Abstract
Subacute ruminal acidosis (SARA) represents one of the most important digestive disorders in intensive dairy farms, and dairy cows are individually different in the severity of SARA risk. The objectives of the current study were to investigate differences in the ruminal bacterial community and metabolome in dairy cattle with different susceptibility to SARA. In the present study, 12 cows were initially enrolled in the experiment. Based on average ruminal pH, 4 cows with the lowest ruminal pH were assigned to the susceptible group (SUS, pH = 5.76, n = 4) and 4 cows with the highest ruminal pH assigned to the tolerant group (TOL, pH = 6.10, n = 4). Rumen contents from susceptible (SUS, n = 4) and tolerant (TOL, n = 4) dairy cows were collected through rumen fistula to systematically reveal the rumen microbial and metabolic alterations of dairy cows with different susceptibility to SARA using multi-omics approaches (16S and 18S rRNA gene sequencing and metabolome). The results showed that despite being fed the same diet, SUS cows had lower ruminal pH and higher concentrations of total volatile fatty acids (VFA) and propionate than TOL cows (P < 0.05). No significant differences were observed in dry matter intake, milk yield, and other milk compositions between the SUS and TOL groups (P > 0.05). The principal coordinates analysis based on the analysis of molecular variance indicated a significant difference in bacterial composition between the two groups (P = 0.01). More specifically, the relative abundance of starch-degrading bacteria (Prevotella spp.) was greater (P < 0.05), while the proportion of fiber-degrading bacteria (unclassified Ruminococcaceae spp., Ruminococcus spp., Papillibacter, and unclassified Family_XIII) was lower in the rumen of SUS cows compared with TOL cows (P < 0.05). Community analysis of protozoa showed that there were no significant differences in the diversity, richness, and community structure (P > 0.05). Metabolomics analysis revealed that the concentrations of organic acids (such as lactic acid), biogenic amines (such as histamine), and bacterial degradation products (such as hypoxanthine) were significantly higher in the SUS group compared to the TOL group (P < 0.05). These findings revealed that the higher proportion of starch-degrading bacteria/lower fiber-degrading bacteria in the rumen of SUS cows resulted in higher VFA-producing capacity, in particular propionate. This caused a disruption in metabolic homeostasis in the rumen which might be the reason for the higher susceptibility to SARA. Overall, these findings enhanced our understanding of the ruminal microbiome and metabolic changes in cows susceptible to SARA.
Keywords: Subacute ruminal acidosis, Microbiome, Metabolome, Dairy cow
1. Introduction
Subacute ruminal acidosis (SARA) is an important metabolic disorders of dairy cattle characterized by low ruminal pH in intensive dairy farms (Colman et al., 2010). Many studies have confirmed that the depressed ruminal pH was mainly caused by the accumulation of volatile fatty acids (VFA) produced from the microbial fermentation of high concentrate diets (Kleen et al., 2003; Mao et al., 2013). Although some preventive strategies such as adding buffer agents (sodium bicarbonate and magnesium oxide) have proven to be effective, several dairy cows under the same diet and management conditions in a herd are still experiencing SARA(Cottee et al., 2004; Penner et al., 2009). Nowadays, the difference in susceptibility of individual dairy cows to SARA has attracted wide attention. However, there is a paucity of information with regard to the underlying mechanisms behind the difference in susceptibility of individual dairy cows to SARA.
The variation in the susceptibility to SARA can be attributed to several factors regulating ruminal pH, such as ruminal acid production by microbiota, epithelial absorption, and buffer neutralization (Allen, 1997; Schlau et al., 2012). Previous studies had demonstrated that SARA susceptible (SUS) cows had lower ruminal pH and higher VFA than the tolerant (TOL) cows (Chen et al., 2012; Gao and Oba, 2014). Moreover, SUS cows appeared to have more active microbial ruminal fermentation, which was supported by a higher amount of total bacterial 16S rRNA genes in the rumen of SUS steers than that of TOL steers (Chen et al., 2012). Thus, the rumen microbial community and its metabolism may play an important role in causing the variation in susceptibility to SARA among dairy cows (Chen et al., 2012). However, some previous studies have only conducted preliminary explorations on rumen bacteria (such as total bacterial population) and metabolites (such as VFA and NH3–N) (Chen et al., 2012; Gao and Oba, 2014); there are no systematic studies on the effects of SARA susceptibility on rumen microbiota (bacteria and protozoa) and their metabolism in dairy cows.
Recently, the application of omics approaches has largely advanced our knowledge of rumen microbiota and its metabolism (Mao et al., 2016). We hypothesize that the variations in rumen microbiota and their metabolites were responsible for the different susceptibility to SARA. Therefore, the present study integrated 16S rRNA genes sequencing, 18S rRNA gene sequencing, and metabolomics to study the responsive changes in rumen microbiota and metabolites in dairy cows with different susceptibility to SARA. Investigating the rumen microbiota and its metabolism in both SUS and TOL cows will further explain the causative mechanism of SARA and provide new insights into its prevention.
2. Materials and methods
All the procedures in the current experiment were conducted according to the Animal Protection Law based on the Guide for the Care and Use of Laboratory Animals approved by the Ethics Committee of Nanjing Agricultural University.
2.1. Animal experimental design
Twelve multiparous ruminally cannulated mid-lactating Holstein cows (days in milk = 114 ± 22 d) were housed in individual tie-stalls during the experimental period. On average, the cows had similar body weight (579.3 ± 53.3 kg) and parities (2 to 3) at the beginning of the experiment and all the cows had never previously received rumen modifiers or been previously exposed to ruminal acidosis challenge studies. The experiment was performed over 35 d, including a 14-d diet adaptation and a 21-d experimental period. The ratio of concentrate to forage in the total mixed ration (TMR) diet was gradually adjusted from 30:70 to 40:60 over the adaption period and kept continuous during the experimental period. Cows were fed twice at 07:00 and 19:00 (5% to 10% orts on an as-fed basis) and milked twice before feeding every day. Detailed dietary composition and nutritional levels are shown in Table 1.
Table 1.
Ingredients and chemical composition of the diet (DM basis, %).
| Ingredient composition | Content | Chemical composition | Content |
|---|---|---|---|
| Alfalfa | 24.00 | DM, % as fresh fed | 46.77 |
| Oat | 24.00 | CP | 16.16 |
| Corn silage | 12.00 | NDF | 36.14 |
| Corn | 19.40 | NFC2 | 38.68 |
| Soybean meal | 13.50 | Ash | 5.97 |
| DDGS | 3.80 | Ca | 1.14 |
| Limstone | 0.80 | P | 0.52 |
| Ca (HCO3)2 | 1.10 | Crude fat | 3.05 |
| NaCl | 0.40 | Starch | 17.96 |
| Premix1 | 1.00 | NEL3, Mcal/kg | 1.57 |
| Total | 100 | NFC/NDF | 1.07 |
CP = crude protein; NDF = neutral detergent fiber; NFC = non-fiber carbohydrate.
Premix contained the following ingredients per kilogram of diet: vitamin A, 22.5 kIU; vitamin D3, 5.0 kIU; vitamin E, 37.5 IU; vitamin K3, 5.0 mg; Mn, 63.5 mg; Zn, 111.9 mg; Cu, 25.6 mg; and Fe, 159.3 mg.
NFC = 100 − [NDF (%) + CP (%) + ether extract (%) + ash (%)].
Calculated based on Ministry of China recommendations (MOA, 2004).
