Abstract
Circular RNAs (circRNAs) have been proven to play key roles in the development and progression of various types of cancers. However, there were no reported studies on the roles of circRNA mediator complex subunit 27 (circMED27) in tumors including hepatocellular carcinoma (HCC). In this study, we found that circMED27 was significantly increased in HCC serum and that higher levels of circMED27 were correlated with bad clinical characteristics and poor prognoses of patients with HCC. Furthermore, upregulated circMED27 promoted HCC resistance to lenvatinib. Our mechanistic investigations revealed that circMED27 functions as a competing endogenous RNA (ceRNA) for miR-655-3p to upregulate ubiquitin-specific peptidase 28 (USP28) expression. Thus, we are led to conclude that circMED27 acts as a potential therapeutic target for HCC patients receiving lenvatinib therapy and may represent a promising molecular biomarker for forecasting lenvatinib-resistant HCC.
Keywords: circMED27, lenvatinib resistance, USP28, miR-655-3p, HCC
Graphical abstract
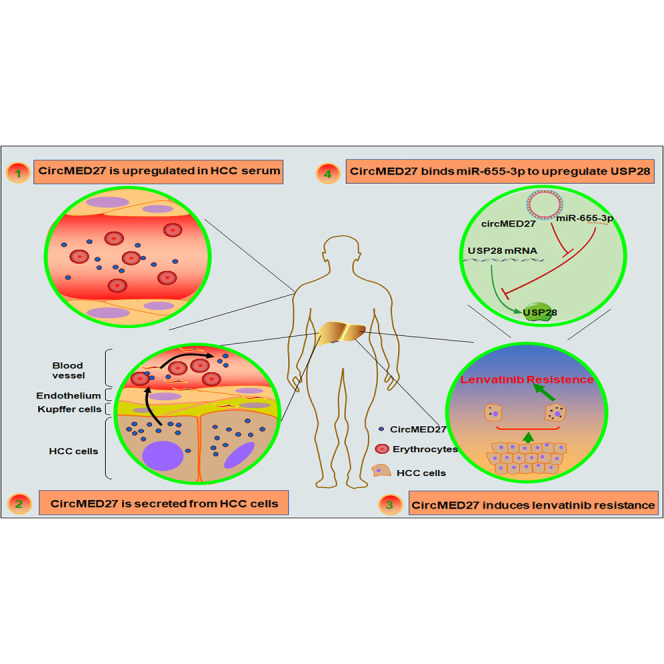
Zhang et al. provide a promising biomarker and therapeutic target for the recovery sensitivity of HCC to lenvatinib. circMED27 was upregulated in both tumor tissues and serum of HCC patients. In addition, circMED27 could bind to miR-655-3p and positively regulate USP28 expression, further inducing lenvatinib resistance in HCC.
Introduction
Hepatocellular carcinoma (HCC) is one of the most frequent malignancies worldwide; it is the third leading cause of cancer-related death.1 Lenvatinib, a multitargeted tyrosine kinase inhibitor (TKI), was approved for the treatment of unresectable HCC with significant inhibitory activity.2 Although lenvatinib has recently demonstrated a survival advantage in HCC, the therapeutic effects of lenvatinib are temporary, and the overall response rate in HCC is only 24%.3 Therefore, it is of critical importance to explore the molecular mechanisms underlying lenvatinib resistance in HCC, which might help develop novel therapeutic drugs for HCC treatment.
Circular RNAs (circRNAs), a class of non-coding RNAs, widely exist in mammalian cells and have been detected in the body fluids, tissues, and exosomes.4,5 Recently, accumulating studies have revealed that tens of thousands of circRNAs have been identified in mammalian genomes, the dysregulation of which has been extensively explored in tumors development and progression.6,7 However, the expression of biological functions of newly identified circRNAs in tumors still needs further investigation. The data of previously published studies have confirmed that “miRNA sponge” is the major role of circRNAs in many tumors. For example, circPSMC3 promotes gastric cancer progression by sponging miRNA-296-5p.8 Our previous study also reported that circFGFR3 promotes non-small cell lung cancer (NSCLC) progression via sponging miR-22-3p to upregulate the galectin-1 expression.9
circRNA mediator complex subunit 27 (circMED27; circBase: hsa_circ_0006825), derived from back-splicing of MED27 mRNA, is located on chromosome 9q34.13 and is 1,304 nucleotides in length. However, the function of circMED27 has not been explored. Here, we reported that circMED27 was significantly upregulated in HCC tissues and was correlated with the bad prognosis of HCC patients. More importantly, our results have shown that circMED27 exerts its oncogenic role in HCC resistance to lenvatinib via sponging miR-655-3p to upregulate ubiquitin-specific peptidase 28 (USP28) expression. Therefore, we concluded that circMED27 can act as a molecular biomarker for lenvatinib-sensitivity predication and as a potential treatment target for HCC patients.
