Abstract
Ovarian cancer is the deadliest gynecologic malignancy in the United States with most patients diagnosed in the advanced stage of the disease. Platinum-based antineoplastic therapeutics is indispensable to treating advanced ovarian serous carcinoma. However, patients have heterogeneous responses to platinum drugs, and it is difficult to predict these interindividual differences before administering medication. In this study, we investigated the tumor proteomic profiles and clinical characteristics of 130 ovarian serous carcinoma patients analyzed by the Clinical Proteomic Tumor Analysis Consortium (CPTAC), predicted the platinum drug response using supervised machine learning methods, and evaluated our prediction models through leave-one-out cross-validation. Our data-driven feature selection approach indicated that tumor proteomics profiles contain information for predicting binarized platinum response (P < 0.0001). We further built a least absolute shrinkage and selection operator (LASSO)-Cox proportional hazards model that stratified patients into early relapse and late relapse groups (P = 0.00013). The top proteomic features indicative of platinum response were involved in ATP synthesis pathways and Ran GTPase binding. Overall, we demonstrated that proteomic profiles of ovarian serous carcinoma patients predicted platinum drug responses as well as provided insights into the biological processes influencing the efficacy of platinum-based therapeutics. Our analytical approach is also extensible to predicting response to other antineoplastic agents or treatment modalities for both ovarian and other cancers.
Keywords: bioinformatics, cancer biomarkers, drug resistance, ovarian cancer, tandem mass spectrometry
Graphical Abstract
INTRODUCTION
Ovarian cancer is the second most common and the deadliest gynecologic malignancy in the United States, causing more than 14,200 deaths per year.1 The lifetime risk for ovarian cancer is approximately one in 73.2 Early detection has been difficult, as most patients in the early stage are either asymptomatic or experience nonspecific symptoms, such as abdominal distention, nausea, anorexia, or early satiety.3,4 For this reason, ovarian cancer was once called the “silent killer”.5 There is no standard screening test for ovarian cancer to date,6 and multimodal screening with serum CA 125 and ultrasonography has not been proven to decrease mortality.7 As such, more than 75% of patients have tumor outside of their ovaries at the time of initial diagnosis.8
Chemotherapy with platinum-based antineoplastic drugs is an indispensible part of treating advanced ovarian serous carcinoma. Common platinum drugs for treating this malignancy include carboplatin and cisplatin.9 Previous studies demonstrated that these platinum-containing drugs have very similar modes of action and mechanisms of resistance.10,11 Despite the initial response to platinum-based therapeutics, some patients experienced tumor relapse shortly after treatment. Researchers proposed several mechanisms that render ovarian tumor cells resistant to platinum chemotherapy,11,12 such as inactivating the drug by glutathione,13 increasing repair of the platinum adducts,14 reducing drug uptake or increasing efflux,15 and increasing the tolerance of the platinum adducts.16,17 Clinicians define platinum-free interval (PFI), the interval between the completion of platinum-based treatment and the detection of tumor relapse, to quantify the response of platinum treatment and stratify patients. Patients with a PFI of less than 6 months are considered platinum resistant, and patients with a PFI of more than or equal to 6 months are considered platinum sensitive.18,19 This stratification informs treatment strategies for relapsed tumors.20–22
Predicting platinum response is of clinical importance. Previous studies investigated the gene expression profiles (using cDNA array)23 and microRNA patterns24 that correlated with the response to chemotherapeutic agents. Other studies proposed using the promoter methylation status of selected genes25 or serum proteins26 to predict platinum response. Although the expression levels of certain proteins or genes were implicated with platinum resistance, most of the proposed biomarkers were never validated or cross-validated, and there is no large-scale study on how the tumor proteome correlated with platinum response. Furthermore, the massive amount of data generated by high-throughput methods posed additional challenges in determing the real associations.27
Previous research showed that a combination of protein markers28 could improve the accuracy of diagnosing ovarian cancer.29 Integrating the serum levels of CA125, β2-micro-globulin, transferrin, apolipoprotein A1, transthyretin, and clinical assessments, the OVA1 immunoassay detected 98.3% of early stage ovarian cancer in postmenopausal patients, outperforming clinical assessments alone or any single protein marker.30 This diagnostic test is the first FDA-cleared in vitro multivariate index assay of proteomic biomarkers, demonstrating the value of protein markers in detecting ovarian cancer.31 However, the utility of tissue protein markers in predicting patient responses to chemotherapy remains to be established.
In this study, we analyzed proteins associated with platinum response using liquid chromatography-tandem mass spectrometry (LC-MS/MS), predicted individual platinum response using robust machine learning models, and discovered proteins and biological processes associated with platinum response. These results contribute to precision medicine32 by building an accurate prediction model for individual chemotherapy responses for ovarian serous carcinoma patients.
MATERIALS AND METHODS
Proteomic Profiling Using LC-MS/MS
Proteomic profiles of 130 ovarian serous carcinoma patients were analyzed by The Cancer Genome Atlas (TCGA) Clinical Proteomic Tumor Analysis Consortium (CPTAC). In short, tumor tissues were collected from newly diagnosed patients with ovarian serous carcinoma who were undergoing surgical resection. No cases received chemotherapy or radiotherapy before surgery. Samples were processed and profiled using LC-MS/MS analysis by TCGA CPTAC. In short, proteins from the tumor tissues were extracted and digested by trypsin. 4-Plex iTRAQ (isobaric tag for relative and absolute quantitation) labeling was performed on the peptides, and peptide fractionation was conducted by basic reversed-phase liquid chromatography (bRPLC). Phosphopeptides were enriched by immobilized metal affinity chromatography (IMAC). Liquid chromatography-tandem mass spectrometry using a high-resolution and high-accuracy Orbitrap mass spectrometer was employed to perform the proteome and phosphoproteome analyses. Peptide identification was conducted through database searching, and the identified peptides were assembled into proteins. iTRAQ reporter ion intensities were computed for protein quantitation. This method has been shown to successfully profile abundant proteins in the tumor tissue samples but may miss proteins at low picomolar concentrations. Detailed descriptions of the experimental and data processing method were described in the TCGA CPTAC paper.33
The abundance of each protein was treated as a feature in the subsequent data analysis. All of the raw proteomics data are deposited to the CPTAC Data Coordinating Center (https://cptac-data-portal.georgetown.edu).
Determining Platinum Response Status of Ovarian Serous Carcinoma Patients
Using the conventional definition, we define platinum-free interval (PFI) as the interval between the completion date of platinum treatment to the date of detection of relapse. For patients who are alive and without relapse, we define PFI to be the interval between the completion date of platinum treatment and the date of the last follow-up. A patient was defined to be platinum resistant if her PFI was less than six months and the tumor had progressed or recurred. Patients with PFI six months or greater were defined as platinum sensitive. If a patient had been followed-up for less than six months after completion of platinum treatment and did not have tumor relapse at the last contact, it would be too early to determine her true platinum response status, and the associated data was excluded from analyses. There were 95 platinum sensitive patients and 35 platinum resistant patients in our data set. Detailed clinical profiles of the patients are shown in Table S-1.
Identifying Differentially Expressed Proteins between Platinum-Sensitive and -Resistant Patients
For each protein, mean fold change between platinum-sensitive and -resistant groups were calculated. Proteins with extreme fold change (fold changes greater than the 99th percentile or less than the 1st percentile of all fold change values) were identified, and the Wilcoxon rank-sum test was used to determine the significance of protein expression differences. The Benjamini-Hochberg method was used to correct for multiple hypothesis tests with a corrected P-value of less than 0.05 deemed as significant. All analysis was conducted with R version 3.2, and heatmaps were generated using the NMF package in R.
GO Analysis and Network Analyses
GO analysis and network analysis were performed to investigate the biological implications of proteins significantly associated with platinum response. STRING version 10 was used to perform network analysis as well as GO and KEGG pathway enrichment analyses. For network analysis, the highest confidence threshold (0.9) was applied, and 20 additional nodes were added to visualize the indirect protein-protein interactions. For GO and KEGG pathway enrichment analyses, a hypergeometric test was performed, and the Benjamini-Hochberg method was used to correct for the multiple hypothesis test. Significance level was set to 0.05 for the corrected P-values.
Supervised Machine Learning Methods for Binarized Platinum Response Prediction
For platinum response from proteomics features to be predicted, multiple supervised machine learning algorithms were utilized and compared, including bootstrap aggregating (bagging), random forests, support vector machines (with radial or polynomial kernels), and naïve Bayes classifiers. The platinum response status was binarized into two classes, where “1” indicate platinum sensitive and “0” platinum resistant. For reducing the risk of overfitting, only the selected proteins based on literature review were used to build the models. All models were built and evaluated with R version 3.2 (with R packages rpart, party, randomForest, ipred, and e1071).