2.2. Feed sampling and analysis
Feed residuals were collected and weighed before feeding for 3 d every week, and the dry matter intake (DMI) of cattle was calculated according to the feeding amount and water content. Samples of the TMR and feed ingredients were collected in the last 3 d of the experimental period and stored at −20 °C until chemical composition analysis. The dry matter (Method: 934.01), crude protein (Method: 98903), starch (Method: 948.03), ash (Method: 942.05), and crude fat (Method: 2003.05) were detected based on the Association of Official Analytical Chemists method (AOAC, 1990). Acid detergent fiber and neutral detergent fiber were measured according to the method found in Van Soest et al. (1991).
2.3. Milk collection and analysis
Milk samples were collected both in the morning and afternoon on d 33, 34, and 35 of the experimental periods and treated with potassium dichromate. Subsequently, the milk collected for each of the 3 d for morning and afternoon were mixed according to the daily milk yield ratio, and transferred to Shanghai DHI Center (China) for milk composition analysis using a near-infrared analyzer (Foss Electric, Denmark) (Luinge et al., 1993). The analyzer detects the composition of milk fat, protein, lactose, total solids, and milk urea nitrogen.
2.4. Rumen fermentation parameters
On d 34 and 35, ruminal pH was monitored at 0 h before the morning feeding and 2, 4, 6, 8, and 12 h later using the pH meter (HANNA HI 99161; Woonsocket, RI, USA). Based on average ruminal pH, 4 cows with the lowest ruminal pH were assigned to the susceptible group (SUS = 5.76, n = 4) and 4 cows with the highest ruminal pH were assigned to the tolerant group (TOL = 6.10, n = 4). The rumen contents were collected from the ventral sac of the rumen via the rumen fistula at 0 h before morning feeding on d 35. Ruminal fluid samples were filtered through four layers of cheesecloth in 2 d (34 and 35 d) at four different time points: 0, 4, 8, and 12 h after the morning feeding. Then, about 50 mL of the filtered rumen fluid was mixed with 25% (wt/vol) metaphosphoric acid (4:1) (Mao et al., 2016), which was immediately stored at −20 °C until the VFA determination. Ruminal fluid samples were analyzed for VFA composition by gas chromatography (GC-2014B, Shimadzu, Japan; capillary column: 30 m × 0.32 mm × 0.25 μm; oven temperature = 140 °C; injector temperature = 180 °C; detector temperature = 180 °C) based on the procedures described by Sun et al. (2016).
2.5. Metabolite profiling of ruminal fluid
The analysis of rumen metabolites profile was performed using the ruminal fluid collected before the morning feeding on d 35. About 10 mL of ruminal fluid was centrifuged at 1,000 × g for 10 min and pipetted into two 5-mL plastic containers before being stored in liquid nitrogen. The samples were analyzed by liquid–chromatography/mass spectrometry (LC/MS). About 100 μL of the samples were transferred into centrifuge tubes (1.5 mL) and mixed with 300 μL methanol and 10 μL internal standard (2.8 mg/mL, DL-o-Chlorophenylalanine). Then, the mixture was vortexed for 30 s (Votex-5, Kylin-Bell Lab Instruments Co, LTD, Haimen, China), kept for 1 h, and centrifuged at 13,000 × g and 4 °C for 15 min. In the end, 200 μL of supernatant was transferred into a vial for LC/MS analysis.
The organic extraction was carried out using liquid chromatography (Thermo, Ultimate 3000 LC, Q Exactive) with a chromatographic column (Hyper gold C18, 100 m × 2.1 mm × 1.9 μm). The automatic injector temperature was set at 4 °C. Exactly 10 μL aliquot was injected into a Thermo 3000 LC system equipped with a capillary column at a constant flow rate of 3.0 mL/min and at 40 °C. Mobile phase A was 0.1% formic acid with 5% acetonitrile in water and mobile phase B was 0.1% formic acid in acetonitrile. The gradient of mobile phase A:B was 95:5 for 1 min, ramped to 60:40 at 2 min, and ramp to 20:80 at 7 min, then to 5:95 at 11 min; this was done to achieve the original state at 15.5 min, which was followed by 4 min of rebalancing.
Mass spectrometry has expressed the result in both positive and negative modes. The spray voltage was 3.0 kV and 3.2 kV for the positive and negative modes, respectively. Other mass detection parameters were set as the same index between positive and negative modes: heater temperature 300 °C, capillary temperature 350 °C, sheath gas flow rate 15 arb, sweep gas flow rate 1 arb, and S-Lens RF level 60%. Data were extracted and pretreated with Compound Discoverer software (Thermo Fisher Scientific, Waltham, MA, USA).
2.6. DNA extraction and 16S rRNA/18S rRNA gene sequencing
The microbial DNA was extracted from the ruminal content (0 h) as described by Mao et al. (2016). In brief, DNA was extracted using a series of wash steps with the phosphate buffered saline (PBS) (0.1 mmoL/L, pH 7.0), which was followed by transferring the detergents to a zirconium bead tube in a bead beater, used for breaking down the microbial cell wall; this was followed by extraction with combinations of phenol/chloroform, and sedimentation with isopropanol. The DNA was acquired after being resuspended in Tris–ethylene diamine tetraacetic acid (EDTA) (1 mol/L Tris HCl and 0.5 mol/L EDTA, pH = 8.0) and after measuring the concentrate and quality on NanoDrop spectrophotometrically (NanoDrop 1000, Thermo Fisher Scientific, Madison, USA).
We investigated the structure of rumen microbial communities by sequencing the region of the bacterial 16S rRNA gene and ciliate protozoal 18S rRNA. For the bacteria, we used universal primers to augment the V3 and V4 regions of the 16S rRNA gene and combed them with an individual 6 bp barcode for each sample. The primer sequences were 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′) (Lin et al., 2019). For ciliate protozoa, we used special primers to amplify the protozoal 18S rRNA gene with a 6 bp barcode unique to each sample. The primers were RP841F (5′-GACTAGGGATTGGARTGG-3′) and Reg1320 (5′-AATTGCAAAGATCTATCC-3′) (Lin et al., 2019). All the polymerase chain reaction (PCR) processes used a 20 μL TransStart Fastpfu DNA polymerase (TransGen Biotech, Beijing, China) reaction system, which included 4 μL 5× FastPfu buffer, 2 μL 2.5 mmol/L dNTPs, 0.8 μL 5 μmol/L Forward primer, 0.8 μL Reverse primer, 0.4 μL FastPfu polymerase, and 10 ng of DNA. Amplification was performed as follows: the first pre-degeneration was performed at 95 °C for 5 min, followed by 27 cycles of denaturation (95 °C, 30 s). Annealing (55 °C, 30 s) and elongation (72 °C, 45 s) were performed before an extension was done for 10 min at 72 °C.
After PCR amplification, all amplicon libraries were sequenced using an Illumina MiSeq PE 250 platform. The raw sequences were processed using QIIME v1.9.0 (Caporaso et al., 2010) and bases with an average quality score more than 20 were retained. Paired-end reads were merged into tags using FLASH v1.2.7 (Magoc and Salzberg, 2011), with a minimum overlap of 10-base sequence. Tags were then clustered into operational taxonomic units (OTU) at a 97% similarity threshold using UPARSE v7.0.1 (Edgar, 2013). The most abundant sequences of the OTU were assigned to a representative sequence, which was identified and marked to the bacterial and protozoa database of SILVA v11.9 (Quast et al., 2013). A rarefaction curve was constructed to ensure sufficient sequencing depth had been achieved. The α diversity, including Chao, Shannon and Simpson index, were used to determine the richness and diversity of the bacterial and protozoa community. The beta-diversity calculations were performed using QIIME. The principal coordinate analysis (PCoA) was based on the unweighted UniFrac distance, and an unweighted distance-based analysis of molecular variance (AMOVA) was conducted to assess the remarkable differences among each sample through MOTHUR v.1.29.0.
