Highlights
-
•
We show that the PAH2 domain of SIN3A is a target when it is inhibited from binding to PF1 results in inhibition of invasive phenotype in TNBC.
-
•
Epigenetic repression of integrins expression and downstream pathways results from enhanced binding of KLF9 /SIN3A repressor complex to their promoters.
-
•
Genome wide transcriptomic analysis showed downregulation of multiple invasion related genes. Tumor growth and lung metastasis were markedly decreased in vivo.
-
•
Our studies highlight that PF1 might serve as a gatekeeper for trafficking SID protein binding to PAH2 of SIN3A and has functional role in presentation of different regulatory complexes.
-
•
Blocking the function of PAH2 offers a promising targeted therapy approach for inhibiting the invasive phenotype in TNBC.
Keywords: SIN3A, PF1, SID, PAH2 domain, KLF9, ITGB1, ITGA6 and TNBC
Abbreviations: TNBC, Triple negative breast cancer; PAH, Paired amphipathic helix; SID, SIN3 Interaction domain; ITGA6, Integrin α6; ITGB1, integrin β1; KLF9, Krüppel-like factor 9; HDAC, Histone deacetylases; EMT, Epithelial to mesenchymal transition; CoIP, Co-immunoprecipitation; ChIP, Chromatin-immunoprecipitation; SA-β-Gal, Senescence associated- β-gal
Abstract
SIN3A, a scaffold protein has regulatory functions in tumor biology. Through its Paired amphipathic helix (PAH2) domain, SIN3A interacts with PHF12 (PF1), a protein with SIN3 interaction domain (SID) that forms a complex with MRG15 and KDM5A/B. These components are often overexpressed in cancer. In the present study, we evaluated the role of SIN3A and its interacting partner PF1 in mediating inhibition of tumor growth and invasion in triple negative breast cancer (TNBC). We found profound inhibition of invasion, migration, and induction of cellular senescence by specific disruption of the PF1/SIN3A PAH2 domain interaction in TNBC cells expressing PF1-SID transcript or peptide treatment. Genome-wide transcriptomic analysis by RNA-seq revealed that PF1-SID downregulates several gene sets and pathways linked to invasion and migration. Integrin α6 (ITGA6) and integrin ß1 (ITGB1) and their downstream target proteins were downregulated in PF1-SID cells. We further determined increased presence of SIN3A and transcriptional repressor, KLF9, on promoters of ITGA6 and ITGB1 in PF1-SID cells. Knockdown of KLF9 leads to re-expression of ITGA6 and ITGB1 and restoration of the invasive phenotype, functionally linking KLF9 to this process. Overall, these data demonstrate that specific disruption of PF1/SIN3A, inhibits tumor growth, migration, and invasion. Also, PF1-SID not only inhibits tumor growth by senescence induction and reduced proliferation, but it also targets cancer stem cell gene expression and blocks mammosphere formation. Overall, these data demonstrate a mechanism whereby invasion and metastasis of TNBC can be suppressed by inhibiting SIN3A-PF1 interaction and enhancing KLF9 mediated suppression of ITGA6 and ITGB1.
Introduction
Dysregulation of epigenetic mechanisms is involved in pathogenesis of cancer, including triple negative breast cancer (TNBC) [1]. Currently, patients with TNBCs lack targeted therapy options. We and others have shown that specific interventions can modify cancer epigenetics and we propose that this approach can be utilized as a potential strategy for anti-cancer therapy [2], [3], [4]. We chose to study SIN3A, a scaffold for chromatin modification which is a global regulator of transcriptional repression and predominant isoform expressed in TNBC cells (Fig. SF1A). In mammals, two paralogs of SIN3 are reported (SIN3A and SIN3B) that can target common as well as unique transcriptional targets [5], [6], [7]. SIN3 interacts with Class I histone deacetylases (HDAC1/2) and accessory proteins forming the SIN3/HDAC chromatin modifying complex [8]. Transcription factors that specifically bind to one or more of the four paired amphipathic helical (PAH1–4) domains of SIN3A provide interface for assembly of multiple proteins [9,10]. These proteins bind to SIN3 via their amino acid sequence specific SIN3-interaction domain (SID) and are known as SID proteins. We previously reported, using a MAD1-SID peptide, that blocking binding of MAD1 to the PAH2 domain of SIN3A, strongly decreases tumor growth and metastasis [4,11]. PF1 (also known as PHF12) is the only SID protein known to form a chromatin modifying complex with the epigenetic modulator MRG15 and H3K4me3 histone lysine 4 demethylase KDM5A/B [12], [13], [14]. Components of the PF1 chromatin modifying complex are overexpressed in breast cancer, thereby highlighting the importance of PF1 as epigenetic modifier [15,16]. Moreover, because PF1 is uniquely part of the SIN3 complex and has been shown to be of low affinity, it might be more amenable to therapeutic disruption. This important background information prompted the current work which aims to find ways to disrupt just the SIN3A/PF1 complex, test the biological consequences of this disruption on TNBC phenotype and identify pathways controlled by PF1/SIN3A interaction that might serve as potential target for combatting TNBC. Our experimental results reveal that among factors that are crucial for maintaining the aggressive aspects of TNBC are elevated PF1/SIN3A interaction , and reduced level and interaction of another SID containing protein, Krüppel-like factor 9 (KLF9) with SIN3A PAH2 domain. Low level of KLF9 has been implicated in poor prognosis of patients with breast cancer lines [17]. KLF9 can also modulate gene expression by directly regulating promoter activity of MMP9 and integrin α6 in breast cancer and glioblastoma cells respectively [18,19]. Aberrant expression of integrins, including α6 and β1 has been linked to poor prognosis of cancer patients and other aspects of cancer, including breast cancer invasiveness [20,21]. Each integrin is a dimer of an α and β subunit, and since each subunit can associate with more than one partner, more than 20 dimers have been described [22,23]. We chose to focus on the α6 and β1 subunits because important functions in breast cancer aggressiveness have been previously ascribed to these subunits [19,24,25]. Therefore, although importance of integrins in regulating cancer behavior is well documented, the epigenetic mechanism of their regulation has not been extensively studied. Moreover, it is an established fact that epigenetic regulation is tissue and/or cell specific. Here, we report that disrupting interaction either by stable overexpression of PF1-SID domain or separation, with a highly specific reagent (PF1-SID peptide), of SIN3A PAH2 complex with PF1 in TNBC models, leads to inhibition of invasion, of cell migration, of tumor growth, and of metastasis. Our genome wide transcriptomic analysis identifies the migration pathway as the most modified by the SIN3A/PF1 disruption. We reveal the mechanism of regulation of α6 and β1 subunits - an integrin that is a laminin receptor and a crucial contributor to the migration/invasion/metastasis processes which we document in culture and in vivo. We also show that blocking the SIN3A/PF1 association induces senescence, a mechanism through which this intervention most likely contributes to inhibition of primary tumor growth. Because the therapeutic intervention works equally well in cells expressing the PF1-SID and with cells treated with the peptide which is engineered to penetrate cell membranes, we propose that the complex and the genes it regulates should be considered as a valid strategy for development of anti-TNBC therapy.
Materials and methods
Cell culture and media
The human MDA-MB-231 (Cat# HTB-26) and MDA-MB-157 (Cat# HTB-24) triple negative breast cancer cell lines were purchased from the American Type Culture Collection (ATCC). MDA-MB-231, MDA-MB-157, and mouse 4T1 cells (Cat# CRL-2539) cell lines were maintained as described in [4]. Cell lines were authenticated by short tandem repeat (STR) profiling in accordance with the standard ASN-0002-2011 in February 2019 (Genetica Cell Lines, case number# CX4006499).
Generation of stable cell lines
Cells were transfected with 1 μg of DNA of empty pCMV-3xFlagvector (Stratagene) or same in-house prepared vector containing PF1-SID sequence followed by SV40 nuclear localization signal (CAGCTGAGGCGGCCCTTTGAGCTGCTGATTGCTGCCGCCATGGAGCGGAACCCCACCCAA or QLRRPFELLIAAAMERNPTQ, CCTCCAAAAAAGAAGAGAAAGGTA or PPKKKRKV, respectively) using TurboFect (Thermo Scientific) transfection reagent according to the manufacturer's recommendations. Stable cell clones were selected in G418-containing medium for at least 15 days. Mutation in PF1-SID sequence (QLRRPFELAIAAAMERNPTQ) was made in the similar way using oligonucleotide containing two nucleotide replacements (CTG codon for L was replaced with GCG codon for A). This mutation blocks interaction of PF1-SID with SIN3A [26]. All plasmids were confirmed by sequencing; successfully transfected clones were verified by qPCR using the transgene specific primers.
siRNA transfection
MDA-MB-231 PCMV and PF1-SID cells were transfected with 1 μg of control (sc-44230) or KLF9 siRNA (sc-37716) in triplicates in 6-well plates according to manufacturer's instructions (Santa Cruz Biotechnology). Briefly, cells were treated with control and KLF9 siRNA for 48 h using standard lipofectamine reagent (Santa Cruz) and later harvested for confirming at protein and transcript level with KLF9 specific primers (37716-PR) and our internally designed primers and KLF9 specific antibody (sc-376422).