Results
circMED27 is upregulated in HCC serum, and increased circMED27 expression positively correlates with a bad prognosis in HCC patients
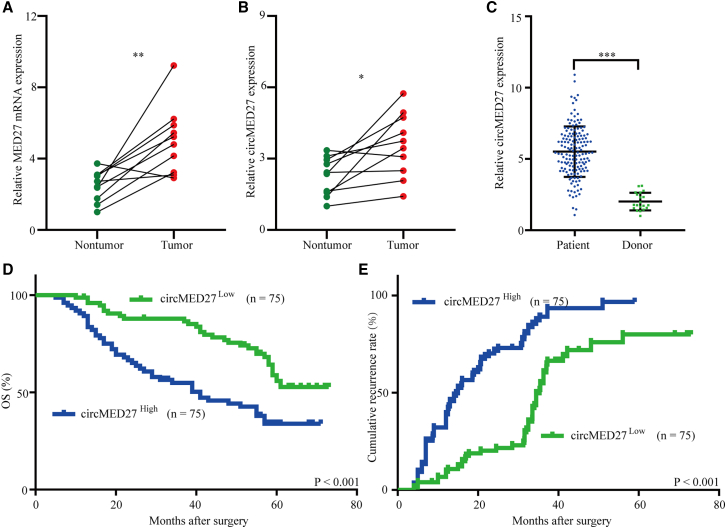
Previous studies have reported that MED27 expression was significantly increased in several cancers.10,11 However, the expression pattern of MED27 in HCC tissues has not been reported. Therefore, we explored the MED27 mRNA expressed in 10 pairs of HCC tissues and adjacent non-tumor tissues (Figure 1A). The results showed that MED27 mRNA expression was significantly upregulated in HCC tissues compared with matched adjacent non-tumor tissues. Interestingly, the data from exoRBase (http://www.exorbase.org/) indicates that med27 gene-derived circRNA circMED27 (circBase: hsa_circ_0006825) expression was detected in human blood and urine. Therefore, we further explored whether circMED27 was expressed in HCC tissues and HCC patient-derived exosomes. Then, we examined circMED27 expression in the above HCC tissues and the matched adjacent non-tumor tissues. The results showed that circMED27 (circBase: hsa_circ_0006825) expression levels were significantly upregulated in 10 of the HCC tumor tissues compared with the expression levels in the matched adjacent non-tumor liver tissues (Figure 1B). Next, we examined the levels of exosomal circMED27 in 150 HCC patients and 20 cases of healthy donor serum by quantitative real-time reverse transcriptase PCR (quantitative real-time RT-PCR). Our results showed that exosomal circMED27 levels in HCC patients were higher than in the healthy donor serum (Figure 1C; p < 0.001). Next, we explored the relationship between serum exosomal circMED27 levels and the clinical characteristics in 150 HCC patients, as listed in Table 1. Results reported that HCC patients with circMED27high had larger tumor sizes (p = 0.009), advanced tumor, node, metastasis (TNM) stages (p = 0.021), and forced alpha-fetoprotein (AFP) (p = 0.005). (The cut-off value is the median of the 150 HCC patients.) Therefore, we investigated the prognostic implication of circMED27 expression. Evidently, our results showed that patients with serum exosomal circMED27high levels had a significantly poorer prognosis than those with serum exosomal with circMED27low expression (Figures 1D and 1E). Multivariate analysis identified higher levels of serum exosomal circMED27 as an independent predictor for overall survival (OS) and postoperative recurrence (Tables 2 and 3). These findings show that circMED27 is likely to have participated in promoting the progression of HCC.
Figure 1.
High circMED27 expression in HCC serum exosomes and prognostic significance
(A) The expression of MED27 mRNA in HCC tissues and matched adjacent non-tumor tissues was verified by quantitative real-time RT-PCR. (B) The expression of circMED27 in HCC tissues and matched adjacent non-tumor tissues was verified by quantitative real-time RT-PCR. (C) The expression of circMED27 in HCC patient serum exosomes and healthy donor serum exosomes was verified by quantitative real-time RT-PCR. (D and E) Prognostic analysis of circMED27 expression in 150 HCC patient serum exosomes.
Table 1.
Correlation between serum circMED27 and clinicopathological characteristics in 150 HCCs
| Variable | circMED27 |
p value | |
|---|---|---|---|
| Low (n = 75) | High (n = 75) | ||
| Age (years) | |||
| ≤50 | 43 | 49 | 0.402 |
| >50 | 32 | 26 | |
| Sex | |||
| Female | 23 | 31 | 0.234 |
| Male | 52 | 44 | |
| HBs Ag | |||
| Negative | 4 | 6 | 0.745 |
| Positive | 71 | 69 | |
| HCV Ab | |||
| Negative | 68 | 70 | 0.765 |
| Positive | 7 | 5 | |
| Liver cirrhosis | |||
| Yes | 47 | 41 | 0.407 |
| No | 28 | 34 | |
| Serum AFP, ng/mL | |||
| ≤20 | 52 | 34 | 0.005 |
| >20 | 23 | 41 | |
| Serum ALT, U/L | |||
| ≤75 | 34 | 37 | 0.744 |
| >75 | 41 | 38 | |
| Tumor size (diameter, cm) | |||
| ≤5 | 47 | 30 | 0.009 |
| >5 | 28 | 45 | |
| Encapsulation invasion | |||
| Absent | 45 | 39 | 0.411 |
| Present | 30 | 36 | |
| Vascular invasion | |||
| No | 32 | 24 | 0.237 |
| Yes | 43 | 51 | |
| Tumor number | |||
| Single | 31 | 27 | 0.615 |
| Multiple | 44 | 48 | |
| TNM stages | |||
| I/II | 42 | 27 | 0.021 |
| III/IV | 33 | 48 | |
HBs Ag, hepatitis B surface antigen; HCV Ab, hepatitis C virus antibody; AFP, alpha-fetoprotein; ALT, alanine transaminase.
Table 2.
Univariate and multivariate analyses of factors associated with overall survival
| Factors | OS |
|||
|---|---|---|---|---|
| Multivariate | ||||
| Univariate, p | HR | 95% CI | p value | |
| Sex (female versus male) | 0.243 | – | – | NA |
| Age (years) (≤50 versus >50) | 0.762 | – | – | NA |
| HBs Ag (positive versus negative) | 0.309 | – | – | NA |
| HCV Ab (positive versus negative) | 0.173 | – | – | NA |
| Liver cirrhosis (yes versus no) | 0.253 | – | – | NA |
| Serum AFP, ng/mL (≤20 versus >20) | 0.075 | – | – | NA |
| Serum ALT, U/L (≤75 versus >75) | 0.413 | – | – | NA |
| Tumor size (diameter, cm) (>5 versus ≤ 5) | 0.007 | 1.021 | 0.932–1.766 | NS |
| Tumor number (multiple versus single) | 0.247 | – | – | NA |
| TNM (III/IV versus I/II) | 0.069 | – | – | NA |
| circMED27 expression (high versus low) | 0.002 | 2.312 | 1.433–2.876 | 0.017 |
OS, overall survival; NA, not adopted; AFP, alpha-fetoprotein; HBs Ag, hepatitis B surface antigen; 95% CI, 95% confidence interval; HR, hazard ratio; NS, not significantly. ∗Cox proportional hazards regression model.
Table 3.
Univariate and multivariate analyses of factors associated with cumulative recurrence
| Factors | Cumulative recurrence |
|||
|---|---|---|---|---|
| Univariate, p | HR | Multivariate |
||
| 95% CI | p value | |||
| Sex (female versus male) | 0.232 | – | – | NA |
| Age (years) (≤50 versus >50) | 0.477 | – | – | NA |
| HBs Ag (positive versus negative) | 0.129 | – | – | NA |
| HCV Ab (positive versus negative) | 0.307 | – | – | NA |
| Liver cirrhosis (yes versus no) | 0.429 | – | – | NA |
| Serum AFP, ng/mL (≤20 versus >20) | 0.672 | – | – | NA |
| Serum ALT, U/L (≤75 versus >75) | 0.351 | – | – | NA |
| Tumor size (diameter, cm) (>5 versus ≤ 5) | 0.003 | 1.347 | 0.917–2.044 | 0.013∗ |
| Tumor number (multiple versus single) | 0.422 | – | – | NA |
| TNM (III/IV versus I/II) | 0.071 | – | – | NA |
| circMED27 expression (high versus low) | 0.006 | 1.502 | 0.814–1.923 | 0.029∗ |
NA, not adopted; AFP, alpha-fetoprotein; HBs Ag, hepatitis B surface antigen; 95% CI, 95% confidence interval; HR, hazard ratio. ∗Cox proportional hazards regression model.
