Abstract
In multicellular organisms, cellular behaviour is tightly regulated to allow proper embryonic development and maintenance of adult tissue. A critical component in this control is the communication between cells via signalling pathways, as errors in intercellular communication can induce developmental defects or diseases such as cancer. It has become clear over the last years that signalling is not static but varies in activity over time. Feedback mechanisms present in every signalling pathway lead to diverse dynamic phenotypes, such as transient activation, signal ramping or oscillations, occurring in a cell type- and stage-dependent manner. In cells, such dynamics can exert various functions that allow organisms to develop in a robust and reproducible way. Here, we focus on Erk, Wnt and Notch signalling pathways, which are dynamic in several tissue types and organisms, including the periodic segmentation of vertebrate embryos, and are often dysregulated in cancer. We will discuss how biochemical processes influence their dynamics and how these impact on cellular behaviour within multicellular systems.
Keywords: embryonic development, Erk signalling, Notch signalling, signalling, signalling dynamics, Wnt signalling
Introduction
Communication between cells coordinates the self-organization process of embryonic development. Cell proliferation, differentiation, migration and death have to be regulated across scales — from single-cell to organism level — to allow the formation of an organism from a single cell. When cells communicate, an extracellular signal often induces an intracellular signal transduction cascade, which results in a cellular response encompassing, for instance, changes in cytoskeleton, metabolism or gene expression. Regulatory and feedback mechanisms at every level of such transduction cascades modulate pathway activity over time [1]. This leads to temporal variations in signalling, e.g. transient, pulsatile, ramping or oscillatory activity, collectively termed signalling dynamics [2]. Signalling oscillations are, for example, found in neural stem cells and the developing pancreas [3–5]. Dynamic changes in signalling can also occur upon changes in the levels of signalling molecules surrounding a cell, for instance when a cell moves through a spatial gradient of signalling molecules. Such gradients, also termed morphogen gradients, have been shown to organize the embryo into its anteroposterior (head to tail), dorsoventral (back to front), proximodistal and left-right axes [6,7].
Function of signalling dynamics
While feedback mechanisms might initially have evolved to keep signalling activity in check and prevent, for instance, overactivation, the resulting signalling dynamics have diverse functions in model systems found today [8,9].
Dynamic signal encoding
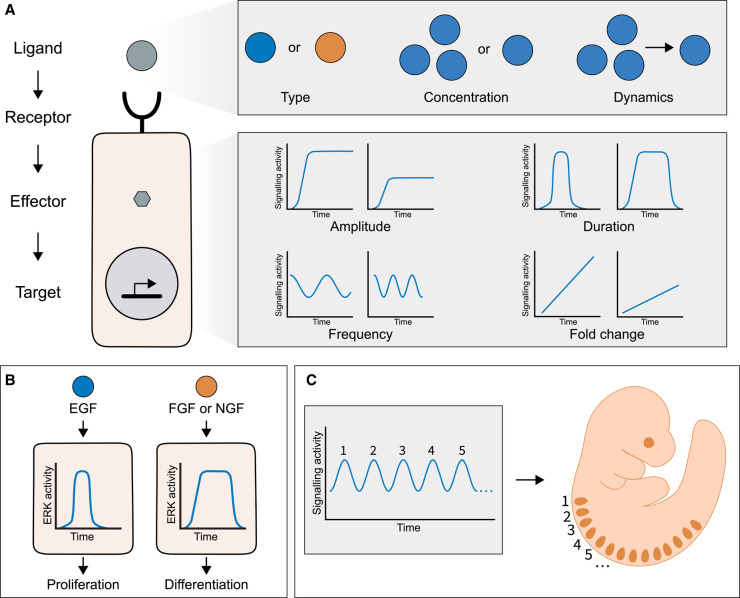
Often, signalling dynamics are used to encode and transmit biological information in the dynamics (Figure 1A). This has two consequences for the signalling process: (A) Encoding information in dynamics can make the signal more robust to noise and ensure proper transmission. Noise naturally occurs in a cell as a result of all biochemical reactions, leading to random fluctuations in, e.g. gene expression, protein–protein interactions, post-translational modifications and metabolic changes. Therefore, information transmission is less accurate and precise when encoding information in the absolute signalling activity. Such information loss can be reduced or prevented by encoding information in the dynamics of ERK activity, Ca2+ concentration or nuclear localization of the transcription factor NFκB [10,11]. In Xenopus embryos, the fold-change in Wnt signalling activity controls development even with varying levels of baseline Wnt signalling [12]. (B) Encoding information in the dynamics increases the versatility of signalling pathways [13]. How a dozen of signalling pathways can transmit diverse information with high specificity in organisms has been a matter of investigation for decades. Besides employing distinct isoforms of pathway components, combinatorial use of multiple pathways and activation of specific transduction cascades, varying the dynamics is another means to increase the repertoire of signalling modalities [14]. A classic example is the stimulation of the rat PC12 (pheochromocytoma) cell line by growth factors. While epidermal growth factor (EGF) induces a transient ERK signal resulting in proliferation, nerve growth factor (NGF) or fibroblast growth factor (FGF) activate a sustained ERK response leading to neuronal differentiation (Figure 1B) [15–17].
Figure 1. Cellular signalling dynamics.
(A) Signal transduction is induced when a ligand interacts with the receptor, which leads to the activation of effector proteins. Effectors exert various functions in cells, among which is the induction of gene expression. The type, concentration and dynamics of ligands can be encoded in signalling dynamics, such as the amplitude of a stable signal, the amplitude and duration of a transient signal and the amplitude, duration and frequency of an oscillatory signal or the fold-change of a ramping signal. (B) An example of dynamic signal encoding is the effect of growth factors on PC12 cells. EGF induces a transient signal of Erk activity, which results in proliferation, while FGF or NGF induce a sustained signal, which leads to differentiation. (C) Oscillatory signals can coordinate periodic events, such as the sequential segment formation in vertebrate embryos.
Regulation of repetitive events
Apart from transmitting information in the dynamics of a signal, oscillations can control periodic events. This way, oscillations of the circadian clock adjust physiological processes of the body to the repeating day-night cycles [18]. In embryonic development, oscillations regulate the sequential segmentation of vertebrate embryos (Figure 1C) [19] and the consecutive steps of larval development in Caenorhabditis elegans [20].
Communication across tissues
Tissue-wide oscillation dynamics can synchronize cells within a multicellular system to regulate collective behaviour. It allows the efficient transfer of information from one side of a tissue to the other. This is exemplified by the coordination of tissue growth with segmentation in vertebrate embryos to ensure the formation of properly sized embryos [19].
How dynamics are decoded to induce specific cellular responses is still largely unknown. For dynamic signal encoding, the change in signal activity over time has to be detected and converted to an absolute signal, such as the expression of differentiation markers. In a ramping signal, information can be encoded in its derivative, while in an oscillatory signal, the frequency, amplitude or number of oscillations can encode information (Figure 1A) [2]. It has been shown that in mouse and human cell cultures, the number of NFκB oscillations is directly converted to a stepwise expression of different sets of target genes [21–23].
Signalling dynamics from a biochemical perspective
Intracellular signalling is initiated when a ligand from a signal-sending cell interacts with a receptor on a signal-receiving cell. Signalling can be autocrine, when signal-sending and -receiving cell coincide, paracrine, when neighbouring cells communicate with each other, or endocrine, when signal-sending and -receiving cells are spatially separated and signalling molecules travel via the circulation as in the case of hormones. An intracellular signalling event is initiated upon ligand–receptor interaction, entailing protein–protein interactions, post-translational modifications, proteolytic cleavages, protein translocations or lysosomal degradation of pathway components. These ultimately lead to changes in enzyme activities, which modulate e.g. metabolism or cytoskeleton, and to changes in gene expression. The dynamics of this signalling process directly depend on the thermodynamics of the system itself, the kinetics of the individual reactions and induced feedback mechanisms (Figures 1, 2A).
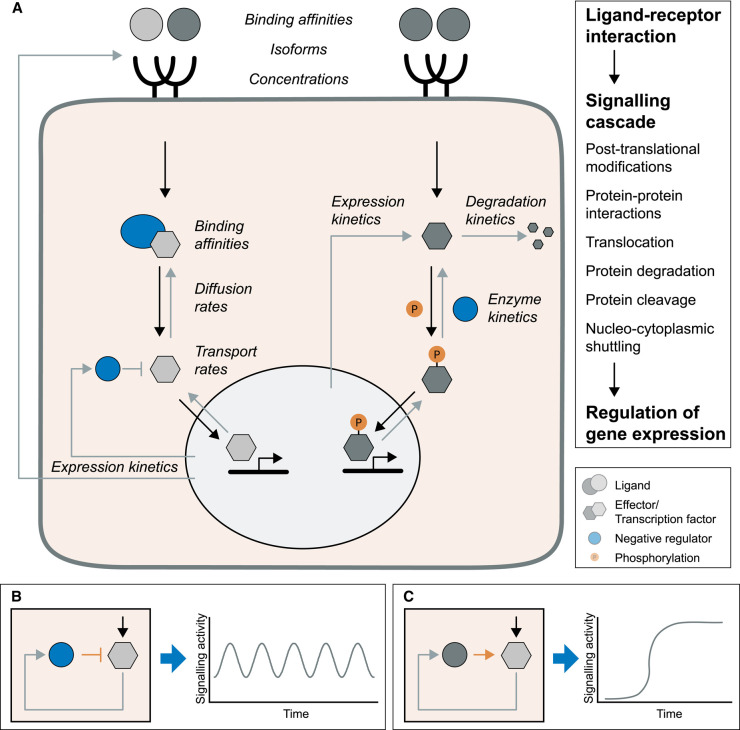
Figure 2. Processes influencing the timing of dynamics.
(A) Signalling is initiated when ligands interact with their receptors. Intracellular signalling can encompass various processes, which ultimately lead to the activation of effector proteins. These effector proteins have diverse functions, often modulating gene expression. Two potential signal transduction cascades and biochemical parameters that can influence the dynamics of a signalling pathway are indicated in the scheme. On the left-hand side, an effector protein is activated by dissociation from an inhibitory interaction partner. On the right-hand side, the effector protein is activated by phosphorylation. (B) A delayed negative feedback system can induce oscillations, for instance, if the pathway induces the expression of a negative regulator. Stable oscillations occur if parameters for activation and delayed inhibition are synchronized. (C) A positive feedback loop can induce a sigmoidal cellular response or even bistability under the right conditions. In developmental biology, such networks can guide cell fate transitions.
The expression levels of all involved signalling components and their isoforms can vary widely in a cell type- and developmental stage-dependent manner [24–27]. The concentrations of receptors on the cell surface and ligands and their respective binding affinities and geometry are critical determinants of the maximal and actual extent of pathway activation. In combination with the stoichiometry, affinity of protein–protein interactions, e.g. the ligand–receptor interaction, the concentration of all components and their interactions collectively influence the equilibrium of steady-state signalling. The intracellular signalling cascades often involve enzymatic reactions, such as post-translational modifications, proteolytic cleavages or degradation of inhibitory components via the proteasome [28] that occur at the second to minute time-scale. In most cases, this culminates in the activation of transcription factors (TFs) that shuttle to the nucleus, where they directly or indirectly modulate gene expression. When labelling these TFs with fluorescent proteins, nuclear accumulation can be detected within minutes after stimulating cells with a ligand [29,30]. In mice, the resulting gene expression, including transcription, splicing, mRNA export and translation, leads to the generation of proteins within at least 20 min in mice [31–33]. Thus, pathway stimulation can result in the appearance of new proteins within 25 to 30 min, but can also take significantly longer.
When ligand stimulation ends, the whole signal transduction cascade is reset to baseline. While post-translational modifications, e.g. protein phosphorylation, are reversible, protein cleavage and degradation cannot be reversed, which has immediate implications for the dynamics of a pathway. For instance, the MAPK (mitogen-activated protein kinase) pathway consists of a series of phosphorylation events. In many cell types, the effector kinase MAPK shows pulsatile cycles of activation/ inactivation with a period of a few minutes [34]. In contrast, dynamics of other pathways building on, for instance, proteolytic cleavage of pathway components take more time to reverse, since the active protein needs to be re-expressed.
Besides pathway inactivation by ligand–receptor dissociation or degradation, signalling activity of most pathways is actively modulated via feedback mechanisms. Negative feedback can result in the inactivation of the pathway at various levels of the cascade. For instance, the binding of ligands to their receptor can trigger the endocytosis of the signalling complex, which is followed either by the recycling of the receptor or degradation of the complex in the lysosome [35]. The extent of Wnt signalling in cells is regulated by receptor degradation mediated by a network of E3 Ubiquitin ligases and regulatory factors [36]. In addition, the expression of negative regulators upon pathway stimulation can reverse post-translational modifications, block protein degradation or repress the expression of target genes of the pathway [37]. Negative feedback can result in adaptation, when continued pathway stimulation results only in a minimal response or baseline activity [1]. If the parameters for pathway activation and induced negative feedback are precisely coordinated regarding levels and timescales of activation, delay and inactivation, oscillatory activity can be induced (Figure 2B) [1]. In addition, network motifs containing positive feedback loops can, for instance, lead to bistability that controls developmental cell fate decisions or cell cycle progression (Figure 2C) [38]. In reality, the regulatory networks are usually more complex than this and can include both positive and negative feedback loops, which modulate signalling activity in complex ways [1].
Since the study of dynamic processes in the context of a multicellular organism is technologically challenging, our understanding of signalling dynamics in multicellular systems is limited, especially at the mechanistic level. With new technologies to analyse, visualize and perturb signalling activity in cells and embryos, signalling dynamics are found in more and more model systems and tissues, and their functions are being revealed. It seems that signalling dynamics is a common characteristic of signalling processes and that there might be general principles underlying the function of dynamic signalling in different tissues and organisms. Here, we review the current knowledge of the role of signalling dynamics in embryonic development and aim to make a conjunction with our understanding of the biochemical properties of the individual signalling pathways. Although pathways such as hedgehog, TGFβ or Hippo signalling are equally important in embryonic development, we will focus on Wnt, Notch and ERK signalling to highlight the complexities of dynamic signalling.
