Abstract
N6-methyladenosine (m6A) RNA modification is a reversible mechanism that regulates eukaryotic gene expression. Growing evidence has demonstrated an association between m6A modification and tumorigenesis and response to immunotherapy. However, the overall influence of m6A regulators on the tumor microenvironment and their effect on the response to immunotherapy in lung adenocarcinoma remains to be explored. Here, we comprehensively analyzed the m6A modification patterns of 936 lung adenocarcinoma samples based on 24 m6A regulators. First, we described the features of genetic variation in these m6A regulators. Many m6A regulators were aberrantly expressed in tumors and negatively correlated with most tumor-infiltrating immune cell types. Furthermore, we identified three m6A modification patterns using a consensus clustering method. m6A cluster B was preferentially associated with a favorable prognosis and enriched in metabolism-associated pathways. In contrast, m6A cluster A was associated with the worst prognosis and was enriched in the process of DNA repair. m6A cluster C was characterized by activation of the immune system and a higher stromal cell score. Surprisingly, patients who received radiotherapy had a better prognosis than patients without radiotherapy only in the m6A cluster C group. Subsequently, we constructed an m6A score model that qualified the m6A modification level of individual samples by using principal component analysis algorithms. Patients with high m6A score were characterized by enhanced immune cell infiltration and prolonged survival time and were associated with lower tumor mutation burden and PD-1/CTLA4 expression. The combination of the m6A score and tumor mutation burden could accurately predict the prognosis of patients with lung adenocarcinoma. Furthermore, patients with high m6A score exhibited greater prognostic benefits from radiotherapy and immunotherapy. This study demonstrates that m6A modification is significantly associated with tumor microenvironment diversity and prognosis. A comprehensive evaluation of m6A modification patterns in single tumors will expand our understanding of the tumor immune landscape. In addition, our m6A score model demonstrated that the level of immune cell infiltration plays a significant role in cancer immunotherapy and provides a basis to increase the efficiency of current immune therapies and promote the clinical success of immunotherapy.
Keywords: m6A, lung adenocarcinoma, tumor microenvironment, tumor-infiltrating immune cells, tumor mutation burden, radiotherapy, immunotherapy
Introduction
Lung cancer remains one of the most difficult-to-treat cancers, and its morbidity and mortality are rising rapidly (1). Lung adenocarcinoma (LUAD) accounts for approximately 40% of all lung cancers (2). Driver genes in LUAD include RTKs (aberrantly expressed), EGFR/KRAS (mutations), ALK (rearrangement), and others (3–6). Despite recent advances in surgery, radiotherapy, chemotherapy, targeted therapy, and immunotherapy, the prognosis of patients with LUAD is still unsatisfactory (7). LUAD is a complicated disease with complex pathogenesis and high heterogeneity (8). Therefore, having a good understanding of the molecular mechanisms underlying LUAD is necessary for the selection of optimal therapeutic strategies.
N6-methyladenosine (m6A) RNA modification has recently been identified as a regulatory mechanism for controlling eukaryotic gene expression (9). As a dynamic reversible epigenetic modification, m6A modification exists in mRNAs, microRNAs, circular RNAs, and long noncoding RNAs, accounting for 80% of all RNA methylation modifications in eukaryotic cells (10). m6A modification is mediated by three subtypes of regulatory proteins: methyltransferases (writers), binding proteins (readers), and demethylases (erasers) (11). The modification is mainly regulated by the following components: writers, which catalyze m6A methylation, such as methyltransferase-like 3/14/16 (METTL3/14/16) (12–14), zinc finger CCCH domain-containing protein 13 (ZC3H13) (15), ELAV-like RNA-binding protein 1 (ELAVL1) (16), Cbl proto-oncogene-like 1 (CBLL1) (17), RNA-binding motif protein 15/15B (RBM15/15B) (17, 18), WT1-associated protein (WTAP) (19), and VIR-like m6A methyltransferase associated (KIAA1429) (20). Erasers are proteins involved in maintaining the balance of the m6A content in the transcriptome and include fat mass and obesity-associated protein (FTO) (21) and AlkB homolog H5 (ALKBH5) (22). Readers are proteins that recognize the m6A consensus motif (DRACH) and promote stimulatory and inhibitory effects on translation dynamics, such as YTH domain family 1/2/3 (YTHDF1/2/3) (23, 24), YTH domain containing 1/2 (YTHDC1/2) (25, 26), IGF2 mRNA-binding proteins 1/2/3 (IGF2BP1/2/3) (27, 28), Fragile X mental retardation 1 (FMR1) (29), leucine-rich pentatricopeptide repeat containing (LRPPRC) (29), heterogeneous nuclear ribonucleoprotein C (HNRNPC) (30), and heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1) (31). m6A regulators posttranscriptionally modify RNA molecules and are associated with many biological processes, including carcinogenesis, immune response, cell differentiation, neurodevelopment, and stress responses (9). In addition, m6A mRNA modification may play a significant role in the occurrence and development of human cancers (32), such as lung cancer, hepatic cell carcinoma, and glioblastoma. METTL3 directly promotes YAP translation and increases its activity, which induces resistance to nonsmall cell lung cancer drugs and metastasis (33). Moreover, the upregulation of WTAP contributes to hepatocellular carcinoma tumorigenesis by repressing ETS1 expression (34). Furthermore, the m6A demethylase ALKBH5 maintains the tumorigenicity of stem-like cells by supporting cell proliferation and FOXM1 expression in glioblastoma (35). However, the relationship between m6A modulators and tumors, especially immunotherapy, remains unclear. Therefore, further elucidation of m6A regulatory factors could provide an attractive perspective for cancer therapy (36).
Immune checkpoint therapy has shown unprecedented efficacy for various malignancies by boosting the immune system to fight cancer (37). Immune checkpoints refer to a plethora of inhibitory or stimulatory molecules that maintain self-tolerance, prevent autoimmunity, and control the duration and extent of immune responses, which are hijacked by cancer cells to evade immune eradication (38–40). Immune checkpoint inhibitors (ICIs) target these checkpoints and show remarkable clinical efficacy in a broad spectrum of tumors. Unfortunately, only a considerable proportion of patients receive clinical benefits from ICIs (41). In recent years, many studies have demonstrated the correlation between tumor-infiltrating immune cells and m6A modification patterns, which cannot be explained by RNA degradation mechanisms. Wang et al. reported that the inhibition of m6A modification could enhance the response to anti-PD-1 therapy in pMMR-MSI-L CRC and melanoma (42). ALKBH5 gene expression and mutation status are correlated with response to immunotherapy in melanoma patients, demonstrating that m6A erasers influence the therapeutic effects of immunotherapy (43). However, the overall impact of all m6A regulators on the immune microenvironment and their effect on the response to immunotherapy are still unclear.
In this study, we analyzed genomic information from 936 patients with LUAD to determine their methylation modification patterns. In addition, we constructed an m6A score model to quantify the m6A modification patterns of individual tumors and predict the clinical response to ICI treatment. Our findings clarify the important role of m6A methylation in LUAD and provide clues for improving the efficiency of current immune therapies, which will contribute to the selection of an effective personalized immunotherapy strategy.
Materials and Methods
Data Source and Processing
The expression matrices and corresponding clinical characteristics of LUAD samples were obtained from the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases. We excluded patients without survival, information, or incomplete clinicopathological characteristics from further assessment. A total of 936 patients were enrolled, including the cohorts GSE68465 (N = 438) and TCGA-LUAD (N = 498). The four validation databases were downloaded from the GEO database, including GSE11969, GSE13213, GSE37745, and GSE50081. The expression matrix data of the TCGA-LUAD cohort (FPKM format) were downloaded from the Genomic Data Commons platform, and FPKM units were converted to transcripts per kilobase million (TPM) units. The “Normalized Between Arrays” function of the R package “Limma” was performed for data standardization. Genome mutation data of TCGA-LUAD (including somatic mutation and copy number variation (CNV)) were downloaded from the UCSC Xena platform. Based on previous studies, we collected 24 m6A regulators, including 10 writers (CBLL1, ELAVL1, METTL3, METTL14, METTL16, KIAA1429, RBM15, RBM15B, WTA, and ZC3H13), two erasers (ALKBH5 and FTO), and 12 readers (YTHDC1, YTHDC2, YTHDF1, YTHDF2, YTHDF3, FMR1, HNRNPA2B1, HNRNPC, IGF2BP1, IGF2BP2, IGF2BP3, LRPPRC). Clinicopathological information and clinical immunotherapy scores (IPS) were obtained from the TCIA database.
m6A Modification Pattern
Based on mRNA expression levels, 19 m6A regulators were extracted from the TCGA-LUAD and GSE68465 cohorts, and the samples were divided into diverse subtypes based on transcriptome data with the R package “Consensus Cluster Plus.” A thousand repetitions were performed to guarantee the stability of the classification (44).
