Abstract
PURPOSE
Clonal hematopoiesis (CH) can be transmitted from a donor to a recipient during allogeneic hematopoietic cell transplantation. Exclusion of candidate donors with CH is controversial since its impact on recipient outcomes and graft alloimmune function is uncertain.
PATIENTS AND METHODS
We performed targeted error-corrected sequencing on samples from 1,727 donors age 40 years or older and assessed the effect of donor CH on recipient clinical outcomes. We measured long-term engraftment of 102 donor clones and cytokine levels in 256 recipients at 3 and 12 months after transplant.
RESULTS
CH was present in 22.5% of donors, with DNMT3A (14.6%) and TET2 (5.2%) mutations being most common; 85% of donor clones showed long-term engraftment in recipients after transplantation, including clones with a variant allele fraction < 0.01. DNMT3A-CH with a variant allele fraction ≥ 0.01, but not smaller clones, was associated with improved recipient overall (hazard ratio [HR], 0.79; P = .042) and progression-free survival (HR, 0.72; P = .003) after adjustment for significant clinical variables. In patients who received calcineurin-based graft-versus-host disease prophylaxis, donor DNMT3A-CH was associated with reduced relapse (subdistribution HR, 0.59; P = .014), increased chronic graft-versus-host disease (subdistribution HR, 1.36; P = .042), and higher interleukin-12p70 levels in recipients. No recipient of sole DNMT3A or TET2-CH developed donor cell leukemia (DCL). In seven of eight cases, DCL evolved from donor CH with rare TP53 or splicing factor mutations or from donors carrying germline DDX41 mutations.
CONCLUSION
Donor CH is closely associated with clinical outcomes in transplant recipients, with differential impact on graft alloimmune function and potential for leukemic transformation related to mutated gene and somatic clonal abundance. Donor DNMT3A-CH is associated with improved recipient survival because of reduced relapse risk and with an augmented network of inflammatory cytokines in recipients. Risk of DCL in allogeneic hematopoietic cell transplantation is driven by somatic myelodysplastic syndrome–associated mutations or germline predisposition in donors.
INTRODUCTION
Allogeneic hematopoietic cell transplantation is the only curative treatment for most high-risk hematologic malignancies. Patients with an available donor have improved overall survival (OS) compared with patients who do not have a donor.1,2 The National Marrow Donor Program (NMDP) prioritizes donors under age 453 years because younger donor age has been associated with improved recipient survival,4 but suitable younger donors are not uniformly available to all transplant candidates. The likelihood of finding a matching donor in the NMDP Be The Match Registry varies widely depending on ethnic background. Although HLA-matched adult donors can be identified in the NMDP registry for 75% of White patients, far fewer patients of color will find matched donors (16%-19% of African Americans, 33%-42% of Asian Americans, and 34%-40% of Hispanic Americans).5 The best available graft for many patients may thus be from an older donor such as an HLA-identical sibling or haploidentical relative,6,7 highlighting the importance of understanding characteristics of older donors that influence recipient outcomes.
CONTEXT
Key Objective
Clonal hematopoiesis (CH) can be transmitted from a donor to a recipient during allogeneic hematopoietic cell transplantation, but its impact on recipient outcomes is unclear. This study assessed the frequency of CH in 1,727 transplant donors age 40 years and older and its effect on survival, relapse, and graft-versus-host disease (GVHD) in recipients.
Knowledge Generated
CH with mutations in DNMT3A was associated with improved overall survival, reduced risk of relapse, and elevated risk of chronic GVHD in patients who received traditional calcineurin inhibitor–based GVHD prophylaxis. Rare cases of donor cell leukemia evolved from CH with less common mutations in TP53 and splicing factor genes.
Relevance
Older individuals with the most common forms of CH need not be excluded from stem-cell donation. More broadly, inhibiting de novo DNA methylation in donor immune cells could reduce risk of relapse in transplant recipients.
Clonal hematopoiesis (CH) is an age-related, asymptomatic condition in which leukemia-associated somatic mutations are detected in the blood of individuals without a hematologic malignancy. In the nontransplant setting, CH is uniformly associated with adverse outcomes, including an elevated risk of developing hematologic malignancies8,9 and an increased risk of nonhematologic outcomes because of altered inflammatory signaling.10 These clinical effects of native CH are apparent with large clones,8,10 but CH at very low clonal abundance with an uncertain biologic effect has been reported to be ubiquitous in adults over age 40 years.11
Exploratory studies have found that CH in transplant donors can engraft in recipients12-15 but have reported conflicting results on the impact of donor CH on transplant-specific clinical outcomes, graft immunologic function, and risk of donor cell leukemia (DCL).12,13,16,17 These studies have been limited by modest sample sizes that affected outcomes analysis, cohort characteristics that restricted generalizability, and a lack of mechanistic rationale. Furthermore, these cohorts have not parsed the clinical impact of different CH mutations or used sequencing technologies that support evaluation of low-abundance clones, which could have unique dynamics in the context of allogeneic transplantation. Current evidence has thus been insufficient to resolve disagreement about whether to screen older candidate donors for CH,18,19 and some transplant centers have begun excluding donors found to have CH on the basis of the assumption that the adverse associations of native CH also apply in the context of transplant.20 In this study, we performed a comprehensive analysis of samples from donors age 40 years or older to determine the impact of CH on overall recipient outcomes, risk of DCL, and measures of graft alloimmune activity.
PATIENTS AND METHODS
Patients
All patients who underwent transplantation with donor age 40 years or older at Dana-Farber Cancer Institute (Cohort 1) or Johns Hopkins University (Cohort 2) between 2000 and 2016 were considered for study inclusion, and 1,727 samples met technical requirements for analysis (Data Supplement, online only). The study was conducted with approval and waivers of consent from both institutional review boards. Additional details are provided in the Data Supplement (online only).
Genetic Studies
We sequenced 46 genes mutated recurrently in CH and myeloid malignancies (Data Supplement).5,6,19-21 We included samples with average unique molecular identifier–deduplicated consensus coverage of at least 1,000× and considered all variants with a variant allele fraction (VAF) ≥ 0.005. We classified variants as pathogenic on the basis of published genetic criteria.20,22 Genetic analysis was blinded to clinical characteristics and locked before merging with clinical data.
Statistical Analysis
The primary end point was progression-free survival (PFS), defined as the time from transplantation until death or relapse of the original disease, whichever occurred first. OS was defined as the time from transplantation until death from any cause or until censoring at the time last known to be alive. OS and PFS were estimated using the Kaplan-Meier method, and the difference was tested using log-rank tests. Cumulative incidences of nonrelapse mortality (NRM), relapse, and graft-versus-host disease (GVHD) were estimated in competing risk frameworks that treated relapse, NRM, and relapse or death without GVHD as competing events, respectively. Cumulative incidences were compared using the Gray test.23 Multivariable analysis was performed using Cox models for OS and PFS and Fine and Gray models for NRM, relapse, and GVHD.23,24 Additional details are provided in the Data Supplement.