Evaluation of Prediction Models for Binarized Platinum Response
For the performance of the two-class (platinum-resistant and -sensitive) classifiers to be evaluated, leave-one-out cross-validation was employed. For each previously reported gene or protein associated with platinum response,34–36 its prediction value for binarized platinum response was assessed by the random forest importance measure (derived from R package FSelector) in the training set. The top four protein candidates with the highest random forest importance scores in the training set were included in the statistical models. Samples in the training set were used to estimate all parameters of the prediction models. After the models were built and all parameters were finalized, the proteomics data of the held-out test case were input into the prediction models, and the predicted platinum status generated was compared against the true response information. The performance of each binary classifier was evaluated using area under receiver operator characteristic curves (AUC). For ensuring the robustness of our machine learning approach, this process was repeated 20 times. We employed the Wilcoxon rank-sum test to evaluate the differences in AUCs between our classifiers and the null classifier.
LASSO-Cox Proportional Hazards Model for Platinum Response Stratification
In addition to predicting the binarized platinum response status (platinum-resistant or -sensitive), a Cox proportional hazards model37 was utilized to exploit the more nuanced interindividual difference in the raw PFI data. The Cox-proportional hazards model assumed that the differences in some protein expression levels were related to the different rates of tumor recurrence observed in our patients and associated the time passed before tumor recurrence with the abundance of proteins. With the aim of regularizing the coefficients in the model and avoiding overfitting, a least absolute shrinkage and selection operator (LASSO)-Cox model (R package glmnet)38 was used to select the most predictive proteomic features in the training set, and the lambda value in the resulting formula was selected as the value that gave the smallest mean cross-validation error in the training data. For each patient, a relapse index (RI) was calculated as a linear combination of selected protein abundance values using the resulting LASSO-Cox model.
Evaluation of LASSO-Cox Proportional Hazards Models
For accommodating the relatively small sample size, performances of the LASSO-Cox proportional hazards models were assessed by leave-one-out cross-validation. As described above, models were built and proteins were selected using the training data. After the parameters were optimized through cross-validation on the training data, the relapse index was first calculated for each patient in the training set, and then the median relapse index (median RI) in the training set was used as the threshold to stratify patients in the test set into the early or late relapse group. If the relapse index of a test case was greater than the median RI (which indicated that this patient has a higher risk of relapse), this patient was categorized as a member of the early relapse group; otherwise, the patient was grouped into the late relapse group. The probability of relapse at each time point for each relapse group was evaluated and compared. The differences between the platinum-free interval of early and late relapse groups were evaluated by log-rank test with the alpha-level set at 0.05. The process of parameter initialization, model building, and model evaluation was also repeated 20 times to ensure the robustness of the prediction models.
RESULTS
Patient Characteristics
To predict response to therapy, as described in Zhang et al.,33 we examined the proteomic profiles of excisional tumor samples from 130 ovarian serous adenocarcinoma patients prior to chemotherapy and radiotherapy. The clinical characteristics of the patients are summarized in Table 1. A total of 3,584 proteins were profiled across the different patients; the abundance of each protein serves as a raw feature that can be used to establish the prediction models of platinum response either individually or in combination.
Table 1.
Clinical Characteristics of Ovarian Serous Carcinoma Patients under Study
| characteristic | summary |
|---|---|
| number of patients | 130 |
| age (mean ± SD) | 60.8 ± 11.6 years |
| Histology Type | |
| serous | 130 (100%) |
| Tumor Stage | |
| I | 2 (1.54%) |
| II | 8 (6.15%) |
| III | 96 (73.85%) |
| IV | 24 (18.46%) |
| Grade | |
| G1 | 0 (0%) |
| G2 | 21 (16.15%) |
| G3 | 107 (82.31%) |
| GX | 2 (1.54%) |
| overall platinum free interval (mean ± SD) | 7.99 ± 14.21 months |
| Platinum Status | |
| platinum sensitive | 95 (73.08%) |
| PFI in platinum-sensitive patients | 3.05 ± 1.83 months |
| platinum resistant | 35 (26.92%) |
| PFI in platinum-resistant patients | 19.39 ± 18.64 months |
| Vital Status | |
| alive | 57 (43.85%) |
| dead | 73 (56.15%) |
Patients with Similar Platinum Responses Shared Certain Proteomic Signatures
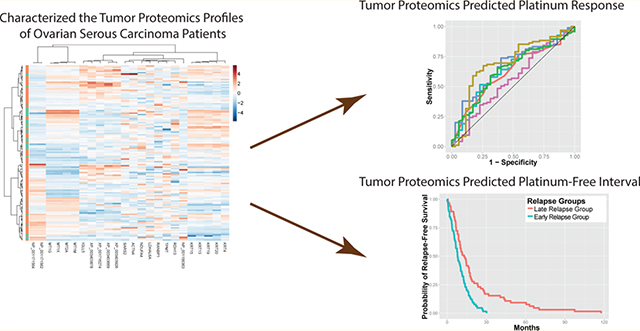
We first attempted to identify proteins whose abundance levels were associated with platinum response in ovarian serous carcinoma patients. Overall, the global proteomic profiles did not predict clinical platinum response (Figure S-1). By examining the distribution of the fold difference between the two platinum responses groups, we selected a subset of proteins differentially expressed in patients with different platinum responses. In this subset of proteins, patients with platinum resistance formed clusters in the protein abundance heatmap (Figure 1A). However, no single protein showed more than 2-fold difference when comparing platinum-sensitive patients with platinum-resistant ones, and the associations between raw expression levels of proteins and platinum response did not perfectly distinguish patients with different platinum responses. Thus, single protein markers might not be sufficient for providing robust prediction of platinum status; integrating the expression levels of multiple proteins is necessary to achieve accurate prediction models.
Figure 1.
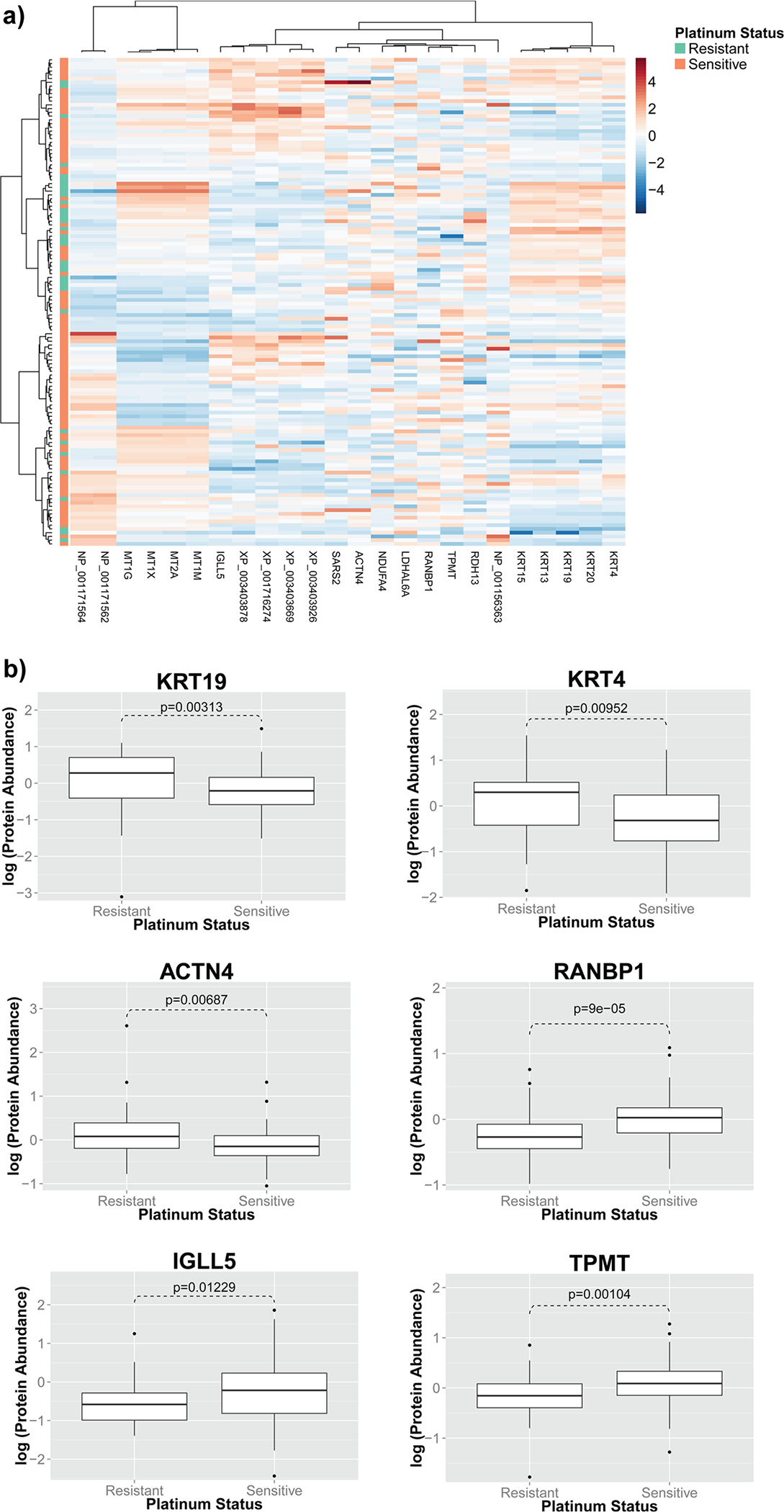
Protein expression levels were associated with platinum response of ovarian serous carcinoma patients. (A) Patients with similar platinum responses shared certain proteomic signatures. In this heatmap, each row is a patient, and each column is a protein. Hierarchical clustering results showed that platinum-resistant patients formed clusters, indicating that these protein signatures are correlated with platinum status of ovarian serous carcinoma patients. (B) Log expression levels of proteins associated with platinum response status. Abundance levels of 24 proteins, including KRT19, KRT4, ACTN4, RANBP1, IGLL5, and TPMT, were strongly associated with the clinical platinum response in ovarian serous carcinoma patients.