Pathway Enrichment Analysis
To investigate the difference between m6A modification patterns in the biological process, we explored the variation in signaling pathways between each of the two subtypes of m6A regulators by using “Gene set variation analysis (GSVA)” R packages (45). We downloaded the gene set file “c2.cp.kegg.v7.4.symbols” from the MSigDB database for the GSVA analysis. An adjusted p-value of less than 0.05 was considered statistically significant.
Analysis of Immune Cell Infiltration
To estimate the relative abundance of 23 immune cell types in the tumor microenvironment of LUAD, single-sample gene set enrichment analysis (ssGSEA) was used to calculate the enrichment scores and represent the relative abundance of each tumor-infiltrating immune cell type in each sample. The set of genes used to label each tumor-infiltrating immune cell type was obtained from the study by Charoentong (46).
Identification of Differentially Expressed Genes Among Subtypes of m6A Regulators
The patients were divided into three subtypes according to the expression level of 19 m6A regulators. We identified differentially expressed genes (DEGs) among different subtypes using the empirical Bayesian method in the R package “Limma,” and selected p-values less than 0.001 as DEG candidates for further analysis (47).
Construction of the m6A Score Model
To quantify the level of m6A modification in a single tumor, we established an m6A score model by performing principal component analysis (PCA). The procedure for the established m6A score model was as follows: first, overlapping DEGs were identified from different m6A clusters, and significant prognosis-related genes were identified by univariate Cox regression analysis; second, PCA was performed on the gene expression profile, and the principal components 1 and 2 were extracted as feature scores. This method minimized the deprivation of information contained in the original index and reduced the indicators to be analyzed, thereby allowing a comprehensive analysis of the collected data; lastly, the m6A score was defined by performing a formula similar to that used in previous studies (48, 49). m6A score = ∑ (PC1i + PC2i), where i is the expression of m6A phenotype-related genes.
Statistical Analysis
All statistical analyses were performed with R software (version 4.0.3). The R package “Limma” was used for differential gene expression analysis. Spearman’s correlation analysis was used to calculate the correlation coefficients between the levels of different tumor-infiltrating immune cell types and the expression of m6A regulators. Kruskal-Wallis test and one-way analysis of variance were used to compare differences between more than two groups. The Kaplan-Meier (KM) method was used to draw the survival curve (5-year survival rate), and univariate Cox regression analysis was used to calculate the hazard ratios (HRs) for m6A regulators and m6A phenotype-related genes. LUAD samples were divided into high and low m6A score subgroups using the “surv-cutpoint” function in the R package “survival.”
Results
Genetic m6A Regulator Variation in LUAD
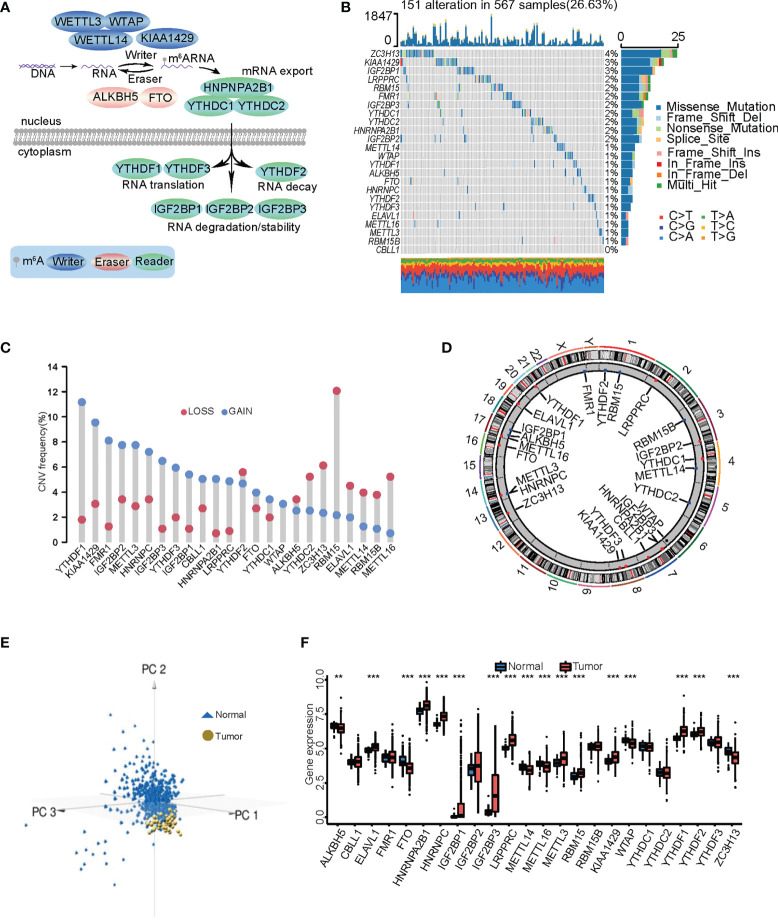
Twenty-four m6A RNA methylation regulators were selected for LUAD according to previous studies, including 10 writers, 12 readers, and two erasers ( Table S1 ). We summarized the dynamic reversible epigenetic modification behavior of these m6A regulators and their biological functions in RNA, including mRNA export, mRNA translation, mRNA decay, and mRNA degradation/stability ( Figure 1A ). Somatic mutations in m6A regulators were found in 151 (26.63%) of 567 samples. ZC3H13 had the highest mutation frequency, followed by KIAA1429 and IGF2BP1, while no CBLL1 mutations were found in the samples ( Figure 1B ). In addition, YTHDC1 was significantly positively correlated with ZC3H13, YTHDC2, FMR1, and HNRNPA2B1 ( Supplementary Figure S1B ). There was widespread CNV in the 24 regulatory factors; METTL16, RBM15B, METTL14, ELAVL1, and RBM15 had copy number losses, while YTHDF1, KIAA1429, FMR1, IGF2BP2, and METTL3 showed numerous copy number gains (gene amplification) ( Figure 1C ). The locations of the CNVs in the 24 m6A regulators were labeled on the chromosomes ( Figure 1D ). Tumor samples were distinguished from normal samples by three-dimensional PCA (3D-PCA) of the 24 m6A regulators. The results showed that the two groups were completely separated from each other ( Figure 1E ). We then compared mRNA expression in normal and tumor samples to explore whether the expression levels of the m6A regulators were affected by the above gene variation. Seventeen of the 24 m6A regulators showed significant overexpression or downregulation in LUAD samples. YTHDF1, KIAA1429, HNRNPC, and METTL3 were most significantly upregulated in tumor samples, and METTL16 and METTL14 were markedly downregulated ( Figure 1F ). These results showed that the CNV was an important factor in controlling the expression of m6A regulators. Most m6A regulators underwent remarkable expression changes in LUAD, suggesting that the abnormal status of m6A regulators is involved in the development of LUAD.
Figure 1.
Gene mutation profile and expression of m6A regulators in lung adenocarcinoma (LUAD). (A) The dynamic and reversible modification process of m6A RNA methylation mediated by 24 m6A regulators and their major biological functions. (B) The mutation frequency of 24 m6A regulators in 567 patients with LUAD. Each column represents each individual patient. The upper bar plot represents TMB. The number on the right represents the mutation frequency in each regulator. The bar graph on the right shows the proportion of each variant type. The stacked bar chart below shows the conversion of each sample. (C) Histogram reflecting the CNV of the m6A regulators. The height of the bar indicates the frequency of variation. Gain, blue; loss, red. (D) The specific location of the CNVs in m6A regulators on 23 chromosomes. (E) Principal component analysis of 24 m6A regulators adapted to distinguishing tumors from normal samples. Tumor was marked with blue and normal with golden. (F) Differences in the expression of the 24 m6A regulators between normal and LUAD tissues. The asterisks show the statistical p-value (** p < 0.01; *** p < 0.001).
