Highlights
-
•
We used resting-state fMRI data from the Philadelphia Neurodevelopmental Cohort to derive a brain age prediction model based on functional brain connectivity.
-
•
The model was able to predict brain age in independent data (the Healthy Brain Network sample).
-
•
Our findings support consistency in functional connectivity patterns between samples and scanners and support the utility of the brain age prediction framework for fMRI based multisite investigations.
Keywords: Machine learning, Brain age, Development, Resting-state fMRI, The Healthy brain network, The Philadelphia neurodevelopmental cohort
Abstract
Objective
Magnetic resonance imaging (MRI) has shown that estimated brain age is deviant from chronological age in various common brain disorders. Brain age estimation could be useful for investigating patterns of brain maturation and integrity, aiding to elucidate brain mechanisms underlying these heterogeneous conditions. Here, we examined functional brain age in two large samples of children and adolescents and its relation to mental health.
Methods
We used resting-state fMRI data from the Philadelphia Neurodevelopmental Cohort (PNC; n = 1126, age range 8–22 years) to estimate functional connectivity between brain networks, and utilized these as features for brain age prediction. We applied the prediction model to 1387 individuals (age range 8–22 years) in the Healthy Brain Network sample (HBN). In addition, we estimated brain age in PNC using a cross-validation framework. Next, we tested for associations between brain age gap and various aspects of psychopathology and cognitive performance.
Results
Our model was able to predict age in the independent test samples, with a model performance of r = 0.54 for the HBN test set, supporting consistency in functional connectivity patterns between samples and scanners. Linear models revealed a significant association between brain age gap and psychopathology in PNC, where individuals with a lower estimated brain age, had a higher overall symptom burden. These associations were not replicated in HBN.
Discussion
Our findings support the use of brain age prediction from fMRI-based connectivity. While requiring further extensions and validations, the approach may be instrumental for detecting brain phenotypes related to intrinsic connectivity and could assist in characterizing risk in non-typically developing populations.
1. Introduction
Psychiatric disorders are complex disorders with substantial heterogeneity in symptoms and prognosis, and comorbidities are usually the rule rather than the exception (Craddock and Owen, 2010, Krueger and Bezdjian, 2009, Nemeroff, 2002). This heterogeneity is also apparent in the brain, and magnetic resonance imaging (MRI) studies suggest greater structural variability among patients compared to healthy controls (Alnaes et al., 2019). This poses substantial challenges for elucidating the brain mechanisms underlying these disorders, and observed effects for neuromarkers are commonly minor in mental health research (Jollans and Whelan, 2018, Linden, 2012, Paulus and Thompson, 2019). This is further exacerbated by the lack of strong mapping between the current symptom-based nosology and the underlying biology (Owen, 2014). Given this multivariate complexity, studies have increasingly turned to machine-learning approaches that can utilize large amounts of data to provide individual-level predictions by the use of dimensionality reduction and pattern recognition (Cao and Schwarz, 2020, Mansourvar et al., 2020). The aim is to provide a better mapping of brain imaging data to symptoms, cognition and behavior.
One avenue provided by machine learning is its utility to map the substantial anatomical and functional changes that the brain undergoes throughout life. Training machine learning models to predict chronological age from brain imaging data allows us to derive the apparent age of the brain - referred to as ‘brain age’ - and its deviation from chronological age, referred to as the ‘brain age gap’ (Franke, Ziegler, Kloppel, Gaser, & Alzheimer's Disease Neuroimaging, 2010). Depending on the modality that is fed into the model, such estimates of apparent brain age can reflect different neurophysiology such as anatomical brain age (Franke et al., 2010) and functional brain age (Dosenbach et al., 2010). It is also possible to study this process for specific parts of the brain to gain insights into region specific alterations (Kaufmann et al., 2019). Studies that have applied brain age prediction to clinical data have shown that adults with psychiatric disorders such as schizophrenia, have a brain that appears older and age faster compared to people without mental disorders (Schnack et al., 2016). Regional differences in brain age gaps between different disorders have also been observed. For instance, individuals with schizophrenia were found to have most pronounced age gap for the frontal lobe, while increased cerebellar-subcortical age gaps was found to be predominant in dementia and multiple sclerosis (Kaufmann et al., 2019).
As the mechanisms of psychiatric disorders are assumed to have a strong neurodevelopmental component (Insel, 2010) and deviant developmental trajectories have been observed in imaging data of youths with early signs of psychiatric disorders (Besenek, 2020, Chung et al., 2018, Collin et al., 2020, Kaufmann et al., 2017, Lian et al., 2018, Saito et al., 2020), estimating brain age gap as a proxy for maturation in young individuals may provide further insights into the early phases of the disorders. Indeed, an increase in anatomical brain age gaps has been observed in association with psychopathology in children and adolescents (Chung et al., 2018, Cropley et al., 2020, Franke et al., 2012). Anatomical brain age during neurodevelopment has also been found to be dependent on sex and to be partly heritable (Brouwer et al., 2020). Additionally, functional measures have been used for brain age prediction in development (Dosenbach et al., 2010, Kassani et al., 2020, Li et al., 2018, Rudolph et al., 2017, Truelove-Hill et al., 2020, Zhai and Li, 2019), but the implications of functional brain age gaps have yet to be further explored across neurodevelopmental disorders in large samples of children and adolescents.
Here, we used functional MRI data from 1126 individuals aged 8–22 years to train a machine learning model to estimate brain age. We applied the resulting model to independent functional imaging data of 1387 youths aged 8 to 22 to derive functional brain age gaps. Next, we used linear models to test for associations between brain age gap and psychopathology in youths, using behavioral measures constructed from diagnostic criterion for selected DSM-5 disorders. Based on previous findings, we expected to observe an association between delayed functional brain development and psychopathology on top of differences related to sex and cognitive test performance.
2. Methods
2.1. Study samples
The Healthy Brain Network (HBN) study sample is an initiative coordinated by the Child Mind Institute, where the aim is to provide a unique understanding of the time period when psychiatric disorders emerge (Alexander et al., 2017). Age range for inclusion of participants is 5–21 years, and consists of both individuals that receive a diagnosis and healthy controls (see supplementary information for overview of diagnosis and demographic information). Individuals are included in the New York area through announcements circulated to community members, educators, and local care providers. In addition, information via email lists was sent out and spread on parent events, where children with clinical concerns were encouraged to take part in this study (Alexander et al., 2017). The participants go through an extensive assessment package where MRI, genetics, electroencephalography (EEG), eye-tracking, biological testing, actigraphy, voice and video interviews are incorporated. In addition, the assessments include a neuropsychological battery and rich information on cognitive, lifestyle, behavioral and psychiatric factors (Alexander et al., 2017). Exclusion criteria comprise serious neurological disorders, neurodegenerative disorders, acute encephalopathy, hearing or visual impairment, lifetime substance abuse that required chemical replacement therapy/acute intoxication at time of study, recent diagnosis of a severe mental disorder or manic/psychotic episode within the last 6 months that did not receive continuing treatment. The onset of suicidality/homicidality where there is no current treatment was also an exclusion criterion (Alexander et al., 2017). Participants over the age of 18 years gave signed informed consent, and legal guardians signed informed consent for participants under the age of 18, in addition to participants giving a written assent (Alexander et al., 2017). The Chesapeake Institutional Review Board approved the study (https://www. chesapeakeirb.com/).
The Philadelphia Neurodevelopmental Cohort (PNC) is a research initiative funded by the National Institutes of Mental Health (NIMH) that aims to describe the interaction between the brain, behavior and genetics (Satterthwaite et al., 2016). The PNC participants were selected after stratification by sex, age and ethnicity (Satterthwaite et al., 2014) from a larger sample of children enrolled in a genetic study at the Center of Applied Genomics, at the Children’s Hospital of Philadelphia (see supplementary material for demographic information). They were included after they had been to a primary care facility that was CHOP-affiliated in the Delaware Valley. The sample includes individuals with different medical conditions, varying from a well-child visit and minor problems to individuals with more complicated illnesses, however, individuals with medical problems that could affect brain function were excluded (Satterthwaite et al., 2016). The inclusion criteria comprised 1) ability to provide signed informed consent (parental consent was acquired for participants under age 18), 2) English language proficiency, and 3) physical and cognitive ability to participate in computerized clinical assessment and neurocognitive testing (Satterthwaite et al., 2014). Participants underwent a structured neuropsychiatric interview in addition to completion of the Computerized Neurocognitive Battery (CNB) (Satterthwaite et al., 2014). The University of Pennsylvania and CHOP Institutional Review Boards approved the study (Satterthwaite et al., 2016).
