Abstract
Coronavirus Disease 2019 (COVID-19), which poses an extremely serious global impact on human public healthcare, represents a high transmission and disease-causing viral infection caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) that is expanding at a rapid pace. Therefore, it is urgent for researchers to establish effective platforms for the assay and treatment of SARS-CoV-2. Functional nucleic acids (FNAs), comprising aptamers and nucleases, are of primary concern within the biological and medical communities owing of the distinctive properties of their target recognition and catalysis. This review will concentrate on the essential aspects of insights regarding FNAs and their technological expertise for the diagnostic and therapeutic utilization against COVID-19. We first offer a historical perspective of the COVID-19 pandemics, its clinical characteristics and potential biomarkers. Then, we briefly discuss the current diagnostic and therapeutic methodology towards COVID-19, highlighting the superiorities and existing shortcomings. After that, we introduce the key features of FNAs, and summarize recent progress of in vitro selection of FNAs for SARS-CoV-2 specific proteins and RNAs, followed by highlighting the general concept of translating FNAs into functional probes for diagnostic and therapeutic purposes. Then, we critically review the emerging FNAs-based diagnostic and therapeutic strategies that are fast, precise, efficient, and highly specific to fight COVID-19. Finally, we identify remaining challenges and offer future outlook of this emerging field.
Keywords: SARS-CoV-2, COVID-19, Diagnosis, Aptamer, DNAzyme, Therapeutics
1. Introduction
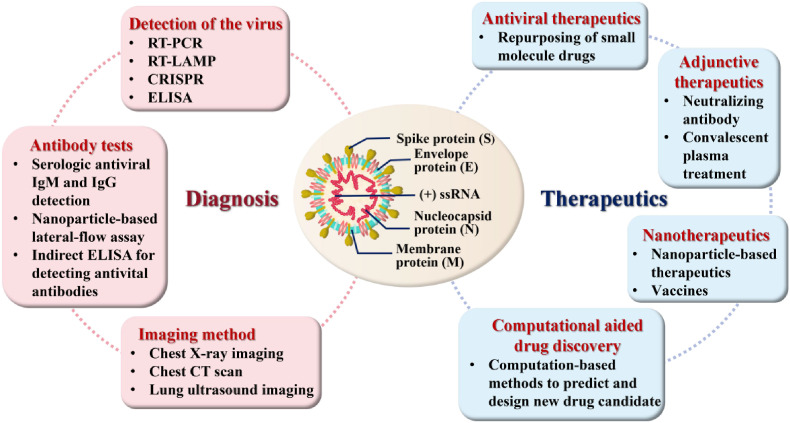
Since December 2019, the pandemic of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), also known as Coronavirus Disease 2019 (COVID-19), the world has been experiencing a huge threat to human health (Zhu et al., 2020; Fernandez et al., 2021; Wang et al., 2020a). As of November 26, 2021, there have been more than 259,502,031 confirmed cases worldwide, including 5,183,003 deaths, from 212 countries and territories (https://covid19.who.int/). The World Health Organization warned that if the current outbreak continues, more than 300 million people worldwide will be diagnosed with COVID-19 by early 2022. SARS-CoV-2 is an enveloped, positive single-stranded RNA virus with a diameter of approximately 50–200 nm (Xu et al., 2020b). SARS-CoV-2 has four major structural proteins, respectively, the spike (S) proteins, membrane (M) proteins, envelope (E) proteins and nucleocapsid (N) proteins (Fig. 1 ), resembling other coronaviruses (Boopathi et al., 2021; Rhouati et al., 2021). Similar to viruses in general, SARS-CoV-2 requires host cells for replication, which commonly comprises attachment, penetration, decapsidation, replication, assembly and release steps (Majumder and Minko, 2021). S proteins are composed of three identical monomers and two distinct subunits (S1 and S2). During the entire infection procedure, subunit S1 is linked towards the angiotensin-converting enzyme 2 (ACE2) receptor, whereas subunit S2 immobilizes the S protein on the membrane of the host cell and manages the fusion of the viral envelope within the host cell membrane. Two fundamental structural domains in the S1 subunit, called the receptor binding domain (RBD) and the N-terminal domain (NTD), are present. The RBD is well-known to be responsible for binding to ACE2 and mediating host cell recognition (Bosso et al., 2020). The N protein, by contrast, encompasses the viral genome and is also involved in the host cell's response to viral infection. Collectively, the S, E and M proteins combine to form a hard outer shell that wraps around and protects the virus (Wrapp et al., 2020).
Fig. 1.
Important diagnostic and therapeutic methods for COVID-19 and structure of SARS-CoV-2.
Physical signs and clinical indications of COVID-19 disease vary from individual to individual (Xu, 2020; Chen et al., 2020a). Clinical manifestations are mostly fever, malaise and cough, but may also present with symptoms such as sore throat, diarrhoea, anorexia, sputum production and shortness of breath (Wu and McGoogan, 2020; Han et al., 2020). Numerous reports have revealed the existence of both asymptomatic (pre-symptomatic) and atypical (asymptomatic) patients (Hu et al., 2020; Wang et al., 2020b; Mizumoto et al., 2020; Pan et al., 2020). Therefore, refinement of the clinical presentation of different clusters is necessary for early further diagnosis and treatment of suspected cases to control the spread of the outbreak. Both the viral protein and RNA can be considered as diagnostic targets for SARS-CoV-2 (Xu et al., 2020a). In addition, COVID-19-specific IgM is mostly positive 3–5 d after the onset of indications, and IgG antibody titres increase 4-fold or more during restoration as compared to the acute-phase, thus detecting IgM and IgG allows for an overview of the patient's infection history. SARS-CoV-2 RNA can be measured up to 3 d before the onset of symptoms and can be retained for 25–50 d after the onset of symptoms (He et al., 2020).
Viral detection, especially rapid testing, before vaccines or effective drugs become accessible is critically essential and a potent approach to pandemic surveillance and management (Xu et al., 2020a). Prompt detection also contributes to the efficient allocation of health care resources in hospitals and time-saving for front-line health workers. Various diagnostic and detection kits/assays have been described for the determination of COVID-19, comprising reverse transcription-polymerase chain reaction (RT-PCR) (Smyrlaki et al., 2020), enzyme-linked immunosorbent assays (ELISA)-based immunoassays (Carter et al., 2020), lateral flow devices (LFDs) (Huang et al., 2020), CT scan (Salehi et al., 2020), etc., which can profile cellular and antibody reactions to SARS-CoV-2 infection. Nevertheless, several disadvantages of these methods exist, which limit their application. These shortcomings include high cost, non-specificity/low specificity, high number of false positive/negative reactions, long testing time, labour intensity and complexity of operation (Iravani, 2020). So far, a number of antiviral drugs and various types of vaccines have been proactively developed. The security and efficacy of these investigational therapies for the treatment of COVID-19 are still being continuously evaluated (Titanji et al., 2021). Treatment of COVID-19 disease typically consists of supportive care in general, ventilatory assistance and nutritional help (Majumder and Minko, 2021). Depending on the current epidemic, rapid, accurate, sustainable, easily manufactured, target-specific and novel treatment techniques for monitoring and therapeutic purposes are still urgently needed. Meanwhile, functional nucleic acids (FNAs) were discovered via in vitro selection with significant concern are of primary concern within the biological and medical communities owing of the distinctive properties of their target recognition and catalysis (Zhang et al., 2019). FNAs possess the following advantages: lower molecular weight and better permeability of FNAs make in vivo applications more possible; it is easy and inexpensive to modify FNAs, which can be produced in large quantities, by solid-phase synthesis; FNAs play a role in immune-related assays due to their lower immunospecificity than antibodies; the library contains a large number of random sequences, and different FNAs for a target can be endowed with a wealth of functionalities (Zhu et al., 2021). FNAs are currently employed for the detection and therapeutic purposes of diseases caused by pathogens. According to these merits, FNAs are ideal candidates diagnosing and treating COVID-19. In this work, we concentrate on the basic comprehension of FNAs and the technical expertise for the diagnostic or therapeutic applications against COVID-19.
2. In vitro selection of functional nucleic acids towards understanding COVID-19
FNAs, mainly comprising aptamers and nucleic acid enzymes (NAEs), are typically isolated through the combined technology termed in vitro selection, and the discovery of new FNAs targeting SARS-CoV-2 has enabled significant advances not only within the realm of viral biology, but has also radically changed the design of functional probes for diagnostic or therapeutic applications against COVID-19 (Zhang et al., 2019). Table 1 summarized brief details of the aptamers that have been selected to date of diagnostic or therapeutic purposes towards COVID-19.
Table 1.