Identification of m6A Modification Patterns in LUAD
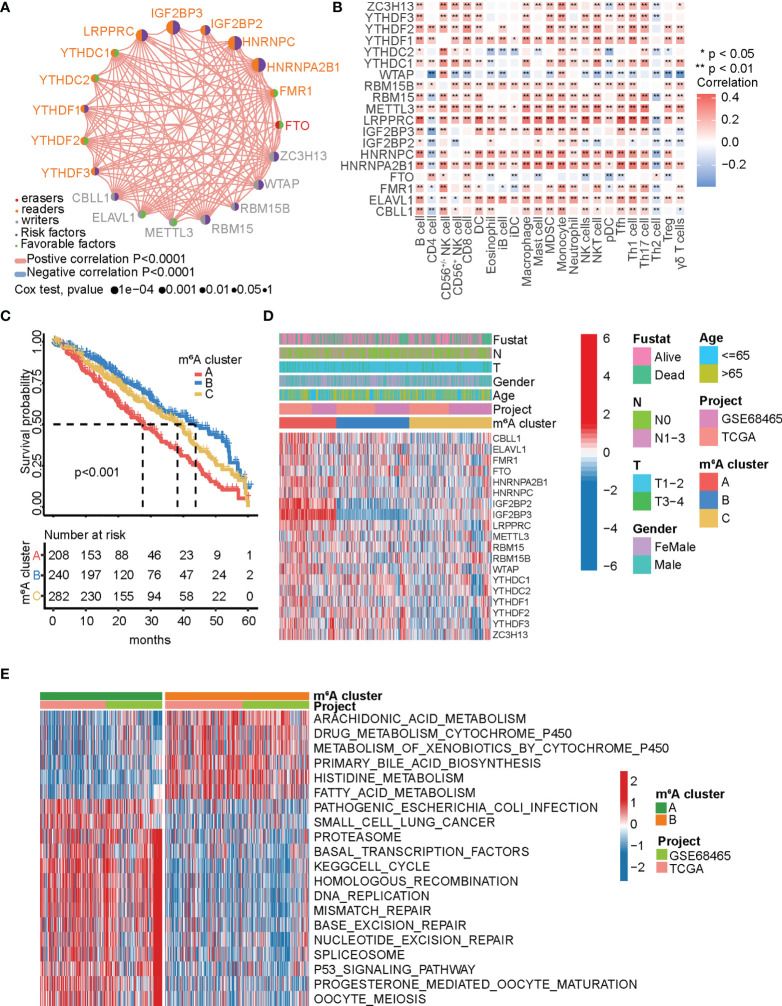
Meta-analysis was performed using TCGA-LUAD and GEO (GSE68465) datasets to further elucidate the modification characteristics of the m6A regulators in LUAD. This algorithm finally identified 19 m6A regulators through data consolidation. To examine whether the 19 m6A regulators could be used as prognostic markers for LUAD, we used univariate Cox regression analysis ( Supplementary Figures S1C and S2A–S ). Eight of the 19 genes were significantly correlated with prognosis, including HNRNPA2B1, HNRNPC, IGF2BP2, IGF2BP3, LRPPRC, RMB15, WTAP, and ZC3H13 (HR >1). The prognostic significance of 19 m6A regulators in LUAD, the connection to the regulator, and the interactions are shown in the m6A regulator network ( Figure 2A ). The results showed a remarkable correlation among writers, readers, and erasers, and this crosstalk was essential for the generation of different m6A modification modes. The landscape also suggested that the occurrence and progression of LUAD are related to m6A regulators. Effective immune infiltration in tumors is considered a key factor in carcinogenesis and prognosis (50, 51). The correlations between individual regulators and each tumor-infiltrating immune cell type were analyzed using Spearman’s correlation analysis ( Figure 2B ). All m6A regulators were significantly correlated with some types of tumor-infiltrating immune cells, suggesting that m6A regulators are critical for immune infiltration in tumors. Most m6A regulators were significantly positively correlated with immune infiltration, while WTAP, an m6A methyltransferase, was negatively correlated.
Figure 2.
m6A methylation patterns and related biological processes. (A) The interplay among the m6A regulators in LUAD. The m6A regulators in three RNA modification types were indicated by the different colors in the circle left. Favorable factors for patients’ survival were indicated by grass green in the circle right and risk factors indicated by blue in the circle right. The circle size indicates the influence of each regulator on prognosis, and the range of values calculated by Log-rank test was represented by the size of each circle. The lines connecting the regulators show their interplay, and the thickness indicates the strength of the association between the regulators. Negative correlation was marked with blue and positive correlation with red. (B) Analysis of the relationship between each tumor-infiltrating immune cell type and each m6A regulator in LUAD using Spearman’s analysis. Red indicates positive correlation; blue indicates negative correlation. * p < 0.05, ** p < 0.01. (C) Survival analyses for the three m6A modification patterns in from TCGA-LUAD and GSE68645, including 208 cases of m6A cluster A, 240 cases of m6A cluster B, and 282 cases of m6A cluster C. Log-rank test, p < 0.0001. (D) Heatmap showing the correlation between the three m6A clusters and the clinicopathological characteristics. Clinicopathological information including age, gender, fustat, and tumor stage, as well as the m6A cluster, is shown in annotations above. Red represents high expression, and blue represents low expression. (E) Heatmap showing the biological processes in different m6A modification patterns obtained by GSVA enrichment analysis. Red shows activated pathways and blue shows inhibited pathways. m6A cluster A vs. m6A cluster B.
Based on the expression of the 19 m6A regulators, the R package “Consensus Cluster Plus” classified patients into various m6A modification patterns. Consequently, the unsupervised consensus clustering algorithm revealed that patients were well defined when k = 3 ( Supplementary Figure S3A ). Thus, three distinct patient clusters were identified based on m6A modification patterns, including 253 cases of cluster A, 324 cases of cluster B, and 359 cases of cluster C. The Kaplan-Meier analysis of the three clusters revealed that cluster A had the worst prognosis, while cluster B had a significant survival advantage ( Figure 2C ). Next, the expression levels of the m6A regulators in the three clusters were analyzed; the expression profiles of most m6A regulators significantly differed among the three clusters. Cluster A was upregulated compared with the other two clusters, including CBLL1, ELAVL1, FMR1, HNRNPA2B1, HNRNPC, IGF2BP2, IGF2BP3, LRPPRC, METTL3, RBM15, RBM15B, WTAP, YTHDF1, and ZC3H13 ( Supplementary Figure S3B ). The expression levels of IGF2BP2 and IGF2BP3 were significantly downregulated in cluster B but upregulated in cluster A ( Figure 2D ). These results indicate that IGF2BP2 and IGF2BP3 are the dominant risk factors for malignant progression. Based on the KEGG gene set, GSVA enrichment analysis was used to explore the biological processes of the two m6A modification patterns ( Figure 2E ; Supplementary Figures S3C, D ). We found that cluster A was clearly enriched in the process of DNA repair, such as nucleotide excision repair, mismatch repair, DNA replication, and base excision repair; cluster B was prominently enriched in metabolism-associated pathways, such as fatty acid metabolism, amino acid metabolism, arachidonic metabolism, drug metabolism cytochrome P450, and primary bile acid biosynthesis. Cluster C was enriched in pathways associated with immune activity, including intestinal immune network production, antigen processing and presentation, and cytokine receptor interaction.
Immune Landscapes With Distinct m6A Modification Patterns
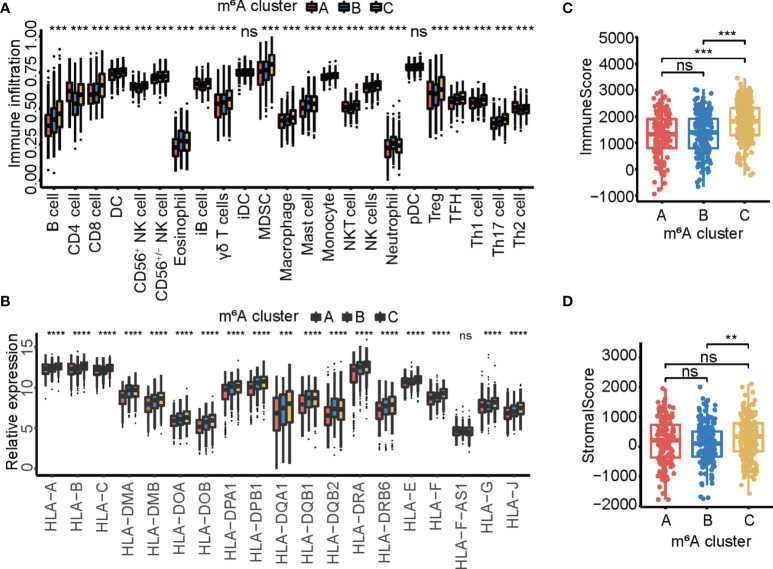
Subsequently, the correlation between 23 tumor-infiltrating immune cell types and three m6A cluster subsets was examined by ssGSEA analysis. The results showed that cluster C was associated with more adaptive and innate immune cell infiltration, including CD8 cells, NK cells, Tregs, MDSCs, macrophages, and B cells ( Figure 3A ). Consistently, cluster C exhibited a comprehensively elevated expression of MHC molecules ( Figure 3B ). Immune cell infiltration in the three m6A cluster subsets was assessed using the ESTIMATE algorithm. The results showed that cluster C exhibited higher immune and stromal scores, indicating that cluster C had significantly increased immune cell infiltration ( Figures 3C, D ), which was mainly due to increased immune infiltration and high MHC expression.
Figure 3.