RESULTS
Clinical and Genetic Characteristics of Donor CH
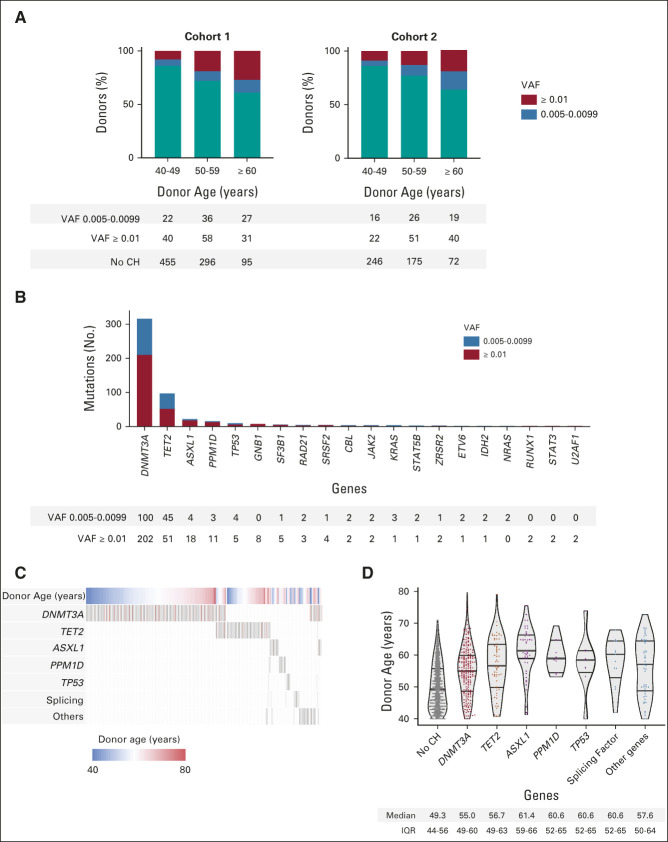
We identified CH with VAF ≥ 0.005 in 388 of 1,727 donors (22.5%, Data Supplement). Its prevalence increased with advancing age: CH was present in 12.6% of donors age 40-49 years, 26.6% of donors age 50-59 years, and 41.2% of donors age 60 years or older. The proportion of donors with CH in each age decade was similar in both cohorts (Fig 1A). In multivariable analysis stratified by cohort and considering donor, recipient, and transplant variables, only older donor age was independently associated with the presence of CH (median 56 v 49 years with and without CH, respectively, P < .001, Table 1).
FIG 1.
Characteristics of CH in transplant donors age 40 years and older. (A) The proportion of donors with and without CH in each cohort, subdivided by donor age decade. CH with VAF 0.005-0.0099 is in blue, and CH with VAF ≥ 0.01 is in red. (B) The number of mutations in each gene mutated in two or more donors, with variants at VAF 0.005-0.0099 again in blue and variants at VAF ≥ 0.01 in red. (C) The patterns of comutation among donors with CH. Each column represents a donor, with rows for donor age and the most commonly mutated genes or gene groups. Donors were hierarchically grouped on the basis of the presence of mutations in genes other than DNMT3A or TET2, DNMT3A, and then TET2. Genes mutated more than once in the same donor are in red. (D) The distribution of donor age on the basis of CH status and mutations in individual genes or groups of genes. Medians and IQRs are reported below each column. CH, clonal hematopoiesis; IQR, interquartile range; VAF, variant allele fraction.
TABLE 1.
Characteristics of Donors and Recipients of Donors With and Without CH
Among 501 total mutations, 324 had VAF ≥ 0.01 and 177 had VAF of 0.005-0.0099. The most frequently mutated genes were DNMT3A (302 mutations in 253 donors), TET2 (96 mutations in 89 donors), ASXL1 (22 mutations in 22 donors), and PPM1D (14 mutations in 14 donors, Fig 1B). No other gene was mutated in more than 10 donors. Most donors with CH (n = 301, 77.5%) had only one mutation. Donors with mutations other than DNMT3A or TET2 were more likely to have mutations in more than one gene (compared with donors who had DNMT3A or TET2 mutations; odds ratio 3.5; 95% CI, 2.0 to 6.1; P < .0001; Fig 1C). Donors with CH were older than those without CH irrespective of the gene mutated (Fig 1D). Using an orthogonal duplex unique molecular identifier–based sequencing platform, we validated 100% (28 of 28) of variants from 20 donors (VAF range 0.0056-0.2159; Data Supplement).
Donor CH and Recipient Outcomes
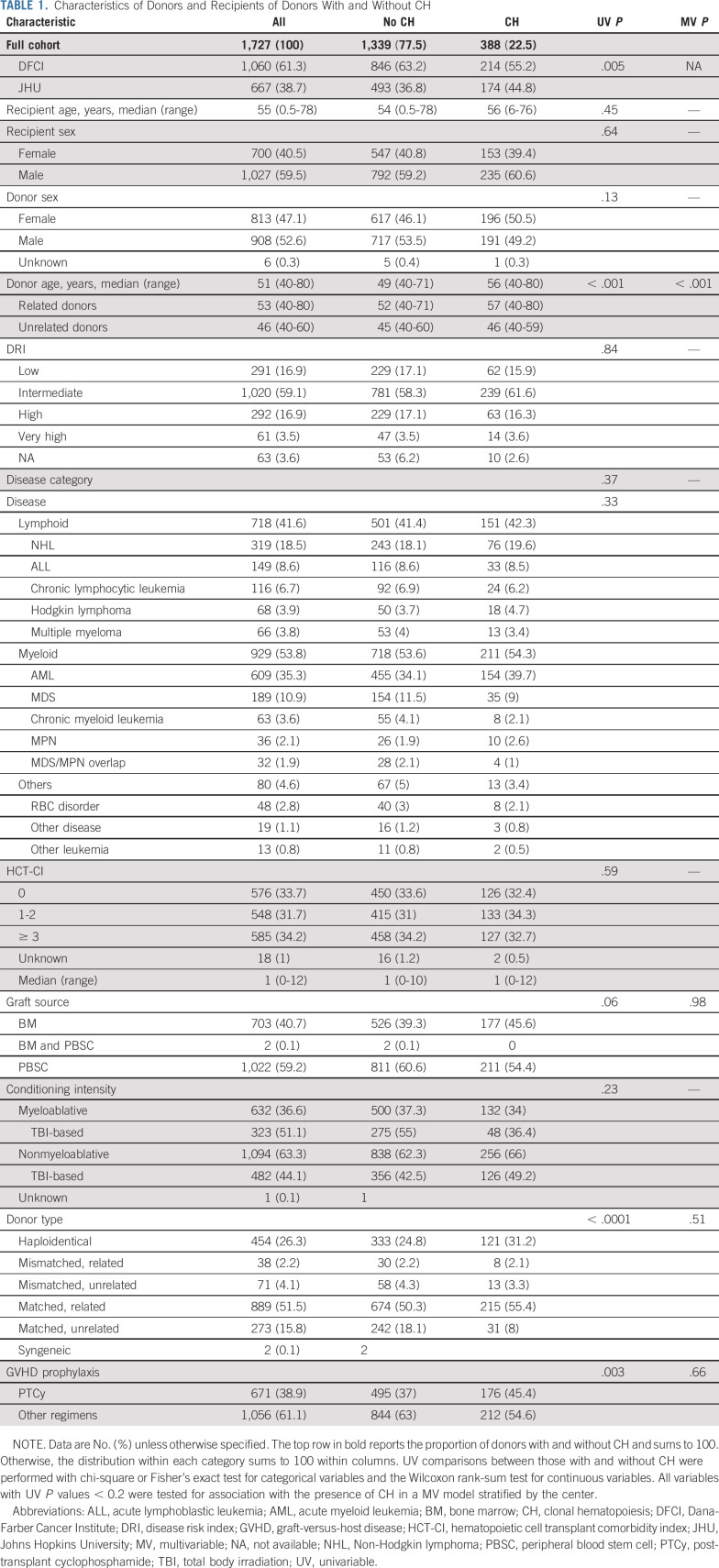
In native CH, larger clones have a greater effect on clinical outcomes than smaller clones,8,10 but no evidence-based VAF cutoff for defining CH has been established. To define a clinically relevant VAF threshold for CH in the setting of transplantation, we examined the relative hazards of PFS, relapse, and NRM across the full range of donor clone sizes and found that the effect of CH was greatest with VAF ≥ 0.01 (Data Supplement). Recipients of donor CH with VAF ≥ 0.01 (n = 241) had improved PFS compared with recipients whose donors did not have CH in a multivariable model that included recipient and donor age, donor-recipient sex mismatch, hematopoietic cell transplantation–specific comorbidity index score,21 conditioning intensity, donor type, disease category, and Disease Risk Index classification22 (hazard ratio [HR], 0.79; 95% CI, 0.66 to 0.95; P = .011; Fig 2A). CH with only smaller donor clones (VAF 0.005-0.0099, n = 147) was not significantly associated with any outcome.
