Supplemental Digital Content is available in the text.
Keywords: C-reactive protein, evidence-based medicine, interleukin-6, neonatal sepsis, neonatology, procalcitonin
Abstract
BACKGROUND:
Late-onset neonatal sepsis is a major complication in preterm neonates. Early identification of the type of infection could help to improve therapy and outcome depending on the suspected microorganism by tailoring antibiotic treatment to the individual patient based on the predicted organism. Results of blood cultures may take up to 2 days or may remain negative in case of clinical sepsis. Chemical biomarkers may show different patterns in response to different type of microorganisms.
OBJECTIVE:
The aim of this study was to develop, as a proof of concept, a simple classification tree algorithm using readily available information from biomarkers to show that biomarkers can potentially be used in discriminating in the type of infection in preterm neonates suspected of late-onset neonatal sepsis.
DERIVATION COHORT:
A total of 509 suspected late-onset neonatal sepsis episodes in neonates born before less than 32 weeks of gestation were analyzed. To examine model performance, 70% of the original dataset was randomly selected as a derivation cohort (n = 356; training dataset).
VALIDATION COHORT:
The remaining 30% of the original dataset was used as a validation cohort (n = 153; test dataset).
PREDICTION MODEL:
A classification tree prediction algorithm was applied to predict type of infection (defined as no/Gram-positive/Gram-negative sepsis).
RESULTS:
Suspected late-onset neonatal sepsis episodes were classified as no sepsis (80.8% [n = 411]), Gram-positive sepsis (13.9% [n = 71]), and Gram-negative sepsis (5.3% [n = 27]). When the derived classification tree was applied to the test cohort, the overall accuracy was 87.6% (95% CI, 81.3–92.4; p = 0.008). The classification tree demonstrates that interleukin-6 is the most important differentiating biomarker and C-reactive protein and procalcitonin help to further differentiate.
CONCLUSION:
We have developed and internally validated a simple, clinically relevant model to discriminate patients with different types of infection at moment of onset. Further research is needed to prospectively validate this in a larger population and assess whether adaptive antibiotic regimens are feasible.
Neonatal sepsis is a major health issue, particularly in preterm neonates, where low gestational age, low birth weight, immature immune system, and other compromising factors make it the primary cause of morbidity and death (1–5). Late-onset neonatal sepsis (LONS) is defined as sepsis that occurs after 3 days of life and may be caused by pathogens acquired at delivery or during the course of hospital care (6). Although, in most cases, the initiation of LONS is often inconspicuous, the clinical course may be alarmingly fulminant leading to septic shock and death within hours of onset (7, 8). Therefore, infected neonates must be promptly identified, and appropriate therapy should be started timely to reduce mortality and morbidity (9).
Current treatment of LONS in neonates consists of antibiotic therapy and supportive care (e.g., respiratory and circulatory support) (10). When LONS is suspected, empirical antibiotic therapy is started since the definitive identification of the bacterial pathogen could take up to 2 days using conventional culture methods (11). Blood cultures could also remain negative due to due to small volumes of blood drawn and low bacterial load in preterm neonates (12). Therefore, culture results are not sufficient to guide specific antibiotic therapy in early stages of LONS. Although necessary at this stage, this approach inevitably leads to unnecessary administration of antibiotics with potential side effects such as nephrotoxicity of aminoglycosides, interference with the microbiome, potential for the development of antibiotic resistance, and in some cases, suboptimal therapy (e.g., delayed start of cephalosporin’s in potential Gram-negative meningitis). Information on the most probable causative microorganism of LONS, available at the moment of first suspicion of sepsis, could guide precise antibiotic and appropriate supportive therapy for the individual patient.
Several chemical biomarkers can be determined to guide clinical care in patients suspected of LONS. Three commonly used pro-inflammatory biomarkers are interleukin (IL)-6, procalcitonin, and C-reactive protein (CRP) (13). It is known that Gram-positive or Gram-negative bacteria activate different Toll-like receptor signaling pathways, resulting in production of different pro-inflammatory cytokines and proteins (14).