The immune landscape in three m6A modification patterns. (A) The relative abundance of 23 tumor-infiltrating immune cell types in three m6A modification patterns. The upper and lower ends of the boxes represented interquartile range of values. The lines in the boxes represented median value, and black dots showed outliers. The asterisks represented the statistical p-value (** p < 0.01; *** p < 0.001). (B) The expression of MHC molecules in three m6A modification patterns. The upper and lower ends of the boxes represented interquartile range of values. The lines in the boxes represented median value, and black dots showed outliers. The asterisks represented the statistical p-value (ns p > 0.05; ** p < 0.01; *** p < 0.001). (C) Box plot showing the immune scores of the three m6A clusters. (D) Box plot showing the stromal score of the three m6A clusters.
m6A Phenotype-Related DEGs in LUAD
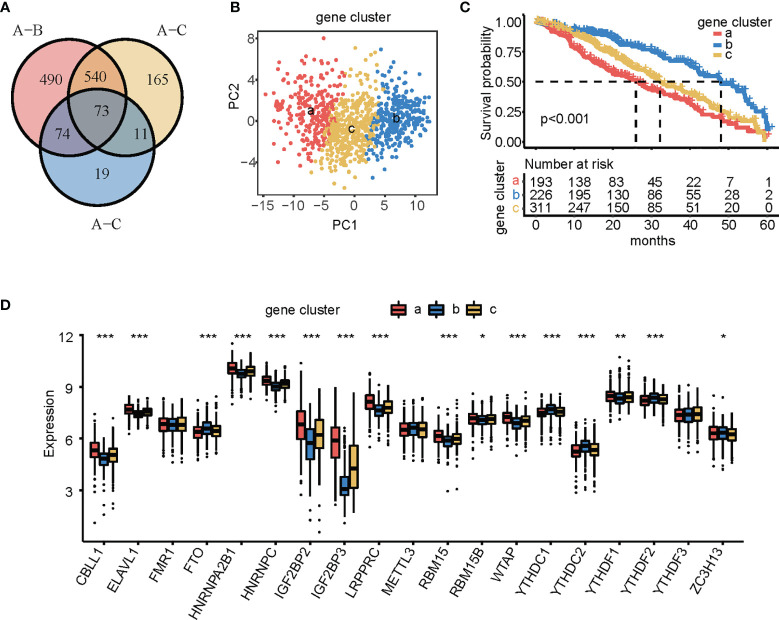
To further elucidate the underlying biological processes and functional annotation of a single m6A modification pattern, seventy-three m6A phenotype-related DEGs were identified using the R package “Limma” and represented on a Venn diagram ( Figure 4A ). GO enrichment analysis for DEGs was performed using the R package “Cluster Profiler.” Surprisingly, these genes were remarkably related to nuclear division and organelle fission ( Supplementary Figure S3E ). To further validate this regulatory mechanism, we obtained 68 prognosis-related DEGs using univariate Cox regression analysis. Unsupervised cluster analysis was performed on the 68 prognosis-related DEGs, which were divided into three subgroups: gene clusters a, b, and c. We performed PCA on the expression of the 68 DEGs, showing that the three groups were completely separated from each other ( Figure 4B ). The results showed 226, 193, and 311 patients in gene clusters a, b, and c, respectively. Patients in gene cluster a were associated with a worse prognosis, patients in gene cluster b had a better prognosis, and patients in gene cluster c had a moderate prognosis ( Figure 4C ). m6A regulator expression was significantly different between the three gene clusters, which was the same as that of the m6A modification patterns ( Figure 4D ).
Figure 4.
Identification of m6A phenotype-related differentially expressed genes (DEGs) and construction of gene clusters. (A) Venn diagram showing 73 m6A phenotype-related DEGs between three m6A clusters. (B) Principal component analysis of three gene cluster patterns. (C) Kaplan-Meier curves showing the overall survival of the patients in the three gene clusters, including 193 cases of gene cluster a, 226 cases of gene cluster b, and 311 cases of gene cluster c. Log-rank test, p < 0.001. (D) Expression of the 24 m6A regulators in the three gene clusters. The upper and lower ends of the boxes represented interquartile range of values. The lines in the boxes represented median value, and black dots showed outliers. The asterisks represented the statistical p-value (* p < 0.05; ** p < 0.01; *** p < 0.001).
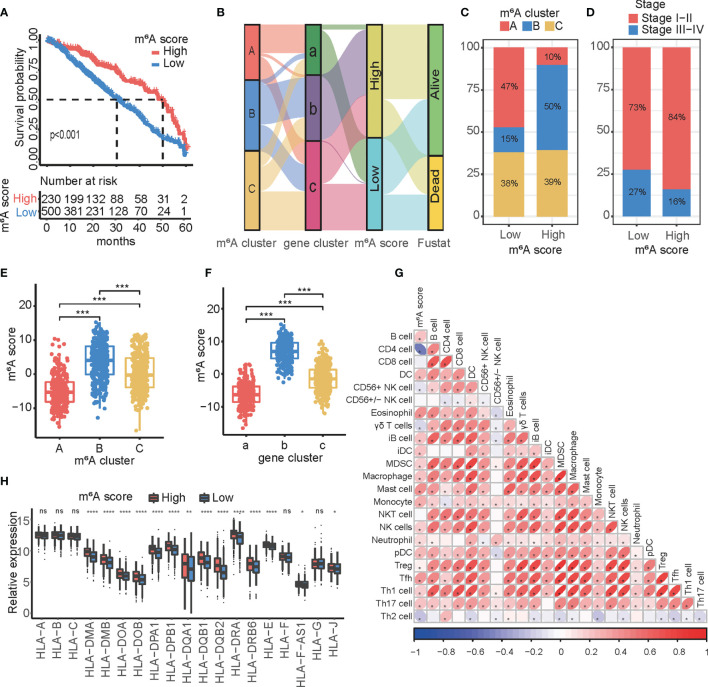
Construction of an m6A Score Model
To predict the immune status and prognosis in a single patient, we sought to develop an m6A score based on the 68 DEGs identified. Therefore, patients were divided into high- or low-m6A score groups based on the cutoff value. The high m6A score group had a better clinical survival profile ( Figure 5A ). In addition, we externally verified m6A score model to predict the prognosis of patients with lung adenocarcinoma from GSE11969, GSE13213, GSE37745, and GSE50081 datasets ( Supplementary Figures S4A–D ). An alluvial diagram was used to illustrate the workflow of the m6A score construction and to visualize the attribute changes in individual patients ( Figure 5B ). The results indicated that gene cluster b was associated with a high m6A score, whereas gene cluster c was associated with a lower m6A score. Notably, most patients who were still alive were included in the high m6A score group. Most patients with m6A cluster A were defined as low m6A, while patients with m6A cluster B had a high m6A score. The m6A cluster C was similarly distributed ( Figure 5C ). The group with a statistically low m6A score had more advanced patients ( Figure 5D ). We also examined the relationship between the above subtypes and the m6A score. Kruskal−Wallis analysis revealed that m6A cluster B and gene cluster b exhibited the highest m6A score, while m6A cluster A and gene cluster a showed the lowest score ( Figures 5E, F ). Multimodel crossvalidation suggested that the m6A score could serve as a prediction model for LUAD. Based on Spearman’s analysis, a heat map demonstrated a positive correlation between the m6A score and tumor-infiltrating immune cells ( Figure 5G ). Furthermore, the high m6A score group also exhibited a comprehensively elevated expression of MHC molecules ( Figure 5H ). These results indicated that the m6A score may be used as a model to predict the immune status in LUAD.
Figure 5.
Construction and characteristics of the m6A score. (A) Kaplan-Meier curves showing the differences in survival of the high (n = 230) and low (n = 500) m6A score groups in LUAD. Log-rank test, p < 0.001. (B) Changes in m6A clusters among groups with different gene clusters, m6A score, and fustat (alive, dead) are shown through an alluvial diagram. (C) The proportion of the three m6A modification patterns in high and low m6A score groups. (D) The proportion of patients with different stages in high and low m6A score groups. (E) Differences in the m6A score between the three m6A clusters. The asterisks represented the statistical p-value (*** p < 0.001). (F) Differences in m6A score between the three gene clusters. The asterisks represented the statistical p-value (*** p < 0.001). (G) Correlations between the m6A score and tumor-infiltrating immune cells using Spearman’s analysis. The positive and negative correlations are marked with red and blue, respectively. (H) Differences in the expression of MHC molecules between the high and low m6A score groups. The upper and lower ends of the boxes represented interquartile range of values. The lines in the boxes represented median value, and black dots showed outliers. The asterisks represented the statistical p value (ns p > 0.05; * p < 0.05; ** p < 0.01; **** p < 0.0001).