Exosomal circMED27 levels positively correlated with HCC tissue circMED27 levels
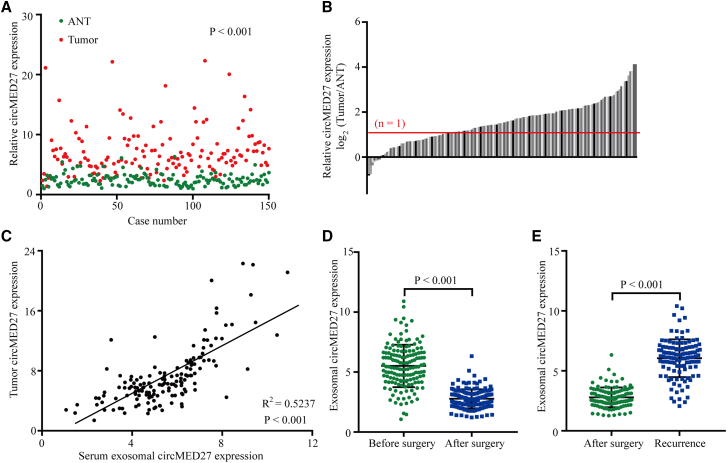
To better understand the biological function of circMED27 on HCC, we explored the levels of circMED27 in 150 pairs of HCC tissues and the corresponding adjacent non-tumor tissues and found that the levels of circMED27 were higher in HCC tissues than in the corresponding adjacent non-tumor tissues (103/150) (Figures 2A and 2B). A previous study reported that tumor cells can secrete exosomes into the circulation; thus, we hypothesized that circMED27 is secreted from HCC cells in an exosome-dependent manner. To verify this hypothesis, we examined the correlation between serum exosomal circMED27 levels and tumor tissue circMED27 levels in 150 HCC patients. Our results reported that there was a positive correlation between serum exosomal circMED27 levels and tumor tissue circMED27 levels in HCC patients (Figure 2C). Importantly, circMED27 expression was significantly reduced in HCC patient serum after surgery (Figure 2D). Moreover, the levels of circMED27 were significantly increased in HCC patients with tumor relapse (Figure 2E).
Figure 2.
High circMED27 expression in HCC tissues and the correlation with the expression in serum exosomes
(A and B) The expression of circMED27 in HCC tumor tissues and matched adjacent non-tumor tissues was verified by quantitative real-time RT-PCR. (C) A positive correlation between circMED27 in HCC tumor tissues and serum exosomes was observed in 150 HCC patient samples. (D) The expression of circMED27 in HCC patient serum exosomes decreased after surgery. (E) The expression of circMED27 in HCC patient serum exosomes increased after the tumor relapsed.
circMED27 expression is negatively correlated with HCC sensitivity to lenvatinib
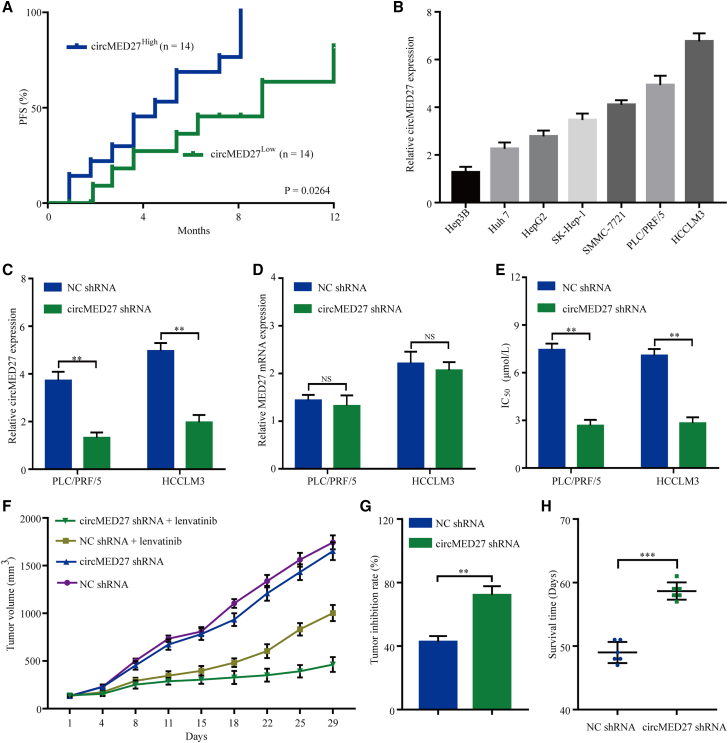
Interestingly, we investigated retrospective data from 28 relapsed HCC patients receiving lenvatinib therapy. All of these patients had received a liver resection 2–60 months before the lenvatinib therapy and were divided into two groups based on circMED27 levels using quantitative real-time RT-PCR. A Kaplan–Meier survival analysis showed that the progression-free survival (PFS) probability for the circMED27low group was much higher than that for the circMED27high group (Figures S1 and 3A). The median PFS was 4.5 months in the circMED27high group and 9 months in the circMED27low group (circMED27high group hazard ratio 2.537; 95% confidence interval, 1.014–6.345; p < 0.05). Thus, we speculated that high levels of circMED27 led to lenvatinib resistance in HCC.
Figure 3.
circMED27 induces lenvatinib resistance in HCC cells
(A) Comparison of progression-free survival curves for patients with high and low circMED27 expressions that were treated with lenvatinib. (B) circMED27 expression in several HCC lines was examined using by quantitative real-time RT-PCR. (C) circMED27 expression in PLC/PRF/5 and HCCLM3 cell lines was modified by shRNA interference transfection. (D) MED27 mRNA expression in circMED27 knockdown HCC cells. (E) Reduced circMED27 expression in PLC/PRF/5 and HCCLM3 cells increased their sensitivity to lenvatinib. (F) Anti-tumor effect of lenvatinib on HCC xenografts in an established model (n = 6). (G) The data were expressed as the percentage of inhibition of tumor growth. (H) The survival time of mice bearing HCC subcutaneous xenografts received lenvatinib therapy. Data are represented as the mean ± SD. ∗∗p < 0.01; ∗∗∗p < 0.001.