Generation of SID decoy peptide
PF1-SID and PF1 scramble (SCR) peptides were commercially synthesized (BioSynthesis, Inc) to a purity level of 95%. SID peptide consists of TAT (underlined) sequences followed by N-terminal SID region of PF1 protein (YGRKKRRQGGGQLRRPFELLIAAAMERNPTQ). SCR peptide in addition to TAT peptide has the following sequence: (YGRKKRRQGGGRLFMQLELRATPAEAPINQR). A similar peptide of MAD-SID sequence was previously shown to be rapidly delivered to nuclei of breast cancer cells [11].
Quantitative real-time PCR
RNA was isolated using RNeasy Plus Mini Kit (Qiagen), and cDNA was prepared using Superscript First Strand Synthesis System for RT-PCR Kit (Biorad) following manufacturers’ instructions. Quantitative real-time PCR was performed using manufacturers’ instructions for SSO advanced SYBR green kit (Biorad) on ABI-qPCR system using equal amount of template (50–100 ng cDNA) per reaction. Obtained values were normalized to housekeeping gene (RPL30) and are presented as fold differences over control using the ∆∆Ct method for relative quantifications. Each comparison was made using triplicate reactions and in at least 3 independent experiments. P-values were calculated using Student's t-test. Primer sequences for different analyzed genes are described in detail in (SI Table 1).
Proximity ligation assay (PLA) assay
PLA assay in MDA-MB-231 PCMV and PF1-SID was carried out as described by Bansal et al[11].
Western blot analysis
Cell lysates were obtained using ice-cold Immunoprecipitation lysis (IP) buffer, supplemented with proteinase inhibitor (Thermo Fischer Scientific Pierce), 10 mg/mL PMSF, phosphatase inhibitors sodium orthovanadate (100 mM) and sodium fluoride (100 mM). Protein concentration was determined using BCA kit (Pierce) and 30 μg of protein from each cell lysate was analyzed according to standard SDS/PAGE and Western immunoblotting protocols with specific primary antibodies. β actin, GAPDH or tubulin antibodies were used as loading control. For KLF9 detection, we used 50 μg of protein for loading due to its low basal expression level in the cell lines used for study.
Coimmunoprecipitation assay
Sub confluent cultures of cells were harvested and lysed in IP lysis buffer supplemented with protease inhibitor cocktail as described above. Total protein (1–2 mg) from each sample was immunoprecipitated with antibody amounts as suggested in the manufacturer's datasheets. Briefly, immunoprecipitated proteins were collected using 40 μl of protein A/G agarose beads and each immune precipitate was washed thrice with ice cold RIPA buffer and eluted with 2X loading dye, fractionated on SDS/PAGE. Immunodetection was done with anti-SIN3A, PF1, KDM5A/B and KLF9 antibodies at a dilution of 1:1000 (listed in SI Table 3). For each immunoprecipitation 10% of input lysate and normal mouse IgG were used as internal control.
Three-dimensional (3D) cell morphology studies
3D cell culture assays were performed in 24-well plates (Thermo Fisher Scientific, Inc.) with 400 µl/well of Matrigel™ Matrix Growth Factor Reduced (BD Biosciences). Cells were suspended in complete medium supplemented with 2% Matrigel™, plated at a density of 1 × 103 for 4T1 and 3 × 103 cells/well for MDA-MB-231 and MDA-MB-157 cells respectively and incubated at 37°C for 5–12 days. A fresh layer of complete medium supplemented with Matrigel™ was added following 2 days of incubation, and the medium was replaced every 48 h with fresh DMEM medium supplemented with 2% Matrigel. Colonies formed after 5–7 days were monitored daily and images were captured under phase contrast microscope (Nikon TS100 inverted phase contrast microscope).
Transwell migration, invasion and cell adhesion assays
Cell migration assays were performed in transwell chambers (Corning Life Sciences, Manassas, VA, USA) as described with some modifications [27]. Cells were seeded (5 × 104 cells/insert) in serum-free medium (SFM) (3 Transwells per treatment). Complete culture medium was added to the lower wells as chemoattractant, treated and after 24 h of incubation processed as described [27]. Images of migrated cells (8 images/insert) were captured with ECLIPSE E600 microscope (Nikon Corporation, Tokyo, Japan). Similar procedure was used for invasion assays except the membranes were coated with a layer of Matrigel™ Basement Membrane Matrix (3D culture RGF reduced from R&D), and the seeding density was 5 × 105 cells/insert. For quantitation of migrated and invaded cells, the bound crystal violet was eluted by adding 400 μL of 33% acetic acid into each insert and shaking for 10 min. The eluent from the lower chamber was transferred to a 96-well clear microplate and the absorbance at 590 nm was measured using SpectraMAX 340 pc (Molecular Devices) plate reader. For cell adhesion assay PCMV, PF1-SID and PF1-SID mut cells were dissociated and plated on laminin-coated wells. Cells were photographed at various time points. At the end of 24 h, adherent cells in PCMV, PF1-SID and PF1-SID mut were stained with crystal violet, dissolved with 2% SDS, and quantified spectrophotometrically at 590 nm using a SpectraMAX 340 pc (Molecular Devices) plate reader. Results show relative adhesion measured after subtracting the background absorbance from all values in three independent biological replicates.
Immunofluorescence
MDA-MB-231 or 4T1 cells were cultured in 8-well chambers (BD Biosciences) and fixed with 4% paraformaldehyde/PBS for 15 min at room temperature. For 3D cultures cells were seeded (4 × 103 cells/well) in quadruplicate onto Matrigel (R&D, cultrex) in 8-well culture slides as described by Farias et al. [4]. Confocal microscopy was performed using a Leica SP5 confocal microscope. Colony morphology was determined by phase contrast microscopy (Nikon TS100 inverted phase contrast microscope) and later captured on ECLIPSE E600 microscope (Nikon Corporation, Tokyo, Japan). Cells were stained with respected antibodies as listed in the figures and the dilutions and antibodies used are listed in the supplementary table (Supplementary table 3 SI Table 3).
SA-β-gal assay
Sub confluent cultures of PCMV and PF1-SID cells were serum starved for 48 h and processed according to manufacturer's protocol (Bio-vision SA-beta gal Kit). The percentage of SA-βGal-positive cells was quantified using ECLIPSE E600 microscope (Nikon Corporation, Tokyo, Japan). At least 200 cells were counted per sample in triplicates and results were plotted from three independent biological replicates.
Immunohistochemistry
IHC was performed on paraffin fixed tumor sections as previously described. The slides were incubated with antibody directed against ITGB1, Ki67, CDH1, γ-H2AX and p16. The slides were processed as described by Farias etal [4].
RNA extraction and RNA-Seq
RNA was extracted in biological triplicates from PCMV and PF1-SID cells (n = 3) as described earlier as per manufacturer's instructions (RNesasy Plus mini kit from Qiagen). Libraries were prepared using the Illumina TruSeq Stranded mRNA Library Kit according to the manufacturer's instructions. Libraries were sequenced (paired end, 75 base reads each) on Illumina Next-Seq High-output platform.
RNA-seq data processing and analysis
Assessment of quality of the raw RNA-seq reads was carried out using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The paired end read were then aligned to the human reference genome (hg38) using STAR aligner (version 2.5.1a) allowing up to 3 mismatches [28]. The mapped reads were then counted using Feature Counts. Raw read counts from three independent biological replicates from PCMV and PF1-SID samples (n = 3) were subjected to differential gene expression analysis using DeSeq2 [29]. For generating heatmap representing top expressed genes, mitochondrial genes were removed, and the reads were normalized to obtain transcript per million (TPM) values and log2(TPM) was plotted.
Chromatin immunoprecipitation
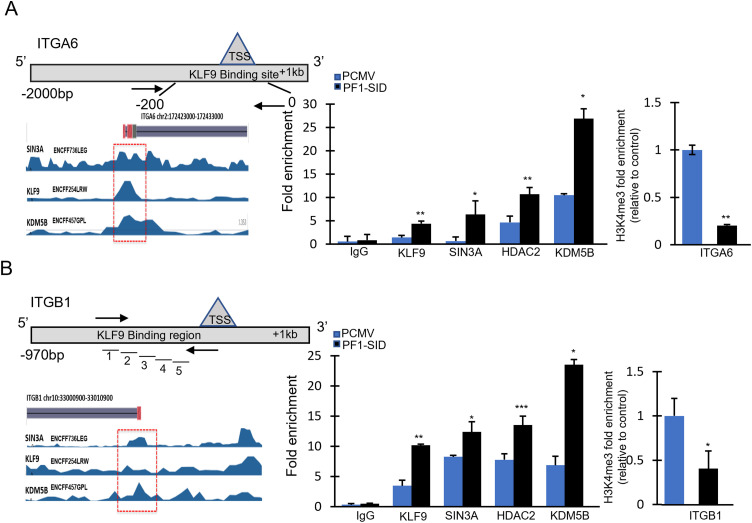
Stably transfected cells or cells treated with the peptide were cross-linked, washed, and lysed according to protocol (EZ-ChIP; Millipore 17–371) followed by chromatin sonication using Qsonica bioruptor with power setting of 50 and 30 cycles of 30 s on/off. Extracted DNA samples from control and test groups (the input sample and ChIP DNA samples) were used for qPCR amplification using primers specific to promoter fragments of the integrin α6 and integrin β1 genes and control primers. Control primers (EZ-ChIP 22–004; Millipore) for the human GAPDH and CDH1 gene were used as an internal and positive control. ITGB1 promoter regions were identified (with data from the University of California, Santa Cruz, Genome Bioinformatics website, http://genome.ucsc.edu); five different primer pairs were designed for the promoter region of the ITGB1 gene using the Primer 3 program (SimGene.com). Primer sequences that were used to amplify the ITGB1 and ITGA6 gene promoter (with position numbers relative to the transcription start site) are shown in schematic of (Fig. 5) and sequences used are listed in (SI Table 2).