Twenty-Four Proteins Were Significantly Differentially Expressed in Tumors with Different Responses to Platinum-Containing Chemotherapy
We next investigated the proteins associated with different clinical platinum responses in ovarian serous carcinoma. After multiple test correction by the Benjamini-Hochberg method, 24 proteins still showed statistically significant differences in their abundance levels between the two clinically defined platinum response groups. Fourteen of the proteins in the list, including KRT19, KRT4, and ACTN4, were higher in abundance in the resistance group, whereas the other 10, including RANBP1, IGLL5, and TPMT, were higher in the platinum-sensitive group. The expression levels of the most dysregulated proteins are visualized in Figure 1B, and the expression levels of all 24 proteins are summarized in Table S-2.
Oxidative Phosphorylation Pathways and Ran GTPase Binding Function Were Highly Associated with Differentially Expressed Proteins
Network analysis revealed that proteins associated with platinum response had indirect protein-protein interactions (Figure 2). For instance, RANBP1 and ACTN4 interacted with RAN-related proteins and alpha-actinins, respectively, and these two modules of proteins both interact with Ubiquitin C (UBC), which is associated with protein degradation, DNA repair, cell cycle regulation, as well as regulation of other cell signaling pathways. In addition, KRT4, KRT19, and BCAT1 also had interaction links with UBC. In contrast, NDUFA4 formed a small interaction module with other NADH dehydrogenase (NDUFA2 and NDUFA6) and proteins involved in ATP synthesis (ATP5L and UQCR10).
Figure 2.
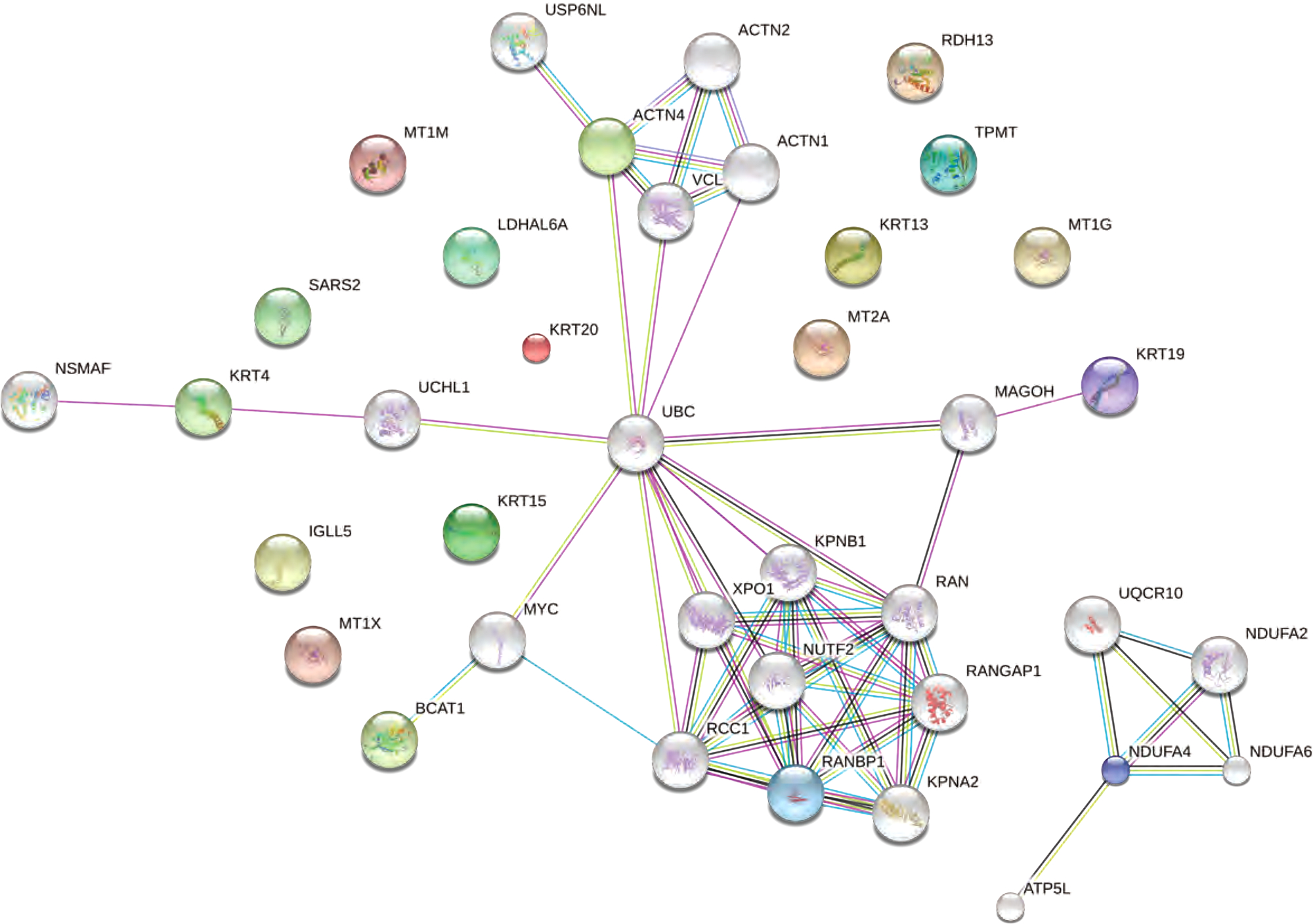
Protein-protein interaction network revealed that proteins indicative of platinum response were significantly enriched in Ran-GTPase binding, ATP synthesis pathways, and regulation mechanisms of the cell cycle. The top proteins associated with platinum response were shown in color, and their interacting proteins were shown in gray.
Pathway analysis revealed that these proteins were highly enriched in mineral absorption, oxidative phosphorylation, and RNA transport pathways (Table 2). GO analysis also showed significant enrichment in Ran GTPase binding, cellular response to zinc ion, and biological processes involving ATP synthesis and metabolism (Table 3).
Table 2.
