Abstract
We previously isolated and sequenced two genomic segments of Mycobacterium avium subsp. paratuberculosis, namely, f57, a species-specific sequence, and the p34 gene, coding for a 34-kDa antigenic protein. Comparison of sequences upstream of the p34 open reading frame (us-p34) from M. avium subsp. paratuberculosis and M. tuberculosis showed a 79-base deletion in M. tuberculosis. Sequence analysis of the p34 genes in another two species, M. bovis (strain BCG) and M. avium (strain D4), confirmed the differences observed between tuberculous and nontuberculous species. A duplex diagnostic PCR strategy based on coamplification of nonhomologous us-p34 and species-specific f57 sequences was therefore developed. Duplex PCR yielded three different patterns, specific either for tuberculous bacilli (M. tuberculosis, M. bovis, and M. africanum), for both nontuberculous mycobacteria M. avium and M. intracellulare, or for M. avium subsp. paratuberculosis. The specificity of this single-step DNA-based assay was assessed on DNA from cultured mycobacterial strains, as well as on a panel of formalin-fixed and paraffin-embedded tissues from cattle. Molecular assay results from tissular DNA were compared to conventional bacteriological and histological test results, including those obtained by Ziehl-Neelsen staining on tissue biopsy specimens. Molecular discrimination was successful and confirmed the value of duplex us-p34 and f57 sequence amplification for differential diagnosis of tuberculosis, paratuberculosis, or infections caused by other members of the M. avium complex.
Bovine paratuberculosis and tuberculosis are still a major concern in many countries of the world. Indeed, they are responsible for heavy economic losses related to decreases in weight, milk production, and fertility. Additional economic costs include increased culling rates, diagnostic testing, and control measures (12). A specific identification of these infectious agents is complicated by the fact that Mycobacterium avium subsp. paratuberculosis and M. bovis are, respectively, part of the M. avium complex (MAC) and the M. tuberculosis complex, each including closely related species (M. avium, M. intracellulare, M. avium subsp. paratuberculosis, and M. scrofulaceum for MAC and M. bovis, M. tuberculosis, M. africanum, and M. microti for M. tuberculosis complex).
Both M. avium and M. avium subsp. paratuberculosis can cause chronic granulomatous enteritis in cattle and other ruminants (3). Paratuberculosis (Johne's disease) is widespread, highly contagious, and usually remains clinically undetectable until the onset of severe clinical symptoms. Contaminated cattle must be isolated and are often sent to slaughter. M. avium infection can sporadically cause chronic granulomatous enteritis in cattle and other ruminants. However, the significance of isolating M. avium, with regard to its contagiousness within a cattle herd, remains unclear. Although better controlled, M. bovis infection remains an important disease in many countries. The presence of M. bovis infection in a population of wild animals may interfere with plans for eradication of bovine tuberculosis (20, 25). Moreover, all animals from infected herds have to be slaughtered.
A presumptive diagnosis of bovine mycobacterial infection in slaughtered animals is made on the basis of the histopathology of lymph nodes and tissue specimens showing tuberculosis-like lesions and acid-fast bacilli. Definitive diagnosis then requires mycobacterial species identification. Culture is considered to be the “gold standard,” but this is a very slow and labor-intensive procedure. Furthermore, culture may become positive only several weeks after inoculation, especially for samples containing low numbers of mycobacteria. Despite continuous methodological improvements, cultures are frequently negative (13, 26). Optimized radiometric techniques have reduced the time taken for detecting mycobacteria but do not solve the crucial issue of sensitivity, while requiring special equipment and facilities and the use of radioisotopes (32). Immunohistological techniques directly applied to clinical specimens are more sensitive than Ziehl-Neelsen staining, but specificity remains controversial and antigenic alteration due to poor fixation or conservation may alter the assay results (4, 29). Diagnosis of paratuberculosis and tuberculosis therefore remains difficult with the tests presently available (18). Altogether, the lack of a rapid, simple, specific, and sensitive diagnostic test for detecting mycobacteria has greatly hampered programs for the control and eradication of these diseases. In this respect, molecular assays appear promising for identification of infected animals, as well as of potential sources of infection, and could bring further insights into epidemiological studies. The need for a specific identification of mycobacteria in cattle also is justified by the potential impact of the bacteria on human health. Although considerably reduced in developed countries, the risk of human exposure to bovine tuberculosis has not yet been totally eradicated (5, 24). Identification of M. avium infections in humans has gained interest with the human immunodeficiency virus (HIV) epidemics (21). M. avium subsp. paratuberculosis is considered a potentially food-borne pathogen (27), and its relationship with Crohn's disease is still debated (3).
Members of our group, along with others, have previously isolated two DNA segments of M. avium subsp. paratuberculosis: the p34 gene, coding for a 34-kDa mycobacterial antigenic protein (3), and the f57 segment (23). The carboxyl-terminal portion (a362) of the p34 protein carries species-specific epitopes and has been used in the development of a serological assay for Johne's disease (30). The f57 sequence is specific for M. avium subsp. paratuberculosis and is not found in any other mycobacterial species or M. avium subspecies (23).
