Abstract
Age-related macular degeneration (AMD) is a leading cause of severe vision loss. With our aging population, it may affect 288 million people globally by the year 2040. AMD progresses from an early and intermediate dry form to an advanced one, which manifests as choroidal neovascularization and geographic atrophy. Conversion to AMD-related exudation is known as progression to neovascular AMD, and presence of geographic atrophy is known as progression to advanced dry AMD. AMD progression predictions could enable timely monitoring, earlier detection and treatment, improving vision outcomes. Machine learning approaches, a subset of artificial intelligence applications, applied on imaging data are showing promising results in predicting progression. Extracted biomarkers, specifically from optical coherence tomography scans, are informative in predicting progression events. The purpose of this mini review is to provide an overview about current machine learning applications in artificial intelligence for predicting AMD progression, and describe the various methods, data-input types, and imaging modalities used to identify high-risk patients. With advances in computational capabilities, artificial intelligence applications are likely to transform patient care and management in AMD. External validation studies that improve generalizability to populations and devices, as well as evaluating systems in real-world clinical settings are needed to improve the clinical translations of artificial intelligence AMD applications.
Keywords: Artificial intelligence, machine learning, deep learning, age-related macular degeneration, disease progression, imaging modalities
Impact statement
Prediction of disease progression from an early or intermediate dry age-related macular degeneration (AMD) stage is crucial, since prompt intervention after conversion to neovascular AMD as well as identification and future therapies for geographic atrophy (GA) can improve visual outcomes. Learning algorithms in artificial intelligence applications, particularly on imaging data, are showing promising results in identifying high-risk patients. The purpose of this mini review is to describe machine and deep learning algorithms for AMD progression, highlighting the methods, data-inputs, findings, and challenges of translating AI AMD systems to clinical applications.
Introduction
Age-related macular degeneration (AMD) is a chronic degenerative disease of the macula, resulting in progressive vision loss. It is the leading cause of blindness among the geriatric population worldwide, with 11 million individuals affected with AMD in the United States alone, and a global prevalence of 170 million. 1 , 2 These estimates are expected to increase to 22 million in the US by the year 2050, and the global prevalence to 288 million by the year 2040. 1 , 2
The pathogenesis of AMD is not clearly understood. AMD is categorized into early, intermediate, or advanced stages based on structural and functional clinical signs and symptoms. One in five patients with AMD may progress to an advanced stage within five years, manifesting as choroidal neovascularization (CNV) and/or geographic atrophy (GA). 3 CNV occurs from the ingrowth of new choriocapillaris vessels through Bruch’s membrane into the sub-pigment epithelial space, which may result in subretinal fluid, intraretinal fluid, hemorrhaging within the retina, sub-retina or sub-RPE layers and RPE detachment and tear. Conversion to AMD-related exudation is known as progression to “neovascular AMD”. Several classes of CNV have been described, based on the site of infiltration of new vessels. 4 End stage of the disease produces a fibrovascular or atrophic macular scar (disciform scar), and results in permanent central vision loss to the patient. Current treatments include intravitreal injections of anti-vascular endothelial growth factor (VEGF) drugs. The inhibition of VEGF, an angiogenic protein which promotes neovascularization, has been shown to maintain or improve visual impairment, with early treatment after a progression event (after exudation) corresponding to better outcomes. 5 GA, on the other hand, presents as well-demarcated lesions representing thinning and loss of outer retina layers. Presence of GA is known as progression to “advanced dry” AMD. The pathogenesis and development of GA from the earlier dry AMD stage remain unclear; however, investigations of the molecular pathways leading to GA have suggested that retinal pigment epithelium death leads to photoreceptor loss and degeneration of choroidal capillaries.6–8 Lesions may occur in single or multiple locations and have been reported to grow at an average rate of 2 mm2/year, although this rate varies considerably from case to case. 9 Lesion size does not correlate with severity of vision loss, as atrophy may or may not involve the fovea. 10 GA is an irreversible disease which leads to progressive loss of vision with no currently available treatment; however, phase II and III clinical trials are ongoing. 11 Most treatments currently under investigation aim to slow progression, rather than inhibit GA development from early-stage AMD, making early diagnosis critical to the future success of the proposed therapies.
Predicting which early and intermediate AMD patients are at progression risk to an advanced form, distinguishing between neovascular and GA forms of the disease, could improve patient screening and enable earlier detection, improving vision outcomes. Traditional, epidemiologic, regression-type modeling in AMD has included biomarkers and risk factors to predict progression.12–17 As more data became available, machine-learning methods became prominent for predictions and in identifying related biomarkers and features. More recently, with advances in computational technology, the use of deep learning-based algorithms has increased significantly for classification and prediction of AMD disease progression, leading to the development of several artificial intelligence (AI) systems. Several studies have used AI systems to predict AMD progression, which have shown promising performance results. These studies have identified imaging biomarkers based on drusen features in color fundus photos and optical coherence tomography (OCT), as well as the identification of genetic and sociodemographic factors for AMD progression.
In this mini review, we provide an overview of AI applications, encompassing machine and deep learning models, for AMD progression to neovascular and/or advanced dry AMD. We will describe the various methods, data-input types, and imaging modalities used to identify high-risk patients.
Artificial intelligence—Statistical and machine learning-based algorithms
AI refers to any hardware or software exhibiting intelligent behavior. 18 For our purposes, AI systems involve learning algorithms for defined medical tasks. With advancements in computational technology, AI methods are facilitating the analysis of medical data and the development of automated algorithms for disease classification and prediction to develop clinical decision support systems. Machine learning (ML) and statistical learning methodologies are a subset of AI methods that have the ability to provide inference for current data and predictions for future data based on learned automated pattern detections from a training dataset. 19 Traditional statistical models such as logistic regression, Poisson regression, and Cox proportional hazards models use statistical learning methods, such as leave one out cross validation and k-fold cross validation to aid in predictive modeling. Cross-validation techniques are used to estimate the performance of the statistical model in a population with the same characteristics as the designated training data. Additionally, when the number of available feature predictors is large, shrinkage methods such as L1, L2, or combination L1, L2 regularization are used to perform variable selection. L1 regularization, also known as Lasso regression, applies a penalty term to regression coefficients. The variable selection process is embedded in that when coefficients get shrunk to zero, unimportant features are removed from the model. Other popular shrinkage methods include Ridge regression, 20 which is also a shrinkage method which applies a penalty term to the regression coefficient, though never shrinking the coefficients exactly to zero, and elastic net, which includes both a ridge and a lasso regression penalty term. Lasso regression and elastic net methods allow researchers to determine which predictors are better suited for the AI task to solve. The inclusion of biomarkers and features is usually task dependent. If the goal is to study the reproducibility of previously identified biomarkers, then one targets those specific features to determine generalizability to populations and devices. If the goal is to identify or uncover new biomarkers, then all image features are analyzed to determine the prognostic relationship between those features and the outcome, followed by modeling.