KEGG Pathway Enrichments for Proteins Associated with Platinum Response in Ovarian Serous Carcinoma
| ID | term | number of genes | corrected P-value |
|---|---|---|---|
| 4978 | mineral absorption | 4 | 4.88 × 10−04 |
| 190 | oxidative phosphorylation | 5 | 4.88 × 10−04 |
| 5012 | Parkinson’s disease | 5 | 4.88 × 10−04 |
| 3013 | RNA transport | 5 | 4.88 × 10−04 |
| 4520 | adherens junction | 4 | 4.88 × 10−04 |
| 5146 | amoebiasis | 4 | 1.95 × 10−03 |
| 4670 | leukocyte transendothelial migration | 4 | 2.31 × 10−03 |
| 4932 | nonalcoholic fatty liver disease (NAFLD) | 4 | 5.60 × 10−03 |
| 5010 | Alzheimer’s disease | 4 | 7.58 × 10−03 |
| 5412 | arrhythmogenic right ventricular cardiomyopathy (ARVC) | 3 | 8.27 × 10−03 |
| 5203 | viral carcinogenesis | 4 | 8.27 × 10−03 |
| 5016 | Huntington’s disease | 4 | 8.27 × 10−03 |
| 4510 | focal adhesion | 4 | 1.16 × 10−02 |
| 4810 | regulation of actin cytoskeleton | 4 | 1.20 × 10−02 |
| 5322 | systemic lupus erythematosus | 3 | 1.35 × 10−02 |
| 5166 | HTLV-I infection | 4 | 2.08 × 10−02 |
| 4530 | tight junction | 3 | 2.84 × 10−02 |
Table 3.
Gene Ontology (GO) Enrichments for Proteins Associated with Platinum Response in Ovarian Serous Carcinoma
| biological processes | |||
|
| |||
| Gene Ontology ID | term | number of genes | corrected P-value |
| GO:0071294 | cellular response to zinc ion | 4 | 9.61 × 10−05 |
| GO:0075733 | intracellular transport of virus | 4 | 1.28 × 10−03 |
| GO:0046794 | transport of virus | 4 | 1.28 × 10−03 |
| GO:1902579 | multiorganism localization | 4 | 1.28 × 10−03 |
| GO:0044766 | multiorganism transport | 4 | 1.28 × 10−03 |
| GO:0010043 | response to zinc ion | 4 | 1.53 × 10−03 |
| GO:0015980 | energy derivation by oxidation of organic compounds | 7 | 1.83 × 10−03 |
| GO:0016032 | viral process | 9 | 2.92 × 10−03 |
| GO:0044764 | multiorganism cellular process | 9 | 2.92 × 10−03 |
| GO:0006091 | generation of precursor metabolites and energy | 7 | 3.08 × 10−03 |
| GO:0022900 | electron transport chain | 5 | 3.08 × 10−03 |
| GO:0022904 | respiratory electron transport chain | 5 | 3.08 × 10−03 |
| GO:1902396 | protein localization to tight junction | 2 | 3.23 × 10−03 |
| GO:0044419 | interspecies interaction between organisms | 9 | 3.90 × 10−03 |
| GO:0044403 | symbiosis, encompassing mutualism through parasitism | 9 | 3.90 × 10−03 |
| GO:0042773 | ATP synthesis coupled electron transport | 4 | 3.90 × 10−03 |
| GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 4 | 3.90 × 10−03 |
| GO:0006119 | oxidative phosphorylation | 4 | 6.26 × 10−03 |
| GO:0045333 | cellular respiration | 5 | 6.26 × 10−03 |
| GO:0000056 | ribosomal small subunit export from nucleus | 2 | 6.45 × 10−03 |
| GO:0051649 | establishment of localization in cell | 12 | 7.57 × 10−03 |
| GO:0002576 | platelet degranulation | 4 | 9.69 × 10−03 |
| GO:0000055 | ribosomal large subunit export from nucleus | 2 | 9.69 × 10−03 |
| GO:0051764 | actin cross-link formation | 2 | 9.69 × 10−03 |
| GO:0036018 | cellular response to erythropoietin | 2 | 9.69 × 10−03 |
| GO:0055114 | oxidation—reduction process | 9 | 9.69 × 10−03 |
| GO:0034333 | adherens junction assembly | 3 | 9.69 × 10−03 |
| GO:0007010 | cytoskeleton organization | 8 | 1.19 × 10−02 |
| GO:0071248 | cellular response to metal ion | 4 | 1.64 × 10−02 |
| GO:0000054 | ribosomal subunit export from nucleus | 2 | 1.87 × 10−02 |
| GO:0071426 | ribonucleoprotein complex export from nucleus | 2 | 1.87 × 10−02 |
| GO:0046034 | ATP metabolic process | 4 | 1.98 × 10−02 |
| GO:0071241 | cellular response to inorganic substance | 4 | 2.21 × 10−02 |
| GO:0019058 | viral life cycle | 5 | 2.48 × 10−02 |
| GO:0051641 | cellular localization | 12 | 2.55 × 10−02 |
| GO:0009205 | purine ribonucleoside triphosphate metabolic process | 4 | 2.72 × 10−02 |
| GO:0043297 | apical junction assembly | 3 | 2.72 × 10−02 |
| GO:1902414 | protein localization to cell junction | 2 | 2.86 × 10−02 |
| GO:0071166 | ribonucleoprotein complex localization | 2 | 2.86 × 10−02 |
| GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | 3 | 2.89 × 10−02 |
| GO:0009199 | ribonucleoside triphosphate metabolic process | 4 | 2.93 × 10−02 |
| GO:0009144 | purine nucleoside triphosphate metabolic process | 4 | 2.95 × 10−02 |
| GO:0044765 | single-organism transport | 13 | 3.96 × 10−02 |
| GO:0009141 | nucleoside triphosphate metabolic process | 4 | 3.99 × 10−02 |
| GO:0009167 | purine ribonucleoside monophosphate metabolic process | 4 | 3.99 × 10−02 |
| molecular functions | |||
|
| |||
| Gene Ontology ID | term | number of genes | corrected P-value |
| GO:0008536 | Ran GTPase binding | 3 | 4.02 × 10−02 |
| cellular components | |||
|
| |||
| Gene Ontology ID | term | number of genes | corrected P-value |
| GO:0031967 | organelle envelope | 13 | 5.31 × 10−06 |
| GO:0031975 | envelope | 13 | 5.31 × 10−06 |
| GO:0005635 | nuclear envelope | 8 | 7.72 × 10−05 |
| GO:0070062 | extracellular exosome | 18 | 7.72 × 10−05 |
| GO:0043230 | extracellular organelle | 18 | 7.72 × 10−05 |
| GO:0065010 | extracellular membrane-bounded organelle | 18 | 7.72 × 10−05 |
| GO:0031143 | pseudopodium | 3 | 4.20 × 10−04 |
| GO:0031988 | membrane-bounded vesicle | 18 | 6.78 × 10−04 |
| GO:0005643 | nuclear pore | 4 | 6.78 × 10−04 |
| GO:0044421 | extracellular region part | 18 | 1.12 × 10−03 |
| GO:0005746 | mitochondrial respiratory chain | 4 | 1.12 × 10−03 |
| GO:0046930 | pore complex | 4 | 1.29 × 10−03 |
| GO:0005829 | cytosol | 16 | 1.58 × 10−03 |
| GO:0070469 | respiratory chain | 4 | 1.58 × 10−03 |
| GO:1990204 | oxidoreductase complex | 4 | 1.58 × 10−03 |
| GO:0031982 | vesicle | 17 | 2.48 × 10−03 |
| GO:0043227 | membrane-bounded organelle | 30 | 2.91 × 10−03 |
| GO:0030018 | Z disc | 4 | 2.91 × 10−03 |
| GO:0031674 | I band | 4 | 4.46 × 10−03 |
| GO:0005747 | mitochondrial respiratory chain complex I | 3 | 5.48 × 10−03 |
| GO:0031093 | platelet alpha granule lumen | 3 | 6.68 × 10−03 |