Fig. 5.
Disruption of interaction of SIN3A with PF1 enhances formation SIN3A/KLF9/HDAC complex near promoter region of ITGA6 and ITGB1 gene.
(A&B; left panels) describe schematic presentations of DNA fragment bound by these proteins in chromatin. Different primer pairs used for scanning the binding site are labelled as (1–5) to span −1000 bp region upstream of T.S.S. (A&B; middle panel) ChIP analysis of ITGA6 (A) and ITGB1 (B) gene promoters showing fold enrichment for occupancy of KLF9, SIN3A, HDAC2 and KDM5B in MDA-MB-231 PCMV and PF1-SID cells. Isotype antibody was used as background control. (A&B; right panel) shown results of qPCR showing fold change in the recruitment of H3K4me3 to the promoter of ITGA6 and ITGB1 corresponding to fold enrichment changes occurred in A&B middle panel. Error bars = mean ± SD. * p < 0.05 and ** p < 0.02. Average fold enrichment from three independent experimental data set is plotted.
In vivo studies
BALB/c and BALB/nu female mice (8 weeks old) were purchased from the Jackson Laboratory/Charles River Laboratories and maintained under specific pathogen-free (SPF) conditions. All experiments were carried out according to IACUC guidelines. 0.5 × 104 4T1 and 2.5 × 106 MDA-MB-231 cells stably transfected with pCMV-3XFlag or pCMV-3XFlag-PF1-SID vectors were resuspended in serum free RPMI medium and were inoculated in interscapular space of BALB/c or BALB/c nude mice respectively. The size of the tumors from each animal were recorded every 2 or 3 days until the tumors reached 1000mm3 in control groups for 4T1 or 500mm3 for MDA-MB-231 cells. At this stage, the tumors were surgically removed and later the animals were followed for another 1 month. Metastatic dissemination was evaluated by dissecting the lungs from sacrificed mice and inspecting the Bouin-fixed (Sigma) lung surface for lesions using a stereoscope (Nikon SMZ800 stereoscope X3 to X5). Tumor volumes were calculated as ellipsoids (Dxd2 /2) by measuring the main diameter (D) and the smaller diameter (d) and plotted versus time (days). In another pilot experiment, 0.5 x 10410 PCMV 4T1 cells and 2.5 × 104 PF1-SID cells were orthotopically injected into the inguinal mammary gland of Balb/c mice (n = 11 for PCMV and n = 14 for PF1-SID). The experiment was stopped when tumors in the control group reached ∼1000 mm3, then, the mice were sacrificed, tumors were isolated for counting tumor volume and lungs were fixed with bouin's to count lung metastatic nodules.
Statistical analysis
To ensure reproducibility, in-vitro experiments were repeated at least three times unless otherwise indicated. Data were expressed as means ± SDs from at least three independent experiments. Unpaired t-test was used when the results from two groups were compared. Where shown, P-values were calculated using the unpaired Student's t-test, Mann–Whitney, or one-way ANOVA analysis as indicated. A P value of < 0.05 was considered significant.
Results
Disrupting the SIN3A/PF1 interaction inhibits EMT, migration and invasive phenotype in TNBC cells
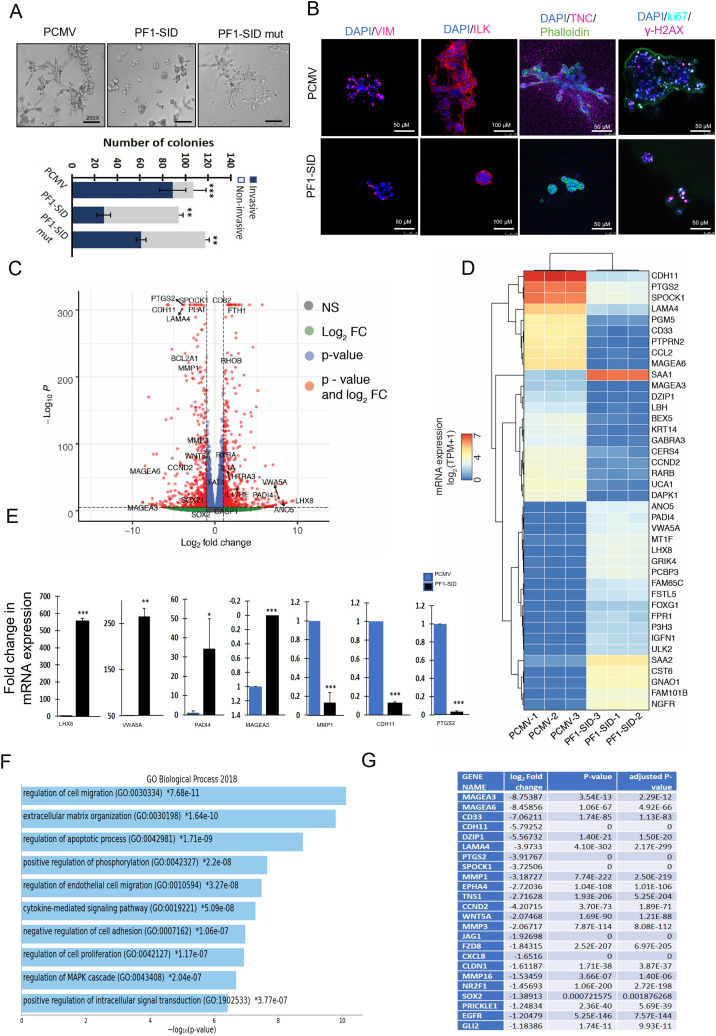
We stably expressed PF1-SID domain of PF1 (PF1-SID) or a mutant form of PF1-SID (PF1-SID mut) in human MDA-MB-231, MDA-MB-157 and mouse 4T1 TNBC cell lines as described in Materials and methods section (2.2). Following generation of stable breast cancer cell lines, we confirmed transgene expression by quantitative PCR (qPCR) and immunofluorescence (IF) analysis of the Flag tag (Fig. SF1B) and showed by proximity ligation assay (PLA) and coimmunoprecipitation the inhibition of SIN3A/PF1 association in MDA-MB-231 cells (Fig. 1A&1B). Coimmunoprecipitation assay also showed reduced association between SIN3A and PF1 complex members KDM5A/B and MRG15 (Fig. 1B). Cells transfected with a PF1-SID single amino acid substitution mutant (PF1-SID mut) that is incapable of interacting with SIN3A PAH2 [26] showed no inhibition of PF1/SIN3A interaction confirming that an intact PF1-SID motif is indispensable for disrupting the interaction (Fig. 1B). Inhibition of SIN3A/PF1 was shown in another human MDA-MB-157 cell line and in mouse 4T1 PF1-SID cell line (Fig. SF1C&D). When grown to low density, MDA-MB-231 cells expressing PF1-SID have cobblestone-like appearance (epithelial phenotype) in comparison to empty vector cells (PCMV) that have elongated fibroblast-like morphology (mesenchymal phenotype) (Fig. 1C). Consistent with these morphological changes we found that expression of CDH1 is upregulated while β-catenin and Vimentin expression is downregulated in PF1-SID cells. (Fig. 1D). These results correspond to our previous observations using MAD1-SID peptide, suggesting that PF1-SID mediated disruption also reverse EMT phenotype [11,30]. These results show that PF1-SID overexpression can disassemble and functionally block the interaction of SIN3A with PF1 and its chromatin modifying complex partners and partially reprogram the EMT phenotype. Consistent with the role of SIN3 paralogs in regulating cell migration [31,32] using Boyden chambers we tested these genetically modified MDA-MB-231 cells for their ability to migrate. Migration of PF1-SID expressing cells was reduced compared to PF1-SID mut cells and PCMV (Fig. 1E; upper panel). The reduced ability of PF1-SID cells to migrate was confirmed in a wound healing assay using MDA-MB-231, MDA-MB-157 and 4T1 PF1-SID cells and MDA-MB-231 cells treated with 2.5 μM of TAT-PF1-SID peptide for 72 h, showing that elevated levels of PF1-SID effectively reduced migration in three different breast cancer cell lines (Fig. SF2A-D). We next tested whether invasion of cells through Matrigel is also impeded by the presence of PF1-SID and found that, like migration, invasion was also inhibited by PF1-SID (Fig. 1E; lower panel). Similar findings were observed with 4T1, MDA-MB-157 cells stably overexpressing PF1-SID and MDA-MB-231 cells treated with TAT-PF1-SID peptide for 72 h (Fig. S2A&B).
Fig. 1.