A brief overview of the aptamers probes against COVID-19.
| Aptamer name | Type | Molecular Target | Sequence | Kd (nM) | SELEX method | Limit of Detection/IC50 | Ref. |
|---|---|---|---|---|---|---|---|
| CoV2-RBD-1C | ssDNA | RBD of S protein | CAGCACCGACCTTGTGCTTTGGGAGTGCTGGTCCAAGGGCGTTAATGGACA | 5.8 | Ni-Beads-SELEX | – | Song et al. (2020) |
| CoV2-RBD-4C | ssDNA | RBD of S protein | ATCCAGAGTGACGCAGCATTTCATCGGGTCCAAAAGGGGCTGCTCGGGATTGCGGATATGGACACGT | 19.9 | Ni-Beads-SELEX | 130 fg/mL (antigen) 8 particles/mL (virus) |
(Pramanik et al., 2021) (Song et al., 2020) |
| cb-CoV2-6C3 | ssDNA | RBD of S protein | CGCAGCACCCAAGAACAAGGACTGCTTAGGATTGCGATAGGTTCGG | 0.13 | Ni-Beads-SELEX | 0.42 ± 0.15 nM (authentic virus) | Sun et al. (2021) |
| Aptamer-1 | ssDNA | RBD of S protein | TCGAGTGGCTTGTTTGTAATGTAGGGTTCCGGTCGTGGGT | 6.05 ± 2.06 | SELEX | 5.2 nM (inhibition of binding) | Liu et al. (2021) |
| Aptamer-2 | ssDNA | RBD of S protein | ATTACCGATGGCTTGTTTGTAATGTAGGGTTCCGTCGGAT | 6.95 ± 1.10 | SELEX | 4.4 nM (inhibition of binding) | Liu et al. (2021) |
| nCoV-S1-Apt1 | ssDNA | S1 | CCGCAGGCAGCTGCCATTAGTCTCTATCCGTGACGGTATG | 0.327 ± 0.016 | CE-based SELEX | 3.125 nM (S1 protein) | Yang et al. (2021) |
| SNAP1 | ssDNA | NTD of S protein | TCGCTCTTTCCGCTTCTTCGCGGTCATTGTGCATCCTGACTGACCCTAAGGTGCGAACATCGCCCGCGTAAGTCCGTGTGTGCGAA | 39.32 ± 0.12 | protein-SELEX | 250 pM (S with LFA) 10 pM (S protein with ELISA) | Kacherovsky et al. (2021) |
| Np-A48 | ssDNA | N protein | GCTGGATGTCGCTTACGACAATATTCCTTAGGGGCACCGCTACATTGACACATCCAGC | 0.49 | SELEX | 1 ng mL−1 (N protein) | (Zhang et al., 2020a) |
| MSA1 | ssDNA | S1 | CACTTTCCGGTTAATTTATGCTCTACCCGTCCACCTACCG | 1.8 ± 0.4 | Bead- and EMSA-based SELEX | 400 fM (pseudo-typed lentivirus) | Li et al. (2021) |
| S14 | ssDNA | NTD of S protein | TGGGAGCCTGGGACATAGTGGGGAAAGAGGGGAAGAGTGGGTCT | 21.8 | Talon-based SELEX | 2 nM (antigen) | Gupta et al. (2021) |
| SP6 | ssDNA | S protein (not RBD) | CCCATGGTAGGTATTGCTTGGTAGGGATAGTGGGCTTGATG | 21 ± 46 | Automated selection procedure | 0.2–1 μM (pseudo-virus) | Schmitz et al. (2021) |
| MAptapro-IR1 | ssRNA | Mpro | GGCGGCGAAACGGAGCUCCCAGGGAAUGGUCCAAAGGCGC | – | MAWS | – | Morena et al. (2021) |
Classical repetitive in vitro selection procedures, called systematic evolution of ligands by exponential enrichment (SELEX) (Ellington and Szostak, 1990), have been employed towards separate out various aptamers (biological receptors composed of single-stranded DNA/RNA), ribozymes (catalytically active RNA molecules) and deoxyribozymes (DNAzymes). Collectively, aptamers, ribozymes and DNAzymes are referred to as FNAs for distinguishing them from the typical functions of DNA and RNA as molecules that store and transmit genetic information (McConnell et al., 2021).
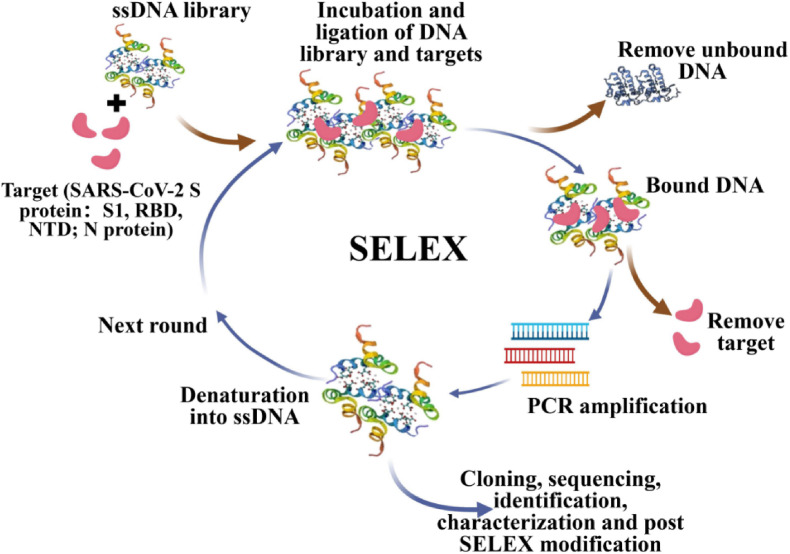
Currently, the majority of exploitation has been devoted to the selection of excellent nucleic-acid aptamers for the diagnosis and therapy of COVID-19. Fig. 2 illustrates the general procedure of in vitro SELEX for COVID-19. An initial random library consisting of short ssDNA sequences, chemically synthesized, is employed and incubated with biomolecular target complexes (e.g., SARS-CoV-2 RBDs modified to interact with A-protein via IgG-tag epitopes to form RBD-protein A-beads) to be ligated. During the initial round, the participating libraries and targets are bound by incubation. After removal of unbound oligonucleotides, an elution of bound target oligonucleotides is performed into amplification by PCR. The pools as selected by the above process serve for the next selection round. During the final round, the enriched pool of aptamers through cloning and sequencing, where the identification and characterization of high affinity aptamers is accomplished by DNA sequencing. Ultimately, multiple distinct high-affinity aptamers are obtained. Overall, the continuous steps of repeated selection (binding, washing and elution), amplification and purification characterizes the process (Kruger et al., 2021).
Fig. 2.
Schematic representation of general procedure of in vitro SELEX for SARS-CoV-2. Reprint from (Kruger et al., 2021).
The in vitro SELEX procedure has been continually refined to obtain aptamers with higher affinity. Cobalt resin (Talon) was incubated with the aptamer library by Gupta et al. and 14 aptamers were obtained after 6 rounds of SELEX (Gupta et al., 2021). Interestingly, from the first round onwards, the aptamers bound to the spiked antigen appeared to be saturated. Structural analysis of the S14 aptamer, the one with the highest affinity for S proteins, revealed a high guanine content and a secondary structure containing a stem-loop-like structure with an abundant GC/GT sequence in the loop region, forming the G-quadruplex. Molecular docking simulations showed that S14 binds to the NTD of S1, but not to the RBD. Similarly in combination with NTD, Kacherovsky et al. (2021) interchanged protein tags and the respective segmentation platforms in a manner that added pressure for selection via the addition of several competitors (e.g. MERS-CoV S1 and SARS-CoV S1). Song et al. (2020) modified RBD as protein A-beads, performed 8 rounds of SELEX and then put in ACE2 to incubate with RBD-beads to obtain sequences that compete with ACE2. Subsequently, 4 more rounds of SELEX were performed and used to simulate the infection process. The enrichment process was monitored based on the RBD-modified Ni-beads. Li et al. (2021) underwent 13 rounds of SELEX on SARS-CoV-2 S1, three of which were combined with magnetic beads and 10 were based on natural gels. Design of two flanking constant regions for the creation of stable pairing units enhanced the opportunity to identify diverse, high-affinity DNA aptamers. The SELEX method based on capillary electrophoresis (CE) has been reported for selection of S1 protein aptamers (Yang et al., 2021). To ensure evolutionary efficiency, a positive, negative and complex background screening process incorporating SELEX strategies with continuously reducing S1 concentrations and adding screening pressure was adopted to achieve high performance aptamers. In addition to aptamers for S proteins, aptamers with high affinity for N proteins have also been selected and optimized (Zhang et al., 2020a).
Besides ssDNA aptamers, ssRNA aptamers have also been explored for binding to major proteases (Morena et al., 2021). As a general matter, DNA aptamers are simpler to manufacture and cheaper to produce than RNA aptamers, stemming from the fact that reverse transcription and in vitro transcription steps with no need for them. Whereas RNA aptamers tend to generate unique, compact and diverse tertiary structures with potentially higher affinity and therapeutic applicability than DNA aptamers, it may allow easier entry into cells (Kruger et al., 2021).