2.2. MRI acquisition
HBN: MRI scans were acquired at 3 sites; Rutgers University Brain Imaging Center (RUBIC), Citigroup Biomedical Imaging Center (CBIC) and a mobile scanner placed in Staten Island. Rutgers used a Siemens 3 T Trim Tio scanner, while CBIC deployed a Siemens 3 T Prisma. For RUBIC and CBIC, structural MRI data was acquired with a repeated 3D T1-weighted sequence (repetition time (TR): 2.5 s, echo time (TE): 3.15 ms, flip angle (FA): 8°, field of view (FOV): 256 mm, slice thickness: 0.8 mm, slices: 224). In addition, CBIC acquired a structural scan based on the ABCD study protocol with the following parameters; TR: 2.5 s, TE: 2.88 ms, FA: 8°, FOV: 256 mm, slice thickness: 1 mm, slices: 176. Resting- state blood-oxygen-level-dependent (BOLD) fMRI data was acquired by means of a T2*-weighted BOLD echo-planar imaging (EPI) sequence with a TR of 800 ms, TE of 30 ms, multiband acceleration factor = 6, number of slices: 60, and 375 repetitions and voxel size = 2.4 × 2.4 × 2.4 mm. Further, the mobile scanner located in Staten Island employed a 1.5 T Siemens Avanto system operational with 45 mT/m gradients (Alexander et al., 2017), using these parameters for T1w data; TR: 2730 ms, TE1: 1.64 ms, TE2: 3.5 ms, TE3: 5.36 ms, TE4: 7.22 ms, multiband acceleration factor = 3, FA: 7°, FOV: 256 mm, slice thickness: 1 mm, slices: 176. And for fMRI; TR: 1.45 s, TE: 40 ms, number of volumes: 420, slices: 54, resolution: 2.5 × 2.5 × 2.5 mm (http://fcon_1000.projects.nitrc.org/indi/cmi_healthy_brain_network/MRI%20Protocol.html).
PNC: A 3D T1-weighted magnetization prepared rapid acquisition gradient echo (MPRAGE) sequence was obtained with a TR of 1.81 s, a TE of 3.5 ms, while FA: 9°, FOV: 240 × 180 mm, slice thickness: 1 mm, and number of slices: 160, and used for structural purposes. MRI data was collected at the hospital of the University of Pennsylvania (Satterthwaite et al., 2014). Resting-state BOLD fMRI data was collected by means of a T2*-weighted BOLD EPI sequence with a TR of 3000 ms, TE of 32 ms, 46 number of slices, 124 repetitions and voxel size = 3 × 3 × 3 mm (Satterthwaite et al., 2014).
2.3. Cognitive and psychiatric measures
HBN: In order to test for associations between brain age and cognition in the test sample, we included the full-scale intelligence quotient (FSIQ) from the Wechsler Intelligence Scale for Children (WISC-V) as a measure of cognitive abilities for HBN participants. This score incorporates visual spatial, verbal comprehension, fluid reasoning, working memory, and processing speed domains (Wechsler, 2003). Furthermore, to test for associations between brain age and mental health we carried out a principal component analysis (PCA) on mental health data, in line with our earlier work (Lund et al., 2020). Specifically, we used the Extended Strengths and Weaknesses Assessment of Normal Behavior (E‐SWAN), where domains include depression, social anxiety, disruptive mood dysregulation disorder (DMDD), and panic disorder (Alexander, Salum, Swanson, & Milham, 2020). This allowed us to assess mental health symptoms on a continuum from healthy to patients to overcome the shortcomings of the typical case-control dichotomy. We excluded three of the items which related to panic disorder, as these questions had a high degree of missing values (90%). We ended up with 62 items for the PCA. Further, excluding individuals with missing scores, we had data for 2626 subjects for the PCA. We utilized the “prcomp” function in R to implement the PCA. The resulting first component, referred to as the p-factor or pF (Caspi et al., 2013) explained most variance across all the symptom domains of the ESWAN questionnaire (43.6%) and was particularly associated with items linked to self-control and depression/anxiety. We also included the second principal component (pF2), which explained 11.3% of the variance and was associated with items describing mood dysregulation. For both, pF and pF2, a high score reflects lower mental health.
PNC: We made use of an already existing delineation of mental health data using PCA (Alnaes et al., 2018) to derive a general psychopathology factor from clinical scores (Calkins et al., 2015, Calkins et al., 2014), and to derive a general cognition component (Alnaes et al., 2018) from various cognitive performance measures (Gur et al., 2014).
2.4. MRI processing and functional connectivity
MR data was collected by the study team of HBN (Alexander et al., 2017) and PNC (Satterthwaite et al., 2016) and we processed both samples with the same pipeline. For details on the preprocessing steps and quality assessment see Lund et al. (2020). Briefly, preprocessing included FSL MCFLIRT (Jenkinson, Bannister, Brady, & Smith, 2002) with spatial smoothing (FWHM:6.0) and a high-pass filter cutoff of 100, non-aggressive ICA-AROMA (Pruim et al., 2015a, Pruim et al., 2015b), followed by ICA FIX with a threshold of 20 (Griffanti et al., 2014, Salimi-Khorshidi et al., 2014), described in earlier work (Kaufmann et al., 2017, Lund et al., 2021). Preprocessing was followed by group level ICA using MELODIC group Independent Component Analysis (Beckmann and Smith, 2004, Hyvärinen, 1999). For each sample and scanning site (1 for PNC; N = 1252, 3 for HBN; N = 1685), we performed one group level ICA, followed by a meta-ICA across all four sites, yielding ICs compliant across samples and sites. Due to the meta-ICA framework, the number of components had to be pre-specified and we chose a model order of 100, as used in prior studies (de Lange et al., 2020, Miller et al., 2016, Smith et al., 2013). After manual quality control through visual inspection of each IC, 53 components were marked as artefactual. Examples of noise that were excluded included physiological noise, movement artifacts, in addition to systematic and scanner artifacts. The rest of the components (47), were included for FSLNets (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSLNets), that was implemented in Matlab, where the time courses from the dual regression analysis were included to estimate partial correlations between each functional network for each subject, giving a 47x47 matrix of direct node connection strengths/partial correlation matrices. In line with earlier work (Kaufmann et al., 2016), we used a Ledoit & Wolf shrinkage estimator procedure for L2 Regularization (Ledoit and Wolf, 2003, Schäfer and Strimmer, 2005), which estimates regularization strength (lambda) at the individual level. Finally, we z-transformed the estimated correlations using Fisher’s transformation and used the upper triangle of the correlation matrix as a feature set for machine learning, giving 1081 unique edges denoting the connection strength between two IC’s at the subject level.
The Shrinkage Estimation of Regression Coefficients (slm) function from the care package (http://strimmerlab.org/software/care) in R was utilized for brain age prediction where default parameters were applied (Schäfer & Strimmer, 2005). Using PNC data, we trained a model to predict age based on the 1081 correlations reflecting connection strengths between the 47 IC’s referred to here as nodes. We excluded the 10% (N = 126) that scored highest on general psychopathology (pF) in order to obtain a model based on healthy individuals, and used the data from the remaining 1126 subjects (age: 8.17–22.9 years, 47.1% males, mean: 15.2 years, sd: 3.54 years, median: 15.4 years) for model training. To validate model performance within PNC data, we performed a 10‐fold cross validation, estimating age for each individual in each of the left-out folds while training the model on the rest. Next, we merged the brain age estimates from the N = 126 individuals with the brain age estimates computed in a cross-validation framework run within the training set, to test for associations between BAG and cognitive and clinical variables.
Afterwards, we tested the PNC model on HBN data (N = 1387, age: 8.01–22.4 years, mean: 12.3 years, sd: 3.24 years, median: 11.5 years, 61.8% males, where information about sex was missing for N = 30, and 298 subjects from the initial meta-ICA were excluded due to age outside the training set range (age < 8) or missing information about age at MRI). For each individual in HBN, we estimated the brain age gap (BAG) by subtracting chronological age from the estimated brain age.
2.5. Statistical analysis
We used brain age gap as a response variable in linear models and tested for associations with psychopathology and cognitive abilities in the HBN sample (N = 941, age: 8.01–17.5 years, 63.5% males, where 446 subjects from the test set was excluded due to missing clinical (N = 325), or cognitive (N = 120) values or information on sex (N = 1)). In addition to the HBN test sample, we performed the same analysis for the 10% of individuals initially excluded from the PNC sample (N = 126; age: 9.5–22.9 years, 39.7% males) and combined the brain age estimates from the N = 126 individuals with brain age estimates obtained through cross-validation in the rest of the PNC sample. We performed analyses using BAG scores and tested for associations between BAG and covariates using linear regression models. HBN models were adjusted for age, age-orthogonalized age squared (age2, using the poly function in R), sex, tSNR, motion and scanning site, while PNC was adjusted for age, age-orthogonalized age squared (age2, using the poly function in R), sex, motion and tSNR.