Readily available information at the moment of LONS suspicion may help clinicians to start or adapt specific antibiotic therapy within a few hours in contrast to waiting for the results of standard blood cultures. We hypothesized that the levels of inflammatory biomarkers differ according to the causative microorganism (Gram-positive vs Gram-negative), and they could be used to predict the type of infection at moment of LONS suspicion. The aim of this study was to develop and validate, as a proof of concept, a simple classification tree algorithm using readily available information from biomarkers to show that biomarkers can potentially be used in discriminating in the type of infection in preterm neonates suspected of LONS.
MATERIALS AND METHODS
Study Design and Population
The study was conducted at the Erasmus MC University Medical Center—Sophia Children’s Hospital Rotterdam, a level III–IV neonatal ICU (NICU). Patient data from January 2018 to June 2020 were available for all neonates with a gestational age below 32 weeks, who were suspected of LONS and in whom blood was taken to determine inflammatory biomarkers.
Ethical Approval
The study was approved by the local ethical board of the Erasmus MC, University Medical Center (MEC-2020-0111).
Biomarkers and Patient Characteristics
The biomarkers included in the analysis were plasma IL-6, procalcitonin, and CRP determined at the time of the initial LONS suspicion. Biomarker levels were measured by the department of Clinical Chemistry and were retrospectively queried from the laboratory information system.
CRP levels were measured using a turbidimetric method (C502, Cobas 8000 system; Roche Diagnostics, Rotkreuz, Switzerland). Procalcitonin and IL-6 were both measured using electrochemiluminescence immunoassay tests (E801, Cobas 8000 system; Roche Diagnostics).
Plasma levels of IL-6, procalcitonin, and CRP were routinely determined as part of a local diagnostic protocol whenever LONS was suspected in neonates. At our center, we use heart rate variability monitoring in preterm neonates as an early warning score for LONS (15). According to the local protocol, clinicians can consider to determine chemical biomarkers when changes in heart rate variability are observed. Blood cultures are drawn and antibiotic therapy is started when the neonate shows evident clinical signs of LONS or when IL-6, procalcitonin, and/or CRP levels are increased.
The following patient characteristics were obtained from clinical charts: sex, gestational age, birth weight, and postnatal age at suspicion.
Outcome: Definition of Type of Infection
The department of Microbiology at the Erasmus MC provided all results of blood cultures, including timing of collection and identified microorganisms. One patient could provide multiple cases of suspected LONS. LONS diagnosis was established by the criteria defined by the National Institute of Child Health and Human Development Neonatal Research Network (16). LONS was defined as a positive blood culture obtained after 72 hours of life and intent to treat with antibiotics for 5 days or more. An episode of culture-proven LONS was defined as a positive blood culture due to an identified bacterial organism (including coagulase-negative Staphylococci), treated with antibiotics for 5 days or more or treated for a shorter duration if death occurred during therapy (16). Culture-negative LONS was defined as 1) CRP level greater than 10 mg/L within 2 days after blood culture, 2) antibiotic therapy longer than 5 days (or intention to treat longer), and 3) clinical symptoms of sepsis assessed by treating physician. We classified suspected LONS episodes as no sepsis (with or without blood culture), Gram-positive sepsis, or Gran-negative sepsis. Culture-negative sepsis episodes were left out of the final analyses, due to the uncertainty regarding the type of infection that hampers classification tree analyses, and the inclusion of CRP in the definition, which would bias the discriminative value of CRP. Also, prediction of culture-negative sepsis is not an outcome of interest for clinicians and does not provide information for specific antibiotic therapy or monitoring.
Statistical Analysis
Statistical analysis was performed using R (R Core Team [2017], Vienna, Austria).
Categorical variables were described using absolute and relative frequencies. Continuous variables were described using medians and range (minimum–maximum) because of non-normal distributions. Baseline biomarker levels between the different types of infection were analyzed with Kruskal-Wallis test. The dataset was split in a train and test dataset, in the ratio 70/30; 70% of the data served to train the model, and 30% to make predictions.