DNAzymes consist of ssDNA molecules from a random DNA library selected in vitro and distinguished from aptamers by the addition of catalytic activity (Zhu et al., 2021). Among these, the commonly available are RNA-cleaved DNAzymes and G-quadruplex DNAzymes. Existing DNAzymes were used in current strategies for the detection and diagnosis of COVID-19 on a DNAzymes basis (Tian et al., 2021; Yang and Chaput, 2021; Ahmad et al., 2021; Zhang et al., 2021a). Although the selection of both aptamers and DNAzymes was done from ssDNA libraries, of which ssDNA libraries randomized and loaded up with RNA sequences serve for the selection of aptamers and DNAzymes, respectively. However, in the case of aptamers, the emphasis of selection was on affinity, whereas DNAzymes evolved mainly on the basis of cleavage activity or peroxidase activity.
3. The general concept of translating functional nucleic acids into functional probes
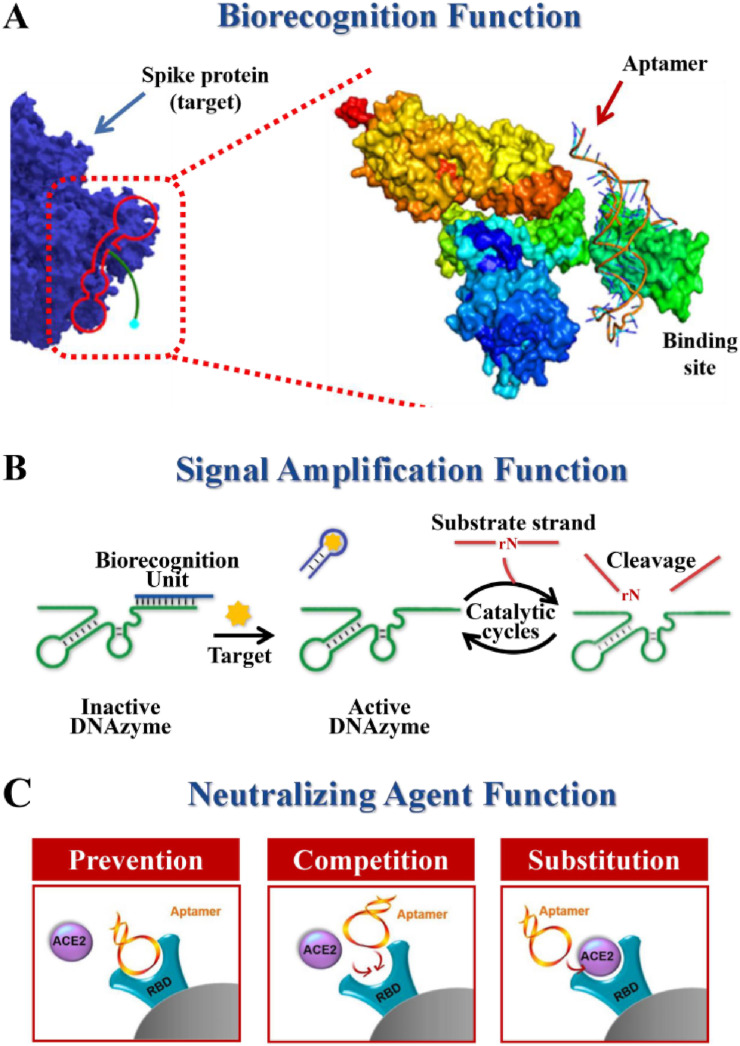
The appealing strengths and multi-functionality of FNAs have led to the emergence of FNA-based diagnostic platforms and therapeutic strategies to combat COVID-19. In these FNAs-based systems, the FNAs act as functional probes, assuming both diagnostic and therapeutic purposes. In general, according to the functions of FNAs, FNAs-based probes can be divided into three types: biorecognition agents, signal amplification agents, and neutralizing agents.
The former two aspects of functionality are beneficial for diagnostic purposes. As biorecognition agents (Fig. 3 A), FNAs can directly sense specific targets, and FNAs-based probes can convert target binding into target-dependent signals, allowing determination by various modalities. High specificity and high affinity are derived from the in vitro screening process. As biocatalysts (Fig. 3B), such as DNAzyme, can sense the target indirectly through signal amplification. DNAzymes are typically embedded in the biorecognition unit in inactive forms. With the existence of the target, the biocognitive unit interacts with the target and activates DNAzyme to cleave the substrate chain. Cleavage of the substrate strand, in general, causes regeneration of the target strand or release of the DNAzyme strand, which subsequently triggers a new round of catalytic cleavage reactions. In brief, a signal input that identifies the target will trigger a large number of catalytic cleavage events, cyclically amplifying the output signal. Apart from these, in the therapeutic context of targeting COVID-19, FNAs, especially aptamers, act as neutralizing agents that specifically bind viral proteins with preventive, competitive and alternative inhibitory effects on the receptors to which they bind, thus inhibiting viral infection for therapeutic purposes (Fig. 3C).
Fig. 3.
A summary of the general concept of translating functional nucleic acids into functional probes. Reprint from (Sun et al., 2021; Chen et al., 2021).
4. Functional nucleic acid-based diagnostic strategies for COVID-19
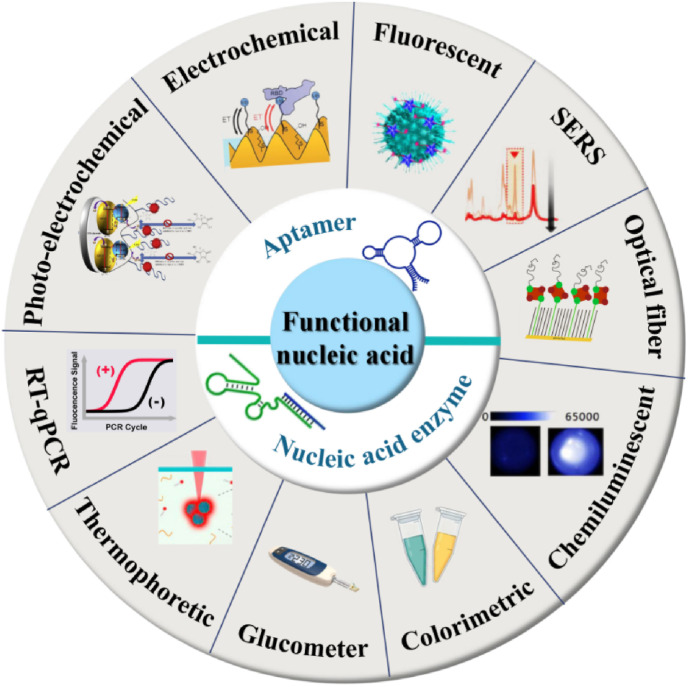
Two major branches of FNAs exist for COVID-19 diagnosis, namely, aptamers and NAEs. As illustrated in Fig. 4 , FNAs on the basis of which diverse signaling strategies can be integrated, including colorimetric, electrochemical, fluorescent, RT-qPCR, glucometer, etc. for detecting a wide range of SARS-CoV-2 biomarkers towards rapid, timely, and precise diagnosis of COVID-19. FNAs have an exciting field diagnostic aspect in early screening, field detection and in situ diagnosis for outbreak control.
Fig. 4.
Schematic of the functional nucleic acid-based diagnostic strategies for COVID-19.
4.1. Aptamers-based diagnostic strategies
Aptamers, termed “chemical antibodies”, represent to be a potent category of recognizing agents that offer a few merits such as high specificity and affinity, fast and dependable synthesis, easiness of conjugation, and excellent feasibility of integration into other DNA-based reactors towards amplification of signals (Liu et al., 2020). All these benefits render the aptamer a prospering bioreceptor with a wide range of applications for COVID-19 diagnosis.