2.7. Quantitative real-time PCR analysis
The copy numbers of the total bacteria and protozoa from each rumen sample were measured by quantitative real-time PCR using specific 16S rRNA (Forward: GTGSTGCAYGGYYGTCGTCA, Reverse: ACGTCRTCCMCNCCTTCCTC) (Maeda et al., 2003) and 18s rRNA (Forward: GCTTTCCGWTGGTAGTGTATT, Reverse: CTTGCCCTCYAATCGTWCT) (Sylvester et al., 2004) primers, respectively. A real-time PCR was performed on the StepOnePlus platform (Applied Biosystems, Foster City, CA, United States) using SYBR Premix Ex Taq dye (Takara, Beijing, China). The PCR was performed in a two-step thermal cycling process that consisted of hot start activation at 95 °C for 30 s, which was followed by 40 cycles at 95 °C for 5 s, and 60 °C for 1 min (Ren et al., 2020). Quantification of the copies of total bacteria and protozoa in each sample was performed in triplicate, and the mean value was calculated. Standard curves were generated using the 10-fold serial dilutions of each standard DNA containing the target gene sequences of the respective microbial group. The absolute abundance of each microbial population was expressed as the log10 gene copies per ng DNA.
2.8. Statistical analysis
Power analyses calculated before the start of the experiment identified a minimum total sample of 8 cows. Each group was to have a power of 82.9% with a type I error of 5% using G∗Power 3.1.9.4 based on F-test of repeated measures between factors ANOVA. The average ruminal pH and VFA were analyzed by One-way repeated measures ANOVA in IBM statistics SPSS V26.0 (Chicago, IL, USA). The models included the main effects of the group (SUS vs TOL), the cow was considered as the random effect, and the hours after morning feeding was considered as the repeated measured. The effects were deemed significant at P < 0.05. The DMI, milk yield, and milk composition were analyzed using an independent sample t-test procedure. The non-parametric Mann–Whitney test was carried out to test the alpha-diversity and relative abundance of the microbiota. P < 0.05 was defined as statistical significance.
A principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) were conducted using the SIMCA 13.0 software (Umetrics, Umea, Sweden). The fold change (FC) was the ratio of the level of the corresponding metabolite obtained from the SUS and TOL groups (SUS/TOL). Differential metabolites were selected by combining the variable importance in the projection (VIP) generated in PLS-DA and the P-value obtained in non-parametric Mann–Whitney test analysis (VIP > 1, P < 0.05). Then, the metabolic pathway and enrichment analyses were conducted on Metabo-Analyst web server (http://www.metaboanalyst.ca) using the differential metabolites. The correlations between rumen metabolites and microbiota were calculated using the Spearman's correlation test implemented in GraphPad Prism version 6.01 (San Diego, CA, United States). The software Gephi 0.8.2 (https://gephi.org/) was used to visualize the correlation network of ruminal pH, VFA, microbiota, and metabolites.
3. Results
3.1. Classification method and rumen fermentation parameters
Based on the individual ruminal pH values (Appendix Fig. 1), dairy cows with relatively higher (pH = 6.10, n = 4) and lower (pH = 5.76, n = 4) mean ruminal pH (Table 2) were assigned to the TOL and SUS groups, respectively. Accordingly, the ruminal pH (mean, minimum, and maximum) in the SUS group was lower (P < 0.05) than that of the TOL group. As shown in Table 3, no significant differences (P > 0.05) were observed in DMI, milk yield, milk fat, milk protein, total solids, and urea nitrogen between the TOL and SUS groups. Compared with the TOL group, the concentrations of total VFA, propionate, butyrate, valerate, and proportions of butyrate (Table 4) were higher in the rumen of the SUS group (P < 0.05), while the proportions of acetate and isobutyrate were low in the SUS group (P < 0.05).
Table 2.
Variation on ruminal pH of lactating cows between the susceptible (SUS) and tolerant (TOL) groups.
| Item | Groups |
SEM | P-value | |
|---|---|---|---|---|
| TOL | SUS | |||
| Mean ruminal pH | 6.10 | 5.76 | 0.04 | <0.001 |
| Minimum ruminal pH | 5.79 | 5.45 | 0.07 | 0.008 |
| Maximum ruminal pH | 6.50 | 6.18 | 0.11 | 0.019 |
Table 3.
The level of DMI and milk production between the susceptible (SUS) and tolerant (TOL) groups.
| Item | Groups |
SEM | P-value | |
|---|---|---|---|---|
| TOL | SUS | |||
| DMI, kg/d | 22.05 | 20.39 | 2.82 | 0.576 |
| Milk yield, kg/d | 19.80 | 18.82 | 1.35 | 0.496 |
| Components, % | ||||
| MF | 4.15 | 4.34 | 0.16 | 0.470 |
| MP | 4.22 | 4.35 | 0.28 | 0.858 |
| Lactose | 4.84 | 5.03 | 0.12 | 0.146 |
| Total solid | 13.16 | 13.50 | 0.22 | 0.154 |
| MUN, mg N/dL | 19.25 | 17.35 | 1.16 | 0.151 |
MF = milk fat; MP = milk protein; MUN = milk urea nitrogen.
Table 4.
The changes of ruminal fermentation parameters between the susceptible (SUS) and tolerant (TOL) groups.
| Item | Groups |
SEM | P-value | |
|---|---|---|---|---|
| TOL | SUS | |||
| Total VFA, mmol/L | 98.43 | 116.56 | 4.97 | 0.042 |
| Acetate, mmol/L | 66.39 | 72.73 | 4.53 | 0.360 |
| Propionate, mmol/L | 19.30 | 26.54 | 1.75 | 0.027 |
| Isobutyrate, mmol/L | 0.83 | 0.80 | 0.06 | 0.722 |
| Butyrate, mmol/L | 9.62 | 13.56 | 0.54 | 0.002 |
| Isovalerate, mmol/L | 1.01 | 1.21 | 0.07 | 0.101 |
| Valerate, mmol/L | 1.28 | 1.72 | 0.12 | 0.029 |
| Acetate-to-propionate ratio | 3.47 | 2.83 | 0.27 | 0.100 |
| Acetate, % | 67.46 | 62.40 | 1.09 | 0.044 |
| Propionate, % | 19.61 | 22.77 | 1.64 | 0.209 |
| Isobutyrate, % | 0.86 | 0.68 | 0.05 | 0.046 |
| Butyrate, % | 9.83 | 11.63 | 0.51 | 0.044 |
| Isovalerate, % | 1.04 | 1.04 | 0.04 | 0.708 |
| Valerate, % | 1.29 | 1.48 | 0.11 | 0.257 |
3.2. The composition of rumen bacteria communities
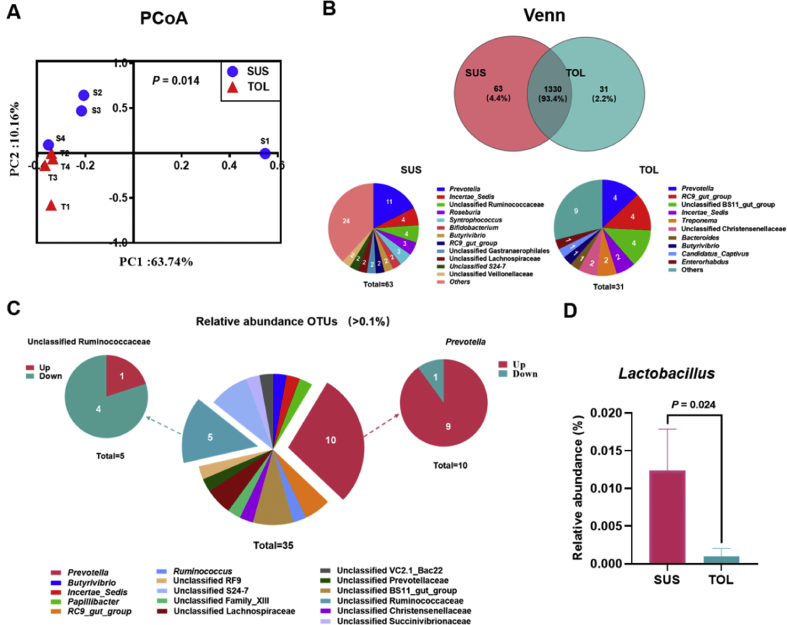
There was an average of 37,671 ± 7,004 reads per sample via 16S rRNA gene sequencing. The rarefaction curves approximately trended to a plateau at 24,349 reads, revealing that the sequencing coverage was saturated (Appendix Fig. 2A). As shown in Appendix Fig. 2B, the Chao1, Shannon, and Simpson index in the SUS group were similar to those in the TOL group (P > 0.05). In addition, we also observed no significant variation between the TOL and SUS cows for total bacteria (Appendix Table 1, P = 0.227). The PCoA accompanied AMOVA analysis showed that the SUS group was significantly separated from the TOL group (AMOVA, P = 0.014; Fig. 1A). The Venn analysis showed (Fig. 1B) that the SUS and TOL groups had 63 and 31 unique OTU, respectively, and shared 1330 OTU. According to the classification, 11 unique OTU belong to Prevotella genus in the SUS group.