Notch signalling
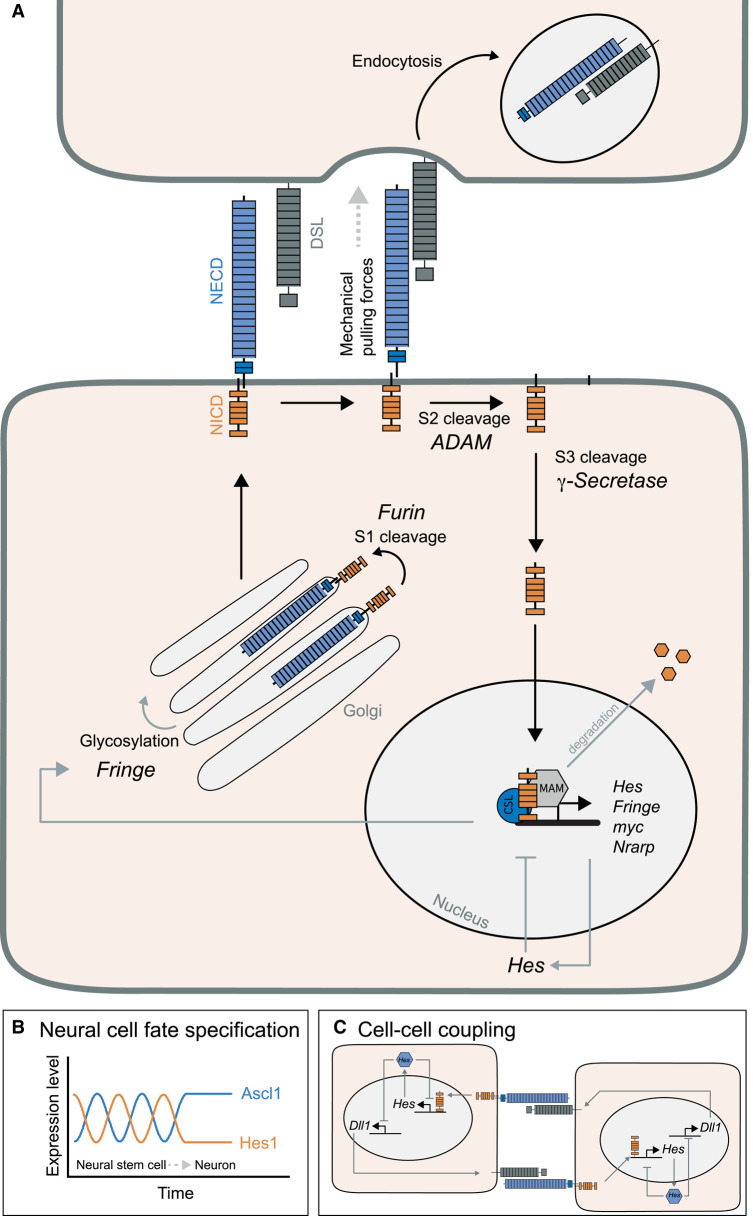
Notch signalling is a highly conserved signalling pathway essential for embryonic development and the maintenance of adult tissues [39]. It mediates the communication between neighbouring cells by cell–cell contact. The ligands Delta/Jagged/Serrate and the receptor Notch are transmembrane proteins, with the ligand being expressed in signal-sending cells and the receptor in neighbouring signal-receiving cells (Figure 3A). Notch activation requires several proteolytic cleavage steps that occur with specific kinetics and therefore influence the kinetics of pathway activation. After translation, Notch is cleaved in the Golgi apparatus by a Furin protease (S1 cleavage), which results in the formation of a heterodimer consisting of the Notch extracellular domain (NECD) bound to a fragment containing a transmembrane domain and the Notch intracellular domain (NICD) [40]. Upon interaction of the ligand with the NECD, an intracellular signal transduction cascade is induced, whereby the Notch intracellular domain (NICD) is released by proteolytic cleavages and travels to the nucleus [41]. These Notch cleavage steps require mechanical pulling forces exerted by the ligand on the receptor, which opens a negative regulatory region (NRR) [42,43]. This allows proteolytic cleavage by ADAM metalloproteases (S2 cleavage) on the extracellular site [44]. This step releases the ligand-bound NECD and enables the final cleavages: γ-Secretase (a multi-subunit complex consisting of Nicastrin, Presenilin, PEN2 and APH1) exerts the S3 and S4 cleavages on the intracellular site, which releases NICD into the cytoplasm [45]. Due to the mechanical forces required for Notch activation, it has so far not been possible to develop a soluble receptor agonist for experimental activation of Notch signalling.
Figure 3. Dynamics of Notch signalling.
(A) Notch is generated in the endoplasmic reticulum and travels to the plasma membrane via the Golgi apparatus. In the Golgi, the first proteolytic cleavage is mediated by Furin, which leads to the formation of a Notch heterodimer. There, Notch binding affinities can be modulated by post-translational modifications, e.g. glycosylation by Fringe. When the ligand from a neighbouring cell interacts with Notch, a mechanical pulling force results in a conformational change in Notch. This allows the proteolytic cleavage of Notch by ADAM and then γ-Secretase, which finally releases NICD (Notch intracellular domain) into the cytoplasm. In the nucleus, NICD interacts with the transcription factor CSL and the co-activator Mam to induce the expression of Notch target genes. Among these are Hes genes, which initiate a delayed negative feedback loop, as well as Fringe and Nrarp, which both feed back onto Notch signalling. Finally, NICD is phosphorylated and degraded by the proteasome. (B) Due to the negative feedback loop of Hes proteins, Hes expression oscillates in many systems. In neural stem cells of the developing brain Hes1 oscillates alternatingly with pro-neural genes. When Hes1 oscillations cease and pro-neural proteins get stabilized, cells differentiate. (C) Neighbouring cells can couple via Notch signalling and synchronize their intracellular oscillations, for instance during periodic segmentation of vertebrate embryos. This is thought to be achieved by reciprocal activation of Notch signalling in neighbouring cells and induction of the ligand Dll1.
Within the nucleus, NICD interacts with the transcription factor CSL (CBF1/RBPJ, Su(H), Lag-1) and the co-activator mastermind (MAM) as well as others to induce expression of Notch target genes [46–48]. Among these are basic helix–loop–helix transcription factors of the Hes family, but also other genes controlling cellular behaviour such as myc, Cyclin D or Nrarp [49–53]. Notch signalling is terminated by phosphorylation of NICD in its PEST domain and degradation by the proteasome [54–56]. In contrast, the ligand-NECD complex is endocytosed by the signal-sending cell [57]. The combination of nuclear translocation, affinity to the CSL complex and the timing until phosphorylation for proteasomal degradation determines the lifetime of activated NICD. In human cell lines, NICD has a half-life of 180 min [54]. This half-life is further modified by the precision of the final Notch cleavage that results in the release of NICD. Depending on the resulting N-terminal amino acid, NICD half-life can vary in accordance with the N-end rule, which states that N-terminal amino acids are recognized by ubiquitin ligases with different efficiencies to mark the protein for degradation [58,59]. Such processes modulate the dynamics of Notch signalling in a cell type- and state-dependent manner, which should be taken into account when investigating the molecular mechanism of signalling dynamics.
Cis- and lateral inhibition
When the Notch receptor and ligands are expressed in the same cell, they interact in cis. Since such an interaction cannot generate the required pulling forces for Notch activation, signalling is essentially blocked, a process termed cis inhibition [60,61]. Thus, within a tissue, Notch signalling is active, if ligand and receptor are expressed in different ratios or if ligand and receptor are spatially separated within a cell [62,63]. This auto-regulatory mechanism, in combination with the fact that Notch ligands themselves are downstream targets of the pathway, can result in lateral inhibition by which neighbouring cells induce each other to express either the ligand or the receptor and thereby assume different cell identities [64–66]. This way, a multicellular tissue can self-organize into a highly ordered structure with alternating, equally spaced cell types. For instance, a combination of cis inhibition and lateral inhibition allows the formation of regularly spaced sensory hairs in separate lines along the back of the fly [67]. In the mammalian gut epithelium, lateral inhibition coordinates the regular spacing of secretory and non-secretory cells along the gut lining. Secretory cells express Notch ligands, which block their neighbours to adopt the same cell fate [68,69]
Cellular Notch signalling oscillations
Several Notch target genes function as negative regulators of the pathway [50]. Prominent among these are members of the Hes transcription factor family, which can function as transcriptional repressors of their own expression as well as other genes [70,71]. Another negative regulator is Nrarp (Notch regulated ankyrin repeat protein), a Notch signalling target in Xenopus embryos [72]. It interacts with the NICD-CSL complex and induces its dissociation, resulting in NICD degradation via the proteasome [50]. Such delayed negative feedback loops induce oscillations of Notch target genes. Indeed, Hes oscillations have been observed in various tissue types, such as embryonic stem cells, neural stem cells and the developing pancreas (Figure 2B) [3,5,73]. In neural stem cells of the developing mouse brain, oscillatory expression of Hes1 and several pro-neural genes such as Ascl1 controls the balance between proliferation and differentiation [5,74]. When pro-neural genes are stabilized and Hes genes down-regulated, neural differentiation is initiated (Figure 3B). Similar oscillation dynamics of Hes5 have been observed in the developing spinal cord [75,76]. To test the functional significance of Hes dynamics in regulating cell behaviour, Imayoshi et al. generated an optogenetic system to modulate Ascl1 expression in cells by light pulses [4,77]. When oscillations of Ascl1 were induced in neural stem cells, cells proliferated and did not differentiate [4]. Conversely, sustained Ascl1 expression resulted in neuronal differentiation. This work highlights the importance of signalling oscillations in controlling the balance between proliferation and differentiation in stem cells; however, the molecular decoding mechanism remains to be investigated.
Spatiotemporal Notch dynamics
Owing to the intrinsic propensity of Notch signalling to oscillate, the fact that expression of both ligand and receptor is induced by the pathway itself as well as its paracrine nature, Notch signalling can generate tissue-wide oscillation or wave dynamics. One example is the periodic formation of segments or somites during vertebrate development, a process termed somitogenesis. Somites are transient embryonic structures and precursors of vertebrae, muscles and dermis [78]. Periodic segmentation of the growing tissue is regulated by signalling oscillations in Notch, Wnt and FGF signalling in the unsegmented tissue, the presomitic mesoderm (PSM) [19]. The network of oscillating signalling pathways is termed the segmentation clock. Each cell is thought to be an autonomous oscillator [79–81], but cell-to-cell communication via Notch signalling synchronizes neighbouring cells (Figure 3C) [82–84]. This way, kinematic waves of signalling activity are observed in tissues travelling from the posterior tip of the tail to the side of segment formation. Since the segmentation clock also induces the expression of the Notch ligand Dll1, a model has been proposed in which oscillatory expression of Dll1 drives Notch oscillations and mediates cell–cell communication to form waves of Notch activity travelling through the segmenting tissue (Figure 3C, also see Figure 4B) [85]. In fact, Notch signalling seems to be central to generating these oscillations. Oscillations of the downstream target Hes7 have been detected in various model organisms such as zebrafish, mouse and human cells [84,86–89]. When the timing of Hes7 transcription was altered by the removal of several introns (see Figure 2), the oscillation period and, consequently, the timing of segmentation were shortened [90]. Interestingly, shortening gene expression timing resulted in damped oscillations, highlighting that the right parameters of the delayed negative feedback loop are essential for stable oscillations (Figure 2A,B). Another set of experiments underscoring the role of Notch signalling for oscillations was performed by Soza-Ried et al. who induced periodic overexpression of the ligand DeltaC in zebrafish, which could control segmentation oscillations [91].
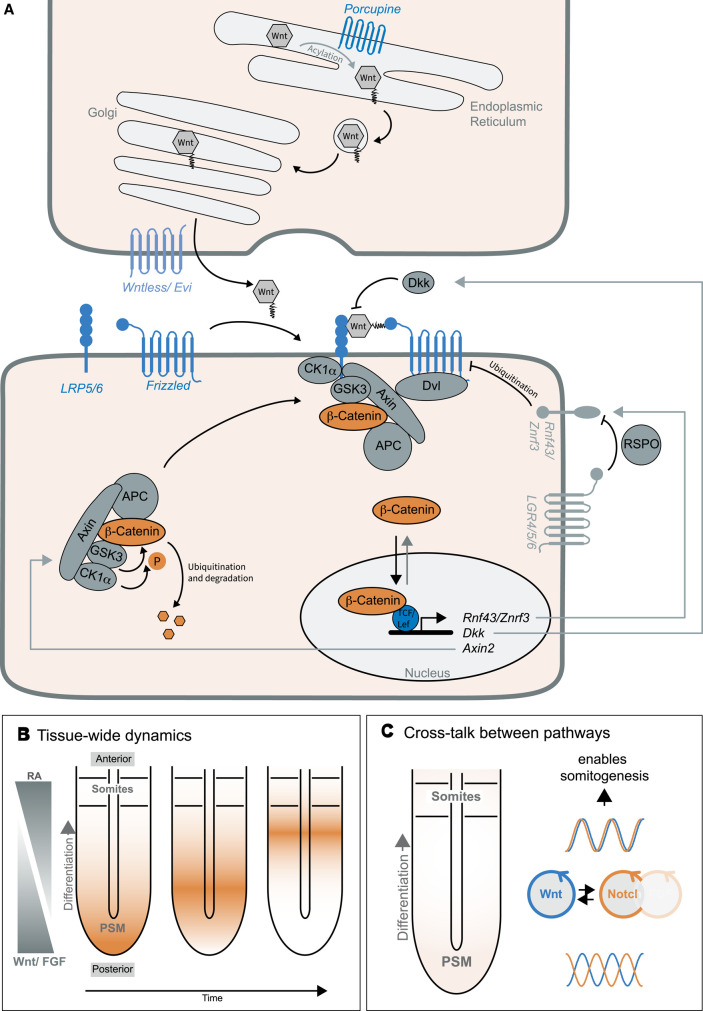
Figure 4. Dynamics of Wnt signalling.
(A) Wnts are secreted proteins that are lipidated in the ER by Porcupine. Wntless/ Evi regulates Wnt secretion. In the signal-receiving cell, many Wnt ligands induce the β-catenin-dependent signalling cascade. In the absence of Wnt, β-catenin is constantly phosphorylated by the destruction complex and marked for degradation by the proteasome. When Wnt binds to its receptor Fzd and the co-receptor LRP5/6, Dvl and the destruction complex are recruited to the plasma membrane. This prevents phosphorylation and degradation of β-catenin, which results in the accumulation of hypo-phosphorylated β-catenin in the cytoplasm. β-Catenin travels to the nucleus to modulate gene expression by interacting with the transcription factor TCF/LEF. Several Wnt target genes function as negative regulators of the pathway, such as Axin2, Dkk and Rnf43/Znrf3. Rnf43/Znrf3 marks Fzd for proteasomal degradation. When RSPO binds to Lgr, this complex recruits and inhibits Rnf43/Znrf3. This double-negative regulation amplifies Wnt signalling activity. (B) Periodic segmentation of mammalian embryos or somitogenesis is regulated by oscillations of Wnt, Notch and Fgf signalling. Coupling between cells via Notch signalling leads to the formation of signalling waves (in orange) travelling from the posterior tip to the anterior side of the presomitic mesoderm (PSM), where segment formation is induced. In addition to signalling waves, the tissue is organized by antagonistic gradients of Wnt/Fgf and retinoic acid (RA). (C) In mouse somitogenesis, Wnt and Notch signalling oscillations are coupled and have different dynamics in the segmenting tissue: In the posterior, Wnt and Notch signalling oscillate out-of-phase, in the anterior they oscillate in phase. This change in phase-relationship is essential for proper segmentation.
Specificity in information transmission
Mammalian genomes encode for 4 Notch isoforms (Notch1 to Notch4) and 5 Notch ligands (Jagged1 (Jag1), Jag2, Delta-like 1 (DLL1), DLL3 and DLL4). However, even if cells express the same Notch receptors, different intracellular dynamics and effects can be activated. How different ligands induce different intracellular dynamics and result in different downstream effects is still a matter of investigation. One potential explanation comes from structural and biochemical analyses revealing that different Notch ligands bind their receptors with different affinities. For instance, DLL4 binds Notch 1 with a higher affinity than DLL1 [3,92–95]. In agreement with this, DLL4 has recently been found to induce sustained Notch activation, which results in expression of the downstream target Hey1, while Dll1 induces Notch pulses and expression of Hes1. It has been suggested that ligand–receptor clustering in the case of Dll4 results in sustained activation, which explains how ligand identity can be encoded in the dynamics of Notch signalling [96].
Ligand–receptor affinity is further fine-tuned by post-translational modifications such as glycosylation of the Notch receptors by several enzymes, including Fringe, POFUT1, POGLUT1 or EOGT1 [97–99]. Local modifications of Notch receptors in Drosophila embryos ensures spatiotemporal specificity of Notch signalling [97,98]. Interestingly, in mammalian cells, Lunatic Fringe (Lfng) is one of the cyclic genes whose expression is periodically induced during vertebrate segmentation [100,101]. Considering that the Notch receptor pool has to be replenished continuously to maintain Notch signalling, oscillatory expression of the glycosylating enzymes might further amplify signalling activity and thereby enhance the oscillation amplitude. It has been shown recently that Hes7 oscillations downstream of Notch signalling do not depend on the presence of Lfng. In contrast, Lfng is essential for coupling between cells, highlighting the importance for glycosylation for proper intercellular signalling [102].
How specific dynamics are decoded to induce a precise, cell type-specific effect is not understood. The architecture and affinity of CSL binding sites and the recruitment of other regulatory proteins seem to play a role, which is discussed in detail elsewhere [41]. In vitro systems, such as periodic induction of ligand expression [91] or the synthetic Notch system syn-Notch [103], allow defined external activation of Notch signalling to analyze the encoding and decoding process in detail.