A classification and regression tree (CART) approach was employed to generate a classification tree prediction algorithm. The CART analysis determines optimal cutoffs for investigated variables and results in a classification tree. All three candidate biomarkers were considered in the CART analysis. The classification tree was built using the R package “RPART” (link: https://cran.r-project.org/package=rpart). Initially, a large tree that contains splits for all input variables is generated. This initial tree is generally too large to be useful as the final subgroups are too small to make sensible statistical inference. A pruning process is then applied to the initial tree with the goal of finding the “subtree” that is most predictive of the outcome of interest and to prevent overfitting. In the “RPART” package, any observation with values for the dependent variable (type of infection) and at least one independent variable (biomarker) is included in the classification process. When an independent variable is missing, a surrogate variable is used (17). This means that for each split observations, where the split variable is missing (in our case, a missing biomarker), the split is based on the best surrogate variable available (another biomarker). Also, bootstrap aggregating (bagging) of the classification tree was used to fit multiple versions of the prediction model. A benefit to creating ensembles via bagging, which is based on resampling with replacement, is that it can provide its own internal estimate of predictive performance of the training dataset with the out-of-bag (OOB) sample. When the OOB misclassification error is similar to the misclassification error rate achieved with cross validation, it suggests that the dataset is sufficiently large.
The resulting classification tree was then used as a prediction algorithm and applied to the test dataset. Additional classification trees were derived with adding baseline characteristics (gestational age, gender, and age at onset) and with different classes for type of infection (no sepsis, coagulase-negative Staphylococci, other Gram-positive and Gram-negative). The model parameters, pruning criteria, the command file for reproducing the classification tree, and the additional classification trees are provided in the supplementary material appendix (see Supplemental File S1, http://links.lww.com/CCX/A855). For all tests, a p value of less than 0.05 was considered statistically significant.
RESULTS
During the study period, 580 suspected LONS episodes occurred in preterm neonates (< 32 wk gestational age at birth), after excluding the culture-negative sepsis (n = 71) episodes, a total number of 509 suspected LONS episodes remained. The demographics and clinical characteristics of the derivation cohort are depicted in Table 1. Median values (and ranges) for IL-6, procalcitonin, and CRP were 39 pg/mL (2–1,396,700 pg/mL), 0.63 ng/mL (0.05–100.00 ng/mL), and 2.7 mg/L (0.30–297 mg/L), respectively. IL-6, procalcitonin, and CRP levels significantly differed between the different types of infection (p < 0.001 for all biomarkers). Patients with a Gram-negative sepsis had the highest levels of IL-6 and procalcitonin levels, while patients with Gram-positive sepsis had the highest CRP levels at onset of suspected LONS (Fig. 1).
TABLE 1.
Demographics and Clinical Characteristics of the Total Cohort
| Variable | All Episodes (n = 509) |
|---|---|
| Median gestational age (wk) | 26.43 (23.71–31.86) |
| Median birth weight (g) | 850 (435–2,100) |
| Number of males (%) | 277 (54.4) |
| Age at onset of suspicion (d) | 13 (3–165) |
| Interleukin-6 levels (pg/mL) | 39 (2–1,396,700) |
| Procalcitonin levels (ng/mL) | 0.63 (0.05–100.00) |
| C-reactive protein levels (mg/L) | 2.7 (0.30–297.00) |
| Intubated patients (%) | 114 (22.4) |
| Number with no sepsis (%) | 411 (80.7) |
| Number with Gram-positive bacteria (%) | 71 (13.9) |
| Number with Gram-negative bacteria (%) | 27 (5.3) |
The demographics and clinical characteristics of the total cohort. Values are medians (minimum–maximum range) or percentages.
Figure 1.