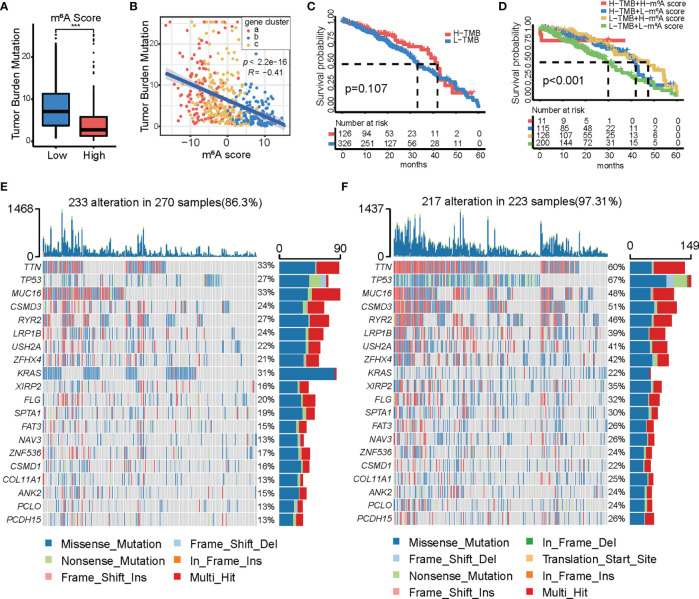
Characteristics of Tumor Somatic Mutations in Patients From the High and Low m6A Score Groups
Several studies have confirmed correlations between tumor somatic mutations, genomic alterations, and immunotherapeutic effects. Therefore, we evaluated the distribution of the tumor mutation burden (TBM) in different m6A score groups. A box and scatter diagram showed that the high m6A score group had a lower TMB and that the m6A score was negatively correlated with TMB ( Figures 6A, B ). K-M survival analysis showed that TMB alone was not sufficient to accurately predict the prognosis of LUAD ( Figure 6C ). To further understand the relationship between TMB, m6A score, and survival outcomes, a K-M survival analysis based on the combination of m6A score and TMB was performed. The results revealed that the low TMB and low m6A score subgroups were associated with poor prognosis. The combination of the m6A score and TMB could accurately predict the quality of life of patients with LUAD ( Figure 6D ). These data emphasize the impact of the m6A score and TMB on cancer development. A list of the somatic mutations in the high- and low-m6A score groups is shown ( Figures 6E, F ). The low m6A score group presented more extensive TMB than the high m6A score group, except for KRAS (22% vs. 31%).
Figure 6.
Characteristics of m6A modification in tumor mutation burden (TMB). (A) Differences in TMB distribution between the high and low m6A score groups. (B) Correlation between TMB and m6A score. (C) Kaplan-Meier curves showing the differences in survival between the high (n = 126) and low (n = 326) TMB groups. Log-rank test, p = 0.107. (D) Survival analyses for subgroup patients stratified by both m6A score and TMB using Kaplan-Meier curves. H, high; L, low. TMB, tumor mutation burden. Log-rank test, p < 0.001. (E, F) Waterfall plot of tumor somatic mutations in patients with high (E) and low (F) m6A score. Each column represents each individual patient. The upper bar plot represents TMB. The number on the right represents the mutation frequency in each regulator. The bar graph on the right shows the proportion of each variant type. The stacked bar chart below shows the conversion of each sample.
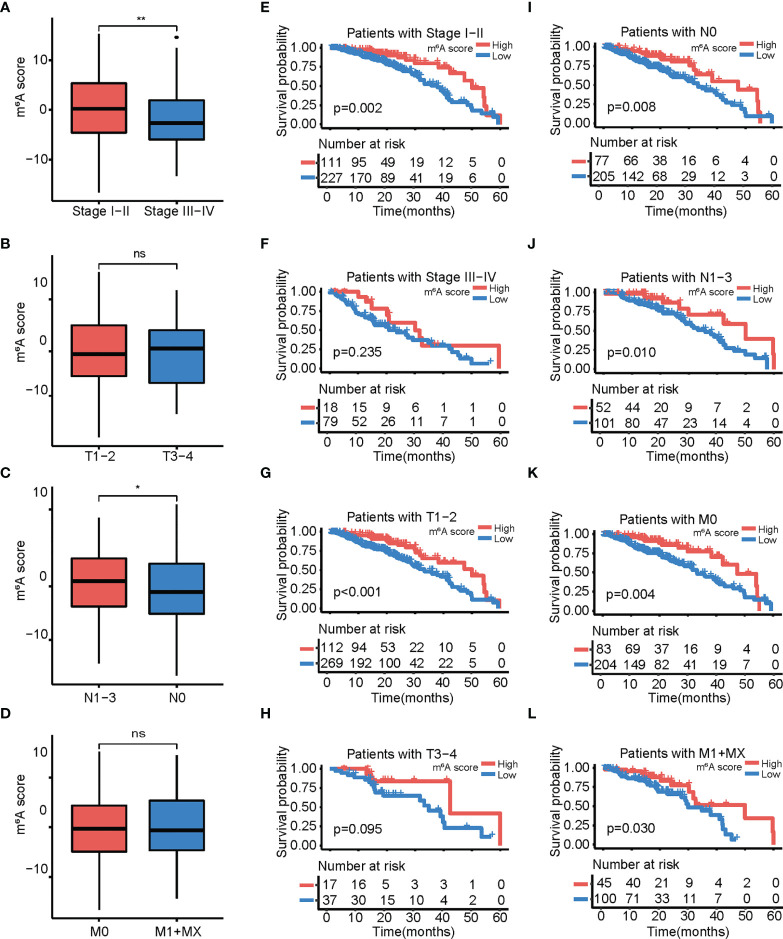
Strong Association of the m6A Score With Clinicopathological Characteristics in LUAD
We examined the relationship between the m6A score and clinicopathological characteristics, and observed that the m6A score significantly differed between patients by stage, node (N), but not tumor (T) and metastasis (M) ( Figures 7A–D ). In this study, the prognostic value of the m6A score was determined for different clinicopathological characteristics, and it was found that patients with high m6A score had significantly longer overall survival than patients with low m6A score in stages I/II, T1/2, N0, and M0 ( Figures 7E–L ). Therefore, we considered the m6A score to be a possible prognostic factor for LUAD.
Figure 7.
Relationship between the m6A score and different clinical characteristics. (A−D) Box plot showing differences in m6A score among patients with different clinical characteristics. (A) Stages I–II vs. stages III–VI; (B) T 1–2 vs. T 3–4; (C) N 1–3 vs. N 0; (D) M 0 vs. M 1+X. (E−I) Kaplan-Meier curves showing the differences in survival depending on the m6A score and different clinical characteristics. (E) Stages I–II. (F) Stages III–VI; (G) T 1–2; (F) T 3–4; (I) N 0; (J) N 1–3; (K) M 0; (L) M 1+X. The asterisks represented the statistical p-value (ns p > 0.05; *p <0.05; **p < 0.01).
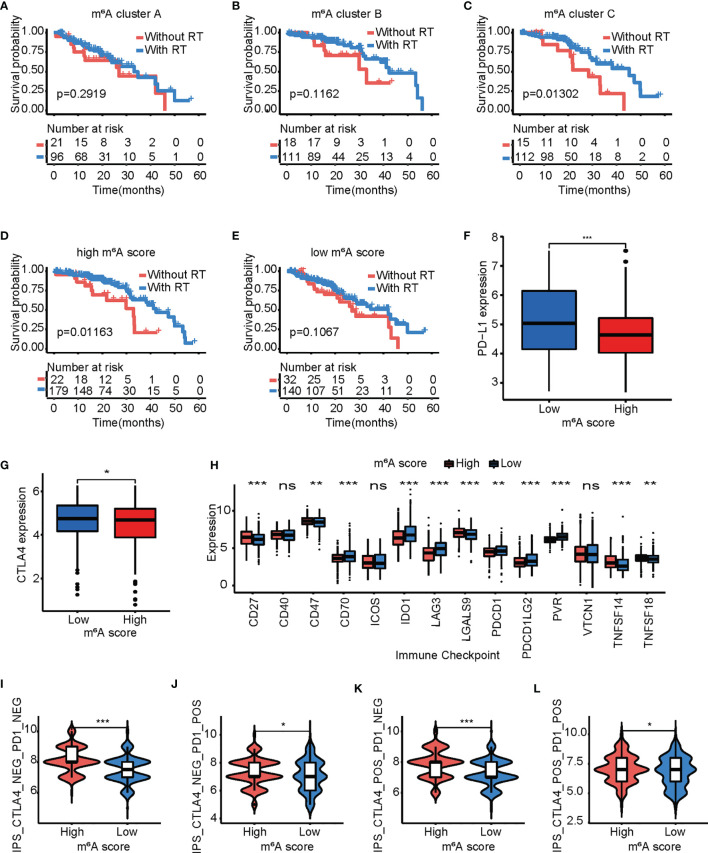
The Role of the m6A Score in Predicting Benefits From Radiotherapy and Immunotherapy
The prognostic value of the m6A score was investigated for patients with LUAD who accepted radiotherapy. Surprisingly, only in the m6A cluster C group did patients with radiotherapy had a better quality of life than those without radiotherapy ( Figures 8A–C ). Interestingly, patients with radiotherapy had a better quality of life than those without radiotherapy in the high m6A score group. The survival advantage from radiotherapy in the low m6A score group was virtually zero ( Figures 8D, E ).
Figure 8.