| GO:0005576 | extracellular region | 18 | 6.68 × 10−03 |
| GO:0032991 | macromolecular complex | 17 | 8.98 × 10−03 |
| GO:0012505 | endomembrane system | 15 | 1.05 × 10−02 |
| GO:0044430 | cytoskeletal part | 9 | 1.05 × 10−02 |
| GO:0044455 | mitochondrial membrane part | 4 | 1.07 × 10−02 |
| GO:0034774 | secretory granule lumen | 3 | 1.09 × 10−02 |
| GO:0030017 | sarcomere | 4 | 1.10 × 10−02 |
| GO:0031091 | platelet alpha granule | 3 | 1.13 × 10−02 |
| GO:0005916 | fascia adherens | 2 | 1.24 × 10−02 |
| GO:0044449 | contractile fiber part | 4 | 1.33 × 10−02 |
| GO:0030863 | cortical cytoskeleton | 3 | 1.49 × 10−02 |
| GO:0030016 | myofibril | 4 | 1.49 × 10−02 |
| GO:0043292 | contractile fiber | 4 | 1.71 × 10−02 |
| GO:0043234 | protein complex | 15 | 1.72 × 10−02 |
| GO:0060205 | cytoplasmic membrane-bounded vesicle lumen | 3 | 1.72 × 10−02 |
| GO:0031983 | vesicle lumen | 3 | 1.73 × 10−02 |
| GO:0070069 | cytochrome complex | 2 | 1.73 × 10−02 |
| GO:0005856 | cytoskeleton | 10 | 2.00 × 10−02 |
| GO:0043034 | costamere | 2 | 2.07 × 10−02 |
| GO:0048471 | perinuclear region of cytoplasm | 6 | 2.16 × 10−02 |
| GO:0044448 | cell cortex part | 3 | 2.89 × 10−02 |
| GO:0005743 | mitochondrial inner membrane | 5 | 3.68 × 10−02 |
| GO:0031965 | nuclear membrane | 4 | 3.74 × 10−02 |
Machine Learning Models Predicted Binarized Platinum Response Status in Ovarian Serous Carcinoma Patients
With an aim to build a robust prediction model for patients’ binarized platinum response (platinum-sensitive or -resistant), we utilized machine learning methods to integrate the expression levels of a panel of proteins into an accurate classifier for patients’ platinum responses. Previous studies revealed that protein expression or gene expression levels of ACTN4, RANBP1, and KRT19 were associated with platinum response.34–36 However, the utility of combining the abundances of these proteins was not explored. To build an unbiased and robust multivariate model for platinum response, we calculated the random forest importance score for every protein in the training set, selected the top proteins, and built our classifiers with the training set. After all parameters in the models were finalized, we evaluated their performance on the held-out test cases. We determined the predicted probability of platinum resistance and calculated the number of true positives and negatives as a function of the probability threshold to obtain receiver operating characteristic curves. Our results showed that when combining the expression levels of the top proteins (the top proteins in different cross-validation folds varied slightly but in most folds they include RANBP1, KRT19, ACTN4, or SNAP29), we attained good prediction performance in the test set. The areas under the curves for different classifiers varied slightly but are generally at the level of 0.58–0.64 (Figure 3). Although these classifiers only weakly predicted binarized platinum response status, the performance is significantly better than random predictors (P = 7.96 × 10−9 for random forest with conditional inference trees, P = 2.10 × 10−7 for SVM with Gaussian kernel, P = 8.01 × 10−9 for SVM with polynomial kernel, P = 4.68 × 10−10 for naïve Bayes classifiers, P = 7.99 × 10−9 for Breiman’s random forest, and P = 7.99 × 10−9 for bagging). These results demonstrated that the expression levels of a panel of proteins provide information to the platinum response status in ovarian serous carcinoma patients. The fact that the prediction is not always accurate may indicate insufficiency in protein markers, tumor heterogeneity, or the arbitrariness of setting six month PFI as the threshold for platinum response, which is the most common practice in clinical settings.39–42
Figure 3.
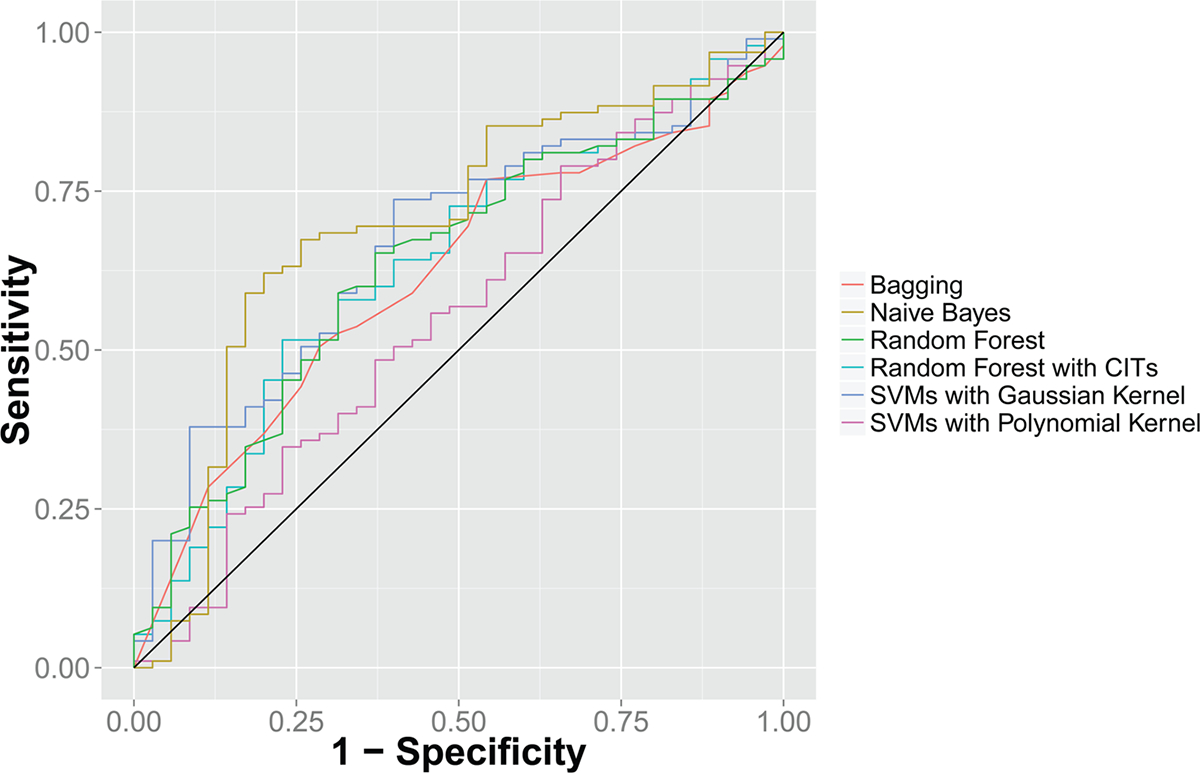
Receiver operating characteristic (ROC) curve for platinum response prediction. Integrating the abundance levels of the top protein features identified in the training set, random forests, support vector machines, bagging, and naïve Bayes classifiers weakly predicted patients with different platinum responses on leave-one-out cross-validation with areas under the curves of approximately 0.58—0.64. These classifiers are significantly better than a null classifier (P = 7.96 × 10−9 for random forest with conditional inference trees, P = 2.10 × 10−7 for SVM with Gaussian kernel, P = 8.01 × 10−9 for SVM with polynomial kernel, P = 4.68 × 10−10 for naïve Bayes classifiers, P = 7.99 × 10−9 for Breiman’s random forest, and P = 7.99 × 10−9 for bagging). The black line indicates the performance of the null classifier. CIT: conditional inference trees.
LASSO-Cox Proportional Hazards Model Successfully Predicted the Observed Platinum-Free Interval
To better correlate the diversity of platinum drug responses in ovarian cancer patients, we also modeled the platinum-free interval as a function of protein expression levels with a Cox proportional hazards model, and used L1 regularization to optimize for the number of proteins included in the model and to avoid overfitting. Applying our LASSO-Cox model, we selected 10–13 proteomic features based on cross-validation on the training set and derived a prognostic index for each patient using the expression levels of the selected proteins. On the basis of the distribution of prognostic indices of patients in the training set, we used the median prognostic index in the training set to stratify patients in the test set into two predicted relapse groups. Cross-validation revealed that patients in the two predicted groups differed significantly in their PFI (P = 0.00013 ± 0.00012) with a median PFI of 14 months in the late relapse group and a median PFI of 7.5 months in the early relapse group (Figure 4). This indicated that the combination of protein markers also predicted the length of time before relapse in ovarian serous carcinoma patients.
Figure 4.