Fig. 1.
Results of 16S rRNA gene sequence of the rumen bacteria in the susceptible (SUS) and tolerant (TOL) cows. (A) Principal coordinate analysis (PCoA) based on the operational taxonomic units (OTU) level of the bacteria in the SUS and TOL groups. (S1-4: SUS1-4; T1-4: TOL1-4) (B) Venn diagram based on the average reads of bacteria community in the rumen. (C) The significantly different (P < 0.05) OTU (>0.1% at least one group) in the rumen bacteria between the SUS and TOL groups. (D) The relative abundance of Lactobacillus between the SUS and TOL groups.
All the rumen bacterial reads were allocated to 16 phyla and 106 genera. As shown in Appendix Fig. 3, the most abundant phyla were Bacteroidetes (53.6% vs 45.5%) and Firmicutes (40.8% vs 49.1%) in the SUS and TOL groups, respectively. At the genus level, we only listed the top 25 bacterial genera whose relative abundance was greater than 0.5% in at least one group (Appendix Fig. 4). Our results showed that the Prevotella (SUS vs TOL: 28.8% vs 18.4%) was the dominant genus in both the SUS and TOL groups. We also observed that the relative abundances of unclassified Family_XIII (P = 0.021) and Papillibacter (P = 0.043) were significantly lower in the SUS group. Furthermore, we screened out 311 OTU whose relative abundance was greater than 0.1% in at least one group. Compared with the TOL group (Appendix Table 2), the SUS group had a higher relative abundance of Prevotella (OTU322, OTU305, OTU239, OTU17, OTU158, OTU156, OTU146, OTU138, and OTU134). However, the abundances of unclassified Ruminococcaceae (OTU4, OTU378, OTU216, and OTU14) and Ruminococcus (OTU141) were lower in the SUS group. In addition, we found the abundance of Lactobacillus was significantly high in the SUS group (Fig. 1D).
3.3. The communities of rumen protozoa
After a quality filter was applied, an average of 40,614 ± 8,541 reads for each sample were observed. The rarefaction curves (Appendix Fig. 5A) approximately trended to stabilize at 30, 365 reads, which revealed that the sequencing coverage was saturated. Based on the unweighted UniFrac distances, the PCoA plot (Appendix Fig. 5B) did not reveal any segregation, and the AMOVA analysis confirmed no significant difference between the two groups (P = 0.69). Similarly, the analysis of protozoa density also showed no significant variation between the TOL and SUS cows (Appendix Table 1, P = 0.626). Since the PCoA chart showed that the S3 sample was obviously an outlier, our subsequent analysis excluded the S3 cow. Estimators of richness and diversity showed that there was no difference (P > 0.05) in the OTU numbers, Chao 1, Simpson, and Shannon index (Appendix Fig. 5C) between the TOL and SUS group. The Venn diagram showed that 20 OTU were shared by the two groups, and two unique OTU were observed in the SUS group (Appendix Fig. 5D). According to the results of subtotals (Appendix Table 3), Entodinium was dominant in the rumen of both groups (78.11% vs. 75.28%). Furthermore, at the genus and OTU levels, ruminal protozoa had a similar relative abundance between the two groups (P > 0.05, Appendix Table 3).
3.4. LC/MS analysis of ruminal fluid
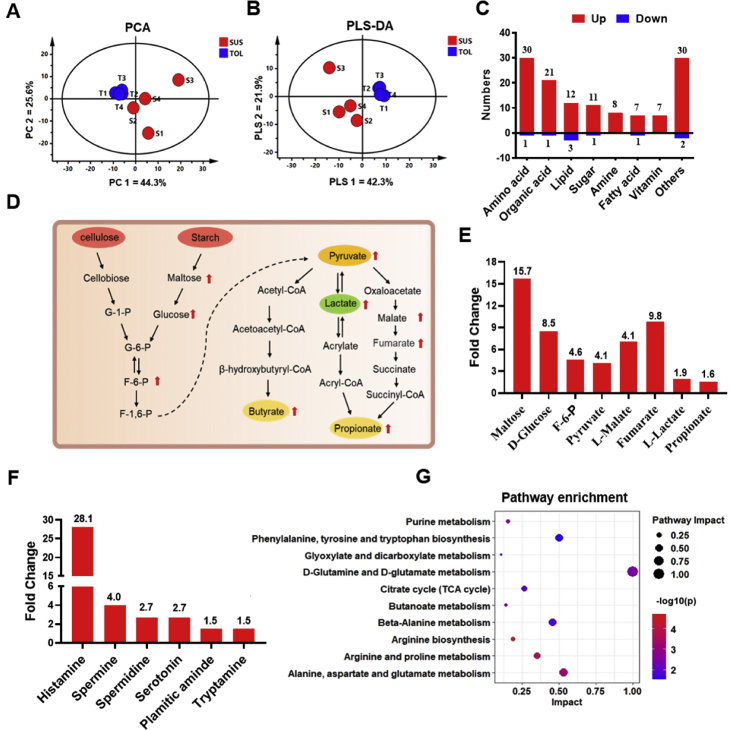
After rigorous quality screening and identification, we obtained 195 reliable metabolites across all samples. As shown in Fig. 2A and B, there was marked clustering between the two groups based on the PCA and PLS-DA models. This result indicates that the distribution of rumen metabolites was significantly different between the TOL and SUS cows. Combined with statistical analysis and the VIP value obtained from the PLS-DA analysis, we identified 135 differential metabolites (P < 0.05 and VIP > 1). These differential rumen metabolites were further classified according to the properties of the compounds (Appendix Table 4), which are mainly distributed in amino acids, organic acids, lipids, sugars, fatty acids, and biogenic amines (Fig. 2C). Compared with the TOL group, 126 differential compounds were increased, while 9 differential compounds were decreased in the SUS group. The metabolite classification indicated that a higher level of amino acids, organic acids, sugars, and biogenic amines were observed in the SUS group than in the TOL group. In the 30 variational amino acids and derivatives, 10 common amino acids, including L-glutamic acid, L-tryptophan, L-lysine, L-phenylalanine, L-threonine, L-histidine, D-leucine acid, L-proline L-arginine, and L-isoleucine, were significantly higher (P < 0.05 and VIP > 1) in the rumen of cows in the SUS group. In addition, 21 organic acids were obviously higher in the SUS group, such as fumarate, L-malate, pyruvate, and L-lactic acid (Fig. 2D). For the sugars and derivatives, the results revealed that the SUS group had higher maltose, D-glucose, beta-D-fructose 6-phosphate, and 6-phospho-2-dehydro-D-gluconate levels but a lower sedoheptulose 7-phosphate level than the TOL group (Fig. 2E). In the case of amines (Fig. 2F), the levels of histamine, spermine spermidine, serotonin, palmitic amide, and tryptamine were higher in the SUS group.