FIG 2.
Association of donor CH with recipient clinical outcomes. (A) The association between donor CH at either VAF 0.005-0.0099 or VAF ≥ 0.01 and recipient PFS in multivariable Cox proportional hazards models. (B) The association between donor CH at VAF ≥ 0.01 and recipient OS, PFS, relapse, and NRM in multivariable models divided by gene group. The groups consist of CH with any mutation other than DNMT3A or TET2 (Other CH), remaining DNMT3A mutations, and then remaining TET2 mutations. (C) The 5-year cumulative incidence of chronic GVHD split by DNMT3A-CH status (VAF ≥ 0.01) and receipt of PTCy for GVHD prophylaxis. (D) The 5-year cumulative incidence of relapse split by DNMT3A-CH status (VAF ≥ 0.01) and receipt of PTCy. (E) The 5-year PFS split by DNMT3A-CH status (VAF ≥ 0.01) and receipt of PTCy. (F) The associations between DNMT3A-CH and outcomes among recipients who did or did not receive PTCy in multivariable Fine-Gray competing risk regressions (chronic GVHD, relapse) and Cox proportional hazards models (PFS and OS). CH, clonal hematopoiesis; GVHD, graft-versus-host disease; HR, hazard ratio; NRM, nonrelapse mortality; OS, overall survival; PFS, progression-free survival; PTCy, post-transplant cyclophosphamide; sHR, subdistribution hazard ratio; VAF, variant allele fraction.
Individual gene mutations in CH have distinct associations with clinical outcomes. In nontransplant settings, DNMT3A and TET2 mutations have driven the bulk of association with inflammatory outcomes like cardiovascular disease,10 whereas mutations other than DNMT3A or TET2 have been associated with a higher risk of progression to hematologic malignancy.10,23,24 We therefore tested the effects of three prespecified, hierarchically defined groups at VAF ≥ 0.01: (1) donors with mutations in any gene other than DNMT3A or TET2 (Other CH, n = 49), (2) remaining donors with DNMT3A mutations (DNMT3A-CH, n = 157), and (3) remaining donors with only TET2 mutations (TET2-CH, n = 35).
Only donor DNMT3A-CH was significantly associated with recipient outcomes. Recipients whose donors had DNMT3A-CH had improved OS and PFS and reduced risk of relapse in the same multivariable model (HR for death 0.78; 95% CI, 0.62 to 0.98; P = .037; HR for death or relapse 0.72; 95% CI, 0.58 to 0.89; P = .003; subdistribution hazard ratio [sHR] for relapse 0.74; 95% CI, 0.57 to 0.96; P = .022, Fig 2B and Data Supplement), when compared with recipients whose donors did not have CH with VAF ≥ 0.01. DNMT3A-CH was not associated with differences in NRM, and causes of death without relapse were similar irrespective of the presence of DNMT3A-CH (Data Supplement). Neither TET2-CH nor Other CH had significant impacts on outcomes.
Alloreactive donor immune cells not only reduce relapse via graft-versus-leukemia (GVL) activity but also mediate a complementary risk of GVHD. Conventional GVHD prophylaxis with calcineurin-based regimens suppresses global T cell function, whereas post-transplant cyclophosphamide (PTCy) is thought to prevent GVHD by selective suppression or elimination of pathogenic alloreactive T cells.25,26 To evaluate the interactions between DNMT3A-CH and immune-modulating therapy, we analyzed GVHD outcomes in patients who did or did not receive PTCy. In multivariable analysis, DNMT3A-CH with VAF ≥ 0.01 was independently associated with an increased risk of chronic GVHD in recipients who did not receive PTCy (Fig 2C; sHR 1.37; 95% CI, 1.02 to 1.84; P = .04 in multivariable analysis). By contrast, we observed no effect of DNMT3A-CH on chronic GVHD in recipients who received PTCy (compared with PTCy and no DNMT3A-CH; sHR 1.15; 95% CI, 0.82 to 1.6; P = .88). The improvements in relapse, PFS, and OS associated with DNMT3A-CH were also confined to recipients who did not receive PTCy (Figs 2D-2F and Data Supplement). Among these patients, donor DNMT3A-CH was associated with reduced risk of relapse (sHR from multivariable model 0.59; 95% CI, 0.39 to 0.9; P = .014) and reduced risk of death (HR 0.65; 95% CI, 0.47 to 0.9; P = .01). This difference was evident for both myeloid and lymphoid diseases (Data Supplement), related and unrelated donors (Data Supplement), and was not evidently influenced by either bone marrow graft source (Data Supplement) or haploidentical donors (Data Supplement), both of which are closely associated with PTCy use. There was no association between DNMT3A-CH and acute GVHD (Data Supplement).