ML methods are generally divided into supervised or unsupervised learning. In supervised learning, the aim is to identify a set of predictors related to a specific outcome of interest. Unsupervised learning is used to discover relationships between available features, without the use of a specific outcome variable. Deep learning (DL) is a further subset of ML, which makes use of deep neural networks to learn underlying features of data. DL provides data representation, rather than function through task-specific algorithms or engineered features, as in traditional ML methods.
AI systems are trained and tested first on internal datasets, often from a single patient population, but may be further tested on an external dataset from a different patient population or source. External validation aims to provide further evidence of an AI algorithm’s accuracy and usefulness. A recent 2019 review published by Kim et al. 21 analyzed over 500 studies presenting AI systems for various disease prediction and classifications, published over a six-month period. The authors reported that only 6% (31 studies) externally validated their algorithms. 21 A similar lack of external validation can be seen in studies proposing AI algorithms for AMD progression predictions. In the absence of external validation sets, many authors apply cross-validation techniques to assess the predictive performance of their machine learning methods. Demographics such as race, age, as well as genetic and environmental factors have been shown to affect AMD incidence and case progression. 17 An algorithm that may show high performance validation within one population may not be translatable to another population with different demographic and environmental characteristics. Performance on external validation datasets is needed to help address translatability of AI systems to clinical environments serving demographically and geographically different populations.
Risk factors for AMD progression have been determined in previous studies, which have included demographic factors, such as age, gender and race; environmental factors, including smoking, sunlight exposure and diet; and genetic predispositions. 17 Analyses of retinal images have also shown associations of progression with drusen-related features. 22 While some studies attempt the inclusion of several feature types in their predictive models, others use imaging data only, or imaging data in combination with genetic or demographic or clinical data. Several imaging modalities are typically used and complement one another in the diagnosis and treatment of AMD, like fundus photography, spectral-domain optical coherence tomography (SD-OCT) or fundus autofluorescence (FAF), and both classic ML and deep learning algorithms have exhibited the most promising results for AMD-related tasks with imaging biomarkers derived from these imaging techniques (Table 1, Figures 1 and 2). SD-OCT has been specifically useful in providing detailed, cross-sectional retinal images with the capability of volumetric evaluation of features for AMD progression (Figure 1). Studies have shown that automatically extracted SD-OCT imaging features are informative in predicting CNV and GA events.23–27
Table 1.
Identified biomarkers for AMD progression by imaging modality.
| Imaging modality | Biomarkers for progression to neovascular AMD | Biomarkers for progression to advanced dry AMD/GA |
|---|---|---|
| Fundus (color fundus and fundus autofluorescence) | Color fundus photography: Large drusen size, increase in total drusen area, non-GA-hyperpigmentation, depigmantation 28 , soft and hard drusen number, reticular pseudodrusen, pigment clumping, atrophy, hemorrhage, and fibrosis. | Fundus autofluorescence: Reticular pseudodrusen presence, hyper- and hypoautofluorescence 29 |
| Spectral domain optical coherence tomography (SD-OCT) | Total and mean drusen volume, total and mean drusen area, drusen density, max drusen height, average (avg) drusen slope, avg. and std. drusen reflectivity, area and volume occupied by druse regions within 3 mm and 5 mm of the fovea center 24 | Hyperreflective foci, retinal pigment epithelium (RPE) layer atrophy/absence, choroid thickness in those with no subretinal drusenoid deposits, photoreceptor outer segment loss, RPE drusen volume, RPE drusen thinning volume 30 |
| OCT-angiographya | CNV flow area, lesion edge complexity, flow index (FI), adjusted flow index, flow void (FV), vascular connectivity,7 and total retinal blood flow (TRBF), CNV location, CNV maturity, the presence of core vessels, and the presence of margin loops.31,32 | GA area, edge complexity, Choriocapillaris flow change, vessel density around GA, regional distribution of flow void in the choriocapillaris. 33 |
OCTA: biomarkers identified to quantify changes in AMD for neovascular and advanced dry AMD.
aThe identification of biomarkers for progression are underway.
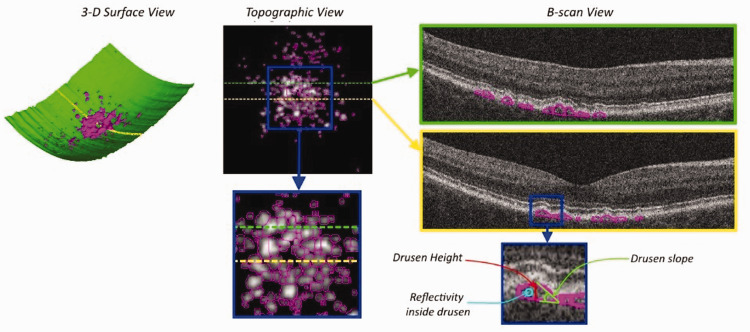
Figure 1.
Spectral domain optical coherence tomography (SD-OCT) drusen segmentation example in different views. 3D surface view with the estimation of Bruch’s membrane surface indicated in green and the detected drusen regions identified in magenta (volumetric properties). Topographic view of a drusen elevation map with individual drusen indicated in magenta (area, extent, and density properties). The dotted green and yellow lines indicate B-scan locations. B-scan view, with generated druse outlines shown in magenta. The blue square identifies a region shown in detail, where indications of drusen height, slope and reflectivity are shown. Adapted from: Hallak JA, de Sisternes L, Osborne A, Yaspan B, Rubin DL, Leng T. Imaging, genetic, and demographic factors associated with conversion to neovascular age-related macular degeneration: secondary analysis of a randomized clinical trial. JAMA Ophthalmol 2019;137:738–44. (A color version of this figure is available in the online journal.)