Fig. 2.
Results of rumen metabolites analysis. (A) Principal component analysis (PCA) of rumen metabolites. (S1-4: SUS1-4, T1-4: TOL1-4). (B) Partial least squares-discriminate analysis (PLS-DA, of rumen metabolites. The x-axis and the y-axis indicate the first and second principal components, respectively. Dots represented samples and the distances among dots demonstrated the similarities among samples according to the rumen metabolites. (C) Classification of rumen differential metabolites (SUS/TOL, variable importance in the projection [VIP] > 1.0, P < 0.05). (D) Metabolic routes for propionate and butyrate production by direct conversion from carbohydrates. G: glucose; P: phosphate; F: fructose; The red arrows indicate a significant higher in rumen metabolites in SUS group. (E) The fold change of different metabolites involved in carbohydrates metabolism (SUS/TOL, VIP > 1.0, P < 0.05). (F) The fold change of amine in rumen metabolites. (G) Metabolic pathways were analyzed based on different metabolites (Impact > 0.1, P < 0.05).
Based on the above results, it is necessary to comprehensively evaluate how the multiple pathways changed in response to the different susceptibility to SARA. Therefore, we further analyzed these differential rumen metabolites. According to the pathway topology analysis, 10 metabolic pathways were significantly enriched by differential metabolites (Fig. 2G, P < 0.05, impact > 0.1). According to the impact value, arginine and proline metabolism, alanine, aspartate and glutamate metabolism, and beta-alanine metabolism were closely related to SARA susceptibility.
3.5. Correlation analysis of rumen microbiota and metabolites
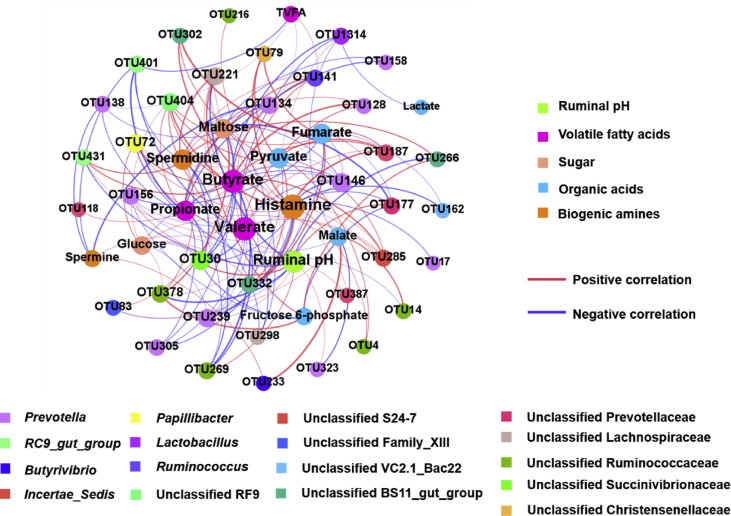
To explore the underlying relationship between rumen microbiota and metabolites related to the variation in susceptibility to SARA, we plotted the correlation network using the affected ruminal OTU, VFA concentration, metabolites, and ruminal pH. The correlation network consisted of 52 nodes and 151 edges (Fig. 3), including 73 positive correlations and 78 negative correlations (|r| > 0.8 and P < 0.05). For instance, ruminal pH was positively correlated with the relative abundance of unclassified Ruminococcaceae (OTU14 and OTU378) and negatively correlated with Prevotella (OTU146, OTU156, OTU239, and OTU323) and unclassified Prevotellaceae (OTU177). Propionate concentration had a positive correlation with Prevotella (OTU134, OTU138, OTU146, OTU158, and OTU305) and a negative correlation with Ruminococcus (OTU141) Unclassified RF9 (OTU404). Moreover, the Prevotella (OTU146) had a significantly positive correlation with maltose, glucose, pyruvate, and fumarate. Based on correlation network analysis, we found that the abundance of Prevotella may be important for the changes of ruminal metabolites.
Fig. 3.
Correlation networks of the rumen volatile fatty acids (VFA), different ruminal metabolites and discrepant rumen bacteria in the susceptible (SUS) and tolerant (TOL) groups based on Spearman's correlation coefficients (|r| > 0.8 and P < 0.05). Node size and color corresponds to the degree and classification, respectively. Red lines denote positive correlations, while green lines denote negative correlations.
4. Discussion
4.1. The same diet and similar DMI resulted in different rumen fermentation profiles between SUS and TOL cows
In dairy farms, high-concentrate diets can easily cause SARA, which is characterized by low ruminal pH, and the duration of ruminal pH < 5.8 has usually been used to define the occurrence of SARA (Hook et al., 2011). However, considerable variability in SARA was observed in dairy cows independent of dietary treatment (Bevans et al., 2005; Penner et al., 2006). In agreement with the studies of Schlau et al. (2012), a significant variation in ruminal pH of twelve experimental dairy cows was observed in the present study. It confirmed the claim that there was discrepant sensitivity to SARA in the dairy cows and indicated the model of different susceptibility to SARA that was established successfully in our work.
As expected, a low ruminal pH and high concentration of total VFA, propionate, and butyrate in SUS cows were observed in the present study. It is generally accepted that a decreased ruminal pH during SARA is associated with the accumulation of VFA (Schlau et al., 2012; Gao and Oba, 2014). However, there were no significant differences in DMI and milk production between the TOL and SUS cows in the present study. Consistently, Gao and Oba (2016) reported that a greater or lower risk of SARA had no significant effects on DMI, milk fat, and milk yield of dairy cows. Hence, this different susceptibility to SARA was not caused by the intake difference of fermentable carbohydrates and did not further affect the milk performance of dairy cows in the present study. Overall, as discussed above, the experimental cows ate the same diets, with similar DMI and milk production, but the SUS cows had lower ruminal pH and higher VFA, suggesting that the rumen microbiome may be responsible for the difference in SARA sensitivity.
4.2. The variation in bacterial communities rather than protozoal communities was partly responsible for the differences in rumen fermentation
To further explore the mechanism that caused this difference, we performed 16S/18S rRNA genes sequencing to reveal the disparity of microbial communities in the rumen between the TOL and SUS groups. Although the TOL and SUS cows had similar bacterial diversity and richness, the PCoA and AMOVA analysis showed a significant difference in bacterial composition between the two groups. More specifically, the phylogenetic analysis of detectable microbial genera showed that Prevotella, a main starch-degrading bacterium, was the dominant genus in the rumen of both the TOL and SUS cows (Pitta et al., 2010). Previous studies have suggested that the degree of SARA risk may be related to the rise of Prevotella populations (Petri et al., 2013; McCann et al., 2016). In order to further reveal the changes in Prevotella abundance, we performed a statistical analysis at the OTU level. Our results showed that the relative abundance of 9 OTU classified as Prevotella was higher in the SUS group than in the TOL group. More interestingly, among these above-mentioned affected OTU, the abundance of Prevotella (OTU134 and OTU138) showed significant positive correlations with TVFA. Moreover, the ruminal pH showed significant negative correlations with Prevotella (OTU146, OTU156, OTU239, and OTU323). This observation confirmed that a high abundance of Prevotella may promote the accumulation of VFA (Khafipour et al., 2009) and reduce the ruminal pH.