PF1-SID overexpression disrupts interaction of SIN3A with the protein complex containing PF1, MRG15 and KDM5A/B and inhibits EMT and invasive phenotype. (A) Representative image of Proximity ligation assay (PLA) of SIN3A/PF1 interaction. Red dots represent the presence of SIN3A:PF1 interactions. Scale bar: 25 μM. Quantification is then reported as relative to PCMV biological controls within a given experiment. Bar graph: nuclear red dots as marked by arrows (sites of interaction) plotted as signal per cell; ** p < 0.05. (B) Protein lysates of MDA-MB-231 cells stably expressing PF1-SID, PF1-SID mutant or vector alone were immunoprecipitated and immunoblotted with indicated antibodies. Input corresponds to 10% of the total protein used for immunoprecipitation. Inhibition of interaction (relative to PCMV) is shown below each blot as analyzed by densitometric analysis (C) Phase contrast images of MDA-MB-231 cells showing morphology of overexpressing PF1-SID compared to vector alone (PCMV) at low density. Magnification: 200x. (D) Western blot analysis of the expression of CDH1, β-catenin and vimentin in PF1-SID cells as compared to vector alone. Tubulin expression was used as loading control. (E) Representative experiment of 24 h migration and invasion assays performed with PCMV, PF1-SID and PF1-SID mut cells. Magnification: 200x. Right panel represents the quantification of migration and invasion assays from at-least 4 different replicates.
PF1-SID overexpression inhibits gene sets and pathway related to migration and invasion
Since SIN3A and PF1 are known global transcriptional regulators, to identify gene sets and specific pathways they regulate upon overexpression of PF1-SID specific domain in MDA-MB-231 cells, we conducted an unbiased test of gene expression profile using RNA sequencing. We chose to extract RNA from 3D colonies in Matrigel because they are the best in vitro representation of malignant growth for transcriptomic analysis. Prior to extracting the RNA, we first tested the effect of PF1-SID on the morphology of the 3D colonies and found that while the majority (85%) of 3D colonies formed by parental (PCMV) cells were large and, as indicated by their star-like shape (invasive), the majority (70%) of colonies formed by cells expressing PF1-SID were small and round with smooth edges (non-invasive). PF1-SID mut also resulted in formation of high invasive colonies as compared to cells expressing intact PF1-SID (Fig. 2A). Similar results were obtained in control and PF1-SID expressing MDA-MB-157 and 4T1 PF1-SID cells respectively (Fig. S3A, left panel) and in MDA-MB-231 cells treated with 2.5 μM of PF1-SID peptide for 72 h (Fig. S3A, right panel). Immunofluorescence analysis on 3D colonies formed by PF1-SID cells showed reduction in the expression vimentin, ILK and TNC (markers related to invasion), Ki67 (marker of proliferation) and γ-H2AX (DNA damage maker) (Fig. 2B). These profound phenotypic changes in 3D culture corresponds to reduced invasion and migration observed under 2D conditions and provided good starting base point for studying gene expression profile signatures perturbed upon PF1-SID overexpression. Differential gene expression analysis of 26,485 profiled transcripts using DESeq2 revealed that 887 and 797 genes were, respectively significantly upregulated and downregulated, in the PF1-SID group (Fig. 2C). Top 50 differentially expressed upregulated and downregulated genes as shown in the heat map revealed that PF1-SID overexpression significantly modulated several genes (Fig. 2D). We further validated by qRT-PCR few top up- (LHX8) and down-regulated genes (MAGEA3) as well as for genes with intermediate fold change differences (± 3–7 log fold) (Fig. 2E). Notably, upregulated genes LHX8, VWA5FA and PADI4 are mainly involved in inhibition of tumorigenesis while downregulated genes MAGEA3, MMP1, CDH11 and PTGS2 are known for promoting invasion and migration (Fig. 2E). Using ENRICHR program for pathway analysis we show that the most significant transcriptomic changes are associated with pathways related to: (i) regulation of cell migration, (ii) cell adhesion, (iii) ECM-receptor interaction, (iv) cytokine mediated signaling and (v) pathways in cancer (Fig. 2F and SI Table 1). Most of the downregulated genes in PF1-SID are associated with positive regulation of invasion and migration (Fig. 2G). Similar gene subset was also investigated in MDA-MB-231 PF1-SID grown under 2D conditions, and we observed similar trend of expression profile except for PADI4 and VWA5A that show downregulation in PF1-SID cells instead of upregulation (Fig. S3B).
Fig. 2.
PF1-SID downregulates invasive phenotype and pathways in 3D culture.
(A) Phase contrast of colony morphogenesis of MDA-MB-231 cells expressing PCMV, PF1-SID and PF1-SID mut cells cultured in 3D Matrigel. Lower panel shows quantification of invasive (blue; long extending protrusion), and non-invasive (gray; smooth and few extending protrusions) colonies formed by PCMV, PF1-SID and PF1-SID mut cells, respectively. Error bar represents ± S.D. ** p <0.01 and * p < 0.05; unpaired t-test. (B) Confocal images of colonies formed by PCMV and PF1-SID cells stained with specific antibodies as listed in the figure. Nucleus is counterstained with DAPI stain (blue). Scale bars: 50 µm except for ILK for which scale bar: 100 µm. (C) Volcano plot of RNAseq results depicting differentially expressed genes between PCMV and PF1-SID as analyzed by DeSeq2. (D) Heatmap showing expression of top 50 differentially expressed upregulated and downregulated genes in each of the individual samples (n = 3) from PCMV and PF1-SID group. The scale represents log2 of transcript per million plus one (TPM +1) expression values. We removed MMP1 from the heatmap due to its extremely high expression values in the PCMV samples. (E) qRT-PCR for validation of genes that were differentially regulated in PF1-SID overexpressing cells cultured on 3D basement membrane. Expression of various genes is plotted as fold change in mRNA expression relative to empty vector control PCMV. RPL30 gene is used as internal loading control. Error bars represent ± S.D from three independent biological replicates. *** represents p-value < 0.001; ** represents p-value <0.01 and * represents p-value < 0.05; unpaired t-test. (F) Distribution of biological processes predicted to be modulated in PF1-SID cells based on gene expression profiling. Processes are arranged according to statistical significance, from lowest to cyan highest as analyzed by Enrichr Appyter program. (G) List of genes with log2 fold change values downregulated in PF1-SID cells are shown known for regulating cell migration processes.
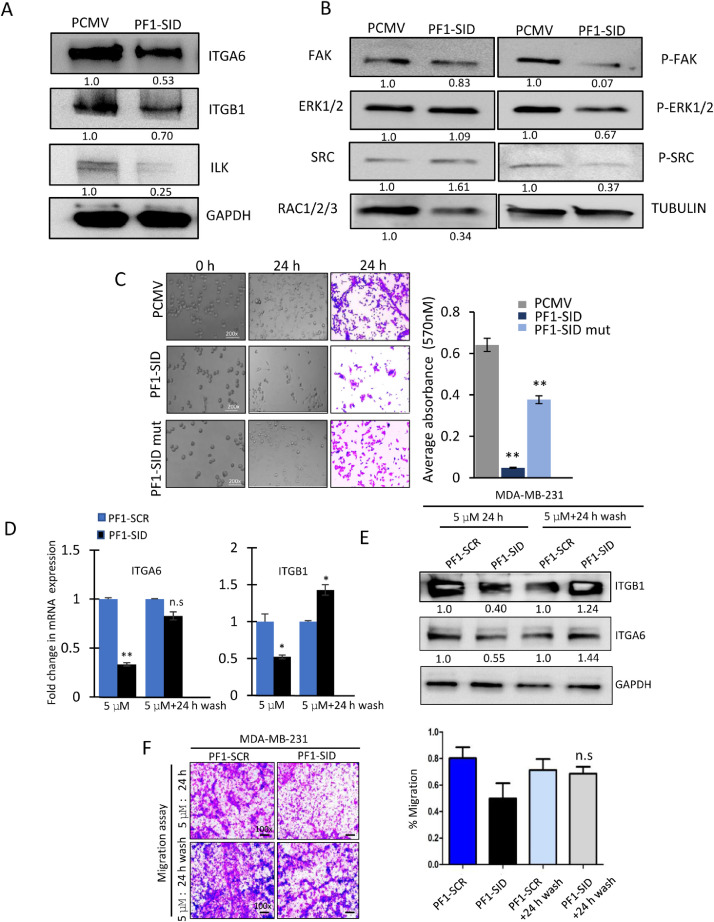
Although pathway analysis revealed several genes of potential importance for migration, for example MAGEA3, MMP3 and PTGS2 our previous work also suggested that the expression of Integrin α6 (ITGA6), an important integrin in breast cancer malignancy, including in migration, might be modulated by the SIN3A-PF1 complex, compelled our current focus. We previously found that in MAD1-SID cells, the H3K4me3 activation signal is decreased on the promoters of ITGA6 and Integrin β1 (ITGB1) [11]. Our unpublished data, in which we used ChIP-seq analysis, showed loss of H3K4me3 activation mark from ITGB1 gene upon MAD1-SID peptide treatment (data not shown). We now tested the expression of IGTA6 and ITGB1 subunits (known to be overexpressed in TNBC cells) and crucial for promoting invasion and metastasis [21] and found reduced level of both proteins in PF1-SID cells (Fig. 3A). We next tested whether reduction in ITGA6 and ITGB1 level causes a reduced activation of the integrin downstream signaling cascade and we found that integrin linked kinase (ILK); a kinase involved in phosphorylation of GSK-3β that regulates β-catenin, as well as p-GSK3β were significantly reduced in PF1-SID cells. Integrin signaling component like phosphorylated FAK, ERK1/2, and SRC, RAC1/2/3, CDC42, RHOA and ITGB4 expression were also inhibited in PF1-SID (Fig. 3B &S2E) suggesting that lowering the integrin level has a profound effect on its function.