3. Results
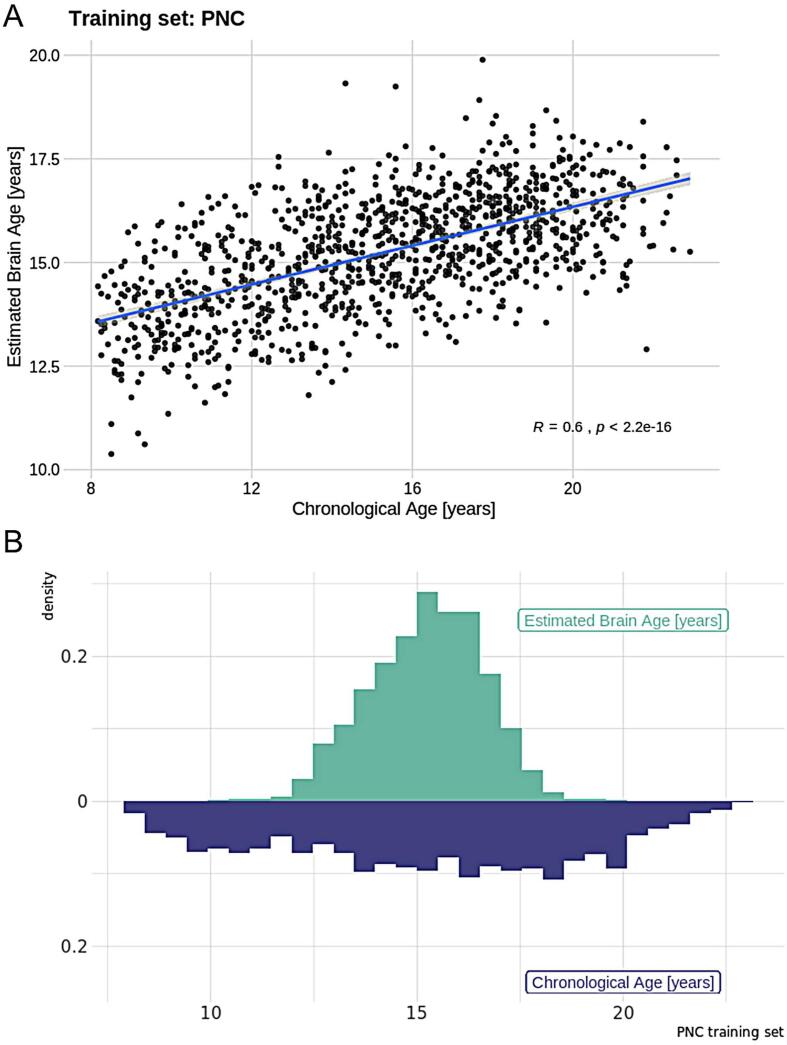
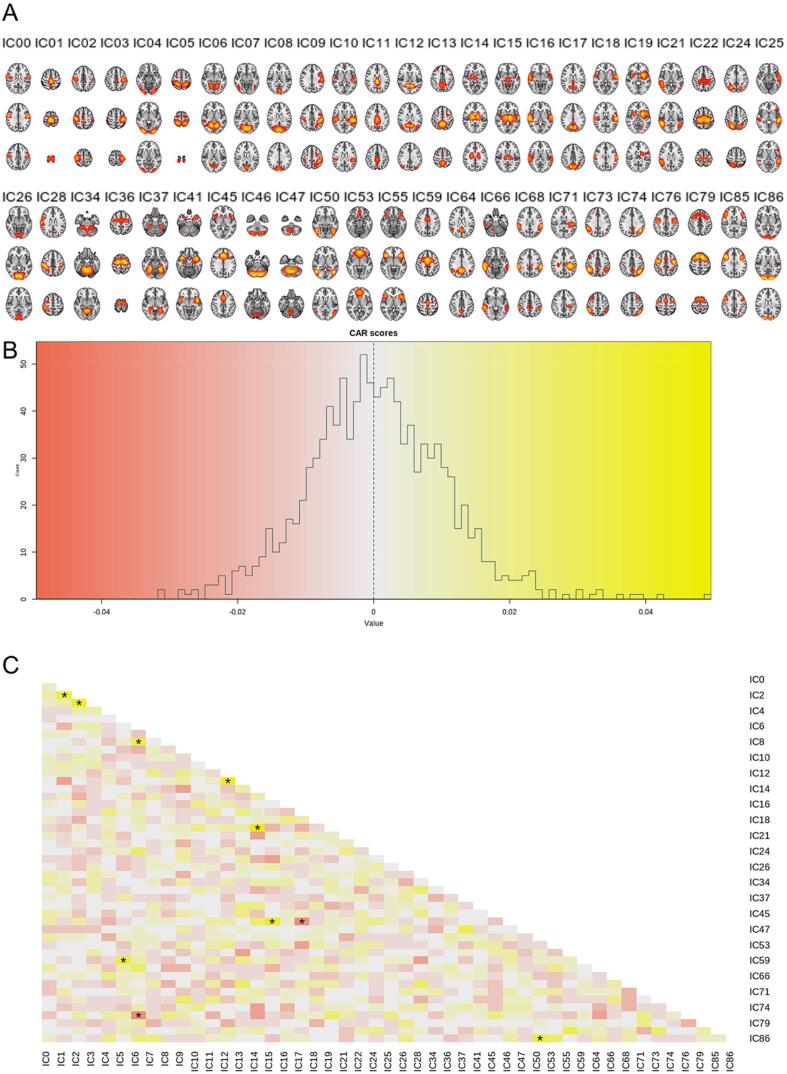
The correlation between the estimated brain age and the chronological age, computed through 10-fold cross-validation in the training set (PNC), was r = 0.60 [95% CI: 0.56, 0.63] (Fig. 1), the mean absolute error (MAE) was 2.43 years and root mean square error (RMSE) was 2.93 years, confirming the utility of the functional connectivity features for predicting age. The slm model explained 11.2% of the variance. Fig. 2 depicts the feature ranking by use of Correlation-Adjusted (marginal) correlation (CAR) scores from the model giving a measure of variable importance. The top 10 most important edges were between a sensorimotor (SM;IC1) and right SM node (IC2), right SM node (IC2) and left SM node (IC3), Visual Medial node (VM;IC6) and VM (IC8), Default Mode Network (DMN;IC12) and precuneus with frontal gyrus (IC13), thalamus (IC14) and putamen/left insular cortex (IC19), thalamus (IC15) and cerebellar node (IC46), precuneus and posterior cingulate (IC17) with cerebellar node (IC46), superior parietal lobe and SM regions (IC5) with Juxtapositional lobule and cingulate gyrus (IC59), VM (IC6), with Juxtapositional lobule, precentral and middle frontal gyrus (IC76), as well as lateral occipital cortex, and pre/postcentral gyrus (IC50) and Visual Occipital node (VO;IC86).
Fig. 1.
A) Model performance for the training set (PNC) where the Pearson correlation between estimated and real age is r = 0.60. B) Density plot showing the distributions of chronological age and estimated brain age for the training set.
Fig. 2.
A) The components from the meta-ICA that were included for the analysis. B) Distribution of CAR scores. As expected, the dependencies are low given that we used a partial correlation framework with regularization in FSLNets to estimate connectivity. C) CAR score matrix showing the dependencies among predictors, where the 10 most important features calculated from the correlation between the response and the Mahalanobis-decorrelated predictors are marked with a star.
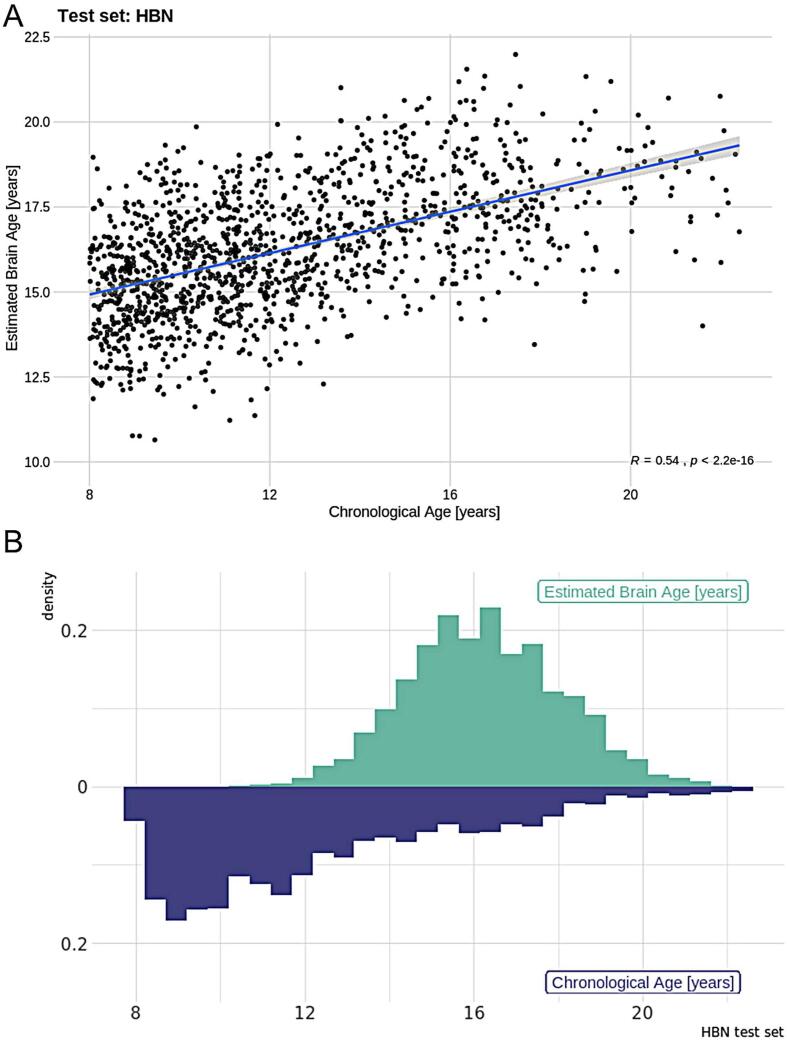
Next, we applied the PNC model to independent test data from the HBN sample. The correlation between chronological age and estimated brain age was r = 0.54 [ 95% CI: 0.50, 0.57] (Fig. 3), MAE was 4.24 years and RMSE = 4.79 years. While these errors were higher than the errors within the PNC sample, considering the test sample comes from different scanners these results nonetheless confirmed the validity of the model, illustrating high consistency between samples and scanners. For further investigation of scanning site effects, we examined model performance for each site. We found that the correlation between the estimated brain age and the chronological age for the scanner located in Staten Island (N = 272) was r = 0.64 [95% CI: 0.56, 0.71], the MAE was 3.46 years and RMSE = 4.03 years. For Rutgers (N = 584), the correlation was r = 0.58 [95% CI: 0.53, 0.64], the MAE was 4.87 years and RMSE = 5.36 years, while for CBIC (N = 531) r = 0.55 [95% CI: 0.49, 0.61], the MAE was 3.95 years and RMSE = 4.47 years. This shows that there is some variability between sites, but that the model overall performs similarly across sites. We therefore dealt with site effects by incorporating it as a fixed factor in the association analyses. The model performance across sites also underlines the feasibility of the meta-ICA framework to derive robust functional networks in data from different scanners, yielding compatible whole-brain networks across sites.