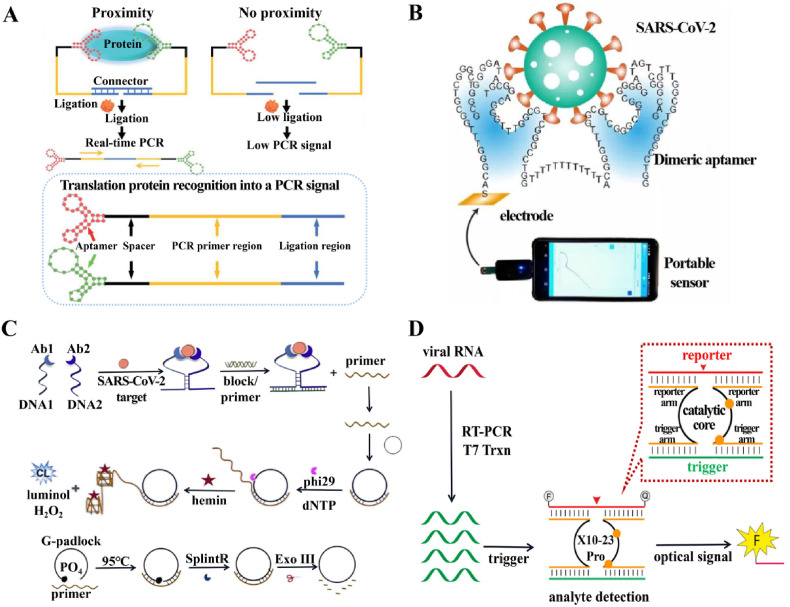
N protein of SARS-CoV has been discovered to be one of the essential biomarkers, and it can be detectable from a diverse range of patient samples, which include nasopharyngeal secretions, urine and faeces. Previously, single-stranded DNA aptamers have been described to bind within specificity of SARS-CoV N protein, while the sequence homology between the SARS-CoV-2 N protein and the SARS-CoV N protein was 91%. Chen et al. (2020b) have further modified the DNA aptamer to perform early diagnosis of COVID-19 by developing a methodology for detecting the SARS-CoV-2 N protein. Subsequently, Liu et al. (2020) presented an N-protein high-affinity aptamer-assisted proximity ligation immunoassay (Apt-PLA) for the ultrasensitive determination of SARS-CoV-2-related antigens. In contrast to the usual RT-qPCR approach, when two adjacent ssDNA aptamers bind to the same protein target in the Apt-PLA strategy, the ligated regions are in close proximity and the templates hybridise to form a complex that initiates ligation-dependent qPCR amplification. The signal is reflected by the level of the cycling threshold (Ct), the higher the Ct value, the lower the ligation efficiency and the lower the viral concentration (Fig. 5 A). This strategy can be used as a facile and versatile platform to provide an essential functionalised component for the diagnosis of coronaviruses.
Fig. 5.
Functional nucleic acid-based diagnostic strategies for COVID-19. (A) Scheme of aptamer-assisted proximity ligation assay for COVID-19 antigens. Probes that bind to the same protein target are in close proximity, initiating ligation-dependent qPCR amplification. Unbounded probes that are not in proximity show low ligation efficiency, resulting in low PCR signal. Reprint from (Liu et al., 2020). (B) Overview of electrochemical assay of wild-type and B.1.1.7 SARS-CoV-2. Reprint from (Li et al., 2021). (C) Schematic Diagrams of CL Imaging for Detection of the SARS-CoV-2 protein and preparation of circular DNA. Reprint from (Zhang et al., 2021a). (D) Overview of REVEALR detection of SARS-CoV-2. Reprint from (Yang and Chaput, 2021).
The RBD of the SARS-CoV-2 S protein has been implicated be associated with the entry of the virus and the pathogenesis of the disease. Pramanik et al. (2021) found that Au nanostars with high affinity ssDNA aptamer attachment of the S protein were available for the detection and inactivation of the virus. The distance-dependent nanoparticle surface energy transfer (NSET) spectroscopy involved in this methodology relies on the superior bursting performance exhibited by Au nanostars. This strategy was employed to test for COVID-19 spike antigen at a concentration level of 130 fg mL−1 and virus at a level of 8 particles mL−1. Furthermore, a rapid and high-sensitivity immunoassay for SARS-CoV-2 has been performed using autogrown gold nanopopcorn as substrate in combination with surface-enhanced Raman scattering (SERS) spectroscopy of the DNA aptamer of the S protein (Chen et al., 2021). The assay allowed the detection of viruses down to 10 PFU mL−1 within 15 min.
In principle, nucleic acid-based assays that specifically target characteristic regions of RNA viruses can deliver a high level of precision and sensibility towards molecular diagnosis. However, the currently utilized detection techniques for nucleic acids have not adequately addressed mutations in SARS-CoV-2. Mutations in SARS-CoV-2 virus raises the uncertainty about prevention and control in the COVID-19 global pandemic. Possible avenues of solution include the availability of a nucleic acid aptamer with exceptional affinity to serve as a recognition factor for the molecule, or coverage of the binding of this aptamer to the virus via extremely sensitive signal transduction strategies. Wang et al. (2021) exploited a CRISPR-Cas13 amplification-based strategy for sensing SARS-CoV-2 and its variants (wild or with D614G mutation) by illuminating specific RNA nucleic acid aptamer signals. The incorporation of RNA aptamers allows CRISPR-Cas13a to detect target RNA sequences label-free. The dual recognition and dual amplification steps enable this sensing platform by ensuring high-level of specificity for resolution of single nucleotide variants and high-level of sensitivity for detection of low abundance viruses. In another case, Zhang et al. (2021b) synthesized the dimeric aptamer DSA1N5 on the basis of previously identified aptamers against the wildtype and B.1.1.7 variants (Li et al., 2021) (Fig. 5B), which significantly improved its affinity through polythymidine (polyT) linkage. DSA1N5 exclusively identified the S protein of the wildtype virus (Kd = 0.12 nM) and its B.1.1.7 Alpha variant (Kd = 0.29 nM) and B.1.617.2 Delta variant (Kd = 0.48 nM), as well as binding to pseudotyped lentiviruses which express wildtype and trimeric S proteins. In conjunction with an electrochemical impedance sensor, virus was detected into less than 10 min straight from a patient saliva sample (1:1 dilution) with a clinically relevant accuracy of 80.5% and a specificity of 100%. The insufficient sensitivity probably stems from the presence of inhibitory factors (neutralizing antibodies) in the complex environment of these saliva samples, preventing recognition by the aptamers.
In addition to the above detection methodology, photoelectrochemical aptasensor have also been described through the determination of the SARS-CoV-2 RBD quantitatively using Chitosan/CdS-gC3N4 nanocomposites as photoactive materials for modified electrodes with sequences of aptamers immobilised on the electrode surface, with a detection limit of 0.12 nM for determination of the SARS-CoV-2 RBD (Amouzadeh Tabrizi et al., 2021). Moreover, Cennamo et al. (2021) demonstrated an aptamer-based platform towards optical assay of SARS-CoV-2 acting on the RBD. The biosensor exploited surface plasmon resonance (SPR) phenomena for highly sensitive and specific protein assay with a detection limit of 37 nM after immobilisation of specific aptamers on short polyethylene glycol (PEG) interfaces of Au nanomembranes deposited on D-shaped plastic optical fibres (POFs) probes. The assay platform has potential value for POC with rapid response and good portability. In another fascinating study, Deng et al. (2021) instead combined a thermoelectrophoretic approach for rapidly quantification of pseudo-SARS-CoV-2 viral particles due to the high-affinity aptamer of SARS-CoV-2 S protein. With the merits of rapidity (15 min), high sensitivity (detection limit of 176 TU μL−1), high accuracy (100%), one-step detection (no pre-treatment) and low cost (US$ 0.4 per test), this method has the capacity to be a validated tool for large-scale population-based screening towards SARS-CoV-2 infection.
The development of portable diagnostic tools for SARS-CoV-2 is not only an innovation in detection methods, but also allows for rapid screening using off-the-shelf portable instruments. Recently, Singh et al. (2021) developed a glucometer-based strategy of detecting SARS-CoV-2 salivary antigen. Using the catalytic properties of the convertase, the biotinylated aptamers of the SARS-CoV-2 S or N protein were pre-coupled into the convertase and, in the presence of the virus, the aptamers conformationally transformed, releasing the convertase and hydrolysing the disaccharide into a glucose signal, completing signal amplification that was captured by the glucometer with signal intensity proportional to the viral antigen concentration. The approach features low cost, high specificity and sensitivity, which is expected to be suitable for repeated population screening and diagnosis on a large scale, nevertheless, prospective clinical trials are required.
In POC detection devices, reliable and reproducible collection of samples is equally critical as in diagnostics. Daniels et al. (2021) improved the sample collection approach and reported an electrochemical biosensor towards directly testing of SARS-CoV-2 virus particles based on an aptamer for the RBD of S protein, collected via a mouthpiece sampling device. The biosensor converts exhaled gas into exhaled breath condensate and can be selective for the detection of SARS-CoV-2 virus particles below 10 pfu mL−1 in cultivated SARS-CoV-2 suspensions, however, a few false-negative problems need to be overcome.
4.2. Nucleic acid enzymes-based diagnostic strategies
NAEs were already discovered for catalyzing numerous chemical reactions, involving DNA or RNA cleavage, ligation and phosphorylation. Such characteristics have been employed with success in a variety of platforms for COVID-19 diagnosis (Table 2 ).
Table 2.