Engraftment and Biologic Characteristics of Donor CH in Recipients
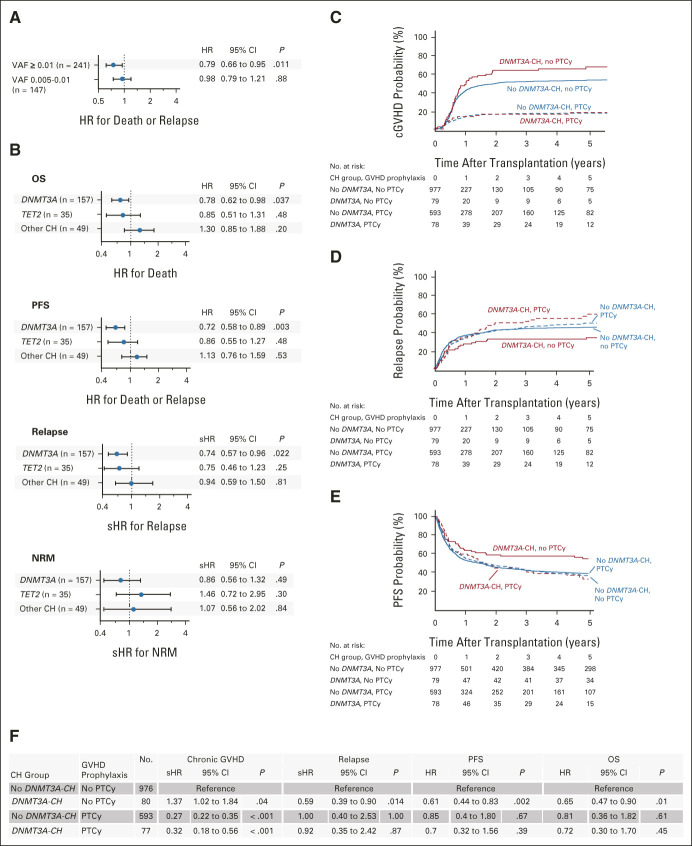
To define the capacity of donor clones to contribute to long-term recipient hematopoiesis, we sequenced samples collected 3 and 12 months after transplant from 69 recipients whose donors had CH and who survived without relapse for at least one year (Data Supplement). In total, 86 of 102 donor mutations were detectable in paired recipients at 12 months (Fig 3A), including 84.9% of DNMT3A mutations, 94.4% of TET2 mutations, and 70.6% of other mutations (Fig 3B). Mutations with VAF ≥ 0.01 engrafted more frequently than mutations with VAF< 0.01 (91.7% v 76.2%, P = .045, Data Supplement). No other donor, recipient, or transplant variables were associated with 12-month engraftment of the clone (Data Supplement).
FIG 3.
Engraftment of donor CH in recipients after transplantation. (A) The VAFs of 102 donor mutations assessed in 69 recipients after transplantation. Each row is an individual mutation, with columns representing the VAFs in the donors at the time of transplant and in the recipients at 3 and 12 months after transplant. VAFs are color-coded from white (lower VAF) to red (higher VAF), with blue indicating that the mutation was not detected. Mutations that were detectable in recipients at 12 months (engrafted) are on top and those that were not engrafted are on bottom. (B) The donor VAFs of DNMT3A, TET2, ASXL1, and other mutations that did or did not engraft in recipients. (C) The proportion of R882 and non-R882 DNMT3A mutations assessed post-transplant (n = 10 and 54, respectively) that did or did not engraft in recipients at 12 months. (D) The VAFs for expansion of non-R882 (left) and R882 DNMT3A mutations (right) in donors and corresponding 12-month samples from recipients. (E) The plasma levels of IL-12p70 at 12 months after transplant in recipients of DNMT3A-CH (n = 21, red) compared with other recipients (n = 241, teal). (F) Correlations between 10 plasma cytokines measured at 12 months after transplant in the same recipients with and without DNMT3A-CH. Correlations are color-coded from blue (negative correlation) to red (positive correlation). CH, clonal hematopoiesis; IFN-γ, interferon gamma; IL, interleukin; n.d., not detected; TNF-α, tumor necrosis factor alpha; VAF, variant allele fraction.
DNMT3A encodes the methyltransferase responsible for de novo DNA methylation in hematopoietic stem cells,27 and a broad spectrum of mutations that variably reduce DNMT3A enzymatic function are frequently identified as early events in myeloid malignancies.28-30 Since DNMT3A R882 hotspot mutations have distinct biochemical function and higher risk of progression to acute myeloid leukemia compared with non-R882 DNMT3A mutations,31-34 we analyzed the long-term engraftment and expansion of 10 R882 and 54 non-R882 clones (median baseline VAF 0.017 v 0.012, P = .20). All 10 donor R882 mutations were detectable in recipients at 12 months, compared with 46 of 54 non-R882 mutations (85.2%, P = .34, Fig 3C). At 12 months, the VAF of R882 mutations was significantly higher than that of non-R882 mutations (median 0.051 v 0.021, P = .004, Fig 3D, Data Supplement).
In native CH, DNMT3A-mutated stem-cell clones invariably contribute to myeloid differentiation and have also been reported to have lymphoid potential.11,35 The lineage potential of allogeneic donor-engrafted clones, however, has not been assessed. To determine whether DNMT3A clones from older donors contribute to the T cell lineage after transplantation, we quantified the representation of 19 DNMT3A mutations from 14 donors in purified recipient CD3-positive blood cells one year after transplantation. We found that 18 of 19 (94.7%) donor DNMT3A mutations were detectable, including 14 of 15 (93.3%) clones from patients treated with PTCy (Data Supplement).
Native CH has been associated with alterations of inflammatory cytokines,10 which could modulate graft immunologic function36,37 or accelerate age-associated phenotypes of hematopoietic stem cells from older donors by altering the bone marrow microenvironment.38 We assessed the association of donor CH with levels of 10 cytokines in 256 recipients with (n = 54) or without (n = 202) donor CH who were alive without relapse at 12 months after transplant. Recipients of donor DNMT3A-CH at VAF ≥ 0.01 (n = 21) had higher median interleukin (IL)-12p70 levels (0.37 pg/mL) than other recipients (n = 241, 0.16 pg/mL, Fig 3E, P = .002). In DNMT3A-CH recipients, IL-12p70 levels were positively correlated with levels of IL-1B, IL-4, IL-5, and interferon gamma and negatively correlated with IL-8, IL-22, tumor necrosis factor alpha, and IL-10 (Fig 3F, Data Supplement).
Evolution to DCL
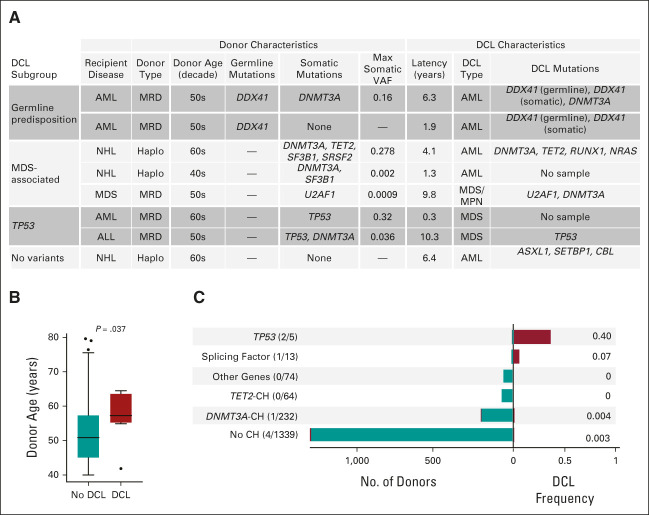
The evolution of donor CH to DCL has been described in individual cases,39-41 but the leukemic risk of donor CH has not been evaluated in a single cohort. We identified eight cases of DCL, for a 10-year cumulative incidence of 0.7% (Fig 4A). The median latency between transplantation and DCL diagnosis was 5.2 years (range 0.3-10.3 years). Donors for recipients who developed DCL were significantly older than donors for recipients who did not develop DCL (median 57.3 v 50.9 years, P = .037, Fig 4B).