To validate the role of circMED27 in the lenvatinib resistance of HCC cells, we carried out quantitative real-time RT-PCR to detect the levels of circMED27 in seven HCC cell lines. As shown in Figure 3B, the levels of circMED27 were significantly higher in PLC/PRF/5 and HCCLM3 cell lines as compared with Hep3B and Huh 7 cell lines. To further confirm the role of circMED27 in HCC cell resistance to lenvatinib, PLC/PRF/5 and HCCLM3, which exhibit high levels of circMED27 expression, were transfected with circMED27 shRNA (Figure 3C). To verify that circMED27 short hairpin RNA (shRNA) did not knock down the other MED27 splicing products, quantitative real-time RT-PCR was carried out in order to examine the MED27 mRNA expression. The result indicated that the levels of MED27 mRNA showed no obvious change after the transfection of circMED27 shRNA, confirming the specificity of circMED27 silencing (Figure 3D). As shown in Figure 3E, circMED27 knockdown significantly enhanced the sensitivity of PLC/PRF/5 and HCCLM3 to lenvatinib. To further investigate the function of circMED27 on lenvatinib resistance, we explored the anti-tumor effects of lenvatinib in nude mice that xenografted with HCCLM3 negative control (NC) shRNA or HCCLM3 circMED27 shRNA cells. Compared with NC shRNA groups, lenvatinib significantly inhibited tumor growth, which derived from HCCLM3 circMED27 shRNA xenografts (Figures 3F and 3G). Moreover, the lenvatinib prolonged the survival time of mice bearing subcutaneous xenograft HCCLM3-circMED27 shRNA tumors as compared with HCCLM3 NC shRNA tumors (Figure 3H).
circMED27 binds miR-655-3p in HCC cells
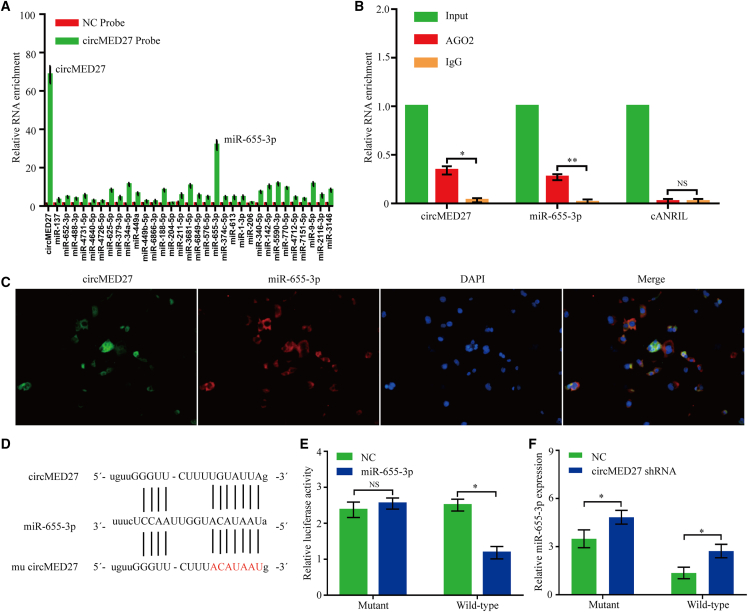
Recently, several studies have confirmed that circRNAs can act as miRNA sponges to inhibit functional miRNAs and then upregulate gene expression. Thus, we tried to detect the potential miRNAs that interacted with circMED27. Through starBase 3.0 prediction, 32 miRNAs were predicted as potential targets of circMED27. To verify the critical functional miRNAs that may interact with circMED27 in HCC cells, we used a circMED27-specific probe to carry out RNA in vivo RNA immunoprecipitation (RIP) and then screened the potential miRNAs. By circRIP experiments, we purified the circMED27-associated RNAs and analyzed the potential 32 candidate miRNAs in the complex. Our results showed a significant enrichment of circMED27 and miR-655-3p as compared with the NCs, whereas the other miRNAs had slight or no enrichment, determining that miR-655-3p is the critical circMED27-associated miRNA in HCC cells (Figure 4A).
Figure 4.
circMED27 may function as a sponge for miR-655-3p
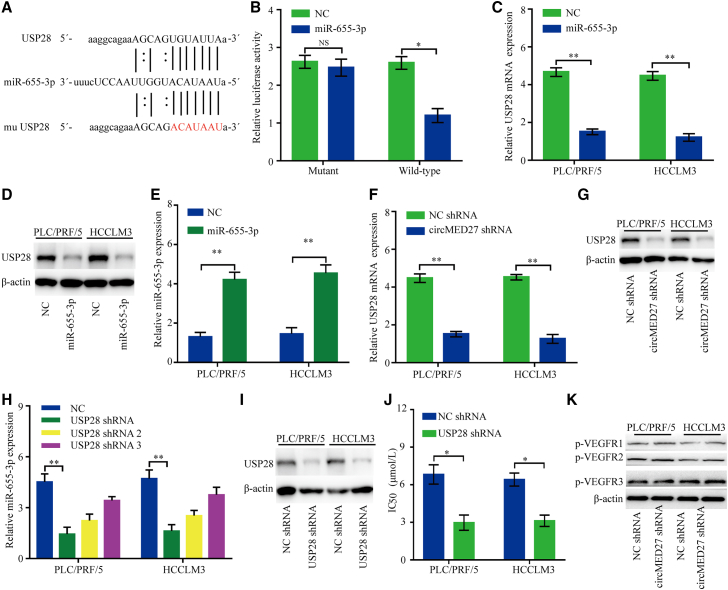
(A) circRIP was carried out in HCCLM3 cells using a circMED27-specific probe and a negative control probe, respectively. The enrichment of circMED27 and microRNAs was detected by quantitative real-time RT-PCR and normalized to the negative control probe. (B) RIP experiments were carried out using an AGO2 antibody on extracts from HCCLM3 cells. (C) Co-localization between circMED27 and miR-655-3p was detected using RNA in situ hybridization in HCCLM3 cells. Nuclei were stained with DAPI. (D) A schematic drawing showing the putative binding sites of miR-655-3p with circMED27. (E) The luciferase activity of luc-circMED27 or mutant luc-circMED27 in HCCLM3 cells after co-transfection with miR-655-3p. (F) miR-655-3p expression in HCC cell lines with decreased circMED27 was examined using quantitative real-time RT-PCR. The data are represented as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01; NS, not significant.