In the current study, species-specific polymorphisms were identified within the nucleotide sequences upstream of the p34 open reading frame (us-p34) in tuberculous and nontuberculous mycobacteria. Accordingly, we developed a us-p34 and f57 sequence-based duplex PCR strategy for discriminating between M. avium subsp. paratuberculosis, other nontuberculous mycobacteria (M. avium subsp. and M. intracellulare), and M. tuberculosis complex, hence providing a rapid differential identification of the major mycobacteria in cattle. This method was assessed on DNA extracts from formol-fixed and paraffin-embedded tissue samples. It compared favorably with conventional testing methods, including culture, histopathology, and Ziehl-Neelsen staining. The current duplex molecular assay could therefore facilitate batch processing of clinical samples in large-scale paratuberculosis and tuberculosis control programs and contribute to confirmation of dubious cases.
(Part of this work was presented at the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Diego, Calif. [poster D-104]).
MATERIALS AND METHODS
Bacterial isolates.
Clinical mycobacterial isolates were obtained from two Belgian Mycobacterium reference laboratories: the Institute of Tropical Medicine (Antwerp, Belgium) and the Pasteur Institute (Brussels, Belgium). The numbers of isolates tested for each species were as follows: M. tuberculosis, 15 (including ATCC 27294); M. bovis, 4 (including ATCC 19210); M. avium, 10 (including M. avium D4, ATCC 10708, ATCC 10719, and ATCC 2529); M. avium subsp. paratuberculosis, 11 (including ATCC 19698); M. africanum, 4 (including ATCC 25420); and M. intracellulare, 6 (including ATCC 13950).
Tissue samples.
Diagnosis of mycobacteriosis was made on the basis of conventional clinical criteria (diarrhea, decreased milk production, emaciation, and anorexia), confirmed in each case by culture and microbiological identification of the etiological agent. Biopsy specimens of intestinal wall and mesenteric lymph nodes from tuberculous (n = 6), nontuberculous (2 infected by M. avium and 15 by M. avium subsp. paratuberculosis), and healthy (n = 3) cows were obtained from two different slaughterhouses. There was one specimen per cow. Samples were formol fixed for 12 h, and paraffin embedded according to current histological techniques.
According to the types of lesions observed in intestinal tissue and mesenteric lymph node biopsy specimens, a distinction was made between pluribacillary and paucibacillary forms of the disease. In the pluribacillary form, tissues contained numerous macrophage-infiltrated granulomas, giant cells and clumps of colored rods (n = 5) or rare (n = 1) or no bacilli (n = 2). Conversely, other tissues disclosed only a few focal lesions with either rare (n = 6) or no (n = 9) bacilli after Ziehl-Neelsen staining.
Preparation of mycobacterial DNA.
Mycobacteria (10 mg [wet weight]) were suspended in 200 μl of lysing solution (0.1 M NaOH, 1 M NaCl, and 5% sodium dodecyl sulfate [SDS]) and heated (100°C) for 20 min. The suspension was then cooled, neutralized with 3 volumes of 0.1 M Tris-HCl (pH 7.4) buffer, and centrifuged (5,000 × g, 5 min). Supernatants were extracted with phenol-chloroform, and DNA was precipitated with ethanol, collected by centrifugation, dissolved in 50 μl of H2O, and stored at −20°C.
DNA extraction from paraffin-embedded tissues.
Three sections (20 μm) of each paraffin-embedded biopsy specimen were dewaxed twice with 1 ml of xylene for 5 min and centrifuged (10,000 × g, 5 min). The supernatant was discarded, and traces of solvent were removed by washing the pellet twice for 5 min with 1 ml of 100% ethanol. After centrifugation (10,000 × g, 5 min), the pellet was air dried. Tissues were digested with 150 μl of 50 mM Tris-HCl buffer (pH 7.4), containing 1 mg/ml proteinase K (Boehringer Mannheim, Mannheim, Germany) and 1% SDS, for 2 h at 37°C. Mycobacteria were then lysed in a final volume of 200 μl, and DNA was purified as previously described.
PCR amplifications.
Based on sequence homology between the M. tuberculosis (GenBank accession no. Z79700) and M. avium subsp. paratuberculosis (GenBank accession no. X68102) regions upstream of the open reading frame of the p34 gene, two sets of oligonucleotides, myc1-myc2 and myc1-myc3, were designed to amplify the regions corresponding to M. bovis BCG and M. avium D4, respectively (Table 1 and Fig. 1). For amplification, an aliquot (10 μl) of the DNA samples was added to 90 μl of PCR mixture consisting of 10 mM Tris-HCl (pH 8.8), 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 0.25 mM (each) deoxynucleoside triphosphates, 10 pmol of each primer and 0.625 U of DyNAzyme DNA polymerase (Finnzymes Inc., Espoo, Finland). After an initial denaturation step (3 min at 96°C), 30 cycles of amplification were performed as follows: denaturation at 96°C for 30 s, annealing at 58°C for 45 s, and DNA extension at 72°C for 30 s, with an increment of 1 s per cycle for the denaturation and extension segments. A final extension was performed at 72°C for 15 min. Amplifications were carried out in a DNA 2400 thermocycler (Perkin-Elmer Applied Biosystems, Foster City, Calif.). After amplification, PCR products were cloned using the TOPO XL PCR cloning kit (Invitrogen, Carlsbad, Calif.), according to the manufacturer's protocol. The clones were further sequenced with the Taq Dye Deoxy Terminator Cycle sequencing kit and an ABI 377 DNA sequencer (Perkin-Elmer Applied Biosystems).
TABLE 1.