In addition, our results also revealed that the relative abundance of 5 OTU belonging to unclassified Ruminococcaceae and Ruminococcus were significantly lower in the SUS group than in the TOL group. These findings are consistent with the results in several studies on SARA or high-concentrate diets (Li et al., 2017; Zhang et al., 2019b), and indicate the abundance of unclassified Ruminococcaceae and Ruminococcus could be used as potential biomarkers of SARA risk. In addition, the abundance of Papillibacter and unclassified Family_XIII were lower in the SUS cows than in the TOL group, and both of them have a fiber-degrading ability (Mao et al., 2013; Zened et al., 2013). The low ruminal pH in SUS cows may be responsible for the decreased relative abundance of these pH-sensitive fiber-degrading bacteria. Overall, the above results demonstrated that the more starch-degrading bacteria and fewer fiber-degrading bacteria in the rumen were partly responsible for the variation in the acidity of the rumen between the SUS and TOL groups.
Besides the bacterial community, ciliate protozoa are also an important part of the rumen microbiome, which could engulf the starch granules and change the digestion products to reserve carbohydrates and slow down starch fermentation (Belzecki et al., 2017). Hence, we further investigated the relationships between SARA susceptibility and ciliate protozoa microbiota. In the present study, there was no difference in the diversity, richness, and community structure of ruminal protozoa. These results reconfirmed that rumen bacteria rather than rumen protozoa mainly caused the changes in the rumen acid environment. Therefore, the rumen ciliate protozoa may be less important than bacteria for the variation in SARA susceptibility.
4.3. Rumen metabolites between SUS and TOL cows exhibited huge differences in response to changes in bacterial communities
In the present study, we used a metabolomics approach to comprehensively reveal the differences in rumen metabolites between TOL and SUS cows. Previously, growing evidence has suggested that rumen metabolites derived from rumen microbiome disorders were important factors for SARA in dairy cows (Mao et al., 2016; Zhang et al., 2017). In the present study, the variation of rumen microorganism and rumen fermentation parameters implied that rumen metabolites might be changed. As expected, a total of 135 metabolites were significantly different between the SUS group and the TOL group. In particular, we observed a significant increase of carbohydrate metabolites in the SUS group, such as maltose, glucose, and β-D-Fructose 6-phosphate, which are important intermediates of carbohydrate metabolism. Carbohydrate metabolites are mainly derived from the degradation of starch and cellulose and they will be digested into glucose by rumen microbial enzymes and then through the glycolytic pathway to produce pyruvate, which is the main precursor of VFA production (Xue et al., 2018). Furthermore, we also discovered a distinctly advantageous level of L-malate, and fumarate in the SUS cows, both of which were important in propionate production (Nisbet and Martin, 1993). Correspondingly, the abundance of Prevotella had a significantly positive correlation with the propionate concentration in the rumen. These observations suggested that the high accumulation of total VFA and propionate in the SUS group was related to the rapid and efficient degradation of soluble carbohydrates by these bacteria. Taken together, the higher risk to SARA in SUS cows was possibly caused by the increased relative abundance of the starch-degrading bacteria, which might be related to a high abundance of Prevotella.
Previous studies have reported that the accumulation of organic acids may promote the risk of SARA in dairy cows (Ewaschuk et al., 2004; Vyas et al., 2015). We found that 21 organic acids had higher levels in the SUS group, including acetoacetic acid, L-lactic acid, and other acids, which contributed to changes in the rumen acid environment. Acetoacetic acid is an intermediate metabolite of the citric acid cycle. In the present study, as compared with the TOL group, a higher concentration of acetoacetic acid in SUS group indicated that the ruminal microbial metabolism in the SUS group might have been more active, and this may have led to the production of more ruminal VFA in the SUS group, which further stimulated SARA development. In ruminal conditions, lactic acid originates from dietary carbohydrates through the glycolytic pathway and pentose phosphate pathway of rumen microorganisms (Sauer et al., 1975; Zhang et al., 2017). In the present study, there was a significantly higher alteration in the relative abundance of Lactobacillus in the SUS group, and it was positively correlated with the concentration of lactic acid, which contributed to the production of lactic acid, and played a significant role in the initiation of SARA (Mao et al., 2013). Moreover, a previous study revealed that pyruvate could be converted to lactic acid by lactic acid dehydrogenase (Chen et al., 2016). Thus, an elevated lactic acid content could potentially improve the concentration of propionate in the SUS group. In general, these observations showed that the degradation of carbohydrates by rumen microbiota was more efficient and promoted the lactic acid and propionate products in the SUS group.
One remarkable alteration in the present study was the increased levels of 30 amino acids and their derivatives in the SUS group. Amino acids are mainly derived from the degradation of proteins and microorganisms by rumen microbiota (Fuller, 2012). Thus, it can be seen that the protein degradation efficiency of rumen microorganisms was different between the two groups. In addition, previous studies have reported that the rumen microorganisms can use VFA or other substances as a carbon source, and use nitrogen compounds, such as ammonia, as a nitrogen source, for the de novo synthesis of amino acids (Sauer et al., 1975; Abdul-Razzaq and Bickerstaffe, 1989; Kajikawa et al., 2002). In the present study, at a lower ruminal pH, higher concentrations of propionate and butyrate may have provided more substrates for amino acid synthesis in the SUS group. Therefore, the above evidence indicated that the ruminal amino acid metabolism was stronger in the SUS group. Interestingly, the levels of biogenic amines (histamine, spermine, spermidine, and tryptamine) in the rumen of the SUS group were significantly higher. Correspondingly, their precursors (L-histidine, L-arginine, L-lysine, and L-tryptophan) were enriched in the rumen of SUS cows. The process of amino acid synthesis and metabolism released biogenic amines from the decarboxylation of certain amino acids. Low concentrations of biogenic amines are essential for normal growth and differentiation of cells (Medina et al., 2003), but those compounds can cause severe damage to the rumen epithelial barrier's tight connection in high concentrations (Wang et al., 2013). Previous studies have revealed that biogenic amines, especially histamine, were responsible for the incidence of SARA because of their pro-inflammatory effects (Aschenbach and Gäbel, 2000; Sun et al., 2017). Collectively, the above evidence indicated that the decarboxylation of amino acids was enriched in the rumen of the SUS group and promoted the release of biogenic amines, which might be an important factor for increasing SARA risk.
The current study also observed an increase of gram-negative bacteria degradation products in the rumen of SUS groups, such as hypoxanthine, uracil, and thymine. The elevated levels of these metabolites reflected that more bacterial nucleic acids were degraded in the SUS group (McAllan and Smith, 1973). Correspondingly, a study by Zhang et al. (2017) showed that an increased level of gram-negative bacteria degradation products (hypoxanthine, uracil, and thymine etc.) were caused by low ruminal pH in dairy cows. The increased bacteria degradation could damage the rumen epithelial barrier (Emmanuel et al., 2007; Zhang et al., 2019a), thereby reducing the absorption rate of VFA and increasing the risk of rumen acidosis (Tao et al., 2014). In general, we speculated that the passaging process of rumen microbes was faster in the SUS group, and it will lead to an increased amount of bacteria degradation products.