Wnt signalling
The Wnt signalling pathway is an evolutionarily conserved signalling pathway essential for stem cell renewal, cell proliferation and cell differentiation during embryogenesis and adult tissue regeneration and repair. Deregulated Wnt signalling is implicated in various diseases, including cancer, developmental disorders, and degenerative diseases [104–106]. Multiple, functionally distinct Wnt signal transduction pathways have been identified [107]. Here, we will focus on the Wnt/β-catenin-dependent pathway, the canonical Wnt pathway, that regulates context-specific gene expression programs (Figure 4A).
The transcriptional co-activator β-catenin is a master regulator of this transduction pathway and is constantly synthesized. In the absence of Wnt ligands, it is rapidly degraded by ubiquitin-dependent proteasomal degradation [108]. This process is initiated by the β-catenin destruction complex, a multiprotein complex composed of the scaffold proteins adenomatous polyposis coli (APC) and Axin1 [109–112] the protein kinases casein kinase 1 (CK1) and glycogen synthase kinase-3 (GSK3), which phosphorylate β-catenin on key serines/threonines [113–115]. The phosphorylated motif is a docking site for the β-TrCP-containing E3 ubiquitin ligase that mediates ubiquitination and subsequent proteasomal degradation [116–118]. Wnt binding to the cell surface receptors Frizzled (Fzd) and co-receptor low-density lipoprotein receptor-related protein 5 (LRP5) or LRP6 triggers the recruitment of Dishevelled (Dvl) to the intracellular regions of Fzd [119–121], and of the Axin-GSK3 complex to the C-terminal tail of LRP5/6, promoting its phosphorylation by GSK3 [122,123]. This results in the inhibition of the β-catenin destruction complex, and the accumulation of hypo-phosphorylated β-catenin in the cytoplasm and the nucleus [124,125]. In the nucleus, β-catenin functions as a co-activator for transcription factors of the TCF/LEF family and modifies the expression of Wnt target genes [126–128], thereby leading to changes in key cellular processes including cell proliferation, cell fate determination and migration depending on cell type and cell state. Besides its function in Wnt signalling, β-catenin stably localizes to the plasma membrane as a component of adherens junctions [129]. The relevance of this β-catenin pool for signalling and the dynamics between the different pools is still controversial and might be context-dependent [130,131].
While the Wnt/β-catenin pathway and the molecular interactions of signalling components are extensively studied, only a few studies investigate the dynamics of Wnt signalling and how cells receive, process, and interpret different features of extracellular ligands, such as molecular identity, concentration, and combinations with other ligands to control specific cellular behaviour. However, many of the ‘communication codes’ identified in other signalling pathways could also be critical determinants for Wnt/β-catenin signalling dynamics and cell fate decisions.
Wnt gradient
Wnts are post-translationally lipidated by the O-acyl transferase porcupine in the ER [132,133] and further shuttled through the Golgi to the plasma membrane by the escort protein Wntless/Evi [134–136] (Figure 4A). After secretion, lipidation is essential for Wnt binding to the Fzd receptor [137] and to limit diffusion in the extracellular environment [132]. As outlined above, the Wnt gradient is critical for coordinated spatial and temporal activity (Figure 4B), but the molecular mechanism of gradient formation remains the subject of many ongoing studies. The current notion suggests that Wnts ‘travel’ away from the secreting cells when incorporated into particles, vesicles or exosomes [138,139] or bound to receptors, i.e. Fzd and Rnf43/Znrf3 through direct cell contact and cell division [140]. Furthermore, in many systems, the Wnt gradient appears to be fine-tuned by negative feedback mechanisms or counteracting gradients of Wnt antagonists. For instance, in the mouse embryo at gastrulation, the expression of Wnt3 in the posterior region opposes the expression of the antagonist dickkopf 1 (Dkk1) to control head morphogenesis [141].
Feedback mechanisms
Wnt pathway activity is carefully controlled in healthy cells by antagonists and potentiators, as well as positive and negative feedback loops that enhance, augment, dampen or terminate Wnt signalling (Figure 4A). The interactions of Wnts with its receptors are negatively regulated by numerous proteins, including Dkk1-4 [142–144], sclerostin/SOST [145], the Wnt inhibitory factor WIF [146] and Notum [147,148], several being Wnt target genes themselves (Figure 4A). In contrast with Axin1, Axin2 is a universal Wnt target gene, and increased Wnt signalling enhances destruction complex formation and β-catenin degradation, thereby suppressing β-catenin-mediated gene expression [12,149,150]. Furthermore, Rnf43 and Znrf3 are Wnt target genes and transmembrane E3-ubiquitin-protein ligases that mediate Fzd degradation [151,152], leading to reduced Fzd cell surface density and decreased sensitivity of cells to Wnt ligands. This results in the attenuation or termination of the Wnt signal necessary for many biological processes. For instance, Znrf3 knockout mouse embryos die around birth and show a lack of lens development as a most obvious phenotype [151]. Conversely, Wnts also trigger the expression of leucine-rich repeat-containing G protein-coupled receptor family 5 (Lgr5) [152]. R-spondin ligands (RSPO) are expressed in many stem cell niches and engage Lgr4-6 and Rnf43/Znrf3, thereby triggering endocytosis of the ternary complex and blocking Rnf43/Znrf3-mediated Fzd degradation [151–153] (Figure 4A). This leads to a higher sensitivity of cells to Wnt ligand, markedly increased Wnt/β-catenin signalling levels and, in the small intestine, to the activation of the transcriptional programs required for stem cell proliferation and self-renewal [154].
Combinatorial use of Wnt ligands and receptors
Many developmental signalling pathways have evolved multiple ligand and receptor variants that interact promiscuously, and the Wnt pathway is no exception. The human genome encodes 19 Wnts, 10 Fzds, and two LRP5/6 co-receptors. Although loss-of-function of most Wnts result in characteristic phenotypes (http://web.stanford.edu/group/nusselab/cgi-bin/Wnt/), observations suggest that these are related to the localized expression, rather than intrinsic activities of different Wnt subtypes. It is generally accepted that Wnts/Fzds are highly cross-reactive. Biochemical and functional approaches that characterize interacting Wnt — Fzd pairs [155,156] have revealed broad Wnt — Fzd cross-reactivity, with some Wnts having broader specificity than others. Consistent with the broad Fzd specificity, Wnt3a is pleiotropic and can exert a broad range of biological activities, such as supporting the growth of organoids from many different epithelial tissues [157], exceeding what could be expected based on the Wnt3a expression profile. Nevertheless, it can be expected that the different Wnts are not functionally equivalent, meaning that their signalling outputs have different characteristics. This notion is supported by the observation that different Wnt subtypes synergistically activate Wnt/β-catenin signalling in multiple cell types [158].
Furthermore, biophysical parameters including stability, geometry and stoichiometry of the Wnt/Fzd/LRP5/6 signalling complex are likely to infer signalling dynamics. Despite some progress in recent years [137,159–162] studies to exploit ligand/receptor-induced Wnt signalling dynamics remain challenging owing to the complex nature of the receptors and the Wnts. To overcome these difficulties, several laboratories have developed Wnt mimetics, consisting of non-lipidated Fzd–LRP5/6 ‘heterodimerizers’ that are structurally distinct from natural Wnts but recapitulate their activities with a tuneable activity [137,159,160,163,164]. Furthermore, ESCs and adult neural stem cells with light-inducible Wnt signalling have been generated by expressing the cytoplasmic tail of LRP6 fused to the blue-light photoreceptor Cryptochrome 2, allowing to probe the effect of dynamic Wnt signalling activation [165,166].
In contrast, the relevance of receptor abundance for tuning Wnt signalling dynamics is better established. RSPO ligands strongly potentiate Wnt signalling by inhibiting the Rnf43/Znrf3-mediated proteasomal degradation of Fzd, leading to increased receptor abundance on the surface of Wnt receiving cells [151,152]. Recently, it has been demonstrated that Wnts and RSPOs exert differential activities in the intestinal crypt stem-cell niche. While Wnts confer a basal competence of stem cells for Wnt signalling by maintaining the expression of Lgr5, RSPOs support the expansion of stem cells [154]. Hence, it would be interesting to elucidate the differential functions of Wnts and RSPOs and their association with diverse signalling dynamics in other systems and models. Besides attenuating or potentiating Wnt signalling, some co-receptors have been found to be required for correct Wnt signalling initiation. For instance, RECK is a selective Wnt7 receptor that, together with GPR124, is required for transducing a Wnt/β-catenin signalling to control embryogenic CNS angiogenesis and blood-brain barrier formation [167,168].
Fold change detection
Intracellularly, β-catenin is the master effector of the Wnt pathway critically involved in regulating gene expression in response to Wnt stimulation. A first mathematical model describing the functioning of the Wnt/β-catenin pathway in time and space was developed by Kirschner, Heinrich, and colleagues and is based on measurements with Xenopus oocyte extracts, and the molecular reactions of the pathways’ core effectors including Axin, APC, and β-catenin [169]. This model was subsequently modified regarding signalling characteristics, refined interactions, functional diversity, compartmentalization and nuclear shuttling of β-catenin, and negative feedback loops [12,169–172]. An important observation was that the fold-change rather than a linear increase in β-catenin is the critical determinant read out by the downstream transcriptional system. While the absolute β-catenin levels induced by Wnt stimulation are very sensitive to relatively small, stochastic pathway perturbations, a fold-change is robustly buffered. Having the output of Wnt signalling sensed in fold-change allows the cells to detect ligand levels while cancelling out fluctuating noise. It requires that at least some Wnt target genes perceive fold-changes, rather than absolute levels, of β-catenin, giving rise to adaptive responses. Detection of the fold change may be accomplished by an incoherent feedforward loop of gene regulation [12,173]. This suggests that Wnt signalling, at least in the absence of Lgr5/RSPO contribution, is optimized for controlling transient events, such as cell fate decisions, rather than continuously transmitting information about extracellular ligand levels.
Adaption
A series of studies used quantitative microscopy to interrogate the dynamics of fluorescently labelled endogenous β-catenin, or β-catenin expressed at near endogenous levels, in response to Wnt signal activation and inhibition [174,175]. Stimulation of HEK293 cells with Wnt3a led to an increase in the total amount of β-catenin. However, the increase was initially faster in the nucleus than in the cytoplasm, resulting in a higher nucleus/cytoplasm protein ratio. The shuttling of β-catenin to the nucleus appears to be mediated by active transport alongside passive diffusion. While the dynamics of ß-catenin nuclear build-up, i.e. rates, levels, wave profile, varied substantially between individual cells, the total levels of accumulated β-catenin were similar in most cells, suggesting that the balance between accumulation and degradation affects the extent of β-catenin accumulation in individual cells. The expression kinetics of the Wnt target gene cyclin D paralleled the kinetics of the initial phase of β-catenin nuclear accumulation. Rate of change, and not actual protein levels, of nuclear β-catenin correlated with the transcription of cyclin D1 mRNA, especially during the early phase of Wnt stimulation [174].
However, β-catenin dynamics appear to be related to cell type, context, and fate. For instance, stimulation of hESCs with exogenous Wnt, resulted in an adaptive β-catenin response. β-catenin accumulated rapidly in both the nucleus and the cytoplasm but started to decline in the nucleus after four hours [176]. The level of adaptation depended on the concentration of the Wnt signal, being complete at a low dose of Wnt3a and partial at saturating concentration of Wnt3a. In contrast, the small-molecule GSK3ß inhibitor CHIR99021, commonly used as a Wnt substitute, induced a nonadaptive dose-dependent increase in nuclear β-catenin, indicating the mechanism that controls adaptation in hESCs is upstream of GSK3β. Interestingly, Wnt target genes showed various response profiles. While some genes were transiently activated in response to Wnt3a, others were sustained, further adding to the complexity of Wnt signalling dynamics and questions to address.
Wnt signalling oscillations
Besides fold-change detection and adaption, Wnt signalling can display complex oscillatory dynamics. The main example is the segmentation of mouse embryos, which is controlled by oscillatory pathway activity (see above, Figure 4B). The first cyclic genes identified in chicken, mouse and zebrafish embryos were downstream targets of Notch signalling [88,100] (Holley et al. 2000; Palmeirim et al. 1997). To identify other oscillatory genes and pathways, a transcriptomic screen was performed, in which one half of the PSM was used for transcriptome analysis [177]. The other half was stained for the cyclic gene Lfng, which allowed the authors to put the transcriptomic samples into temporal order based on the Lfng staining pattern. This revealed Wnt signalling as a new oscillatory component of the segmentation clock. Subsequently, staining of nascent Axin2 mRNA as downstream target of Wnt signalling and the quantification of dynamic Wnt signalling reporters confirmed oscillatory Wnt activity in models of mouse and human somitogenesis [87,178,179]. The regulatory network inducing Wnt signalling oscillations is not understood in detail. Modelling of the Wnt signalling network indicated that a delayed negative feedback loop via the negative regulator Axin2 could account for the oscillations [180,181]. However, other negative regulators such as Dkk are induced likewise (Figure 4A) and their function should be tested experimentally.
The cross-talk between and hierarchy of Wnt and Notch signalling in somitogenesis has been investigated and has remained inconclusive [178,182–185]. This stems in part from the presence of a Wnt signalling gradient in the tissue that adds to the complexity and makes the experimental dissection difficult [178,182]. When knocking out the transcriptional repressor Hes7, Notch signalling oscillations were not detected anymore, indicating Hes7 as part of the oscillatory mechanism [90,184]. In contrast, Wnt signalling oscillations persisted in Hes7 knockout mice. To analyse a potential coupling between Wnt and Notch signalling oscillations, a microfluidic system was applied that allows the subtle modulation of signalling oscillations in segmenting tissue [179,186]. This showed that Wnt signalling oscillations followed, when Notch signalling oscillations were entrained, and vice versa, implying that Wnt and Notch signalling oscillations are indeed coupled (Figure 4C) [179]. Interestingly, Wnt and Notch signalling oscillations change phase-relationship from out-of-phase in the posterior tissue to in-phase in the anterior part of the PSM, where segments form [177–179]. When changing this phase relationship using microfluidics, segment formation was impaired, which highlights that phase shift carries critical information for somitogenesis (Figure 4C). How this phase-shift changes and how it controls segment formation is not understood. Furthermore, whether and how Wnt signalling oscillations regulate the development of other tissues has to be investigated in future studies.