Oligonucleotide sequences used in PCR amplifications
| Primers | Sequenced | Descriptiona |
|---|---|---|
| myc1 | GAGTAGGTCATGGCTCCTCC | R, nt 344–364 of M. tuberculosis us-p34 sequenceb and nt 423–443 of M. avium subsp. paratuberculosis us-p34 sequenceb |
| myc2 | GTGCGCATATAGCGGTCGTC | F, nt 1–20 of M. tuberculosis us-p34 sequenceb |
| myc3 | CATGCACCGAATTAGAACGT | F, nt 186–205 of M. tuberculosis us-p34 sequenceb |
| f57a | GGTCGCGTCATTCAGAATC | F, nt 179–197 of M. avium subsp. paratuberculosis f57 genec |
| f57b | TCTCAGACAGTGGCAGGTG | R, nt 599–617 of M. avium subsp. paratuberculosis f57 genec |
FIG. 1.
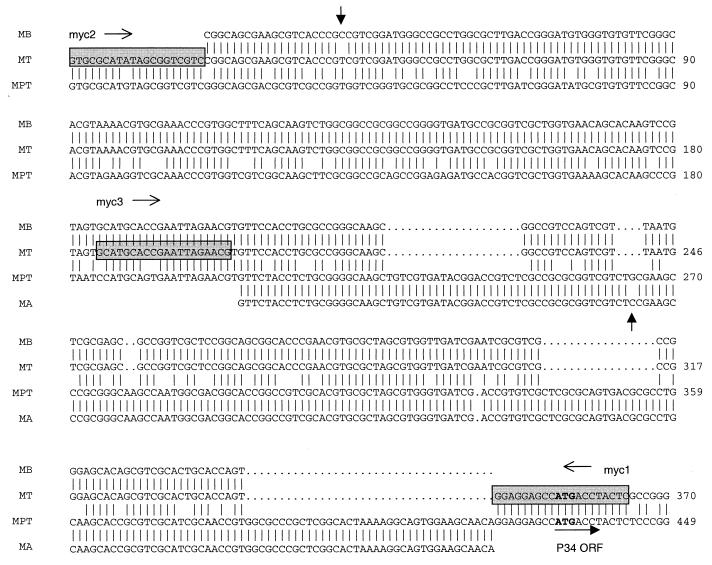
Multiple nucleotidic sequence alignment of M. bovis (MB), M. tuberculosis (MT), M. avium subsp. paratuberculosis (MPT), and M. avium subsp. (MA) us-p34 genes. Gaps between sequences are indicated by dots. Vertical bars indicate identity across sequences. The start codon (ATG) for the p34 open reading frame is in bold. Primer sequences are indicated by shaded boxes. The vertical arrows indicate point mutations between M. tuberculosis and M. bovis and between M. avium subsp. paratuberculosis and M. avium subsp. The horizontal arrows indicate directions of transcription.
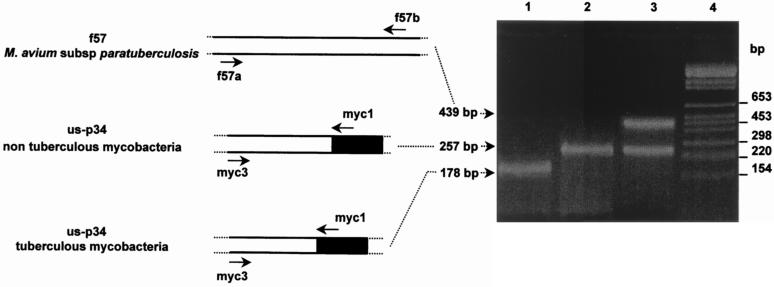
For the duplex PCR amplification, oligonucleotides were designed on the basis of the f57 (Genbank accession no. X70277) and us-p34 sequences (Fig. 1 and Table 1). Primers f57a and f57b amplify a 439-bp fragment from the f57 gene, and primers myc3 and myc1 generate a 178-bp amplicon from us-p34 in M. tuberculosis and M. bovis while amplifying a 257-bp fragment in M. avium subsp. paratuberculosis and M. avium.
A strict procedure was followed to avoid cross-contamination between samples or carryover of PCR products. DNA extraction was carried out sample by sample. In any series of reactions, contamination at the DNA level was ruled out by performing PCR analysis without any DNA template. As positive controls, DNA samples from ATCC reference strains of M. bovis, M. avium subsp. avium, and M. avium subsp. paratuberculosis were included in each test.
DNA sequence analysis was performed with the Genetics Computer Group software obtained from the University of Wisconsin, through the use of the Belgian EMBnet Node facility. Sequences were aligned by the Pileup program.
Hybridization analysis.
Amplified DNA segments were transferred onto nylon membranes (Hybond-N+; Amersham, Little Chalfont, United Kingdom) according to the Southern blot method. After 2 h of prehybridization, membranes were hybridized with f57 and us-p34 digoxigenin-labeled probes for 4 h at 50°C. f57 and us-p34 probes were obtained by PCR amplification of 10 ng of M. avium subsp. paratuberculosis DNA, with primers sets f57a-f57b and myc3-myc1, respectively, in the presence of DIG-11-dUTP. Filters were then washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 37°C for 5 min, and twice with 0.2× SSC–0.1% SDS at 50°C for 5 min. Hybridized digoxigenin-labeled DNA fragments were detected through alkaline-phosphatase-labeled anti-DIG Fab fragments. Colorimetric detection was performed with Nitro Blue Tetrazolium Chlorure and 5,3-bromo-4-indolylphosphate (BCIP) (Boerhinger Mannheim) according to the manufacturer's instructions.
RESULTS
Analysis of the upstream p34 mycobacterial DNAs.
