Abstract
Background:
The available evidence has increasingly demonstrated that a combination of genetic and epigenetic factors, such as DNA methylation, could be considered as causing leukemia. Epigenetic changes and methylation of the suppressor of the cytokine signaling 1 promoter (SOCS1) CpG region silence SOCS1 expression in cancer. In the current study, we evaluated the impact of epigallocatechin gallate (EGCG) and RG108 on SOCS1 promoter methylation and expression in U937 cells.
Methods:
In the current study, U937 leukemic cells were treated with EGCG and RG108 for 12, 24, 48, and 72 h and SOCS1 promoter methylation and its expression were measured by methylation-specific PCR (MSP) and quantitative real-time PCR, respectively.
Results:
The outcomes indicated that the SOCS1 promoter is methylated in U937 cells, and treatment of these cells with either EGCG or RG108 reduced its methylation. Moreover, we observed that SOCS1 expression was significantly upregulated in a time-dependent manner by both EGCG and RG108 in U937 cells compared with control cells. In the RG108-treated group at 12, 24, 48, and 72 h, SOCS1 expression was upregulated by 1, 4.2, 16.6, and 32.6 -fold respectively, and in the EGCG-treated group, by 0.5, 3.2, 10.8, and 22.3 -fold, respectively.
Conclusion:
Treatment with either EGCG or RG108 reduced SOCS1 promoter methylation and increased SOCS1 expression in U937 cells in a time-dependent manner, which may play a role in leukemia therapy.
Key Words: DNA Methylation, EGCG, Leukemia, RG108, SOCS1
Introduction
Leukemia is associated with genetic and epigenetic changes in genes responsible for the inhibition of tumors and oncogenes (1). Myeloid leukemia is the most prevalent form of leukemia, and most of its sufferers are adults. Moreover, the likelihood of developing leukemia increases with age (2, 3). Epigenetic changes alter gene expression without changing DNA sequences. DNA methylation is one of the most substantial epigenetic changes, and is frequently detected at the onset of leukemia (4-7). DNA methylation occurs exclusively at CpG nucleotides, and plays a significant role in the regulation of gene expression and in silencing repetitive elements in the genome (8).
Signaling occurs through the stimulation of cytokine receptors in both acquired and intrinsic immunity. The Janus kinases-signal transducer and activator of transcription proteins (JAK/STAT) signaling pathway is one of the most studied signaling pathways (9). Recent studies have elucidated that JAKs play fundamental roles in carcinogenesis in humans and other species (10). Suppressors of cytokine signaling proteins (SOCS) are considered as one of the most significant attenuators of cytokine signaling (11).
The SOCS family consists of eight of proteins; SOCS1-7 and CIS (12). SOCS1 inhibits JAKs significantly and acts as a tumor suppressor. SOCS1 also acts as a regulator that can influence cell proliferation, survival, differentiation, and cellular transformation signaling (13). Altered SOCS1 expression has been widely reported in human cancers and is associated with prolonged activity of the JAK/STAT signaling pathway (14). Various experiments have acknowledged the potential role of SOCS1 and proposed it as a potent cancer therapy target. Recent experiments have elucidated that CpG island methylation is responsible for SOCS1 silencing in various carcinomas including multiple myeloma, human hepatocellular carcinoma (HCC), acute myeloid lymphoma, and pancreatic ductal neoplasm (15-18).
The relationship between methylated SOCS1 and cancer was first observed in HCC (15). SOCS1 is abundantly upregulated in organs responsible for blood cell synthesis, and lack of SOCS1 expression not only causes changes in lymphoid cells, but also in myeloid cells (19). Previous studies have demonstrated that the SOCS1 gene is both hypermethylated and downregulated in acute myeloid leukemia (AML) patients (20).
Epigallocatechin gallate (EGCG) is the most effective polyphenol in green tea and its antitumor effects have been widely examined in various human and animal cancerous cell lines. It has been demonstrated that EGCG inhibits tumorigenesis in various organs including lung, bladder, skin, intestine, prostate, and breast (21-24). It is believed that EGCG impedes the hypermethylation of new DNA strands through DNA methyltransferase (DNMT) inhibition, which reverses the methylation process and leads to the re-expression of silenced genes (25). According to these studies, it appears that EGCG inhibits tumor suppressor gene methylation. Furthermore, analysis of gene expression in cell models affected by hypermethylation and treated with EGCG may facilitate cancer treatment. Considering the significance of this issue and previous experiments, in this study we evaluated the effects of EGCG and RG108 on the methylation and expression of SOCS1 in U937 cells.