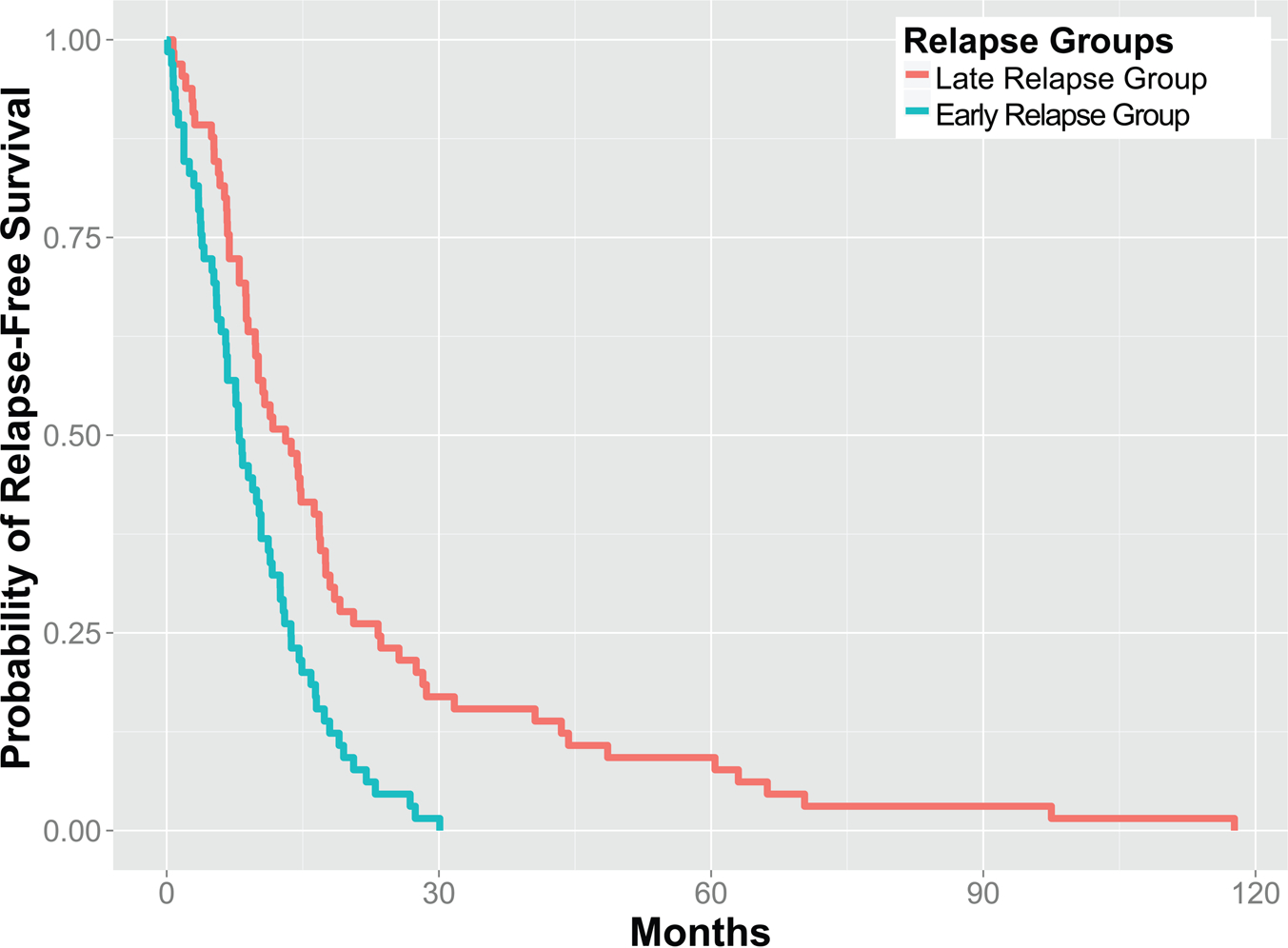
Proteomic signatures informed the platinum-free interval of patients. A LASSO-Cox proportional hazards model stratified ovarian serous carcinoma patients in the test set into two predicted relapse groups with a statistically significant difference in their platinum free interval (P = 0.00013 ± 0.00012).
DISCUSSION
Clinical response to platinum-based chemotherapy is a major determinant of the survival outcomes for ovarian serous carcinoma patients. We utilized LC-MS/MS analysis to quantify proteins from the tumor tissue, established prediction models for platinum response through machine learning methods, and provided insights into the biological mechanisms correlated with the efficacy of platinum drugs. Our results showed that tissue expression levels of 24 proteins are highly associated with platinum response and revealed the biological processes underpinning platinum resistance. In addition, we successfully predicted the binarized platinum response by utilizing a panel of proteomic markers and characterized the nuanced difference in PFIs by the relapse indices derived from the abundances of selected proteins.
Enrichment analysis demonstrated that several proteins involved in Ran (Ras-related nuclear protein) GTPase binding were associated with platinum response. Ran GTPase binding is the most enriched molecular function on our protein list. It plays a significant role in the control of DNA synthesis and cell cycle progression. Studies also suggested that Ran could be a key factor in regulating microtubule polymerization during mitosis. Previous studies showed that platinum-based antineoplastic drugs release platinum-containing ions when dissolved in water. These ions could bind to nucleotide bases, causing DNA to cross-link,43,44 which inhibits the transcription and duplication of DNA. Thus, Ran GTPase binding function might be involved in modulating DNA synthesis in the tumor cells treated by platinum drugs. In addition, decreased RANBP1 expression in platinum-resistant tumor might facilitate DNA replication and cell proliferation, causing tumor relapse despite platinum-based chemotherapy. Moreover, results also revealed that ATP synthesis and biological processes related to the electron transport chain were highly associated with the dysregulated proteins, suggesting a novel role for altered metabolism in developing platinum resistance. Our bioinformatics analysis generated insights into the mechanisms of chemotherapy resistance, which might inspire ways to increase drug sensitivity by combination chemotherapy.
Investigating individual proteins associated with platinum resistance also suggested the molecular mechanisms by which ovarian serous carcinoma interacts with chemotherapeutics. For instance, we found that increased ACTN4 and KRT19 protein expression is associated with resistance to platinum-based drugs. ACTN4 is an isoform of nonmuscular α-actinin, which is concentrated in the cytoplasm and enhances cell motility by bundling the actin cytoskeleton. This isoform is also thought to be involved in metastatic processes.45 A higher copy number of this gene was associated with resistance to first-line chemotherapies and poor survival outcome in ovarian clear cell carcinoma.34 KRT19 is a member of type I cytokeratins,46 which are involved in cell adhesion, spreading, locomotion, and potentially tumor invasion and metastasis processes.47 KRT19 protein is highly expressed in a number of malignancies, including breast cancer, endometrial cancer, lung cancer, ovarian cancer, pancreatic cancer, thyroid cancer, and urothelial cancer.48,49 Upregulation of KRT19 was also associated with increased invasion, invadopodia formation, early postoperative recurrence,50 as well as resistance to doxorubicin, 5-fluorouracil, and sorafenib in human hepatocellular carcinoma.51 It is possible that KRT19 is associated with invasion, and thus, clinically undetectable microinvasion might contribute to resistance and lethality. KRT20, another related cytokeratin, is also highly expressed in platinum-resistant tumor samples and is implicated in the development of bladder cancer,52 gastric cancer,53 and colorectal cancer.54 Further research is needed to identify the exact molecular mechanism linking this cytokeratin to platinum chemoresistance in ovarian carcinoma.
We also identified proteins with lower expression levels in platinum-resistant patients and their roles in chemoresistance. As an illustration, RANBP1, a protein that forms a complex with Ran, showed a lower abundance level in platinum-resistant patients. RANBP1 not only participated in the Ran pathway but also formed a complex that controls the transport of proteins and nucleic acids into the nucleus.55 Thus, its downregulation could potentially affect the binding of platinum drugs to DNA in the nucleus, thus contributing to the development of platinum resistance. Dysregulation of RANBP1 was also associated with activation of apoptosis and taxol sensitivity in several cancer cell lines.56,57 We discovered that, although the protein expression level of RANBP1 was associated with platinum resistance in ovarian serous carcinoma, the prediction value of this single protein was modest. However, when combined with other proteins, we were able to build a robust panel of markers for platinum status.
We further identified a few proteins previously unknown to be correlated with platinum resistance in cancer. For instance, the abundance of the TPMT protein is significant higher in the platinum-sensitive group. TPMT is an enzyme that metabolizes thiopurine chemotherapeutic agents, such as 6-mercaptopurine. Previous studies identified the correlation between the genetic polymorphisms of the gene encoding this protein and variations in sensitivity and toxicity to thiopurine drugs within individuals.58 However, the association between TPMT and platinum drug response was not described.59 In addition, lactate dehydrogenase A-like 6A (LDHAL6A) is highly expressed in platinum-resistant patients and also participates in sulfur amino acid metabolism.60 The connection between LDHAL6A and platinum response was not previously recognized. Thus, overall these proteins point to potential mechanisms affecting patient’s platinum response and can be used for prediction purposes. However, we need to validate the prediction values of these newly characterized proteins in other cohorts.
A combination of proteins is better than a single biomarker in predicting the platinum-free interval of ovarian cancer patients. This might be due to the fact that there are multiple mechanisms of developing drug resistance to platinum drugs11,12 and/or that individual effects of the identified proteins are modest. As an illustration, in addition to halting DNA duplication by cross-linking, one study suggests that cross-linked adducts generated by platinum drugs may activate several signal transduction pathways, resulting in apoptosis.61 Several other mechanisms of actions, such as inducing cytotoxic effects by binding to proteins, have also been proposed.62 As such, a variety of proteins would be affected by the chemotherapeutic agent, and any single biomarker might not reflect the entirety of drug actions. However, a panel of biomarkers captures the diverse biological pathways associated with platinum resistance and thus better predicts the response of such chemotherapy.