Comparison of the homologous p34 gene and its regulatory sequences (us-p34 sequences) for M. tuberculosis and M. avium subsp. paratuberculosis showed the presence of deletions in the region upstream of the p34 protein start codon in M. tuberculosis (Fig. 1). To confirm and extend these findings, the us-p34 sequences of M. bovis BCG and M. avium D4 were amplified and sequenced. Alignment of multiple sequences of this region revealed interspecies polymorphisms specific for both tuberculous (M. tuberculosis and M. bovis BCG) and nontuberculous (M. avium sp. and M. avium subsp. paratuberculosis) mycobacteria, the us-p34 fragment being 79 bases shorter in tuberculous species. Conversely, the us-p34 region appeared to be highly conserved within each group: differentiation between M. tuberculosis and M. bovis relied on a single T-to-C transition at position 41 of the us-p34 sequence and a single C-to-G transversion at position 264 of the us-p34 sequence in M. avium subsp. and M. avium subsp. paratuberculosis (Fig. 1).
Development of duplex PCR diagnostic assay for mycobacterioses.
Based on the sequences of the us-p34 region, a PCR assay was developed to discriminate tuberculous from nontuberculous mycobacteria complexes. Primers matching conserved sequences of the polymorphic us-p34 region allowed to amplify a 178-bp fragment in tuberculous mycobacteria versus a 257-bp fragment in nontuberculous mycobacteria, irrespective of the species. In a next step, amplification of a 439-bp product from the genomic f57 sequence allowed a specific identification of M. avium subsp. paratuberculosis within the nontuberculous group. Duplex amplification generated a distinct amplification pattern in three of the four mycobacterial species: DNA from M. avium subsp. paratuberculosis produced two fragments of 439 and 257 bp, respectively, M. avium subsp. produced the 257-bp fragment only, and M. tuberculosis and M. bovis were characterized by a 178-bp amplicon (Fig. 2).
FIG. 2.
Duplex PCR strategy. Amplified DNA fragments from M. bovis (lane 1), M. avium subsp. (lane 2), and M. avium subsp. paratuberculosis (lane 3) were separated by electrophoresis in a 2% agarose gel. The lengths (in base pairs) of the amplified fragments, as well as of the BglI- and HinfI-cleaved pBR328 DNA fragments (molecular weight marker) (lane 4), are indicated on the left- and righthand sides of the gel, respectively.
Application of duplex PCR procedure to identification of mycobacteria.
The specificity of the duplex assay was confirmed by the analysis of DNA originating from a panel of 50 clinical and reference mycobacterial strains. Our results demonstrate that us-p34 sequence polymorphism is conserved for both nontuberculous and tuberculous mycobacteria, while the f57 sequence is fully specific for M. avium subsp. paratuberculosis. In all the samples, amplicons of the expected size were found after amplification: the 439- and 257-bp fragments were present in all nontuberculous M. avium subsp. paratuberculosis strains (n = 11), the single 257-bp fragment was found in each of the nontuberculous M. avium (n = 10) and M. intracellulare (n = 6) strains, whereas strains of tuberculous mycobacteria, including M. tuberculosis, M. bovis, and M. africanum, were all (n = 23) characterized by the 178-bp amplicon (Table 2).
TABLE 2.
Results of the duplex PCR procedure on reference mycobacterial strains
| Strain | No. of strains tested | No. of strains characterized by amplicon lengtha of:
|
||
|---|---|---|---|---|
| 178 bp | 257 bp | 439 bp | ||
| M. tuberculosis | 15 | 15 | ||
| M. bovis | 4 | 4 | ||
| M. africanum | 4 | 4 | ||
| M. avium | 10 | 10 | ||
| M. avium subsp. paratuberculosis | 11 | 11 | 11 | |
| M. intracellulare | 6 | 6 | ||
The 439-bp fragment is amplified from the f57 gene sequence (primers f57a and f57b), whereas the 178- and 257-bp amplicons are distinctly amplified from the us-p34 sequence (primers myc3 and myc1), depending on the mycobacterial species.
Validation of duplex PCR procedure on embedded tissues.
The duplex assay was also assessed on DNA extracted from formalin-fixed, paraffin-embedded tissues, i.e., intestinal wall and mesenteric lymph nodes, from 26 cattle (Table 3).
TABLE 3.