Nucleic acid enzymes-based biosensors for COVID-19 diagnosis.
| Detection Strategy | Target Genes | LOD | Time | Ref. |
|---|---|---|---|---|
| Fluorescence Lateral flow readout | S gene | 20 aM | 60 min | Yang and Chaput (2021) |
| RT-PCR Colorimetric readout | N gene | 1000 Copies of RNA | ∼75 min | Anantharaj et al. (2020) |
| RT-PCR Colorimetric readout | N gene | 100 Copies of RNA | <1 h | Ahmad et al. (2021) |
| Chemiluminescence Immunoassay | SARS-CoV-2 antibodies | 6.46 fg mL−1 | ∼140 min | (Zhang et al., 2021a) |
| Logic DNA circuit Fluorescent | nucleotide 3562–3581 and 32442–32461 of the COVID-19 complete genome | 3.3 fM | 75 min | Pan et al. (2021) |
| Electrochemical | N protein | 8.33 pg mL −1 | Unknown | Tian et al. (2021) |
The colorimetric method holds promise for large-scale testing by minimally trained personnel in resource-limited settings. The horseradish mimicking peroxidase DNAzyme (HRPzyme) as a short DNA sequence comprised within a guanine (G)-rich sequence that folds into a G-quadruplex structure and is stable in complex with hemin (Travascio et al., 1998). Anantharaj et al. (2020) developed a simple manual visualization method to SARS-CoV-2 RNA assay via PCR induction and conventional thermal cycler, detection limit of which was 1000 copies of viral RNA using HRPzyme. Subsequently, Ahmad et al. (2021) optimized the detection scheme with a modified HRPzyme sequence, resulting in an order of magnitude higher sensitivity.
Zhang et al. (2021a) reported a chemiluminescence (CL)-based high-throughput immunoassay for SARS-CoV-2, triggering the formation of a circular amplified G-quadruplex/hemin DNAzyme via proximity hybridization (Fig. 5C). Concurrently triggering proximity hybridization, strand replacement reactions and rolling circle amplification reactions by mixing samples with DNA antibody couples, cyclic DNA, block/primer complex and rolling circle amplification reaction solution, a homologous CL imaging procedure can be simply undertaken. With a broad detection range and a detection limit of 6.46 fg mL−1, this strategy has been potentially indicated for early screening and rapid diagnosis of COVID-19.
DNA molecular logic systems with recognition specificity, synthetic simplicity and predictability have been used for diverse input activation and amplification strategies(Zhang et al., 2020b; Wang et al., 2019; Gao et al., 2017). Nevertheless, the challenge of developing multiplexed signal amplification for COVID-19 logic systems remains. Pan et al. (2021) reported a logical DNA detector circuit based on a co-amplifying element controlled by exonuclease III and DNAzyme for highly sensitive COVID-19 assays. The logic circuit was applied successfully to the assay of real samples with a high degree of selectivity and robustness.
Combining the strengths of aptamers and NAEs, Tian et al. (2021) constructed an aptamer-protein-nanoprobe intercalated electrochemical sensing platform for highly sensitive and selective assay of COVID-19 on the electrode surface using Au@Pt NPs, G-quadruplex/hemin DNAzymes and HRP for co-catalytic, sulfhydryl-modified SARS-CoV-2 N protein double aptamer recognition.
XNAzyme, an artificial, catalytically active, multi-component nucleic-acid enzyme (Taylor and Holliger, 2015), has modular properties and separation of input and output functions, with potential for integration into a variety of diagnostic biosensors (Mokany et al., 2010). Yang et al. (Yang and Chaput, 2021) developed the RNA-Encoded Viral Nucleic Acid Analyte Reporter (REVEALR) on the basis of the novel discovery of XNAzyme 10–23 (Fig. 5D). The combination of viral pre-amplification and XNAzyme 10–23 pro-mediated RNA inactivation reporter enzymatic specific nucleic acid detection is termed REVEALR. This strategy used optical or visual readout systems for COVID-19 assay with a detection limit of 20 aM. REVEALR has a low cost, extensive sequence targeting, and process simplicity, therefore, offers a broad applicability in the immediate diagnosis of diseases.
5. Functional nucleic acid-based therapeutic strategies for COVID-19
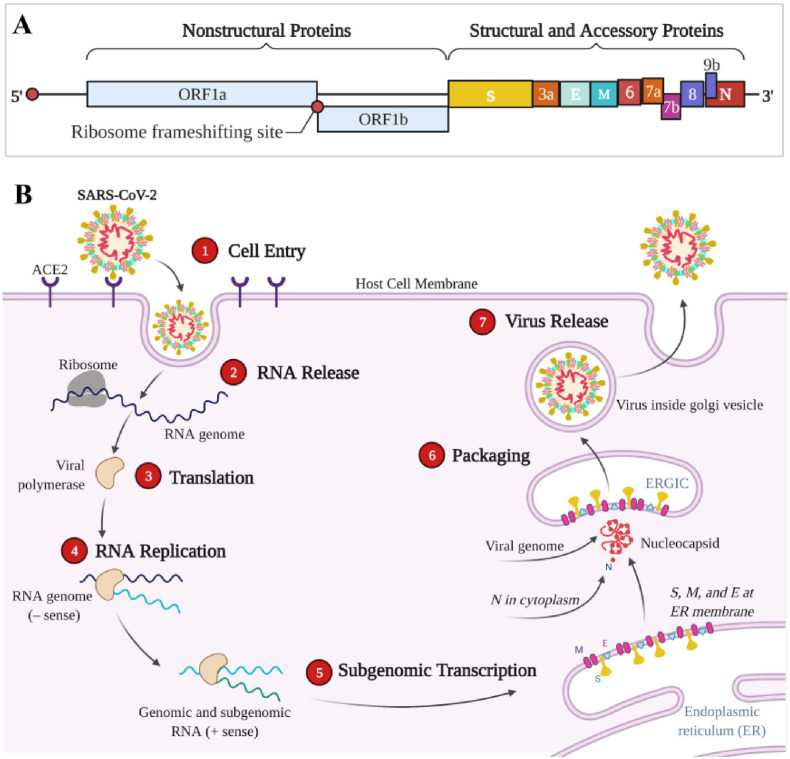
Approximately 29,900 nucleotides and 12 fictitious open reading frames (ORFs) are present in the ssRNA of SARS-CoV-2 (Manfredonia et al., 2020). In Fig. 6 A, the viral genome of SARS-CoV-2 comprises ORF1a and ORF1b encoding 16 nonstructural proteins, S, E, M, and N encoding four structural proteins, and ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF9 encoding six accessory proteins (Brant et al., 2021). The replicase (R), protease (Pro), S, M, N (including RdRp) and E proteins encoded by these genomes play key roles in virus entry into host cells, replication and survival (Zeng et al., 2021).
Fig. 6.
(A) Genomic organization and (B) replication cycle of SARS-CoV-2. ① The SARS-CoV-2 surface spike (S) protein interacts with the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell membrane, binds tightly to the serine protease (TMPRSS211), promotes viral fusion, and subsequently enters the host cell by endocytosis; ② SARS-CoV-2 enters the cell and releases the viral genome; ③ Using the host ribosome, viral RNA is translated to produce polyproteins, and the major protease (M pro) cleaves the polyproteins to form non-structural proteins; ④ RNA-dependent RNA polymerase (RdRp) in turn synthesizes a full-length negative-strand RNA template for making more viral genomic RNA; ⑤ Viral RNA is transcribed to produce a variety of mRNAs, which are subsequently translated to produce four important viral structural proteins, including S, M (membrane), E (envelope) and N (nucleocapsid) proteins; ⑥ The N protein binds genomic RNA, and the S, M, and E proteins integrate into the endoplasmic reticulum membrane to form the ERGIC endoplasmic reticulum-Golgi intermediate compartment (also called vesicle-tube cluster), which assembles the helically twisted RNA into a nucleocapsid that is wrapped within the endoplasmic cavity; ⑦ Vesicles carrying new viral particles fuse with the host cell membrane via ERGIC to release new virus. Created with BioRender.com (BJ2327CX1N).
A schematic diagram of viral infection of host cells and viral replication of SARS-CoV-2 was displayed in Fig. 6B. With respect to COVID-19 therapy, nucleic acids-based therapy is an attractive emerging strategy to combat the COVID-19 pandemic. One strategy is mainly involved the neutralization of SARS-CoV-2 through binding to spike RBD protein, which inhibits the SARS-CoV-2 entry into the human host. Another promising strategy is targeting replication crucial proteins to interfere with viral replication.
5.1. Aptamer-based therapeutic strategies
Current work indicates that one of the aptamer-based therapeutic strategies for COVID-19 is to inhibit viral infection by blocking the RBD of the S protein of SARS-CoV-2 to prevent it from interacting with the ACE2 receptor. Neutralizing antibodies have been reported to function as potentially effective agents in the prophylaxis and therapy against COVID-19 by acting on the RBD of the S protein (Ju et al., 2020; Cao et al., 2020; Pinto et al., 2020). Unfortunately, the large size of neutralizing antibodies significantly restricts the variety of delivery modes, while the antibody-dependent enhancement effect of the virus increases infectivity and virulence (Shi et al., 2020; DiLillo et al., 2014). By contrast, aptamers feature small size, high stability, low antibody-dependent enhancement effects and high programmability (Sun et al., 2021). Based on these advantages, Song et al. (2020) firstly selected and optimized two high-affinity aptamers for the RBD of S proteins, namely CoV2-RBD-1C and CoV2-RBD-4C, utilizing a machine learning screening algorithm. Both screened aptamers were capable of binding to several amino acid residues of the viral RBD, both in competition with ACE2. Small in size, as well as easy to modify and use, they serve as ideal RBD recognition probes for S proteins, both diagnostic and therapeutic for COVID-19.