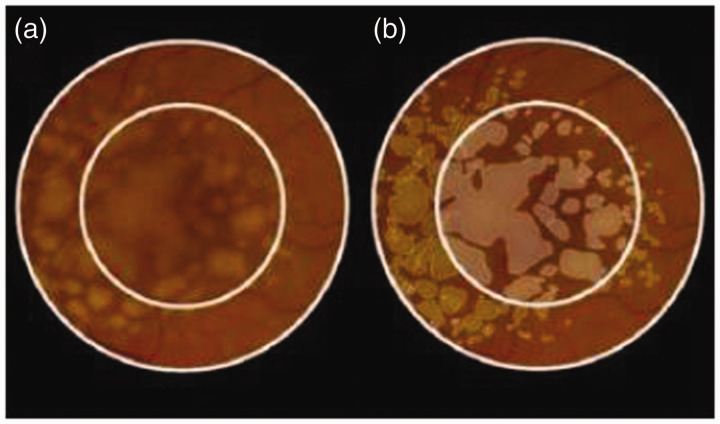
Figure 2.
(a) Color fundus photo with 3- and 5-mm circles. (b) Manual drusen outlines performed by the reading center (drusen area 3.46 mm2 (3 mm), 5.65 mm2 (5 mm)). Adapted from: Yehoshua Z, Gregori G, Sadda SR, Penha FM, Goldhardt R, Nittala MG, Konduru RK, Feuer WJ, Gupta P, Li Y, Rosenfeld PJ. Comparison of drusen area detected by spectral domain optical coherence tomography and color fundus imaging. Invest Ophthalmol Vis Sci 2013;54:2429–34.50 (A color version of this figure is available in the online journal.)
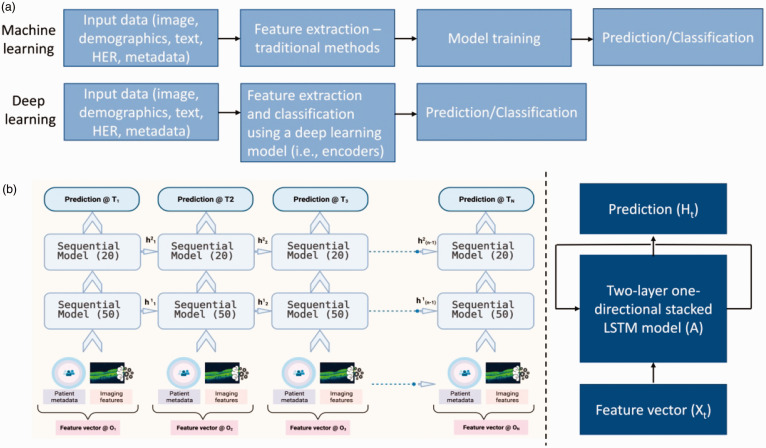
Figure 3.
(a) Simple machine learning (ML) and deep learning (DL) pipeline illustration; (b) Prediction pipeline for models with sequential multi-modal data. An array of patient metadata and imaging features are used as input to a sequential model (recursive neural network) for predictions at different time frames. (A color version of this figure is available in the online journal.)
Machine learning and deep learning algorithms for AMD progression
The inclusion criteria for studies utilizing learning algorithms for AMD progression were as follows: (i) studies published in the last decade, (ii) use of machine and/or deep learning methods for predictive modeling, (iii) an end point of progression to neovascular AMD or advanced dry AMD, and (iv) the inclusion of imaging data/imaging biomarkers. Of the 15 studies included, 7 used color fundus photos,12,15,34,36,37,41,43 7 used SD-OCT scans,23–25,38–40,42 and 1 used both image modalities 35 as input to ML and DL systems for AMD progression prediction (Table 2). Wu et al., 35 Yim et al., 38 and Burlina et al. 41 used imaging only, while others included features representing demographic, environmental, genetics, clinical and temporal characteristics of patients. Many studies reported area under the operating curves (AUCs) as metrics for model performance. Four studies included sensitivity (Sn) and specificity (Sp) metrics in their results 23 , 34 , 42 , 43 and five reported validation on external datasets. 15 , 23 , 34 , 37 , 43 Of those studies that did report AUC metrics, performance ranged from 0.68 to 0.97 (Table 2). We highlighted six studies that looked at progression to neovascular AMD,15,23–25, 38 , 40 one that looked at progression to GA only, 39 and eight that included both.12,34–37,41–43 Progression predictions at different timepoints were reported, and were as early as three months and up to 12 years (Table 2). For studies that had one- to two-year follow-up, performance metrics were reported at three-month intervals, 23 six-month intervals, 39 or yearly. 34 For studies with more than five-year follow-up, performance was reported at every year 37 or every five years. 25 Short-term predictions (such as three months) are particularly important for early identification of high-risk patients. 23
Table 2.
Learning algorithms for age-related macular degeneration progression.
| Study | Description | Dataset | Results |
|---|---|---|---|
| Banerjee et al.23 a | Hybrid sequential prediction model "Deep Sequence" integrating OCT imaging features, demographic and visual variables with an RNN model to predict risk of exudation (progression to neovascular AMD) within 3 to 21 months | HARBOR: 671 fellow eyes | AUC:0.96± 0.02 (3 months)0.97± 0.02 (21 months) |
| Bhuiyan et al. 34 a | Color fundus photos and 12-step severity scale class used with demographic data in logistic tree model to predict progression to advanced dry, neovascular, or all late AMD in 1 or 2 years (yrs). | AREDS >4600 color fundus images | ALL advanced AMDSe: 0.91 (1 yr), 0.92 (2 yr)Sp: 0.85 (1 yr), 0.84 (2 yr)ACC: 0.86 (1 yr), 0.86 (2 yr) |
| Wu et al. 35 | Cox proportional hazards models with LOOCV to compare fundus versus OCT versus both inputs for neovascular and advanced dry AMD progression in 36 months | LEAD: 280 eyes from 140 participants. OCT B-scans, fundus photos. | AUC: 0.85 |
| Wu et al. 36 | Cox proportional hazards model with LOOCV to examine added predictive value of PSD and LLD to fundus data for progression to GA, nascent GA and neovascular AMD within 36 months | LEAD: 280 eyes from 140 participants. fundus photos. | AUC: 0.80 |
| Yan et al.37 a | Genotypes and color fundus images used to predict progression to advanced dry or neovascular AMD within range of 2–7 years with a modified deep convolutional neural network | AREDS > 31,000 fundus images from 1351 subjects | AUC: 0.85–0.86 |
| Yim et al. 38 | DL model based on three-dimensional SD-OCT images and automatic tissue maps combined for neovascular AMD prediction within 6 months | Internal retrospective cohort 2795 patients. OCT | AUC: 0.745 (conversion scan ground truth), 0.886 (first injection ground truth) |