Fig. 3.
A) Model performance for the test set (HBN) where the Pearson correlation between estimated and real age is r = 0.54. B) Density plot showing the distribution between chronological age and estimated brain age for the training sample.
Linear models revealed no significant associations for BAG and mental health (pF: t = -0.11, P = .91, pF2: t = -0.55, P = .58) or cognitive abilities (t = 0.95, P = .34) in the test set, when accounting for age, age-orthogonalized age squared, sex, tSNR, motion and scanning site. In line with earlier research (Kaufmann et al., 2019), there were significant associations between BAG and age, motion and scanning site (see supplementary table 1). We tested for interaction effects of cognitive abilities and chronological age, but we did not find that cognitive abilities depend on age, or influences the association with the brain age gap (FSIQ*Chronological Age: t = 0.1, P = .92). We also investigated if there were interaction effects between age and psychopathology on BAG but there were no significant associations for HBN: (pF*Chronological Age: t = 1.47, P = .14, pF2*Chronological Age: t = 0.07, P = .94), or for PNC (pF*Chronological Age: t = 0.09, P = .93). Additionally, we performed a supplemental analysis where we included a measure of puberty (Peterson Puberty Scale) for the HBN sample to examine if maturation could be associated with BAG, however we did not find a significant effect of adolescent maturation on BAG (t = -0.67, P = .5) (see supplemental information, page 8).
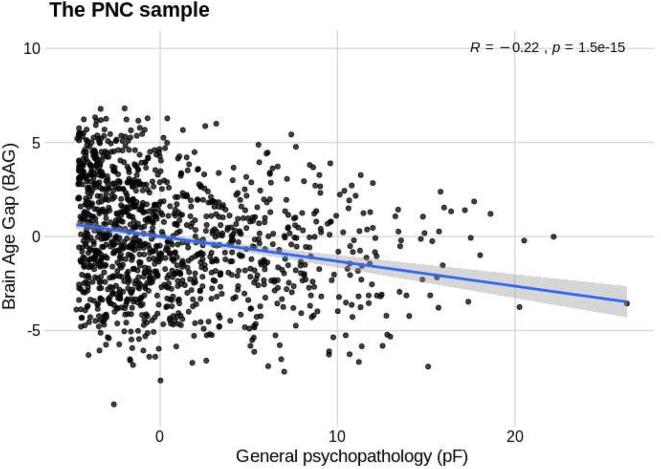
Additionally, using the existing PNC model, we estimated brain age for the 10% of the PNC participants (N = 126) with the highest mental health burden that were excluded from the training set. The correlation between chronological age and estimated brain age in this small sample was r = 0.42 [95% CI: 0.27, 0.56], MAE was 2.26 years and RMSE = 2.81 years. Next, Using the full PNC sample (N = 1252), we tested for associations with pF and a g factor (gF for PNC was estimated based on the instruments given here; Alnaes et al. (2018)), accounting for age, age-orthogonalized age squared, sex, motion and tSNR, using a linear model. We observed a significant association between mental health and BAG (t = -2.8, P < .01). Specifically, higher symptom burden was associated with lower BAG, indicating that individuals with a younger (estimated) brain age compared to chronological age had a higher level of psychiatric symptoms (Fig. 4). We found no significant associations for BAG and cognitive abilities (t = 1.59, P > .1). Further, to elucidate what aspects of mental health that could be relevant for BAG, we tested for effects of clinical ICs on BAG derived from the published work of Alnaes et al. (2018), and found significant associations for clinical IC1 that relates to attention-deficit/hyperactivity disorder (ADHD) (t = -2.04, P = .04), and clinical IC2 (t = -1.98, P < .05) that loaded high on agoraphobia, specific phobias, separation anxiety and social anxiety (see supplementary material, page 4). Additionally, we tested if the association between mental health symptoms and BAG was present in the PNC-90% sample and in the PNC-10% samples separately (see supplemental information page 7).
Fig. 4.
Plot showing the significant negative association of psychopathology (pF) and Brain Age Gap (BAG) for the full PNC sample (N = 1252).
4. Discussion
Here, we performed age prediction using machine learning on functional connectivity derived from resting state data (rsfMRI), allowing us to reduce a sizeable amount of data to a single measure of estimated functional brain age per individual. As rsfMRI does not include a cognitively demanding task or require a long scan duration, this is an appealing method for collecting data across individuals with different disorders. The absence of a specific task protocol also makes it ideally suited to combine data from different samples, thereby meeting the requirements for large samples to obtain robust machine learning models. Indeed, we here show that brain age models trained on functional connectivity of one sample can be successfully applied to other samples, even though predictions regress towards the mean of the training set (Fig. 3b), including those with data obtained from different scanners and protocols. A correlation of around 0.6 between chronological age and estimated brain age was consistent with other PNC studies estimating brain age using functional connectivity as feature input (Li et al., 2018, Truelove-Hill et al., 2020). The moderate correlation is expected as the functional connectome is highly dynamic over the lifespan and across contexts. However, a developmental pattern encompassing a core functional connectome backbone in rsfMRI data has shown high test–retest reliability (Thomason et al., 2011, Zuo and Xing, 2014), and reproducible inter- and intra-subject rsfMRI measures (Damoiseaux et al., 2006, Shehzad et al., 2009). As such, despite moderate correlations the models are likely to capture biologically relevant information and may be used to indicate trajectories for typical and non-typical brain development and neural restructuring important for susceptibility to brain disorders.
The brain age approach allowed us to assess the relationship between an estimated brain age gap and cognition and mental health in independent data, while our dimensional approach to mental health data enabled us to characterize both healthy subjects and individuals with a psychiatric disorder along a symptom dimension, rather than applying a binary distinction as cases and controls. Such dimensional approaches may more aptly identify phenotypes which map to brain biology and the neuronal mechanisms underlying the symptoms than diagnostic categories (Hengartner and Lehmann, 2017, Krueger and Bezdjian, 2009).
Our hypothesis of a link between functional brain age gap, a proxy for brain maturation, and psychopathology was supported in PNC but not in the HBN sample. The strength of the association between BAG and psychopathology for the PNC sample was moderate but of reasonable magnitude, as we are measuring shared variance across symptom domains in relation to distributed connectivity patterns across large-scale brain networks. The significant association between general psychopathology and BAG in the PNC sample, where a higher symptom burden was associated with a lower BAG, is in accordance with studies showing a delay in brain maturation being linked to poorer mental health in the same sample (Kaufmann et al., 2017) and also in young patients with schizophrenia (Douaud et al., 2009). In addition, studies utilizing morphometry and diffusion data for BAG prediction have found that HBN participants that had a lower BAG also had more symptoms on the Child Behavior Checklist (CBCL), and decreased global functioning (Luna et al., 2021). While work in youths’ has pointed to a lower BAG with increased symptoms (interpreted as a delayed development), elevated structural brain age (interpreted as apparent aging) have also been shown for frontal and subcortical regions in PNC individuals with symptoms of psychosis and obsessive–compulsive disorder (Cropley et al., 2020). In addition, elevated BAG has been observed in youths at high-risk, with BAG interacting with age and elevated for individuals at high-risk under the age of 17 years, but not for high- risk participants from 18 to 21 years (Chung et al., 2018). Moreover, studies estimating structural brain age in 16–22 years old individuals has shown higher brain age for schizophrenia patients compared to controls (Truelove-Hill et al., 2020), consistent with studies including older samples (Koutsouleris et al., 2013, Kaufmann et al., 2019). Taken together, these studies illustrate that both structural and functional brain age models capture important patterns, and that whether mental health is associated with a higher or lower BAG may largely depend on its timing.