Log (10) transformed interleukin-6 (IL-6) (pg/mL), procalcitonin (PCT) (ng/mL), and C-reactive protein (CRP) (mg/L) levels at moment of onset according to type of infection. Median levels with their range for the no sepsis group: IL-6 30 pg/mL (44 pg/mL); PCT 0.56 ng/mL (0.62 ng/mL); and CRP 1.8 mg/L (4.7 mg/L). For the Gram-positive sepsis group: IL-6 383 pg/mL (1,433 pg/mL); PCT 1.58 ng/mL (2.68 ng/mL); and CRP 25 mg/L (53 mg/L). For the Gram-negative sepsis group: IL-6 918 pg/mL (25,194 pg/mL); PCT 4.8 ng/mL (33.55 ng/mL); and CRP 17 mg/L (50 mg/L).
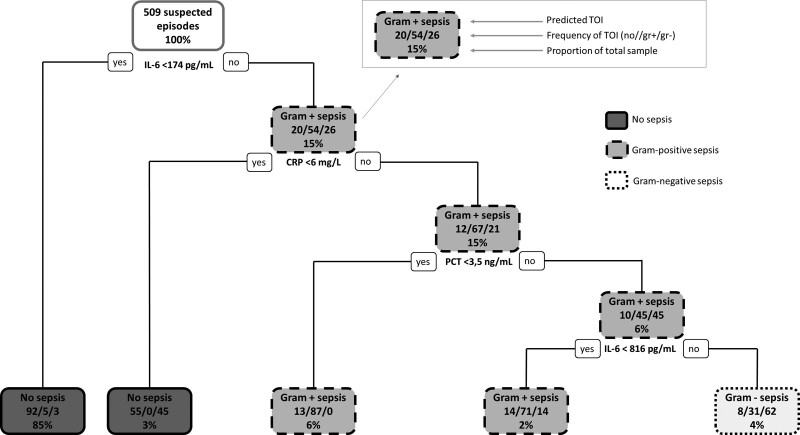
The pruned classification tree is shown in Figure 2 and is derived from the training set (356 suspected LONS episodes, 70% of total episodes). The classification tree shows five terminal nodes: two for no sepsis, two Gram-positive sepsis, and one for Gram-negative sepsis. The classification tree demonstrates that IL-6 is the most important differentiating biomarker and CRP and procalcitonin help to further differentiate.
Figure 2.
Classification tree resulting from RPART analysis of all 2018–2020 data. For each branch, to the left indicates that the patient meets the condition, and to the right, the patient does not meet the condition. The probabilities for the types of infections are depicted next to each other in every node (no infection–Gram-positive sepsis–Gram-negative sepsis). The frequency depicted at the bottom of the node is the proportion of the total sample that is in this category. CRP = C-reactive protein, gr– = Gram-negative sepsis, gr+ = Gram-positive sepsis, IL-6 = interleukin-6, no = no sepsis, PCT = procalcitonin, TOI = type of infection.
The test cohort consisted of 153 suspected LONS episodes (30% of total episodes). When the derived classification tree was applied to the test cohort, the overall accuracy was 87.6% (95% CI, 81.3–92.4%; p = 0.008). The sensitivity values for the different type of infections were 97.5%, 60.0%, and 50% for no, Gram-positive, and Gram-negative sepsis, respectively. The specificity values were 64.5%, 97.6%, and 96.6% for no, Gram-positive, and Gram-negative sepsis, respectively. Additional classification trees were derived with adding baseline characteristics (gestational age, gender, and age at onset) (Supplemental Fig. S1, http://links.lww.com/CCX/A856) and with different classes for type of infection (no sepsis, coagulase-negative Staphylococci, other Gram-positive and Gram-negative) (Supplemental Fig. S2, http://links.lww.com/CCX/A857). The classification trees did not change materially and overall accuracy was similar.
By aggregating 100 bootstrap samples and trees, the OOB estimate of misclassification error was 0.1297, which is similar to the misclassification error in the test cohort, suggesting that the predicted misclassification error in the test cohort is unbiased.
DISCUSSION
We have demonstrated that a simple algorithm that uses routine laboratory values shows a high capability of predicting the type of infection at the onset of LONS suspicion in preterm NICU patients. The generated classification tree with the combination of IL-6, CRP, and procalcitonin showed an overall accuracy rate in the test cohort of 88%.