miR-655-3p was frequently found to be downregulated in HCC cells, and its reduced expression was significantly correlated with the poor prognosis of HCC patients.12,13 Then, the interaction between miR-655-3p and circMED27 was investigated in depth. We conducted RIP in HCC cells using anti-AGO2 antibodies or controlled immunoglobulin G (IgG), followed by quantitative real-time RT-PCR analysis for circMED27 and miR-655-3p levels. The results showed that circMED27 and miR-655-3p, but not cANRIL (a circRNA that has been reported to not bind to AGO2 protein14), were significantly enriched by the anti-AGO2 antibody, indicating that circMED27 may act as a binding platform for AGO2 protein and miR-655-3p (Figure 4B). Using the fluorescence in situ hybridization (FISH) analysis in HCC HCCLM3 cells, we found that miR-655-3p and circMED27 were mainly localized in the cytoplasm (Figure 4C). Furthermore, the results of the luciferase reporter assays confirmed that miR-655-3p significantly inhibited the luciferase activity of the reporter containing the wild-type (WT) circMED27 sequence compared with the reporter containing circMED27 with the mutated miR-655-3p binding site (mu circMED27) (Figures 4D and 4E). Moreover, the miR-655-3p expression shows a significant decrease after the knock down of circMED27 in HCC cells (Figure 4F).
USP28 is the target of miR-655-3p and promotes lenvatinib resistance of HCC
We used starBase 3.0 to predict the potential target mRNAs of miR-655-3p. The results showed that the 3′ UTR of USP28 mRNA bears a potential miR-655-3p binding site (Figure 5A). The effect of miR-655-3p on USP28 mRNA was evaluated by a luciferase reporter assay. Forced miR-655-3p expression significantly decreased luciferase activity of the reporter with the WT USP28 mRNA 3′ UTR as compared to the NC (p < 0.05) (Figure 5B). Conversely, this regulatory effect of miR-655-3p was not inhibited, while the predicted miR-655-3p-binding site in the 3′ UTR of USP28 mRNA was mutated (Figure 5B). Consistent with our expectations, our results indicated that miR-655-3p overexpression significantly decreased the USP28 mRNA and protein levels in HCC cells (Figures 5C–5E). Furthermore, the results showed that knockdown circMED27 expression significantly reduced the USP28 mRNA and protein levels in HCC cells (Figures 5F and 5G). To further evaluate the role of USP28 in HCC cells, three shRNAs against USP28, including shRNA, shRNA-2, and shRNA-3, were designed to silence the USP28 level in PLC/PRF/5 and HCCLM3 cells, and finally, USP28 shRNA was chosen for the following experiment with its high silencing efficiency (Figures 5H and 5I). As shown in Figure 5J, USP28 knockdown significantly enhanced the sensitivity of PLC/PRF/5 and HCCLM3 to lenvatinib. Additionally, circMED27 knockdown does not change the expression of phosphorylated vascular endothelial growth factor receptors 1–3 (pVEGFR1–3) (Figure 5K). Taken together, our results indicate that circMED27-mediated resistance to lenvatinib of HCC is independent of VEGFR-related pathways.
Figure 5.
USP28 binds to miR-655-3p and promotes lenvatinib resistance of HCC
(A) A schematic drawing showing the putative binding sites of miR-655-3p with USP28. (B) The luciferase activity of luc-USP28 or mutant luc-USP28 in HCCLM3 cells after co-transfection with miR-655-3p. (C) USP28 mRNA expression in PLC/PRF/5 and HCCLM3 cells was modified by miR-655-3p mimic transfection. (D) USP28 protein expression in PLC/PRF/5 and HCCLM3 cells was modified by miR-655-3p mimic transfection. (E) miR-655-3p expression in PLC/PRF/5 and HCCLM3 cells was modified by miR-655-3p mimic transfection. (F) USP28 mRNA expression in PLC/PRF/5 and HCCLM3 cells was modified by circMED27 shRNA transfection. (G) USP28 protein expression in PLC/PRF/5 and HCCLM3 cells was modified by circMED27 shRNA transfection. (H) USP28 mRNA expression in PLC/PRF/5 and HCCLM3 cells was modified by shRNA transfection. (I) USP28 protein expression in PLC/PRF/5 and HCCLM3 cells was modified by shRNA transfection. (J) Reduced USP28 expression in PLC/PRF/5 and HCCLM3 cells increased their sensitivity to lenvatinib. (K) pVEGFR1–3 protein expression in PLC/PRF/5 and HCCLM3 cells was modified by circMED27 shRNA transfection. The data are represented as the mean ± SD. ∗p < 0.05; ∗∗p < 0.01; NS, not significant.