A limitation with general trans-diagnostic symptom measures derived within samples are their validity across cohorts. The pF measure utilized as part of the PNC sample was constructed using 129 clinical items (Calkins et al., 2015, Calkins et al., 2014), and included a wide range of psychiatric domains pertaining to depression and mania/hypomania as well as anxiety, attention deficits, conduct disorder, phobias, eating disorder, and obsessive–compulsive disorder. For the HBN sample, the ESWAN questionnaire was utilized to perform a PCA, that was based on 62 items, pertaining to depression, social anxiety, panic disorder, disruptive mood and dysregulation disorder (Alexander et al., 2020). One possibility is that we did not include enough items or wide enough of a range for delineating psychopathology for it to be sensitive for brain measures in the HBN sample. As such, the lack of significant associations in HBN could be related to a number of factors, including that the pF captures different aspects of mental health in the two samples, that HBN samples from the point of transition between decreased BAG (childhood) and increased BAG (adulthood), or that there is no difference due to mental health. Alternatively, it is possible that the linear machine learning model did not have enough flexibility to detect non-linear effects in development. It will be interesting in future research to see if models that do better at capturing non-linear maturation trajectories may reveal associations with psychopathology.
Contrary to our expectations, our analysis revealed no significant associations between BAG and cognitive test performance. In contrast, previous work utilizing the PNC sample have observed that individual differences in working memory performance is linked with the centrality of the cingulo-opercular network (Kolskår et al., 2018) while others have found resting-state modularity to be associated with cognitive performance in PNC (Gu et al., 2015). Also, BAG has been found to be associated with cognitive test performance when investigating cognition in the PNC sample, with elevated BAG being related to better processing speed (Erus et al., 2015). In addition, covariation patterns have shown to have a positive association with cognition when using the HBN sample as a training set for deriving brain age estimates and testing on the Nathan Kline Institute - Rockland Sample (NKI-RS) (Zhao, Klein, Castellanos, & Milham, 2019). Moreover, for developmental trajectories in youths, it has been illustrated that networks associated with cognition and emotion have locally increased functional connectivity compared to adults, indicating fine-tuning and specialization occurring during the first years of adolescence, principally in networks characterized for higher-order cognitive functioning (Hoff, Van den Heuvel, Benders, Kersbergen, & De Vries, 2013).
Studies examining the PNC cohort have also reported there to be sex differences in cognitive styles that are also reflected in resting-state brain connectivity (Satterthwaite et al., 2015). Also, for adults, permutation tests showed above chance-level prediction accuracy for trait-level educational attainment and fluid intelligence in rsfMRI data from UK Biobank, where both variables were negatively linked with functional connectivity in frontal and default mode networks (Maglanoc et al., 2020). In relation to disorders, differences have been found in functional connectivity for the putamen, dorsal and default-mode regions in Alzheimer’s disease in comparison with mild cognitive and subjective cognitive impairment (Córdova-Palomera et al., 2017). Also, anatomical BAG has been associated with sex, with females developing earlier than males (Brouwer et al., 2020), yet there was no significant association with sex for the functional BAGs in our current study. This could be due to not training the model separately for females and males, owing to limitations in number of features versus participants. Still, the model performance showed that shared variability for both sexes was captured.
Although our PNC-trained brain age model generalized well to the HBN data, sample differences such as different acquisition protocols, scanning hardware or age distributions may have factored into reduced generalizability of the PNC-trained model to HBN participants. In addition, it is important to note that HBN comprises almost entirely of individuals with diagnosed disorders and therefore their estimated BAG is not expected to be at zero. Subsequently, when evaluating model performance, we need to take into consideration that accuracy may partly be 'confounded' with biological variance as part of the HBN sample. To examine if the lack of an association between BAG and mental health in the HBN sample may be attributed to model performance, we performed a supplemental analysis in which we estimated BAG within the HBN sample using a cross-validation framework. However, in line with our main results, we did not find an association for the within-HBN estimated BAGs (see supplementary information, page 2). We also tested if accuracy would improve if we had used a subset of HBN individuals, making the two samples more alike in age distributions (see supplementary information, page 2 for age distribution plots and details). We found that performance improved, yielding a MAE of 3.4 for the HBN subsample, yet we still did not find a significant association with mental health in this subsample.
Apart from the investigations into BAG, we identified a set of specific connections important for modeling brain age. Central features for the brain age prediction included sensorimotor, visual, insular, DMN, cerebellar and language processing regions, which is coherent with changes in sensory, motor and cognitive abilities observed across this age span (Casey et al., 2005, de Bie et al., 2012). Our findings are in line with reviews (Power et al., 2010, Uddin et al., 2010) showing alterations in these functional connectivity patterns in development. Specifically, studies have shown that brain maturation in children and adolescence may involve a decrease in connectivity of short-range connections and an increase of long-range connections in functional networks. This has been observed for instance in a reduction in short-length connections for the SM and anterior cingulate cortex in young children (Kelly et al., 2008, Supekar et al., 2009), and for segregation of frontal regions and the DMN in late childhood (Fair et al., 2009).
5. Conclusions
In the present study, we estimated functional brain connectivity from rsfMRI data from children and adolescents, and used connectivity strengths as features for brain age prediction. Our model showed reasonable performance, and consistency across samples and scanning sites. The most important connections for age prediction were related to sensorimotor, visual, insular, DMN, cerebellar and language areas, indicating that these neural circuits are central in adolescent development. While we found mixed results for behavioral and psychopathology measures with the brain age gap, the applicability of models to data from different sites supports the utility of the brain age prediction framework for multisite investigations.
Funding
The authors were funded by the Research Council of Norway #276082 (LifespanHealth), #223273 (NORMENT), #249795, #298646, #300767, #283798; H2020 European Research Council #802998 (BRAINMINT); The South-East Norway Regional Health Authority #2019101, #2019107, #2020086; the Swiss National Science Foundation #PZ00P3_193658.
Financial disclosures
OAA is a consultant for HealthLytix.
Data and code availability
The data incorporated in this work were gathered from the open access Healthy Brain Network and The Philadelphia Neurodevelopmental Cohort resource.
CRediT authorship contribution statement
Martina J. Lund: Conceptualization, Data curation, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. Dag Alnæs: Supervision, Writing – review & editing. Ann-Marie G. de Lange: Investigation, Writing – review & editing. Ole A. Andreassen: Resources, Writing – review & editing. Lars T. Westlye: Supervision, Writing – review & editing. Tobias Kaufmann: Conceptualization, Formal analysis, Investigation, Supervision, Funding acquisition, Writing – original draft, Writing – review & editing.
Acknowledgments
Acknowledgements
This manuscript was prepared using a limited access dataset obtained from the Child Mind Institute Biobank, the HBN resource (http://www.healthybrainnetwork.org, and the publicly available Philadelphia Neurodevelopmental Cohort (PNC,http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id = phs000607.v1.p1) with permission no.8642. Support for the collection of the PNC data set was provided by grant #RC2MH089983 awarded to Raquel Gur, MD, PhD, and #RC2MH089924 awarded to Hakon Hakonarson, MD, PhD. All PNC participants were recruited through the Center for Applied Genomics at The Children’s Hospital in Philadelphia.
This manuscript reflects the views of the authors and does not necessarily reflect the opinions or views of any other agency, organization, employer or company. This work was performed on the TSD (Tjeneste for Sensitive Data) facilities, owned by the University of Oslo, operated and developed by the TSD service group at the University of Oslo, IT-Department (USIT) (tsd-drift@usit.uio.no).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2021.102921.
Contributor Information
Martina J. Lund, Email: m.j.lund@medisin.uio.no.