Fig. 3.
Continuous blocking of SIN3A-PF1 interaction prerequisite for inhibiting invasive and migratory phenotype. (A&B) Western blot showing expression of integrin α6, β1 and ILK in PCMV and PF1-SID MDA-MB-231 cells.and value written at the bottom indicates the fold activation or repression in PF1-SID cells compared to PCMV analyzed by densitometric analysis. (B) Western blot of indicated proteins regulated by integrin signaling in PCMV and PF1-SID cells. GAPDH and tubulin were used as loading control. (C) PCMV, PF1-SID and PF1-SID mut cells were coated onto laminin-coated culture substrata for 24 h. Cell adhesion was evaluated by phase-contrast microscopy by taking photographs at 0 h and 24 h time point; Magnification 200x. At the end of 24 h, cells are dissolved in acetic acid and absorbance was measured for crystal violet-stained cells. Bar graph represents the quantification of average absorbance for PCMV, PF1-SID and PF1-SID mut cells. (D&E) qRT-PCR and western blot analysis for quantification of transcript and protein levels of ITGA6 and ITGB1 in cells treated with 5 μM of PF1-SID peptide for 24 h followed by 24 h wash. RPL30 and GAPDH were used as internal control for qRT-PCR and western blot assay, respectively. (F) Migration assay of cells treated with PF1-SID peptide for 24 h followed by wash. Magnification: 200x. Right panel represents bar graph for quantification of migration. All experiments were performed in triplicates. Error bars represents mean ± SD. * p < 0.05 and ** p < 0.02.
Since α6β1 is one of at-least 3 recognized laminin receptors, which participate in cell adhesion and spreading [25] we tested whether this function is also affected by PF1-SID overexpression. We found that while the PCMV cells were able to attach and spread within 24 h of cell seeding onto laminin coated plates, the PF1-SID expressing cells seeded onto laminin coated plates hardly adhere and spread poorly, such that 24 h post plating only ∼13% of the cells remains attached to the plate as compared to PCMV. The mutant PF1-SID cells (negative control) spread efficiently as the compare to cells with intact PF1-SID. (Fig. 3C). We also found that treatment of the parental MDA-MB-231 cells for 24 h with the membrane penetrating TAT-PF1-SID peptide, reduced both the transcript and the protein level of the α6 and β1 integrin subunits, while control peptide (TAT-PF1-SCR) was ineffective (Fig. 3D&E). When PF1-SID peptide was washed out, the level of integrins rose and the migratory phenotype was also reversed (Fig. 3F), indicating that the level of Integrin α6 and β1 is controlled by the intact and continuously present SIN3A-PF1 complex.
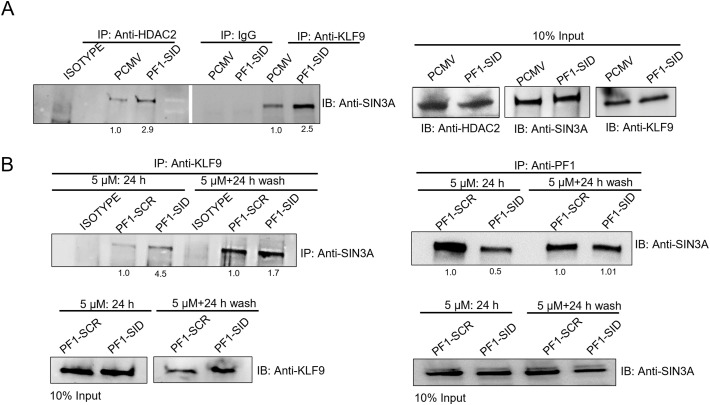
KLF9-SIN3A interaction is enhanced in PF1-SID overexpressing cells
A genome wide analysis of genes regulated by KLF9 identified KLF9 as a transcriptional repressor and regulator of important oncogene related pathways [17,33]. The level of KLF9 in breast cancer tissue was shown to be lower than in normal tissue and its overexpression was shown to block invasion [34]. The genome wide analysis revealed ITGA6 as a gene regulated by KLF9 [19]. Since KLF9 is a SID containing protein and is known to interact with SIN3A via its PAH2 domain, we examined the status of interaction of SIN3A with KLF9 in PF1-SID expressing cells. Using CoIP assay we showed that MDA-MB-231 cells expressing the PF1-SID peptide show a 2.5-fold increase in association between KLF9, SIN3A and HDAC2 (Fig. 4A). A short, 24 h treatment with 5 μM of PF1-SID peptide, but not with scrambled peptide, produced a 4.6-fold increase in KLF9 and SIN3A association, which was lost upon peptide washout (Fig. 4B). The peptide treatment/washout results indicate that the KLF9/SIN3A interaction is reversible and that persistent elevation of KLF9 binding with SIN3A requires continuous disruption of PF1-SIN3A complex.
Fig. 4.
KLF9-SIN3A interaction is enhanced in PF1-SID overexpressing cells.
(A) IP-immunoblot analysis of MDA-MB-231 cells expressing PF1-SID or vector alone were, immunoprecipitated with indicated antibody and probed with anti-SIN3A antibody. Input corresponds to 10% of the total protein used for immunoprecipitation. (B) CoIP assays in MDA-MB-231 cells treated for 24 h with PF1-SID peptide and washed for 24 h and immunoprecipitated with anti-KLF9 and anti-PF1 antibodies and probed with anti-SIN3A. 10% of input was used as internal control and IgG antibody was used as negative control in all experiments. Values at the bottom of IP blots corresponds to densitometric analysis and represent fold activation or repression in case of PF1-SID as compared to PCMV.
Increased recruitment of KLF9/SIN3A/HDAC complex onto ITAG6 and ITGB1 promoter in PF1-SID cells
To query a possible role of SIN3A PAH2 domain function in mediating repression of ITGA6 and ITGB1 we used ChIP assay and found that expression of PF1-SID in breast cancer cells results in concomitant recruitment of KLF9, SIN3A and HDAC2 to the promoter regions of ITGA6 and ITGB1 genes, as shown in the schematic (Fig. 5A&B). The recruitment of KDM5B, a histone demethylase and component of PF1 complex was increased in PF1-SID cells while the recruitment of the activation mark H3K4 trimethylation (H3K4me3) was decreased to ∼80% and ∼60% in the promoters of ITGA6 and ITGB1, respectively (Fig. 5A&B right panel). A promoter of a CDH1 gene, which we previously shown to be activated by the disruption of the SIN3A complex, an effect which we showed to be linked to reversal of EMT [4] showed increased H3K4me3 binding (Fig. S4A). PF1-SID peptide treated MDA-MB-231 cells had similar enrichment for KLF9, SIN3A, HDAC2 and KDM5B on the ITGA6 promoter (Fig. S4B).
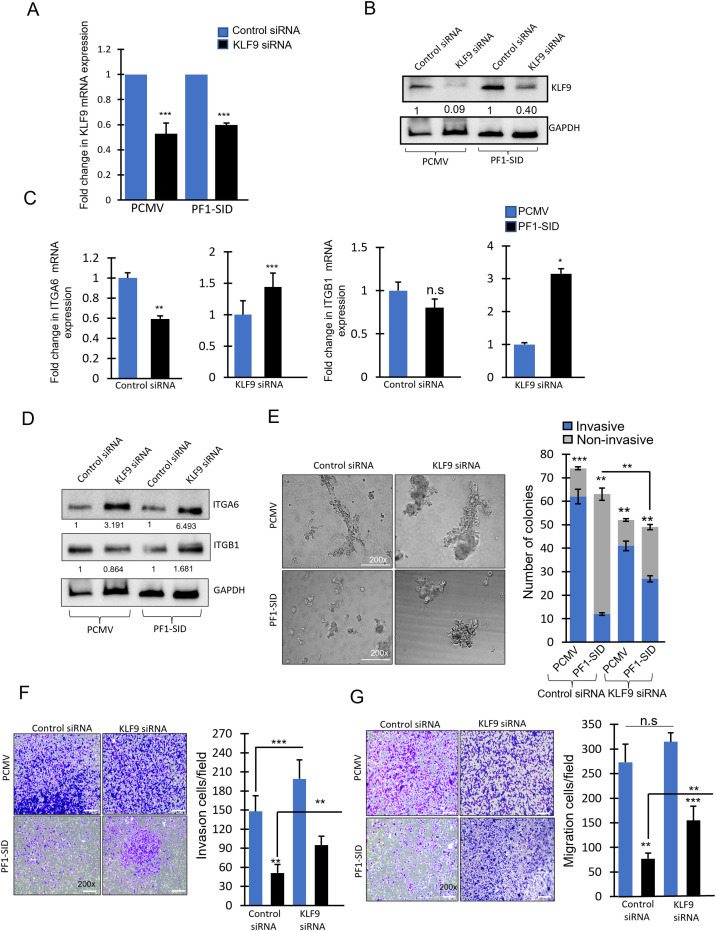
Knockdown of KLF9 relieves repression of integrins genes (ITGA6 and ITGB1)
To directly link KLF9 to downregulation of ITGA6 and ITGB1 expression, we transiently knocked down KLF9. Knockdown of KLF9 was confirmed at both transcript and protein level in MDA-MB-231 PCMV and PF1-SID cells (Fig. 6A &B). Reduction of KLF9 expression by siRNA coincided with significant increase in ITGA6 and ITGB1 expression in PF1-SID cells. As expected, control siRNA did not significantly modify the PF1-SID effect on ITGA6 and ITGB1 (Fig. 6C). A similar effect was observed at the level of protein; KLF9 knockdown increased the integrin protein level in PF1-SID cells while no significant change was observed in PCMV KLF9-siRNA transfected cells (Fig. 6D).