circMED27 induces lenvatinib resistance in HCC cell lines by sponging miR-655-3p
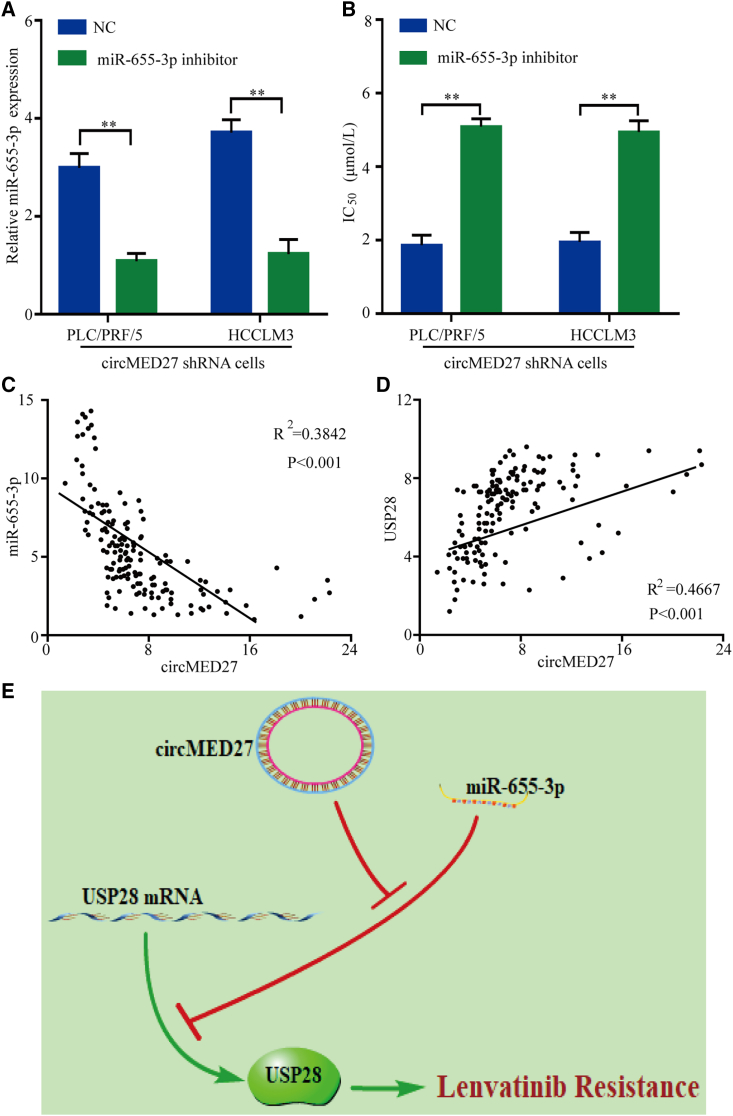
For further verify the biological molecular mechanism of circMED27 in HCC, we first transfected circMED27 shRNA HCC cells with a miR-655-3p inhibitor to verify whether the miR-655-3p inhibitor would recover the original lenvatinib-resistant phenotype (Figure 6A). Indeed, the lenvatinib-resistant capacity was reinstated in circMED27 shRNA HCC cells with the miR-655-3p inhibitor (Figure 6B). Furthermore, a scatterplot analysis showed a negative relationship between circMED27 and miR-655-3p expressions in HCC tissues (R2 = 0.3842; p < 0.001; Figure 6C). Importantly, a scatterplot analysis confirmed a positive relationship between circMED27 and USP28 expressions in HCC tissues (R2 = 0.4667; p < 0.001; Figure 6D). Collectively, these findings indicate that circMED27 induces HCC cell resistance to lenvatinib through the circMED27/miR-655-3p/USP28 axis (Figure 6E).
Figure 6.
circMED27-knockdown-mediated biological function was restored by miR-655-3p inhibition
(A) Relative levels of miR-655-3p were detected in PLC/PRF/5 and HCCLM3 cells after being transfected with a miR-655-3p inhibitor or negative control (NC) using quantitative real-time RT-PCR. (B) Reduced circMED27 expression in PLC/PRF/5 and HCCLM3 cells increased their sensitivity to lenvatinib. (C) The expression of miR-655-3p in 150 cases of HCC tissues was detected by quantitative real-time RT-PCR. A negative correlation between circMED27 and miR-655-3p was observed in tumor tissues (R2 = 0.3842; p < 0.001). (D) The expression of USP28 in 150 cases of HCC tissues was detected by immunohistochemistry. A positive correlation between circMED27 and USP28 was observed in tumor tissues (R2 = 0.4667; p < 0.001). (E) Models of high-level circMED27 on lenvatinib resistance. The data are represented as the mean ± SD. ∗∗p < 0.01.
Discussion
Lenvatinib is a multitargeted TKI with anti-tumor activities via the inhibition of fibroblast growth factor receptors (FGFRs) 1–4, VEGFRs 1–3, ret proto-oncogene, platelet-derived growth factor receptor alpha (PDGFRα), and KIT proto-oncogene receptor tyrosine kinase (KIT).15,16 A phase III REFLECT trial has reported that lenvatinib demonstrated promising anti-tumor activity in HCC patients.17 However, the overall response rate was only 40.7% in HCC patients who received lenvatinib.18 There is scarcely any knowledge about the molecular mechanisms of lenvatinib resistance in HCC, and primary and acquired lenvatinib resistance is very common, which is a major problem in the current HCC-targeting therapy.
Here, we first reported that circMED27 was a critical circRNA frequently overexpressed in HCC serum and tissue and that higher levels of circMED27 expression were positively correlated with a worse prognosis of HCC. Second, from a molecular mechanistic study, we found that the overexpression of circMED27 acted as the competing endogenous RNA (ceRNA) of miR-655-3p and that it upregulated the USP28-related pathway to promote HCC cell resistance to lenvatinib by competing for miR-655-3p. Third, our results showed that increased circMED27 was secreted into the serum of HCC patients by cancer cells in an exosome-dependent manner, predicting a shorter OS. Given the life-threatening nature of USP28-related signaling in the hepatocarcinogenesis,19 our results, for the first time, verified the critical role of circMED27 as a promising treatment target in HCC.
Recently, circRNAs have been validated to function as miRNA sponges.20 For example, circRNA FGFR3 (circFGFR3) was reported to sponge miR-22-3p, leading to the upregulation of galectin-1 and the activation of AKT and ERK signaling in NSCLC.9 Yu reported that circRNA cSMARCA5 could bind to miR-17-3p/miR-181b-5p and positively regulate tissue inhibitor of metalloproteinase 3 (TIMP3) expression, further inhibiting the growth and migration abilities of HCC.21 Here, we found that the overexpression of circMED27 could promote the lenvatinib resistance of HCC cells by sponging miR-655-3p then upregulating USP28 expression. To date, it has been reported that in HCC cells, miR-655-3p functions as a tumor suppressor by directly targeting a disintegrin and metallopeptidase domain 10 and then inhibiting the β-catenin pathway.13 Furthermore, a low level of miR-655-3p was associated with worse clinical characteristics of patients with HCC.12 In this study, we further confirmed USP28 as a novel target regulated by miR-655-3p. These data, collectively, suggested an important role of circMED27-sponged miR-655-3p in the regulation of HCC cells’ resistance to lenvatinib. USP28 was identified as a deubiquitinase that plays a vital role in the multiple biological processes of cancer progression.22 For example, USP28 is a prerequisite for MYC protein stability in human tumor cells.23 Higher levels of USP28 promoted progression and were associated with shorter OS in NSCLC patients.24 Recently, the clinical implications of forced USP28 expression have been documented for various cancers, including HCC.19,22 The importance of targeting USP28 for cancer therapy has attracted more attention than before.22 In this study, we concluded that circMED27 induces the lenvatinib resistance of HCC by upregulating the expression of USP28 and sponging miR-655-3p.
In summary, we demonstrate that, for the first time, a circMED27 was upregulated in HCC serum and tissues. circMED27 is capable of functioning as a ceRNA for sponging miR-655-3p to upregulate the expression of USP28. Furthermore, our results showed that circMED27 boosts HCC cells’ resistance to lenvatinib via increasing the USP28 expression. Moreover, the outcomes of our study provided a novel circMED27/miR-665-3p/USP28 signaling regulatory axis in HCC, which may deliver a promising biomarker and therapeutic target for the recovery of the sensitivity of HCC to lenvatinib.