MAPK signalling
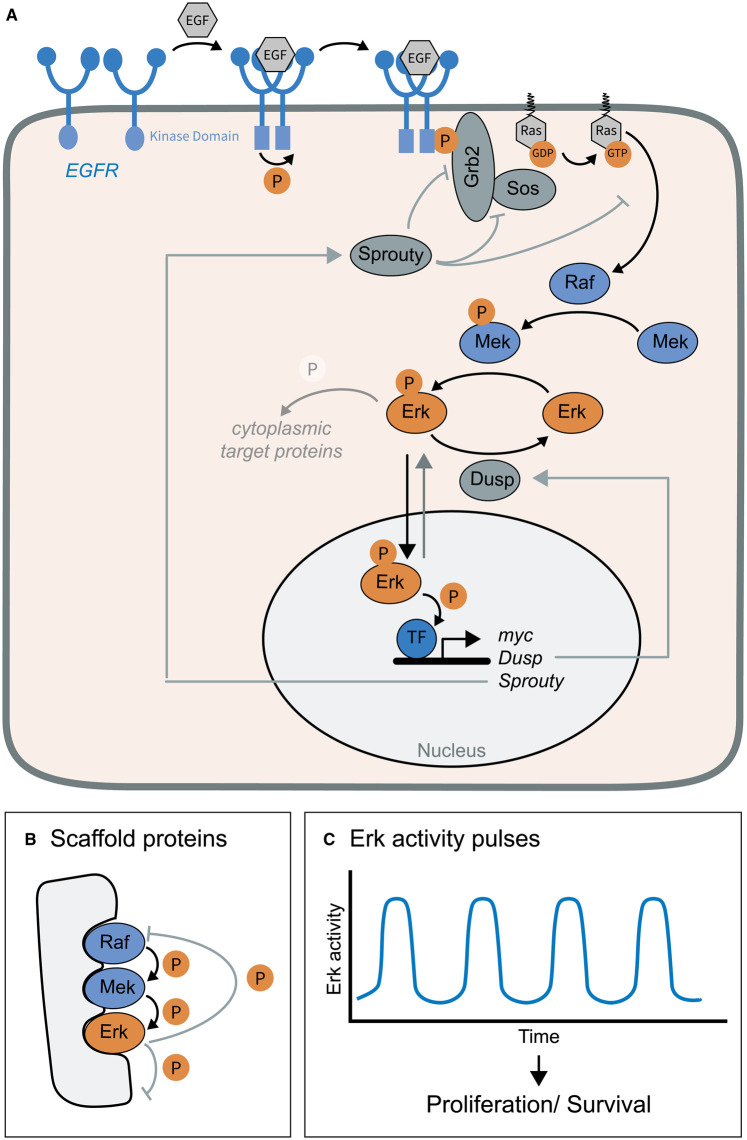
One of the main signal transduction pathways activated by growth factors is MAPK signalling. Growth factors such as EGF or FGF are soluble ligands that bind to receptor tyrosine kinases (RTKs) to mediate diverse functions within organisms, e.g. cell proliferation, survival or modification of cell metabolism [187,188]. Upon ligand–receptor interaction and receptor dimerization, the intracellular RTK kinase domain is activated and several tyrosine residues of the intracellular domain are phosphorylated in trans (Figure 5A) [189–191]. Phosphorylated tyrosines serve as docking sites for the recruitment of Shc, Grb2 and Sos [192]. Sos functions as a guanine nucleotide exchange factor to activate the small G protein Ras by exchanging GDP with GTP [192,193]. Ras-GTP then initiates a kinase cascade by activating the kinase Raf. Raf (a MAPK kinase kinase) phosphorylates and thereby activates the kinase MEK (a MAPK kinase), which in turn activates ERK (a MAPK) by phosphorylation. ERK as effector kinase of the pathway phosphorylates diverse proteins within the cell to change cellular behaviour. It also travels to the nucleus to mediate gene expression of target genes by phosphorylating transcription factors [188]. The activation process of Raf and its dynamics are modulated by the combinatorial usage of ligands and receptors, post-translational modifications, subcellular localization and endocytosis of the receptor complex, all of which is dependent on cell type and developmental stage, discussed in detail elsewhere [187,188]. Here we focus on the kinase cascade itself and its implications for signalling dynamics.
Figure 5. Erk signalling dynamics.
(A) When EGF interacts with a dimer of its receptor EGFR, a conformational change is induced, which results in the activation of the intracellular kinase domain. Phosphorylation in trans allows the recruitment of the adaptor Grb2 and the GEF Sos. Sos promotes the exchange of GDP with GTP in the small G protein Ras, which then activates the kinase Raf. Raf then phosphorylates and activates the kinase Mek, which in turn phosphorylates and activates the effector kinase Erk. Erk phosphorylates various cytoplasmic proteins and travels to the nucleus to activate the expression of target genes. Among these are the negative regulators Sprouty and the phosphatase Dusp. (B) Erk signalling is modulated by scaffold proteins, which make the kinase cascade more specific and efficient. In addition, feedback mechanisms involving the scaffold protein regulate the cascade further. For instance, the activated effector kinase phosphorylates and both the scaffold protein and Raf, which results in dissociation of the two proteins. (C) Erk activity pulses have been observed in various tissue types ranging from ∼15 to 90 min in period. In these tissues, pulses induce cell proliferation and promote cell survival.
Scaffold proteins in ERK signalling
Mechanistic details of Erk signalling have been studied extensively in human cell lines and in vitro. One regulatory component of MAPK signalling is the use of scaffold proteins, which consist of modular interaction domains that coordinate the three kinases of the MAPK cascade (Figure 5B) [194,195]. This has several consequences for ERK signalling: (1) By recruiting the kinases to the plasma membrane, in the vicinity to the activator Ras, the phosphorylation cascade can be activated efficiently. Furthermore, the cascade itself gets more specific and more efficient by bringing the right proteins in close proximity, as long as the ratio between scaffold and kinases is not too high. For instance, the scaffold protein KSR (kinase suppressor of Ras) binds Raf, MEK and ERK, all components of the ERK activation cascade [195]. Despite these advantages for cells, too stable interactions might limit the amplification of the MAPK cascade. (2) The scaffold has an allosteric effect on the phosphorylation process, for instance, on Raf kinase activation [196]. In addition, in a ternary complex consisting of KSR, B-Raf and MEK1 a conformational change in MEK1 promotes phosphorylation by a second B-Raf molecule [197]. (3) Scaffold proteins influence feedback mechanisms of the signalling pathway. For instance, when ERK bound to KSR gets activated, ERK phosphorylates both KSR and Raf, which induces the dissociation of KSR and Raf thereby inhibiting further amplification of the signal within short timeframes [198]. How scaffold proteins contribute to ERK signalling dynamics is not known. It is however conceivable that such proteins modulate the parameters of dynamics, such as timing, amplitude and duration dependent on the cell type and developmental stage. Their expression and dynamics should therefore be studied in the cellular context of ERK signalling dynamics.
ERK signalling dynamics
ERK signalling is driven and regulated by post-translational modifications. Not only phosphorylation, but also others such as ubiquitination, modify activity and stability of the involved proteins [199,200]. These modifications are largely reversible, since both modifying and reversing enzymes are expressed in the same cells, giving rise to complex signalling dynamics. In addition, the expression of activating and inactivating enzymes is induced by the ERK pathway, which generates positive and negative feedback loops [201]. Dynamics in ERK signalling have first been described in the PC12 cell line, in which both growth factors EGF and NGF activate ERK signalling, but with different dynamics and lead to different cellular outcomes, i.e. cell proliferation or differentiation, respectively (see above, Figure 1B) [17,202]. Using an optogenetic tool, Toettcher et al. induced ERK signalling by light with different dynamics, which confirmed that sustained signalling resulted in neuronal differentiation [203]. It has been suggested that EGF and NGF activate different signalling network topologies, thereby leading to different dynamics: EGF binds to the EGF receptor EGFR, while NGF binds to TrkA (Tropomyosine receptor kinase A). This way, EGF mainly induces a negative feedback loop. NGF induces a positive feedback loop via additional activation of PKC (protein kinase C), leading to sustained ERK signalling [204,205]. In fact, single-cell analyses indicated that the relationship between activated ERK and PKB (protein kinase B)/AKT determines cellular outcome [206–208].
ERK activity pulses
Over the last decade, several highly versatile ERK activity reporters have been generated that are based on the phosphorylation of fluorescent reporter constructs, which mediate a change in Foerster resonance energy transfer (FRET) or subcellular localization [209–211]. Such reporters have allowed the identification of ERK dynamics in various organisms and tissue types within their multicellular context. It turns out that ERK signalling is often pulsatile within a range of several minutes, presumably owing to the periodic phosphorylation and dephosphorylation dynamics (Figure 5C) [212]. In skin cells the period of ERK pulses varied between ∼30 min and 1.5 h in both mice and human cells, which was dependent on cell type and cultivation time [212]. The generation of pulses was dependent on the dual specificity phosphatases (DUSP) Dusp6 and Dusp10 [212]. A screen using more than 400 kinase inhibitors indicated the complex network and crosstalk between different signalling pathways influencing ERK dynamics [203,213]. Pulsatile ERK activity correlates with cell proliferation in skin cells [212]. In the context of blocked endogenous EGF signalling with an EGFR inhibitor, induced ERK pulses using optogenetics could recapitulate this proliferative effect [213]. Since continuous activation of ERK activity had a similar effect, it has to be investigated in future studies whether the dynamics or the cumulative activity of ERK determine cellular behaviour. Indeed, similar optogenetic modulation of ERK activity in Drosophila embryos showed that the cumulative dose of ERK activity defines the induced cell fate [214,215]. Moreover, in a two-dimensional layer of the canine kidney cell line MDCK, a wave of ERK activity is initiated around apoptotic cells, which promotes survival of surrounding cells, a phenomenon also found in Drosophila [216,217]. Optogenetic induction of ERK activity pulses showed that pulses with a period of at least 3–4 h could prevent apoptosis [217], which indicates that maintaining the cellular effect of ERK constantly at a sufficiently high level has a pro-survival effect. Similar ERK dynamics have been detected in other tissues such as intestinal organoids and lung cells [209,218,219], indicating that this might be a common phenomenon in epithelial tissue. The functional significance for embryonic development and tissue maintenance as well as the decoding mechanism have to be investigated in future studies.
ERK signalling oscillations
The periodic segmentation of mouse embryos is mediated by a network of oscillatory Wnt, Notch and FGF signalling (see above, Figure 4B,C). Several downstream targets of FGF/ERK signalling have been shown to be oscillatory with a period of 2.5 h in mice and 5 h in humans [87,177]. Periodic absence of FGF signalling in the region of segmentation with the concomitant presence of Notch activity allows the formation of a new somite [183]. Using a luciferase reporter based on the downstream target DUSP4, signalling waves of FGF signalling in the developing mouse embryo were observed [183]. In contrast, the dynamics of ERK activity itself have not been quantified using live reporters yet. However, oscillations of phosphorylated ERK (P-ERK) were detected by immunohistochemistry [183]. In the future, it will be important to analyse ERK dynamics within single cells at high temporal resolution using dynamic ERK activity reporters to determine whether ERK activity is also pulsatile in the segmenting embryo. This will allow the investigation of how ERK dynamics are translated into an oscillatory expression of ERK target genes.
Species-specific differences in signalling dynamics
Even though embryonic development of different mammalian organisms is strikingly similar in its molecular processes, developmental time greatly differs between species [220]. In two recent studies, this has been attributed to differences in biochemical parameters of the molecular processes and signalling events [33,221]. Pluripotent stem cells (PSCs) were either differentiated along neuromesodermal progenitors towards neural tube cells [221] or along oscillating PSM towards somitic cells [33]. In vitro formation of oscillating PSM cells confirmed previous findings that mouse cells oscillate with a period of ∼2.5 h, while in human cells the period is ∼5 h (Figure 6A) [33,80,84,87,222]. By swapping the human and mouse version of the cyclic gene Hes7, it was ruled out that the difference lay in the gene locus. In contrast, it turned out that several biochemical parameters of the delayed negative feedback loop generating oscillations differed (Figure 6B). Transcription, splicing and translation as well as protein degradation take approximately double the amount of time in human compared with mouse cells [33]. Equivalent differences in protein stability were found when differentiating PSCs towards neural tube cells [221]. The cause of this divergence in the timing of cellular processes remains elusive. The question of whether all biochemical reactions are equally changed — or only a subset of processes — will have direct implications on signalling dynamics in the different model organisms. For instance, ERK dynamics are mediated by periodic post-translational modifications on the one hand and gene expression of negative regulators on the other hand. A direct comparison of ERK dynamics can be performed by quantifying equivalent reporters for both ERK activity [209,211] and target gene expression [183] in equivalent mouse and human cells.
Figure 6. Species-specific differences in the timing of dynamics.
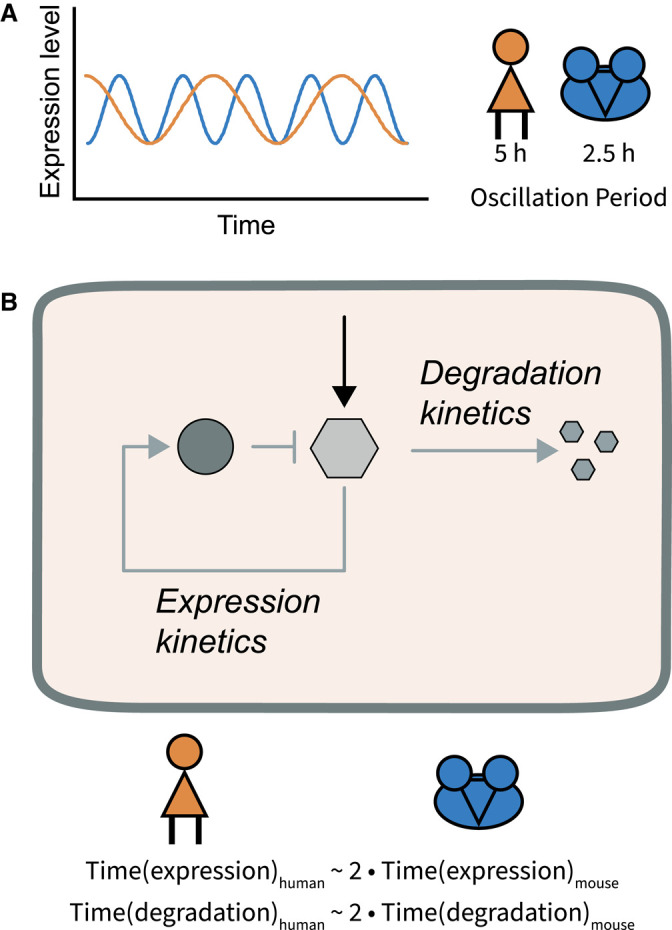
(A) Oscillations of the segmentation clock have a period of ∼5 h in humans and 2.5 h in mice. (B) Segmentation clock oscillations are thought to be generated by a delayed negative feedback loop, involving the induction of the expression of a negative regulator. It has been shown that the timing of expression (transcription, splicing and translation) and protein degradation take 2× longer in humans than in mice.
Perspective
Signalling pathways are intrinsically dynamic. Such dynamics have diverse functions in organisms, ranging from the control of cell proliferation and differentiation to the coordination of periodic events during embryonic development. Here, we have discussed the relevance and mechanisms of dynamics in Notch, Wnt and MAPK signalling. Other signalling pathways are equally important in embryonic development and dynamics have been detected in various contexts [10,223,224]. This indicates that signalling dynamics might be a common characteristic of cellular signalling.
Oscillatory signalling activity occurs in various pathways, model systems and tissue types. These oscillations seem to maintain cells in an undifferentiated and proliferative state. This suggests that signalling oscillations might keep cells in an undecided, but responsive state and allows cells to quickly adapt to changes. Only upon sustained expression of transcription factors, differentiation seems to be initiated. Some questions that emerge from this are the following: Are Erk pulses a general property of epithelial cells to control proliferation? How do dynamics change in the process of epithelial-to-mesenchymal transition and vice versa? How do tumorigenic mutations affect these dynamics [218,219]? During embryogenesis periodic segmentation is a common process often occurring in both vertebrates and invertebrates. Dynamic and oscillatory activity are found in several other segmenting species, the best-studied invertebrate being the beetle Tribolium [225–227]. Whether the mechanism of tissue-wide communication and induction of segments are similar remains to be determined.
With new technological developments, the mechanistic investigation of the role of signalling dynamics becomes amenable. (1) In vitro models of embryonic development and adult tissue homeostasis, including stem cell-based embryo-like structures and organoid models, that can be generated in high numbers, enable the study of signalling dynamics in accessible and simplified systems [228,229]. (2) Signalling reporters allow the spatiotemporally resolved quantification of signalling dynamics. Due to the functionally critical cellular heterogeneity within tissues and organisms, it is essential to analyse signalling within single cells. Signalling reporters either report on the transduction cascade itself, e.g. nuclear translocation or change in abundance of the effector protein [203], or are based on the expression of target genes [179]. To get a more comprehensive understanding of the signalling complexity, the different components of a pathway should be analysed in the same model system. This will allow the analysis of ligand–receptor interaction, the signal transduction cascade, cellular effects, target gene expression and feedback loops. For visualizing newly generated proteins, fast maturing fluorescent proteins or tagging systems should be used that are not dependent on fluorescent protein maturation, such as SNAP-tag or Llama-tag [230,231]. In addition, the correlation between mRNA and protein expression of target genes will be critical to account for species-to-species and isoform-to-isoform variations. In combination with advanced fluorescence microscopes that induce only low phototoxicity, signalling can be quantified at a high spatiotemporal resolution within multicellular systems [232,233]. (3) To reveal the complete picture of cellular signalling dynamics, it is essential to understand the abundance of various signalling components within a cell, as well as their interaction partners. With advances in omics approaches, this indeed becomes feasible. Bulk and single-cell transcriptomics have revealed cyclic genes in vertebrate segmentation [87,177,234]. However, since the transcriptome does not necessarily reflect protein expression, the proteome and phosphoproteome, ideally at a single-cell resolution, should be analysed in addition to revealing the signalling components in cells [24,235–237]. Such approaches will start to give a clear picture of how signalling dynamics are generated and how they control cellular behaviour. (4) To understand the function of signalling dynamics, we need tools to specifically modulate dynamics without altering overall signalling activity. The establishment of optogenetic and microfluidic approaches have enabled changing dynamics with high precision [179,203,238]. Systems to externally induce the degradation of selected proteins [239–241] will allow the dissection of how specific proteins, for instance negative feedback regulators, affect signalling dynamics. (5) Synthetic biology and bioengineering approaches then allow researchers to build intercellular signalling networks in vitro, modulate them and test predictions on the functionality of these networks [159,160,242].