Fig. 6.
Knockdown of KLF9 relieves repression of Integrins genes (ITGA6 and ITGB1). (A) qRT-PCR showing transcript levels of KLF9 in PCMV, and PF1-SID cells transfected with control siRNA and KLF9 siRNA. RPL30 was used as internal loading control. (B) Western blot analysis showing KLF9 expression in PCMV and PF1-SID cells transfected with control or KLF9 (pool of 3 siRNA) siRNA. GAPDH was used as internal loading control. (C) Fold change in mRNA expression of ITGA6 and ITGB1 in PCMV or PF1-SID cells transfected with control or KLF9 siRNA. (D) Western blotting indicating ITGA6 and ITGB1 protein expression levels in PCMV and PF1-SID expressing cells with either control or KLF9 siRNA. GAPDH protein expression was used as loading control. (E) Phase contrast images of colonies formed on the basement membrane by MDA-MB-231 cells stably transfected with PCMV or PF1-SID cells treated with control siRNA or KLF9 siRNA. Bar graph on the right represent the quantification of the invasive (blue) and non-invasive colonies (gray) formed by each group as mentioned in the graph. Magnification: 200x. (F&G) Transwell invasion and migration assays evaluating the effects of KLF9 knockdown on the cellular motility and invasion ability of MDA-MB-231 cells after 24 h. Magnification: 200x. The bar graphs show the number of migrating and invading cells for each category of cells (right). Error bars represent the mean ± SD from at least 3 experiments. Statistically significant data are indicated by * for significance at p value < 0.05, ** and ***p < 0.01; unpaired t-test.
We next tested whether knockdown of KLF9, possibly via its upregulation of integrin expression, will increase invasion. We chose to examine it in 3D colony formation assay in matrigel because we previously showed (Fig. 2A) that PF1-SID is very effective in blocking invasion under these conditions. KLF9-siRNA strongly increased the number and size of invasive colonies formed by PF1-SID cells in matrigel; control siRNA has no effect on colonies grown in 3D matrigel (Fig. 6E). Knockdown of KLF9 with siRNA in PF1-SID cells increased invasion and migration while control siRNA in PF1-SID expressing cells did not interfere with the PF1-SID mediated inhibition of migration while PCMV cells treated with KLF9 siRNA showed increased invasion (Fig. 6F&G). This observation is consistent with the role of KLF9 in inhibiting invasion.
Anti-tumor effect of PF1-SID overexpression- role of cellular senescence?
The anti-tumor effect of PF1-SID was measured in vivo by subcutaneous implantation of MDA-MB-231 PCMV and PF1-SID expressing cells in Balb/c and Balb/c nude mice respectively. Compared to PCMV cells, which in 45 days produced tumors with mean volume of 400mm3, the PF1-SID expressing cells , during the same time formed barely detectable tumor (∼95% volume reduction) (Fig. 7A). A similar tumor growth inhibition (more than 90%) was observed when 4T1 control cells were compared to PF1-SID expressing cells inoculated into immunocompetent Balb/c mice (Fig. 7B). Concordant with the reduced tumor growth we observed reduction in lung metastasis (Fig. 7A&B; right panel). However, we could not conclude if this observed reduction is due to poor formation of tumors by PF1-SID cells or real inhibition of cancer cell dissemination and requires detailed investigation. Additionally, orthotopic injection of 4T1 cellsexpressing PCMV (n = 11) and PF1-SID (n = 14) showed significant reduction in tumor growth and incidence of lung metastasis indicating that PF1-SID overexpression indeed leads to reduction in tumor growth and lung metastasis irrespective of primary injection site (Fig. 7C). In accordance with our findings, that PF1-SID, via KLF9 regulation, controls integrin α6 and β1 expression, we also found that the expression of integrin β1 is significantly reduced in PF1-SID tumor sections in MDA-MB-231 PF1-SID cells (Fig. 7D and Fig. SF5A). The 4T1 PF1-SID tumor sections had increased CDH1 (E-cadherin) protein while Ki67 expression was decreased and show reduction in size of metastatic foci formed in lungs (Fig. 7E&F and Fig SF5B&C).
Fig. 7.
PF1-SID inhibits primary tumor growth and lung metastatisin vivo.
(A&B Left graphs) Tumor progression in Balb/nu and immunocompetent mice implanted with human MDA-MB-231 and mouse 4T1 cells expressing PF1-SID (red) or PCMV (black). Tumor volumes were quantitated at the indicated time points. ***, p = 0.0005; student's t-test. (A&B; right graphs) Quantification of lung metastasis recovered from Balb/nu mice injected with MDA-MB-231 or 4T1 PF1-SID cells as described in A&B. (C) Tumor progression in Balb/c mice orthotopically injected with 4T1 cells expressing PF1-SID and PCMV. Tumor volumes were quantitated at the indicated time points. Day 18, ***, p = 0.0001; p, unpaired t-test. Lungs from sacrificed Balb/c mice inoculated as described in (C) were isolated and quantified for the number of metastasis observed. ***, p > 0.001; p, unpaired t-test =. (D) ITGB1 expression was detected by IHC in tumor sections of animals implanted with MDA-MB-231 PCMV or PF1-SID cells. (E) Same for CDH1 and Ki67 expression for animals implanted with 4T1 PCMV and PF1-SID cells. (F) Representative image of lungs isolated from sacrificed animals injected with 4T1 cells expressing PCMV and PF1-SID showing the presence of metastatic nodules.
Reduction of proliferation (reduced Ki67) and increased CDH1 might affect tumor mass but as previous reports [38,39] suggested that SIN3 and PF1 might be regulating senescence, we tested whether senescence, as defined by increased β-gal (SA-β-gal) expression, was responsible for the observed reduction in tumor growth. We found that MDA-MB-231 PF1-SID cells grown in 2D and 3D conditions showed elevated SA-β-gal expression both conditions (Fig. 8A). MDA-MB-157 and 4T1 PF1-SID cells or MDA-MB-231 cells treated with 2.5 μM of PF1-SID peptide for 72 h also showed a marked increase in SA-β-gal expression (Fig. S6A). Moreover, senescence associated genes (IL-6, IL-1α, IL-1β and Gro-α) were also upregulated in MDA-MB-231 PF1-SID cells (Fig. 8B). Nucleolar protein NPM1 implicated in suppression of senescence was also downregulated in PF1-SID cells at both transcript and protein level (Fig. 8C and Fig SF5D).
Fig. 8.
PF1-SID overexpression promotes cellular senescence.
(A) Representative pictures of SA-β-gal staining for the PCMV and PF1-SID cells in MDA-MB-231 cells under 2D and 3D culture conditions. Magnification: 200x. (B) Fold change in mRNA expression of indicated senescence associated gene markers in MDA-MB-231 serum starved PCMV and PF1-SID cells. (C; left panel) qRT-PCR, measuring fold change in mRNA expression levels of NPM1 in PF1-SID relative to vector control. RPL30 gene expression was used as housekeeping control. Right panel show Immunofluorescence analysis in MDA-MB-231 PCMV and PF1-SID cells for staining of NPM1 (nucleolar staining) in PCMV and PF1-SID cells in 2D conditions. Nucleus was counterstained with DAPI. Scale bar: 50 μM. (D) IHC for p16 and γ-H2AX expression in tumor sections of animals inoculated with PCMV of PF1-SID stably overexpressing MDA-MB-231 cells. Scale bar: 10 µM.
Because it is difficult to reliably stain fixed tumor section for β-gal, we used 2 accepted markers of senescence, p16INK4a (a tumor suppressor) and γ-H2AX (a phosphorylated form of histone H2AX isoform which marks DNA damage). The expression of both markers was increased in tumors formed by MDA-MB-231 PF1-SID expressing cells suggesting that senescence might contribute to tumor growth inhibition (Fig. 8D and Fig. SF5E&F).
Mechanism of anti-tumor effect of PF1-SID; the role of cancer stem cells (CSCs)
Induction of senescence and/or apoptosis and inhibition of proliferation are important mechanisms in reducing overall tumor mass, but they cannot eliminate the tumor formation and prevent it from regrowing. It is currently believed that to accomplish this, cancer stem cells (CSCs) need to be eliminated. To test whether in vivo PF1-SID might target stem cells, we focused our gene expression analysis of tumor sections on stemness and invasion-related genes. qRT-PCR of RNA from MDA-MB-231 PCMV and PF1-SID xenografts tumors showed downregulation of invasion and stemness related genes in PF1-SID tumors while expression of CDH1, linked to reversion of EMT, was increased (Fig. 9A). Gene expression profile of transcript from 4T1 PF1-SID cells formed tumors also showed that CDH1 was upregulated, while stemness, invasion and WNT target genes were significantly downregulated, except for Tenascin C (Fig. 9B). We examined the functional significance of the decreased expression of stemness genes by analyzing mammosphere formation ability of MDA-MB-231 and 4T1 PF1-SID cells that were used for in vivo inoculation. Both MDA-MB-231 and 4T1 PF1-SID cells produced fewer and small size mammospheres (Fig. 9C). We then tested whether PF1-SID affects the protein level of cancer stem cell markers and we found that the expression of cancer stem cell markers Nanog, Sox-2 and Oct-4 proteins linked to stem cell self-renewal, pluripotency, and cancer malignancy [35,36] were downregulated in mammospheres formed by MDA-MB-231 PF1-SID cells (Fig. 9D).