| Hallak et al. 24 | Mixed methods to determine associations between variables and progression to neovascular AMD. Bivariate analysis for genetic variants and LASSO regression for OCT imaging features decided the variables included alongside demographic data in survival analysis and Cox proportional hazards regression. | HARBOR: 686 fellow eyes with non neovascular AMD at baseline | Female sex (HR, 1.57; 95% CI, 1.11–2.20)Drusen area within 3 mm of the fovea (HR, 1.45; 95% CI, 1.24–1.69)Mean drusen reflectivity (HR, 3.97; 95% CI, 1.11–14.18 |
| Rivail et al. 39 | Deep Siamese network capturing time-specific features on longitudinal data to predict advanced dry AMD in 6, 12, and 18 months. | Internal: 3308 OCT B-scans from 221 patients (420 eyes) | AUC:6 months: 0.75312 months: 0.78418 months: 0.773 |
| Russakoff et al. 40 | Comparison of two deep convolutional neural networks for neovascular AMD risk prediction over 2 years (17–27-months follow-up). | Internal: 71 eyes, 71 subjects (progressors =31). 9088 OCT B-scans from two devices. | AUC:VGG16: 0.87AMDnet: 0.91 |
| Burlina et al. 41 | 3 DCNN models developed to estimate 5-year risk of progression to neovascular and GA based on 9-step AREDS severity scale | AREDS< 6000 fundus images across 9 classifications | PE:Soft model: 0.038Hard model: 0.035Regressed: 0.053 |
| Schmidt-Erfurth et al. 42 | Demographic, genetic, and image features input to Cox proportional hazards models with 10-fold cross validation to predict neovascular AMD or GA in 2 years | HARBOR: 495 eyes (progressors = 159 eyes) SD-OCT | Se: 0.8 (CNV), 0.8 (GA)Sp: 0.46 (CNV), 0.69 (GA)AUC: 0.68 (CNV), 0.8 (GA) |
| Seddon et al. 12 | Genetic, demographic, environmental, and image features input to Cox proportional hazards models to predict neovascular AMD and/or GA at any follow-up visit within 12 years | AREDS: fundus images, 2951 subjects (834 progressors) | AUC:0.911 (All AMD), 0.923 (GA), 0.896 (neovascular AMD) |
| Chiu et al.43 a | Demographic and environmental features included in logistic model producing a risk scoring system for neovascularization or central GA by end of study timeframe (12 years) | AREDS: fundus images from 4507 participants (1185 progressors) | Se: 0.876Sp: 0.736 |
| de Sisternes et al. 25 | Automated pipeline for segmentation and extraction of longitudinal image features used in L1-penalized Poisson model predicting neovascular AMD progression within 5 years | Private: 2146 SD-OCT scans of 330 eyes of 244 patients (36 eyes progressed) | AUC: 5 yr: 0.7411 months:0.9216 months:0.8618 months:0.748 months:0.79 |
| Seddon et al.15 a | Output from Cox proportional hazards regression, including demographic, environmental and genetic features, used for predictive algorithm of neovascular AMD and GA within 5 or 10 years | AREDS: fundus photos from 2937 individuals (819 progressors) | AUC:5 yr: 0.88510 yr: 0.915 |
Se: sensitivity; Sp: specificity; Acc: accuracy; AUC: area under the receiver operating curve; PE: prediction error.
aValidation performed on external dataset.
Machine learning models
Eight of the included studies employed supervised models to predict progression to neovascular AMD and/or advanced dry AMD and one used machine learning as a part of mixed methods to select features for explanatory statistical modeling. 24 Hallak et al., investigated the progression to neovascular AMD during a two-year period, LASSO regression for SD-OCT imaging feature selection and bivariate analysis of genetic variants were performed to determine which variables to include alongside demographic data in a survival Cox proportional hazards regression model. 24 After controlling for demographic and treatment effects, drusen area within 3 mm of the fovea (HR, 1.45; 95% CI, 1.24–1.69; HR for 1-SD increase, 1.36 [95% CI, 1.20–1.54]) and mean drusen reflectivity (HR, 3.97; 95% CI, 1.11–14.18; HR for 1-SD increase, 1.32 [95% CI, 1.02–1.71]) were significantly associated with future progression with an exudation event within two years. In addition, one genetic variant (rs61941274) was found to be potentially associated with future exudations during a two-year follow-up 24 (Table 2).
Some ML techniques were preceded by DL data processing and classification of images, as was the case with Buhiyan et al.’s 34 study, which used a logistic model tree technique on pre-classified fundus photo data, along with sociodemographic and clinical data to predict progression to neovascular and/or advanced dry AMD (GA). 34 In all reported metrics, including accuracy via AUC, Sn, and Sp, their model performed better predicting any advanced AMD, versus only neovascular or advanced dry AMD in both one- and two-year predictions. For example, while AUC reached 0.8619 and 0.8636 for one- and two-year predictions of all advanced AMD development, advanced dry AMD only predictions achieved 0.6679 and 0.6688 accuracies for one- and two-year predictions and neovascular AMD only predictions achieved 0.6815 and 0.6715 accuracies for one- and two-year predictions, respectively. Similar discrepancies were found among Sn and Sp metrics 34 (Table 2).