Tobias Kaufmann, Email: tobias.kaufmann@medisin.uio.no.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Alexander L.M., Escalera J., Ai L., Andreotti C., Febre K., Mangone A., Vega-Potler N., Langer N., Alexander A., Kovacs M., Litke S., O'Hagan B., Andersen J., Bronstein B., Bui A., Bushey M., Butler H., Castagna V., Camacho N., Chan E., Citera D., Clucas J., Cohen S., Dufek S., Eaves M., Fradera B., Gardner J., Grant-Villegas N., Green G., Gregory C., Hart E., Harris S., Horton M., Kahn D., Kabotyanski K., Karmel B., Kelly S.P., Kleinman K., Koo B., Kramer E., Lennon E., Lord C., Mantello G., Margolis A., Merikangas K.R., Milham J., Minniti G., Neuhaus R., Levine A., Osman Y., Parra L.C., Pugh K.R., Racanello A., Restrepo A., Saltzman T., Septimus B., Tobe R., Waltz R., Williams A., Yeo A., Castellanos F.X., Klein A., Paus T., Leventhal B.L., Craddock R.C., Koplewicz H.S., Milham M.P. An open resource for transdiagnostic research in pediatric mental health and learning disorders. Sci. Data. 2017;4(1) doi: 10.1038/sdata.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander L.M., Salum G.A., Swanson J.M., Milham M.P. Measuring strengths and weaknesses in dimensional psychiatry. J. Child Psychol. Psychiatry. 2020;61(1):40–50. doi: 10.1111/jcpp.v61.110.1111/jcpp.13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnæs D., Kaufmann T., Doan N.T., Córdova-Palomera A., Wang Y., Bettella F., Moberget T., Andreassen O.A., Westlye L.T. Association of Heritable Cognitive Ability and Psychopathology With White Matter Properties in Children and Adolescents. JAMA Psychiatry. 2018;75(3):287. doi: 10.1001/jamapsychiatry.2017.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnæs D., Kaufmann T., van der Meer D., Córdova-Palomera A., Rokicki J., Moberget T., Bettella F., Agartz I., Barch D.M., Bertolino A., Brandt C.L., Cervenka S., Djurovic S., Doan N.T., Eisenacher S., Fatouros-Bergman H., Flyckt L., Di Giorgio A., Haatveit B., Jönsson E.G., Kirsch P., Lund M.J., Meyer-Lindenberg A., Pergola G., Schwarz E., Smeland O.B., Quarto T., Zink M., Andreassen O.A., Westlye L.T. Brain Heterogeneity in Schizophrenia and Its Association With Polygenic Risk. JAMA Psychiatry. 2019;76(7):739. doi: 10.1001/jamapsychiatry.2019.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann C.F., Smith S.M. Probabilistic Independent Component Analysis for Functional Magnetic Resonance Imaging. IEEE Trans. Med. Imaging. 2004;23(2):137–152. doi: 10.1109/tmi.2003.822821. [DOI] [PubMed] [Google Scholar]
- Besenek M. Anterior cingulate cortex disconnectivity in high-risk offspring of bipolar patients: a preliminary DTI study. Dusunen Adam: J. Psychiat. Neurol. Sci. 2020 doi: 10.14744/dajpns.2019.00030. [DOI] [Google Scholar]
- Brouwer, R. M., Schutte, J., Janssen, R., Boomsma, D. I., Hulshoff Pol, H. E., & Schnack, H. G. (2020). The Speed of Development of Adolescent Brain Age Depends on Sex and Is Genetically Determined. Cereb Cortex. doi:10.1093/cercor/bhaa296. [DOI] [PMC free article] [PubMed]
- Calkins M.E., Merikangas K.R., Moore T.M., Burstein M., Behr M.A., Satterthwaite T.D., Ruparel K., Wolf D.H., Roalf D.R., Mentch F.D., Qiu H., Chiavacci R., Connolly J.J., Sleiman P.M.A., Gur R.C., Hakonarson H., Gur R.E. The Philadelphia Neurodevelopmental Cohort: constructing a deep phenotyping collaborative. J. Child Psychol. Psychiatry. 2015;56(12):1356–1369. doi: 10.1111/jcpp.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins M.E., Moore T.M., Merikangas K.R., Burstein M., Satterthwaite T.D., Bilker W.B., Ruparel K., Chiavacci R., Wolf D.H., Mentch F., Qiu H., Connolly J.J., Sleiman P.A., Hakonarson H., Gur R.C., Gur R.E. The psychosis spectrum in a young US community sample: findings from the Philadelphia Neurodevelopmental Cohort. World Psychiatry. 2014;13(3):296–305. doi: 10.1002/wps.20152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., & Schwarz, E. (2020). Opportunities and challenges of machine learning approaches for biomarker signature identification in psychiatry. In Personalized Psychiatry (pp. 117-126).
- Casey B.J., Tottenham N., Liston C., Durston S. Imaging the developing brain: what have we learned about cognitive development? Trends Cogn Sci. 2005;9(3):104–110. doi: 10.1016/j.tics.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Caspi A., Houts R.M., Belsky D.W., Goldman-Mellor S.J., Harrington HonaLee, Israel S., Meier M.H., Ramrakha S., Shalev I., Poulton R., Moffitt T.E. The p Factor. Clinical. Psychol. Sci. 2013;2(2):119–137. doi: 10.1177/2167702613497473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y., Addington J., Bearden C.E., Cadenhead K., Cornblatt B., Mathalon D.H., McGlashan T., Perkins D., Seidman L.J., Tsuang M., Walker E., Woods S.W., McEwen S., van Erp T.G.M., Cannon T.D. Use of Machine Learning to Determine Deviance in Neuroanatomical Maturity Associated With Future Psychosis in Youths at Clinically High Risk. JAMA Psychiatry. 2018;75(9):960. doi: 10.1001/jamapsychiatry.2018.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin G., Seidman L.J., Keshavan M.S., Stone W.S., Qi Z., Zhang T., Tang Y., Li H., Anteraper S.A., Niznikiewicz M.A., McCarley R.W., Shenton M.E., Wang J., Whitfield-Gabrieli S. Functional connectome organization predicts conversion to psychosis in clinical high-risk youth from the SHARP program. Mol. Psychiatry. 2020;25(10):2431–2440. doi: 10.1038/s41380-018-0288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Córdova-Palomera A., Kaufmann T., Persson K., Alnæs D., Doan N.T., Moberget T., Lund M.J., Barca M.L., Engvig A., Brækhus A., Engedal K., Andreassen O.A., Selbæk G., Westlye L.T. Disrupted global metastability and static and dynamic brain connectivity across individuals in the Alzheimer's disease continuum. Sci. Rep. 2017;7(1) doi: 10.1038/srep40268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N., Owen M.J. The Kraepelinian dichotomy - going, going... but still not gone. Br. J. Psychiatry. 2010;196(2):92–95. doi: 10.1192/bjp.bp.109.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cropley V.L., Tian Y.e., Fernando K., Mansour L.S., Pantelis C., Cocchi L., Zalesky A. Brain-Predicted Age Associates With Psychopathology Dimensions in Youths. Biol. Psychiatry Cogn Neurosci Neuroimaging. 2020;6(4):410–419. doi: 10.1016/j.bpsc.2020.07.014. [DOI] [PubMed] [Google Scholar]
- Damoiseaux J.S., Rombouts S.A., Barkhof F., Scheltens P., Stam C.J., Smith S.M., Beckmann C.F. Consistent resting-state networks across healthy subjects. Proc. Natl. Acad. Sci. U.S.A. 2006;103(37):13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bie H.M.A., Boersma M., Adriaanse S., Veltman D.J., Wink A.M., Roosendaal S.D., Barkhof F., Stam C.J., Oostrom K.J., Delemarre-van de Waal H.A., Sanz-Arigita E.J. Resting-state networks in awake five- to eight-year old children. Hum. Brain Mapp. 2012;33(5):1189–1201. doi: 10.1002/hbm.v33.510.1002/hbm.21280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange A.-M., Anatürk M., Suri S., Kaufmann T., Cole J.H., Griffanti L., Zsoldos E., Jensen D.E.A., Filippini N., Singh-Manoux A., Kivimäki M., Westlye L.T., Ebmeier K.P. Multimodal brain-age prediction and cardiovascular risk: The Whitehall II MRI sub-study. NeuroImage. 2020;222:117292. doi: 10.1016/j.neuroimage.2020.117292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach N.U.F., Nardos B., Cohen A.L., Fair D.A., Power J.D., Church J.A., Nelson S.M., Wig G.S., Vogel A.C., Lessov-Schlaggar C.N., Barnes K.A., Dubis J.W., Feczko E., Coalson R.S., Pruett J.R., Barch D.M., Petersen S.E., Schlaggar B.L. Prediction of Individual Brain Maturity Using fMRI. Science. 2010;329(5997):1358–1361. doi: 10.1126/science:1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douaud, G., Mackay, C., Andersson, J., James, S., Quested, D., Ray, M. K., . . . James, A. (2009). Schizophrenia delays and alters maturation of the brain in adolescence. Brain, 132(Pt 9), 2437-2448. doi:10.1093/brain/awp126. [DOI] [PubMed]