In current clinical practice in our center, when LONS seems likely (either due to clinical symptoms or based on early inflammatory signals in combination with heart rate variability monitoring), empirical broad spectrum antibiotics are administered to the preterm neonate, without knowledge or probability assessment of the causative microorganism. From a clinical point of view, the prediction of causative microorganism could reduce the time required to initiating the most precise antimicrobial therapy and could help select high-risk patients with a likely Gram-negative sepsis. This risk assessment increases awareness and may lead to personalized monitoring and treatment. For example, more intensive monitoring of hemodynamics (near-infrared spectroscopy, echocardiography, peripheral arterial blood pressure measurement) could be considered in an identified high-risk patient to monitor possible hemodynamic deterioration. Additionally, the classification tree might be applied for antibiotic stewardship in combination with an early warning system, such as heart rate monitoring (15, 18). After a rise in the heart rate characteristic scores, the classification tree may be useful for reducing antibiotic exposure in patients with predicted no sepsis and also for identifying the highest-risk patients that would benefit from starting antibiotics (18).
Furthermore, this classification tree could enable personalized treatment, such as starting immunomodulatory compounds, like pentoxifylline in high-risk patients (19). We recently showed that IL-6 and procalcitonin levels were positively associated with the risk of mortality in preterm neonates suspected of LONS and can also be used for initiating specific therapies or intensifying monitoring (20). This association may partly be explained by the underlying causative microorganism in line with the current study findings. In adults sepsis patients, it has already been shown that procalcitonin levels might differ according to causative pathogens (21, 22). Similar to these studies, in the classification tree developed in this cohort, procalcitonin levels helped to discriminate Gram-negative from Gram-positive pathogens together with IL-6. Also, IL-6 was previously shown to have substantial discriminatory value for Gram-negative versus Gram-positive sepsis in neonates, compared with other cytokines such as CRP, in line with our study findings (18, 21). In line with this study, the developed classification tree showed that IL-6 levels were the most discriminating factor and that CRP levels only helped differentiating further. Furthermore, in adult patients that were suspected of sepsis, it was shown that procalcitonin levels could help select patients at high risk of sepsis and which patients would need a blood culture, thereby reducing the number of unnecessary blood cultures and antibiotic therapy (23). In this study, we determined inflammatory biomarkers based on changes in heart rate characteristics (15). Our findings highlight that biomarker testing can be applied as a tool to guide decisions when an early warning score, such as heart rate variability monitoring, is elevated but clinical signs remain equivocal, as was also suggested by Raynor et al (18). This includes not initiating antibiotic therapy when biomarkers are reassuring, which could reduce antibiotic exposure in patients without sepsis, important for antibiotic stewardship.
The purpose of this clinical classification tree was to develop a model capable of recognizing patterns in common laboratory data that indicate toward a causative microorganism that is otherwise unidentified within the first few days of LONS suspicion (11). Inclusion of baseline variables did not improve the accuracy of the classification tree, indicating that biomarkers levels are most informative for predicting the type of infection. In the future, when the model is further developed, we envisage it would be best implemented as an automated tool into an electronic medical record system. This tool would then enable to practice personalized medicine; when a neonate is suspected of LONS, the classification tree will predict a causative organism and a visual feedback system will send a notification to the clinicians caring for the patient and could suggest specific antibiotic therapy to the clinician. In adult patients, this has already been shown to be feasible, where Paul et al (24, 25) showed that a computerized model including local signs, symptoms, laboratory, and radiological findings can predict individual pathogens causing infection and thereby can advise physicians on appropriate empirical antibiotic treatment within the first hours of infection.