FIG 4.
Clinical and genomic features of DCLs. (A) The clinical and genomic features of eight DCLs that developed in the cohort during the study period. Recipient diseases: AML, NHL, MDS, ALL. DCL type: AML, MDS, or MDS/MPN overlap syndrome. Somatic mutations reported include both those meeting criteria for CH and lower abundance mutations identified by sequencing the donor cell leukemia sample. For donor samples with multiple somatic mutations, the maximum VAF is reported. Mutations present in the donor cell leukemia are reported for the six of eight DCLs with available samples. (B) The distribution of donor age for recipients who did or did not develop DCL. (C) The total number of DCLs in each genetic subgroup of CH on the left and the corresponding proportion of recipients in each group who developed DCL on the right. Note that CH is defined here as VAF ≥ 0.005, meaning that two of the donors with low-abundance splicing factor mutations reported in (A) are classified as no CH. ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CH, clonal hematopoiesis; DCL, donor cell leukemia; MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasm; NHL, non-Hodgkin lymphoma; VAF, variant allele fraction.
To determine the proportion of DCLs that evolved directly from donor CH, we sequenced six DCL samples and analyzed the correlation between DCL mutations and the mutations present in the donor. In five of six cases, DCL mutations were detectable in the donor sample. In both DCLs without available samples, the donors had detectable CH. Donor clones that progressed to leukemia were genetically distinct from more common DNMT3A- or TET2-mutated CH: three had mutations in myelodysplastic syndrome (MDS)–associated splicing factors (SF3B1, SRSF2, and U2AF1),42 two had TP53 mutations, and in two cases, we identified germline mutations in the leukemia predisposition gene DDX41 that were shared between the recipient and their sibling donors (Data Supplement).43 Although donor CH with splicing factor or TP53 mutations was infrequent overall, relatively large proportions developed DCL (Fig 4C).
DISCUSSION
CH in older stem-cell donors may influence recipient outcomes after allogeneic transplantation,13 but previous studies have not shown an association between donor CH and overall recipient survival, examined its genetic heterogeneity and interaction with immune-modulating therapies, or evaluated the impact of low-abundance clones. Here, we paired sensitive detection of CH in 1,727 healthy older transplant donors with comprehensive clinical annotation of recipient outcomes. Our findings directly inform screening and selection of older donors and have implications for our fundamental understanding of GVL. We find that the presence of DNMT3A-CH or TET2-CH in stem-cell donors does not adversely affect recipient outcomes and that DNMT3A-CH is independently associated with improved survival in recipients as a consequence of reduced relapse. By contrast, rare mutations in MDS-associated genes pose a high risk of leukemic transformation after transplant.
Concerns about the risks of donor CH have stemmed largely from extrapolating results of nontransplant studies linking CH to increased risks of hematologic malignancies8 and nonhematologic diseases.10 These effects are reported to be caused by pathologic dysregulation of inflammatory cytokine signaling from differentiated clonal myeloid cells. In this study, we found that recipients of donor CH had evidence of altered inflammatory cytokine signaling, but paradoxically had improved survival mediated by a lower risk of disease relapse, consistent with the critical importance of graft-versus-tumor immunity in allogeneic transplant efficacy. Increased inflammatory signaling from DNMT3A-mutant myeloid cells could augment graft alloimmune activity by effects on either T-cell function44 or malignant cells, where inflammatory signaling positively regulates major histocompatibility complex Class II expression that is required for maintaining tumor immunogenicity after transplantation.45 IL-12p70, which was significantly higher in recipients of DNMT3A-CH than others and was correlated with other proinflammatory cytokines, has been implicated in the development of GVHD and GVL through its positive effects on Th1 polarization and interferon gamma production in CD4+ T cells.44,46-48 Cross talk with donor-engrafted myeloid cells might explain why DNMT3A-CH is associated with an increased risk of chronic GVHD, but not acute GVHD, which is mediated by alloreactive T cells present in the graft at the time of transplantation.49 Loss-of-function DNMT3A mutations in donor-engrafted T cells could also potentiate alloimmune activity in a cell autonomous manner by limiting exhaustion programs50 or augmenting development of memory CD8+ T-cell populations,51 both of which are mediated by de novo DNA methylation in T cells. Together, our results provide a mechanistic rationale for exploring therapeutic modulation of DNA methyltransferase activity to augment efficacy of cell-based immune therapies.
We empirically defined a VAF cutoff for CH clinical significance. We found that even small clones engraft reliably in recipients, but that the biologic and clinical consequences of donor DNMT3A-CH are only meaningful above VAF 0.01. Notably, 246 of 388 donors with CH had VAF ≤ 0.02 and thus would not have been evaluated in studies relying on the provisional definition of CH of indeterminate potential.13 This is particularly important for clinical applications, as small clones are expected to be identified more frequently as ever more sensitive sequencing technologies are deployed in the clinical setting.
Administration of PTCy is an alternative to traditional calcineurin inhibitor (CNI)–based GVHD prophylaxis because it potently reduces the risk of chronic GVHD.52,53 In our study, the type of GVHD prophylaxis regimen had no overall impact on survival or relapse, but the effects of DNMT3A-CH were only observed in those who received CNI-based regimens. If confirmed by additional studies, this observation may have immediate clinical implications in cases where the best available donor has DNMT3A-CH. In such cases, recipients with high-risk hematologic malignancies who could benefit from enhanced GVL might opt for CNI-based regimens, whereas recipients with nonmalignant hematologic disease in whom minimizing GVHD is a priority might opt for PTCy-based prophylaxis.
The potential for leukemic evolution of pre-existing donor clones has been proposed as a basis for excluding candidate donors with CH,18,20,39 but the magnitude of risk on the basis of the CH genotype has not been defined. In our study, no recipients of DNMT3A- or TET2-mutated donor CH developed DCL without concurrent MDS-associated mutations or germline risk alleles. Instead, DCLs arose from donor gene mutations that were rare in the cohort overall, such as TP53 and MDS-associated splicing factors, supporting findings of published case reports.39,54,55 High-risk mutations were associated with older donor age, raising the possibility that age alone also contributes to risk of transformation. The frequency of such high-risk gene mutations was lower in this healthy donor cohort than in cross-sectional studies,8,9 which could either reflect their association with clinical abnormalities that would lead to exclusion from the donor pool or a difference in the age structure of donors compared with population-based studies of CH.24
By exonerating donors with the most common form of CH, this study expands the donor pool for patients unable to find matched younger donors in unrelated registries or in whom sibling donors are preferred. By contrast, our results do not engage the question of whether an older sibling donor with CH is equivalent to a younger matched unrelated donor when both choices are available. This question is of interest given studies suggesting that, for both unrelated and haploidentical donors, younger donor age is associated with improved recipient outcomes.4,56 Although 80% of transplants in our study were from related donors, we nevertheless observed improved outcomes among recipients of DNMT3A-CH in the unrelated donor subset. Dedicated analysis of donor registries may definitively determine the extent to which donor CH modulates relapse and survival in recipients of grafts from unrelated donors.