One limitation of this study is that the LC-MS/MS method employed by CPTAC could not identify proteins with very low abundance. As the concentration of clinically important proteins could be in the low picomolar range,63 proteins not quantified by our method might provide additional information for platinum response prediction. Another limitation is that the number of platinum resistant patients is relatively small (n = 35), and several patients participating in the CPTAC analysis had missing platinum response. Because of the limited sample size, we used very stringent criteria to select the proteins, regularize the models, and avoid overfitting. Nonetheless, because of the small sample size, proteins with moderate correlations with platinum response might not be identified. Future studies with larger cohorts might be able to identify such proteins and further validate the statistical models described here.
In summary, through deep proteomics profiling and applying tailored data mining approaches, we successfully established novel prediction models of platinum-based chemotherapy response for ovarian serous carcinoma patients and provided insights into the biological processes influencing the drug efficacy. Our analytical pipeline is likely extensible to determine the interindividual variability of the response to other antineoplastic agents or treatment modalities. This approach can also be extended to other types of cancers. Such studies are expected to contribute to precision medicine and better guide treatment for these deadly diseases.
Supplementary Material
ACKNOWLEDGMENTS
The authors express their appreciation to Dr. Karin D. Rodland for her efforts to coordinate the proteomics analysis, and to Drs. Henry Rodriguez, Christopher Kinsinger, and Emily S. Boja for organizing the Clinical Proteomic Tumor Analysis Consortium (CPTAC), and to all CPTAC investigators for generating the tumor proteomic data and their valuable feedback on the manuscript. The authors thank the anonymous reviewers for their constructive comments. K.-H.Y. is a Winston Chen Stanford Graduate Fellow and Howard Hughes Medical Institute (HHMI) International Student Research Fellow. This research was supported by National Institutes of Health Grant 5U24CA160036-05.
ABBREVIATIONS
- AUC
area under receiver operating characteristic curve
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- PFI
platinum-free interval
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.5b01129.
(Figure S-1) Global proteomic profiles of ovarian serous carcinoma patients, (Table S-1) clinical profiles of the participating ovarian serous carcinoma patients, and (Table S-2) proteins with significant abundance differences between platinum-sensitive and -resistant patients (PDF)
REFERENCES
- (1).Siegel R; Ma J; Zou Z; Jemal A Cancer statistics, 2014. Ca-Cancer J. Clin. 2014, 64,9–29. [DOI] [PubMed] [Google Scholar]
- (2).What are the key statistics about ovarian cancer? 2014. http://www.cancer.org/cancer/ovariancancer/detailedguide/ovarian-cancer-keystatistics (accessed December 11, 2014).
- (3).Goff BA; Mandel LS; Melancon CH; Muntz HG Frequency of symptoms of ovarian cancer in women presenting to primary care clinics. Jama 2004, 291, 2705–12. [DOI] [PubMed] [Google Scholar]
- (4).Olson SH; Mignone L; Nakraseive C; Caputo TA; Barakat RR; Harlap S Symptoms of ovarian cancer. Obstet. Gynecol. 2001, 98, 212–7. [DOI] [PubMed] [Google Scholar]
- (5).Goff BA; Mandel L; Muntz HG; Melancon CH Ovarian carcinoma diagnosis. Cancer 2000, 89, 2068–75. [DOI] [PubMed] [Google Scholar]
- (6).Rosenthal AN; Menon U; Jacobs IJ Screening for ovarian cancer. Clin. Obstet. Gynecol. 2006, 49, 433–47. [DOI] [PubMed] [Google Scholar]
- (7).Buys SS; Partridge E; Black A; et al. Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. Jama 2011, 305, 2295–303. [DOI] [PubMed] [Google Scholar]
- (8).Barnholtz-Sloan JS; Schwartz AG; Qureshi F; Jacques S; Malone J; Munkarah AR Ovarian cancer: changes in patterns at diagnosis and relative survival over the last three decades. Am. J. Obstet. Gynecol. 2003, 189, 1120–7. [DOI] [PubMed] [Google Scholar]
- (9).du Bois A; Luck HJ; Meier W; et al. A randomized clinical trial of cisplatin/paclitaxel versus carboplatin/paclitaxel as first-line treatment of ovarian cancer. J. Natl. Cancer I 2003, 95, 1320–1329. [DOI] [PubMed] [Google Scholar]
- (10).Eckstein N Platinum resistance in breast and ovarian cancer cell lines. J. Exp. Clin. Cancer Res. 2011, 30, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Stewart DJ Mechanisms of resistance to cisplatin and carboplatin. Critical reviews in oncology/hematology 2007, 63, 12–31. [DOI] [PubMed] [Google Scholar]
- (12).Wang D; Lippard SJ Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discovery 2005, 4, 307–20. [DOI] [PubMed] [Google Scholar]
- (13).Lai GM; Ozols RF; Young RC; Hamilton TC Effect of glutathione on DNA repair in cisplatin-resistant human ovarian cancer cell lines. J. Natl. Cancer Inst 1989, 81, 535–9. [DOI] [PubMed] [Google Scholar]
- (14).Lai GM; Ozols RF; Smyth JF; Young RC; Hamilton TC Enhanced DNA repair and resistance to cisplatin in human ovarian cancer. Biochem. Pharmacol. 1988, 37, 4597–600. [DOI] [PubMed] [Google Scholar]
- (15).Nakayama K; Kanzaki A; Ogawa K; Miyazaki K; Neamati N; Takebayashi Y Copper-transporting P-type adenosine triphosphatase (ATP7B) as a cisplatin based chemoresistance marker in ovarian carcinoma: comparative analysis with expression of MDR1, MRP1, MRP2, LRP and BCRP. Int. J. Cancer 2002, 101, 488–95. [DOI] [PubMed] [Google Scholar]
- (16).Mamenta EL; Poma EE; Kaufmann WK; Delmastro DA; Grady HL; Chaney SG Enhanced replicative bypass of platinum-DNA adducts in cisplatin-resistant human ovarian carcinoma cell lines. Cancer Research 1994, 54, 3500–5. [PubMed] [Google Scholar]
- (17).Eliopoulos AG; Kerr DJ; Herod J; et al. The control of apoptosis and drug resistance in ovarian cancer: influence of p53 and Bcl-2. Oncogene 1995, 11, 1217–28. [PubMed] [Google Scholar]
- (18).Pujade-Lauraine E How to approach patients in relapse. Annals of oncology: official journal of the European Society for Medical Oncology/ ESMO 2012, 23 (Suppl 10), x128–31. [DOI] [PubMed] [Google Scholar]
- (19).Friedlander M; Trimble E; Tinker A; et al. Clinical trials in recurrent ovarian cancer. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society 2011, 21, 771–5. [DOI] [PubMed] [Google Scholar]
- (20).Parmar MK; Ledermann JA; Colombo N; et al. Paclitaxel plus platinum-based chemotherapy versus conventional platinum-based chemotherapy in women with relapsed ovarian cancer: the ICON4/AGO-OVAR-2.2 trial. Lancet 2003, 361, 2099–106. [DOI] [PubMed] [Google Scholar]
- (21).Ledermann JA; Kristeleit RS Optimal treatment for relapsing ovarian cancer. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO 2010, 21 (Suppl 7), vii218–22. [DOI] [PubMed] [Google Scholar]
- (22).Gynecologic Oncology G; Markman M; Blessing J; et al. Phase II trial of weekly paclitaxel (80 mg/m2) in platinum and paclitaxel-resistant ovarian and primary peritoneal cancers: a Gynecologic Oncology Group study. Gynecol. Oncol. 2006, 101, 436–40. [DOI] [PubMed] [Google Scholar]
- (23).Hartmann LC; Lu KH; Linette GP; et al. Gene expression profiles predict early relapse in ovarian cancer after platinum-paclitaxel chemotherapy. Clin. Cancer Res. 2005, 11, 2149–55. [DOI] [PubMed] [Google Scholar]
- (24).Eitan R; Kushnir M; Lithwick-Yanai G; et al. Tumor microRNA expression patterns associated with resistance to platinum based chemotherapy and survival in ovarian cancer patients. Gynecol. Oncol. 2009, 114, 253–9. [DOI] [PubMed] [Google Scholar]