The m6A score model predicts the benefits of radiotherapy and immunotherapy. (A–C) Kaplan-Meier curves of overall survival for patients in m6A cluster A (A), m6A cluster B (B), and m6A cluster C (C) based on acceptance/rejection of radiotherapy. Log-rank test. (D, E) Kaplan-Meier curves of overall survival for patients with a high (D)/low (E) m6A score based on acceptance/rejection of radiotherapy. Log-rank test. (F, G) Comparison of the PD-1 (F)/CTLA4 (G) expression levels between the high and low m6A score groups. Log-rank test. (ns p > 0.05;* p < 0.05; ** p < 0.01; *** p < 0.001). (H) Differences in the expression of other immune checkpoints between the high and low m6A score groups. Log-rank test. (* p < 0.05; ** p < 0.01; *** p < 0.001). (I–L) Box plot representing the relative distribution of the immunophenoscore between the low and high m6A score groups in LUAD patients based on TCIA database, (I) CTLA4− PD1−; (J) CTLA4− PD1+; (K) CTLA4+ PD1−; and (L) CTLA4+ PD1+. The asterisks represented the statistical p-value (*p < 0.05; ***p < 0.001).
Furthermore, the expression levels of PD-1 and CTLA4 were examined, and a remarkable elevation was observed in the low m6A score group ( Figures 8F, G ). The levels of other immune checkpoints were then compared in the high and low m6A score groups. The high m6A score group exhibited higher expression of CD27, CD28, LGALS9, TNFSF14, and TNFSF18, while the low m6A score group had higher expression of CD70, IDO1, LAG3, PDCD1LG2, and PVR ( Figure 8H ). In addition to the well-known TMB and checkpoint, IPS is one of the newly discovered predictive factors that is widely used and is strongly suggested to assess patients’ reaction to immunotherapy (52, 53). Compared with patients with low m6A score, patients with high m6A score exhibited significant clinical benefits from anti-PD-1/CTLA4 immunotherapy ( Figures 8I–L ). These findings confirmed that the levels of tumor m6A modification modes play a significant role in the regulation of the expression of immune molecules.
Discussion
m6A methylation, the most common form of mRNA modification, plays an indispensable role in posttranscriptional regulation. In recent years, increasing studies have demonstrated the importance of m6A modification in congenital immunity and inflammation, and its antitumor effects through coaction with unequal m6A regulators (54–56). Many studies have focused on single m6A regulators or tumor-infiltrating immune cell types; however, the association between overall tumor microenvironment characteristics and integrated m6A regulators remains poorly understood. Therefore, the distinction of inverse m6A modification modes in tumors will contribute to understanding the relationship between m6A regulators and the antitumor immune response. Here, we constructed a prognostic model for effective therapeutic strategies.
Based on the presentation level of 21 m6A regulators, this study revealed three m6A modification modes with different characteristics. The m6A cluster A was characterized by a poor prognosis and enrichment in the process of DNA repair. The m6A cluster B was characterized by a favorable prognosis and enrichment in metabolism-associated pathways. The m6A cluster C was characterized by activation of the immune system and a higher stromal cell score. Previous studies have reported that the immune microenvironment plays a key role in tumor evolution and immunotherapy (57). The characteristics of tumor immune infiltration, including the activity of CD4 and CD8 T cells, macrophages, and natural killer cells, are associated with immunotherapeutic efficacy (58–60). Here, we verified that the m6A cluster C was significantly associated with elevated immune cell permeation and high stromal cell infiltration. Previous studies demonstrated that tumors with immune-excluded phenotype showed the presence of abundant immune cells, whereas these immune cells do not penetrate the parenchyma but instead are retained in the stroma that surrounds nests of tumor cells (61). Moreover, stromal cells affect the killing effect of IL-12 delivery, empowering CAR-T immune infiltrating cells (62). Thus, it was not surprising to find that m6A cluster C aroused congenital immunity but a poor prognosis.
DEGs that discerned disparate m6A modification patterns were deemed to be m6A phenotype-related gene signatures. Parallel to the m6A clustering construction, three gene cluster types were constructed according to the m6A-related DEGs, which were significantly related to distinct clinical outcomes and landscapes of immune infiltration. These findings suggest that m6A modification is involved in tumorigenesis, tumor development, and immune cell infiltration. To qualify the m6A modification patterns of individual samples, a quantitative model named “the m6A score” was constructed. As a result, m6A cluster B and gene cluster b exhibited higher m6A score, while m6A cluster A and gene cluster a exhibited a lower m6A score. We found that the m6A score was positively associated with immune cell infiltration. Surprisingly, the high m6A score group also exhibited elevated expression of MHC molecules and lower expression of PD-1 and CTLA4.
Our analysis also demonstrated an obvious subtractive association between the m6A score and TMB. Unlike the results of previous studies, there was no disparity in survival between the high and low TMB groups in LUAD, while the incorporation of the m6A score and TMB level could refine the clinical outcomes of patients with LUAD. We also confirmed that the m6A score could be used to evaluate the clinicopathological characteristics of patients involving the clinical stage. Exhaustive associations between the m6A score and clinicopathological characteristics could be discovered in this study. Similarly, the m6A score could play a role as a stand-alone prognostic biomarker for prognosis. A high m6A score and the m6A cluster C subgroup were beneficial to radiotherapy and anti-PD-1/CTLA4 immunotherapy, which was due to increased immune cell infiltration and immunocompetence. The m6A score model could predict the power of adjuvant radiotherapy and the clinical effect of the patient type on the response to anti-PD-1/CTLA4 immunotherapy. These findings provide new insights into the relationship between tumor-infiltrating immune cells and cancer immunotherapy, and increase our capacity to select clinical immunotherapy strategies.
We compiled a list of 24 identified m6A regulators; however, newly recognized regulators must be integrated into the model to achieve the highest precision with the m6A modification patterns. Furthermore, the m6A modification patterns and m6A score were distinguished by employing retrospective data collection; therefore, a future cohort of patients with LUAD accepting immunotherapy is required to confirm our findings. In addition, since immunotherapy showed strong clinical advantages in a fraction of patients with high m6A score, more clinical cases and tumor types should be introduced into the predicted models to increase precision.
Conclusion
In conclusion, we systematically analyzed m6A modification patterns among 936 LUAD samples considering 24 m6A regulators, and comprehensively evaluated their prognostic value and correlation with tumor-infiltration immune cell characteristics. The comprehensive analysis of individual tumor m6A modification patterns will greatly enhance our understanding of the tumor microenvironment and the characterization of immune cell infiltration. This study provides a basis for improving current immune therapies and promoting the clinical success of immunotherapy. However, due to the small number of samples and the clinical heterogeneity of the study cohort, large-scale cohort studies and prospective studies are necessary to verify the predictive value of the m6A score in the clinical treatment and prognosis of LUAD.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Ethics Statement
The studies involving human participants were reviewed and approved by the Medical Ethics Committee of Guangzhou Medical University Tumor Hospital. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Medical Ethics Committee of Guangzhou Medical University Tumor Hospital. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
MZ and YC have contributed equally to this work. MZ, YL, XZ, and QY designed the experiments. MZ, YL, and QY wrote the paper. MZ, YC, QM, JZ, TZ, YX, ZW, and DS analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (grant numbers 81870409, to QY) and Guangzhou Key Medical Discipline Construction Project (to QY).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
- m6A
N6-methyladenosine
- LUAD
lung adenocarcinoma
- METTL3
methyltransferase-like 3
- METTL14
methyltransferase-like 14
- METTL16
methyltransferase-like16
- ZC3H13
zinc finger CCCH domain-containing protein 13
- ELAVL1
ELAV-like RNA binding protein 1
- CBLL1
Cbl proto-oncogene-like 1
- RBM15
RNA-binding motif protein 15
- RBM15B
RNA-binding motif protein 15B
- WTAP
WT1-associated protein
- KIAA1429
VIR-like m6A methyltransferase associated
- FTO
fat mass and obesity-associated protein
- ALKBH5
AlkB homolog H5
- YTHDF1
YTH domain family 1
- YTHDF2
YTH domain family 2
- YTHDF3
YTH domain family 3
- YTHDC1
YTH domain containing 1
- YTHDC2
YTH domain containing 2
- IGF2BP1
IGF2 mRNA-binding proteins 1
- IGF2BP2
IGF2 mRNA-binding proteins 2
- IGF2BP3
IGF2 mRNA-binding proteins 3
- FMR1
Fragile X mental retardation 1
- LRPPRC
leucine-rich pentatricopeptide repeat containing
- HNRNPC
heterogeneous nuclear ribonucleoprotein C
- HNRNPA2B1
heterogeneous nuclear ribonucleoprotein A2/B1
- NMF
nonnegative matrix factorization
- ssGESA
single sample GSEA
- PCA
principal component analysis
- TME
tumor microenvironment
- TMB
tumor mutation burden
- ICIs
immune checkpoint inhibitors
- TCGA
The Cancer Genome Atlas
- GEO
Gene Expression Omnibus
- GSVA
gene set variation analysis
- DEGs
differentially expressed genes
- HR
hazard ratios
- CNV
copy number variation
- 3D-PCA
three-dimensional principal component analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MHC
major histocompatibility complex
- MSI
microsatellite instability
- IPS
immunophenoscore
- B cell
activated B cell
- CD4 cell
activated CD4 T cell
- CD8 cell
activated CD8 T cell
- DC
activated dendritic cell
- CD56+ NK cell
CD56bright natural killer cell
- CD56+/− NK cell
CD56dim natural killer cell
- eosinophil
eosinophil
- γδ T cells
gamma delta T cell
- iB cell
immature B cell
- iDC
immature dendritic cell
- NKT cell
natural killer T cell
- NK cells
natural killer cell
- pDC
plasmacytoid dendritic cell
- Treg
regulatory T cell
- Tfh
T follicular helper cell
- Th1 cell
T helper 1 cell
- Th2 cell
T helper 2 cell
- Th17 cell
T helper 17 cell
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.782551/full#supplementary-material
The correlation and prognostic analysis of 19 m6A regulators. (A) Overview of this analysis. (B) Correlation analyses for 19 m6A regulators. Red indicates positive correlation; blue indicates negative correlation. (*P < 0.05; **P < 0.01; ***P < 0.001). (C) Prognostic analyses for 19 m6A regulators using univariate Cox regression analysis. Hazard ratio >1 indicated risk factors for survival and represent by red. hazard ratio <1 indicated risk factors for survival and represented by blue.