Wu et al. in two studies in 2020 applied leave one out cross validation (LOOCV) on Cox proportional hazards models on the sham study arm of the LEAD dataset, which included intermediate AMD eyes to study interventions aimed at slowing disease progression, to predict progression to neovascular and/or advanced dry AMD (GA) in a 36-month time period. 35 , 36 In one study, Wu et al. 35 compared inputs of fundus photography, versus OCT scans versus both imaging modalities to investigate the optimum input for their system, finding the best results with the combination of both modalities (Table 2). In the second study, Wu et al. 36 examined the added predictive value of microperimetric sensitivity (PSD) and low luminance deficit (LLD) to fundus photography data, finding no improvement in predictive performance with the added data types (Table 2). Schmidt-Erfurth et al. 42 used a similar modeling strategy to predict progression to CNV or GA in two years. They developed two Cox proportional hazards models with 10-fold cross validation to predict either CNV or GA with SD-OCT, genetic, and demographic data, finding better results for GA prediction (GA: AUC 0.8, CNV: AUC 0.68). Their model learned from a four-month observational period to predict conversion during the two years of a clinical trial study. 42 Using Cox proportional hazards regression, Seddon et al. 15 used demographic, environmental, genetic, and fundus images from the AREDS dataset to produce a risk score for conversion to neovascular or dry advanced AMD within 5 or 10 years. The ultimate goal was to develop an online clinical decision-making tool and to determine the association between genetic variants and conversion with results for combined neovascular and advanced dry AMD prediction (AUC: 0.911). They also used the same modeling method to predict conversion by the end of the study timeframe at 12 years. 12
Other studies employing ML methods include Chiu et al.’s 43 use of demographic, environmental, and imaging features to create a risk scoring system with Bayes’ theorem in a logistic model for progression to neovascularization or central GA by the end of a 12-year clinical trial study (AREDS). Their reported metrics included Sn and Sp only and achieved similar results in their internal validation (Sn: 0.876, Sp: 0.736) compared to validation of a 10-year prediction on an external validation dataset (BMES) (Sn:0.899, Sp: 0.729). 43 de Sisternes et al. 25 used a fully automated pipeline to segment and extract image features of drusen and longitudinal evolution in drusen characteristics from SD-OCT scans. Their ML prediction model used these features in an L1-penalized Poisson predictive model to predict exudation events at arbitrary future time intervals, where time to prediction is a variable in the model. This work was able to produce reliable predictions in the short term (within three months) and in longer time intervals, up to five years. Their results showed very good predictions in the short term (0.92 AUC predictions within 11 months) but a decrease in AUC in the longer time frames (Table 2). 25
Deep learning models
As mentioned above, deep learning is a subset of machine learning that uses multiple layers of algorithms, each providing different interpretations of the data presented to it. DL models have been primarily used in the past decade, and studies including these methods are increasing in number. 15 This mini review includes six studies using DL to predict AMD progression. Banerjee et al. 23 proposed a hybrid sequential prediction model (termed “Deep Sequence”) that utilized multi-modal features (imaging, patient meta-data, and visual factors) in a recursive neural network (RNN) to predict risk of exudation in AMD patients within a short (3 months) and long-term (21 months) timeframe. A representative pipeline for deep-sequence-like sequential models is shown in Figure 3. This study was conducted on the fellow eyes of the HARBOR clinical trial data (671 AMD eyes with 13,954 observations), and the deep sequence model achieved a cross validation AUC of 0.96 and 0.97 for the prediction of exudation within 3 months and 21 months, respectively. This model was further validated on an external real-world dataset from the Bascom Palmer Eye Institute (BPEI). The prediction performance decreased on the validation data as expected due to variability and unstructured characteristics of real-world data with three-month predictions on internal and external data achieving AUCs of 0.96 and 0.82, and 21-month predictions achieving AUCs of 0.97 and 0.68, respectively (Table 3). 23 In another study, Yan et al. 37 predicted progression to advanced dry and late stage neovascular AMD over two, three, four, five, six, and seven years using AREDS data with genotypes and fundus image data as input to their multi-layer convolutional neural network (CNN) and achieved similar performance metrics across years (AUC: 0.85–0.86). Their external validation, predicting three-year conversion using data from a UK biobank (200 participants) produced an even higher AUC of 0.9. 37 Burlina et al. 41 also used AREDS data, first employing a CNN to classify images based on a 4 and 9-step severity scale, and then estimate five-year risk of progression through creating three different deep convolutional neural to produce the following three predictions (DCNN): (i) a soft prediction which defined risk as the expected value of class risk, (ii) a hard prediction where risk was defined as the maximum value of class risk, and (iii) a regressed prediction which skipped classification and used the DCNN directly to map image input to risk prediction. Comparisons of prediction error showed the hard prediction as providing the most accurate results 41 (Table 2).
Table 3.
Studies with AMD progression model validation using external datasets.
| Study | Training and internal validation dataset | Internal results | External testing dataset | External validation results |
|---|---|---|---|---|
| Banerjee et al. 23 | HARBOR: 671 fellow eyes SD-OCT | AUC for neovascular AMD:0.96 ± 0.02 (3 mo)0.97 ± 0.02 (21 mo) | Validation on real-world dataset: 719 eyes from 507 patients, 12,288 OCT volumes | AUC:0.82 (3 mo)0.68 (21 mo) |
| Bhuiyan et al. 34 | AREDS >4600 color fundus images | ALL advanced AMD (neovascular or advanced dry)Se: 1 yr (0.91) 2 yr (0.92)Sp: 1 yr (0.85) 2 yr (0.84)Acc: 1 yr (0.86) 2 yr (0.86) | NAT-2 dataset (88 eyes)prediction to 3-year conversion | ALL advanced AMD (neovascular or advanced dry)Se: 0.84Sp: 0.9Acc: 0.81 |
| Yan et al. 37 | AREDS > 31,000 fundus images from 1351 subjects | AUC for late AMD (neovascular or advanced dry AMD: 0.85–0.86 (2,3,4,5,6 and 7-year predictions) | UK Biobank (200 participants) prediction to 3-year conversion | AUC: 0.9 |
| Chiu et al. 43 | AREDS: fundus photos from 4507 participants (1185 progressors) | Predictions for neovascularization or central GASe: 0.876Sp: 0.736 | Blue Mountains Eye Study (BMES) followed for 10 years 2169 participants (69 progressors) | Se: 0.899Sp: 0.729 |
Se: sensitivity; Sp: specificity; Acc: accuracy; AUC: area under the receiver operating curve.
In the remaining three studies, internal institutional data were collected to train and test DL systems. Yim et al. 38 tested their network’s ability to predict six-month progression to CNV using SD-OCT images and automatic tissue maps. A two-stage segmentation and prediction network was combined with a model trained on raw OCT images and reported AUCs were provided for both conversion scan and first injection ground truths (Table 2). While first injection ground truth showed better results (AUC: 0.886), the researchers chose conversion scan ground truth as the basis for their analysis, which included choosing liberal (higher Sn of 0.8) and conservative (higher Sp of 0.9) cutoff points. While the liberal selection of higher Sn provided a Sp of 0.55, the conservative selection of higher Sp provided a poor Sn of 0.34. 38 Rivail et al. 39 predicted GA conversion in 6, 12, and 18 months with their deep Siamese network, a self-supervised learning system of spatiotemporal representations with 6-fold cross validation. AUC and precision were reported with the former remaining consistent across time and the latter increasing across time points 39 (Table 2). Russakoff et al. 40 compared the ability of two different CNNs to predict two-year (17–27 month) conversion to wet AMD. VGG16, a popular CNN for image recognition was tested against AMDnet, a novel, simplified CNN architecture trained from scratch on a privately acquired set of OCT B-scans using two different OCT devices. AMDnet outperformed VGG16 with an AUC of 0.91 vs. 0.87 respectively. 40
Model validation
Of the 15 selected studies, only four reported validation on separate datasets (Table 3). The ability of any AI system to function well on one trained dataset (internal dataset) does not necessarily foreshadow the successful application of that system to external datasets and real-world clinical data. Attempting and reporting on external validation, specifically on real-world clinical data, will contribute to the knowledge base of the research community attempting these types of predictions for future developments with the overall aim of real-world clinical application.