5. Conclusions
The integrated analysis of ruminal microbiome and metabolome demonstrated that SUS cows have different ruminal bacterial communities including more starch-degrading bacteria (Prevotella spp.) and fewer fiber-degrading bacteria (unclassified Ruminococcaceae spp., Ruminococcus spp., Papillibacter, and unclassified Family_XIII) and showed increased acidic substances and biogenic amines in the rumen compared with the TOL cows. The high abundance of starch-degrading bacteria will promote carbohydrate metabolism and increase VFA production. In addition, the increased biogenic amines and organic acid disrupted the ruminal metabolic homeostasis and possibly resulted in a high susceptibility to SARA. Overall, our findings contribute to the further exploration of the mechanisms of susceptibility to SARA in cows, warranting future studies about the microbial–host interactions under this susceptibility.
Author contributions
Shengyong Mao designed the experiments. Tao Zhang, Yingyu Mu, Wangpan Qi, Changzheng Guo and Jiyou Zhang performed the experiments. Tao Zhang analyzed the experimental data and wrote the draft manuscript, Ruiyang Zhang, Yanfeng Xue, and Shengyong Mao reviewed and edited the manuscript. The authors read and approved the final manuscript.
Availability of data and materials
Raw sequence reads for all samples described above were deposited into the NCBI Sequence Read Archive (SRA) database (NO. PRJNA678648).
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgments
This research was funded by Natural Science Foundation of China (32072755) and the Fundamental Research Funds for the Central Universities (JCQY201905).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
The appendix to this article can be found online at https://doi.org/10.1016/j.aninu.2021.10.009.
Appendix.
The following is the supplementary data to this article:
References
- Abdul-Razzaq H A, Bickerstaffe R. The influence of rumen volatile fatty acids on protein metabolism in growing lambs. Br J Nutr,1989 62(2):297-310. [DOI] [PubMed]
- Allen M.S. Relationship between fermentation acid production in the rumen and the requirement for physically effective fiber. J Dairy Sci. 1997;80(7):1447–1462. doi: 10.3168/jds.S0022-0302(97)76074-0. [DOI] [PubMed] [Google Scholar]
- AOAC . 15th ed. vol. I. Association of Official Analytical Chemists; Arlington, VA: 1990. (Official methods of analysis). [Google Scholar]
- Aschenbach J.R., Gäbel G. Effect and absorption of histamine in sheep rumen: significance of acidotic epithelial damage. J Anim Sci. 2000;78:464–470. doi: 10.2527/2000.782464x. [DOI] [PubMed] [Google Scholar]
- Belzecki G., McEwan N.R., Kowalik B., Michalowski T., Miltko R. Effect of Entodinium caudatum on starch intake and glycogen formation by Eudiplodinium maggii in the rumen and reticulum. Eur J Protistol. 2017;57:38–49. doi: 10.1016/j.ejop.2016.09.007. [DOI] [PubMed] [Google Scholar]
- Bevans D.W., Beauchemin K.A., Schwartzkopf-Genswein K.S., McKinnon J.J., McAllister T.A. Effect of rapid or gradual grain adaptation on subacute acidosis and feed intake by feedlot cattle. J Anim Sci. 2005;83:1116–1132. doi: 10.2527/2005.8351116x. [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Tumbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Luo Y., Wang H., Liu S., Shen Y., Wang M. Effects of glucose and starch on lactate production by newly isolated Streptococcus bovis S1 from Saanen goats. Appl Environ Microbiol. 2016;82:5982–5989. doi: 10.1128/AEM.01994-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Oba M., Guan L.L. Variation of bacterial communities and expression of Toll-like receptor genes in the rumen of steers differing in susceptibility to subacute ruminal acidosis. Vet Microbiol. 2012;159:451–459. doi: 10.1016/j.vetmic.2012.04.032. [DOI] [PubMed] [Google Scholar]
- Colman E., Fokkink W.B., Craninx M., Newbold J.R., De Baets B., Fievez V. Effect of induction of subacute ruminal acidosis on milk fat profile and rumen parameters. J Dairy Sci. 2010;93:4759–4773. doi: 10.3168/jds.2010-3158. [DOI] [PubMed] [Google Scholar]
- Cottee G., Kyriazakis I., Widowski T.M., Lindinger M.I., Cant J.P., Duffield T.F., Osborne V.R., McBride B.W. The effects of subacute ruminal acidosis on sodium bicarbonate-supplemented water intake for lactating dairy cows. J Dairy Sci. 2004;87:2248–2253. doi: 10.3168/jds.S0022-0302(04)70045-4. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Emmanuel D.G., Madsen K.L., Churchill T.A., Dunn S.M., Ametaj B.N. Acidosis and lipopolysaccharide from Escherichia coli B:055 cause hyperpermeability of rumen and colon tissues. J Dairy Sci. 2007;90:5552–5557. doi: 10.3168/jds.2007-0257. [DOI] [PubMed] [Google Scholar]
- Ewaschuk J.B., Naylor J.M., Barabash W.A., Zello G.A. High-performance liquid chromatographic assay of lactic, pyruvic and acetic acids and lactic acid stereoisomers in calf feces, rumen fluid and urine. J Chromatogr B Anal Technol Biomed Life Sci. 2004;805:347–351. doi: 10.1016/j.jchromb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Fuller M. Determination of protein and amino acid digestibility in foods including implications of gut microbial amino acid synthesis. Br J Nutr. 2012;108(Suppl. 2):S238–S246. doi: 10.1017/S0007114512002279. [DOI] [PubMed] [Google Scholar]
- Gao X., Oba M. Relationship of severity of subacute ruminal acidosis to rumen fermentation, chewing activities, sorting behavior, and milk production in lactating dairy cows fed a high-grain diet. J Dairy Sci. 2014;97:3006–3016. doi: 10.3168/jds.2013-7472. [DOI] [PubMed] [Google Scholar]
- Gao X., Oba M. Characteristics of dairy cows with a greater or lower risk of subacute ruminal acidosis: volatile fatty acid absorption, rumen digestion, and expression of genes in rumen epithelial cells. J Dairy Sci. 2016;99:8733–8745. doi: 10.3168/jds.2016-11570. [DOI] [PubMed] [Google Scholar]
- Hook S.E., Steele M.A., Northwood K.S., Dijkstra J., France J., Wright A.D., McBride B.W. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol Ecol. 2011;78:275–284. doi: 10.1111/j.1574-6941.2011.01154.x. [DOI] [PubMed] [Google Scholar]
- Kajikawa H., Mitsumori M., Ohmomo S. Stimulatory and inhibitory effects of protein amino acids on growth rate and efficiency of mixed ruminal bacteria. J Dairy Sci. 2002;85:2015–2022. doi: 10.3168/jds.S0022-0302(02)74278-1. [DOI] [PubMed] [Google Scholar]
- Khafipour E., Li S., Plaizier J.C., Krause D.O. Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl Environ Microbiol. 2009;75:7115–7124. doi: 10.1128/AEM.00739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleen J.L., Hooijer G.A., Rehage J., Noordhuizen J.P. Subacute ruminal acidosis (SARA): a review. J Vet Med A Physiol Pathol Clin Med. 2003;50:406–414. doi: 10.1046/j.1439-0442.2003.00569.x. [DOI] [PubMed] [Google Scholar]
- Li F., Wang Z., Dong C., Li F., Wang W., Yuan Z., Mo F., Weng X. Rumen bacteria communities and performances of fattening lambs with a lower or greater subacute ruminal acidosis risk. Front Microbiol. 2017;8:2506. doi: 10.3389/fmicb.2017.02506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Xie F., Sun D., Liu J., Zhu W., Mao S. Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome. 2019;7:83. doi: 10.1186/s40168-019-0701-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luinge H.J., Hop E., Lutz E.T.G., Vanhemert J.A., Dejong E.A.M. Determination of the fat, protein and lactose content of milk using Fourier transform infrared spectrometry. Anal Chim Acta. 1993;284:419–433. [Google Scholar]