To conclude, dynamic signalling is a common phenomenon in the animal kingdom. With technological advances we have started to reveal the functional significance of signalling dynamics in multicellular systems. This will allow us to study the role in different model systems to reveal general principles of its functioning. To dissect the molecular mechanism of how dynamics are generated and how they control cellular behaviour, it will be essential to transfer our understanding of signal transduction cascades at sub-cellular and molecular levels to multicellular systems and model systems of embryonic development. Moreover, the same signalling pathways, governing embryonic development, also control adult tissue homeostasis and mutations in signalling pathways induce and promote pathological conditions such as cancer [209,219,243,244]. It will be intriguing to compare how signalling dynamics modulate embryonic development, tissue homeostasis and cancer biology.
Acknowledgements
We thank Klaas Mulder, members of the Sonnen lab, in particular Sonja Weterings, Yasmine el Azhar and Marek van Oostrom, and the reviewers for feedback on the manuscript.
Abbreviations
- APC
adenomatous polyposis coli
- CSL
CBF1/RBPJ, Su(H), Lag-1
- DUSP
dual specificity phosphatases
- EGF
epidermal growth factor
- FGF
fibroblast growth factor
- GSK3
glycogen synthase kinase-3
- KSR
kinase suppressor of Ras
- LRP5
lipoprotein receptor-related protein 5
- MAM
mastermind
- MAPK
mitogen-activated protein kinase
- NECD
Notch extracellular domain
- NGF
nerve growth factor
- NICD
Notch intracellular domain
- PSCs
pluripotent stem cells
- PSM
presomitic mesoderm
- RSPO
R-spondin ligands
- TFs
transcription factors
Competing Interests
C.Y.J is a cofounder of Surrozen, Inc. K.F.S. has no conflict of interest to declare.
Funding
This work was partially funded through an ERC starting grant (no. 850554) to K.F.S.
References
- 1.Alon, U. (2007) Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450–461 10.1038/nrg2102 [DOI] [PubMed] [Google Scholar]
- 2.Sonnen, K.F. and Aulehla, A. (2014) Dynamic signal encoding–from cells to organisms. Semin. Cell Dev. Biol. 34, 91–98 10.1016/j.semcdb.2014.06.019 [DOI] [PubMed] [Google Scholar]
- 3.Seymour, P.A., Collin, C.A., Egeskov-Madsen, A.R., Jorgensen, M.C., Shimojo, H., Imayoshi, I.et al. (2020) Jag1 modulates an oscillatory Dll1-Notch-Hes1 signaling module to coordinate growth and fate of pancreatic progenitors. Dev. Cell 52, 731–47.e8 10.1016/j.devcel.2020.01.015 [DOI] [PubMed] [Google Scholar]
- 4.Imayoshi, I., Isomura, A., Harima, Y., Kawaguchi, K., Kori, H., Miyachi, H.et al. (2013) Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science 342, 1203–1208 10.1126/science.1242366 [DOI] [PubMed] [Google Scholar]
- 5.Shimojo, H., Ohtsuka, T. and Kageyama, R. (2008) Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58, 52–64 10.1016/j.neuron.2008.02.014 [DOI] [PubMed] [Google Scholar]
- 6.Wartlick, O., Kicheva, A. and González-Gaitán, M. (2009) Morphogen gradient formation. Cold Spring Harb. Perspect. Biol. 1, a001255 10.1101/cshperspect.a001255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stapornwongkul, K.S. and Vincent, J.-P. (2021) Generation of extracellular morphogen gradients: the case for diffusion. Nat. Rev. Genet. 22, 393–411 10.1038/s41576-021-00342-y [DOI] [PubMed] [Google Scholar]
- 8.Pires-daSilva, A. and Sommer, R.J. (2003) The evolution of signalling pathways in animal development. Nat. Rev. Genet. 4, 39–49 10.1038/nrg977 [DOI] [PubMed] [Google Scholar]
- 9.Freeman, M. (2000) Feedback control of intercellular signalling in development. Nature 408, 313–319 10.1038/35042500 [DOI] [PubMed] [Google Scholar]
- 10.Selimkhanov, J., Taylor, B., Yao, J., Pilko, A., Albeck, J., Hoffmann, A.et al. (2014) Accurate information transmission through dynamic biochemical signaling networks. Science 346, 1370–1373 10.1126/science.1254933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uda, S., Saito, T.H., Kudo, T., Kokaji, T., Tsuchiya, T., Kubota, H.et al. (2013) Robustness and compensation of information transmission of signaling pathways. Science 341, 558–561 10.1126/science.1234511 [DOI] [PubMed] [Google Scholar]
- 12.Goentoro, L. and Kirschner, M.W. (2009) Evidence that fold-change, and not absolute level, of beta-catenin dictates Wnt signaling. Mol. Cell 36, 872–884 10.1016/j.molcel.2009.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purvis, J.E. and Lahav, G. (2013) Encoding and decoding cellular information through signaling dynamics. Cell 152, 945–956 10.1016/j.cell.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, P. and Elowitz, M.B. (2019) Communication codes in developmental signaling pathways. Development 146, dev170977 10.1242/dev.170977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall, C.J. (1995) Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80, 179–185 10.1016/0092-8674(95)90401-8 [DOI] [PubMed] [Google Scholar]
- 16.Traverse, S., Gomez, N., Paterson, H., Marshall, C. and Cohen, P. (1992) Sustained activation of the mitogen-activated protein (MAP) kinase cascade may be required for differentiation of PC12 cells. Comparison of the effects of nerve growth factor and epidermal growth factor. Biochem. J. 288, 351–355 10.1042/bj2880351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greene, L.A. and Tischler, A.S. (1976) Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl Acad. Sci. U.S.A. 73, 2424–2428 10.1073/pnas.73.7.2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patke, A., Young, M.W. and Axelrod, S. (2020) Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 21, 67–84 10.1038/s41580-019-0179-2 [DOI] [PubMed] [Google Scholar]
- 19.Hubaud, A. and Pourquie, O. (2014) Signalling dynamics in vertebrate segmentation. Nat. Rev. Mol. Cell Biol. 15, 709–721 10.1038/nrm3891 [DOI] [PubMed] [Google Scholar]
- 20.Tsiairis, C. and Großhans, H. (2021) Gene expression oscillations in C. elegans underlie a new developmental clock. Curr. Top. Dev. Biol. 144, 19–43 10.1016/bs.ctdb.2020.11.001 [DOI] [PubMed] [Google Scholar]
- 21.Nelson, D.E., Ihekwaba, A.E.C., Elliott, M., Johnson, J.R., Gibney, C.A., Foreman, B.E.et al. (2004) Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science 306, 704–708 10.1126/science.1099962 [DOI] [PubMed] [Google Scholar]
- 22.Zambrano, S., De Toma, I., Piffer, A., Bianchi, M.E. and Agresti, A. (2016) NF-κB oscillations translate into functionally related patterns of gene expression. eLife 5, e09100 10.7554/eLife.09100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tay, S., Hughey, J.J., Lee, T.K., Lipniacki, T., Quake, S.R. and Covert, M.W. (2010) Single-cell NF-κB dynamics reveal digital activation and analog information processing in cells. Nature 466, 267–271 10.1038/nature09145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao, Y., Liu, X., Tang, B., Li, C., Kou, Z., Li, L.et al. (2017) Protein expression landscape of mouse embryos during pre-implantation development. Cell Rep. 21, 3957–3969 10.1016/j.celrep.2017.11.111 [DOI] [PubMed] [Google Scholar]
- 25.Israel, S., Ernst, M., Psathaki, O.E., Drexler, H.C.A., Casser, E., Suzuki, Y.et al. (2019) An integrated genome-wide multi-omics analysis of gene expression dynamics in the preimplantation mouse embryo. Sci. Rep. 9, 13356 10.1038/s41598-019-49817-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochsner, S.A., Abraham, D., Martin, K., Ding, W., McOwiti, A., Kankanamge, W.et al. (2019) The signaling pathways project, an integrated ‘omics knowledgebase for mammalian cellular signaling pathways. Sci. Data 6, 252 10.1038/s41597-019-0193-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramilowski, J.A., Goldberg, T., Harshbarger, J., Kloppmann, E., Lizio, M., Satagopam, V.P.et al. (2015) A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 6, 7866 10.1038/ncomms8866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kramer, I.M. (2015) Signal Transduction, Academic Press, 1104 p [Google Scholar]
- 29.Pranada, A.L., Metz, S., Herrmann, A., Heinrich, P.C. and Müller-Newen, G. (2004) Real time analysis of STAT3 nucleocytoplasmic shuttling. J. Biol. Chem. 279, 15114–15123 10.1074/jbc.M312530200 [DOI] [PubMed] [Google Scholar]
- 30.Lidke, D.S., Huang, F., Post, J.N., Rieger, B., Wilsbacher, J., Thomas, J.L.et al. (2010) ERK nuclear translocation is dimerization-independent but controlled by the rate of phosphorylation. J. Biol. Chem. 285, 3092–3102 10.1074/jbc.M109.064972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shamir, M., Bar-On, Y., Phillips, R. and Milo, R. (2016) Snapshot: timescales in cell biology. Cell 164, 1302.e1 10.1016/j.cell.2016.02.058 [DOI] [PubMed] [Google Scholar]
- 32.Harima, Y., Takashima, Y., Ueda, Y., Ohtsuka, T. and Kageyama, R. (2013) Accelerating the tempo of the segmentation clock by reducing the number of introns in the Hes7 gene. Cell Rep. 3, 1–7 10.1016/j.celrep.2012.11.012 [DOI] [PubMed] [Google Scholar]
- 33.Matsuda, M., Hayashi, H., Garcia-Ojalvo, J., Yoshioka-Kobayashi, K., Kageyama, R., Yamanaka, Y.et al. (2020) Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science 369, 1450–1455 10.1126/science.aba7668 [DOI] [PubMed] [Google Scholar]
- 34.Albeck, J.G., Mills, G.B. and Brugge, J.S. (2013) Frequency-Modulated pulses of ERK activity transmit quantitative proliferation signals. Mol. Cell 49, 249–261 10.1016/j.molcel.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cullen, P.J. and Steinberg, F. (2018) To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 19, 679–696 10.1038/s41580-018-0053-7 [DOI] [PubMed] [Google Scholar]
- 36.de Lau, W., Peng, W.C., Gros, P. and Clevers, H. (2014) The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 28, 305–316 10.1101/gad.235473.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandman, O. and Meyer, T. (2008) Feedback loops shape cellular signals in space and time. Science 322, 390–395 10.1126/science.1160617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pomerening, J.R., Kim, S.Y. and Ferrell, Jr, J.E. (2005) Systems-level dissection of the cell-cycle oscillator: bypassing positive feedback produces damped oscillations. Cell 122, 565–578 10.1016/j.cell.2005.06.016 [DOI] [PubMed] [Google Scholar]
- 39.Bray, S.J. (2016) Notch signalling in context. Nat. Rev. Mol. Cell Biol. 17, 722–735 10.1038/nrm.2016.94 [DOI] [PubMed] [Google Scholar]
- 40.Logeat, F., Bessia, C., Brou, C., LeBail, O., Jarriault, S., Seidah, N.G.et al. (1998) The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl Acad. Sci. U.S.A. 95, 8108–8112 10.1073/pnas.95.14.8108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovall, R.A., Gebelein, B., Sprinzak, D. and Kopan, R. (2017) The canonical Notch signaling pathway: structural and biochemical insights into shape, sugar, and force. Dev. Cell 41, 228–241 10.1016/j.devcel.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon, W.R., Zimmerman, B., He, L., Miles, L.J., Huang, J., Tiyanont, K.et al. (2015) Mechanical allostery: evidence for a force requirement in the proteolytic activation of Notch. Dev. Cell 33, 729–736 10.1016/j.devcel.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parks, A.L., Klueg, K.M., Stout, J.R. and Muskavitch, M.A. (2000) Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development 127, 1373–1385 10.1242/dev.127.7.1373 [DOI] [PubMed] [Google Scholar]
- 44.Brou, C., Logeat, F., Gupta, N., Bessia, C., LeBail, O., Doedens, J.R.et al. (2000) A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216 10.1016/S1097-2765(00)80417-7 [DOI] [PubMed] [Google Scholar]
- 45.De Strooper, B., Annaert, W., Cupers, P., Saftig, P., Craessaerts, K., Mumm, J.S.et al. (1999) A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 10.1038/19083 [DOI] [PubMed] [Google Scholar]
- 46.Petcherski, A.G. and Kimble, J. (2000) Mastermind is a putative activator for Notch. Curr. Biol. 10, R471–R473 10.1016/S0960-9822(00)00577-7 [DOI] [PubMed] [Google Scholar]
- 47.Wu, L., Aster, J.C., Blacklow, S.C., Lake, R., Artavanis-Tsakonas, S. and Griffin, J.D. (2000) MAML1, a human homologue of Drosophila Mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26, 484–489 10.1038/82644 [DOI] [PubMed] [Google Scholar]
- 48.Schweisguth, F. and Posakony, J.W. (1992) Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell 69, 1199–1212 10.1016/0092-8674(92)90641-O [DOI] [PubMed] [Google Scholar]
- 49.Weng, A.P. (2006) c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 15, 2096–2109 10.1101/gad.1450406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamar, E., Deblandre, G., Wettstein, D., Gawantka, V., Pollet, N., Niehrs, C.et al. (2001) Nrarp is a novel intracellular component of the Notch signaling pathway. Genes Dev. 15, 1885–1899 10.1101/gad.908101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jarriault, S., Le Bail, O., Hirsinger, E., Pourquié, O., Logeat, F., Strong, C.F.et al. (1998) Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol. Cell. Biol. 18, 7423–7431 10.1128/MCB.18.12.7423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palomero, T., Lim, W.K., Odom, D.T., Sulis, M.L., Real, P.J., Margolin, A.et al. (2006) NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl Acad. Sci. U.S.A. 103, 18261–6 10.1073/pnas.0606108103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronchini, C. and Capobianco, A.J. (2001) Induction of cyclin D1 transcription and CDK2 activity by Notch ic: implication for cell cycle disruption in transformation by Notch ic. Mol. Cell. Biol. 21, 5925–5934 10.1128/MCB.21.17.5925-5934.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fryer, C.J., White, J.B. and Jones, K.A. (2004) Mastermind recruits cycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16, 509–520 10.1016/j.molcel.2004.10.014 [DOI] [PubMed] [Google Scholar]
- 55.Oberg, C., Li, J., Pauley, A., Wolf, E., Gurney, M. and Lendahl, U. (2001) The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 276, 35847–35853 10.1074/jbc.M103992200 [DOI] [PubMed] [Google Scholar]
- 56.Carrieri, F.A., Murray, P.J., Ditsova, D., Ferris, M.A., Davies, P. and Dale, J.K. (2019) CDK 1 and CDK 2 regulate NICD 1 turnover and the periodicity of the segmentation clock. EMBO Rep. 20, e46436 10.15252/embr.201846436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langridge, P.D. and Struhl, G. (2017) Epsin-dependent ligand endocytosis activates Notch by force. Cell 171, 1383–96.e12 10.1016/j.cell.2017.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tagami, S., Okochi, M., Yanagida, K., Ikuta, A., Fukumori, A., Matsumoto, N.et al. (2008) Regulation of Notch signaling by dynamic changes in the precision of S3 cleavage of Notch-1. Mol. Cell. Biol. 28, 165–176 10.1128/MCB.00863-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bachmair, A., Finley, D. and Varshavsky, A. (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 10.1126/science.3018930 [DOI] [PubMed] [Google Scholar]