A recommendation to incorporate screening for CH into the standard evaluation of transplant donor candidates would require synthesis of scientific evidence, technical feasibility, cost-effectiveness, and ethical considerations.18,19 Our results provide scientific evidence for such a policy discussion by showing that individuals age 40 years or older should not be excluded from stem-cell donation on the basis of identification of CH involving sole DNMT3A or TET2 mutations. By contrast, clinicians may consider excluding individuals with splicing factor or TP53-mutated CH, or with germline predisposition alleles, from donation because of an apparently elevated risk of DCL.
Alana Ogata
Patents, Royalties, Other Intellectual Property: AFO reports royalty payments from Brigham and Women’s Hospital for the intellectual property of the SARS-CoV-2 antibody assays that was licensed to Quanterix Inc.
Mark Fleharty
Patents, Royalties, Other Intellectual Property: I am listed as an inventor for a patent, DNA/RNA sequencing using a semiconducting nanopore. Patent Number(s): 9,988,677. https://www.osti.gov/biblio/1456913. I do not think this is relevant to my recent coauthorship, but I am including it for completeness.
Christopher D. Gocke
Leadership: OncoMEDX, Inc
Stock and Other Ownership Interests: OncoMEDx, Inc
Patents, Royalties, Other Intellectual Property: Intellectual property licensed from Penn State to my company, OncoMEDx, Inc
Joseph H. Antin
Consulting or Advisory Role: CSL Behring, Janssen, Pharmacosmos
Sarah Nikiforow
Consulting or Advisory Role: Kite/Gilead, Iovance Biotherapeutics, GlaxoSmithKline
Amy E. DeZern
Consulting or Advisory Role: Gilead Sciences, Taiho Pharmaceutical, Novartis, Jasper Therapeutics
Research Funding: Celgene (Inst), Astex Pharmaceuticals (Inst)
Travel, Accommodations, Expenses: AbbVie
Yi-Bin Chen
Consulting or Advisory Role: Magenta Therapeutics, Incyte, AbbVie, Equillium, Daiichi Sankyo/Lilly, Celularity, Actinium Pharmaceuticals
Vincent T. Ho
Consulting or Advisory Role: Jazz Pharmaceuticals, Janssen, Alexion Pharmaceuticals, Omeros
Research Funding: Jazz Pharmaceuticals
Niall J. Lennon
Expert Testimony: ArcherDx
David R. Walt
Employment: Partners Healthcare
Leadership: Exicure, Quanterix
Stock and Other Ownership Interests: Quanterix, Exicure, Illumina
Consulting or Advisory Role: Danaher
Research Funding: Canon Medical System (Inst)
Patents, Royalties, Other Intellectual Property: My laboratory has licensed technology to both Illumina and Quanterix. I receive royalties on the patents from my previous academic employer, Tufts University
Jerome Ritz
Stock and Other Ownership Interests: LifeVault Bio, Tscan Therapeutics, Clade Therapeutics
Consulting or Advisory Role: Draper Labs, Avrobio, TScan Therapeutics, Talaris, Rheos Medicines, Infinity Pharmaceuticals, Akron Biotech, Garuda Therapeutics, Novartis Institutes for BioMedical Research
Research Funding: Kite/Gilead, Equillium, Amgen, Novartis Institutes for BioMedical Research
Robert J. Soiffer
Leadership: Kiadis Pharma, Be the Match/NMDP
Consulting or Advisory Role: Juno Therapeutics, Gilead Sciences, Rheos Medicines, Cugene, Jazz Pharmaceuticals, Precision Biosciences, Takeda, Jasper Therapeutics, Alexion Pharmaceuticals
Expert Testimony: Pfizer
Travel, Accommodations, Expenses: Gilead Sciences
Lukasz P. Gondek
Consulting or Advisory Role: Bristol Myers Squibb/Celgene
R. Coleman Lindsley
Consulting or Advisory Role: Bluebird Bio, Takeda
Patents, Royalties, Other Intellectual Property: International Patent Application No. PCT/US2020/049257. Title: CRISPR Effector System Based Multiplex Cancer Diagnostics. International Filing Date: September 3, 2020. Inventors: Jonathan Gootenberg, Omar Abudayyeh, Jeremy Koob, Rahul Vedula, Coleman Lindsley, Feng Zhang. Publication No/Date: WO 2021/046257, March 11, 2021. Applicants: The Broad Institute, Inc Massachusetts Institute of Technology, and Dana-Farber Cancer Institute, Inc. Broad Ref: BI-10578 MIT Ref: 21822JR DFCI Ref.: DFCI 2775.010 JMIN Ref: BROD-4630WP.
No other potential conflicts of interest were reported.
DISCLAIMER
The authors attest that all clinical and genetic data required for replication are contained in the article and Data Supplement.
SUPPORT
Supported by the National Institutes of Health [K08CA204734 (R.C.L.), P01CA229092 (R.C.L., J.R., and R.J.S.), K08HL136894 (L.P.G.), R01HL156144 (L.P.G.)], the Damon Runyon Cancer Research Foundation (C.J.G.), the Alan and Lisa Dynner Fund (R.J.S. and R.C.L.), the James A. and Lois J. Champy Family Fund (R.C.L.), the Jock and Bunny Adams Education and Research Fund (J.H.A.), the Ted and Eileen Pasquarello Tissue Bank in Hematologic Malignancies, and the Connell and O'Reilly Families Cell Manipulation Core Facility.
L.P.G. and R.C.L. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Christopher J. Gibson, Robert J. Soiffer, Lukasz P. Gondek, R. Coleman Lindsley
Financial support: Robert J. Soiffer, Lukasz P. Gondek, R. Coleman Lindsley
Provision of study materials or patients: Christopher D. Gocke, Sarah Nikiforow, Amy E. DeZern, Richard J. Jones, Robert J. Soiffer
Collection and assembly of data: Christopher J. Gibson, Lin Zhao, H. Moses Murdock, Bryan Hambley, Alana Ogata, Rafael Madero-Marroquin, Shiyu Wang, Lisa Green, Tyler Dougan, Chi-An Cheng, Brendan Blumenstiel, Carrie Cibulskis, Christopher D. Gocke, Sarah Nikiforow, Amy E. DeZern, Yi-Bin Chen, Vincent T. Ho, Niall J. Lennon, David R. Walt, Jerome Ritz, Lukasz P. Gondek, R. Coleman Lindsley
Data analysis and interpretation: Haesook T. Kim, Alana Ogata, Mark Fleharty, Tyler Dougan, Junko Tsuji, Madeleine Duran, Joseph H. Antin, Amy E. DeZern, Yi-Bin Chen, Richard J. Jones, David R. Walt, Jerome Ritz, Robert J. Soiffer, Lukasz P. Gondek, R. Coleman Lindsley
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Donor Clonal Hematopoiesis and Recipient Outcomes After Transplantation
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Alana Ogata
Patents, Royalties, Other Intellectual Property: AFO reports royalty payments from Brigham and Women’s Hospital for the intellectual property of the SARS-CoV-2 antibody assays that was licensed to Quanterix Inc.