Validation of the duplex PCR procedure on paraffin-embedded tissues
| Specimen | Mycobacterial species | Histological observationa | Result of:
|
|
|---|---|---|---|---|
| Ziehl-Neelsen stainingb | PCR analysisc | |||
| 98/264 | Negative control | No lesion | − | − |
| 98/265 | Negative control | No lesion | − | − |
| 96/278a | Negative control | No lesion | − | − |
| 96/5a | M. avium subsp. paratuberculosis | Pluribacillary | ++ | ++ |
| 96/8d | M. avium subsp. paratuberculosis | Pluribacillary | ++ | ++ |
| N96/445 | M. avium subsp. paratuberculosis | Pluribacillary | ++ | ++ |
| 98/266 | M. avium subsp. paratuberculosis | Pluribacillary | ++ | ++ |
| 98/267 | M. avium subsp. paratuberculosis | Pluribacillary | ++ | ++ |
| N95/558E | M. avium subsp. paratuberculosis | Paucibacillary | + | + |
| N95/558A | M. avium subsp. paratuberculosis | Paucibacillary | + | + |
| 96/230a | M. avium subsp. paratuberculosis | Paucibacillary | + | + |
| N96/670B | M. avium subsp. paratuberculosis | Paucibacillary | + | + |
| W95/3A | M. avium subsp. paratuberculosis | Paucibacillary | − | + |
| W95/635F | M. avium subsp. paratuberculosis | Paucibacillary | + | − |
| 96/14d | M. avium subsp. paratuberculosis | Paucibacillary | − | − |
| 96/17a | M. avium subsp. paratuberculosis | Paucibacillary | − | − |
| 96/229b | M. avium subsp. paratuberculosis | Paucibacillary | − | − |
| N95/600A | M. avium subsp. paratuberculosis | Paucibacillary | − | − |
| N86/498A | M. avium | Paucibacillary | + | + |
| N86/498B | M. avium | Paucibacillary | − | − |
| 95P3006 | M. bovis | Pluribacillary | + | + |
| 93/00/214/2 | M. bovis | Pluribacillary | − | + |
| 93/00/483/14 | M. bovis | Pluribacillary | − | − |
| 96/277a | M. bovis | Paucibacillary | − | + |
| 96/276b | M. bovis | Paucibacillary | − | − |
| 96/279b | M. bovis | Paucibacillary | − | − |
Histological classification based on either the presence of marked lesions containing numerous macrophage-infiltrated granulomas and giant cells (pluribacillary form) or scarce lesions (paucibacillary form).
Ziehl-Neelsen staining determining the presence of clumps of colored rods (++) and either rare (+) or no (−) bacilli in the tissue.
Species-specific amplicons are either not detected (−) or detected directly by visualization of ethidium bromide-stained DNA (++) or following Southern blotting and hybridization (+).
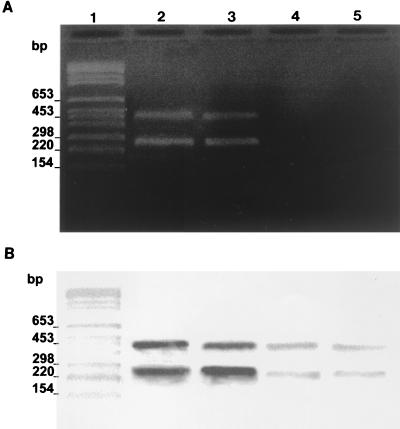
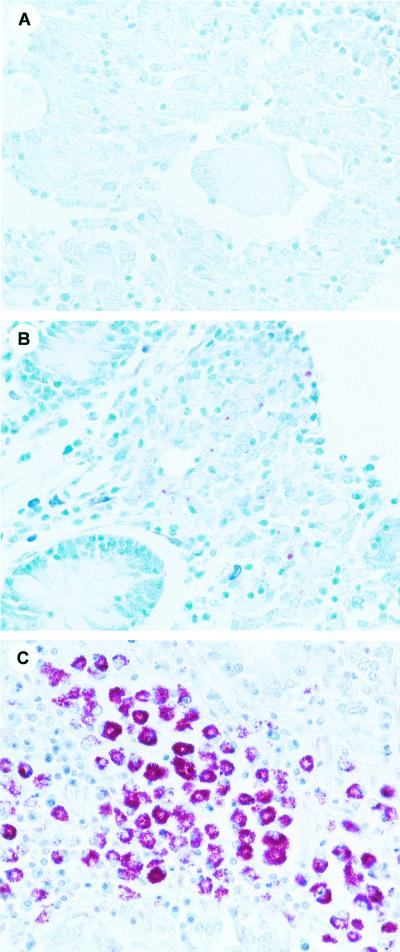
The result of duplex PCR assay was positive in 7 of 8 pluribacillary tissues, 5 after direct visualization of the amplified DNA on agarose gel, and 2 after Southern blotting of the gel followed by hybridization with biotinylated probes, including one of the two Ziehl-Neelsen-negative specimens. Although no amplification product was directly observed with the 15 paucibacillary tissues, a positive signal was obtained for 7 of them, including two Ziehl-Neelsen-negative tissues, after Southern blotting and hybridization. Altogether, a full agreement between culture and molecular assay results was observed for the 14 tissue samples for which amplification was successful. Figure 3 illustrates a positive detection from two pluribacillary specimens infected by M. avium subsp. paratuberculosis and presenting marked histological lesions (Fig. 3A, lanes 2 and 3) and a positive signal obtained after Southern blotting and hybridization on DNA from two paucibacillary forms containing scarce histological lesions (Figure 3B, lanes 3 and 4). Three distinct histological samples following Ziehl-Neelsen staining are shown (Fig. 4). They illustrate the histological difference between negative (Fig. 4A) paucibacillary tissues disclosing rare acid-fast bacilli (Fig. 4B) and pluribacillary tissues containing clumps of acid-fast organisms (Fig. 4C).
FIG. 3.
PCR duplex analysis of DNA purified from formalin-fixed and paraffin-embedded paratuberculosis tissues. DNA samples (100 ng) isolated from mesenteric lymph node biopsy specimens and were used in a duplex PCR procedure. (A) Aliquots of PCR-amplified DNA (50 μl of each amplified sample) were analyzed by 2% agarose gel electrophoresis. (B) Hybridization analysis of the same amplified products with the DIG-labeled f57 and us-p34 probes. Lanes 1, BglI- and HinfI-cleaved pBR328 weight marker; lanes 2 and 3, specimens from pluribacillary cows; lanes 4 and 5, specimens from paratuberculous cows (lanes of panel B are as labeled in panel A).
FIG. 4.