- 60.Micchelli, C.A., Rulifson, E.J. and Blair, S.S. (1997) The function and regulation of cut expression on the wing margin of Drosophila: Notch, wingless and a dominant negative role for delta and serrate. Development 124, 1485–1495 10.1242/dev.124.8.1485 [DOI] [PubMed] [Google Scholar]
- 61.de Celis, J.F, Celis de, J.F. and Bray, S. (1997) Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development 124, 3241–3251 10.1242/dev.124.17.3241 [DOI] [PubMed] [Google Scholar]
- 62.Couturier, L., Mazouni, K. and Schweisguth, F. (2013) Numb localizes at endosomes and controls the endosomal sorting of notch after asymmetric division in Drosophila. Curr. Biol. 23, 588–593 10.1016/j.cub.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 63.Couturier, L., Vodovar, N. and Schweisguth, F. (2012) Endocytosis by Numb breaks Notch symmetry at cytokinesis. Nat. Cell Biol. 14, 131–139 10.1038/ncb2419 [DOI] [PubMed] [Google Scholar]
- 64.Sprinzak, D., Lakhanpal, A., Lebon, L., Santat, L.A., Fontes, M.E., Anderson, G.A.et al. (2010) Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 465, 86–90 10.1038/nature08959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cubas, P., de Celis, J.F., Campuzano, S. and Modolell, J. (1991) Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal wing disc. Genes Dev. 5, 996–1008 10.1101/gad.5.6.996 [DOI] [PubMed] [Google Scholar]
- 66.Skeath, J.B. and Carroll, S.B. (1991) Regulation of achaete-scute gene expression and sensory organ pattern formation in the Drosophila wing. Genes Dev. 5, 984–995 10.1101/gad.5.6.984 [DOI] [PubMed] [Google Scholar]
- 67.Corson, F., Couturier, L., Rouault, H., Mazouni, K. and Schweisguth, F. (2017) Self-organized Notch dynamics generate stereotyped sensory organ patterns in Drosophila. Mech. Dev. 145, S10 10.1016/j.mod.2017.04.549 [DOI] [PubMed] [Google Scholar]
- 68.Crosnier, C., Vargesson, N., Gschmeissner, S., Ariza-McNaughton, L., Morrison, A. and Lewis, J. (2005) Delta-Notch signalling controls commitment to a secretory fate in the zebrafish intestine. Development 132, 1093–1104 10.1242/dev.01644 [DOI] [PubMed] [Google Scholar]
- 69.van Es, J.H., van Gijn, M.E., Riccio, O., van den Born, M., Vooijs, M., Begthel, H.et al. (2005) Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963 10.1038/nature03659 [DOI] [PubMed] [Google Scholar]
- 70.Krejcí, A. and Bray, S. (2007) Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 21, 1322–1327 10.1101/gad.424607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weerkamp, F., Luis, T.C., Naber, B.A.E., Koster, E.E.L., Jeannotte, L., van Dongen, J.J.M.et al. (2006) Identification of Notch target genes in uncommitted T-cell progenitors: no direct induction of a T-cell specific gene program. Leukemia 20, 1967–1977 10.1038/sj.leu.2404396 [DOI] [PubMed] [Google Scholar]
- 72.Gawantka, V., Pollet, N., Delius, H., Vingron, M., Pfister, R., Nitsch, R.et al. (1998) Gene expression screening in Xenopus identifies molecular pathways, predicts gene function and provides a global view of embryonic patterning. Mech. Dev. 77, 95–141 10.1016/S0925-4773(98)00115-4 [DOI] [PubMed] [Google Scholar]
- 73.Kobayashi, T. and Kageyama, R. (2010) Hes1 regulates embryonic stem cell differentiation by suppressing Notch signaling. Genes Cells 15, 689–698 10.1111/j.1365-2443.2010.01413.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ochi, S., Imaizumi, Y., Shimojo, H., Miyachi, H. and Kageyama, R. (2020) Oscillatory expression of Hes1 regulates cell proliferation and neuronal differentiation in the embryonic brain. Development 147, dev182204 10.1242/dev.182204 [DOI] [PubMed] [Google Scholar]
- 75.Manning, C.S., Biga, V., Boyd, J., Kursawe, J., Ymisson, B., Spiller, D.G.et al. (2019) Quantitative single-cell live imaging links HES5 dynamics with cell-state and fate in murine neurogenesis. Nat. Commun. 10, 1–19 10.1038/s41467-018-07882-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Biga, V., Hawley, J., Soto, X., Johns, E., Han, D., Bennett, H.et al. (2021) A dynamic, spatially periodic, micro-pattern of HES5 underlies neurogenesis in the mouse spinal cord. Mol. Syst. Biol. 17, e9902 10.15252/msb.20209902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson, H.E. and Toettcher, J.E. (2018) Illuminating developmental biology with cellular optogenetics. Curr. Opin. Biotechnol. 52, 42–48 10.1016/j.copbio.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pourquié, O. (2011) Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell 145, 650–663 10.1016/j.cell.2011.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hubaud, A., Regev, I., Mahadevan, L. and Pourquié, O. (2017) Excitable dynamics and Yap-dependent mechanical cues drive the segmentation clock. Cell 171, 668–682.e11 10.1016/j.cell.2017.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Masamizu, Y., Ohtsuka, T., Takashima, Y., Nagahara, H., Takenaka, Y., Yoshikawa, K.et al. (2006) Real-time imaging of the somite segmentation clock: revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc. Natl Acad. Sci. U.S.A. 103, 1313–1318 10.1073/pnas.0508658103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Webb, A.B., Lengyel, I.M., Jörg, D.J., Valentin, G., Jülicher, F., Morelli, L.G.et al. (2016) Persistence, period and precision of autonomous cellular oscillators from the zebrafish segmentation clock. eLife 5, e08438 10.7554/eLife.08438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Riedel-Kruse, I.H., Müller, C. and Oates, A.C. (2007) Synchrony dynamics during initiation, failure, and rescue of the segmentation clock. Science 317, 1911–1915 10.1126/science.1142538 [DOI] [PubMed] [Google Scholar]
- 83.Tsiairis, C.D. and Aulehla, A. (2016) Self-organization of embryonic genetic oscillators into spatiotemporal wave patterns. Cell 164, 656–667 10.1016/j.cell.2016.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Diaz-Cuadros, M., Wagner, D.E., Budjan, C., Hubaud, A., Tarazona, O.A., Donelly, S.et al. (2020) In vitro characterization of the human segmentation clock. Nature 580, 113–118 10.1038/s41586-019-1885-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shimojo, H., Isomura, A., Ohtsuka, T., Kori, H., Miyachi, H. and Kageyama, R. (2016) Oscillatory control of Delta-like1 in cell interactions regulates dynamic gene expression and tissue morphogenesis. Genes Dev. 30, 102–116 10.1101/gad.270785.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bessho, Y. (2003) Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 17, 1451–1456 10.1101/gad.1092303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matsuda, M., Yamanaka, Y., Uemura, M., Osawa, M., Saito, M.K., Nagahashi, A.et al. (2020) Recapitulating the human segmentation clock with pluripotent stem cells. Nature 580, 124–129 10.1038/s41586-020-2144-9 [DOI] [PubMed] [Google Scholar]
- 88.Oates, A.C. and Ho, R.K. (2002) Hairy/E(spl)-related (Her) genes are central components of the segmentation oscillator and display redundancy with the Delta/Notch signaling pathway in the formation of anterior segmental boundaries in the zebrafish. Development 129, 2929–2946 10.1242/dev.129.12.2929 [DOI] [PubMed] [Google Scholar]
- 89.Henry, C.A., Urban, M.K., Dill, K.K., Merlie, J.P., Page, M.F., Kimmel, C.B.et al. (2002) Two linked hairy/Enhancer of split-related zebrafish genes, her1 and her7, function together to refine alternating somite boundaries. Development 129, 3693–3704 10.1242/dev.129.15.3693 [DOI] [PubMed] [Google Scholar]
- 90.Hirata, H., Bessho, Y., Kokubu, H., Masamizu, Y., Yamada, S., Lewis, J.et al. (2004) Instability of Hes7 protein is crucial for the somite segmentation clock. Nat. Genet. 36, 750–754 10.1038/ng1372 [DOI] [PubMed] [Google Scholar]
- 91.Soza-Ried, C., Ozturk, E., Ish-Horowicz, D. and Lewis, J. (2014) Pulses of Notch activation synchronise oscillating somite cells and entrain the zebrafish segmentation clock. Development 141, 1780–1788 10.1242/dev.102111 [DOI] [PubMed] [Google Scholar]
- 92.Luca, V.C., Jude, K.M., Pierce, N.W., Nachury, M.V., Fischer, S. and Garcia, K.C. (2015) Structural biology. Structural basis for Notch1 engagement of Delta-like 4. Science 347, 847–853 10.1126/science.1261093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Luca, V.C., Kim, B.C., Ge, C., Kakuda, S., Wu, D., Roein-Peikar, M.et al. (2017) Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science 355, 1320–1324 10.1126/science.aaf9739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weisshuhn, P.C., Sheppard, D., Taylor, P., Whiteman, P., Lea, S.M., Handford, P.A.et al. (2016) Non-linear and flexible regions of the human Notch1 extracellular domain revealed by high-resolution structural studies. Structure 24, 555–566 10.1016/j.str.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Andrawes, M.B., Xu, X., Liu, H., Ficarro, S.B., Marto, J.A., Aster, J.C.et al. (2013) Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1. J. Biol. Chem. 288, 25477–25489 10.1074/jbc.M113.454850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nandagopal, N., Santat, L.A., LeBon, L., Sprinzak, D., Bronner, M.E. and Elowitz, M.B. (2018) Dynamic ligand discrimination in the Notch signaling pathway. Cell 172, 869–880.e19 10.1016/j.cell.2018.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harvey, B.M., Rana, N.A., Moss, H., Leonardi, J., Jafar-Nejad, H. and Haltiwanger, R.S. (2016) Mapping sites of O-Glycosylation and fringe elongation on Drosophila Notch. J. Biol. Chem. 291, 16348–16360 10.1074/jbc.M116.732537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pandey, A., Harvey, B.M., Lopez, M.F., Ito, A., Haltiwanger, R.S. and Jafar-Nejad, H. (2019) Glycosylation of specific Notch EGF repeats by O-Fut1 and fringe regulates Notch signaling in Drosophila. Cell Rep. 29, 2054–2066.e6 10.1016/j.celrep.2019.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kakuda, S. and Haltiwanger, R.S. (2017) Deciphering the fringe-Mediated Notch code: identification of activating and inhibiting sites allowing discrimination between ligands. Dev. Cell 40, 193–201 10.1016/j.devcel.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aulehla, A. and Johnson, R.L. (1999) Dynamic expression of lunatic fringe suggests a link between notch signaling and an autonomous cellular oscillator driving somite segmentation. Dev. Biol. 207, 49–61 10.1006/dbio.1998.9164 [DOI] [PubMed] [Google Scholar]
- 101.Evrard, Y.A., Lun, Y., Aulehla, A. and Gan, L. (1998) Johnson RL. lunatic fringe is an essential mediator of somite segmentation and patterning. Nature 394, 377–381 10.1038/28632 [DOI] [PubMed] [Google Scholar]
- 102.Yoshioka-Kobayashi, K., Matsumiya, M., Niino, Y., Isomura, A., Kori, H., Miyawaki, A.et al. (2020) Coupling delay controls synchronized oscillation in the segmentation clock. Nature 580, 119–123 10.1038/s41586-019-1882-z [DOI] [PubMed] [Google Scholar]
- 103.Morsut, L., Roybal, K.T., Xiong, X., Gordley, R.M., Coyle, S.M., Thomson, M.et al. (2016) Engineering customized cell sensing and response behaviors using synthetic Notch receptors. Cell 164, 780–791 10.1016/j.cell.2016.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nusse, R. and Clevers, H. (2017) Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 105.Steinhart, Z. and Angers, S. (2018) Wnt signaling in development and tissue homeostasis. Development 145, dev146589 10.1242/dev.146589 [DOI] [PubMed] [Google Scholar]
- 106.Clevers, H., Loh, K.M. and Nusse, R. (2014) Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012 10.1126/science.1248012 [DOI] [PubMed] [Google Scholar]
- 107.van Amerongen, R. (2012) Alternative Wnt pathways and receptors. Cold Spring Harb. Perspect. Biol. 4, a007914 10.1101/cshperspect.a007914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stamos, J.L. and Weis, W.I. (2013) The β-catenin destruction complex. Cold Spring Harb. Perspect. Biol. 5, a007898 10.1101/cshperspect.a007898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Munemitsu, S., Albert, I., Souza, B., Rubinfeld, B. and Polakis, P. (1995) Regulation of intracellular beta-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc. Natl Acad. Sci. U.S.A. 92, 3046–3050 10.1073/pnas.92.7.3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rubinfeld, B., Souza, B., Albert, I., Müller, O., Chamberlain, S.H., Masiarz, F.R.et al. (1993) Association of the APC gene product with beta-catenin. Science 262, 1731–1734 10.1126/science.8259518 [DOI] [PubMed] [Google Scholar]
- 111.Behrens, J., Jerchow, B.A., Würtele, M., Grimm, J., Asbrand, C., Wirtz, R.et al. (1998) Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science 280, 596–599 10.1126/science.280.5363.596 [DOI] [PubMed] [Google Scholar]
- 112.Hart, M.J., de los Santos, R., Albert, I.N., Rubinfeld, B. and Polakis, P. (1998) Downregulation of beta-catenin by human axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr. Biol. 8, 573–581 10.1016/S0960-9822(98)70226-X [DOI] [PubMed] [Google Scholar]
- 113.Amit, S., Hatzubai, A., Birman, Y., Andersen, J.S., Ben-Shushan, E., Mann, M.et al. (2002) Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 16, 1066–1076 10.1101/gad.230302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rubinfeld, B., Albert, I., Porfiri, E., Fiol, C., Munemitsu, S. and Polakis, P. (1996) Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science 272, 1023–1026 10.1126/science.272.5264.1023 [DOI] [PubMed] [Google Scholar]
- 115.Liu, C., Li, Y., Semenov, M., Han, C., Baeg, G.H., Tan, Y.et al. (2002) Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108, 837–847 10.1016/S0092-8674(02)00685-2 [DOI] [PubMed] [Google Scholar]
- 116.Winston, J.T., Strack, P., Beer-Romero, P., Chu, C.Y., Elledge, S.J. and Harper, J.W. (1999) The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in ikappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 13, 270–283 10.1101/gad.13.3.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aberle, H., Bauer, A., Stappert, J., Kispert, A. and Kemler, R. (1997) beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 16, 3797–3804 10.1093/emboj/16.13.3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hart, M., Concordet, J.P., Lassot, I., Albert, I., del los Santos, R., Durand, H.et al. (1999) The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr. Biol. 9, 207–210 10.1016/S0960-9822(99)80091-8 [DOI] [PubMed] [Google Scholar]
- 119.Klingensmith, J., Nusse, R. and Perrimon, N. (1994) The Drosophila segment polarity gene dishevelled encodes a novel protein required for response to the wingless signal. Genes Dev. 8, 118–130 10.1101/gad.8.1.118 [DOI] [PubMed] [Google Scholar]
- 120.Cong, F., Schweizer, L. and Varmus, H. (2004) Wnt signals across the plasma membrane to activate the beta-catenin pathway by forming oligomers containing its receptors, frizzled and LRP. Development 131, 5103–5115 10.1242/dev.01318 [DOI] [PubMed] [Google Scholar]
- 121.Tauriello, D.V.F., Jordens, I., Kirchner, K., Slootstra, J.W., Kruitwagen, T., Bouwman, B.A.M.et al. (2012) Wnt/β-catenin signaling requires interaction of the dishevelled DEP domain and C terminus with a discontinuous motif in frizzled. Proc. Natl Acad. Sci. U.S.A. 109, E812–E820 10.1073/pnas.1114802109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tamai, K., Zeng, X., Liu, C., Zhang, X., Harada, Y., Chang, Z.et al. (2004) A mechanism for Wnt coreceptor activation. Mol. Cell 13, 149–156 10.1016/S1097-2765(03)00484-2 [DOI] [PubMed] [Google Scholar]