- Maeda H., Fujimoto C., Haruki Y., Maeda T., Kokeguchi S., Petelin M., Arai H., Tanimoto I., Nishimura F., Takashiba S. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol Med Microbiol. 2003;39:81–86. doi: 10.1016/S0928-8244(03)00224-4. [DOI] [PubMed] [Google Scholar]
- Magoc T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S.Y., Huo W.J., Zhu W.Y. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ Microbiol. 2016;18:525–541. doi: 10.1111/1462-2920.12724. [DOI] [PubMed] [Google Scholar]
- Mao S.Y., Zhang R.Y., Wang D.S., Zhu W.Y. Impact of subacute ruminal acidosis (SARA) adaptation on rumen microbiota in dairy cattle using pyrosequencing. Anaerobe. 2013;24:12–19. doi: 10.1016/j.anaerobe.2013.08.003. [DOI] [PubMed] [Google Scholar]
- McAllan A.B., Smith R.H. Degradation of nucleic acids in the rumen. Br J Nutr. 1973;29:331–345. doi: 10.1079/bjn19730107. [DOI] [PubMed] [Google Scholar]
- McCann J.C., Luan S., Cardoso F.C., Derakhshani H., Khafipour E., Loor J.J. Induction of subacute ruminal acidosis affects the ruminal microbiome and epithelium. Front Microbiol. 2016;7:701. doi: 10.3389/fmicb.2016.00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M.A., Urdiales J.L., Rodriguez-Caso C., Ramirez F.J., Sanchez-Jimenez F. Biogenic amines and polyamines: similar biochemistry for different physiological missions and biomedical applications. Crit Rev Biochem Mol Biol. 2003;38:23–59. doi: 10.1080/713609209. [DOI] [PubMed] [Google Scholar]
- MOA . MOA; Beijing, China: 2004. Feeding standard of dairy cattle (NY/T 34–2004) [Google Scholar]
- Nisbet D.J., Martin S.A. Effects of fumarate, L-malate, and an Aspergillus oryzae fermentation extract on D-lactate utilization by the ruminal bacterium Selenomonas ruminantium. Curr Microbiol. 1993;26(3):133–136. [Google Scholar]
- Penner G.B., Aschenbach J.R., Gabel G., et al. Epithelial capacity for apical uptake of short chain fatty acids is a key determinant for intraruminal pH and the susceptibility to subacute ruminal acidosis in sheep. J Nutr. 2009;139(9):1714–1720. doi: 10.3945/jn.109.108506. [DOI] [PubMed] [Google Scholar]
- Penner G.B., Beauchemin K.A., Mutsvangwa T. An evaluation of the accuracy and precision of a stand-alone submersible continuous ruminal pH measurement system. J Dairy Sci. 2006;89:2132–2140. doi: 10.3168/jds.S0022-0302(06)72284-6. [DOI] [PubMed] [Google Scholar]
- Petri R.M., Schwaiger T., Penner G.B., Beauchemin K.A., Forster R.J., McKinnon J.J., McAllister T.A. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS One. 2013;8(12):e83424. doi: 10.1371/journal.pone.0083424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitta D.W., Pinchak E., Dowd S.E., Osterstock J., Gontcharova V., Youn E., Dorton K., Yoon I., Min B.R., Fulford J.D., Wickersham T.A., Malinowski D.P. Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets. Microb Ecol. 2010;59:511–522. doi: 10.1007/s00248-009-9609-6. [DOI] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glockner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q., Si H., Yan X., Liu C., Ding L., Long R., Li Z., Qiu Q. Bacterial communities in the solid, liquid, dorsal, and ventral epithelium fractions of yak (Bos grunniens) rumen. Microbiologyopen. 2020;9:e963. doi: 10.1002/mbo3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer F.D., Erfle J.D., Mahadevan S. Amino acid biosynthesis in mixed rumen cultures. Biochem J. 1975;150:357–372. doi: 10.1042/bj1500357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlau N., Guan L., Oba M. The relationship between rumen acidosis resistance and expression of genes involved in regulation of intracellular pH and butyrate metabolism of ruminal epithelial cells in steers. J Dairy Sci. 2012;95:5866–5875. doi: 10.3168/jds.2011-5167. [DOI] [PubMed] [Google Scholar]
- Sun X., Yuan X., Chen L., Wang T., Wang Z., Sun G., Li X., Li X., Liu G. Histamine induces bovine rumen epithelial cell inflammatory response via NF-kappaB pathway. Cell Physiol Biochem. 2017;42:1109–1119. doi: 10.1159/000478765. [DOI] [PubMed] [Google Scholar]
- Sun Y., Yu K., Zhou L., Fang L., Su Y., Zhu W. Metabolomic and transcriptomic responses induced in the livers of pigs by the long-term intake of resistant starch. J Anim Sci. 2016;94:1083–1094. doi: 10.2527/jas.2015-9715. [DOI] [PubMed] [Google Scholar]
- Sylvester J.T., Karnati S.K., Yu Z., Morrison M., Firkins J.L. Development of an assay to quantify rumen ciliate protozoal biomass in cows using real-time PCR. J Nutr. 2004;134:3378–3384. doi: 10.1093/jn/134.12.3378. [DOI] [PubMed] [Google Scholar]
- Tao S., Duanmu Y., Dong H., Tian J., Ni Y., Zhao R. A high-concentrate diet induced colonic epithelial barrier disruption is associated with the activating of cell apoptosis in lactating goats. BMC Vet Res. 2014;10:235. doi: 10.1186/s12917-014-0235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soest P.J., Robertson J.B., Lewis B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci. 1991;74:3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- Vyas D., Beauchemin K.A., Koenig K.M. Using organic acids to control subacute ruminal acidosis and fermentation in feedlot cattle fed a high-grain diet. J Anim Sci. 2015;93:3950–3958. doi: 10.2527/jas.2015-9009. [DOI] [PubMed] [Google Scholar]
- Wang D.S., Zhang R.Y., Zhu W.Y., Mao S.Y. Effects of subacute ruminal acidosis challenges on fermentation and biogenic amines in the rumen of dairy cows. Livest Sci. 2013;155:262–272. [Google Scholar]
- Xue F., Pan X., Jiang L., Guo Y., Xiong B. GC-MS analysis of the ruminal metabolome response to thiamine supplementation during high grain feeding in dairy cows. Metabolomics. 2018;14:67. doi: 10.1007/s11306-018-1362-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zened A., Combes S., Cauquil L., Mariette J., Klopp C., Bouchez O., Troegeler-Meynadier A., Enjalbert F. Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol Ecol. 2013;83:504–514. doi: 10.1111/1574-6941.12011. [DOI] [PubMed] [Google Scholar]
- Zhang K., Meng M., Gao L., Tu Y., Bai Y. Rumen-derived lipopolysaccharide induced ruminal epithelium barrier damage in goats fed a high-concentrate diet. Microb Pathog. 2019;131:81–86. doi: 10.1016/j.micpath.2019.02.007. [DOI] [PubMed] [Google Scholar]
- Zhang R., Zhu W., Jiang L., Mao S. Comparative metabolome analysis of ruminal changes in Holstein dairy cows fed low- or high-concentrate diets. Metabolomics. 2017;13:74. [Google Scholar]
- Zhang R.Y., Liu Y.J., Yin Y.Y., Jin W., Mao S.Y., Liu J.H. Response of rumen microbiota, and metabolic profiles of rumen fluid, liver and serum of goats to high-grain diets. Animal. 2019;13:1855–1864. doi: 10.1017/S1751731118003671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence reads for all samples described above were deposited into the NCBI Sequence Read Archive (SRA) database (NO. PRJNA678648).