Ziehl-Neelsen stained, formalin-fixed, paraffin-embedded tissues (intestinal mucosa). (A) Negatively stained tissue; (B) paucibacillary tissue disclosing rare acid-fast bacilli; (C) pluribacillary tissue containing clumps of acid-fast organisms.
DISCUSSION
Conventional methods for identification of mycobacteria, involving evaluation of phenotypic and biochemical growth characteristics, are time-consuming and tedious. Difficulties in identifying mycobacteria also arise with other conventional methods, including serological assay and fecal culture (18, 26). Accordingly, molecular techniques based on hybridization or amplification of species-specific genomic regions have recently been developed. Amplification of specific target DNA by PCR has the potential for early detection of subclinical disease in slaughtered animals and, hence, for better identifying the source of infection (2). This also appears to be of paramount importance, considering the high contagiousness of M. bovis and M. avium subsp. paratuberculosis, their potential role as food-borne pathogens contributing to human pathology, and the interspecies transmission suspected to occur through wildlife (20, 25). However, appropriate control measures require a clear differentiation between M. avium subsp. paratuberculosis, tuberculous mycobacteria, and ubiquitous mycobacteria present in soil, water, and the intestinal tract.
In this respect, various genetic targets have been used, including 16S rRNA sequences (14), multiple-copy insertion elements like IS900 (10), IS901 (15), and IS6110 (28), and unique species-specific determinants (7, 23). Over the last few years, multiplex PCR-based assays have been designed to coidentify distinct mycobacterial strains in the same PCR tube (6, 11, 14). However, it is worth noting that none of these assays has so far allowed the discrimination of M. avium subsp. paratuberculosis from other M. avium subspecies and tuberculous species by direct analysis and in a single reaction. Current assays are mostly based on restriction endonuclease analysis (17) or restriction fragment length polymorphism (8, 31) of DNA amplified from mycobacterial cultures. Commercial reference tests, such as the AccuProbe (Gen-Probe) or the Amplicor (Roche) tests, are available only for M. bovis and M. tuberculosis or M. avium identification. They provide results restricted to a single species per testing, and require prior mycobacterial culture. In contrast, the duplex PCR procedure offers an easy, rapid, and inexpensive way for identifying several mycobacterial species in a single experiment (J.-L. Gala, B. Vandercam, P. Vannufel, M. Reynaert, and P.-F. Laterre, 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. O-14d, 1998). Moreover, the presence of deletions in the DNA sequence upstream of the start signal of the p34 gene in tuberculous bacteria allows the differentiation of M. tuberculosis complex from MAC with a single pair of primers. Simultaneous amplification of the f57 sequence during the same analytical procedure allows the distinguishing of M. avium subsp. paratuberculosis from other M. avium subspecies.
The interest in PCR-based assays is also based on their potential for detecting and identifying mycobacteria directly from clinical samples. However, the sensitivity is not as high as that reported for molecular analysis on short-term cultures. Such a difference can be explained by the presence of inhibitors to the PCR reaction as well as in the difficulty in extracting mycobacterial DNA when the mycobacterial load tends to be low (1). Nonetheless, there have been a few studies reporting a specific molecular identification of either M. bovis (16, 19) or M. avium subsp. paratuberculosis (22, 33), directly from animal tissue samples at slaughter. While a presumptive diagnosis of tuberculosis can be made if a tissue has characteristic histopathologic changes and contains acid-fast bacilli, definitive diagnosis based on culture and species identification of mycobacteria may require several weeks to complete. Accordingly, the clinical performance of our assay was assessed on a series of embedded tissues, all of them disclosing histological evidence of granulomatous lesions, albeit sometimes scarcely.
Compared with these reference methods, molecular results indicate that the duplex PCR assay based on f57 and us-p34 sequences was sensitive enough to detect and correctly identify mycobacterioses associated with marked histological lesions. In the pluribacillary group, direct staining of amplified fragments on agarose gel was indeed successful in five of eight samples, whereas another two samples became positive after Southern blotting and hybridization. Failure in this group was only observed with one Ziehl-Neelsen-negative specimen. Altogether, a positive molecular result was obtained in the majority of Ziehl-Neelsen-positive samples (n = 11 of 12 samples) and amplification followed by Southern blotting was also successful in three Ziehl-Neelsen-negative specimens (one classified as pluribacillary and two classified as paucibacillary).
The p34 and f57 sequence-based duplex assay appears to quickly differentiate visible tuberculous lesions found at postmortem examination of slaughtered animals. Its ability to detect paucibacillary lesions suggests a potential for detecting subclinical diseases, for which diagnosis remains difficult with present tests (13). These features are thought to make it a relevant assay for mycobacterial diagnosis in veterinary medicine. In the case of paratuberculous animals, new infections must indeed be prevented and infected animals should be isolated from the flock, whereas herd slaughtering is compulsory for tuberculous cattle. Conversely, no special measures are taken to isolate cattle infected with other M. avium mycobacteria. In the panel of available detection techniques, the duplex assay could be a useful adjunct, able to confirm dubious cases and suitable for multiple processing of clinical samples in large-scale paratuberculosis control programs. This may contribute to better eradication measures in herds suspected of mycobacterial infection and to limitating of interherd spread.
Although not the topic of this work, the p34 and f57 sequence-based duplex assay may also contribute to rapid differential diagnosis between tuberculosis and opportunistic nontuberculous infections in human diseases, with particular application for HIV type 1 infected patients, among whom the prevalence of nontuberculous mycobacteria is high (9, 21), and for Crohn's disease where a pathogenic role for M. avium subsp. paratuberculosis is still questioned (3).