- 123.Mao, J., Wang, J., Liu, B., Pan, W., Farr, III, G.H., Flynn, C.et al. (2001) Low-density lipoprotein receptor-related protein-5 binds to axin and regulates the canonical Wnt signaling pathway. Mol. Cell 7, 801–809 10.1016/S1097-2765(01)00224-6 [DOI] [PubMed] [Google Scholar]
- 124.Stamos, J.L., Chu, M.L.-H., Enos, M.D., Shah, N. and Weis, W.I. (2014) Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6. eLife 3, e01998 10.7554/eLife.01998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Piao, S., Lee, S.-H., Kim, H., Yum, S., Stamos, J.L., Xu, Y.et al. (2008) Direct inhibition of GSK3beta by the phosphorylated cytoplasmic domain of LRP6 in Wnt/beta-catenin signaling. PLoS ONE 3, e4046 10.1371/journal.pone.0004046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Molenaar, M., van de Wetering, M., Oosterwegel, M., Peterson-Maduro, J., Godsave, S., Korinek, V.et al. (1996) XTcf-3 transcription factor mediates beta-catenin-induced axis formation in xenopus embryos. Cell 86, 391–399 10.1016/S0092-8674(00)80112-9 [DOI] [PubMed] [Google Scholar]
- 127.Behrens, J., von Kries, J.P., Kühl, M., Bruhn, L., Wedlich, D., Grosschedl, R.et al. (1996) Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382, 638–642 10.1038/382638a0 [DOI] [PubMed] [Google Scholar]
- 128.van de Wetering, M., Cavallo, R., Dooijes, D., van Beest, M., van Es, J., Loureiro, J.et al. (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88, 789–799 10.1016/S0092-8674(00)81925-X [DOI] [PubMed] [Google Scholar]
- 129.Kemler, R. (1993) From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends Genet. 9, 317–321 10.1016/0168-9525(93)90250-L [DOI] [PubMed] [Google Scholar]
- 130.van der Wal, T. and van Amerongen, R. (2020) Walking the tight wire between cell adhesion and WNT signalling: a balancing act for beta-catenin. Open Biol. 10, 200267 10.1098/rsob.200267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sieiro, D., Rios, A.C., Hirst, C.E. and Marcelle, C. (2016) Cytoplasmic NOTCH and membrane-derived beta-catenin link cell fate choice to epithelial-mesenchymal transition during myogenesis. eLife 5, e14847 10.7554/eLife.14847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Willert, K., Brown, J.D., Danenberg, E., Duncan, A.W., Weissman, I.L., Reya, T.et al. (2003) Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423, 448–452 10.1038/nature01611 [DOI] [PubMed] [Google Scholar]
- 133.Takada, R., Satomi, Y., Kurata, T., Ueno, N., Norioka, S., Kondoh, H.et al. (2006) Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev. Cell 11, 791–801 10.1016/j.devcel.2006.10.003 [DOI] [PubMed] [Google Scholar]
- 134.Bänziger, C., Soldini, D., Schütt, C., Zipperlen, P., Hausmann, G. and Basler, K. (2006) Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 125, 509–522 10.1016/j.cell.2006.02.049 [DOI] [PubMed] [Google Scholar]
- 135.Bartscherer, K., Pelte, N., Ingelfinger, D. and Boutros, M. (2006) Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell 125, 523–533 10.1016/j.cell.2006.04.009 [DOI] [PubMed] [Google Scholar]
- 136.Goodman, R.M., Thombre, S., Firtina, Z., Gray, D., Betts, D., Roebuck, J.et al. (2006) Sprinter: a novel transmembrane protein required for Wg secretion and signaling. Development 133, 4901–4911 10.1242/dev.02674 [DOI] [PubMed] [Google Scholar]
- 137.Janda, C.Y., Waghray, D., Levin, A.M., Thomas, C. and Garcia, K.C. (2012) Structural basis of Wnt recognition by frizzled. Science 337, 59–64 10.1126/science.1222879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Langton, P.F., Kakugawa, S. and Vincent, J.P. (2016) Making, exporting, and modulating wnts. Trends Cell Biol. 26, 756–765 10.1016/j.tcb.2016.05.011 [DOI] [PubMed] [Google Scholar]
- 139.Gross, J.C. and Boutros, M. (2013) Secretion and extracellular space travel of Wnt proteins. Curr. Opin. Genet. Dev. 23, 385–390 10.1016/j.gde.2013.02.017 [DOI] [PubMed] [Google Scholar]
- 140.Farin, H.F., Jordens, I., Mosa, M.H., Basak, O., Korving, J., Tauriello, D.V.F.et al. (2016) Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 10.1038/nature16937 [DOI] [PubMed] [Google Scholar]
- 141.Lewis, S.L., Khoo, P.-L., De Young, R.A., Steiner, K., Wilcock, C., Mukhopadhyay, M.et al. (2008) Dkk1 and Wnt3 interact to control head morphogenesis in the mouse. Development 135, 1791–1801 10.1242/dev.018853 [DOI] [PubMed] [Google Scholar]
- 142.Glinka, A., Wu, W., Delius, H., Monaghan, A.P., Blumenstock, C. and Niehrs, C. (1998) Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 391, 357–362 10.1038/34848 [DOI] [PubMed] [Google Scholar]
- 143.Bafico, A., Liu, G., Yaniv, A., Gazit, A. and Aaronson, S.A. (2001) Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat. Cell Biol. 3, 683–686 10.1038/35083081 [DOI] [PubMed] [Google Scholar]
- 144.Semënov, M.V., Tamai, K., Brott, B.K., Kühl, M., Sokol, S. and He, X. (2001) Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr. Biol. 11, 951–961 10.1016/S0960-9822(01)00290-1 [DOI] [PubMed] [Google Scholar]
- 145.Semënov, M., Tamai, K. and He, X. (2005) SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J. Biol. Chem. 280, 26770–26775 10.1074/jbc.M504308200 [DOI] [PubMed] [Google Scholar]
- 146.Hsieh, J.C., Kodjabachian, L., Rebbert, M.L., Rattner, A., Smallwood, P.M., Samos, C.H.et al. (1999) A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature 398, 431–436 10.1038/18899 [DOI] [PubMed] [Google Scholar]
- 147.Kakugawa, S., Langton, P.F., Zebisch, M., Howell, S., Chang, T.-H., Liu, Y.et al. (2015) Notum deacylates Wnt proteins to suppress signalling activity. Nature 519, 187–192 10.1038/nature14259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang, X., Cheong, S.-M., Amado, N.G., Reis, A.H., MacDonald, B.T., Zebisch, M.et al. (2015) Notum is required for neural and head induction via Wnt deacylation, oxidation, and inactivation. Dev. Cell 32, 719–730 10.1016/j.devcel.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jho, E.-H., Zhang, T., Domon, C., Joo, C.-K., Freund, J.-N. and Costantini, F. (2002) Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22, 1172–1183 10.1128/MCB.22.4.1172-1183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lustig, B., Jerchow, B., Sachs, M., Weiler, S., Pietsch, T., Karsten, U.et al. (2002) Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 22, 1184–1193 10.1128/MCB.22.4.1184-1193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hao, H.-X., Xie, Y., Zhang, Y., Charlat, O., Oster, E., Avello, M.et al. (2012) ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–200 10.1038/nature11019 [DOI] [PubMed] [Google Scholar]
- 152.Koo, B.-K., Spit, M., Jordens, I., Low, T.Y., Stange, D.E., van de Wetering, M.et al. (2012) Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488, 665–669 10.1038/nature11308 [DOI] [PubMed] [Google Scholar]
- 153.de Lau, W., Barker, N., Low, T.Y., Koo, B.-K., Li, V.S.W., Teunissen, H.et al. (2011) Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 10.1038/nature10337 [DOI] [PubMed] [Google Scholar]
- 154.Yan, K.S., Janda, C.Y., Chang, J., Zheng, G.X.Y., Larkin, K.A., Luca, V.C.et al. (2017) Non-equivalence of Wnt and R-spondin ligands during Lgr5 intestinal stem-cell self-renewal. Nature 545, 238–242 10.1038/nature22313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dijksterhuis, J.P., Baljinnyam, B., Stanger, K., Sercan, H.O., Ji, Y., Andres, O.et al. (2015) Systematic mapping of WNT-FZD protein interactions reveals functional selectivity by distinct WNT-FZD pairs. J. Biol. Chem. 290, 6789–6798 10.1074/jbc.M114.612648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Voloshanenko, O., Gmach, P., Winter, J., Kranz, D. and Boutros, M. (2017) Mapping of Wnt-Frizzled interactions by multiplex CRISPR targeting of receptor gene families. FASEB J. 31, 4832–4844 10.1096/fj.201700144R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Clevers, H. (2016) Modeling development and disease with organoids. Cell 165, 1586–1597 10.1016/j.cell.2016.05.082 [DOI] [PubMed] [Google Scholar]
- 158.Alok, A., Lei, Z., Jagannathan, N.S., Kaur, S., Harmston, N., Rozen, S.G.et al. (2017) Wnt proteins synergize to activate β-catenin signaling. J. Cell Sci. 130, 1532–1544 10.1242/jcs.198093 [DOI] [PubMed] [Google Scholar]
- 159.Janda, C.Y., Dang, L.T., You, C., Chang, J., de Lau, W., Zhong, Z.A.et al. (2017) Surrogate Wnt agonists that phenocopy canonical Wnt and β-catenin signalling. Nature 545, 234–237 10.1038/nature22306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Miao, Y., Ha, A., de Lau, W., Yuki, K., Santos, A.J.M., You, C.et al. (2020) Next-Generation surrogate wnts support organoid growth and deconvolute frizzled pleiotropy in vivo. Cell Stem Cell 27, 840–851.e6 10.1016/j.stem.2020.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tsutsumi, N., Mukherjee, S., Waghray, D., Janda, C.Y., Jude, K.M., Miao, Y.et al. (2020) Structure of human Frizzled5 by fiducial-assisted cryo-EM supports a heterodimeric mechanism of canonical Wnt signaling. eLife 9, e58464 10.7554/eLife.58464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wesslowski, J., Kozielewicz, P., Wang, X., Cui, H., Schihada, H., Kranz, D.et al. (2020) eGFP-tagged Wnt-3a enables functional analysis of Wnt trafficking and signaling and kinetic assessment of Wnt binding to full-length Frizzled. J. Biol. Chem. 295, 8759–8774 10.1074/jbc.RA120.012892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen, H., Lu, C., Ouyang, B., Zhang, H., Huang, Z., Bhatia, D.et al. (2020) Development of potent, selective surrogate WNT molecules and their application in defining frizzled requirements. Cell Chem. Biol. 27, 598–609.e4 10.1016/j.chembiol.2020.02.009 [DOI] [PubMed] [Google Scholar]
- 164.Mukherjee, A., Stathos, M.E., Varner, C., Arsiwala, A., Frey, S., Hu, Y.et al. (2020) One-pot synthesis of heterodimeric agonists that activate the canonical Wnt signaling pathway. Chem. Commun. 56, 3685–3688 10.1039/D0CC00920B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Repina, N.A., McClave, T., Johnson, H.J., Bao, X., Kane, R.S. and Schaffer, D.V. (2020) Engineered illumination devices for optogenetic control of cellular signaling dynamics. Cell Rep. 31, 107737 10.1016/j.celrep.2020.107737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rosenbloom, A.B., Tarczyński, M., Lam, N., Kane, R.S., Bugaj, L.J. and Schaffer, D.V. (2020) β-Catenin signaling dynamics regulate cell fate in differentiating neural stem cells. Proc. Natl Acad. Sci. U.S.A. 117, 28828–28837 10.1073/pnas.2008509117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhou, Y. and Nathans, J. (2014) Gpr124 controls CNS angiogenesis and blood-brain barrier integrity by promoting ligand-specific canonical wnt signaling. Dev. Cell 31, 248–256 10.1016/j.devcel.2014.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Vallon, M., Yuki, K., Nguyen, T.D., Chang, J., Yuan, J., Siepe, D.et al. (2018) A RECK-WNT7 receptor-ligand interaction enables isoform-Specific regulation of Wnt bioavailability. Cell Rep. 25, 339–349.e9 10.1016/j.celrep.2018.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lee, E., Salic, A., Krüger, R., Heinrich, R. and Kirschner, M.W. (2003) The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 1, E10 10.1371/journal.pbio.0000010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.van Leeuwen, I.M.M., Byrne, H.M., Jensen, O.E. and King, J.R. (2007) Elucidating the interactions between the adhesive and transcriptional functions of beta-catenin in normal and cancerous cells. J. Theor. Biol. 247, 77–102 10.1016/j.jtbi.2007.01.019 [DOI] [PubMed] [Google Scholar]
- 171.Mirams, G.R., Byrne, H.M. and King, J.R. (2010) A multiple timescale analysis of a mathematical model of the Wnt/beta-catenin signalling pathway. J. Math. Biol. 60, 131–160 10.1007/s00285-009-0262-y [DOI] [PubMed] [Google Scholar]
- 172.Schmitz, Y., Rateitschak, K. and Wolkenhauer, O. (2013) Analysing the impact of nucleo-cytoplasmic shuttling of β-catenin and its antagonists APC, axin and GSK3 on Wnt/β-catenin signalling. Cell Signal. 25, 2210–2221 10.1016/j.cellsig.2013.07.005 [DOI] [PubMed] [Google Scholar]
- 173.Goentoro, L., Shoval, O., Kirschner, M.W. and Alon, U. (2009) The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol. Cell 36, 894–899 10.1016/j.molcel.2009.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kafri, P., Hasenson, S.E., Kanter, I., Sheinberger, J., Kinor, N., Yunger, S.et al. (2016) Quantifying β-catenin subcellular dynamics and cyclin D1 mRNA transcription during Wnt signaling in single living cells. eLife 5, e16748 10.7554/eLife.16748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tan, C.W., Gardiner, B.S., Hirokawa, Y., Smith, D.W. and Burgess, A.W. (2014) Analysis of Wnt signaling β-catenin spatial dynamics in HEK293T cells. BMC Syst. Biol. 8, 44 10.1186/1752-0509-8-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Massey, J., Liu, Y., Alvarenga, O., Saez, T., Schmerer, M. and Warmflash, A. (2019) Synergy with TGFβ ligands switches WNT pathway dynamics from transient to sustained during human pluripotent cell differentiation. Proc. Natl Acad. Sci. U.S.A. 116, 4989–4998 10.1073/pnas.1815363116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dequéant, M.L., Glynn, E., Gaudenz, K., Wahl, M., Chen, J., Mushegian, A.et al. (2006) A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science 314, 1595–1598 10.1126/science.1133141 [DOI] [PubMed] [Google Scholar]
- 178.Aulehla, A., Wehrle, C., Brand-Saberi, B., Kemler, R., Gossler, A., Kanzler, B.et al. (2003) Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell 4, 395–406 10.1016/S1534-5807(03)00055-8 [DOI] [PubMed] [Google Scholar]
- 179.Sonnen, K.F., Lauschke, V.M., Uraji, J., Falk, H.J., Petersen, Y., Funk, M.C.et al. (2018) Modulation of phase shift between Wnt and Notch signaling oscillations controls mesoderm segmentation. Cell 172, 1079–1090 e12 10.1016/j.cell.2018.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cavallo, J.C., Scholpp, S. and Flegg, M.B. (2020) Delay-driven oscillations via Axin2 feedback in the Wnt/β-catenin signalling pathway. J. Theor. Biol. 507, 110458 10.1016/j.jtbi.2020.110458 [DOI] [PubMed] [Google Scholar]
- 181.Jensen, P.B., Pedersen, L., Krishna, S. and Jensen, M.H. (2010) A Wnt oscillator model for somitogenesis. Biophys. J. 98, 943–950 10.1016/j.bpj.2009.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Aulehla, A., Wiegraebe, W., Baubet, V., Wahl, M.B., Deng, C., Taketo, M.et al. (2008) A beta-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat. Cell Biol. 10, 186–193 10.1038/ncb1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Niwa, Y., Shimojo, H., Isomura, A., González, A., Miyachi, H. and Kageyama, R. (2011) Different types of oscillations in Notch and Fgf signaling regulate the spatiotemporal periodicity of somitogenesis. Genes Dev. 25, 1115–1120 10.1101/gad.2035311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Niwa, Y., Masamizu, Y., Liu, T., Nakayama, R., Deng, C.-X. and Kageyama, R. (2007) The initiation and propagation of Hes7 oscillation are cooperatively regulated by Fgf and notch signaling in the somite segmentation clock. Dev. Cell 13, 298–304 10.1016/j.devcel.2007.07.013 [DOI] [PubMed] [Google Scholar]