Fig. 9.
PF1-SID overexpression inhibits cancer stem cell phenotype.
(A) Expression of stemness and invasion genes in MDA-MB-231 cell originated tumors as measured by qRT-PCR. (B) Expression of stemness and invasion genes in 4T1 cell originated tumors as measured by qRT-PCR. Error bars represent the mean ± SD from at least 3 experiments. Statistically significant data are indicated by * for significance at p value < 0.05, ** and ***p < 0.01; unpaired t-test. (C) Phase contrast images of mammospheres formed by MDA-MB-231 and 4T1 PCMV and PF1-SID cells. Magnification: 200x. Right panel represents quantification of the mammospheres in MDA-MB-231 and 4T1 cells respectively. (D) Western blot showing expression of Nanog, Sox-2 and Oct-4 protein in MDA-MB-231, PCMV and PF1-SID cells. Tubulin was used as loading control.
Discussion
Our study shows that disrupting the SIN3A/PF1 interaction in TNBC cells, modulates the chromatin modifying complex containing MRG15 and KDM5A/B, an event that leads to epigenetic reprogramming of TNBC cells culminating in reduced cell invasion, diminished tumor growth and increased cellular senescence. We further show that in PF1-SID cells, KLF9 and components of the PF1 complex (SIN3A/HDAC2 and KDM5B), which reside in the regulatory region of ITGA6 and ITGB1 in PF1-SID cells (Fig. 5) identify components of pathways responsible for these biological effects. Together, these results contribute to a potentially new approach to more effective targeted therapies, which might extend to the treatment strategies for poor survival of TNBC patients [37]. In this regard, epigenetic modifiers are currently being investigated as inducers of phenotypic reprogramming of cancer cells and ultimately as cancer therapies [38,39]. SIN3A chromatin scaffold protein has been previously identified as a target that can epigenetically reprogram TNBC cells from more basal to a luminal phenotype [4,11].
Histone demethylase KDM5B, which is part of the PF1 complex, inhibits genes involved in inflammatory response, cell proliferation and cell adhesion in MDA-MB-231 cell line [40,41]. We previously reported that disrupting the interaction of KDM5B with SIN3A results in decrease in H3K4me3 signal and enhances binding of KDM5B to the promoter of ITGA6 gene [11]. Here, we found that in PF1-SID cells, the H3K4me3 activation signal is decreased on the promoter of ITGA6 and ITGB1 genes, critical for regulating invasion and migration pathways. In a related study, using ChIP-seq analysis we showed loss of H3K4me3 activation mark from ITGB1 gene upon MAD-SID peptide treatment (data not shown). KLF9, a SIN3A interacting partner, negatively regulates ITGA6 expression leading to reduced cell spreading and invasive phenotype in the glioblastoma cells [19]. Our ChIP results indicate increased recruitment of KLF9 and components of the PF1 complex (SIN3A/HDAC2 and KDM5B) upstream of ITGA6 and ITGB1 in PF1-SID cells (Fig. 5). Analysis of genome wide binding of proteins using ENCODE identified that both ITGA6 and ITGB1 gene show overlapping peaks for KLF9, SIN3A and KDMB5B indicating their combined role in regulation of these integrins. Further, PF1-SID cells when coated on laminin adhered poorly compared to PCMV and mut PF1-SID cells (Fig. 3C).
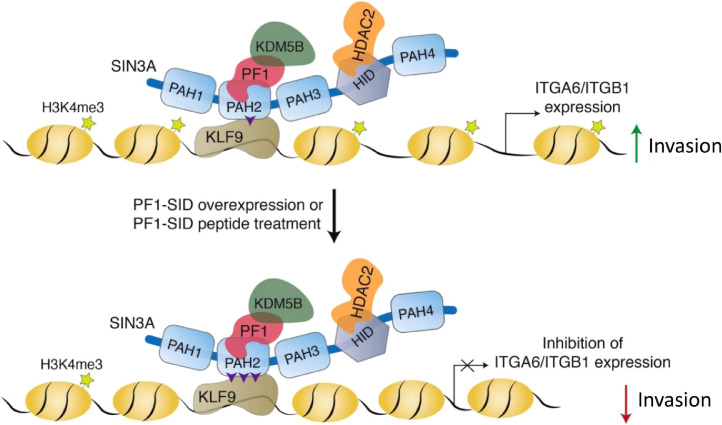
Oncomine database analysis shows that downregulation of KLF9 expression in breast cancer is correlated with poor prognosis and increased metastasis [17]. In our study, knockdown of KLF9 resulted in increased expression of ITGA6 and ITGB1 with subsequent increase in migration and invasion in PF1-SID cells (Fig. 6). The ChIP and transient knockdown assay showed that KLF9 association with SIN3A is upregulated upon PF1-SID overexpression thereby repressing ITGA6 and ITGB1 at mRNA level as shown in the schematic model (Fig. 10) and on protein level.
Fig. 10.
PF1 as a gatekeeper for repression of ITGA6 and ITGB1 via SIN3A PAH2 and KLF9: PF1-SID overexpression or PF1-SID peptide treatment increases the interaction between PAH2 of SIN3A and KLF9 and decreases PAH2 interaction with PF1 as indicated in Fig. 4A&B (for clarity increased interaction is shown by three purple arrows heads and low interaction is shown by one arrowhead). This results in recruitment of KLF9/SIN3A/HDAC2 and histone H3K4me3 demethylase KDM5B near the promoter (KLF9 response element) of ITGA6 or ITGB1 genes. Binding of this chromatin modifying complex induced chromatin changes through increased HDAC2, KDM5B and reduction in H3K4me3 activation mark thus causing gene repression. The ultimate effect of repression of integrins is reflected as decreased Invasion and migratory phenotype.
In conjunction, we found their downstream pathway proteins are also affected in PF1-SID cells. PF1-SID peptide treatment also inhibits ITGA6 and ITGB1 expression and increased recruitment of KLF9/SIN3A/HDAC2 and KDM5B onto the promoter of ITGA6. Diminished tumor growth and lung metastasis correlated with decreased integrin β1 expression in PF1-SID xenografts (Fig. 7). These observations are in agreement with reduced invasion and migration following inactivation of integrins and also with reduced cell spreading in PF1-SID cells.
Structural studies of the mammalian PAH2 domain show conformational heterogeneity enabling SIN3A to regulate its interaction with diverse protein binding partners in a context dependent manner [9,26,42,43]. We found increased physical association between KLF9 and SIN3A in PF1-SID cells whereas short-term PF1-SID peptide treatment increased interaction but was reversed upon peptide removal (Fig. 4). This suggests that the low affinity interactions between SIN3A PAH2 and SID domain proteins are highly dynamic in nature and might allow diverse interaction of partner proteins with SIN3A in a range of repressor proteins. In this context, the SID domain of KLF9 and KLF11 are quite different from the SID of MAD, TGIF1, HBP1 and PF1 in both binding affinity to PAH2 and binding orientation [26,44]. Perturbation of SIN3B PAH2 interactions with a range of SID containing proteins was studied using 3D simulation models. Small molecule, sulfatide mediated confirmational changes that reduced the interaction of MAD1 and HDAC2 with PAH2 domain of SINB and induced the expression of Integrin αV suggesting interaction affinity can be modulated by changing orientation of alpha helix of PAH domains [45]. Importantly, PF1 contains two binding sites for SIN3A; one interacts with PAH1 via PF1-SID2, while the other interacts with PAH2 through PF1-SID1 [14]. Additionally, it has independent binding site for MRG15, a known epigenetic modifier [46]. PF1-SID1 binds PAH2 directly via its amphipathic alpha helix with nanomolar affinity and is known to induce structural changes in PAH2 [14]. The blocking of PF1 interaction with SIN3A PAH2 domain may facilitate the binding of SIN3A PAH1 to PF1-SID2 and enhanced interaction of KLF9 with SIN3A. However, further structural studies are needed to validate this hypothesis and to address the kinetics of various SID proteins binding to PAH1 and PAH2 domains.