Materials and Methods
Cell culture
The U937 monocyte cell line was purchased from the cell bank of the Pasteur Institute of Tehran, Tehran, Iran. The cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS), 100 IU/mL penicillin, and 100 IU/mL streptomycin. The incubator temperature was 37 °C with a humid atmosphere containing 5% CO2. After determining the concentrations of EGCG and RG108 needed for to produce LD50, RPMI 1640 media containing 48 µM RG108 or 25 µM EGCG were prepared. Cells grown in untreated cell culture medium for 12, 24, 48, and 72 h were used as the control. Then, 106 cells were cultured in the prepared culture media containing EGCG and RG108 for 12, 24, 48, and 72 h. The cells were harvested and RNA and DNA were extracted. This study was approved by Lorestan University of Medical Sciences with code 467.
Chemicals and kits
EGCG, RG108, and thiazolyl blue tetrazolium bromide (MTT) with cat. nos. 93894, R8279, and M2128, respectively, were purchased from Sigma-Aldrich; MERCK, Inc., St. Louis, MO, USA. Methylated DNA by M.SssI enzyme with cat. no. D5014 was purchased from Zymo Research, Germany. DNA extracted from healthy donor lymphocytes was utilized for positive and negative controls of methylation-specific PCR (MSP). To compare the samples, the methylation control kit of Zymo Research, Germany, which contains methylated DNA as a positive control and non-methylated DNA as negative control, was used for positive control and negative control.
MTT colorimetric assay
MTT is a colorimetric assay used to evaluate cell activity and viability. After seeding 2 × 104 cells to each well of an MTT-specific plate, cells were incubated with EGCG at 0.76, 1.53, 3.06, 6.125, 12.5, 25, 50, and 100 μM and RG108 at 1.95, 3.9, 7.81, 15.6, 31.25, 62.5, 125, and 250 μM for 72 h. Then, 100 µL of complete culture media containing 0.5 mg MTT powder was added to each well and incubated at 37 °C with 5% CO2 for 4 h. MTT is absorbed by viable and activated cells and produces formazan crystals through reduction by the mitochondrial succinate dehydrogenase enzyme. Then, 100 µL of dimethyl sulfoxide (DMSO) was added to each well and plates were incubated at 37 °C with 5% CO2 with shaking for 30 minutes. Absorbance was measured at 570 nm. DMSO (100 μL/well) was employed to dissolve the formazan. The following formula was applied to achieve sufficient concentrations of EGCG and RG108.
‰Cytotoxicity=1-mean absorbance of toxicant / mean absorbance of negative control ×100
RNA extraction and quantitative real-time PCR
Total RNA was isolated using a kit from Jena Bioscience, Germany, according to the manufacturer's instructions. After adding recombinant RNase-Free DNase I (Roche, Mannheim, Germany) to RNA samples, cDNA was obtained using a cDNA synthesis kit (Zistroyesh, Tehran, Iran) according to the manufacturer’s instructions. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an inner control. Real-time PCR was set up with 1 µL of cDNA and specific primers (Zistroyesh, Tehran, Iran). Thermal cycling conditions were as follows: initial denaturation at 95 °C for 10 min; 40 cycles of denaturation at 95 °C for 20 seconds, and annealing and elongation at 60 °C for 60 seconds (26, 27).
Methylation-specific PCR
Whole genomic DNA was isolated from the U937 mononuclear cells with a DNA extraction kit (CinnaGen Co, Tehran, Iran) according to its protocol. MSP was applied to evaluate the methylation levels of methylated promoters. First, DNA was subjected to sodium bisulfite with the CpGenome DNA Modification Kit (Intergen, New York, NY, USA) according to the kit’s instructions. Sodium bisulfite converts unmethylated cytosines to uracil but does not alter methylated cytosines. Primer sequences for methylated and unmethylated SOCS1 promoters are presented in Table 1.
Table 1.