- Erus G., Battapady H., Satterthwaite T.D., Hakonarson H., Gur R.E., Davatzikos C., Gur R.C. Imaging patterns of brain development and their relationship to cognition. Cereb. Cortex. 2015;25(6):1676–1684. doi: 10.1093/cercor/bht425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair D.A., Cohen A.L., Power J.D., Dosenbach N.U.F., Church J.A., Miezin F.M., Schlaggar B.L., Petersen S.E., Sporns O. Functional brain networks develop from a “local to distributed” organization. PLoS Comput. Biol. 2009;5(5):e1000381. doi: 10.1371/journal.pcbi.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K., Luders E., May A., Wilke M., Gaser C. Brain maturation: predicting individual BrainAGE in children and adolescents using structural MRI. NeuroImage. 2012;63(3):1305–1312. doi: 10.1016/j.neuroimage.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Franke K., Ziegler G., Kloppel S., Gaser C., Alzheimer's Disease Neuroimaging I. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. NeuroImage. 2010;50(3):883–892. doi: 10.1016/j.neuroimage.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Griffanti Ludovica, Salimi-Khorshidi Gholamreza, Beckmann Christian F., Auerbach Edward J., Douaud Gwenaëlle, Sexton Claire E., Zsoldos Enikő, Ebmeier Klaus P., Filippini Nicola, Mackay Clare E., Moeller Steen, Xu Junqian, Yacoub Essa, Baselli Giuseppe, Ugurbil Kamil, Miller Karla L., Smith Stephen M. ICA-based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. NeuroImage. 2014;95:232–247. doi: 10.1016/j.neuroimage.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S., Satterthwaite T.D., Medaglia J.D., Yang M., Gur R.E., Gur R.C., Bassett D.S. Emergence of system roles in normative neurodevelopment. Proc. Natl. Acad. Sci. U.S.A. 2015;112(44):13681–13686. doi: 10.1073/pnas.1502829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur Ruben C., Calkins Monica E., Satterthwaite Theodore D., Ruparel Kosha, Bilker Warren B., Moore Tyler M., Savitt Adam P., Hakonarson Hakon, Gur Raquel E. Neurocognitive growth charting in psychosis spectrum youths. JAMA Psychiatry. 2014;71(4):366. doi: 10.1001/jamapsychiatry.2013.4190. [DOI] [PubMed] [Google Scholar]
- Hengartner M.P., Lehmann S.N. Why Psychiatric Research Must Abandon Traditional Diagnostic Classification and Adopt a Fully Dimensional Scope: Two Solutions to a Persistent Problem. Front. Psychiatry. 2017;8:101. doi: 10.3389/fpsyt.2017.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff G.E., Van den Heuvel M.P., Benders M.J., Kersbergen K.J., De Vries L.S. On development of functional brain connectivity in the young brain. Front. Hum. Neurosci. 2013;7:650. doi: 10.3389/fnhum.2013.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvärinen, A. (1999). Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw., 10(3), 626–634. [DOI] [PubMed]
- Insel T.R. Rethinking schizophrenia. Nature. 2010;468(7321):187–193. doi: 10.1038/nature09552. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Bannister P., Brady M., Smith S. Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. NeuroImage. 2002;17(2):825–841. doi: 10.1006/nimg.2002.1132. [DOI] [PubMed] [Google Scholar]
- Jollans L., Whelan R. Neuromarkers for Mental Disorders: Harnessing Population Neuroscience. Front. Psychiatry. 2018;9:242. doi: 10.3389/fpsyt.2018.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassani Peyman Hosseinzadeh, Gossmann Alexej, Wang Yu-Ping. Multimodal Sparse Classifier for Adolescent Brain Age Prediction. IEEE J Biomed Health Inform. 2020;24(2):336–344. doi: 10.1109/JBHI.622102010.1109/JBHI.2019.2925710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann T., Alnaes D., Doan N.T., Brandt C.L., Andreassen O.A., Westlye L.T. Delayed stabilization and individualization in connectome development are related to psychiatric disorders. Nat. Neurosci. 2017;20(4):513–515. doi: 10.1038/nn.4511. [DOI] [PubMed] [Google Scholar]
- Kaufmann Tobias, Elvsåshagen Torbjørn, Alnæs Dag, Zak Nathalia, Pedersen Per Ø., Norbom Linn B., Quraishi Sophia H., Tagliazucchi Enzo, Laufs Helmut, Bjørnerud Atle, Malt Ulrik F., Andreassen Ole A., Roussos Evangelos, Duff Eugene P., Smith Stephen M., Groote Inge R., Westlye Lars T. The brain functional connectome is robustly altered by lack of sleep. NeuroImage. 2016;127:324–332. doi: 10.1016/j.neuroimage.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, T., van der Meer, D., Doan, N. T., Schwarz, E., Lund, M. J., Agartz, I., . . . Westlye, L. T. (2019). Common brain disorders are associated with heritable patterns of apparent aging of the brain. Nature Neuroscience, 22(10), 1617-1623. doi:10.1038/s41593-019-0471-7. [DOI] [PMC free article] [PubMed]
- Kelly A.M.C., Di Martino A., Uddin L.Q., Shehzad Z., Gee D.G., Reiss P.T., Margulies D.S., Castellanos F.X., Milham M.P. Development of Anterior Cingulate Functional Connectivity from Late Childhood to Early Adulthood. Cereb. Cortex. 2008;19(3):640–657. doi: 10.1093/cercor/bhn117. [DOI] [PubMed] [Google Scholar]
- Kolskar, K. K., Alnaes, D., Kaufmann, T., Richard, G., Sanders, A. M., Ulrichsen, K. M., . . . Westlye, L. T. (2018). Key Brain Network Nodes Show Differential Cognitive Relevance and Developmental Trajectories during Childhood and Adolescence. eNeuro, 5(4). doi:10.1523/ENEURO.0092-18.2018. [DOI] [PMC free article] [PubMed]
- Koutsouleris N., Davatzikos C., Borgwardt S., Gaser C., Bottlender R., Frodl T., Falkai P., Riecher-Rossler A., Moller H.-J., Reiser M., Pantelis C., Meisenzahl E. Accelerated Brain Aging in Schizophrenia and Beyond: A Neuroanatomical Marker of Psychiatric Disorders. Schizophr. Bull. 2013;40(5):1140–1153. doi: 10.1093/schbul/sbt142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger, R. F., & Bezdjian, S. (2009). Enhancing research and treatment of mental disorders with dimensional concepts: toward DSM-V and ICD-11. . World Psychiatry, 8(1)(3). [DOI] [PMC free article] [PubMed]
- Ledoit O., Wolf M. Improved estimation of the covariance matrix of stock returns with an application to portfolio selection. J. Empir. Fin. 2003;10(5):603–621. doi: 10.1016/s0927-5398(03)00007-0. [DOI] [Google Scholar]
- Li H., Satterthwaite T.D., Fan Y. In 2018 IEEE 15th International Symposium on Biomedical Imaging. 2018. Brain age prediction based on resting-state functional connectivity patterns using convolutional neural networks; pp. 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Nan, Lv Hailong, Guo Wenbing, Shen Yidong, Wu Renlong, Liu Yong, Zhu Furong, Hu Maorong, Ou Jianjun, Zhao Jingping. A comparative study of magnetic resonance imaging on the gray matter and resting-state function in prodromal and first-episode schizophrenia. Am. J. Med. Genet. B Neuropsychiatr Genet. 2018;177(6):537–545. doi: 10.1002/ajmg.b.v177.610.1002/ajmg.b.32644. [DOI] [PubMed] [Google Scholar]
- Linden D.E. The challenges and promise of neuroimaging in psychiatry. Neuron. 2012;73(1):8–22. doi: 10.1016/j.neuron.2011.12.014. [DOI] [PubMed] [Google Scholar]
- Luna Alex, Bernanke Joel, Kim Kakyeong, Aw Natalie, Dworkin Jordan D., Cha Jiook, Posner Jonathan. Maturity of gray matter structures and white matter connectomes, and their relationship with psychiatric symptoms in youth. Hum. Brain Mapp. 2021;42(14):4568–4579. doi: 10.1002/hbm.v42.1410.1002/hbm.25565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund Martina J., Alnæs Dag, Schwab Simon, Meer Dennis, Andreassen Ole A., Westlye Lars T., Kaufmann Tobias. Differences in directed functional brain connectivity related to age, sex and mental health. Hum. Brain Mapp. 2020;41(15):4173–4186. doi: 10.1002/hbm.v41.1510.1002/hbm.25116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, M. J., Alnæs, D., Rokicki, J., Schwab, S., Andreassen, O. A., Westlye, L. T., & Kaufmann, T. (2021). Functional connectivity directionality between large-scale resting-state networks in children and adolescence from the Healthy Brain Network sample. medRxiv, 2020-10. [DOI] [PMC free article] [PubMed]
- Maglanoc Luigi A., Kaufmann Tobias, van der Meer Dennis, Marquand Andre F., Wolfers Thomas, Jonassen Rune, Hilland Eva, Andreassen Ole A., Landrø Nils Inge, Westlye Lars T. Brain Connectome Mapping of Complex Human Traits and Their Polygenic Architecture Using Machine Learning. Biol. Psychiatry. 2020;87(8):717–726. doi: 10.1016/j.biopsych.2019.10.011. [DOI] [PubMed] [Google Scholar]
- Mansourvar M., Wiil U.K., & C., N. (2020). Big Data Analytics in Healthcare: A Review of Opportunities and Challenges. Paper presented at the In International Conference for Emerging Technologies in Computing.