Prognostic analysis of 19 m6A regulators. (A–S). Kaplan-Meier survival curves are used to analyze the survival difference between high and low gene expression groups of 19 m6A regulators. High gene expression groups indicated by red. Low gene expression groups indicated by blue.
Consensus clustering of 24 m6A regulators in lung adenocarcinoma. (A) Consensus clustering matrix for k = 3. (B) Expression of the 24 m6A regulators in the three m6A clusters. The upper and lower ends of the boxes represented interquartile range of values. The lines in the boxes represented median value, and black dots showed outliers. The asterisks represented the statistical p value (*P< 0.05; **P< 0.01; ***P< 0.001). (C, D) Heatmap showing the biological processes in different m6A modification patterns obtained by GSVA enrichment analysis. Red shows activated pathways and blue shows inhibited pathways. B m6A cluster A vs m6A cluster C; C m6A cluster B vs m6A cluster C. (E) GO enrichment analysis of the 73 m6A phenotype-related DEGs.
External validation of the m6A score model in lung adenocarcinoma patients from GSE11969, GSE13213, GSE37745, and GSE50081 datasets. (A) Kaplan-Meier curves showing the differences in survival of the high and low m6A score groups in GSE11969 (P = 0.002, Log-rank test). (B) Kaplan-Meier curves showing the differences in survival of the high and low m6A score groups in GSE13213 (P = 0.002, Log-rank test). (C) Kaplan-Meier curves showing the differences in survival of the high and low m6A score groups in GSE37745 (P = 0.032, Log-rank test). (D) Kaplan-Meier curves showing the differences in survival of the high and low m6A score groups in GSE50081 (P < 0.001, Log-rank test).
References
- 1. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Jr, Wu YL, et al. Lung Cancer: Current Therapies and New Targeted Treatments. Lancet (2017) 389:299–311. doi: 10.1016/S0140-6736(16)30958-8 [DOI] [PubMed] [Google Scholar]
- 2. Denisenko TV, Budkevich IN, Zhivotovsky B. Cell Death-Based Treatment of Lung Adenocarcinoma. Cell Death Dis (2018) 9:017–0063. doi: 10.1038/s41419-017-0063-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen Z, Chen J, Ren D, Zhang J, Yang Y, Zhang H, et al. EPHA5 Mutations Predict Survival After Immunotherapy in Lung Adenocarcinoma. Aging (2020) 13:598–618. doi: 10.18632/aging.202169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, et al. The Transcriptional Landscape and Mutational Profile of Lung Adenocarcinoma. Genome Res (2012) 22:2109–19. doi: 10.1101/gr.145144.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saito M, Suzuki H, Kono K, Takenoshita S, Kohno T. Treatment of Lung Adenocarcinoma by Molecular-Targeted Therapy and Immunotherapy. Surg Today (2018) 48:1–8. doi: 10.1007/s00595-017-1497-7 [DOI] [PubMed] [Google Scholar]
- 6. Giroux Leprieur E, Dumenil C, Julie C, Giraud V, Dumoulin J, Labrune S, et al. Immunotherapy Revolutionises Non-Small-Cell Lung Cancer Therapy: Results, Perspectives and New Challenges. Eur J Cancer (2017) 78:16–23. doi: 10.1016/j.ejca.2016.12.041 [DOI] [PubMed] [Google Scholar]
- 7. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2019. CA Cancer J Clin (2019) 69:7–34. doi: 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 8. Luo W, Tian P, Wang Y, Xu H, Chen L, Tang C, et al. Characteristics of Genomic Alterations of Lung Adenocarcinoma in Young Never-Smokers. Int J Cancer (2018) 143:1696–705. doi: 10.1002/ijc.31542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao BS, Roundtree IA, He C. Post-Transcriptional Gene Regulation by mRNA Modifications. Nat Rev Mol Cell Biol (2017) 18:31–42. doi: 10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yi YC, Chen XY, Zhang J, Zhu JS. Novel Insights Into the Interplay Between m(6)A Modification and Noncoding RNAs in Cancer. Mol Cancer (2020) 19:020–01233. doi: 10.1186/s12943-020-01233-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-Methyladenosine and Its Role in Cancer. Mol Cancer (2019) 18:019–1109. doi: 10.1186/s12943-019-1109-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schumann U, Shafik A, Preiss T. METTL3 Gains R/W Access to the Epitranscriptome. Mol Cell (2016) 62:323–4. doi: 10.1016/j.molcel.2016.04.024 [DOI] [PubMed] [Google Scholar]
- 13. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat Chem Biol (2014) 10:93–5. doi: 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell (2017) 169:824–35. doi: 10.1016/j.cell.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell (2018) 69:1028–38. doi: 10.1016/j.molcel.2018.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, et al. METTL3-Mediated N6-Methyladenosine Modification Is Critical for Epithelial-Mesenchymal Transition and Metastasis of Gastric Cancer. Mol Cancer (2019) 18:019–1065. doi: 10.1186/s12943-019-1065-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The Role of m6A Modification in the Biological Functions and Diseases. Signal Transduct Target Ther (2021) 6:020–00450. doi: 10.1038/s41392-020-00450-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. M(6)A RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature (2016) 537:369–73. doi: 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ. Mammalian WTAP Is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res (2014) 24:177–89. doi: 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA Mediates Preferential m(6)A mRNA Methylation in 3’UTR and Near Stop Codon and Associates With Alternative Polyadenylation. Cell Discov (2018) 4:018–0019. doi: 10.1038/s41421-018-0019-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-Methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat Chem Biol (2011) 7:885–7. doi: 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 Is a Mammalian RNA Demethylase That Impacts RNA Metabolism and Mouse Fertility. Mol Cell (2013) 49:18–29. doi: 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-Methyladenosine Modulates Messenger RNA Translation Efficiency. Cell (2015) 161:1388–99. doi: 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ. YTHDF3 Facilitates Translation and Decay of N(6)-Methyladenosine-Modified RNA. Cell Res (2017) 27:315–28. doi: 10.1038/cr.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell (2016) 61:507–19. doi: 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 26. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 Is an N(6)-Methyladenosine Binding Protein That Regulates Mammalian Spermatogenesis. Cell Res (2017) 27:1115–27. doi: 10.1038/cr.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Müller S, Glaß M, Singh AK, Haase J, Bley N, Fuchs T, et al. IGF2BP1 Promotes SRF-Dependent Transcription in Cancer in a m6a- and miRNA-Dependent Manner. Nucleic Acids Res (2019) 47:375–90. doi: 10.1093/nar/gky1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H. Recognition of RNA N(6)-Methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation. Nat Cell Biol (2018) 20:285–95. doi: 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arguello AE, DeLiberto AN, Kleiner RE. RNA Chemical Proteomics Reveals the N(6)-Methyladenosine (m(6)A)-Regulated Protein-RNA Interactome. J Am Chem Soc (2017) 139:17249–52. doi: 10.1021/jacs.7b09213 [DOI] [PubMed] [Google Scholar]
- 30. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-Methyladenosine-Dependent RNA Structural Switches Regulate RNA-Protein Interactions. Nature (2015) 518:560–4. doi: 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF, et al. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell (2015) 162:1299–308. doi: 10.1016/j.cell.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang S, Sun C, Li J, Zhang E, Ma Z, Xu W, et al. Roles of RNA Methylation by Means of N(6)-Methyladenosine (m(6)A) in Human Cancers. Cancer Lett (2017) 408:112–20. doi: 10.1016/j.canlet.2017.08.030 [DOI] [PubMed] [Google Scholar]
- 33. Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. Correction to: m6A mRNA Methylation Initiated by METTL3 Directly Promotes YAP Translation and Increases YAP Activity by Regulating the MALAT1-miR-1914-3p-YAP Axis to Induce NSCLC Drug Resistance and Metastasis. J Hematol Oncol (2020) 13:020–00942. doi: 10.1186/s13045-020-00942-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Y, Peng C, Chen J, Chen D, Yang B, He B, et al. WTAP Facilitates Progression of Hepatocellular Carcinoma via M6a-HuR-Dependent Epigenetic Silencing of ETS1. Mol Cancer (2019) 18:127. doi: 10.1186/s12943-019-1053-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z. M(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-Like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell (2017) 31:591–606. doi: 10.1016/j.ccell.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the Tumor Immune Microenvironment (TIME) for Effective Therapy. Nat Med (2018) 24:541–50. doi: 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Galluzzi L, Humeau J, Buqué A, Zitvogel L, Kroemer G. Immunostimulation With Chemotherapy in the Era of Immune Checkpoint Inhibitors. Nat Rev Clin Oncol (2020) 17:725–41. doi: 10.1038/s41571-020-0413-z [DOI] [PubMed] [Google Scholar]