Methylated and unmethylated SOCS1 promoter sequences.
| Gene | Forward primer (5-3) | Reverse primer | Annealing T (°C) | Reference |
|---|---|---|---|---|
| SOCS1 (U) | TTATGAGTATTTGTGTGTATTTTTAGGTTGGTT | CACTAACAACACAACTCCTACAACAACCA | 63 | (14) |
| SOCS1(M): | TTCGCGTGTATTTTTAGGTCGGTC | CGACACAACTCCTAC AACGACCG | 63 | (14) |
The MSP procedure was as follows: 95 °C for 12 min followed by 35-40 cycles of 95 °C for 35 sec, annealing at 72 °C for 30 sec, and final extension for 10 min at 72 °C. Then, 10 µL of the MSP product was electrophoresed on a 2% agarose gel. The gel was stained with DNA-safe stain and bands were analyzed on a GelDoc system under UV light.
Statistical analysis
The data obtained from real-time PCR were analyzed by the ΔΔCt method. Significant differences between groups were determined by one-way analysis of variance (ANOVA) and by the post-hoc Tukey test after verification of normal distribution of the data by the Shapiro–Wilk and equality of variances tests. The data are presented as means±SEMs. All experiments were performed in triplicate. Statistics were analyzed using SPSS (version 22.0, SPSS Inc., Chicago, IL, USA).
Results
LD50 concentrations of RG108 and EGCG
According to the MTT assays, the approximate LD50 concentrations of EGCG and RG108 are 25 and 48 µL, respectively (Fig. 1).
Fig. 1.
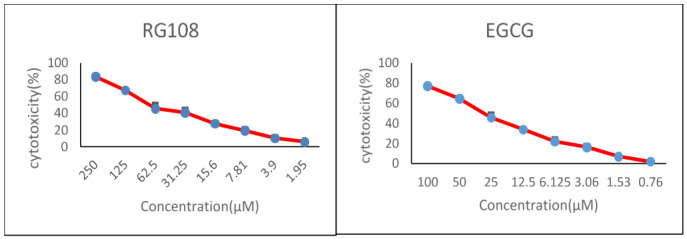
MTT results of U937 cell cytotoxicity in the presence of EGCG and RG108. A. The percentage of U937 cell cytotoxicity in the presence of 1.95-250 µM RG108 at 72 h. B. The percentage of U937 cell cytotoxicity in the presence of 0.76-100 µM EGCG at 72 h.
SOCS1 expression in the leukemic U937 cell line
SOCS1 expression in U937 cells increased over time in the presence of both RG108 and EGCG relative to the controls. SOCS1 expression was normalized to GAPDH expression. SOCS1 expression was 1, 4.2, 16.6, and 32.6 -fold greater at 12, 24, 48, and 72 h in the RG108 group, respectively, and 0.5, 3.2, 10.8, and 22.3 -fold greater at 12, 24, 48, and 72 h in the ECGC group, respectively, than that of the control cells at 72h (Fig. 2).
Fig. 2.
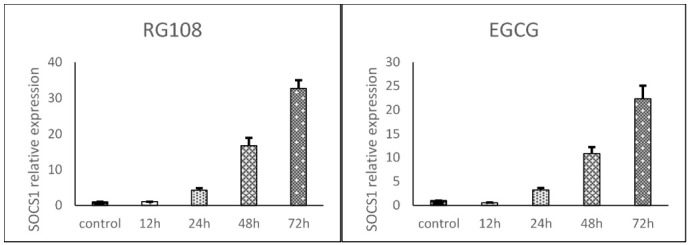
SOCS1 expression in RG108 and EGCG -treated U937 cells relative to controls at 12, 24, 48, and 72 h as determined by real-time PCR. SOCS1 expression was normalized to GAPDH. Statistical significance was determined by ANOVA and the post-hoc Tukey test. Data are reported as means±SEMs.
SOCS1 promoter methylation in U937 cells
MSP results indicate that the SOCS1 promoter is hypermethylated in U937 cells and that both RG108 and EGCG reduced this promoter methylation most efficiently at 72 h (Fig. 3).
Fig. 3.