- Miller Karla L, Alfaro-Almagro Fidel, Bangerter Neal K, Thomas David L, Yacoub Essa, Xu Junqian, Bartsch Andreas J, Jbabdi Saad, Sotiropoulos Stamatios N, Andersson Jesper L R, Griffanti Ludovica, Douaud Gwenaëlle, Okell Thomas W, Weale Peter, Dragonu Iulius, Garratt Steve, Hudson Sarah, Collins Rory, Jenkinson Mark, Matthews Paul M, Smith Stephen M. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 2016;19(11):1523–1536. doi: 10.1038/nn.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff Charles B. Comorbidity of mood and anxiety disorders: the rule, not the exception? Am. J. Psychiatry. 2002;159(1):3–4. doi: 10.1176/appi.ajp.159.1.3. [DOI] [PubMed] [Google Scholar]
- Owen M.J. New approaches to psychiatric diagnostic classification. Neuron. 2014;84(3):564–571. doi: 10.1016/j.neuron.2014.10.028. [DOI] [PubMed] [Google Scholar]
- Paulus M.P., Thompson W.K. The Challenges and Opportunities of Small Effects: The New Normal in Academic Psychiatry. JAMA Psychiatry. 2019;76(4):353–354. doi: 10.1001/jamapsychiatry.2018.4540. [DOI] [PubMed] [Google Scholar]
- Power J.D., Fair D.A., Schlaggar B.L., Petersen S.E. The Development of Human Functional Brain Networks. Neuron. 2010;67(5):735–748. doi: 10.1016/j.neuron.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim R.H.R., Mennes M., Buitelaar J.K., Beckmann C.F. Evaluation of ICA-AROMA and alternative strategies for motion artifact removal in resting state fMRI. NeuroImage. 2015;112:278–287. doi: 10.1016/j.neuroimage.2015.02.063. [DOI] [PubMed] [Google Scholar]
- Pruim R.H.R., Mennes M., van Rooij D., Llera A., Buitelaar J.K., Beckmann C.F. ICA-AROMA: A robust ICA-based strategy for removing motion artifacts from fMRI data. NeuroImage. 2015;112:267–277. doi: 10.1016/j.neuroimage.2015.02.064. [DOI] [PubMed] [Google Scholar]
- Rudolph Marc D., Miranda-Domínguez Oscar, Cohen Alexandra O., Breiner Kaitlyn, Steinberg Laurence, Bonnie Richard J., Scott Elizabeth S., Taylor-Thompson Kim, Chein Jason, Fettich Karla C., Richeson Jennifer A., Dellarco Danielle V., Galván Adriana, Casey B.J., Fair Damien A. At risk of being risky: The relationship between “brain age” under emotional states and risk preference. Dev. Cogn. Neurosci. 2017;24:93–106. doi: 10.1016/j.dcn.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Junichi, Nemoto Takahiro, Katagiri Naoyuki, Hori Masaaki, Tagata Hiromi, Funatogawa Tomoyuki, Yamaguchi Taiju, Tsujino Naohisa, Mizuno Masafumi. Can reduced leftward asymmetry of white matter integrity be a marker of transition to psychosis in at-risk mental state? Asian J. Psychiatr. 2020;54:102450. doi: 10.1016/j.ajp.2020.102450. [DOI] [PubMed] [Google Scholar]
- Salimi-Khorshidi G., Douaud G., Beckmann C.F., Glasser M.F., Griffanti L., Smith S.M. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. NeuroImage. 2014;90:449–468. doi: 10.1016/j.neuroimage.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite Theodore D., Connolly John J., Ruparel Kosha, Calkins Monica E., Jackson Chad, Elliott Mark A., Roalf David R., Hopson Ryan, Prabhakaran Karthik, Behr Meckenzie, Qiu Haijun, Mentch Frank D., Chiavacci Rosetta, Sleiman Patrick M.A., Gur Ruben C., Hakonarson Hakon, Gur Raquel E. The Philadelphia Neurodevelopmental Cohort: A publicly available resource for the study of normal and abnormal brain development in youth. NeuroImage. 2016;124:1115–1119. doi: 10.1016/j.neuroimage.2015.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite Theodore D., Elliott Mark A., Ruparel Kosha, Loughead James, Prabhakaran Karthik, Calkins Monica E., Hopson Ryan, Jackson Chad, Keefe Jack, Riley Marisa, Mentch Frank D., Sleiman Patrick, Verma Ragini, Davatzikos Christos, Hakonarson Hakon, Gur Ruben C., Gur Raquel E. Neuroimaging of the Philadelphia neurodevelopmental cohort. NeuroImage. 2014;86:544–553. doi: 10.1016/j.neuroimage.2013.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite Theodore D., Wolf Daniel H., Roalf David R., Ruparel Kosha, Erus Guray, Vandekar Simon, Gennatas Efstathios D., Elliott Mark A., Smith Alex, Hakonarson Hakon, Verma Ragini, Davatzikos Christos, Gur Raquel E., Gur Ruben C. Linked Sex Differences in Cognition and Functional Connectivity in Youth. Cereb. Cortex. 2015;25(9):2383–2394. doi: 10.1093/cercor/bhu036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnack H.G., van Haren N.E.M., Nieuwenhuis M., Hulshoff Pol H.E., Cahn W., Kahn R.S. Accelerated Brain Aging in Schizophrenia: A Longitudinal Pattern Recognition Study. Am. J. Psychiatry. 2016;173(6):607–616. doi: 10.1176/appi.ajp.2015.15070922. [DOI] [PubMed] [Google Scholar]
- Schäfer, J., & Strimmer, K. (2005). A shrinkage approach to large-scale covariance estimation and implications for functional genomics. Statistical applications in genetics and molecular biology, 4(1). doi:DOI:10.2202/1544-6115.1175. [DOI] [PubMed]
- Shehzad, Z., Kelly, A. M. C., Reiss, P. T., Gee, D. G., Gotimer, K., Uddin, L. Q., . . . Milham, M. P. (2009). The Resting Brain: Unconstrained yet Reliable. Cerebral Cortex, 19(10), 2209-2229. doi:10.1093/cercor/bhn256. [DOI] [PMC free article] [PubMed]
- Smith Stephen M., Beckmann Christian F., Andersson Jesper, Auerbach Edward J., Bijsterbosch Janine, Douaud Gwenaëlle, Duff Eugene, Feinberg David A., Griffanti Ludovica, Harms Michael P., Kelly Michael, Laumann Timothy, Miller Karla L., Moeller Steen, Petersen Steve, Power Jonathan, Salimi-Khorshidi Gholamreza, Snyder Abraham Z., Vu An T., Woolrich Mark W., Xu Junqian, Yacoub Essa, Uğurbil Kamil, Van Essen David C., Glasser Matthew F. Resting-state fMRI in the Human Connectome Project. NeuroImage. 2013;80:144–168. doi: 10.1016/j.neuroimage.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar Kaustubh, Musen Mark, Menon Vinod, Friston Karl J. Development of large-scale functional brain networks in children. PLoS Biol. 2009;7(7):e1000157. doi: 10.1371/journal.pbio.1000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason Moriah E., Dennis Emily L., Joshi Anand A., Joshi Shantanu H., Dinov Ivo D., Chang Catie, Henry Melissa L., Johnson Rebecca F., Thompson Paul M., Toga Arthur W., Glover Gary H., Van Horn John D., Gotlib Ian H. Resting-state fMRI can reliably map neural networks in children. NeuroImage. 2011;55(1):165–175. doi: 10.1016/j.neuroimage.2010.11.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truelove-Hill Monica, Erus Guray, Bashyam Vishnu, Varol Erdem, Sako Chiharu, Gur Ruben C., Gur Raquel E., Koutsouleris Nikolaos, Zhuo Chuanjun, Fan Yong, Wolf Daniel H., Satterthwaite Theodore D., Davatzikos Christos. A Multidimensional Neural Maturation Index Reveals Reproducible Developmental Patterns in Children and Adolescents. J. Neurosci. 2020;40(6):1265–1275. doi: 10.1523/JNEUROSCI.2092-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin L.Q., Supekar K., Menon V. Typical and atypical development of functional human brain networks: insights from resting-state FMRI. Front. Syst. Neurosci. 2010;4:21. doi: 10.3389/fnsys.2010.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler, D. (2003). Wechsler intelligence scale for children--Fourth Edition (WISC-IV). [DOI] [PubMed]
- Zhai Jian, Li Ke. Predicting Brain Age Based on Spatial and Temporal Features of Human Brain Functional Networks. Front. Hum. Neurosci. 2019;13 doi: 10.3389/fnhum.2019.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Yihong, Klein Arno, Castellanos F. Xavier, Milham Michael P. Brain age prediction: Cortical and subcortical shape covariation in the developing human brain. NeuroImage. 2019;202:116149. doi: 10.1016/j.neuroimage.2019.116149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X.N., Xing X.X. Test-retest reliabilities of resting-state FMRI measurements in human brain functional connectomics: a systems neuroscience perspective. Neurosci. Biobehav. Rev. 2014;45:100–118. doi: 10.1016/j.neubiorev.2014.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data incorporated in this work were gathered from the open access Healthy Brain Network and The Philadelphia Neurodevelopmental Cohort resource.