Materials and methods
Human tissue microarray and immunohistochemistry
Clinical tissue samples and healthy donor samples were collected from the Fudan University Shanghai Cancer Center. We received informed consent from all subjects. All of the procedures were approved by the Institutional Review Board of Fudan University Shanghai Cancer Center and were conducted in accordance with the Declaration of Helsinki. All tumor tissues were histologically characterized by two pathologists in accordance with the criteria established by the World Health Organization and were stored in liquid nitrogen or formalin after surgery. For tissue microarray construction and immunohistochemistry, paraffin-embedded tissue sections (4 μm) were prepared using classical methods, and USP28 expression was detected using an immunoperoxidase method. USP28 expression levels were classified as either high (>20% of tumor section) or low (<20% of tumor section) using an integrated imaging system (MetaMorph Imaging System version 3.0; Universal Imaging, Buckinghamshire, UK).
Exosome isolation
Exosome isolation was performed as described in our previous study. In brief, exosomes from the serum of HCC patients and healthy donors were isolated using ExoQuick Exosome Precipitation Solution (SBI System Biosciences, catalogue no.: EXOQ5A-1) according to the manufacturer's instructions.
Cells and transfection
From the Chinese Academy of Science Cell Bank (Shanghai, China), the human HCC cell lines Hep3B, Huh7, HepG2, SK-Hep-1, and PLC/PRF/5 were obtained for the study. Furthermore, from the Liver Cancer Institute, Zhongshan Hospital, Fudan University, HCCLM3 was obtained, and from the Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences (Shanghai, China) SMMC-7721 was obtained for the study. The miR-655-3p mimic, the NC, and the miR-655-3p inhibitor (a chemically synthesized siRNA targeting miR-655-3p) were purchased from Shanghai GeneChem. The USP28 shRNA plasmids were purchased from Shanghai Yeasen. The transfection of the miR-655-3p mimic, NC, and USP28 shRNA plasmids was carried out with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Cell Counting Kit-8 (CCK-8) assay
HCC cells were seeded onto 96-well plates at 1 × 103 cells per well. 24 h later, the cells were treated with lenvatinib (1, 3, 9, 27, and 81 μM) and cultured for 72 h. Cell viability was then measured by using CCK-8 (Dojindo Molecular Technologies, Kumamoto, Japan), and half-maximal inhibitory concentration (IC50) values were measured by using GraphPad Prism (GraphPad Software, La Jolla, CA, USA).
circRNAs in vivo precipitation (circRIP)
A biotin-labeled circMED27-specific probe (bound to the back splice site of circMED27) and NC probes were synthesized by Sangon Biotech. For the circRIP assay, HCCLM3 cells were washed with ice-cold phosphate-buffered saline and fixed with 1% formaldehyde. The cells were subsequently lysed in 500 μL co-immunoprecipitation (coIP) buffer, sonicated, and centrifuged. The supernatant was added to a probes-M280 streptavidin dynabeads (Invitrogen) mixture and was then incubated at 30°C for 12 h. The probes-dynabeads-circRNAs mixture was washed and incubated with 200 μL lysis buffer and proteinase K to reverse the formaldehyde cross-linking. Finally, the RNA was extracted from the mixture using TRIzol Reagent (Invitrogen).
Quantitative real-time RT-PCR
Total RNA was extracted using the TRIzol kit (Invitrogen, Carlsbad, CA, USA) and quantitative real-time RT-PCR was performed using the SYBR Green PCR kit (TaKaRa, Otsu, Japan). The sequences of primers for quantitative real-time RT-PCR were as follows: circMED27 sense 5′-AGAAGCCCTAGTAGTGCCCAG-3′ and antisense 5′-TTTCTAATCCTAATAGATGCC-3′; USP28 sense 5′-TGGAGGAGAGTTGATGGTTG-3′ and antisense 5′-ATAGATCCAGGGCTGCAGAC-3′. GAPDH was used as an internal control reference: sense 5′-GGGGCTCTCCAGAACATCATCC-3′, antisense 5′-ACGCCTGCTTCACCACCTCTT-3′.
Western blotting and RIP
For western blotting, an equal amount of total cellular protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto 0.22 μm polyvinylidene difluoride (PVDF) membranes, and incubated with the corresponding antibodies. USP28, pVEGFR1-3, and β-actin antibodies were from Abcam. The membranes were subsequently developed using the enhanced chemiluminescence method (Pierce, Rockford, IL, USA). The RIP assays were conducted using a Magna RIP RNA-binding protein immunoprecipitation kit (Millipore). AGO2 and IgG antibodies were from Abcam.
FISH
Alexa Fluor 555-labeled circMED27 probes and Alexa Fluor 488-labeled miR-655-3p probes were designed and synthesized by RiboBio (Guangzhou, China). The probe signals were determined with the fluorescent In Situ Hybridization kit (RiboBio, Guangzhou, China) according to the manufacturer’s guidelines, and all fluorescence images were acquired by fluorescence microscope (Eclipse E600; Nikon Corporation, Tokyo, Japan).
Luciferase reporter assay
The mutant luciferase reporter vectors were generated using a Mutagenesis kit (QIAGEN, Valencia, CA, USA). HCCLM3 cells were seeded into 96-well plates. The cells were then co-transfected with a luciferase reporter vector and miR-655-3p mimic or the NC using the lipofectamine 2000 transfection reagent and incubated for 48 h. Finally, the firefly and Renilla luciferase activities were quantified with a dual-luciferase reporter assay (Promega, USA).
Subcutaneous xenograft tumor models
5 × 105 HCC cells were subcutaneously injected into 4- to 6-week-old nude mice. The mice were randomly assigned to treatment groups and vehicle solution groups when the tumor volumes reached an average of approximately 150 mm3. For treatment groups, a daily oral dose of lenvatinib was given at 10 mg/kg for 15 days. The length and width of each tumor were measured twice every week with calipers, and the volume was calculated using the following formula: length × width2 × 0.5. The mice were considered dead until the tumor volumes reached 2,000 mm3. Mice were manipulated and housed according to protocols approved by the Medical Experimental Animal Care Commission of Fudan University Shanghai Cancer Center.