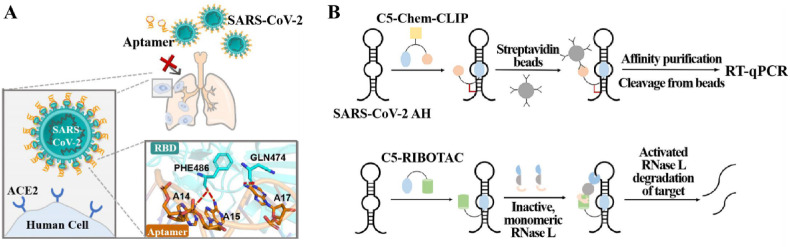
Subsequently, Sun et al. (2021) identified the CoV2-6 aptamer, also functioned the RBD of the viral S protein, through screening and molecular docking (Fig. 7 A). The screened aptamer was further optimized to a circular divalent nucleosome, cb-CoV2-6C3, which improved its stability and inhibitory effect with a Kd value of 0.13 nM and a half-maximal inhibitory concentration (IC50) value of 0.42 nM against SARS-CoV-2 virus. Yang et al. (2021) selected six aptamers with high affinity for SARS-CoV-2 S1 protein based on capillary electrophoresis (CE) SELEX method, among which nCoV-S1-Apt1 possesses excellent neutralizing activity. Two nucleic acid aptamers with the functionality to inhibit SARS-CoV-2 virus infection were screened through the work of Liu et al. (2021). The high affinity ssDNA aptamer-attached Au nanostars of SARS-CoV-2 S protein reported by Pramanik et al. (2021) was utilized for the assay of SARS-CoV-2 virus in addition to the inhibition of virus replication and inactivation. The effect was primarily due to two factors, on the one hand, the virus was unable to bind to ACE2 after binding to the Au nanostars attached to the S protein aptamer, and on the other hand, the Au nanostars adhering to the aptamer destroyed the lipid membrane of the pseudobaculovirus, causing the virus particles to collapse and inactivate.
Fig. 7.
Functional nucleic acid-based therapeutic strategies for COVID-19. (A) The mechanism of aptamer blocking strategy to inhibit SARS-CoV-2 infection. Reprint from (Sun et al., 2021). (B) Schematic of direct target engagement of SARS-CoV-2 FSE by C5-Chem-CLIP in cells and C5-RIBOTAC degradation of the SARS-CoV-2 RNA. Reprint from (Haniff et al., 2020). AH: attenuator hairpin; C5-Chem-CLIP: a covalent small molecule, directly engages the SARS-CoV-2 frameshifting element, validating C5's target site.
Owing to mutations in the RBD of the S protein of SARS-CoV-2 leads to ineffective RBD-targeting antibodies, but the interaction of SARS-CoV-2 with ACE2 remains unchanged or strengthened (Li et al., 2020). As a result, additional inhibitors of viral infection in diverse modes of action require development. The aptamer SP6 developed by Schmitz et al. (2021) is non-dependent on the RBD targeting S proteins but suppresses viral infection. Nevertheless, the authors were unclear about the exact mechanism of action of SP6 to suppress infection with the virus, but speculated that it would hinder the link within the virus and the host cell after binding, which may relate to prevention from S2 cleavage or disrupting the pre-fusion conformation of the S protein. The work suggests that inhibition is indeed possible despite the fact that the virus has already conjugated to the cells.
Alternatively, another promising therapeutic strategy is the binding of the aptamer to the major protease to inhibit viral replication. Mpro, a major protease in the non-structural protein of SARS-CoV-2, is also termed 3-chymotrypsin-like cysteine protease (Mousavizadeh and Ghasemi, 2021). Primarily involved in viral multiprotein processing and in the generation of RNA polymerase and decapping enzymes, Mpro is the key viral protease participating in the viral replication process (Koulgi et al., 2020). As a noteworthy fact, Mpro with a gene that is highly conserved, SARS-CoV-2 bears 96% sequence identities to SARS-CoV, and as such, is an efficacious target in developing viral variants against SARS-CoV-2 (Yang et al., 2003; Abian et al., 2020). The ssRNA aptamers for SARS-CoV-2 Mpro reported by Morena et al. (2021) were screened by modeling through an entropic fragment-based strategy followed by optimization of the initial sequence and three-dimensional structure using an elite genetic algorithm and an RNA inverse process. These specific aptamers were located in the SARS-CoV-2 Mpro dimer region key to enzyme activity, with the MAptapro-IR1 aptamer interacting most strongly and stably with the SARS-CoV-2 Mpro enzyme and being a prospective drug for the inhibition of SARS-CoV-2 virus replication and the treatment of COVID-19.
Current aptamer-based COVID-19 therapeutic strategies have been designed to target the viral entry mechanism and replication cycle, respectively. However, with little validation using real SARS-CoV-2 viruses and lack of in vivo experiments, the above process faces numerous challenges. Firstly, aptamers screened by the in vitro SELEX procedure may ignore some in vivo conditions, thus affecting the specificity for real samples. Secondly, therapeutic aptamers suffered from high renal clearance, safety and drug delivery issues when entering the treatment regimen. Finally, mutant proteins on the SARS-CoV-2 spectrum evade therapeutic regimens, requiring reselection of aptamer strategies to accommodate the mutations.
5.2. Other therapeutic strategies that targeting the SARS-CoV-2 RNA genome
While NAEs have not been directly used as therapeutic agents for COVID-19, other alternative therapeutic strategies that target the SARS-CoV-2 RNA genome have been developed. By research on the SARS-CoV-2 genome, frame-shifting, an essential process to suppress viruses via targeting small molecules with conserved, functional structures within the viral genome, was identified (Haniff et al., 2020). Both the coding and non-coding regions of the RNA genome of viruses are structurally convergent, and structural elements of this region may serve as potential novel targets for small molecule therapeutics (Brant et al., 2021). The frame-shifting element of SARS-CoV-2 engages the translation of two multiprotein critical for viral replication, where the existence of a structural convergence site suspends the ribosome and initiates frame-shifting, thereby altering protein coding (Manfredonia et al., 2020). Haniff et al. (2020) inhibited SARS-CoV-2 infection by suppressing frame shifting through two different mechanisms of action: simple binding and recruitment of endogenous nucleases to degrade the viral genome. The development of a targeted small molecule, C5, selectively binds and stabilizes frame-shifting elements, reducing their frame-shifting efficiency in cells (Fig. 7B). Optimized with the Kd value of 11 nM, the further refined recruitment of ribonuclease (C5-RIBOTAC) can disrupt the viral genome. The subsequent development of the drug holds more challenges, but the small molecule can indeed engage a functional position in the SARS-CoV-2 genome and act as a viral inhibitor.
Other therapeutic strategies that targeting the SARS-CoV 2 RNA genome, such as RNA G-Quadruplex (Zhao et al., 2021), targeting bulge structures in the 5’ UTR (Melidis et al., 2021), and siRNA-nanoparticle-based therapeutic against SARS-CoV-2 infection (Idris et al., 2021), have also been reported for therapeutic applications. Despite these advances, however, several critical issues should be considered when proceeding with the development of NAEs-based antiviral therapies. First, the target regions of the SARS-CoV-2 RNA genome should be highly conserved and critical for viral replication as well as, for more importantly, affinity for the binding and subsequent efficient shearing of NAEs. Second, the effective delivery methodology is the essential factor to enhance the effectiveness of the treatment. The capability of nanocarriers for rapid design and delivery of NAEs opens up a promising strategy for future NAEs-based anti-SARS-CoV-2 pharmacotherapy. Finally, the as-prepared NAEs have a limiting effect in some cases. For example, ribozymes can stimulate the immune system leading to certain off-target effects, saturation of natural RNAi in RNAi-based actions, and other conditions. It is possible to overcome these limitations by changing vectors, NAEs, target genes or via chemical modification of NAEs.
6. Conclusions and outlook
To summarize, we have performed a review of excited advancements in the molecular engineering of FNAs, including aptamers and NAEs, to fight COVID-19. These FNAs display several distinctive characteristics which render them hopeful candidates for the designs of diagnostic and therapeutic strategies. For example, different signal transduction strategies, such as electrochemical, fluorescent, SERS, thermophoretic, and glucose meter, have been integrated with aptamers and NAEs for detecting a wide range of SARS-CoV-2 biomarkers. Additionally, aptamers which have both targeting and therapeutic capabilities have been designed in order to improve therapy efficiency. Finally, targeting the SARS-CoV-2 RNA genome with RIBOTAC, RNA G-Quadruplex inhibitor and siRNA-nanoparticle also yielded new therapeutic strategies to interfere with viral replication. These emerging FNAs-based diagnostic and therapeutic strategies provide a fast, precise, efficient, and highly specific toolbox to fight COVID-19.
