–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Collins MA; et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J. Clin. Invest 2012, 122, 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Tom Dieck S; et al. Metabolic labeling with noncanonical amino acids and visualization by chemoselective fluorescent tagging. Curr. Protoc. Cell. Biol 2012, 7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Calve S; Witten AJ; Ocken AR; Kinzer-Ursem TL Incorporation of non-canonical amino acids into the developing murine proteome. Sci. Rep 2016, 6, No. 32377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Glenn WS; et al. Bioorthogonal Noncanonical Amino Acid Tagging (BONCAT) Enables Time-Resolved Analysis of Protein Synthesis in Native Plant Tissue. Plant Physiol 2017, 173, 1543–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Searle BC; et al. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat. Commun 2018, 9, No. 5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Gessulat S; et al. Prosit: proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat. Methods 2019, 16, 509–518. [DOI] [PubMed] [Google Scholar]
- (29).Ritchie ME; et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015, 43, No. e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kuleshov MV; et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016, 44, W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Newman AM; et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ren B; et al. Tumor microenvironment participates in metastasis of pancreatic cancer. Mol. Cancer 2018, 17, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Crystal AS; et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 2014, 346, 1480–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Bertotti A; et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discovery 2011, 1, 508–523. [DOI] [PubMed] [Google Scholar]
- (35).Zhang X; et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res 2013, 73, 4885–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Hanna C; Kwok L; Finlay-Schultz J; Sartorius CA; Cittelly DM Labeling of Breast Cancer Patient-derived Xenografts with Traceable Reporters for Tumor Growth and Metastasis Studies. J. Vis. Exp 2016, DOI: 10.3791/54944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Liu JF; et al. Establishment of Patient-Derived Tumor Xenograft Models of Epithelial Ovarian Cancer for Preclinical Evaluation of Novel Therapeutics. Clin. Cancer Res 2017, 23, 1263–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Yang AC; et al. Multiple Click-Selective tRNA Synthetases Expand Mammalian Cell-Specific Proteomics. J. Am. Chem. Soc 2018, 140, 7046–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