ACKNOWLEDGMENTS
We are grateful to F. Portaels and M. Fauville-Dufaux for providing mycobacterial isolates.
This work was supported by grant 3.4506.96 from the Fund for Medical Scientific Research (Belgium) and by JSM-R&D-T2, the Joint Staff section of the Belgian Army supporting research and development. The work was started in the ISTO department under the supervision of C. Cocito and J.-F. Denef. ISTO and LBCM university departments contributed equally to it. P.V. was supported by grants 9713655 and 9813902 from the Région wallonne.
REFERENCES
- 1.Catanzano A, Davidson B L, Fujiwara P I, Coldberger M J, Gordin F, Salfinger M, Spodoro J, Schluger N W, Sierra M F, Woods G L. Rapid diagnostic tests for tuberculosis: what is the appropriate use? American Thoracic Society Workshop. Am J Respir Crit Care Med. 1997;155:1804–1814. doi: 10.1164/ajrccm.155.5.9154896. [DOI] [PubMed] [Google Scholar]
- 2.Cetinkaya B, Egan K, Harbour D A, Morgan K L. An abattoir-based study of the prevalence of subclinical Johne's disease in adult cattle in south west England. Epidemiol Infect. 1996;116:373–379. doi: 10.1017/s0950268800052705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cocito C, Gilot P, Coene M, De Kesel M, Poupart P, Vannuffel P. Paratuberculosis. Clin Microbiol Rev. 1994;7:328–345. doi: 10.1128/cmr.7.3.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coetsier C, Havaux X, Mattelard F, Sadatte S, Cormont F, Buergelt K, Limbourg B, Latinne D, Bazin H, Denef J-F, Cocito C. Detection of Mycobacterium avium subsp. paratuberculosis in infected tissues by new species-specific immunohistological procedures. Clin Diagn Lab Immunol. 1998;5:446–451. doi: 10.1128/cdli.5.4.446-451.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cousins D V, Dawson D J. Tuberculosis due to Mycobacterium bovis in the Australian population: cases recorded during 1970-1994. Int J Tuberc Lung Dis. 1999;3:715–721. [PubMed] [Google Scholar]
- 6.Del Portillo P, Thomas M C, Martinez E, Maranon C, Valladares B, Patarroyo M E, Lopez M C. Multiprimer PCR system for differential identification of mycobacteria in clinical samples. J Clin Microbiol. 1996;34:324–328. doi: 10.1128/jcm.34.2.324-328.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellingson J L E, Bolin C A, Stabel J R. Identification of a gene unique to Mycobacterium avium subspecies paratuberculosis and application to diagnosis of paratuberculosis. Mol Cell Probes. 1998;12:133–142. doi: 10.1006/mcpr.1998.0167. [DOI] [PubMed] [Google Scholar]
- 8.Eriks I S, Munck K T, Besser T E, Cantor G H, Kapur V. Rapid differentiation of Mycobacterium avium and M. paratuberculosis by PCR and restriction enzyme analysis. J Clin Microbiol. 1996;34:734–737. doi: 10.1128/jcm.34.3.734-737.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falkinham J O. Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev. 1996;9:177–215. doi: 10.1128/cmr.9.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green E P, Tizard M L, Moss M T, Thompson J, Winterbourne D J, McFadden J J, Hermon-Taylor J. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 1989;17:9063–9073. doi: 10.1093/nar/17.22.9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrera E A, Perez O, Segovia M. Differentiation between Mycobacterium tuberculosis and Mycobacterium bovis by a multiplex-polymerase chain reaction. J Appl Bacteriol. 1996;80:596–604. doi: 10.1111/j.1365-2672.1996.tb03263.x. [DOI] [PubMed] [Google Scholar]
- 12.Johnson-Ifearulundu Y, Kaneene J B, Lloyd J W. Herd-level economic analysis of the impact of paratuberculosis on dairy herds. J Am Vet Med Assoc. 1999;214:822–825. [PubMed] [Google Scholar]
- 13.Kalis C H J, Hesselink J W, Russchen E W, Barkema H W, Collins M T, Visser I J R. Factors influencing the isolation of Mycobacterium avium subsp. paratuberculosis from bovine fecal samples. J Vet Diagn Investig. 1999;11:345–351. doi: 10.1177/104063879901100409. [DOI] [PubMed] [Google Scholar]
- 14.Kox L F F, Jansen H M, Kuijper S, Kolk A H J. Multiplex PCR assay for immediate identification of the infecting species in patients with mycobacterial disease. J Clin Microbiol. 1997;35:1492–1498. doi: 10.1128/jcm.35.6.1492-1498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunze Z M, Wall S, Appelberg R, Silva M T, Portaels F, McFadden J J. IS901, a new member of a wide-spread class of atypical insertion sequences, is associated with pathogenicity in Mycobacterium avium. Mol Microbiol. 1991;5:2265–2272. doi: 10.1111/j.1365-2958.1991.tb02157.x. [DOI] [PubMed] [Google Scholar]