Statistical analysis
Statistical analyses were performed using SPSS 21.0 software. The comparisons of quantitative data between two groups were carried out by the Student’s t test. The relationship between two proteins was calculated with the Pearson correlation coefficient. The OS and cumulative recurrence rates of patients were explored by the Kaplan–Meier method, and differences between the two groups were evaluated by the log rank test. p < 0.05 was considered statistically significant.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (no. 82072575 and 81702868) and the Nature Science Foundation of Jiangxi province (no. 20192BAB205064).
Author contributions
P.Z., J.W., Y.W., and Y.C. conceived and designed the experiments; P.Z., H.S., and P.W. performed the experiments; J.W. and Y.W. analyzed the data; P.Z. and Y.C. wrote the paper. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.12.001.
Contributor Information
Yilin Wang, Email: linglingwangyi@126.com.
Yuehong Cui, Email: cui.yuehong@zs-hospital.sh.cn.
Jing Wu, Email: wu.jing3@zs-hospital.sh.cn.
Supplemental information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Hiraoka A., Kumada T., Kariyama K., Takaguchi K., Atsukawa M., Itobayashi E., Tsuji K., Tajiri K., Hirooka M., Shimada N., et al. Clinical features of lenvatinib for unresectable hepatocellular carcinoma in real-world conditions: multicenter analysis. Cancer Med. 2019;8:137–146. doi: 10.1002/cam4.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Personeni N., Pressiani T., Rimassa L. Lenvatinib for the treatment of unresectable hepatocellular carcinoma: evidence to date. J. Hepatocell. Carcinoma. 2019;6:31–39. doi: 10.2147/JHC.S168953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beermann J., Piccoli M.T., Viereck J., Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 5.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang X., Li Z., Zhang Q., Wang W., Li B., Wang L., Xu Z., Zeng A., Zhang X., Zhang X., et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol. Cancer. 2019;18:71. doi: 10.1186/s12943-019-0969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bach D.H., Lee S.K., Sood A.K. Circular RNAs in cancer. Mol. Ther. Nucleic Acids. 2019;16:118–129. doi: 10.1016/j.omtn.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rong D., Lu C., Zhang B., Fu K., Zhao S., Tang W., Cao H. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol. Cancer. 2019;18:25. doi: 10.1186/s12943-019-0958-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Qiu B.Q., Zhang P.F., Xiong D., Xu J.J., Long X., Zhu S.Q., Ye X.D., Wu Y., Pei X., Zhang X.M., Wu Y.B. CircRNA fibroblast growth factor receptor 3 promotes tumor progression in non-small cell lung cancer by regulating Galectin-1-AKT/ERK1/2 signaling. J. Cell. Physiol. 2019;234:11256–11264. doi: 10.1002/jcp.27783. [DOI] [PubMed] [Google Scholar]
- 10.Tang R., Xu X., Yang W., Yu W., Hou S., Xuan Y., Tang Z., Zhao S., Chen Y., Xiao X., et al. MED27 promotes melanoma growth by targeting AKT/MAPK and NF-kappaB/iNOS signaling pathways. Cancer Lett. 2016;373:77–87. doi: 10.1016/j.canlet.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y.H., Huang J.H., Tu J.F., Wu M.H. MED27 promotes malignant behavior of cells by affecting Sp1 in breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2020;24:6802–6808. doi: 10.26355/eurrev_202006_21669. [DOI] [PubMed] [Google Scholar]
- 12.Zhao X.Q., Liang B., Jiang K., Zhang H.Y. Down-regulation of miR-655-3p predicts worse clinical outcome in patients suffering from hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2017;21:748–752. [PubMed] [Google Scholar]
- 13.Wu G., Zheng K., Xia S., Wang Y., Meng X., Qin X., Cheng Y. MicroRNA-655-3p functions as a tumor suppressor by regulating ADAM10 and beta-catenin pathway in Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2016;35:89. doi: 10.1186/s13046-016-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A., et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tohyama O., Matsui J., Kodama K., Hata-Sugi N., Kimura T., Okamoto K., Minoshima Y., Iwata M., Funahashi Y. Antitumor activity of lenvatinib (e7080): an angiogenesis inhibitor that targets multiple receptor tyrosine kinases in preclinical human thyroid cancer models. J. Thyroid Res. 2014;2014:638747. doi: 10.1155/2014/638747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto Y., Matsui J., Matsushima T., Obaishi H., Miyazaki K., Nakamura K., Tohyama O., Semba T., Yamaguchi A., Hoshi S.S., et al. Lenvatinib, an angiogenesis inhibitor targeting VEGFR/FGFR, shows broad antitumor activity in human tumor xenograft models associated with microvessel density and pericyte coverage. Vasc. Cell. 2014;6:18. doi: 10.1186/2045-824X-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Salama Z.T., Syed Y.Y., Scott L.J. Lenvatinib: a review in hepatocellular carcinoma. Drugs. 2019;79:665–674. doi: 10.1007/s40265-019-01116-x. [DOI] [PubMed] [Google Scholar]
- 18.Hiraoka A., Kumada T., Kariyama K., Takaguchi K., Itobayashi E., Shimada N., Tajiri K., Tsuji K., Ishikawa T., Ochi H., et al. Therapeutic potential of lenvatinib for unresectable hepatocellular carcinoma in clinical practice: multicenter analysis. Hepatol. Res. 2019;49:111–117. doi: 10.1111/hepr.13243. [DOI] [PubMed] [Google Scholar]
- 19.Han H., Sun D., Li W., Shen H., Zhu Y., Li C., Chen Y., Lu L., Li W., Zhang J., et al. A c-Myc-MicroRNA functional feedback loop affects hepatocarcinogenesis. Hepatology. 2013;57:2378–2389. doi: 10.1002/hep.26302. [DOI] [PubMed] [Google Scholar]
- 20.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 21.Yu J., Xu Q.G., Wang Z.G., Yang Y., Zhang L., Ma J.Z., Sun S.H., Yang F., Zhou W.P. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 2018;68:1214–1227. doi: 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Liu Z., Zhang L., Yang Z., Chen X., Luo J., Zhou Z., Mei X., Yu X., Shao Z., et al. Targeting deubiquitinase USP28 for cancer therapy. Cell Death Dis. 2018;9:186. doi: 10.1038/s41419-017-0208-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popov N., Wanzel M., Madiredjo M., Zhang D., Beijersbergen R., Bernards R., Moll R., Elledge S.J., Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat. Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L., Xu B., Qiang Y., Huang H., Wang C., Li D., Qian J. Overexpression of deubiquitinating enzyme USP28 promoted non-small cell lung cancer growth. J. Cell. Mol. Med. 2015;19:799–805. doi: 10.1111/jcmm.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.