- 185.Wahl, M.B., Deng, C., Lewandoski, M. and Pourquié, O. (2007) FGF signaling acts upstream of the NOTCH and WNT signaling pathways to control segmentation clock oscillations in mouse somitogenesis. Development 134, 4033–4041 10.1242/dev.009167 [DOI] [PubMed] [Google Scholar]
- 186.van Oostrom, M.J., Meijer, W.H.M. and Sonnen, K.F. (2021) A microfluidics approach for the functional investigation of signaling oscillations governing somitogenesis. J. Vis. Exp. 169, 1–17 10.3791/62318 [DOI] [PubMed] [Google Scholar]
- 187.Casaletto, J.B. and McClatchey, A.I. (2012) Spatial regulation of receptor tyrosine kinases in development and cancer. Nat. Rev. Cancer 12, 387–400 10.1038/nrc3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lavoie, H., Gagnon, J. and Therrien, M. (2020) ERK signalling: a master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell. Biol. 21, 607–632 10.1038/s41580-020-0255-7 [DOI] [PubMed] [Google Scholar]
- 189.Ferguson, K.M., Berger, M.B., Mendrola, J.M., Cho, H.S., Leahy, D.J. and Lemmon, M.A. (2003) EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell 11, 507–517 10.1016/S1097-2765(03)00047-9 [DOI] [PubMed] [Google Scholar]
- 190.Ogiso, H., Ishitani, R., Nureki, O., Fukai, S., Yamanaka, M., Kim, J.-H.et al. (2002) Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110, 775–787 10.1016/S0092-8674(02)00963-7 [DOI] [PubMed] [Google Scholar]
- 191.Zhang, X., Gureasko, J., Shen, K., Cole, P.A. and Kuriyan, J. (2006) An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137–1149 10.1016/j.cell.2006.05.013 [DOI] [PubMed] [Google Scholar]
- 192.Margolis, B.L., Lax, I., Kris, R., Dombalagian, M., Honegger, A.M., Howk, R.et al. (1989) All autophosphorylation sites of epidermal growth factor (EGF) receptor and HER2/neu are located in their carboxyl-terminal tails. identification of a novel site in EGF receptor. J. Biol. Chem. 264, 10667–10671 10.1016/S0021-9258(18)81674-X [DOI] [PubMed] [Google Scholar]
- 193.Rogge, R.D., Karlovich, C.A. and Banerjee, U. (1991) Genetic dissection of a neurodevelopmental pathway: son of sevenless functions downstream of the sevenless and EGF receptor tyrosine kinases. Cell 64, 39–48 10.1016/0092-8674(91)90207-F [DOI] [PubMed] [Google Scholar]
- 194.Chol, K.-Y., Satterberg, B., Lyons, D.M. and Elion, E.A. (1994) Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell 78, 499–512 10.1016/0092-8674(94)90427-8 [DOI] [PubMed] [Google Scholar]
- 195.Therrien, M., Michaud, N.R., Rubin, G.M. and Morrison, D.K. (1996) KSR modulates signal propagation within the MAPK cascade. Genes Dev. 10, 2684–2695 10.1101/gad.10.21.2684 [DOI] [PubMed] [Google Scholar]
- 196.Rajakulendran, T., Sahmi, M., Lefrançois, M., Sicheri, F. and Therrien, M. (2009) A dimerization-dependent mechanism drives RAF catalytic activation. Nature 461, 542–545 10.1038/nature08314 [DOI] [PubMed] [Google Scholar]
- 197.Brennan, D.F., Dar, A.C., Hertz, N.T., Chao, W.C.H., Burlingame, A.L., Shokat, K.M.et al. (2011) A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature 472, 366–369 10.1038/nature09860 [DOI] [PubMed] [Google Scholar]
- 198.McKay, M.M., Ritt, D.A. and Morrison, D.K. (2009) Signaling dynamics of the KSR1 scaffold complex. Proc. Natl Acad. Sci. U.S.A. 106, 11022–11027 10.1073/pnas.0901590106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chen, Z.J. and Sun, L.J. (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33, 275–286 10.1016/j.molcel.2009.01.014 [DOI] [PubMed] [Google Scholar]
- 200.Nguyen, L.K., Kolch, W. and Kholodenko, B.N. (2013) When ubiquitination meets phosphorylation: a systems biology perspective of EGFR/MAPK signalling. Cell Commun. Signal. 11, 52 10.1186/1478-811X-11-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wilson, M.Z., Ravindran, P.T., Lim, W.A. and Toettcher, J.E. (2017) Tracing information flow from Erk to target gene induction reveals mechanisms of dynamic and combinatorial control. Mol. Cell 67, 757–769.e5 10.1016/j.molcel.2017.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Huff, K., End, D. and Guroff, G. (1981) Nerve growth factor-induced alteration in the response of PC12 pheochromocytoma cells to epidermal growth factor. J. Cell Biol. 88, 189–198 10.1083/jcb.88.1.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Toettcher, J.E., Weiner, O.D. and Lim, W.A. (2013) Using optogenetics to interrogate the dynamic control of signal transmission by the ras/erk module. Cell 155, 1422–1434 10.1016/j.cell.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ryu, H., Chung, M., Song, J., Lee, S.S., Pertz, O. and Jeon, N.L. (2018) Integrated platform for monitoring single-cell MAPK kinetics in computer-controlled temporal stimulations. Sci. Rep. 8, 11126 10.1038/s41598-018-28873-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Santos, S.D.M., Verveer, P.J. and Bastiaens, P.I.H. (2007) Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat. Cell Biol. 9, 324–330 10.1038/ncb1543 [DOI] [PubMed] [Google Scholar]
- 206.Chen, J.-Y., Lin, J.-R., Cimprich, K.A. and Meyer, T. (2012) A Two-Dimensional ERK-AKT signaling code for an NGF-Triggered cell-Fate decision. Mol. Cell 45, 196–209 10.1016/j.molcel.2011.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ryu, H., Chung, M., Dobrzyński, M., Fey, D., Blum, Y., Sik Lee, S.et al. (2016) Frequency modulation of ERK activation dynamics rewires cell fate. Mol. Syst. Biol. 12, 866 10.15252/msb.20166982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.von Kriegsheim, A., Baiocchi, D., Birtwistle, M., Sumpton, D., Bienvenut, W., Morrice, N.et al. (2009) Cell fate decisions are specified by the dynamic ERK interactome. Nat. Cell Biol. 11, 1458–1464 10.1038/ncb1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ponsioen, B., Post, J.B., Buissant des Amorie, J.R., Laskaris, D., van Ineveld, R.L., Kersten, S.et al. (2021) Quantifying single-cell ERK dynamics in colorectal cancer organoids reveals EGFR as an amplifier of oncogenic MAPK pathway signalling. Nat. Cell Biol. 23, 377–390 10.1038/s41556-021-00654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.de la Cova, C., Townley, R., Regot, S. and Greenwald, I. (2017) A real-Time biosensor for ERK activity reveals signaling dynamics during C. elegans cell fate specification. Dev. Cell 42, 542–553 e4 10.1016/j.devcel.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Harvey, C.D., Ehrhardt, A.G., Cellurale, C., Zhong, H., Yasuda, R., Davis, R.J.et al. (2008) A genetically encoded fluorescent sensor of ERK activity. Proc. Natl Acad. Sci. U.S.A. 105, 19264–19269 10.1073/pnas.0804598105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hiratsuka, T., Bordeu, I., Pruessner, G. and Watt, F.M. (2020) Regulation of ERK basal and pulsatile activity control proliferation and exit from the stem cell compartment in mammalian epidermis. Proc. Natl Acad. Sci. U.S.A. 117, 17796–17807 10.1073/pnas.2006965117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Goglia, A.G., Wilson, M.Z., Jena, S.G., Silbert, J., Basta, L.P., Devenport, D.et al. (2020) A live-cell screen for altered Erk dynamics reveals principles of proliferative control. Cell Syst. 10, 240–253.e6 10.1016/j.cels.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lim, B., Dsilva, C.J., Levario, T.J., Lu, H., Schupbach, T., Kevrekidis, I.G.et al. (2015) Dynamics of inductive ERK signaling in the Drosophila embryo. Curr. Biol. 25, 1784–1790 10.1016/j.cub.2015.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Johnson, H.E. and Toettcher, J.E. (2019) Signaling dynamics control cell fate in the early Drosophila embryo. Dev. Cell 48, 361–370.e3 10.1016/j.devcel.2019.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Valon, L., Davidović, A., Levillayer, F., Villars, A., Chouly, M., Cerqueira-Campos, F.et al. (2021) Robustness of epithelial sealing is an emerging property of local ERK feedback driven by cell elimination. Dev. Cell 56, 1700–1711.e8 10.1016/j.devcel.2021.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Gagliardi, P.A., Dobrzyński, M., Jacques, M.-A., Dessauges, C., Ender, P., Blum, Y.et al. (2021) Collective ERK/Akt activity waves orchestrate epithelial homeostasis by driving apoptosis-induced survival. Dev. Cell 56, 1712–1726.e6 10.1016/j.devcel.2021.05.007 [DOI] [PubMed] [Google Scholar]
- 218.Muta, Y., Fujita, Y., Sumiyama, K., Sakurai, A., Taketo, M.M., Chiba, T.et al. (2018) Composite regulation of ERK activity dynamics underlying tumour-specific traits in the intestine. Nat. Commun. 9, 2174 10.1038/s41467-018-04527-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Bugaj, L.J., Sabnis, A.J., Mitchell, A., Garbarino, J.E., Toettcher, J.E., Bivona, T.G.et al. (2018) Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science 361, eaao3048 10.1126/science.aao3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rayon, T. and Briscoe, J. (2021) Cross-species comparisons and models to study tempo in development and homeostasis. Interface Focus 11, 20200069 10.1098/rsfs.2020.0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Rayon, T., Stamataki, D., Perez-Carrasco, R., Garcia-Perez, L., Barrington, C., Melchionda, M.et al. (2020) Species-specific pace of development is associated with differences in protein stability. Science 369, eaba7667 10.1126/science.aba7667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Turnpenny, P.D., Alman, B., Cornier, A.S., Giampietro, P.F., Offiah, A., Tassy, O.et al. (2007) Abnormal vertebral segmentation and the notch signaling pathway in man. Dev. Dyn. 236, 1456–1474 10.1002/dvdy.21182 [DOI] [PubMed] [Google Scholar]
- 223.Balaskas, N., Ribeiro, A., Panovska, J., Dessaud, E., Sasai, N., Page, K.M.et al. (2012) Gene regulatory logic for reading the Sonic hedgehog signaling gradient in the vertebrate neural tube. Cell 148, 273–284 10.1016/j.cell.2011.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zagorski, M., Tabata, Y., Brandenberg, N., Lutolf, M.P., Tkačik, G., Bollenbach, T.et al. (2017) Decoding of position in the developing neural tube from antiparallel morphogen gradients. Science 356, 1379–1383 10.1126/science.aam5887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.El-Sherif, E., Averof, M. and Brown, S.J. (2012) A segmentation clock operating in blastoderm and germband stages of Tribolium development. Development 139, 4341–4346 10.1242/dev.085126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sarrazin, A.F., Peel, A.D. and Averof, M. (2012) A segmentation clock with two-segment periodicity in insects. Science 336, 338–341 10.1126/science.1218256 [DOI] [PubMed] [Google Scholar]
- 227.Liao, B.-K. and Oates, A.C. (2017) Delta-Notch signalling in segmentation. Arthropod. Struct. Dev. 46, 429–447 10.1016/j.asd.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gupta, A., Lutolf, M.P., Hughes, A.J. and Sonnen, K.F. (2021) Bioengineering in vitro models of embryonic development. Stem Cell Rep. 16, 1104–1116 10.1016/j.stemcr.2021.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.El Azhar, Y. and Sonnen, K.F. (2021) Development in a dish- models of mammalian embryonic development. Front Cell Dev Biol. 9, 655993 10.3389/fcell.2021.655993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Gautier, A., Juillerat, A., Heinis, C., Corrêa, Jr, I.R., Kindermann, M., Beaufils, F.et al. (2008) An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 15, 128–136 10.1016/j.chembiol.2008.01.007 [DOI] [PubMed] [Google Scholar]
- 231.Bothma, J.P., Norstad, M.R., Alamos, S. and Garcia, H.G. (2018) Llamatags: a versatile tool to image transcription factor dynamics in live embryos. Cell 173, 1810–1822 10.1016/j.cell.2018.03.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Serra, D., Mayr, U., Boni, A., Lukonin, I., Rempfler, M., Challet Meylan, L.et al. (2019) Self-organization and symmetry breaking in intestinal organoid development. Nature 569, 66–72 10.1038/s41586-019-1146-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Strnad, P., Gunther, S., Reichmann, J., Krzic, U., Balazs, B., de Medeiros, G.et al. (2016) Inverted light-sheet microscope for imaging mouse pre-implantation development. Nat. Methods 13, 139–142 10.1038/nmeth.3690 [DOI] [PubMed] [Google Scholar]
- 234.Krol, A.J., Roellig, D., Dequeant, M.L., Tassy, O., Glynn, E., Hattem, G.et al. (2011) Evolutionary plasticity of segmentation clock networks. Development 138, 2783–2792 10.1242/dev.063834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Rudolph, J.D., de Graauw, M., van de Water, B., Geiger, T. and Sharan, R. (2016) Elucidation of signaling pathways from large-scale phosphoproteomic data using protein interaction networks. Cell Syst. 3, 585–593.e3 10.1016/j.cels.2016.11.005 [DOI] [PubMed] [Google Scholar]
- 236.Gerlach, J.P., van Buggenum, J.A.G., Tanis, S.E.J., Hogeweg, M., Heuts, B.M.H., Muraro, M.J.et al. (2019) Combined quantification of intracellular (phospho-)proteins and transcriptomics from fixed single cells. Sci. Rep. 9, 1469 10.1038/s41598-018-37977-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.van Eijl, R., van Buggenum, J., Tanis, S.E.J., Hendriks, J. and Mulder, K.W. (2018) Single-Cell ID-seq reveals dynamic BMP pathway activation upstream of the MAF/MAFB-Program in epidermal differentiation. iScience 9, 412–422 10.1016/j.isci.2018.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sonnen, K.F. and Merten, C.A. (2019) Microfluidics as an emerging precision tool in developmental biology. Dev. Cell 48, 293–311 10.1016/j.devcel.2019.01.015 [DOI] [PubMed] [Google Scholar]
- 239.Nabet, B., Roberts, J.M., Buckley, D.L., Paulk, J., Dastjerdi, S., Yang, A.et al. (2018) The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 14, 431–441 10.1038/s41589-018-0021-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Caussinus, E. and Affolter, M. (2016) deGradFP: a system to knockdown GFP-tagged proteins. Methods Mol. Biol. 1478, 177–187 10.1007/978-1-4939-6371-3_9 [DOI] [PubMed] [Google Scholar]
- 241.Zhang, L., Ward, J.D., Cheng, Z. and Dernburg, A.F. (2015) The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374–4384 10.1242/dev.125393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ho, C. and Morsut, L. (2021) Novel synthetic biology approaches for developmental systems. Stem Cell Rep. 16, 1051–1064 10.1016/j.stemcr.2021.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hanahan, D. and Weinberg, R.A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 244.Weterings, S.D.C., Oostrom, M.J. and Sonnen, K.F. (2021) Building bridges between fields: bringing together development and homeostasis. Development 148, dev193268 10.1242/dev.200265 [DOI] [PMC free article] [PubMed] [Google Scholar]