- 16.Liébana E, Aranaz A, Mateos A, Vilafranca M, Gomez-Mampaso E, Tercero J C, Alemany J, Suarez G, Domingo M, Dominguez L. Simple and rapid detection of Mycobacterium tuberculosis complex organisms in bovine tissue samples by PCR. J Clin Microbiol. 1995;33:33–36. doi: 10.1128/jcm.33.1.33-36.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh I, Whittington R, Cousins D. PCR-restriction endonuclease analysis for identification and strain typing of Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. avium based on polymorphisms in IS1311. Mol Cell Probes. 1999;13:115–126. doi: 10.1006/mcpr.1999.0227. [DOI] [PubMed] [Google Scholar]
- 18.McDonald W L, Ridge S E, Hope A F, Condron R J. Evaluation of diagnostic tests for Johne's disease in young cattle. Aust Vet J. 1999;77:113–119. doi: 10.1111/j.1751-0813.1999.tb11679.x. [DOI] [PubMed] [Google Scholar]
- 19.Miller J, Jenny A, Rhyan J, Saari C, Suarez D. Detection of Mycobacterium bovis in formalin-fixed, paraffin-embedded tissues of cattle and elk by PCR amplification of an IS6110 sequence specific for Mycobacterium tuberculosis complex organisms. J Vet Diagn Investig. 1997;9:244–249. doi: 10.1177/104063879700900304. [DOI] [PubMed] [Google Scholar]
- 20.Munroe F A, Dohoo I R, McNab W B, Spangler L. Risk factors for the between-herd spread of Mycobacterium bovis in Canadian cattle and cervids between 1985 and 1994. Prev Vet Med. 1999;41:119–133. doi: 10.1016/s0167-5877(99)00051-3. [DOI] [PubMed] [Google Scholar]
- 21.Naser S A, Felix J, Liping H, Romero C, Naser N, Walsh A, Safranek W. Occurrence of the IS900 gene in Mycobacterium avium complex derived from HIV patients. Mol Cell Probes. 1999;13:367–372. doi: 10.1006/mcpr.1999.0261. [DOI] [PubMed] [Google Scholar]
- 22.Plante Y, Remenda B W, Chelack B J, Haines D M. Detection of Mycobacterium paratuberculosis in formalin-fixed paraffin embedded tissues by the polymerase chain reaction. Can J Vet Res. 1996;60:115–120. [PMC free article] [PubMed] [Google Scholar]
- 23.Poupart P, Coene M, Van Heuverswyn H, Cocito C. Preparation of a specific RNA probe for detection of M. paratuberculosis and diagnosis of Johne's disease. J Clin Microbiol. 1993;31:1601–1605. doi: 10.1128/jcm.31.6.1601-1605.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robert J, Boulahbal F, Trystram D, Truffot-Pernot C, de Benoist A C, Vincent V, Jarlier V, Grosset J. A national survey of human Mycobacterium bovis infection in France. Int J Tuberc Lung Dis. 1999;3:711–714. [PubMed] [Google Scholar]
- 25.Serraino A, Marchetti G, Sanguinetti V, Rossi M C, Zanoni R C, Catozzi L, Bandera A, Dini W, Mignone W, Franzetti F, Gori A. Monitoring of transmission of tuberculosis between wild boars and cattle: genotypical analysis of strains by molecular epidemiology techniques. J Clin Microbiol. 1999;37:2766–2771. doi: 10.1128/jcm.37.9.2766-2771.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stabel J R. An improved method for cultivation of Mycobacterium paratuberculosis from bovine fecal samples and comparison to three other methods. J Vet Diagn Investig. 1997;9:375–380. doi: 10.1177/104063879700900406. [DOI] [PubMed] [Google Scholar]
- 27.Stabel J R. Johne's disease: a hidden threat. J Dairy Sci. 1998;81:283–288. doi: 10.3168/jds.S0022-0302(98)75577-8. [DOI] [PubMed] [Google Scholar]
- 28.Thierry D, Brisson-Noel A, Vincent-Lévy-Frébault V, Nguyen S, Guesdon J L, Gicquel B. Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J Clin Microbiol. 1990;28:2668–2673. doi: 10.1128/jcm.28.12.2668-2673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thoresen O F, Falk K, Evensen O. Comparison of immunohistochemistry, acid-fast staining, and cultivation for detection of Mycobacterium paratuberculosis in goats. J Vet Diagn Investig. 1994;6:195–199. doi: 10.1177/104063879400600210. [DOI] [PubMed] [Google Scholar]
- 30.Vannuffel P, Limbourg B, Dieterich C, Naerhuyzen B, Gilot P, Coene M, Cocito C. Development of species-specific enzyme-linked immunosorbent assay for diagnosis of Johne's disease in cattle. J Clin Microbiol. 1994;32:1211–1216. doi: 10.1128/jcm.32.5.1211-1216.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Soolingen D, Bauer J, Ritacco V, Leão S C, Pavlik I, Vincent V, Rastogi N, Gori A, Bodmer T, Garzelli C, Garcia M J. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: proposal for standardization. J Clin Microbiol. 1998;36:3051–3054. doi: 10.1128/jcm.36.10.3051-3054.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whittington R J, Marsh I, McAllister S, Turner M J, Marshall D J, Fraser C A. Evaluation of modified BACTEC 12B radiometric medium and solid media for culture of Mycobacterium avium subsp. paratuberculosis from sheep. J Clin Microbiol. 1999;37:1077–1083. doi: 10.1128/jcm.37.4.1077-1083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whittington R J, Reddacliff L, Marsh I, Saunders V. Detection of Mycobacterium avium subsp. paratuberculosis in formalin-fixed paraffin-embedded intestinal tissue by IS900 polymerase chain reaction. Aust Vet J. 1999;77:392–397. doi: 10.1111/j.1751-0813.1999.tb10315.x. [DOI] [PubMed] [Google Scholar]