Our study also has limitations, with the most important one being possible instability of the generated algorithm. The algorithm depends on binary splits to classify LONS episodes and the algorithm selects cutoff values that result in the optimal classification tree. These cutoff values may not immediately seem logical from a clinical standpoint and may vary with differences in practice between hospitals and even between departments within a hospital. In our center, we use heart rate observation variability monitoring as an early warning system of LONS (26). Based on changes in heart rate characteristics, we consider determining inflammatory biomarkers, possibly early during LONS course, when clinical signs are absent or nonspecific. In centers without such monitoring, biomarkers might be determined later in the course of LONS, which could affect cutoff values we determined. For instance, after a rapid increase of IL-6, the levels also decrease rapidly and could provide less information compared with IL-6 levels early in the suspected LONS episode (8).
From a clinical point of view, it would be most important for the prediction model to be able to identify high-risk patients. This means that it is particularly important that the prediction model has a good accuracy in predicting the Gram-negative sepsis cases. One way to incorporate this in the model is by adding weights to observations. By default, the model gives equal weights for each class. However, the model can be trained in a manner that will penalize incorrectly labeled Gram-negative episodes more than it penalizes incorrectly labeled no sepsis episodes (17). Unfortunately, our analysis was restricted by the limited numbers of Gram-negative sepsis.
However, once the method has been developed, the process of updating the algorithm could be essentially automated. As the number of patients to develop the model will grow, some of the biomarker cutoff values that drive the classification tree will change. Since research in neonatal sepsis is growing rapidly, it might also be possible that new biomarkers are identified that might contribute to the model. Some promising new biomarkers that could be added to the model might include the pro-inflammatory biomarkers serum amyloid A, tumor necrosis factor-α, IL-8, the anti-inflammatory IL-10, IL-1 receptor antagonist, and cell surface antigen CD64 (27–30). As many factors, such as mechanical ventilation, can induce inflammation without infection, pure sepsis-related biomarkers are needed. Since neonatal sepsis is a complex disorder involving different organs, which leads to wide variations in the organism’s metabolites, analysis of the entire metabolome or proteome is a promising method (31). These “omics” approaches may enable the identification of new disease-specific biomarkers. These biomarkers are determined by liquid chromatography-mass spectrometry, which allows the simultaneous detection and quantification of up to several hundred metabolites in one sample in a relatively short time (32, 33). Incorporating a (combination of) metabolite and/or protein levels in the classification tree might thereby increase its accuracy. Additionally, traditional hematological biomarkers in adults sepsis, such as WBC count and immature to total neutrophil count ratio, might also be of value in the classification of LONS (34, 35).
Evolution of the classification tree could improve the predictive performance and its generalizability if additional biomarkers are widely available. Our methodological approach might be a valuable tool for future studies investigating the potential of predicting type of infection at moment of suspicion using readily available information.
CONCLUSIONS
In conclusion, as a proof of concept, we generated a classification tree that requires few resources and can already indicate the type of infection before the result of the blood culture. When applied in an automated fashion with the capability to alert clinicians, the method demonstrated here could allow for personalized antibiotic therapy in the treatment of preterm neonates suspected of LONS.
Supplementary Material
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
Ms. Kurul and Dr. Taal conceptualized and designed the study. Dr. Taal supervised data collection and reviewed and revised the article. Ms. Kurul collected data, carried out the initial analyses, and drafted, reviewed, and revised the article. Dr. Ramakers collected data and reviewed and revised the article. Drs. Simons, De Rijke, Kornelisse, Kroon, and Reiss critically reviewed the article for important intellectual content and revised the article. All authors approved the final article as submitted and agree to be accountable for all aspects of the work.
The authors have disclosed that they do not have any potential conflicts of interest.
This work was performed at Erasmus MC, University Medical Center-Sophia Children’s Hospital, Rotterdam, The Netherlands.