Mark Fleharty
Patents, Royalties, Other Intellectual Property: I am listed as an inventor for a patent, DNA/RNA sequencing using a semiconducting nanopore. Patent Number(s): 9,988,677. https://www.osti.gov/biblio/1456913. I do not think this is relevant to my recent coauthorship, but I am including it for completeness.
Christopher D. Gocke
Leadership: OncoMEDX, Inc
Stock and Other Ownership Interests: OncoMEDx, Inc
Patents, Royalties, Other Intellectual Property: Intellectual property licensed from Penn State to my company, OncoMEDx, Inc
Joseph H. Antin
Consulting or Advisory Role: CSL Behring, Janssen, Pharmacosmos
Sarah Nikiforow
Consulting or Advisory Role: Kite/Gilead, Iovance Biotherapeutics, GlaxoSmithKline
Amy E. DeZern
Consulting or Advisory Role: Gilead Sciences, Taiho Pharmaceutical, Novartis, Jasper Therapeutics
Research Funding: Celgene (Inst), Astex Pharmaceuticals (Inst)
Travel, Accommodations, Expenses: AbbVie
Yi-Bin Chen
Consulting or Advisory Role: Magenta Therapeutics, Incyte, AbbVie, Equillium, Daiichi Sankyo/Lilly, Celularity, Actinium Pharmaceuticals
Vincent T. Ho
Consulting or Advisory Role: Jazz Pharmaceuticals, Janssen, Alexion Pharmaceuticals, Omeros
Research Funding: Jazz Pharmaceuticals
Niall J. Lennon
Expert Testimony: ArcherDx
David R. Walt
Employment: Partners Healthcare
Leadership: Exicure, Quanterix
Stock and Other Ownership Interests: Quanterix, Exicure, Illumina
Consulting or Advisory Role: Danaher
Research Funding: Canon Medical System (Inst)
Patents, Royalties, Other Intellectual Property: My laboratory has licensed technology to both Illumina and Quanterix. I receive royalties on the patents from my previous academic employer, Tufts University
Jerome Ritz
Stock and Other Ownership Interests: LifeVault Bio, Tscan Therapeutics, Clade Therapeutics
Consulting or Advisory Role: Draper Labs, Avrobio, TScan Therapeutics, Talaris, Rheos Medicines, Infinity Pharmaceuticals, Akron Biotech, Garuda Therapeutics, Novartis Institutes for BioMedical Research
Research Funding: Kite/Gilead, Equillium, Amgen, Novartis Institutes for BioMedical Research
Robert J. Soiffer
Leadership: Kiadis Pharma, Be the Match/NMDP
Consulting or Advisory Role: Juno Therapeutics, Gilead Sciences, Rheos Medicines, Cugene, Jazz Pharmaceuticals, Precision Biosciences, Takeda, Jasper Therapeutics, Alexion Pharmaceuticals
Expert Testimony: Pfizer
Travel, Accommodations, Expenses: Gilead Sciences
Lukasz P. Gondek
Consulting or Advisory Role: Bristol Myers Squibb/Celgene
R. Coleman Lindsley
Consulting or Advisory Role: Bluebird Bio, Takeda
Patents, Royalties, Other Intellectual Property: International Patent Application No. PCT/US2020/049257. Title: CRISPR Effector System Based Multiplex Cancer Diagnostics. International Filing Date: September 3, 2020. Inventors: Jonathan Gootenberg, Omar Abudayyeh, Jeremy Koob, Rahul Vedula, Coleman Lindsley, Feng Zhang. Publication No/Date: WO 2021/046257, March 11, 2021. Applicants: The Broad Institute, Inc Massachusetts Institute of Technology, and Dana-Farber Cancer Institute, Inc. Broad Ref: BI-10578 MIT Ref: 21822JR DFCI Ref.: DFCI 2775.010 JMIN Ref: BROD-4630WP.
No other potential conflicts of interest were reported.
REFERENCES
- 1.Schlenk RF, Döhner K, Krauter J, et al. : Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med 358:1909-1918, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Nakamura R, Saber W, Martens MJ, et al. : A multi-center biologic assignment trial comparing reduced intensity allogeneic hematopoietic cell transplantation to hypomethylating therapy or best supportive care in patients aged 50-75 with advanced myelodysplastic syndrome: Blood and Marrow Transplant Clinical Trials Network study 1102. Blood 136:19-21, 2020 [Google Scholar]
- 3.Be The Match : Age Requirements and Limits for Donating Bone Marrow. https://bethematch.org/transplant-basics/matching-patients-with-donors/why-donor-age-matters/ [Google Scholar]
- 4.Kollman C, Spellman SR, Zhang M-J, et al. : The effect of donor characteristics on survival after unrelated donor transplantation for hematologic malignancy. Blood 127:260-267, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gragert L, Eapen M, Williams E, et al. : HLA match likelihoods for hematopoietic stem-cell grafts in the U.S. registry. N Engl J Med 371:339-348, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pulsipher MA, Logan BR, Kiefer DM, et al. : Related peripheral blood stem cell donors experience more severe symptoms and less complete recovery at one year compared to unrelated donors. Haematologica 104:844-854, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCurdy SR, Zhang M-J, St Martin A, et al. : Effect of donor characteristics on haploidentical transplantation with posttransplantation cyclophosphamide. Blood Adv 2:299-307, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaiswal S, Fontanillas P, Flannick J, et al. : Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371:2488-2498, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genovese G, Kähler AK, Handsaker RE, et al. : Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371:2477-2487, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaiswal S, Natarajan P, Silver AJ, et al. : Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 377:111-121, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young AL, Challen GA, Birmann BM, et al. : Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun 7:12484, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson CJ, Kennedy JA, Nikiforow S, et al. : Donor-engrafted CHIP is common among stem cell transplant recipients with unexplained cytopenias. Blood 130:91-94, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frick M, Chan W, Arends CM, et al. : Role of donor clonal hematopoiesis in allogeneic hematopoietic stem-cell transplantation. J Clin Oncol 37:375-385, 2019 [DOI] [PubMed] [Google Scholar]
- 14.Wong WH, Bhatt S, Trinkaus K, et al. : Engraftment of rare, pathogenic donor hematopoietic mutations in unrelated hematopoietic stem cell transplantation. Sci Transl Med 12:eaax6249, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boettcher S, Wilk CM, Singer J, et al. : Clonal hematopoiesis in donors and long-term survivors of related allogeneic hematopoietic stem cell transplantation. Blood 135:1548-1559, 2020 [DOI] [PubMed] [Google Scholar]
- 16.Oran B, Champlin RE, Wang F, et al. : Donor clonal hematopoiesis increases risk of acute graft versus host disease after matched sibling transplantation. Leukemia 10.1038/s41375-021-01312-3 [epub ahead of print on June 16, 2021] [DOI] [PubMed] [Google Scholar]
- 17.Newell LF, Williams T, Liu J, et al. : Engrafted donor-derived clonal hematopoiesis after allogenic hematopoietic cell transplantation is associated with chronic graft-versus-host disease requiring immunosuppressive therapy, but no adverse impact on overall survival or relapse. Transplant Cell Ther 27:662.e1-662.e9, 2021 [DOI] [PubMed] [Google Scholar]