- (25).Chaudhry P; Srinivasan R; Patel FD Utility of gene promoter methylation in prediction of response to platinum-based chemotherapy in epithelial ovarian cancer (EOC). Cancer Invest. 2009, 27, 877–84. [DOI] [PubMed] [Google Scholar]
- (26).Oh JH; Gao J; Nandi A; Gurnani P; Knowles L; Schorge J Diagnosis of early relapse in ovarian cancer using serum proteomic profiling. Genome Informatics International Conference on Genome Informatics 2005, 16, 195–204. [PubMed] [Google Scholar]
- (27).Yu KH; Snyder M Omics profiling in precision oncology. Mol. Cell. Proteomics 2016, mcp.O116.059253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Han CL; Chen JS; Chan EC; et al. An informatics-assisted label-free approach for personalized tissue membrane proteomics: case study on colorectal cancer. Mol. Cell. Proteomics 2011, 10, M110 003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Li D; Chan DW Proteomic cancer biomarkers from discovery to approval: it’s worth the effort. Expert Rev. Proteomics 2014, 11, 135–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Longoria TC; Ueland FR; Zhang Z; et al. Clinical performance of a multivariate index assay for detecting early-stage ovarian cancer. Am. J. Obstet. Gynecol. 2014, 210 (78), e1–9. [DOI] [PubMed] [Google Scholar]
- (31).Zhang Z; Chan DW The road from discovery to clinical diagnostics: lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol., Biomarkers Prev. 2010, 19, 2995–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Snyder M Genomics and Personalized Medicine: What Everyone Needs to Know; Oxford University Press, 2016. [Google Scholar]
- (33).Zhang H; Liu T; Zhang Z; et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, DOI: 10.1016/j.cell.2016.05.069, (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Yamamoto S; Tsuda H; Honda K; et al. Actinin-4 gene amplification in ovarian cancer: a candidate oncogene associated with poor patient prognosis and tumor chemoresistance. Mod. Pathol. 2009, 22, 499–507. [DOI] [PubMed] [Google Scholar]
- (35).Macleod K; Mullen P; Sewell J; et al. Altered ErbB receptor signaling and gene expression in cisplatin-resistant ovarian cancer. Cancer Res. 2005, 65, 6789–800. [DOI] [PubMed] [Google Scholar]
- (36).Gong F; Peng X; Zeng Z; Yu M; Zhao Y; Tong A Proteomic analysis of cisplatin resistance in human ovarian cancer using 2-DE method. Mol. Cell. Biochem. 2011, 348, 141–7. [DOI] [PubMed] [Google Scholar]
- (37).Simon N; Friedman J; Hastie T; Tibshirani R Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. J. Stat. Soft. 2011, 39, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Tibshirani R Regression shrinkage and selection via the Lasso. J. Roy. Stat. Soc. B Met 1996, 58, 267–88. [Google Scholar]
- (39).Fong PC; Yap TA; Boss DS; et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J. Clin. Oncol. 2010, 28, 2512–9. [DOI] [PubMed] [Google Scholar]
- (40).Agarwal R; Kaye SB Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat. Rev. Cancer 2003, 3, 502–16. [DOI] [PubMed] [Google Scholar]
- (41).Chuang YT; Chang CL Extending platinum-free interval in partially platinum-sensitive recurrent ovarian cancer by a non-platinum regimen: its possible clinical significance. Taiwanese journal of obstetrics & gynecology 2012, 51, 336–41. [DOI] [PubMed] [Google Scholar]
- (42).Bookman MA Extending the platinum-free interval in recurrent ovarian cancer: the role of topotecan in second-line chemotherapy. Oncologist 1999, 4, 87–94. [PubMed] [Google Scholar]
- (43).Reedijk J; Lohman PH Cisplatin: synthesis, antitumour activity and mechanism of action. Pharm. Weekbl., Sci. Ed. 1985, 7, 173–80. [DOI] [PubMed] [Google Scholar]
- (44).Reedijk J The Mechanism of Action of Platinum Antitumor Drugs. Pure Appl. Chem. 1987, 59, 181–92. [Google Scholar]
- (45).Honda K; Yamada T; Endo R; et al. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J. Cell Biol. 1998, 140, 1383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zhou X; Liao J; Hu L; Feng L; Omary MB Characterization of the major physiologic phosphorylation site of human keratin 19 and its role in filament organization. J. Biol. Chem. 1999, 274, 12861–6. [DOI] [PubMed] [Google Scholar]
- (47).Bernal SD; Stahel RA Cytoskeleton-associated proteins: their role as cellular integrators in the neoplastic process. Critical reviews in oncology/hematology 1985, 3, 191–204. [DOI] [PubMed] [Google Scholar]
- (48).Berglund L; Bjorling E; Oksvold P; et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol. Cell. Proteomics 2008, 7, 2019–27. [DOI] [PubMed] [Google Scholar]
- (49).Schelfhout LJ; Van Muijen GN; Fleuren GJ Expression of keratin 19 distinguishes papillary thyroid carcinoma from follicular carcinomas and follicular thyroid adenoma. Am. J. Clin. Pathol. 1989, 92, 654–8. [DOI] [PubMed] [Google Scholar]
- (50).Uenishi T; Kubo S; Yamamoto T; et al. Cytokeratin 19 expression in hepatocellular carcinoma predicts early postoperative recurrence. Cancer Sci. 2003, 94, 851–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Govaere O; Komuta M; Berkers J; et al. Keratin 19: a key role player in the invasion of human hepatocellular carcinomas. Gut 2014, 63, 674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Mengual L; Burset M; Ars E; et al. Partially degraded RNA from bladder washing is a suitable sample for studying gene expression profiles in bladder cancer. Eur. Urol. 2006, 50, 1347–55, discussion 55–6.. [DOI] [PubMed] [Google Scholar]
- (53).Lee HJ; Nam KT; Park HS; et al. Gene expression profiling of metaplastic lineages identifies CDH17 as a prognostic marker in early stage gastric cancer. Gastroenterology 2010, 139, 213–25, e3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Merlos-Suarez A; Barriga FM; Jung P; et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell stem cell 2011, 8, 511–24. [DOI] [PubMed] [Google Scholar]
- (55).Hayashi N; Yokoyama N; Seki T; Azuma Y; Ohba T; Nishimoto T RanBP1, a Ras-like nuclear G protein binding to Ran/ TC4, inhibits RCC1 via Ran/TC4. Mol. Gen. Genet. 1995, 247, 661–9. [DOI] [PubMed] [Google Scholar]
- (56).Rensen WM; Roscioli E; Tedeschi A; et al. RanBP1 downregulation sensitizes cancer cells to taxol in a caspase-3-dependent manner. Oncogene 2009, 28, 1748–58. [DOI] [PubMed] [Google Scholar]
- (57).Amato R; Scumaci D; D’Antona L; et al. Sgk1 enhances RANBP1 transcript levels and decreases taxol sensitivity in RKO colon carcinoma cells. Oncogene 2013, 32, 4572–8. [DOI] [PubMed] [Google Scholar]
- (58).Stanulla M; Schaeffeler E; Flohr T; et al. Thiopurine methyltransferase (TPMT) genotype and early treatment response to mercaptopurine in childhood acute lymphoblastic leukemia. Jama 2005, 293, 1485–9. [DOI] [PubMed] [Google Scholar]
- (59).Khrunin AV; Khokhrin DV; Moisseev AA; Gorbunova VA; Limborska SA Pharmacogenomic assessment of cisplatin-based chemotherapy outcomes in ovarian cancer. Pharmacogenomics 2014, 15, 329–37. [DOI] [PubMed] [Google Scholar]
- (60).Chen X; Gu X; Shan Y; et al. Identification of a novel human lactate dehydrogenase gene LDHAL6A, which activates transcriptional activities of AP1(PMA). Mol. Biol. Rep. 2009, 36, 669–76. [DOI] [PubMed] [Google Scholar]
- (61).Siddik ZH Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 2003, 22, 7265–79. [DOI] [PubMed] [Google Scholar]
- (62).Fuertes MA; Castilla J; Alonso C; Perez JM Cisplatin biochemical mechanism of action: from cytotoxicity to induction of cell death through interconnections between apoptotic and necrotic pathways. Curr. Med. Chem. 2003, 10, 257–66. [DOI] [PubMed] [Google Scholar]
- (63).Grebe SK; Singh RJ LC-MS/MS in the Clinical Laboratory Where to From Here? Clinical Biochemist Reviews/Australian Association of Clinical Biochemists 2011, 32,5–31. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