REFERENCES
- 1.Ganatra HA, Stoll BJ, Zaidi AK: International perspective on early-onset neonatal sepsis. Clin Perinatol. 2010; 37:501–523 [DOI] [PubMed] [Google Scholar]
- 2.Stoll BJ, Hansen N, Fanaroff AA, et al. : Late-onset sepsis in very low birth weight neonates: The experience of the NICHD Neonatal Research Network. Pediatrics. 2002; 110:285–291 [DOI] [PubMed] [Google Scholar]
- 3.Lawn JE, Wilczynska-Ketende K, Cousens SN: Estimating the causes of 4 million neonatal deaths in the year 2000. Int J Epidemiol. 2006; 35:706–718 [DOI] [PubMed] [Google Scholar]
- 4.Stoll BJ, Hansen NI, Adams-Chapman I, et al. ; National Institute of Child Health and Human Development Neonatal Research Network: Neurodevelopmental and growth impairment among extremely low-birth-weight infants with neonatal infection. JAMA. 2004; 292:2357–2365 [DOI] [PubMed] [Google Scholar]
- 5.Rudd KE, Johnson SC, Agesa KM, et al. : Global, regional, and national sepsis incidence and mortality, 1990-2017: Analysis for the Global Burden of Disease Study. Lancet. 2020; 395:200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hornik CP, Fort P, Clark RH, et al. : Early and late onset sepsis in very-low-birth-weight infants from a large group of neonatal intensive care units. Early Hum Dev. 2012; 88(Suppl 2):S69–S74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng PC: Diagnostic markers of infection in neonates. Arch Dis Child Fetal Neonatal Ed. 2004; 89:F229–F235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng PC, Cheng SH, Chui KM, et al. : Diagnosis of late onset neonatal sepsis with cytokines, adhesion molecule, and C-reactive protein in preterm very low birthweight infants. Arch Dis Child Fetal Neonatal Ed. 1997; 77:F221–F227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu VX, Fielding-Singh V, Greene JD, et al. : The timing of early antibiotics and hospital mortality in sepsis. Am J Respir Crit Care Med. 2017; 196:856–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pammi M, Weisman LE: Late-onset sepsis in preterm infants: Update on strategies for therapy and prevention. Expert Rev Anti Infect Ther. 2015; 13:487–504 [DOI] [PubMed] [Google Scholar]
- 11.Jardine L, Davies MW, Faoagali J: Incubation time required for neonatal blood cultures to become positive. J Paediatr Child Health. 2006; 42:797–802 [DOI] [PubMed] [Google Scholar]
- 12.Connell TG, Rele M, Cowley D, et al. : How reliable is a negative blood culture result? Volume of blood submitted for culture in routine practice in a children’s hospital. Pediatrics. 2007; 119:891–896 [DOI] [PubMed] [Google Scholar]
- 13.Meem M, Modak JK, Mortuza R, et al. : Biomarkers for diagnosis of neonatal infections: A systematic analysis of their potential as a point-of-care diagnostics. J Glob Health. 2011; 1:201–209 [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Ingle H, Prasad DV, et al. : Recognition of bacterial infection by innate immune sensors. Crit Rev Microbiol. 2013; 39:229–246 [DOI] [PubMed] [Google Scholar]
- 15.Moorman JR, Carlo WA, Kattwinkel J, et al. : Mortality reduction by heart rate characteristic monitoring in very low birth weight neonates: A randomized trial. J Pediatr. 2011; 159:900–9006.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boghossian NS, Page GP, Bell EF, et al. ; Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network: Late-onset sepsis in very low birth weight infants from singleton and multiple-gestation births. J Pediatr. 2013; 162:1120–1124, 1124.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Therneau TM, Atkinson EJ: An Introduction to Recursive Partitioning Using the RPART Routines. 2019. Available at: https://cran.r-project org/web/packages/rpart/vignettes/longintro.pdf. Accessed November 21, 2018