- 18.DeZern AE, Gondek LP: Stem cell donors should be screened for CHIP. Blood Adv 4:784-788, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson CJ, Lindsley RC: Stem cell donors should not be screened for clonal hematopoiesis. Blood Adv 4:789-792, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seftel MD, Kuxhausen M, Burns L, et al. : Clonal hematopoiesis in related allogeneic transplant donors: Implications for screening and management. Biol Blood Marrow Transplant 26:e142-e144, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorror ML, Maris MB, Storb R, et al. : Hematopoietic cell transplantation (HCT)-specific comorbidity index: A new tool for risk assessment before allogeneic HCT. Blood 106:2912-2919, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armand P, Gibson CJ, Cutler C, et al. : A disease risk index for patients undergoing allogeneic stem cell transplantation. Blood 120:905-913, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abelson S, Collord G, Ng SWK, et al. : Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559:400-404, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malcovati L, Gallì A, Travaglino E, et al. : Clinical significance of somatic mutation in unexplained blood cytopenia. Blood 129:3371-3378, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eto M, Mayumi H, Tomita Y, et al. : Specific destruction of host-reactive mature T cells of donor origin prevents graft-versus-host disease in cyclophosphamide-induced tolerant mice. J Immunol 146:1402-1409, 1991 [PubMed] [Google Scholar]
- 26.Wachsmuth LP, Patterson MT, Eckhaus MA, et al. : Post-transplantation cyclophosphamide prevents graft-versus-host disease by inducing alloreactive T cell dysfunction and suppression. J Clin Invest 129:2357-2373, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong M, Sun D, Luo M, et al. : Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat Genet 46:17-23, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ley TJ, Ding L, Walter MJ, et al. : DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363:2424-2433, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter MJ, Ding L, Shen D, et al. : Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia 25:1153-1158, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel JP, Gönen M, Figueroa ME, et al. : Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 366:1079-1089, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russler-Germain DA, Spencer DH, Young MA, et al. : The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 25:442-454, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miles LA, Bowman RL, Merlinsky TR, et al. : Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 587:477-482, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young AL, Tong RS, Birmann BM, et al. : Clonal hematopoiesis and risk of acute myeloid leukemia. Haematologica 104:2410-2417, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SJ, Zhao H, Hardikar S, et al. : A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood 122:4086-4089, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buscarlet M, Provost S, Feroz Zada Y, et al. : Lineage restriction analyses in CHIP indicate myeloid bias for TET2 and multipotent stem cell origin for DNMT3A. Blood 132:277-288, 2018 [DOI] [PubMed] [Google Scholar]
- 36.Nisticò A, Young NS: Gamma-interferon gene expression in the bone marrow of patients with aplastic anemia. Ann Intern Med 120:463-469, 1994 [DOI] [PubMed] [Google Scholar]
- 37.Baldridge MT, King KY, Boles NC, et al. : Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature 465:793-797, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young K, Eudy E, Bell R, et al. : Decline in IGF1 in the bone marrow microenvironment initiates hematopoietic stem cell aging. Cell Stem Cell 28:1473-1482.e7, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gondek LP, Zheng G, Ghiaur G, et al. : Donor cell leukemia arising from clonal hematopoiesis after bone marrow transplantation. Leukemia 30:1916-1920, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi S, Kobayashi A, Osawa Y, et al. : Donor cell leukemia arising from preleukemic clones with a novel germline DDX41 mutation after allogenic hematopoietic stem cell transplantation. Leukemia 31:1020-1022, 2017 [DOI] [PubMed] [Google Scholar]
- 41.Nevejan L, Nollet F, Devos H, et al. : Malignant progression of donor-engrafted clonal hematopoiesis in sibling recipients after stem cell transplantation. Blood Adv 4:5631-5634, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida K, Sanada M, Shiraishi Y, et al. : Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478:64-69, 2011 [DOI] [PubMed] [Google Scholar]
- 43.Polprasert C, Schulze I, Sekeres MA, et al. : Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell 27:658-670, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macatonia SE, Hosken NA, Litton M, et al. : Dendritic cells produce IL-12 and direct the development of Th1 cells from naive CD4+ T cells. J Immunol 154:5071-5079, 1995 [PubMed] [Google Scholar]
- 45.Christopher MJ, Petti AA, Rettig MP, et al. : Immune escape of relapsed AML cells after allogeneic transplantation. N Engl J Med 379:2330-2341, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy EE, Terres G, Macatonia SE, et al. : B7 and interleukin 12 cooperate for proliferation and interferon gamma production by mouse T helper clones that are unresponsive to B7 costimulation. J Exp Med 180:223-231, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reddy V, Winer AG, Eksioglu E, et al. : Interleukin 12 is associated with reduced relapse without increased incidence of graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 11:1014-1021, 2005 [DOI] [PubMed] [Google Scholar]
- 48.Darlak KA, Wang Y, Li J-M, et al. : Enrichment of IL-12-producing plasmacytoid dendritic cells in donor bone marrow grafts enhances graft-versus-leukemia activity in allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 19:1331-1339, 2013 [DOI] [PubMed] [Google Scholar]
- 49.Kernan NA, Collins NH, Juliano L, et al. : Clonable T lymphocytes in T cell-depleted bone marrow transplants correlate with development of graft-v-host disease. Blood 68:770-773, 1986 [PubMed] [Google Scholar]
- 50.Ghoneim HE, Fan Y, Moustaki A, et al. : De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 170:142-157.e19, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Youngblood B, Hale JS, Kissick HT, et al. : Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 552:404-409, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanakry CG, O’Donnell PV, Furlong T, et al. : Multi-institutional study of post-transplantation cyclophosphamide as single-agent graft-versus-host disease prophylaxis after allogeneic bone marrow transplantation using myeloablative busulfan and fludarabine conditioning. J Clin Oncol 32:3497-3505, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kasamon YL, Bolanos-Meade J, Prince GT, et al. : Outcomes of nonmyeloablative HLA-haploidentical blood or marrow transplantation with high-dose post-transplantation cyclophosphamide in older adults. J Clin Oncol 33:3152-3161, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herold S, Kuhn M, Bonin MV, et al. : Donor cell leukemia: Evidence for multiple preleukemic clones and parallel long term clonal evolution in donor and recipient. Leukemia 31:1637-1640, 2017 [DOI] [PubMed] [Google Scholar]
- 55.Rojek K, Nickels E, Neistadt B, et al. : Identifying inherited and acquired genetic factors involved in poor stem cell mobilization and donor-derived malignancy. Biol Blood Marrow Transplant 22:2100-2103, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeZern AE, Franklin C, Tsai H-L, et al. : Relationship of donor age and relationship to outcomes of haploidentical transplantation with posttransplant cyclophosphamide. Blood Adv 5:1360-1368, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]