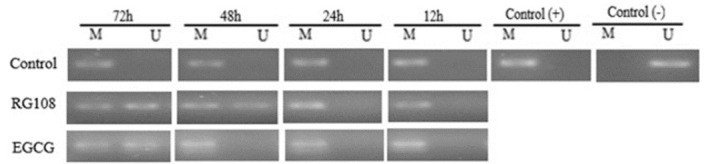
MSP of SOCS1 promoter methylation in U937 cells. Cells were treated with RG108 (48 µM) or EGCG (25 µM) for 12, 24, 48, and 72 h. Control cells received neither. Results were obtained by methylation-specific PCR and agarose gel electrophoresis. M: methylated, U: unmethylated. To compare the samples, the methylation control kit of Zymo Research, Germany, which contains methylated DNA as a positive control and non-methylated DNA as negative control, was used for positive control and negative control. Concentration(µM)
Discussion
Recent epigenetic advances have revealed that cancerous cells are affected by global epigenetic disorders in addition to various genetic changes (28). Previous studies have demonstrated that epigenetic changes and methylation of the SOCS1 promoter CPG region result in SOCS1 silencing in cancer (15-18). One effective treatment for AML is to reduce CPG methylation. DNMT inhibition reduces newly-synthesized DNA hypermethylation and leads to the re-expression of silenced genes (29). EGCG induces re-expression of the silenced genes in cancerous cells by attaching to the DNMT active site and suppressing DNMT activities (30, 31). Molecular modeling of the DNMT/EGCG interaction revealed that EGCG binds to the active site of DNMT, and RG108 attaches to almost the same site (32). Thus, the discovery and development of drugs that bind the DNMT active site should be considered in the design of epigenetic-modulating drugs.
Our results illustrate that the effects of EGCG and RG108 on SOCS1 expression are time dependent, that SOCS1 methylation was less, and expression significantly greater, in the EGCG and RG108 -treated U937 cells than in the controls. These results indicate that EGCG and RG108 probably upregulate SOCS1 expression by inhibiting promoter methylation. Previous studies illustrated that EGCG and RG108 inhibit gene promoter methylation by occupying the DNMT active site, leading to gene upregulation (32). Furthermore, it has been indicated that EGCG causes the re-expression of tumor suppressor genes, such as p161NK4a and Cip1/p21 (33), and also decreases methylation of the tumor suppressor gene, RECK (34).
Meng et al. demonstrated that EGCG leads to time-dependent demethylation of the P16 gene in ECa109 cells (35). Nandakumar et al. demonstrated that the general methylation of DNA is diminished dose-dependently by EGCG in A431 cells, resulting in re-expression of the tumor suppressor genes p161NK4a and Cip1/p21 (33).
Our results are consistent with the mentioned outcomes and reveal that SOCS1 expression is influenced by EGCG and RG108 through reducing methylation of the SOCS1 promoter. In contrast to the mentioned studies, Remely et al. reported that EGCG increased DNMT1 methylation in C57BL/6J cells in male mice (36). This contradictory result may be due to the different cell lines tested, cell sensitivity, or their unique genetic essences.
To our knowledge, this is the first study to investigate the effects of EGCG and RG108 on SOCS1 promoter methylation and expression in U937 cells. Because this study was conducted in vitro, the data may not be applicable to the in vivo situation; therefore, these results alone cannot confirm the effectiveness of these compounds against leukemia; further research using different methods than those employed here in living tissues is required.
Our results demonstrated that EGCG and RG108 reduced SOCS1 gene promoter methylation and upregulated SOCS1 expression in U937 cells in a time-dependent manner. We hypothesize that green tea consumption may be utilized for cancer therapy and prevention due to its polyphenolic substance, EGCG.
Acknowledgements
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Kim Y, Jekarl DW, Kim J, Kwon A, Choi H, Lee S, et al. Genetic and epigenetic alterations of bone marrow stromal cells in myelodysplastic syndrome and acute myeloid leukemia patients. . Stem cell research. . 2015;14(2):177, 84. doi: 10.1016/j.scr.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Zahedpanah M, Shaiegan M, Ghaffari SH, Nikbakht M, Nikugoftar M, Mohammadi S. Parthenolide induces apoptosis in committed progenitor AML cell line U937 via reduction in osteopontin. Rep Biochem Mol Biol. . 2016;4(2):82, 8. [PMC free article] [PubMed] [Google Scholar]
- 3.Gholami M, Bayat S, Manoochehrabadi S, Pashaiefar H, Omrani MD, Jalaeikhoo H, et al. Investigation of CEBPA and CEBPA-AS genes expression in acute myeloid leukemia. . Rep Biochem Mol Biol. . 2019;7(2):136, 141. [PMC free article] [PubMed] [Google Scholar]
- 4.Oakes CC, Seifert M, Assenov Y, Gu L, Przekopowitz M, Ruppert AS, et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat Genet. . 2016;48(3):253, 64. doi: 10.1038/ng.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. . Cancer Res. 2016;76(12):3446–50. doi: 10.1158/0008-5472.CAN-15-3278. [DOI] [PubMed] [Google Scholar]
- 6.Ghaznavi H, Kiani AA, Soltanpour MS. Association study between DNA methylation and genetic variation of APOE gene with the risk of coronary artery disease. . Mol Biol Res Commun. . 2018;7(4):173–179. doi: 10.22099/mbrc.2018.30955.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soltanpour MS, Soheili Z, Pourfathollah AA, Samiei S, Meshkani R, Kiani AA, et al. The A1298C mutation in methylenetetrahydrofolate reductase gene and its association with idiopathic venous thrombosis in an Iranian population. . Laboratory Medicine. 2011;42(4):213–6. [Google Scholar]
- 8.Schübeler D. Function and information content of DNA methylation. . Nature. 2015;517(7534):321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 9.Villarino AV, Kanno Y, Ferdinand JR, O’Shea JJ. Mechanisms of Jak/STAT signaling in immunity and disease. . J Immunol. 2015;194(1):21–7. doi: 10.4049/jimmunol.1401867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas S, Snowden J, Zeidler M, Danson S. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. . Br J Cancer. 2015;113(3):365–71. doi: 10.1038/bjc.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen L, Evel-Kabler K, Strube R, Chen S-Y. Silencing of SOCS1 enhances antigen presentation by dendritic cells and antigen-specific anti-tumor immunity. . Nat Biotechnol. . 2004;22(12):1546–53. doi: 10.1038/nbt1035. [DOI] [PubMed] [Google Scholar]
- 12.Jiang M, Zhang W-w, Liu P, Yu W, Liu T, Yu J. Dysregulation of SOCS-mediated negative feedback of cytokine signaling in carcinogenesis and its significance in cancer treatment. . Front Immunol. 2017;8:70. doi: 10.3389/fimmu.2017.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liau NP, Laktyushin A, Lucet IS, Murphy JM, Yao S, Whitlock E, et al. The molecular basis of JAK/STAT inhibition by SOCS1. . Nature communications. 2018;9(1):1558. doi: 10.1038/s41467-018-04013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beaurivage C, Champagne A, Tobelaim WS, Pomerleau V, Menendez A, Saucier C. SOCS1 in cancer: An oncogene and a tumor suppressor. Cytokine. . 2016;82:87–94. doi: 10.1016/j.cyto.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Yoshikawa H, Matsubara K, Qian G-S, Jackson P, Groopman JD, Manning JE, et al. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. . Nature genetics. . 2001;28(1):29–35. doi: 10.1038/ng0501-29. [DOI] [PubMed] [Google Scholar]
- 16.Nagai H, Naka T, Terada Y, Komazaki T, Yabe A, Jin E, et al. Hypermethylation associated with inactivation of the SOCS-1 gene, a JAK/STAT inhibitor, in human hepatoblastomas. . Journal of human genetics. 2003;48(2):65–69. doi: 10.1007/s100380300008. [DOI] [PubMed] [Google Scholar]
- 17.Galm O, Yoshikawa H, Esteller M, Osieka R, Herman JG. SOCS-1, a negative regulator of cytokine signaling, is frequently silenced by methylation in multiple myeloma. Blood. 2003;101(7):2784–8. doi: 10.1182/blood-2002-06-1735. [DOI] [PubMed] [Google Scholar]
- 18.Chen CY, Tsay W, Tang JL, Shen HL, Lin SW, Huang SY, et al. SOCS1 methylation in patients with newly diagnosed acute myeloid leukemia. . Genes Chromosomes Cancer. . 2003;37(3):300–5. doi: 10.1002/gcc.10222. [DOI] [PubMed] [Google Scholar]
- 19.Liu TC, Lin SF, Chang JG, Yang MY, Hung SY, Chang CS, editors. Epigenetic alteration of the SOCS1 gene in chronic myeloid leukaemia. . Br J Haematol. 2003;123(4):654–61. doi: 10.1046/j.1365-2141.2003.04660.x. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe D, Ezoe S, Fujimoto M, Kimura A, Saito Y, Nagai H, et al. Suppressor of cytokine signalling-1 gene silencing in acute myeloid leukaemia and human haematopoietic cell lines. Br J Haematol. 2004;126(5):726–35. doi: 10.1111/j.1365-2141.2004.05107.x. [DOI] [PubMed] [Google Scholar]
- 21.Mimoto J, Kiura K, Matsuo K, Yoshino T, Takata I, Ueoka H, et al. (–)-Epigallocatechin gallate can prevent cisplatin-induced lung tumorigenesis in A/J mice. . Carcinogenesis. . 2000;21(5):915–9. doi: 10.1093/carcin/21.5.915. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Ye ZQ, Koo M. Growth inhibition and cell cycle arrest effects of epigallocatechin gallate in the NBT-II bladder tumour cell line. . BJU Int. . 2004;93(7):1082–6. doi: 10.1111/j.1464-410X.2004.04785.x. [DOI] [PubMed] [Google Scholar]
- 23.Mantena SK, Roy AM, Katiyar SK. Retracted: Epigallocatechin-3-Gallate Inhibits Photocarcinogenesis Through Inhibition of Angiogenic Factors and Activation of CD8+ T Cells in Tumors. . Photochem Photobiol. . 2005;81(5):1174–9. doi: 10.1562/2005-04-11-RA-487. [DOI] [PubMed] [Google Scholar]
- 24.Stuart EC, Scandlyn MJ, Rosengren RJ. Role of epigallocatechin gallate (EGCG) in the treatment of breast and prostate cancer. . Life Sci. . 2006;79(25):2329–36. doi: 10.1016/j.lfs.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Wang X, Han L, Zhou Y, Sun S. Green tea polyphenol EGCG reverse cisplatin resistance of A549/DDP cell line through candidate genes demethylation. . Biomed Pharmacother. 2015;69:285–90. doi: 10.1016/j.biopha.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 26.Tarhriz V, Wagner KD, Masoumi Z, Molavi O, Hejazi MS, Ghanbarian H. CDK9 regulates apoptosis of myoblast cells by modulation of microRNA-1 expression. . J Cell Biochem. . 2018;119(1):547–554. doi: 10.1002/jcb.26213. [DOI] [PubMed] [Google Scholar]
- 27.Kohram F, Fallah P, Shamsara M, Bolandi Z, Rassoulzadegan M, Soleimani M, et al. Cell type-dependent functions of microRNA-92a. . Journal of cellular biochemistry. . 2018;119(7):5798–804. doi: 10.1002/jcb.26765. [DOI] [PubMed] [Google Scholar]
- 28.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. . N Engl J Med. 2010;363(25):2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Tollefsbol TO. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr Med Chem. . 2010;17(20):2141–51. doi: 10.2174/092986710791299966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong J, Xu C, Reece EA, Yang P. The green tea polyphenol EGCG alleviates maternal diabetes–induced neural tube defects by inhibiting DNA hypermethylation. Am J Obstet Gynecol. . 2016;215(3):368.e1–368.e10. doi: 10.1016/j.ajog.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo J, Medina-Franco JL. Homology modeling, docking and structure-based pharmacophore of inhibitors of DNA methyltransferase. . J Comput Aided Mol Des. . 2011;25(6):555–67. doi: 10.1007/s10822-011-9441-1. [DOI] [PubMed] [Google Scholar]
- 33.Nandakumar V, Vaid M, Katiyar SK. (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p 16 INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. . Carcinogenesis. . 2011;32(4):537–44. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato K, Long N, Makita H, Toida M, Yamashita T, Hatakeyama D, et al. Effects of green tea polyphenol on methylation status of RECK gene and cancer cell invasion in oral squamous cell carcinoma cells. . Br J Cancer. . 2008;99(4):647–654. doi: 10.1038/sj.bjc.6604521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng J, Tong Q, Liu X, Yu Z, Zhang J, Gao B. Epigallocatechin-3-gallate inhibits growth and induces apoptosis in esophageal cancer cells through the demethylation and reactivation of the p16 gene. . Oncol Lett. . 2017;14(1):1152–1156. doi: 10.3892/ol.2017.6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Remely M, Ferk F, Sterneder S, Setayesh T, Roth S, Kepcija T, et al. EGCG prevents high fat diet-induced changes in gut microbiota, decreases of DNA strand breaks, and changes in expression and DNA methylation of Dnmt1 and MLH1 in C57BL/6J male mice. . Oxid Med Cell Longev. . 2017;2017:3079148. doi: 10.1155/2017/3079148. [DOI] [PMC free article] [PubMed] [Google Scholar]