- 18.Raynor LL, Saucerman JJ, Akinola MO, et al. : Cytokine screening identifies NICU patients with Gram-negative bacteremia. Pediatr Res. 2012; 71:261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schüller SS, Kempf K, Unterasinger L, et al. : Intravenous pentoxifylline is well tolerated in critically ill preterm infants with sepsis or necrotizing enterocolitis. Eur J Pediatr. 2020; 179:1325–1330 [DOI] [PubMed] [Google Scholar]
- 20.Kurul Ş, Simons SHP, Ramakers CRB, et al. : Association of inflammatory biomarkers with subsequent clinical course in suspected late onset sepsis in preterm neonates. Crit Care. 2021; 25:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brodská H, Malíčková K, Adámková V, et al. : Significantly higher procalcitonin levels could differentiate Gram-negative sepsis from Gram-positive and fungal sepsis. Clin Exp Med. 2013; 13:165–170 [DOI] [PubMed] [Google Scholar]
- 22.Li S, Rong H, Guo Q, et al. : Serum procalcitonin levels distinguish Gram-negative bacterial sepsis from Gram-positive bacterial and fungal sepsis. J Res Med Sci. 2016; 21:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laukemann S, Kasper N, Kulkarni P, et al. : Can we reduce negative blood cultures with clinical scores and blood markers? Results from an observational cohort study. Medicine (Baltimore). 2015; 94:e2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul M, Andreassen S, Tacconelli E, et al. ; TREAT Study Group: Improving empirical antibiotic treatment using TREAT, a computerized decision support system: Cluster randomized trial. J Antimicrob Chemother. 2006; 58:1238–1245 [DOI] [PubMed] [Google Scholar]
- 25.Paul M, Nielsen AD, Goldberg E, et al. ; TREAT Study Group: Prediction of specific pathogens in patients with sepsis: Evaluation of TREAT, a computerized decision support system. J Antimicrob Chemother. 2007; 59:1204–1207 [DOI] [PubMed] [Google Scholar]
- 26.Griffin MP, Moorman JR: Toward the early diagnosis of neonatal sepsis and sepsis-like illness using novel heart rate analysis. Pediatrics. 2001; 107:97–104 [DOI] [PubMed] [Google Scholar]
- 27.Hedegaard SS, Wisborg K, Hvas A-M: Diagnostic utility of biomarkers for neonatal sepsis–a systematic review. Infect Dis. 2015; 47:117–124 [DOI] [PubMed] [Google Scholar]
- 28.Zeitoun AA, Gad SS, Attia FM, et al. : Evaluation of neutrophilic CD64, interleukin 10 and procalcitonin as diagnostic markers of early- and late-onset neonatal sepsis. Scand J Infect Dis. 2010; 42:299–305 [DOI] [PubMed] [Google Scholar]
- 29.Romagnoli C, Frezza S, Cingolani A, et al. : Plasma levels of interleukin-6 and interleukin-10 in preterm neonates evaluated for sepsis. Eur J Pediatr. 2001; 160:345–350 [DOI] [PubMed] [Google Scholar]
- 30.Küster H, Weiss M, Willeitner AE, et al. : Interleukin-1 receptor antagonist and interleukin-6 for early diagnosis of neonatal sepsis 2 days before clinical manifestation. Lancet. 1998; 352:1271–1277 [DOI] [PubMed] [Google Scholar]
- 31.Dessì A, Corsello G, Stronati M, et al. : New diagnostic possibilities in systemic neonatal infections: Metabolomics. Early Hum Dev. 2014; 90(Suppl 1):S19–S21 [DOI] [PubMed] [Google Scholar]
- 32.Schmerler D, Neugebauer S, Ludewig K, et al. : Targeted metabolomics for discrimination of systemic inflammatory disorders in critically ill patients. J Lipid Res. 2012; 53:1369–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart CJ, Nelson A, Treumann A, et al. : Metabolomic and proteomic analysis of serum from preterm infants with necrotising entercolitis and late-onset sepsis. Pediatr Res. 2016; 79:425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodwell RL, Leslie AL, Tudehope DI: Early diagnosis of neonatal sepsis using a hematologic scoring system. J Pediatr. 1988; 112:761–767 [DOI] [PubMed] [Google Scholar]
- 35.Manucha V, Rusia U, Sikka M, et al. : Utility of haematological parameters and C-reactive protein in the detection of neonatal sepsis. J Paediatr Child Health. 2002; 38:459–464 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.