- 38. Xu F, Jin T, Zhu Y, Dai C. Immune Checkpoint Therapy in Liver Cancer. J Exp Clin Cancer Res (2018) 37(1):018–0777.doi: 10.1186/s13046-018-0777-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. ElTanbouly MA, Noelle RJ. Rethinking Peripheral T Cell Tolerance: Checkpoints Across a T Cell’s Journey. Nat Rev Immunol (2021) 21:257–267. doi: 10.1038/s41577-020-00454-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marin-Acevedo JA, Dholaria B, Soyano AE, Knutson KL, Chumsri S, Lou Y. Next Generation of Immune Checkpoint Therapy in Cancer: New Developments and Challenges. J Hematol Oncol (2018) 11:018–0582. doi: 10.1186/s13045-018-0582-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, Activity, and Immune Correlates of Anti-PD-1 Antibody in Cancer. N Engl J Med (2012) 366:2443–54. doi: 10.15252/embj.2020104514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang L, Hui H, Agrawal K, Kang Y, Li N, Tang R, et al. M(6) A RNA Methyltransferases METTL3/14 Regulate Immune Responses to Anti-PD-1 Therapy. EMBO J (2020) 39:e104514. doi: 10.15252/embj.2020104514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li N, Kang Y, Wang L, Huff S, Tang R, Hui H, et al. ALKBH5 Regulates Anti-PD-1 Therapy Response by Modulating Lactate and Suppressive Immune Cell Accumulation in Tumor Microenvironment. Proc Natl Acad Sci USA (2020) 117:20159–70. doi: 10.1073/pnas.1918986117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilkerson MD, Hayes DN. ConsensusClusterPlus: A Class Discovery Tool With Confidence Assessments and Item Tracking. Bioinformatics (2010) 26:1572–3. doi: 10.1093/bioinformatics/btq170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hänzelmann S, Castelo R, Guinney J. GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data. BMC Bioinformatics (2013) 14:7. doi: 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-Cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep (2017) 18:248–62. doi: 10.1016/j.celrep.2016.12.019 [DOI] [PubMed] [Google Scholar]
- 47. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res (2015) 43:20. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zeng D, Li M, Zhou R, Zhang J, Sun H, Shi M, et al. Tumor Microenvironment Characterization in Gastric Cancer Identifies Prognostic and Immunotherapeutically Relevant Gene Signatures. Cancer Immunol Res (2019) 7:737–50. doi: 10.1158/2326-6066.CIR-18-0436 [DOI] [PubMed] [Google Scholar]
- 49. Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and Microenvironment Evolution During Immunotherapy With Nivolumab. Cell (2017) 171:934–49. doi: 10.1016/j.cell.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res (2019) 79:4557–66. doi: 10.1158/0008-5472.CAN-18-3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural Innate and Adaptive Immunity to Cancer. Annu Rev Immunol (2011) 29:235–71. doi: 10.1146/annurev-immunol-031210-101324 [DOI] [PubMed] [Google Scholar]
- 52. Chen H, Yang M, Wang Q, Song F, Li X, Chen K. The New Identified Biomarkers Determine Sensitivity to Immune Check-Point Blockade Therapies in Melanoma. Oncoimmunology (2019) 8:1608132. doi: 10.1080/2162402X.2019.1608132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen H, Chong W, Wu Q, Yao Y, Mao M, Wang X. Association of LRP1B Mutation With Tumor Mutation Burden and Outcomes in Melanoma and Non-Small Cell Lung Cancer Patients Treated With Immune Check-Point Blockades. Front Immunol (2019) 10:1113. doi: 10.3389/fimmu.2019.01113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen XY, Zhang J, Zhu JS. The Role of m(6)A RNA Methylation in Human Cancer. Mol Cancer (2019) 18:019–1033. doi: 10.1186/s12943-019-1033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shulman Z, Stern-Ginossar N. The RNA Modification N(6)-Methyladenosine as a Novel Regulator of the Immune System. Nat Immunol (2020) 21:501–12. doi: 10.1038/s41590-020-0650-4 [DOI] [PubMed] [Google Scholar]
- 56. Jian D, Wang Y, Jian L, Tang H, Rao L, Chen K, et al. METTL14 Aggravates Endothelial Inflammation and Atherosclerosis by Increasing FOXO1 N6-Methyladeosine Modifications. Theranostics (2020) 10:8939–56. doi: 10.7150/thno.45178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang Y, Zhang Z. The History and Advances in Cancer Immunotherapy: Understanding the Characteristics of Tumor-Infiltrating Immune Cells and Their Therapeutic Implications. Cell Mol Immunol (2020) 17:807–21. doi: 10.1038/s41423-020-0488-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. DeRogatis JM, Viramontes KM, Neubert EN, Tinoco R. PSGL-1 Immune Checkpoint Inhibition for CD4(+) T Cell Cancer Immunotherapy. Front Immunol (2021) 12. doi: 10.3389/fimmu.2021.636238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kumai T, Mizukoshi E, Hashiba T, Nakagawa H, Kitahara M, Miyashita T, et al. Effect of Adoptive T-Cell Immunotherapy on Immunological Parameters and Prognosis in Patients With Advanced Pancreatic Cancer. Cytotherapy (2021) 23:137–45. doi: 10.1016/j.jcyt.2020.08.001 [DOI] [PubMed] [Google Scholar]
- 60. Barkal AA, Brewer RE, Markovic M, Kowarsky M, Barkal SA, Zaro BW, et al. CD24 Signalling Through Macrophage Siglec-10 Is a Target for Cancer Immunotherapy. Nature (2019) 572:392–6. doi: 10.1038/s41586-019-1456-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen DS, Mellman I. Elements of Cancer Immunity and the Cancer-Immune Set Point. Nature (2017) 541:321–30. doi: 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- 62. Agliardi G, Liuzzi AR, Hotblack A, De Feo D, Núñez N, Stowe CL, et al. Intratumoral IL-12 Delivery Empowers ÊR-T Cell Immunotherapy in a Pre-Clinical Model of Glioblastoma. Nat Commun (2021) 12:020–20599. doi: 10.1038/s41467-020-20599-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The correlation and prognostic analysis of 19 m6A regulators. (A) Overview of this analysis. (B) Correlation analyses for 19 m6A regulators. Red indicates positive correlation; blue indicates negative correlation. (*P < 0.05; **P < 0.01; ***P < 0.001). (C) Prognostic analyses for 19 m6A regulators using univariate Cox regression analysis. Hazard ratio >1 indicated risk factors for survival and represent by red. hazard ratio <1 indicated risk factors for survival and represented by blue.
Prognostic analysis of 19 m6A regulators. (A–S). Kaplan-Meier survival curves are used to analyze the survival difference between high and low gene expression groups of 19 m6A regulators. High gene expression groups indicated by red. Low gene expression groups indicated by blue.
Consensus clustering of 24 m6A regulators in lung adenocarcinoma. (A) Consensus clustering matrix for k = 3. (B) Expression of the 24 m6A regulators in the three m6A clusters. The upper and lower ends of the boxes represented interquartile range of values. The lines in the boxes represented median value, and black dots showed outliers. The asterisks represented the statistical p value (*P< 0.05; **P< 0.01; ***P< 0.001). (C, D) Heatmap showing the biological processes in different m6A modification patterns obtained by GSVA enrichment analysis. Red shows activated pathways and blue shows inhibited pathways. B m6A cluster A vs m6A cluster C; C m6A cluster B vs m6A cluster C. (E) GO enrichment analysis of the 73 m6A phenotype-related DEGs.
External validation of the m6A score model in lung adenocarcinoma patients from GSE11969, GSE13213, GSE37745, and GSE50081 datasets. (A) Kaplan-Meier curves showing the differences in survival of the high and low m6A score groups in GSE11969 (P = 0.002, Log-rank test). (B) Kaplan-Meier curves showing the differences in survival of the high and low m6A score groups in GSE13213 (P = 0.002, Log-rank test). (C) Kaplan-Meier curves showing the differences in survival of the high and low m6A score groups in GSE37745 (P = 0.032, Log-rank test). (D) Kaplan-Meier curves showing the differences in survival of the high and low m6A score groups in GSE50081 (P < 0.001, Log-rank test).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .