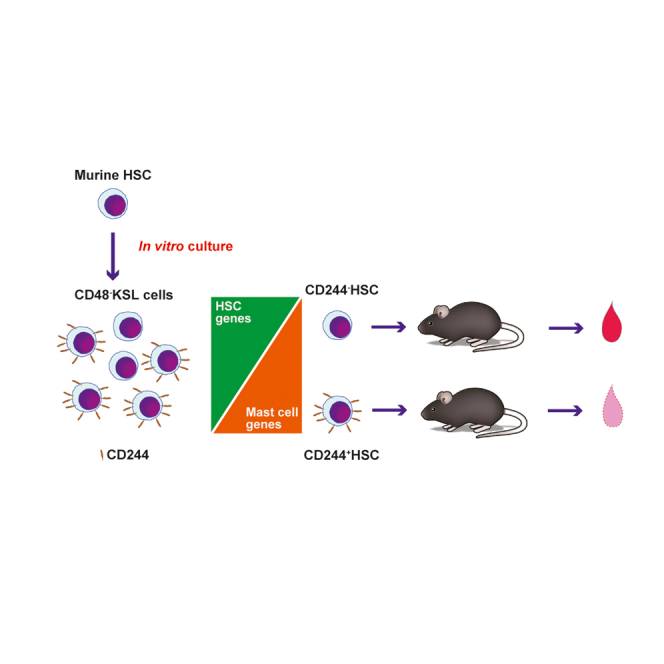
Summary
Isolation of long-term hematopoietic stem cell (HSC) is possible by utilizing flow cytometry with multiple cell surface markers. However, those cell surface phenotypes do not represent functional HSCs after in vitro culture. Here we show that cultured HSCs express mast cell-related genes including Cd244. After in vitro culture, phenotypic HSCs were divided into CD244- and CD244+ subpopulations, and only CD244- cells that have low mast cell gene expression and maintain HSC-related genes sustain reconstitution potential. The result was same when HSCs were cultured in an efficient expansion medium containing polyvinyl alcohol. Chemically induced endoplasmic reticulum (ER) stress signal increased the CD244+ subpopulation, whereas ER stress suppression using a molecular chaperone, TUDCA, decreased CD244+ population, which was correlated to improved reconstitution output. These data suggest CD244 is a potent marker to exclude non-functional HSCs after in vitro culture thereby useful to elucidate mechanism of functional decline of HSCs during ex vivo treatment.
Subject areas: Biological sciences, Immunology, Cell biology, Stem cells research
Graphical Abstract
Highlights
-
•
Murine HSCs up-regulate mast cell-related genes including Cd244 during in vitro culture
-
•
Long-term HSCs after in vitro culture are enriched in CD244−CD48−KSL population
-
•
Induction of unfolded protein response is involved in the increase of CD244+HSC
Biological sciences; Immunology; Cell biology; Stem cells research
Introduction
Hematopoietic stem cells (HSCs) replenish the entire blood system when needed, and transplantation of HSCs remains as one of the most effective treatments for patients with genetic diseases and hematopoietic malignancies (Seita and Weissman, 2010). Identification of multiple cell surface markers, e.g., c-Kit, Sca-I, CD34, SLAM family members, EPCR, CD45RA, CD49f, and CD90 (Balazs et al., 2006; Kiel et al., 2005; Notta et al., 2011; Oguro et al., 2013; Osawa et al., 1996), and utilization of unique cellular properties, e.g., dye-efflux and high aldehyde dehydrogenase 1-A1 activity (Goodell et al., 1996; Storms et al., 1999), have enabled prospective isolation of HSCs from bone marrow (BM) and umbilical cord blood. To this day, long-term reconstitution into recipient mice remains as the only reliable test of HSC functionality. In steady state (dormant) conditions, HSCs can be successfully enriched; however, surface marker phenotypes change and become distorted upon multitude of hematopoietic stress, e.g., inflammation and ex vivo culture (Zhang and Lodish, 2005). Functionality of HSCs is especially difficult to predict after in vitro cell culture, solely based on phenotype, even though we rely heavily on it to study normal and malignant HSC biology. One of the few markers that have been proposed to predict reconstitution potential after in vitro culture is CD48, as functional HSCs are enriched in the CD48- fraction within c-kit+Sca-I+Lineage− (KSL) cell population after culture (Noda et al., 2008). Yet, CD48 negativity cannot always represent HSC functionality. For instance, we and others have previously demonstrated that HSCs in culture are vulnerable to endoplasmic reticulum (ER) stress responses fueled by accumulation of unfolded/misfolded proteins (Miharada et al., 2014; van Galen et al., 2014; Walter and Ron, 2011). In our previous work we could ameliorate ER stress pathways using bile acids (BAs) and significantly enhance reconstitution potential of HSCs after in vitro culture (Miharada et al., 2014; Sigurdsson et al., 2016). However, a standard HSC staining including CD48 was not able to point out noticeable differences between samples cultured with or without BA, indicating increased functionality (Miharada et al., 2014). The discovery of additional cell surface markers that correlate with reconstitution capacity after in vitro culture would be highly beneficial and reduce dependency on the transplantation assay in HSC biology.
In this study our aim was to discover key signatures and cell surface markers representing the functional decline of HSCs during ex vivo culture. We performed gene expression profiling to compare fresh and cultured HSCs and discovered up-regulation of genes that are highly expressed in mast cells. In addition, we identified Cd244 as one of the top up-regulated genes. After in vitro culture, HSC population represented by CD48−KSL was subdivided into CD244+ and CD244- populations. CD244−CD48−KSL expressed high levels of HSC-related genes, whereas CD244+CD48−KSL expressed mast cell-related genes, and only CD244−CD48−KSL cells exhibited long-term reconstitution potential. ER stress induction is correlated to CD244 expression, as treatment with an ER stress inducer, tunicamycin, reduced the ratio of CD244−CD48−KSL cells, whereas addition of the BA, TUDCA, increased the ratio of CD244- HSCs. Furthermore, this was also the case when HSCs were cultured in the efficient expansion medium containing polyvinyl alcohol (PVA) (Wilkinson et al., 2019).
Our data show that CD244 is a potent marker to represent functionally impaired HSCs enabling prospective evaluation of HSC after extensive in vitro culture.
Results
CD48 is unable to distinguish functional HSCs after in vitro culture
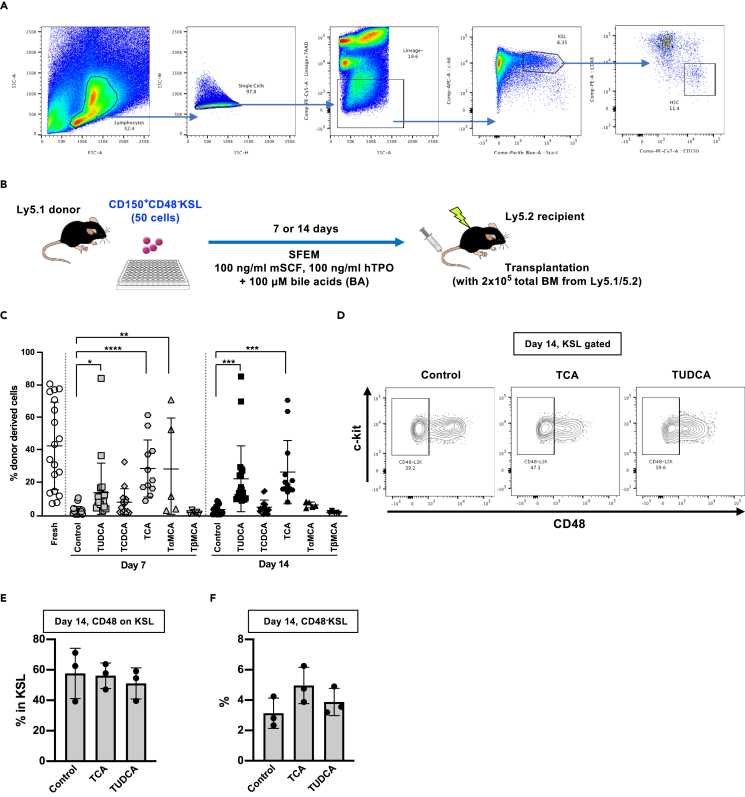
We have previously reported that two specific types of BAs, TUDCA and taurocholic acid (TCA), can maintain functionality of HSCs in vitro (Miharada et al., 2014; Sigurdsson et al., 2016); however, no clear difference was observed in the frequency of CD48−KSL cells after in vitro culture of fetal liver HSCs (Sigurdsson et al., 2016). To reproduce the result using adult HSCs from murine BM, CD150+CD48−KSL cells were cultured with different BA, analyzed, and transplanted into lethally irradiated mice (Figures 1A and 1B). Peripheral blood (PB) analyses showed that long-term engraftment of HSCs cultured with TUDCA and TCA was significantly higher than under other conditions (Figure 1C). Flow cytometry analyses of cultured HSCs using KSL and CD48 staining did not show significant differences between TUDCA/TCA and control condition, despite showing improvement in transplantation chimerism (Figures 1D–1F). These results suggest that CD48 negativity is not sufficient to detect HSC functionality after in vitro culture.
Figure 1.
Transplantation of HSCs cultured with various bile acids
(A) The gating strategy for isolating CD150+CD48−KSL cells from murine bone marrow.
(B) Experimental design of the competitive reconstitution assay. Fifty (50) CD150+CD48−KSL cells were isolated from BM of Ly5.1 mice (donor, two mice per experiment were used) and cultured in Stemspan SFEM medium supplemented with 100 ng/mL mSCF and 100 ng/mL hTPO with or without 100 μM of different types of bile acids (BA) for 7 or 14 days. Cells were then transplanted into lethally irradiated Ly5.2 mice (recipient, 5–18 mice) along with 2 × 105 total BM cells derived from Ly5.1/5.2 (F1) mice (competitor). Donor contribution (chimerism) was monitored by analyzing peripheral blood (PB) every month.
(C) Peripheral blood analysis of the transplanted mice. Chimerism in PB at 16 weeks of transplanted mice are shown. Mean ± SD from three independent experiments (n = 5–18) are displayed. Significance was calculated using one-way ANOVA within the same culture days. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(D–F) Cell surface expression of CD48 on KSL fraction of HSCs cultured with or without BA. Representative FACS plots (C) and summary of the flow cytometry analyses (D and E) are shown. Mean ± SD (n = 3) are displayed.
Induction of mast cell-related genes in HSCs after in vitro culture
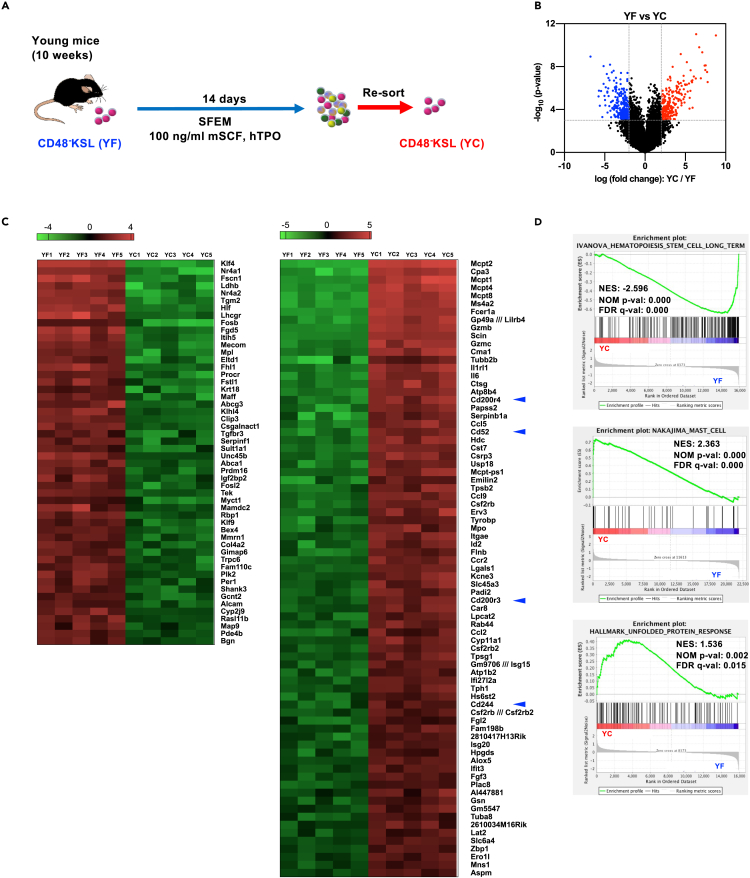
To discover transcriptional changes occurring during the in vitro-cultured HSCs and to identify additional markers that represent functionality/impairment of HSCs after in vitro culture, we performed a gene expression profiling of freshly isolated HSCs compared with in vitro-cultured HSCs. CD48−KSL cells were freshly isolated from 10-week-old mice (YF) and then cultured in serum-free medium supplemented with 100 ng/mL of murine stem cell factor (mSCF) and human thrombopoietin (hTPO) for 14 days, and then CD48−KSL cells were re-isolated (YC) (Figure 2A). Comparison of transcriptional profiles of these samples revealed significant changes of gene expression patterns (Figure 2B). Genes down-regulated upon in vitro culture mainly included genes that are known for their functions in HSC regulations or as markers of HSCs, e.g., Klf4, Procr, Fgd5, Hlf, Nr4a1, and Nr4a2 (Balazs et al., 2006; Calvanese et al., 2019; Freire and Conneely, 2018; Gazit et al., 2014; Komorowska et al., 2017; Park et al., 2019). This might suggest that the CD48−KSL population contains only a small fraction of functional HSCs and/or cells with profoundly reduced HSC function after in vitro culture. In contrast, up-regulated genes included genes that are normally highly expressed in mast cells, such as Mcpt1/2/4/8, Cpa3, Cma1, Ccl2, and Gzmb (Dwyer et al., 2016) (Figure 2C). Using gene set enrichment analysis (GSEA) (Subramanian et al., 2005) we could identify a variety of gene signatures that changed between samples (Table S1) including loss of a long-term HSC signature and induction of the mast cell signature (Figure 2D). In addition, signature of unfolded protein response (UPR) induction was found in the cultured HSCs (Figure 2D), in line with our previous study (Miharada et al., 2014).
Figure 2.
Gene expression changes in CD48−KSL cells upon in vitro culture and aging
(A) Experimental design of the gene expression analysis. Five hundred CD48−KSL cells were sorted from BM of young (10 weeks old, YF) mice. Cells were also cultured in Stemspan SFEM medium supplemented with 100 ng/mL mSCF and 100 ng/mL hTPO for 14 days, and CD48−KSL cells were re-sorted.
(B) Volcano plot showing differentially expressed genes between YF and YC. Significant difference was defined as p < 0.001 and log2 fold change < −2 (blue) or >2 (red).
(C) Heatmaps showing a list of differentially expressed genes. Significant difference was defined as p < 0.0001 and log2 fold change < −3 or >3. Blue arrowheads indicate CD markers.
(D) Gene set enrichment analysis (GSEA) of the microarray data comparing YF and YC. NES, normalized enrichment score; NOM p-val, nominal p value; FDR q-val, false discovery rate q-value. See also Table S1.
Mast cells are immune cells playing major roles in pathogen surveillance as well as promoting host defense through cellular communications with other immune cell types (Abraham and St John, 2010). Mast cells are characterized by existence of histamine-rich granules that are released upon various stimulations, particularly immunoglobulin E (IgE) binding to their cell surface FC receptor, which is a key reaction during allergic responses (Galli and Tsai, 2012). Expression of multiple mast cell-related genes including mature mast cell genes such as Fcer1a, which codes FcεRIα, were observed in the YC cells. However, flow cytometry analysis was not able to detect FcεRI protein on the cell surface and morphological analyses of re-isolated YC cells did not observe the typical granule-rich morphology (data not shown), suggesting that YC cells were not fully differentiated toward mast cells. Mast cells are known to share crucial genes with basophils (Dwyer et al., 2016), but GSEA did not observe enrichment of a basophil signature in both cultured CD48−KSL cells and CD244+CD48−KSL cells (data not shown). SCF is a strong inducer of mast cell differentiation, whereas TPO signaling is considered to inhibit mast cell differentiation (Martelli et al., 2008). Both SCF and TPO are used as standard cytokines in mouse and human HSC culture. However, our gene expression analysis showed that expression levels of Mpl, TPO receptor, were significantly down-regulated (Figure 2C), presumably failing to suppress induction of mast cell genes during the in vitro culture.
Aging is also known as a trigger of functional decline of HSC (Sudo et al., 2000). Therefore, we also analyzed if aging stress also induces mast cell signature. Gene expression profiles of HSCs isolated from young mice (YF) and from aged (18 months) mice (AF) were compared (Figure S1A). GSEA revealed that the mast cell signature was rather enriched in YF (Figure S1B). This finding indicates that induction of UPR genes and the mast cell signature might be specific for ex vivo culture conditions.
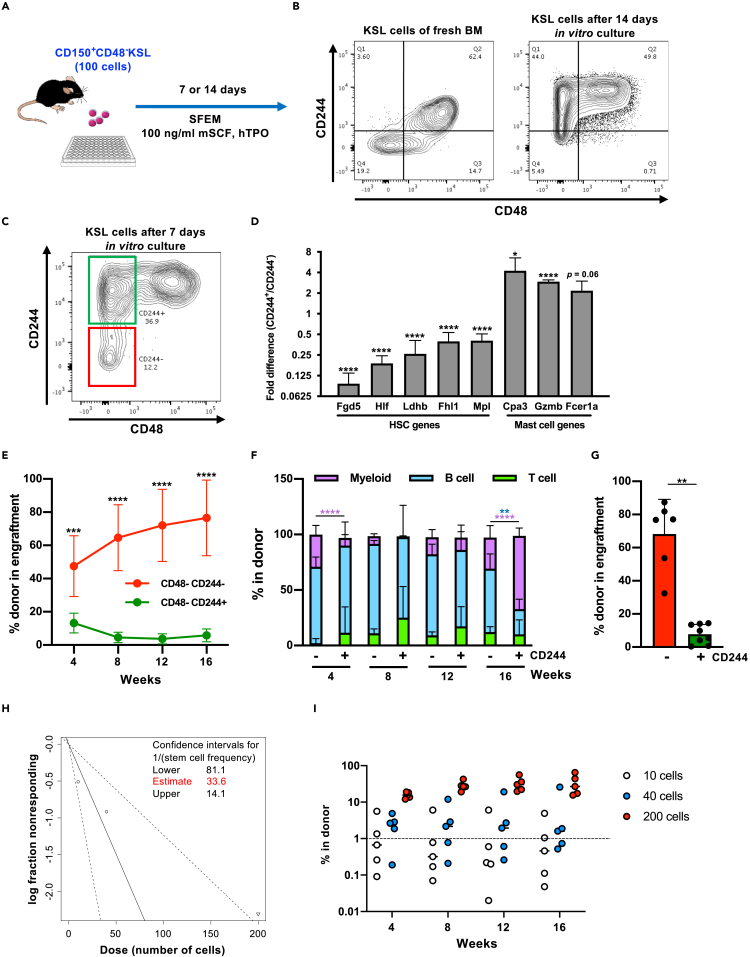
CD244 can functionally subdivide the CD48−KSL population after in vitro culture
We identified that the cell surface marker Cd244 was one of the significantly up-regulated genes in the in vitro-cultured CD48−KSL cells (Figure 2C). Cd244 is a member of the SLAM family of genes and is expressed in various immune cells including NK cells, γδT cells, CD8+ αβT cells, monocytes, basophils, eosinophils, dendritic cells, and mast cells (McArdel et al., 2016). The CD244 molecule plays roles in immune responses partially through its binding to CD48 (Agresta et al., 2018; Elishmereni et al., 2014; Waggoner and Kumar, 2012). CD244 has been reported as a negative marker for HSC isolation but is currently considered to be redundant with other SLAM markers when purifying fresh HSCs from steady-state mice (Kiel et al., 2005; Oguro et al., 2013). Freshly isolated CD150+CD48−LSK cells were mostly negative for CD244, whereas after in vitro culture for 14 days CD48−KSL cells were subdivided into CD244+ and CD244- populations and a vast majority of CD48+ cells were CD244+ (Figures 3A and 3B). The expression pattern of two frequently used HSC markers, CD34 and CD150 (Kiel et al., 2005; Osawa et al., 1996), was abnormal compared with freshly isolated cells (Figure S2A). We also checked the expression of EPCR (Procr, CD201) on the cultured cells, since EPCR is known as a potent marker of human HSCs both before and after in vitro culture and to enrich for functional murine HSC after 5-FU treatment (Fares et al., 2017; Umemoto et al., 2018). Flow cytometry analyses of cultured HSCs showed that a vast majority of CD244−KSL cells were positive for EPCR while EPCR+KSL cells contained both CD244+ and CD244- fractions (Figure S2A). To compare the proliferative status of CD244−CD48−KSL cells (CD244−HSCs) and CD244−CD48−KSL cells (CD244+HSCs), cell cycle profiles of these subpopulations were analyzed using Ki-67 staining after 7 days’ culture. CD244−HSCs contained the highest proportion of Ki-67-positive cells, indicating least cycling of this population (Figures S2B and S2C). In addition, we measured protein synthesis rates in different subpopulations using L-homopropargylglycine (L-HPG) incorporation into newly synthesized proteins. The results indicate that CD244−CD48−KSL cells have the lowest protein synthesis levels among the different subgroups. However, between CD244−HSCs and CD244+HSCs there was not a significant difference (Figures S2D and S2E). To further characterize these subpopulations, CD244−HSCs and CD244- CD244+HSCs were isolated from 7-day cultured HSC samples (Figure 3C) and expression levels of representative genes in HSCs or mast cell regulation were analyzed using quantitative RT-PCR. Significantly higher expression levels of HSC-related genes (Fgd5, Hlf, Fhl1 and Mpl) were observed in CD244−HSCs, whereas higher expression of mast cell-related genes (Cpa3, Gzmb, and Mcpt8) was detected in CD244+HSCs (Figure 3D). However, morphologies of these two types of HSCs are similar (Figure S2F).
Figure 3.
CD244 expression divides CD48−KSL cells into functionally distinct subpopulations after in vitro culture
(A) Experimental design of the in vitro culture experiment. One hundred CD48−KSL cells were sorted from BM of young mice and cultured in Stemspan SFEM medium supplemented with 100 ng/mL mSCF and 100 ng/mL hTPO for 7 days.
(B) Expression patterns of CD244 and CD48 on the cell surface of fresh and 14 days cultured KSL cells. Representative FACS plots on KSL population are shown.
(C and D) qRT-PCR analysis for HSC-related genes and mast cell-related genes in CD244+CD48−KSL cells compared with the CD244+CD48−KSL counterpart. Representative FACS plot and gating of CD244+CD48−KSL cells (green) and CD244−CD48−KSL cells (red) on 7 days cultured KSL cells are shown in (C). Relative expression levels of genes in CD244+CD48−KSL cells to CD244−CD48−KSL are shown in (D).
(E) Competitive reconstitution assay. After 7 days’ culture, two subpopulations were sorted and 1,000 of CD244- or 1,500 of CD244+CD48−KSL cells were separately transplanted into lethally irradiated recipient mice (seven mice) with 2 × 105 total BM cells (competitor). Chimerism was monitored by analyzing PB every month. Significance was calculated using Student's t test at each time point. Mean ± SD from two independent experiments (n = 7) are displayed. ∗∗∗∗p < 0.0001.
(F) Lineage balance of donor-derived cells in the PB of recipient mice. Mean ± SD from two independent experiments (n = 6) are displayed. ∗∗p < 0.01, ∗∗∗∗p < 0.0001. Each color represents different lineages.
(G) Analysis of BM from engrafted mice after 16 weeks. Chimerism in each cell fraction is shown. Significance was calculated using Student's t test. Mean ± SD from two independent experiments (n = 6) are displayed. ∗∗p < 0.01.
(H and I) Limiting dilution assay for CD244−CD48−KSL cells. CD244−CD48−KSL cells were re-sorted after 7 days’ in vitro culture and 200, 40, or 10 cells were transplanted to recipient mice in a competitive manner. Chimerism above 1% was judged as successful engraftment. The frequency of functional HSC was calculated using ELDA (http://bioinf.wehi.edu.au/software/elda/). Chimerism of individual mice is shown in (I).
To compare long-term reconstitution potential of these two populations, CD244- and CD244+ HSCs were separately isolated from 7-day cultured HSC samples and transplanted into lethally irradiated mice. Monthly PB analysis showed that CD244−HSCs showed long-term reconstitution of the hematopoietic system of transplanted mice while engraftment levels of CD244+HSCs was low (Figure 3E). Both CD244- and CD244+HSCs reconstituted Myeloid, B, and T cells with a modest difference; however, the PB profile of CD244+HSC-engrafted mice was not accurate because of their low engraftment levels (Figure 3F). Analyses of BM of the transplanted mice revealed that the reconstitution level of CD244−HSCs was significantly higher than that of CD244+HSCs (Figure 3G). In order to estimate the frequency of functional HSC within the CD244−CD48−KSL fraction, limiting dilution assay was performed. After 7 days of culture, CD244−CD48−KSL cells were re-sorted and 10, 40, and 200 cells were transplanted into lethally irradiated recipient mice in a competitive manner. From this experiment we estimate the HSC frequency in the CD244−CD48−KSL fraction to be 1/33.6 cells (Figures 3H and 3I).
In summary, our experiments indicate that the CD244−CD48−KSL fraction in culture is distinct from CD244+ cells and highly enriched for functional HSCs.
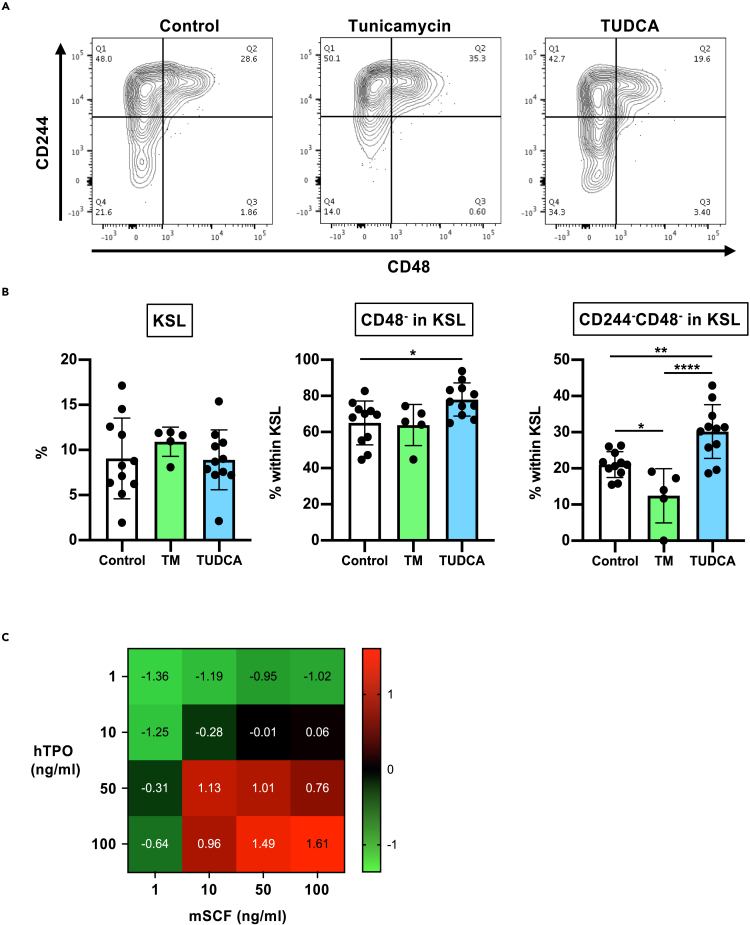
ER stress induction and cytokine signals affect CD244 expression
We and others have previously demonstrated that induction of UPR and ER stress signals impair the potential of mouse and human HSCs (Miharada et al., 2014; van Galen et al., 2014) during in vitro culture. These results were also confirmed with our gene expression analysis in this study (Figure 2D). We therefore asked if ER stress induction would lead to the elevation of CD244 expression. Addition of a chemical ER stress inducer, tunicamycin, increased the ratio of CD244+HSCs within CD48−KSL cells (Figures 4A and 4B). Conversely, addition of TUDCA, a BA known to suppress ER stress (Miharada et al., 2014; Özcan et al., 2006; Sigurdsson et al., 2020), resulted in increased frequency of CD244−HSCs (Figures 4A and 4B). Of note, the frequency of KSL cells did not significantly differ in both cases and CD48 negativity was marginally affected while the frequency of CD244−CD48- population in KSL was significantly changed (Figure 4B).
Figure 4.
ER stress induction and cytokine signals affect CD244 expression
(A) Expression patterns of CD244 and CD48 on KSL cells after in vitro culture in Stemspan SFEM medium supplemented with 100 ng/mL mSCF and 100 ng/mL hTPO for 7 days, and with or without 0.5 μg/mL Tunicamycin or 100 μM TUDCA. Representative FACS plots are shown.
(B) Summary of the flow cytometry analyses. Frequencies of KSL cells, CD48- fraction in KSL population, and CD244−CD48- fraction in KSL population are shown. Mean ± SD from two independent experiments (n = 5–11) are displayed. Significance was calculated using one-way ANOVA within each population. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
(C) A heatmap showing frequencies of CD244−CD48−KSL cells after 7 days’ culture with various SCF/TPO concentrations. Mean values of Z score (n = 3) in each fraction are displayed.
Since TPO signaling is known to inhibit mast cell differentiation (Martelli et al., 2008), we next analyzed the influence of cytokine concentration during the in vitro culture on the frequency of the CD244−CD48−KSL population. High TPO and low SCF concentrations have recently been reported as the optimal culture condition for HSCs (Kobayashi et al., 2019; Wilkinson et al., 2019). In contrast, our results showed that a combination of high concentration (100 ng/mL) of both SCF and TPO maintained the highest CD244−CD48−KSL population in culture. Although lower concentrations of SCF and TPO both resulted in lower frequency of the CD244−CD48−KSL fraction, there was a tendency that a lower concentration of TPO reduced the CD244−CD48−KSL fraction more than the same SCF concentration (Figures 4C and S3).
These findings indicate that in vitro culture-induced ER stress and cytokine signals could be one of the critical causes of functional impairment of HSCs in cell culture that can be measured by CD244 elevation.
Gene expression profiling of CD244- and CD244+ HSCs
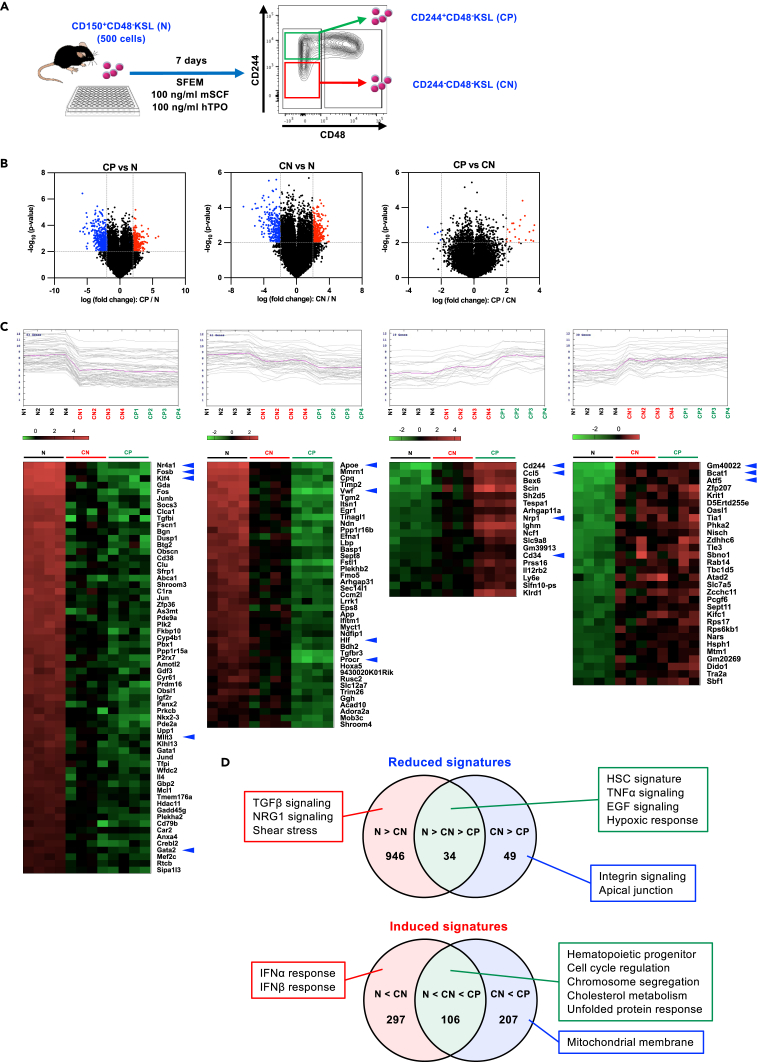
To explore the key molecular changes that impair HSC functionality, we compared gene expression profiles between fresh HSCs and CD244+/CD244−HSCs after 7 days’ culture in the conventional culture condition. CD244−CD48−KSL cells were freshly isolated from young (8 weeks old) mice (N) and cultured for 7 days, and then CD244−CD48−KSL cells (CN) or CD244+CD48−KSL cells (CP) were re-isolated (Figure 5A). Gene expression analysis revealed that there were more differentially expressed genes between N and CN cells than between CN and CP cells, although critical functional difference was seen between CN and CP (Figure 5B). Among significantly differentially expressed genes between three types of cells, multiple expression patterns were detected; in both up- and down-regulated genes, some of the genes showed significant changes between N and CN that were at the same degree in CP cells, which were presumably more directly affected by in vitro culture. This group contained Nr4a1, Fosb, Klf4, Mllt3, Gata2 (down-regulated) and Gm40022, Bcat1, Atf5 (up-regulated) (Figure 5C). Other than these, a part of genes exhibited gradual changes in expression levels, meaning that the expression levels of those genes are more linearly correlated to the reconstitution potential of the cells. This group contained Apoe, Vwf, Hlf, Procr (down-regulated) and Cd244, Ccl5, Nrp1, Cd34 (up-regulated) (Figure 5C). Similarly, GSEA on both N versus CN and CN versus CP observed significant enrichment of gene signatures in various fashions. For instance, tumor necrosis factor alpha (TNFα) signaling, epidermal growth factor (EGF) signaling, and hypoxic response signatures, which have been implicated in HSC function (Doan et al., 2013; Miharada et al., 2011; Suda et al., 2011; Yamashita and Passegué, 2019), were gradually lost in accordance with loss of reconstitution capacity (N > CN > CP), whereas cell cycle-related hallmarks, chromosome segregation, and UPR signatures were gradually induced (N < CN < CP) (Figure 5D and Table S2). In contrast, signature of transforming growth factor β (TGFβ) signaling (Yamazaki et al., 2009) was uniquely enriched in N (N > CN = CP) and interferon alpha (IFNα) signaling pathway was induced in CN (N < CN = CP), suggesting that these pathways might more directly reflect environmental changes upon in vitro culture (Figure 5D and Table S2). Thus, including CD244 as a marker enables one to dissect cellular and molecular changes during the transformation of HSCs upon in vitro culture.
Figure 5.
Gene expression profiling of CD244- and CD244+ KSL cells
(A) Experimental design of the gene expression analysis. Five hundred CD244-48−KSL cells were sorted from BM of 10-week-old mice (N) and were also cultured in Stemspan SFEM medium supplemented with 100 ng/mL mSCF and 100 ng/mL hTPO for 7 days, and CD244−KSL (CN) and CD244+KSL (CP) cells were re-sorted.
(B) Volcano plot showing differentially expressed genes between CP versus N (left), CN versus N (center), and CP versus CN (right). Significant difference was defined as p < 0.01 and log2 fold change < −2 (blue) or >2 (red).
(C) K-mean clustering of genes showing different expression patterns between the three cohorts and heatmaps of selected genes. Significant difference was defined as p < 0.001 and log2 fold change < −2 or >2.
(D) Overlap of hallmarks and GO terms between the three groups based on GSEA. See also Table S2.
CD244 enriches functional HSCs after in vitro culture with PVA
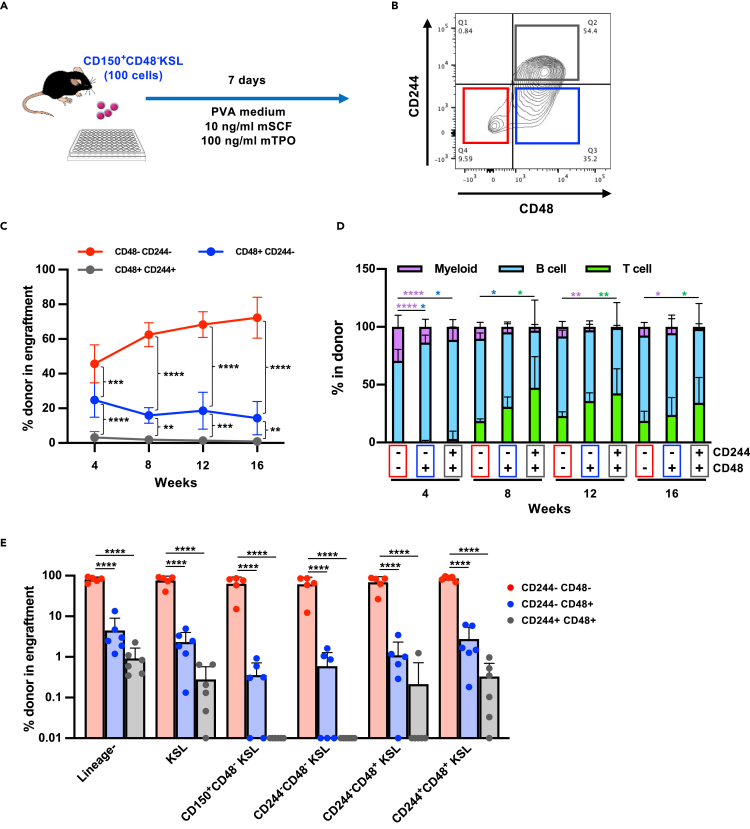
High-efficiency in vitro expansion of phenotypic HSCs has been described using a culture medium containing polyvinyl alcohol (PVA) and a low concentration of SCF (Wilkinson et al., 2019). We used the CD244 marker to ask if there is functional heterogeneity in expanded HSCs after culture in PVA medium. Purified CD150+CD48−KSL cells were cultured for 7 days in PVA medium supplemented with 10 ng/mL of mSCF and 100 ng/mL of mTPO according to the original report (Figure 6A). Unlike the conventional culture condition, CD48+KSL population contained CD244+ and CD244- fractions, whereas all CD244+KSL cells were CD48+ (Figure 6B). A small fraction of CD244−CD48−KSL cells (<10%) was identified in the PVA culture, and these cells were also positive for EPCR (Figure S4). Alternative analysis of the PVA data showed that CD244−CD48+KSL cells contained more EPCR− cells, whereas EPCR+CD48−KSL cells clearly contained CD244+ cells (Figure S4). Based on our flow cytometry analysis we decided to transplant CD244−CD48−KSL, CD244−CD48+KSL, and CD244+CD48+KSL cells from the PVA culture. The transplantation experiment revealed that the highest reconstitution in PB was found within the CD244−CD48−KSL cells and significantly lower but stable long-term reconstitution was seen in CD244−CD48+KSL cells, whereas CD244+CD48+KSL failed to engraft (Figure 6C). The balance of three lineages in the PB of the long-term (16 weeks) reconstituted mice was similar between CD244−CD48−KSL and CD244−CD48+KSL cells (Figure 6D). However, analyses of BM of the transplanted mice revealed that contribution of donor cells in the HSC population was mostly observed in the CD244−CD48−HSCs-engrafted mice (Figure 6E).
Figure 6.
CD244 expression distinguishes functionally distinct subpopulations after in vitro culture in PVA medium
(A) Experimental design of the in vitro culture experiment. One hundred CD48−KSL cells were sorted from BM of young mice and cultured in PVA containing medium (Wilkinson et al., 2019) supplemented with 10 ng/mL mSCF and 100 ng/mL hTPO for 7 days.
(B) Expression patterns of CD244 and CD48 on the cell surface of KSL cells after in vitro culture in PVA medium. A representative FACS plot on KSL population is shown.
(C) Competitive reconstitution assay. After 7 days’ culture, three subpopulations were sorted and 500 of CD244−CD48−KSL cells, CD244−CD48+KSL cells, or CD244+CD48+KSL cells were separately transplanted into lethally irradiated recipient mice with 2 × 105 total BM cells. Chimerism was monitored by analyzing PB every month. Significance was calculated using one-way ANOVA at each time point. Mean ± SD from two independent experiments (n = 12) are displayed. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001
(D) Lineage balance of donor-derived cells in the PB of recipient mice. Significance was calculated using one-way ANOVA at each time point. Mean ± SD from two independent experiments (n = 12) are displayed. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. Each color represents different lineages.
(E) Analysis of BM from engrafted mice after 16 weeks. Chimerism in each cell fraction is shown. Significance was calculated using one-way ANOVA within each population. Mean ± SD from two independent experiments (n = 5) are displayed. ∗∗∗∗p < 0.0001.
These findings reveal that cells equipped with long-term reconstitution potential are limited, even when using PVA expansion medium, and that CD244 negativity is a potent marker to efficiently enrich functional HSCs after ex vivo expansion.
Discussion
The long-term reconstitution assay has been recognized as an exclusive and conclusive method to prove the functional potential of HSCs. However, transplantation assays require a large number of recipient animals, long time periods of monitoring, and high costs. Therefore, developing methods that allow for the prospective evaluation of reconstitution potential of HSCs is of great benefit. The biggest challenge is to identify reliable markers that reflect reconstitution potential of HSCs under non-steady-state conditions, e.g., in vitro culture. This study has revealed that phenotypic murine HSCs after in vitro culture have quite distinct gene expression patterns, particularly the reduction of HSC genes and the induction of mast cell-related genes (Figures 2C and 2D). Although decreased expression levels of HSC-related genes may be a major part of the functional retardation, unexpected elevation of some genes is also considered a critical reason of failed reconstitution. For instance, granzyme B (Gzmb) has been reported to have negative impact on the long-term reconstitution potential of HSCs (Carnevalli et al., 2014).
Among them, we particularly identified CD244 as a robust marker that represents the decline of reconstitution potential of murine HSCs after in vitro culture. CD244 expression is low on freshly isolated HSCs (Kiel et al., 2005; Oguro et al., 2013); however, in vitro culture clearly induced CD244 expression (Figures 3B and 3C). Of note, CD244+CD48−KSL (CD244+HSCs) did not efficiently engraft to transplanted recipient mice, whereas CD244- cells still sustained reconstitution potential (Figure 3E). The potential of CD244 negativity in combination with CD48 was further supported by experiments using a new HSC expansion system with PVA (Wilkinson et al., 2019). After culturing with the PVA system, long-term reconstitution ability was mainly found in the CD244−CD48−KSL population (Figures 6C and 6E) that comprised only 10% of the total culture. Including CD244 as a marker for in vitro cultured HSC is thus highly beneficial and allows for a higher-resolution comparison of gene expression profiles between freshly isolated HSCs and in vitro cultured HSCs.
Although functional HSCs are highly enriched in the CD244−CD48−KSL fraction after in vitro culture, the frequency is still lower than of fresh BM cells. This might be explained by a lack of yet-unidentified additional marker(s) to capture all functional HSC. From the gene expression analysis data (Figures 2C and 5C) we have tested some additional cell surface molecules (e.g., CD53 and Nrp1) but so far not found additional markers to further subdivide CD244−HSCs. Alternatively, such markers might not be cell surface molecules but cellular characteristics reflecting metabolic conditions.
In summary, we conclude that CD244 is an important marker to distinguish functional versus non-functional HSCs after culture. Including CD244 as a marker to evaluate in vitro-cultured HSCs may help to prospectively estimate reconstitution potential and therefore contribute to further improvement of in vitro HSC expansion systems.
Limitations of the study
This study particularly targeted murine HSC, and therefore the ability of CD244 negativity as a marker for HSC of other species, e.g., human, is unknown. In addition, whether CD244 expression is implicated in different stress conditions is unclear at this moment.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-mouse B220 (RA3-6B2), APC | BioLegend | Cat# 103212; RRID: AB_312997 |
| Rat anti-mouse B220 (RA3-6B2), PE/Cy5 | BioLegend | Cat# 103210; RRID: AB_312995 |
| Armenian Hamster anti-mouse CD3ε (145-2C11), APC | BioLegend | Cat# 100312; RRID: AB_ 312677 |
| Armenian Hamster anti-mouse CD3ε (145-2C11), Biotin | BioLegend | Cat# 100304; RRID: AB_ 312669 |
| Armenian Hamster anti-mouse CD3ε (145-2C11), PE/Cy5 | BioLegend | Cat# 100310; RRID: AB_312675 |
| Rat anti-mouse CD4 (L3T4), APC/Cy7 | BioLegend | Cat# 100526; RRID: AB_312727 |
| Rat anti-mouse CD4 (GK1.5), Biotin | BioLegend | Cat# 100404; RRID: AB_312689 |
| Rat anti-mouse CD4 (H129.19), FITC | BioLegend | Cat# 130308; RRID: AB_1279237 |
| Rat anti-mouse CD8a (53-6.7), APC/Cy7 | BioLegend | Cat# 100714; RRID: AB_ 312747 |
| Rat anti-mouse CD8a (53-6.7), PE | BioLegend | Cat# 100708; RRID: AB_ 312747 |
| Rat anti-mouse CD11b (M1/70), Biotin | BioLegend | Cat# 101204; RRID: AB_312787 |
| Rat anti-mouse CD11b (M1/70), PE | BioLegend | Cat# 101208; RRID: AB_312791 |
| Rat anti-mouse CD11b (M1/70), PE/Cy5 | BioLegend | Cat# 101210; RRID: AB_312793 |
| Mouse anti-mouse CD45.1 (A20), Brilliant Violet 510™ | BioLegend | Cat# 110741; RRID: AB_2563378 |
| Rat anti-mouse CD45.1 (A20), PE/Cy7 | BioLegend | Cat# 110730; RRID: AB_1134168 |
| Rat anti-mouse CD45.1 (A20), PerCP | BioLegend | Cat# 110726; RRID: AB_893345 |
| Mouse anti-mouse CD45.2 (104), APC | BioLegend | Cat# 109814; RRID: AB_389211 |
| Mouse anti-mouse CD45.2 (104), FITC | BioLegend | Cat# 109806; RRID: AB_313443 |
| Mouse anti-mouse CD45.2 (104), Brilliant Violet 421™ | BioLegend | Cat# 109832; RRID: AB_2565511 |
| Mouse anti-mouse CD45.2 (104), Brilliant Violet 785™ | BioLegend | Cat# 109839; RRID: AB_2562604 |
| Mouse anti-mouse CD45.2 (104), PE | BioLegend | Cat# 109808; RRID: AB_313445 |
| Rat anti-mouse CD48 (HM48-1), APC | BioLegend | Cat# 103412; RRID: AB_571997 |
| Rat anti-mouse CD48 (HM48-1), FITC | BioLegend | Cat# 103404; RRID: AB_313019 |
| Rat anti-mouse CD150 (TC15-12F12.2), PE/Cy7 | BioLegend | Cat# 115914; RRID: AB_439797 |
| Rat anti-mouse CD244.2 (m2B4), PE | BioLegend | Cat# 133508; RRID: AB_2072855 |
| Rat anti-mouse c-kit (2B8), APC | BioLegend | Cat# 105812; RRID: AB_313221 |
| Rat anti-mouse c-kit (2B8), APC/Cy7 | BioLegend | Cat# 105826; RRID: AB_1626278 |
| Armenian Hamster anti-mouse FcεRIα (MAR-1), FITC | BioLegend | Cat# 134306; RRID: AB_1626108 |
| Rat anti-mouse Ly-6G/Ly-6C (Gr-1) (RB6-8C5), PE | BioLegend | Cat# 108408; RRID: AB_313373 |
| Rat anti-mouse Ly-6G/Ly-6C (Gr-1) (RB6-8C5), PE/Cy5 | BioLegend | Cat# 108410; RRID: AB_313375 |
| Rat anti-mouse Ly-6A/E (Sca-1) (D7), FITC | BioLegend | Cat# 108106; RRID: AB_313343 |
| Rat anti-mouse Ly-6A/E (Sca-1) (D7), Brilliant Violet 421™ | BioLegend | Cat# 108127; RRID: AB_2563064 |
| Rat anti-mouse Ter119 (TER119), Biotin | BioLegend | Cat# 116204; RRID: AB_313705 |
| Rat anti-mouse Ter119 (TER119), PE/Cy5 | BioLegend | Cat# 116210; RRID: AB_313711 |
| Streptavidin-APC | BioLegend | Cat# 405207 |
| Streptavidin-Brilliant Violet 605™ | BD | Cat# 563260 |
| Rat anti-mouse CD34 (RAM34), FITC | eBioscience | Cat# 11-0341-85; RRID: AB_465021 |
| Rat anti-mouse CD201 (eBio1560), APC | eBioscience | Cat# 17-2012-82; RRID: AB_10717805 |
| Rat anti-mouse c-kit (2B8), APC-eFluor 780 | Thermo Fisher Scientific | Cat# 47-1171-82; RRID: AB_1272177 |
| Rat anti-mouse CD8a (53-6.7), Biotin | TONBO | Cat# 20-0081-U100 |
| Chemicals, peptides, and recombinant proteins | ||
| 7-Amino-Actinomycin-D (7AAD) | Sigma-aldrich | Cat# A9400-1MG |
| CD117 MicroBeads, mouse | Miltenyi Biotec | Cat# 130-091-224 |
| Recombinant Murine stem cell factor (mSCF) | PEPROTECH | Cat# 250-03-10 μg |
| Recombinant Mouse thrombopoietin (mTPO) | PEPROTECH | Cat# AF-315-14-10 μg |
| Recombinant Human thrombopoietin (hTPO) | PEPROTECH | Cat# 300-18-10μg |
| 2-Mercaptoethanol | Sigma-aldrich | Cat# M6250-100ML |
| Ham’s F-12 Nutrient Mix | Gibco | Cat# 11765054 |
| StemSpan™ SFEM | Stem Cell Technologies | Cat# 09650 |
| HEPES | Thermo Fisher Scientific | Cat# 15630080 |
| Penicillin–Streptomycin–Glutamine | Thermo Fisher Scientific | Cat# 10378016 |
| Insulin–Transferrin–Selenium–Ethanolamine (ITS-X) | Thermo Fisher Scientific | Cat# 51500056 |
| Polyvinyl alcohol (87–90%-hydrolyzed) | Sigma-Aldrich | Cat# P8136-250G |
| Taurochenodeoxycholic acid | Sigma-Aldrich | Cat# T6260 |
| Taurocholic acid | Sigma-Aldrich | Cat# T9034 |
| Tauro-α-muricholic acid | Toronto Research Chemicals | Cat# T009130 |
| Tauro-β-muricholic acid | Santa Cruz | Cat# sc-361829 |
| Tauroursodeoxycholic acid | Sigma-Aldrich | Cat# T0266 |
| Critical commercial assays | ||
| RNeasy Micro Kit | QIAGEN | Cat# 74004 |
| Ki-67 staining Kit | BD | Cat# 556026 |
| Click-IT™ L-Homopropargylglycine (HPG) | Thermo Fisher Scientific | Cat# C10186 |
| Click-iT™ Plus Alexa Fluor™ 488 Picolyl Azide Toolkit | Thermo Fisher Scientific | Cat# C10641 |
| BD Cytofix/Cytoperm™ Fixation/Permeabilization Solution Kit | Thermo Fisher Scientific | Cat# BDB554714 |
| Deposited data | ||
| Microarray analysis | NCBI Gene Expression Omnibus (GEO) database | Accession number: GSE162408 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 (CD45.2) | Taconic | N/A |
| Mouse: B6.SJL (CD45.1) | In-house breeding | N/A |
| Oligonucleotides | ||
| Quantitative real-time PCR probe for Cpa3 | Thermo Fisher Scientific | Mm00483940_m1 |
| Quantitative real-time PCR probe for Fcer1a | Thermo Fisher Scientific | Mm00438867_m1 |
| Quantitative real-time PCR probe for Fgd5 | Thermo Fisher Scientific | Mm01189735_g1 |
| Quantitative real-time PCR probe for Fhl1 | Thermo Fisher Scientific | Mm04204611_g1 |
| Quantitative real-time PCR probe for Gzmb | Thermo Fisher Scientific | Mm00442834_m1 |
| Quantitative real-time PCR probe for Hlf | Thermo Fisher Scientific | Mm00723157_m1 |
| Quantitative real-time PCR probe for Hprt | Thermo Fisher Scientific | Mm03024075_m1 |
| Quantitative real-time PCR probe for Ldhb | Thermo Fisher Scientific | Mm01267402_m1 |
| Quantitative real-time PCR probe for Mpl | Thermo Fisher Scientific | Mm00440310_m1 |
| Software and algorithms | ||
| Gene Expression Commons | RIKEN | https://gexc.riken.jp/ |
| FlowJo | Tree Star | N/A |
| ELDA | WEHI | https://bioinf.wehi.edu.au/software/elda/ |
| Prism | Graphpad | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kenichi Miharada (kenmiharada@kumamoto-u.ac.jp).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Mice
B6.SJL (Ly-5.1) male mice (8-10 weeks old) were obtained from The Jackson Laboratory. Young (10 weeks old) and aged (18 months old) C57BL/6J (Ly-5.2) male mice were obtained from Janvier Labs. All animals were maintained in individually ventilated racks and given autoclaved food and water ad libitum. All experiments were approved by the Lund University Animal Ethical Committee, Swedish Board of Agriculture and Animal Research Facility of The University of Tokyo guidelines.
Method details
Flow cytometry
Adult BM cells were isolated by crushing tibias, femurs and iliac bones of 8 to 10 weeks old mice with a mortar and pestle in PBS. c-Kit positive (c-Kit+) cells were enriched using magnetic separation system (MACS) with anti-c-Kit magnetic beads (Miltenyi Biotec). The enriched cells were stained with a combination of antibodies (please see STAR Methods for detail) All antibodies were used at a ceoncentration of 1:250. Anti-CD3, -B220, -CD11b, -Gr-I and Ter119 antibodies were used as lineage antibody mix. Dead cells were excluded using 7-Amino-Actinomycin-D (7AAD) staining. Cells were sorted on FACS Aria III or analyzed on FACS LSRII or LSR Fortessa (BD). Collected data were analyzed on the FlowJo software (Tree Star).
In vitro culture of HSCs
In conventional setting, CD150+CD48-KSL cells were sorted from 10 weeks old mice and cultured in StemSpan™ SFEM medium (STEMCELL Technologies) supplemented with 100 ng/mL of mouse stem cell factor (mSCF) and 100 ng/mL of human thrombopoietin (hTPO) for 7 or 14 days. Alternatively, cells were cultured in PVA containing medium (Wilkinson et al., 2019) for 7 days. Briefly, 10 mM HEPES, 1 × Penicillin–streptomycin–glutamine, 1 × Insulin–transferrin–selenium–ethanolamine (all Thermo Fischer) and 1 mg/mL polyvinyl alcohol (87–90%-hydrolyzed, Sigma-Aldrich) were added to F12 medium, and conditioned medium was supplemented with 10 ng/mL of mSCF and 100 ng/mL of mouse thrombopoietin (mTPO).
HSC culture with chemical compounds
Different types of primary bile acids (tauroursodeoxycholic acid: TUDCA, taurochenoceoxycholic acid: TCDCA, taurocholic acid: TCA, tauro-α-muricholic acid: TαMCA, tauro-β-muricholic acid: TβMCA) were dissolved in water or ethanol, and separately added to the cell culture with the concentration of 100 μM. Tunicamycin was dissolved in water and added to the culture with the concentration of 0.5 μg/mL.
Cell cycle analysis
Freshly isolated total BM cells or in vitro cultured cells were stained with HSC markers including CD244, and then fixed and permeabilized using BD Cytofix/Cytoperm Fixation and Permeabilization Kit (BD). Fixed cells were then stained with anti-Ki67 antibody and DAPI. Cell cycle status was determined based on the Ki-67 expression and DNA replication on FACS Fortessa.
Protein synthesis rate analysis
Protein synthesis rate was measured using L-homopropargylglycine (L-HPG) incorporation into newly synthesized proteins. HSCs were culture in standard conditions as described above. 50 μM L-HPG in fresh medium was added to cells on day 7 and incubated for 30 min. After surface staining for HSC markers, cells were fixed and permeabilized using BD Cytofix/cytoperm kit. L-HPG was detected with an Alexa Fluor 488 azide using Click-iT plus. MFI measurements were done in different populations of cultured cells using FACS Aria III.
Microarray analysis
To compare gene expression profiles of freshly isolated HSCs and cultured HSCs, CD48−KSL cells were sorted from BM of 10 weeks old or 18 months old C57BL/6 SJL mice. A part of young HSCs were then cultured in the conventional condition, and 14-days later CD48−KSL fraction was re-sorted. Total RNA was isolated from the sorted cells using RNeasy® Micro Kit (Qiagen) according to the manufacturer's protocol. After the quality/quantity determination of the extracted RNA, cDNA was synthesized and amplified using Ovation® Pico WTA System V2 (NuGEN).
In order to compare gene expression profiles of CD244- and CD244+HSCs, CD244−CD48−KSL cells were sorted from BM of 10 weeks old mice, and a part of cells were cultured in the conventional condition, and 7 days later CD244−CD48−KSL and CD244+CD48−KSL fractions were re-sorted. Total RNA was isolated as mentioned above. After the quality/quantity determination, extracted RNA was amplified and converted to cDNA using GeneChip® 3′ IVT Pico Reagent Kit (Affymetrix).
Fragmented and labeled double-strand cDNA were hybridized to Affymetrix Mouse Genome 430 PM Array Plates using an Affymetrix GeneTitan® system controlled by the Affymetrix GeneChip® Command Console® software v4.2 or v4.3.3. The fluorescent signals were measured with an Affymetrix GeneTitan® system controlled by the Affymetrix GeneChip® Command Console® software v4.2 or v4.3.3. Gene level summarized probe set signals in log2 scale were calculated from Affymetrix CEL files by using the RMA algorithm as implemented in the Affymetrix GeneChip® Expression Console® v1.4 Software. Sample processing was performed at a Genomics Core Facility, “KFB - Center of Excellence for Fluorescent Bioanalytics” (Regensburg, Germany; www.kfb-regensburg.de). Differential genes were called using the Limma R package using a p value cut off of 0.01 or 0.05. Gene clustering was performed using k-means/medians clustering on MeV v4.8. Overlap of differentially expressed genes between multiple cohorts were defined using an online tool Venny v2.1 (https://bioinfogp.cnb.csic.es/tools/venny/). The microarray data are available at the GEO database under the accession number GSE162408.
Long-term competitive repopulation assay
Competitive repopulation assay was performed using the CD45 congenic mouse system. Ten to fifteen hundred (1,000-1,500) freshly isolated various HSC populations (donor) were mixed with 2 × 105 total BM cells (competitor), and then transplanted into mice irradiated with 900 cGy (recipient). Every 4 weeks after transplantation, PB from tail vein of recipient mice was collected and stained with anti-CD45.1, -CD45.2, -CD3, -Gr-1, -CD11b, and -B220 antibodies after red blood cell lysis using NH4Cl. After long-term (16 weeks) monitoring, BM of the engrafted primary recipient mice was analyzed. Donor contribution (chimerism) was determined as a formula of % donor/(% donor + % competitor) × 100.
Limiting dilution assay
Different number (200, 40, 10) of CD244−CD48−KSL cells re-isolated from 7 days cultured HSC were transplanted with 2 × 105 total BM cells as competitor cells into lethally irradiated recipient mice. Every 4 weeks after transplantation, PB was analyzed. Chimerism exceeding 1% in PB at 16-weeks was judged as a successful engraftment.
Quantitative RT-PCR
CD244- or CD244+ CD48−KSL cells were directly sorted into lysis buffer, and total RNA was isolated using RNeasy Micro Kit. CT values were averaged, and relative expression compared to HPRT was calculated using 2−ΔCT formula. For the information about used probes, please see STAR methods.
Quantification and statistical analysis
Statistical significance was determined using the Bon-ferroni method for comparison of multiple groups or the two-tailed Student's t-test for comparison of two groups. Details of used method are described in Figure Legends. All statistical analyses were performed on Prism (GraphPad).
The frequency of HSCs in the limiting dilution assay was calculated using ELDA (Hu and Smyth, 2009).
Acknowledgements
We thank Mattias Magnusson, Mark van der Garde, Terumasa Umemoto, and Satoshi Yamazaki for scientific discussions. This work was supported by JSPS Grant-in-Aid for Young Scientists (20K17370) (S.K.), Uehara Memorial Foundation (S.K.), the Swedish Child Cancer Foundation (V.S.), Åke-Wibergs foundation (V.S., K.M.), the Crafoord foundation (V.S., K.M.), the Swedish Cancer Society (V.S., V.R., K.M.), Olle Engkvist Foundation (K.S.), Grants-in-Aid for Scientific Research (JP19H05653) (A.I.), Scientific Research on Innovative Areas “Replication of Non-Genomic Codes” (JP19H05746) from MEXT, Japan (A.I.), The Swedish Research Council (K.M.), and Knut and Alice Wallenberg Foundation (K.M.). This study was also partly supported by a Grant from International Joint Usage/Research Center, the Institute of Medical Science, The University of Tokyo. K.M. was funded by StemTherapy program at Lund University. The Lund Stem Cell Center was supported by a Center of Excellence grant in life sciences from the Swedish Foundation for Strategic Research.
Author contributions
K.M. designed the project. S.K., V.S., A.I., and K.M. planned experiments. S.K., V.S., V.R., K.S., Z.Z., and K.M. performed experiments. S.K., S.L., S.S., and K.M. analyzed gene expression data. S.K., V.S., A.I., and K.M. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103603.
Supplemental information
Summary of GSEA comparing YF and YC
Summary of GSEA comparing N, CN, and CP cells
Data and code availability
-
•
Microarray data have been deposited at GEO and a re publicly available as of the date of publication. Accession number is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abraham S.N., St John A.L. Mast cell-orchestrated immunity to pathogens. Nat. Rev. Immunol. 2010;10:440–452. doi: 10.1038/nri2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agresta L., Hoebe K.H.N., Janssen E.M. The emerging role of CD244 signaling in immune cells of the tumor microenvironment. Front. Immunol. 2018;9:2809. doi: 10.3389/fimmu.2018.02809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazs A.B., Fabian A.J., Esmon C.T., Mulligan R.C. Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood. 2006;107:2317–2321. doi: 10.1182/blood-2005-06-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevalli L.S., Scognamiglio R., Cabezas-Wallscheid N., Rahmig S., Laurenti E., Masuda K., Jöckel L., Kuck A., Sujer S., Polykratis A., et al. Improved HSC reconstitution and protection from inflammatory stress and chemotherapy in mice lacking granzyme B. J. Exp. Med. 2014;211:769–779. doi: 10.1084/jem.20131072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvanese V., Nguyen A.T., Bolan T.J., Vavilina A., Su T., Lee L.K., Wang Y., Lay F.D., Magnusson M., Crooks G.M., et al. MLLT3 governs human haematopoietic stem-cell self-renewal and engraftment. Nature. 2019;576:281–286. doi: 10.1038/s41586-019-1790-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan P.L., Himburg H.A., Helms K., Russell J.L., Fixsen E., Quarmyne M., Harris J.R., Deoliviera D., Sullivan J.M., Chao N.J., et al. Epidermal growth factor regulates hematopoietic regeneration after radiation injury. Nat. Med. 2013;19:295–304. doi: 10.1038/nm.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer D.F., Barrett N.A., Austen K.F., Immunological Genome Project Consortium Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat. Immunol. 2016;17:878–887. doi: 10.1038/ni.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elishmereni M., Fyhrquist N., Gangwar R.S., Lehtimäki S., Alenius H., Levi-Schaffer F. Complex 2B4 regulation of mast cells and eosinophils in murine allergic inflammation. J. Invest. Dermatol. 2014;134:2928–2937. doi: 10.1038/jid.2014.280. [DOI] [PubMed] [Google Scholar]
- Fares I., Chagraoui J., Lehnertz B., MacRae T., Mayotte N., Tomellini E., Aubert L., Roux P.P., Sauvageau G. EPCR expression marks UM171-expanded CD34+ cord blood stem cells. Blood. 2017;129:3344–3351. doi: 10.1182/blood-2016-11-750729. [DOI] [PubMed] [Google Scholar]
- Freire P.R., Conneely O.M. NR4A1 and NR4A3 restrict HSC proliferation via reciprocal regulation of C/EBPα and inflammatory signaling. Blood. 2018;131:1081–1093. doi: 10.1182/blood-2017-07-795757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit R., Mandal P.K., Ebina W., Ben-Zvi A., Nombela-Arrieta C., Silberstein L.E., Rossi D.J. Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J. Exp. Med. 2014;211:1315–1331. doi: 10.1084/jem.20130428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli S.J., Tsai M. IgE and mast cells in allergic disease. Nat. Med. 2012;18:693–704. doi: 10.1038/nm.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell M.A., Brose K., Paradis G., Conner A.S., Mulligan R.C. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz O.H., Iwashita T., Yilmaz O.H., Terhorst C., Morrison S.J. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Morikawa T., Okinaga A., Hamano F., Hashidate-Yoshida T., Watanuki S., Hishikawa D., Shindou H., Arai F., Kabe Y., et al. Environmental optimization enables maintenance of quiescent hematopoietic stem cells ex vivo. Cell Rep. 2019;28:145–158. doi: 10.1016/j.celrep.2019.06.008. [DOI] [PubMed] [Google Scholar]
- Komorowska K., Doyle A., Wahlestedt M., Subramaniam A., Debnath S., Chen J., Soneji S., Van Handel B., Mikkola H.K.A., Miharada K., et al. Hepatic leukemia factor maintains quiescence of hematopoietic stem cells and protects the stem cell pool during regeneration. Cell Rep. 2017;21:3514–3523. doi: 10.1016/j.celrep.2017.11.084. [DOI] [PubMed] [Google Scholar]
- McArdel S.L., Terhorst C., Sharpea A.H. Roles of CD48 in regulating immunity and tolerance. Clin. Immunol. 2016;164:10–20. doi: 10.1016/j.clim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli F., Ghinassi B., Lorenzini R., Vannucchi A.M., Rana R.A., Nishikawa M., Partamian S., Migliaccio G., Migliaccio A.R. Thrombopoietin inhibits murine mast cell differentiation. Stem Cells. 2008;26:912–919. doi: 10.1634/stemcells.2007-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miharada K., Karlsson G., Rehn M., Rörby E., Siva K., Cammenga J., Karlsson S. Cripto regulates hematopoietic stem cells as a hypoxic-niche-related factor through cell surface receptor GRP78. Cell Stem Cell. 2011;9:330–344. doi: 10.1016/j.stem.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Miharada K., Sigurdsson V., Karlsson S. Dppa5 improves hematopoietic stem cell activity by reducing endoplasmic reticulum stress. Cell Rep. 2014;9:330–344. doi: 10.1016/j.celrep.2014.04.056. [DOI] [PubMed] [Google Scholar]
- Noda S., Horiguchi K., Ichikawa H., Miyoshi H. Repopulating activity of ex vivo-expanded murine hematopoietic stem cells resides in the CD48-c-Kit+Sca-1+lineage marker- cell population. Stem Cells. 2008;26:646–655. doi: 10.1634/stemcells.2007-0623. [DOI] [PubMed] [Google Scholar]
- Notta F., Doulatov S., Laurenti E., Poeppl A., Jurisica I., Dick J.E. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- Oguro H., Ding L., Morrison S.J. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M., Hanada K., Hamada H., Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Özcan U., Yilmaz E., Özcan L., Furuhashi M., Vaillancourt E., Smith R.O., Görgün C.Z., Hotamisligil G.S. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Lewis A., Chen T., Lacorazza D. Concise review: Regulation of self-renewal in normal and malignant hematopoietic stem cells by Krüppel-Like Factor 4. Stem Cells Transl. Med. 2019;8:568–574. doi: 10.1002/sctm.18-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seita J., Weissman I.L. Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson V., Haga Y., Takei H., Mansell E., Okamatsu-Haga C., Suzuki M., Radulovic V., van der Garde M., Koide S., Soboleva S., et al. Induction of blood-circulating bile acids supports recovery from myelosuppressive chemotherapy. Blood Adv. 2020;4:1833–1843. doi: 10.1182/bloodadvances.2019000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson V., Takei H., Soboleva S., Radulovic V., Galeev R., Siva K., Leeb-Lundberg L.M., Iida T., Nittono H., Miharada K. Bile acids protect expanding hematopoietic stem cells from unfolded protein stress in fetal liver. Cell Stem Cell. 2016;18:522–532. doi: 10.1016/j.stem.2016.01.002. [DOI] [PubMed] [Google Scholar]
- Storms R.W., Trujillo A.P., Springer J.B., Shah L., Colvin O.M., Ludeman S.M., Smith C. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc. Natl. Acad. Sci. U S A. 1999;96:9118–9123. doi: 10.1073/pnas.96.16.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T., Takubo K., Semenza G.L. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9:298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Sudo K., Ema H., Morita Y., Nakauchi H. Age-associated characteristics of murine hematopoietic stem cells. J. Exp. Med. 2000;192:1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto T., Hashimoto M., Matsumura T., Nakamura-Ishizu A., Suda T. Ca2+-mitochondria axis drives cell division in hematopoietic stem cells. J. Exp. Med. 2018;215:2097–2113. doi: 10.1084/jem.20180421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Galen P., Kreso A., Mbong N., Kent D.G., Fitzmaurice T., Chambers J.E., Xie S., Laurenti E., Hermans K., Eppert K., et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510:268–272. doi: 10.1038/nature13228. [DOI] [PubMed] [Google Scholar]
- Waggoner S.N., Kumar K. Evolving role of 2B4/CD244 in T and NK cell responses during virus infection. Front. Immunol. 2012;3:377. doi: 10.3389/fimmu.2012.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wilkinson A.C., Ishida R., Kikuchi M., Sudo K., Morita M., Crisostomo R.V., Yamamoto R., Loh K.M., Nakamura Y., Watanabe M., et al. Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature. 2019;571:117–121. doi: 10.1038/s41586-019-1244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M., Passegué E. TNF-α coordinates hematopoietic stem cell survival and myeloid regeneration. Cell Stem Cell. 2019;25:357–372. doi: 10.1016/j.stem.2019.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S., Iwama A., Takayanagi S., Eto K., Ema H., Nakauchi H. TGF-beta as a candidate bone marrow niche signal to induce hematopoietic stem cell hibernation. Blood. 2009;113:1250–1256. doi: 10.1182/blood-2008-04-146480. [DOI] [PubMed] [Google Scholar]
- Zhang C.C., Lodish H.F. Murine hematopoietic stem cells change their surface phenotype during ex vivo expansion. Blood. 2005;105:4314–4320. doi: 10.1182/blood-2004-11-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of GSEA comparing YF and YC
Summary of GSEA comparing N, CN, and CP cells
Data Availability Statement
-
•
Microarray data have been deposited at GEO and a re publicly available as of the date of publication. Accession number is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.