Clinical trial datasets were used for initial training and internal validation in all of the studies that used an external dataset for testing (Table 3). Banerjee et al. 23 and Yan et al. 37 used a DL system for their predictions, while Bhuiyan et al. 34 and Chiu et al. 43 used ML approaches. Banerjee et al. 23 tested their deep sequence model on a real-world clinical dataset for the prediction of exudation in AMD at different time points. The deep sequence model generalized well for making the short-term predictions (within three, six, and nine months, with AUCs of 0.82, 0.77, and 0.71, respectively). However, a decrease was observed for the longer time intervals compared to the performance of the deep sequence model on the HARBOR clinical trial data. This difference is mainly due to population characteristics, the lack of socio-demographic factors in the external dataset, and the fact that the external dataset was from a real-world clinical setting. 23 Yan et al. 37 showed an improvement in AUC when they tested their model on a real-world biobank dataset. While AUCs for two, three, four, five, six, and seven-year predictions to neovascular or advanced dry AMD ranged between 0.85 and 0.86, a three-year conversion prediction on the new dataset achieved an AUC of 0.9. 37 Bhuiyan et al. 34 predicted three-year conversion to advanced AMD using the NAT-2 dataset for external testing. They improved specificity in their external validation results while slightly reducing sensitivity and accuracy. 34 Chiu et al. 43 validated on the BMES dataset and on a random derivation of the original dataset, respectively, and were able to achieve very similar results.
Machine learning versus deep learning methods
As we report different approaches utilizing ML and DL methods for AMD detection and prediction, it must be noted that the use of ML or DL approaches is generally contingent upon the use case and several parameters. The decision to use these computation tools is dependent on different scenarios, available data, and access to compute resources. Traditionally, DL has performed better than ML when the number of data is comparatively larger with a good representation of test use cases in the training data. Traditional ML approaches will work better when the data are smaller, and engineered features are correlated better with subtle changes in the diseases. Oftentimes, it takes the DL model a long time to extract the subtle intuition found in local features, which might require large computational resources. Moreover, for both ML and DL models, it can be challenging to limit overfitting and make the model generalized for widespread deployment. The general trend currently is moving towards hybrid modeling and using multi-modality (different imaging modality, text, metadata etc.) for a more holistic approach for disease detection and progression prediction.
Conclusions
With advances in computational capabilities, AI applications, machine learning, and particularly deep learning are likely to transform patient care and management in AMD, through accessibility, accuracy and timeliness of diagnoses, monitoring, and treatments. This mini review highlighted current work utilizing machine and deep learning applications to develop AI systems for AMD progression predictions. These systems, particularly deep learning algorithms, with imaging biomarkers are showing promising results. Notably, there is some variation across systems. This variation may be due to the type of data inputted, population characteristics, and the prediction methodology utilized. Authors of the reviewed studies reported limitations including small sample sizes (or a desire for larger study populations) 23 , 24 , 35 , 40 , 42 and limited numbers of endpoints reached.35–37 Additional limitations included use of only one imaging modality for analysis, 34 , 38 , 40 lack of diversity in the study populations, 12 , 37 , 38 , 41 and datasets not representing real-world patient data. 23 , 24 , 34 , 36 , 38 As the number of studies continue to increase, the ability to pool results and make comparisons regarding these aspects of predictive model creation may be of use to researchers to improve on their methods. In addition to identifying at-risk patients for AMD progression as defined by conversion to CNV or GA development, AI applications can be used to predict the growth rate and direction of biomarkers, specifically for GA.44–46
Four of the 15 studies in this review 15 , 34 , 37 , 43 reported performances on external data. Validating on real-world clinical data remains limited. For successful clinical translations and deployment of any AI system, research efforts towards building the needed infrastructures for developing databases for AI applications in ophthalmology as well as the ability to share diverse data across health systems will improve the validity and generalizability of AI applications. 47 Solutions to data access problems, such as federated learning, are very valuable in enhancing validation studies. 48 , 49 Federated learning allows the training and testing of algorithms collaboratively without the need to exchange data. 48 , 49 Machine learning models are developed locally at institutions and only model characteristics (parameters and gradients) are shared. 48 , 49 By allowing multiple institutions to train without the need for data centralization, federated learning may improve model generalizability and address data sensitivity. In addition to validation studies, algorithms need to be more interpretable and explainable to ensure targeted representation and to identify potential bias in training data, all while protecting data safety and privacy. 47 Finally, a critical aspect for successful deployment is evaluating the integration of AI systems into clinical workflows. Collectively, building algorithms that are valid, generalizable, interpretable, and that integrate well into clinical workflows in real-world settings will bring us closer to the delivery of personalized care for improved patient outcomes.
Footnotes
AUTHORS’ CONTRIBUTIONS: All authors participated in the writing and editing of the manuscript; KR, MA, SK, and JAH conducted study retrieval, review, and analyses; KR, MA, and JAH wrote the manuscript, and LdS, TL, JIL, and DL reviewed and edited the manuscript.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Luis de Sisternes: Carl Zeiss Meditec (Employee).
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by a BrightFocus Foundation grant M2019155 (JAH), an Unrestricted Grant for Research to Prevent Blindness, Department of Ophthalmology and Visual Sciences, University of Illinois at Chicago, Chicago, IL (RK, SK, JIL, JAH), and the Core grant for Vision Research (2P30 EY001792 41), Department of Ophthalmology and Visual Sciences, University of Illinois at Chicago, Chicago, IL, (RK, SK, JIL, JAH).