The programmability of FNAs, especially in the early stages of acquisition, improves the accuracy and specificity of diagnosis for SARS-CoV-2. To further enhance diagnostic capabilities and accuracy, a combination of various biomarkers can be diagnosed/detected by employing multiplexed identification and biosensing techniques. Furthermore, the details related to mechanical learning-based programming and acquisition of signals are critical fields of interest for scientists in order to enhance the reliability and reproducibility of such sensors. To tackle such provocative challenges, more detailed academic analysis and research in particular regarding sensitivity, false negative/positive outcomes, selectivity, verification procedures, testing velocity, simplicity, affordability and general usability should be systematically undertaken. FNAs are capable of making notable advances in the manufacture of novel diagnostic sensing platforms, incorporation of innovative devices, improved modifications/validation, and enhancement of POC sensing performance. Further research directions need to be mapped out to seek and design novel next-generation non-invasive, specifically tailored, cost-effective and rapid biological sensing approaches and techniques that could be applied to the diagnosis of COVID-19.
In spite of these significant advances in the engineering of FNAs against SARS-CoV2, a number of challenges still limit their clinical utility. Firstly, understanding the intracellular or in vivo behaviours of these FNAs, such as stability, affinity, and specificity, is still in its infancy. In addition, for therapeutic purpose, the potential challenges of in vivo circulation, clearance, and biodistribution should be taken into consideration. Finally, the delivery of FNAs agents in a precisely controlled manner also requires more innovative approaches. Therefore, surmounting these challenges will further expand the range of FNAs-based diagnostic and therapeutic strategies and hopefully promote their conversion for clinical adoptions to combat COVID-19.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We greatly acknowledge the financial support from the National Natural Science Foundation of China (no. 22004063), Natural Science Foundation of Jiangsu Province (no. 20200303).
References
- Abian O., Ortega-Alarcon D., Jimenez-Alesanco A., Ceballos-Laita L., Vega S., Reyburn H.T., Rizzuti B., Velazquez-Campoy A. Int. J. Biol. Macromol. 2020;164:1693–1703. doi: 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M., Sharma P., Kamai A., Agrawal A., Faruq M., Kulshreshtha A. Biosens. Bioelectron. 2021;187 doi: 10.1016/j.bios.2021.113280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amouzadeh Tabrizi M., Nazari L., Acedo P. Sensor. Actuator. B Chem. 2021;345 doi: 10.1016/j.snb.2021.130377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaj A., Das S.J., Sharanabasava P., Lodha R., Kabra S.K., Sharma T.K., Medigeshi G.R. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.586254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi S., Poma A.B., Kolandaivel P. J. Biomol. Struct. Dynam. 2021;39(9):3409–3418. doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosso M., Thanaraj T.A., Abu-Farha M., Alanbaei M., Abubaker J., Al-Mulla F. Mol. Ther. Methods Clin. Dev. 2020;18:321–327. doi: 10.1016/j.omtm.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brant A.C., Tian W., Majerciak V., Yang W., Zheng Z.-M. Cell Biosci. 2021;11(1) doi: 10.1186/s13578-021-00643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.L., Su B., Guo X.H., Sun W.J., Deng Y.Q., Bao L.L., Zhu Q.Y., Zhang X., Zheng Y.H., Geng C.Y., Chai X.R., He R.S., Li X.F., Lv Q., Zhu H., Deng W., Xu Y.F., Wang Y.J., Qiao L.X., Tan Y.F., Song L.Y., Wang G.P., Du X.X., Gao N., Liu J.N., Xiao J.Y., Su X.D., Du Z.M., Feng Y.M., Qin C., Qin C.F., Jin R.H., Xie X.S. Cell. 2020;182(1):73. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. ACS Cent. Sci. 2020;6(5):591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cennamo N., Pasquardini L., Arcadio F., Lunelli L., Vanzetti L., Carafa V., Altucci L., Zeni L. Talanta. 2021:233. doi: 10.1016/j.talanta.2021.122532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Park S.-G., Choi N., Kwon H.-J., Kang T., Lee M.-K., Choo J. ACS Sens. 2021;6(6):2378–2385. doi: 10.1021/acssensors.1c00596. [DOI] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J.a., Yu T., Zhang X., Zhang L. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Wu Q., Chen J., Ni X., Dai J. Virol. Sin. 2020;35(3):351–354. doi: 10.1007/s12250-020-00236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels J., Wadekar S., DeCubellis K., Jackson G.W., Chiu A.S., Pagneux Q., Saada H., Engelmann I., Ogiez J., Loze-Warot D., Boukherroub R., Szunerits S. Biosens. Bioelectron. 2021;192 doi: 10.1016/j.bios.2021.113486. 113486-113486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Tian F., Liu C., Liu Y., Zhao S., Fu T., Sun J., Tan W. J. Am. Chem. Soc. 2021;143(19):7261–7266. doi: 10.1021/jacs.1c02929. [DOI] [PubMed] [Google Scholar]
- DiLillo D.J., Tan G.S., Palese P., Ravetch J.V. Nat. Med. 2014;20(2):143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A.D., Szostak J.W. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Fernandez N.B., Caceres D.H., Beer K.D., Irrazabal C., Delgado G., Farias L., Chiller T.M., Verweij P.E., Stecher D. Med. Mycol. Case Rep. 2021;31:19–23. doi: 10.1016/j.mmcr.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R.-R., Yao T.-M., Lv X.-Y., Zhu Y.-Y., Zhang Y.-W., Shi S. Chem. Sci. 2017;8(6):4211–4222. doi: 10.1039/c7sc00361g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Anand A., Jain N., Goswami S., Ananthraj A., Patil S., Singh R., Kumar A., Shrivastava T., Bhatnagar S., Medigeshi G.R., Sharma T.K., Research, D.B.T.I.C.f.C Molecular Therapy-Nucleic Acids. 2021;26:321–332. doi: 10.1016/j.omtn.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Duan C., Zhang S., Spiegel B., Shi H., Wang W., Zhang L., Lin R., Liu J., Ding Z., Hou X. Am. J. Gastroenterol. 2020;115(6):916–923. doi: 10.14309/ajg.0000000000000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniff H.S., Tong Y., Liu X., Chen J.L., Suresh B.M., Andrews R.J., Peterson J.M., O'Leary C.A., Benhamou R.I., Moss W.N., Disney M.D. ACS Cent. Sci. 2020;6(10):1713–1721. doi: 10.1021/acscentsci.0c00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Nat. Med. 2020;26(5) doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Hu Z., Song C., Xu C., Jin G., Chen Y., Xu X., Ma H., Chen W., Lin Y., Zheng Y., Wang J., Hu Z., Yi Y., Shen H. Sci. China Life Sci. 2020;63(5):706–711. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wen T., Shi F.-J., Zeng X.-Y., Jiao Y.-J. ACS Omega. 2020;5(21):12550–12556. doi: 10.1021/acsomega.0c01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris A., Davis A., Supramaniam A., Acharya D., Kelly G., Tayyar Y., West N., Zhang P., McMillan C.L.D., Soemardy C., Ray R., O'Meally D., Scott T.A., McMillan N.A.J., Morris K.V. 2021. bioRxiv : the Preprint Server for Biology. [Google Scholar]
- Iravani S. Mater. Adv. 2020;1(9):3092–3103. [Google Scholar]