ORCID iD: Joelle A Hallak https://orcid.org/0000-0002-4243-9930
References
- 1.Rein DB, Wittenborn JS, Zhang X, Honeycutt AA, Lesesne SB, Saaddine J, for the Vision Health Cost-Effectiveness Study Group. Forecasting age-related macular degeneration through the year 2050 – the potential impact of new treatments. Arch Ophthalmol 2009; 127:533–40 [DOI] [PubMed] [Google Scholar]
- 2.Wong WL, Su X, Li X, Cheung CMG, Klein R, Cheng CY, Wong TY. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health 2014; 2:e106–e116 [DOI] [PubMed] [Google Scholar]
- 3.Gehrs KM, Anderson DH, Johnson LV, Hageman GS. Age-related macular degeneration–emerging pathogenetic and therapeutic concepts. Ann Med 2006; 38:450–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gass JDM. Stereoscopic atlas of macular diseases. 4th ed. Maryland Heights, MO: Mosby, 1998 [Google Scholar]
- 5.Mitchell P, Liew G, Gopinath B, Wong TY. Age-related macular degeneration. Lancet 2018; 392:1147–59 [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Stratton C, Francis PJ, Kleinman ME, Tan PL, Gibbs D, Tong Z, Chen H, Constantine R, Yang X, Chen Y, Zeng J, Davey L, Ma X, Hau VS, Wang C, Harmon J, Buehler J, Pearson E, Patel S, Kaminoh Y, Watkins S, Luo L, Zabriskie NA, Bernstein PS, Cho W, Schwager A, Hinton DR, Klein ML, Hamon SC, Simmons E, Yu B, Campochiaro B, Sunness JS, Campochiaro P, Jorde L, Parmigiani G, Zack DJ, Katsanis N, Ambati J, Zhang K. Toll-like receptor 3 and geographic atrophy in age-related macular degeneration. N Engl J Med 2008; 359:1456–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaw PX, Stiles T, Douglas C, Ho D, Fan W, Du H, Xiao X. Oxidative stress, innate immunity, and age-related macular degeneration. AIMS Mol Sci 2016; 3:196–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLeod DS, Grebe R, Bhutto I, Merges C, Baba T, Lutty GA. Relationship between RPE and choriocapillaris in age-related macular degeneration. Invest Ophthalmol Vis Sci 2009; 50:4982–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joachim N, Mitchell P, Kifley A, Rochtchina E, Hong T, Wang JJ. Incidence and progression of geographic atrophy: observations from a population-based cohort. Ophthalmology 2013; 120:2042–50 [DOI] [PubMed] [Google Scholar]
- 10.Liew G, Joachim N, Mitchell P, Burlutsky G, Wang JJ. Validating the AREDS simplified severity scale of age-related macular degeneration with 5- and 10-year incident data in a population-based sample. Ophthalmology 2016; 123:1874–8 [DOI] [PubMed] [Google Scholar]
- 11.Sacconi R, Corbelli E, Querques L, Bandello F, Querques G. A review of current and future management of geographic atrophy. Ophthalmol Ther 2017; 6:69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seddon J, Silver R, Kwong M, Rosner B. Risk prediction for progression of macular degeneration: 10 common and rare genetic variants, demographic, environmental, and macular covariates. Invest Ophthalmol Vis Sci 2015; 56:2192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sobrin L, Seddon JM. Nature and nurture- genes and environment- predict onset and progression of macular degeneration. Prog Retin Eye Res 2014; 40:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seddon JM, Reynolds R, Maller J, Fagerness JA, Daly MJ, Rosner B. Prediction model for prevalence and incidence of advanced age-related macular degeneration based on genetic, demographic, and environmental variables. Invest Ophthalmol Vis Sci 2009; 50:2044–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seddon JM, Reynolds R, Yu Y, Daly M, Rosner B. Risk models for progression to advanced age-related macular degeneration using demographic, environmental, genetic, and ocular factors. Ophthalmology 2011; 118:2203–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feigl B, Morris CP. The challenge of predicting macular degeneration. Curr Med Res Opin 2011; 27:1745–8 [DOI] [PubMed] [Google Scholar]
- 17.Heesterbeek TJ, Lorés-Motta L, Hoyng CB, Lechanteur YTE, den Hollander AI. Risk factors for progression of age-related macular degeneration. Ophthalmic Physiol Opt 2020; 40:140–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham N. Artificial intelligence. Vol. 1076. Blue Ridge Summit: Tab Books, 1979 [Google Scholar]
- 19.Lu W, Tong Y, Yu Y, Xing Y, Chen C, Shen Y. Applications of artificial intelligence in ophthalmology: general overview. J Ophthalmol 2018; 2018:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoerl AE, Kennard RW. Ridge regression: biased estimation for nonorthogonal problems. Technometrics 1970; 12:55–67 [Google Scholar]
- 21.Kim DW, Jang HY, Kim KW, Shin Y, Park SH. Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol 2019; 20:405–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanagasingam Y, Bhuiyan A, Abràmoff MD, Smith RT, Goldschmidt L, Wong TY. Progress on retinal image analysis for age related macular degeneration. Prog Retin Eye Res 2014; 38:20–42 [DOI] [PubMed] [Google Scholar]
- 23.Banerjee I, de Sisternes L, Hallak JA, Leng T, Osborne A, Rosenfeld PJ, Gregori G, Durbin M, Rubin D. Prediction of age-related macular degeneration disease using a sequential deep learning approach on longitudinal SD-OCT imaging biomarkers. Sci Rep 2020; 10:15434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallak JA, de Sisternes L, Osborne A, Yaspan B, Rubin DL, Leng T. Imaging, genetic, and demographic factors associated with conversion to neovascular age-related macular degeneration: secondary analysis of a randomized clinical trial. JAMA Ophthalmol 2019; 137:738–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Sisternes L, Simon N, Tibshirani R, Leng T, Rubin DL. Quantitative SD-OCT imaging biomarkers as indicators of age-related macular degeneration progression. Invest Ophthalmol Vis Sci 2014; 55:7093–103 [DOI] [PubMed] [Google Scholar]
- 26.de Sisternes L, Jonna G, Greven MA, Chen Q, Leng T, Rubin DL. Individual drusen segmentation and repeatability and reproducibility of their automated quantification in optical coherence tomography images. Transl Vis Sci Technol 2017; 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Sisternes L, Hu J, Rubin DL, Leng T. Visual prognosis of eyes recovering from macular hole surgery through automated quantitative analysis of Spectral-Domain optical coherence tomography (SD-OCT) scans. Invest Ophthalmol Vis Sci 2015; 56:4631–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saha S, Nassisi M, Wang M, Lindenberg S, Kanagasingam Y, Sadda S, Hu ZJ. Automated detection and classification of early AMD biomarkers using deep learning. Sci Rep 2019; 9:10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleckenstein M, Mitchell P, Freund KB, Sadda S, Holz FG, Brittain C, Henry EC, Ferrara D. The progression of geographic atrophy secondary to Age-Related macular degeneration. Ophthalmology 2018; 125:369–90 [DOI] [PubMed] [Google Scholar]
- 30.Sleiman K, Veerappan M, Winter KP, McCall MN, Yiu G, Farsiu S, Chew EY, Clemons T, Toth CA. Age-Related eye disease study 2 ancillary spectral domain optical coherence tomography study group. Optical coherence tomography predictors of risk for progression to non-neovascular atrophic age-related macular degeneration. Ophthalmology 2017; 124:1764–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao X, Alam MN, Le D, Toslak D. Quantitative optical coherence tomography angiography: a review. Exp Biol Med 2020; 245:301–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma J, Desai R, Nesper P, Gill M, Fawzi A, Skondra D. Optical coherence tomographic angiography imaging in age-related macular degeneration. Ophthalmol Eye Dis 2017; 9:1179172116686075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khashayar N, Zhou H, Rinella N, Zhang Q, Dai Y, Foote KG, Keiner C, Deiner M, Duncan JL, Porco TC, Wang RK, Schwartz DM. OCT angiography to predict geographic atrophy progression using choriocapillaris flow void as a biomarker. Transl Vis Sci Technol 2020; 9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhuiyan A, Wong TY, Ting DSW, Govindaiah A, Souied EH, Smith RT. Artificial intelligence to stratify severity of Age-Related macular degeneration (AMD) and predict risk of progression to late AMD. Transl Vis Sci Technol 2020; 9:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Z, Bogunović H, Asgari R, Schmidt-Erfurth U, Guymer RH. Predicting progression of age-related macular degeneration using OCT and fundus photography. Ophthalmol Retina 2021; 5:118–25 [DOI] [PubMed] [Google Scholar]
- 36.Wu Z, Luu CD, Hodgson LA, Caruso E, Chen FK, Chakravarthy U, Arnold JJ, Heriot WJ, Runciman J, Guymer RH. Examining the added value of microperimetry and low luminance deficit for predicting progression in age-related macular degeneration. Br J Ophthalmol 2020; 105:711–5 [DOI] [PubMed] [Google Scholar]
- 37.Yan Q, Weeks DE, Xin H, Swaroop A, Chew EY, Huang H, Ding Y, Chen W. Deep-learning-based prediction of late age-related macular degeneration progression. Nat Mach Intell 2020; 2:141–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yim J, Chopra R, Spitz T, Winkens J, Obika A, Kelly C, Askham H, Lukic M, Huemer J, Fasler K, Moraes G, Meyer C, Wilson M, Dixon J, Hughes C, Rees G, Khaw PT, Karthikesalingam A, King D, Hassabis D, Suleyman M, Back T, Ledsam JR, Keane PA, De Fauw J. Predicting conversion to wet age-related macular degeneration using deep learning. Nat Med 2020; 26:892–9 [DOI] [PubMed] [Google Scholar]
- 39.Rivail A, Schmidt-Erfurth U, Vogl W, Waldstein S, Riedl S, Grechenig C, Wu Z, Bogunovic H. Modeling disease progression in retinal OCTs with longitudinal self-supervised learning. Int Workshop Predict Intell Med 2019; 11843:44–52 [Google Scholar]
- 40.Russakoff DB, Lamin A, Oakley JD, Dubis AM, Sivaprasad S. Deep learning for prediction of AMD progression: a pilot study. Invest Ophthalmol Vis Sci 2019; 60:712–22 [DOI] [PubMed] [Google Scholar]
- 41.Burlina PM, Joshi N, Pacheco KD, Freund DE, Kong J, Bressler NM. Use of deep learning for detailed severity characterization and estimation of 5-year risk among patients with age-related macular degeneration. JAMA Ophthalmol 2018; 136:1359–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt-Erfurth U, Waldstein SM, Klimscha S, Sadeghipour A, Hu X, Gerendas BS, Osborne A, Bogunovic H. Prediction of individual disease conversion in early AMD using artificial intelligence. Invest Ophthalmol Vis Sci 2018; 59:3199–208 2 [DOI] [PubMed] [Google Scholar]
- 43.Chiu CJ, Mitchell P, Klein R, Klein BE, Chang ML, Gensler G, Taylor A. A risk score for the prediction of advanced age-related macular degeneration: development and validation in 2 prospective cohorts. Ophthalmology 2014; 121:1421–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu S, de Sisternes L, Chen Q, Rubin DL, Leng T. Fully automated prediction of geographic atrophy growth using quantitative spectral-domain optical coherence tomography biomarkers. Ophthalmology 2016; 123:1737–50 [DOI] [PubMed] [Google Scholar]
- 45.Pfau M, Möller PT, Künzel SH, von der Emde L, Lindner M, Thiele S, Dysli C, Nadal J, Schmid M, Schmitz-Valckenberg S, Holz FG, Fleckenstein M. Type 1 choroidal neovascularization is associated with reduced localized progression of atrophy in age-related macular degeneration. Ophthalmol Retina 2020; 4:238–48 [DOI] [PubMed] [Google Scholar]
- 46.Schmidt-Erfurth U, Bogunovic H, Grechenig C, Bui P, Fabianska M, Waldstein S, Reiter GS. Role of deep learning quantified hyperreflective foci for the prediction of geographic atrophy progression. Am J Ophthalmol 2020; 206:257–70 [DOI] [PubMed] [Google Scholar]
- 47.Hallak JA, Azar DT. The AI revolution and how to prepare for it. Transl Vis Sci Technol 2020; 9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheller MJ, Edwards B, Reina GA, Martin J, Pati S, Kotrotsou A, Milchenko M, Xu W, Marcus D, Colen RR, Bakas S. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci Rep 2020; 10:12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rieke N, Hancox J, Li W, Milletarì F, Roth HR, Albarqouni S, Bakas S, Galtier MN, Landman BA, Maier-Hein K, Ourselin S, Sheller M, Summers RM, Trask A, Xu D, Baust M, Cardoso MJ. The future of digital health with federated learning. NPJ Digit Med 2020; 3:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yehoshua Z, Gregori G, Sadda SR, Penha FM, Goldhardt R, Nittala MG, Konduru RK, Feuer WJ, Gupta P, Li Y, Rosenfeld PJ. Comparison of drusen area detected by spectral domain optical coherence tomography and color fundus imaging. Invest Ophthalmol Vis Sci 2013; 54:2429–34 [DOI] [PMC free article] [PubMed] [Google Scholar]