- Ju B., Zhang Q., Ge J.W., Wang R.K., Sun J., Ge X.Y., Yu J.Z., Shan S.S., Zhou B., Song S., Tang X., Yu J.F., Lan J., Yuan J., Wang H.Y., Zhao J.N., Zhang S.Y., Wang Y.C., Shi X.L., Liu L., Zhao J.C., Wang X.Q., Zhang Z., Zhang L.Q. Nature. 2020;584(7819):115. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kacherovsky N., Yang L.F., Dang H.V., Cheng E.L., Cardle I.I., Walls A.C., McCallum M., Sellers D.L., DiMaio F., Salipante S.J., Corti D., Veesler D., Pun S. Angewandte Chemie (International. 2021. in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulgi S., Jani V., Uppuladinne M., Sonavane U., Nath A.K., Darbari H., Joshi R. J. Biomol. Struct. Dynam. 2020;39(15):5735–5755. doi: 10.1080/07391102.2020.1792344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger A., de Jesus Santos A.P., de Sa V., Ulrich H., Wrenger C. Pharmaceuticals. 2021;14(7) doi: 10.3390/ph14070622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang Z., Gu J., Stacey H.D., Ang J.C., Capretta A., Filipe C.D.M., Mossman K.L., Balion C., Salena B.J., Yamamura D., Soleymani L., Miller M.S., Brennan J.D., Li Y. Nucleic Acids Res. 2021;49(13):7267–7279. doi: 10.1093/nar/gkab574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.Q., Wu J.J., Nie J.H., Zhang L., Hao H., Liu S., Zhao C.Y., Zhang Q., Liu H., Nie L.L., Qin H.Y., Wang M., Lu Q., Li X.Y., Sun Q.Y., Liu J.K., Zhang L.Q., Li X.G., Huang W.J., Wang Y.C. Cell. 2020;182(5):1284. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., He L., Hu Y., Luo Z., Zhang J. Chem. Sci. 2020;11(44):12157–12164. doi: 10.1039/d0sc03920a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang Y.-l., Wu J., Qi J., Zeng Z., Wan Q., Chen Z., Manandhar P., Cavener V.S., Boyle N.R., Fu X., Salazar E., Kuchipudi S.V., Kapur V., Zhang X., Umetani M., Sen M., Willson R.C., Chen S.-h., Zu Y. Angew. Chem. Int. Ed. 2021;60(18):10273–10278. doi: 10.1002/anie.202100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder J., Minko T. AAPS J. 2021;23(1) doi: 10.1208/s12248-020-00532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredonia I., Nithin C., Ponce-Salvatierra A., Ghosh P., Wirecki T.K., Marinus T., Ogando N.S., Snijder E.J., van Hemert M.J., Bujnicki J.M., Incarnato D. Nucleic Acids Res. 2020;48(22):12436–12452. doi: 10.1093/nar/gkaa1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell E.M., Cozma I., Mou Q., Brennan J.D., Lu Y., Li Y. Chem. Soc. Rev. 2021;50(16):8954–8994. doi: 10.1039/d1cs00240f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melidis L., Hill H.J., Coltman N.J., Davies S.P., Winczura K., Chauhan T., Craig J.S., Garai A., Hooper C.A.J., Egan R.T., McKeating J.A., Hodges N.J., Stamataki Z., Grzechnik P., Hannon M.J. Angew. Chem. Int. Ed. 2021;60(33):18144–18151. doi: 10.1002/anie.202104179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K., Kagaya K., Zarebski A., Chowell G. Euro Surveill. 2020;25(10):2–6. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokany E., Bone S.M., Young P.E., Doan T.B., Todd A.V. J. Am. Chem. Soc. 2010;132(3):1051–1059. doi: 10.1021/ja9076777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morena F., Argentati C., Tortorella I., Emiliani C., Martino S. Int. J. Mol. Sci. 2021;22(13) doi: 10.3390/ijms22136874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavizadeh L., Ghasemi S. J. Microbiol. Immunol. Infect. 2021;54(2):159–163. doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., He Y., Liu Z., Chen J. Chem. Commun. 2021;57(9):1125–1128. doi: 10.1039/d0cc06799g. [DOI] [PubMed] [Google Scholar]
- Pan X., Chen D., Xia Y., Wu X., Li T., Ou X., Zhou L., Liu J. Lancet Infect. Dis. 2020;20(4):410–411. doi: 10.1016/S1473-3099(20)30114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J.B., Chen R.T.E., Havenar-Daughton C., Snell G., Telenti A., Virgin H.W., Lanzavecchia A., Diamond M.S., Fink K., Veesler D., Corti D. Nature. 2020;583(7815):290. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Pramanik A., Gao Y., Patibandla S., Mitra D., McCandless M.G., Fassero L.A., Gates K., Tandon R., Ray P.C. J. Phys. Chem. Lett. 2021;12(8):2166–2171. doi: 10.1021/acs.jpclett.0c03570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhouati A., Teniou A., Badea M., Marty J.L. Sensors. 2021;21(4) doi: 10.3390/s21041485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Am. J. Roentgenol. 2020;215(1):87–93. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- Schmitz A., Weber A., Bayin M., Breuers S., Fieberg V., Famulok M., Mayer G. Angew. Chem.(Weinheim an der Bergstrasse, Germany) 2021;133(18):10367–10373. doi: 10.1002/ange.202100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X.M., Chen Z.H., Liu P.P., Song J.W., Song T., Bi X.S., Han C., Wu L.A., Gao G., Hu X., Zhang Y.A., Tong Z., Huang W.J., Liu W.J., Wu G.Z., Zhang B., Wang L., Qi J.X., Feng H., Wang F.S., Wang Q.H., Gao G.F., Yuan Z.M., Yan J.H. Nature. 2020;584(7819):120. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Singh N.K., Ray P., Carlin A.F., Magallanes C., Morgan S.C., Laurent L.C., Aronoff-Spencer E.S., Hall D.A. Biosens. Bioelectron. 2021;180 doi: 10.1016/j.bios.2021.113111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyrlaki I., Ekman M., Lentini A., de Sousa N.R., Papanicolaou N., Vondracek M., Aarum J., Safari H., Muradrasoli S., Rothfuchs A.G., Albert J., Hoegberg B., Reinius B. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-18611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C. Anal. Chem. 2020;92(14):9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- Sun M., Liu S., Wei X., Wan S., Huang M., Song T., Lu Y., Weng X., Lin Z., Chen H., Song Y., Yang C. Angew. Chem. Int. Ed. 2021;60(18):10266–10272. doi: 10.1002/anie.202100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.I., Holliger P. Nat. Protoc. 2015;10(10):1625–1642. doi: 10.1038/nprot.2015.104. [DOI] [PubMed] [Google Scholar]
- Tian J., Liang Z., Hu O., He Q., Sun D., Chen Z. Electrochim. Acta. 2021;387 [Google Scholar]
- Titanji B.K., Farley M.M., Mehta A., Connor-Schuler R., Moanna A., Cribbs S.K., O'Shea J., DeSilva K., Chan B., Edwards A., Gavegnano C., Schinazi R.F., Marconi V.C. Clin. Infect. Dis. 2021;72(7):1247–1250. doi: 10.1093/cid/ciaa879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travascio P., Li Y.F., Sen D. Chem. Biol. 1998;5(9):505–517. doi: 10.1016/s1074-5521(98)90006-0. [DOI] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu Y., Liu L., Wang X., Luo N., Li L. JID (J. Infect. Dis.) 2020;221(11):1770–1774. doi: 10.1093/infdis/jiaa119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zhao J., Lu S., Huang J., Yang X. Anal. Chem. 2019;91(11):6971–6975. doi: 10.1021/acs.analchem.9b01319. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang Y., Chen J., Wang M., Zhang T., Luo W., Li Y., Wu Y., Zeng B., Zhang K., Deng R., Li W. Anal. Chem. 2021;93(7):3393–3402. doi: 10.1021/acs.analchem.0c04303. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Science. 2020;367(6483):1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Jama-J. Am. Med. Assoc. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xu BMJ Br. Med. J. (Clin. Res. Ed.) 2020;368 [Google Scholar]
- Xu L., Li D., Ramadan S., Li Y., Klein N. Biosens. Bioelectron. 2020;170 doi: 10.1016/j.bios.2020.112673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Sci. China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Li Z., Mohammed I., Zhao L., Wei W., Xiao H., Guo W., Zhao Y., Qu F., Huang Y. Signal Transduct. Target. Ther. 2021;6(1) doi: 10.1038/s41392-021-00649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.T., Yang M.J., Ding Y., Liu Y.W., Lou Z.Y., Zhou Z., Sun L., Mo L.J., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z.H. Proc. Natl. Acad. Sci. U.S.A. 2003;100(23):13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Chaput J.C. J. Am. Chem. Soc. 2021;143(24):8957–8961. doi: 10.1021/jacs.1c02664. [DOI] [PubMed] [Google Scholar]
- Zeng L., Li D., Tong W., Shi T., Ning B. Biochem. Pharmacol. 2021;189 doi: 10.1016/j.bcp.2021.114424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Lan T., Lu Y. Adv. Healthcare Mater. 2019;8(6) doi: 10.1002/adhm.201801158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Fang X., Liu X., Ou H., Zhang H., Wang J., Li Q., Cheng H., Zhang W., Luo Z. Chem. Commun. 2020;56(70):10235–10238. doi: 10.1039/d0cc03993d. [DOI] [PubMed] [Google Scholar]
- Zhang S., Cheng J., Shi W., Li K.-B., Han D.-M., Xu J.-J. Anal. Chem. 2020;92(8):5952–5959. doi: 10.1021/acs.analchem.0c00147. [DOI] [PubMed] [Google Scholar]
- Zhang R., Wu J., Ao H., Fu J., Qiao B., Wu Q., Ju H. Anal. Chem. 2021;93(28):9933–9938. doi: 10.1021/acs.analchem.1c02229. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Pandey R., Li J., Gu J., White D., Stacey H.D., Ang J.C., Steinberg C.-J., Capretta A., Filipe C.D.M., Mossman K., Balion C., Miller M.S., Salena B.J., Yamamura D., Soleymani L., Brennan J.D., Li Y. Angewandte Chemie (International. 2021. in English) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Qin G., Niu J., Wang Z., Wang C., Ren J., Qu X. Angew. Chem. Int. Ed. 2021;60(1):432–438. doi: 10.1002/anie.202011419. [DOI] [PubMed] [Google Scholar]
- Zhu L., Ling J., Zhu Z., Tian T., Song Y., Yang C. Anal. Bioanal. Chem. 2021;413(18):4563–4579. doi: 10.1007/s00216-020-03124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel C. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]