In Drosophila, knockout of Sin3 is shown to promote invasion and cell motility [31]. In mammals, Hurst and colleagues described SIN3A as a suppressor of breast cancer progression and metastasis. Knockdown of SIN3A caused an increase in invasive phenotype and increased metastatic potential of breast cancer cells [32]. Interestingly, our study shows that specific targeting of SIN3A (PAH2) domain results in suppression of invasion and metastasis related genes. Our transcriptomic analysis upon SIN3A-PF1 perturbation in 3D culture system showed significant alteration of several genes. Matrix metalloproteases proteins (MMP1 & MMP3) known enhancers of invasion and MAGEA3, CDH11, SPOCK, PTGS2 and WNT5A, overexpressed in high grade of invasive cancer, are all downregulated in PF1-SID cells [47], [48], [49], [50], [51], [52], [53], [54]. LHX8 and its family member LHX6, known to inhibit breast cancer cell proliferation by repressing WNT signaling, is the topmost upregulated gene in PF1-SID cells [55,56]. Knockdown of TGIF1, another SID containing protein showed modulation of EMT, translocation of beta-catenin from nucleus to the cytoplasm and inhibition of WNT and invasion target genes [30]. Consistent with this, our GO term analysis showed that most biological processes, modulated in PF1-SID cells, include regulation of cell migration, cell adhesion, cell proliferation and extracellular matrix organization (SI Table 1). Further, in 2D, we found pFAK, ERK1/2 kinase upstream in the integrin signaling are downregulated in PF1-SID. This is consistent with the activation of these kinase proteins in cohort of invasive breast cancer that confirmed high FAK expression is correlated with increased risk of recurrence and reduced survival in invasive breast cancer [57]. Modulation of these biological processes is also linked with EMT in concordance with previously shown role of PF1 in blocking EMT and established role of SID decoys in suppressing invasion [11,58]. Here, we report that disruption in interaction of SIN3A and PF1 results in reversal of EMT, marked by the increased expression of CDH1 and reduction in the levels of vimentin and β-catenin (Fig. 1D).
Senescence can serve as a barrier against cancer progression or may promote tumorigenesis in a context dependent manner [59]. Like in PF1-SID cells or PF1 SID peptide treated cells (Fig. 8A), knockout of PF1 in MEFs cells increases senescence-associated β-galactosidase (SA-β-gal) activity and nucleolar changes [60]. Sin3B overexpression triggers senescence and underscores the contribution of chromatin modifiers in tumor progression [61]. As we showed increased SA-β-gal, p16 and H2AX in tumor sections produced by implantation of PF1-SID cells, we conclude that the reduction in primary tumor growth could be at least partially attributed to increased senescence. David and coworkers reported that PF1 associates with proteins involved in maintenance of nucleolar integrity and ribosomal biogenesis. PF1 has been shown to interact with NPM1 protein which is overexpressed in tumor cells [60]. In addition, increased level of NPM1 is reported to inhibit senescence and induce cellular proliferation whereas NPM1 knockdown causes structural changes in the nucleolus [62,63]. We found that PF1-SID cells have decreased NPM1 expression at both transcript and protein level (Fig. 8C). The increase in senescence and decrease in NPM1 expression is associated with change in nucleolar morphology as observed in PF1-SID cells (Fig. S6B).
In summary, our findings demonstrated that disruption of PF1/SIN3A-PAH2 interaction suppresses breast cancer progression and metastasis. This is in part mediated via inhibition of expression of ITGA6 and ITGB1 by KLF9 transcription factor that is recruited to their promoters along with KDM5B and SIN3A/HDAC complex. Our studies highlight that PF1 might serve as a gatekeeper for trafficking SID protein binding to PAH2 of SIN3A and has functional role in presentation of different regulatory complexes. Blocking the function of PAH2 offers a promising targeted therapy approach for inhibiting the invasive phenotype in TNBC.
Conclusion
In summary we describe the role of SIN3A-PF1-KLF9 interaction axis in regulating invasive phenotype in breast cancer. We show that the PAH2 domain of SIN3A is a target when it is inhibited from binding to sequence specific proteins such as PF1, often overexpressed in TNBC. This results in inhibition of tumor growth by inducing senescence and blocking cancer cell stemness,invasion and potentially preventing metastasis. Epigenetic repression of integrins expression and downstream pathways results from enhanced binding of KLF9 /SIN3A repressor complex to their promoters is demonstrated as a mechanism for this profound loss of the invasive phenotype. The precise mechanisms that underlie the disruption of interaction of PAH2-SIN3A and PF1 could be further assessed to develop specific peptides and small molecule inhibitors that offer a therapeutic target for preventing TNBC recurrence.
Declaration of Competing Interest
No competing and conflict of interests exists.
Acknowledgments
Acknowledgement
We thank Prof. Liliana Ossowski (Professor Emeritus, Icahn School of Medicine at Mount Sinai) and Dr. Yongkui Jing from Shenyang Pharmaceuticals university for doing the in-depth reading and providing valuable comments for the manuscript. We would also like to thank Almudena Bosch and Prof. Martin Walsh at the Tisch Cancer Institute for providing instrumental support to carry out ChIP experiments. We express our gratitude to Prof. Ming- Ming Zhou at the Department of Pharmacological Sciences, The Tisch Cancer Institute for his critical comments and suggestions throughout the course of study. We acknowledge the help from Karen Triff and Maria Ken Figueroa from University of Miami for providing ChIP-seq data that was used to identify some targets related to integrin signalling.
Declaration
Ethics approval and consent to participate: All animal procedures were performed according to institutional IACUC guidelines, and no human consent was required for the study.
Consent for publication: Not applicable.
Availability of data and materials:
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors upon request. All processed data for RNA-seq are available in the main text or supplementary file. Additional data needed to evaluate the conclusions in the paper are available under BioProject ID PRJNA790640.
Funding
This study was supported by grants from Samuel Waxman Cancer Research Foundation and National Institute of Health-National Cancer Institute grant (R01CA158121).
Authors’ Contribution: Rama Kadamb designed and performed the experiments, analyzed the data, and wrote the manuscript. Boris A Leibovitch was involved in experimental designing and preparation of clones used in the study, performed qRT-PCR, analyzed the data and in writing of the manuscript. Eduardo F. Farias was involved in designing animal experiments and conceived the project. Hemant Suryawanshi carried out RNA-sequencing and analysis. Nidhi Bansal was involved in experiments during early phase of this work and for Proximity ligation assay. Nisha Rani Dahiya carried out western blot experiment to Check SIN3B paralog expression (Fig. S1A). Samuel Waxman conceived the project, designed the experiments, supervised, and wrote the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101320.
Contributor Information
Rama Kadamb, Email: rama.kadamb@einsteinmed.edu.
Samuel Waxman, Email: Samuel.waxman@mssm.edu.
Appendix. Supplementary materials
References
- 1.Berdasco M., Esteller M. Clinical epigenetics: seizing opportunities for translation. Nat. Rev. Genet. 2019;20:109–127. doi: 10.1038/s41576-018-0074-2. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 2.Arkin M.R., Tang Y., Wells J.A. Small-Molecule Inhibitors of Protein-Protein Interactions: progressing toward the Reality. Chem. Biol. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fry D.C., Vassilev L.T. Targeting protein–protein interactions for cancer therapy. J. Mol. Med. 2005;83:955–963. doi: 10.1007/s00109-005-0705-x. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 4.Farias E.F., Petrie K., Leibovitch B., Murtagh J., Chornet M.B., Schenk T., Zelent A., Waxman S. Interference with Sin3 function induces epigenetic reprogramming and differentiation in breast cancer cells. Proc. Natl. Acad. Sci. 2010;107:11811–11816. doi: 10.1073/pnas.1006737107. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadamb R., Mittal S., Bansal N., Batra H., Saluja D. Sin3: insight into its transcription regulatory functions. Eur. J. Cell Biol. 2013;92:237–246. doi: 10.1016/j.ejcb.2013.09.001. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 6.Silverstein R.A., Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 7.Bansal N., Kadamb R., Mittal S., Vig L., Sharma R., Dwarakanath B.S., Saluja D. Tumor Suppressor Protein p53 Recruits Human Sin3B/HDAC1 Complex for Down-Regulation of Its Target Promoters in Response to Genotoxic Stress. PLoS One. 2011;6:e26156. doi: 10.1371/journal.pone.0026156. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grzenda A., Lomberk G., Zhang J.-.S., Urrutia R. Sin3: master scaffold and transcriptional corepressor. Biochimica Et Biophysica Acta Bba - Gene Regul Mech. 2009;1789:443–450. doi: 10.1016/j.bbagrm.2009.05.007. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swanson K.A., Knoepfler P.S., Huang K., Kang R.S., Cowley S.M., Laherty C.D., Eisenman R.N., Radhakrishnan I. HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat. Struct. Mol. Biol. 2004;11:738–746. doi: 10.1038/nsmb798. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 10.Kadamb R., Mittal S., Bansal N., Saluja D. Stress-mediated Sin3B activation leads to negative regulation of subset of p53 target genes. Biosci. Rep. 2015;35 doi: 10.1042/bsr20150122. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Bansal N., Petrie K., Christova R., Chung C.-.Y., Leibovitch B.A., Howell L., Gil V., Sbirkov Y., Lee E., Wexler J., Ariztia E.V., Sharma R., Zhu J., Bernstein E., Zhou M.-.M., Zelent A., Farias E., Waxman S. Targeting the SIN3A-PF1 interaction inhibits epithelial to mesenchymal transition and maintenance of a stem cell phenotype in triple negative breast cancer. Oncotarget. 2015;6:34087–34105. doi: 10.18632/oncotarget.6048. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar G.S., Chang W., Xie T., Patel A., Zhang Y., Wang G.G., David G., Radhakrishnan I. Sequence requirements for combinatorial recognition of histone H3 by the MRG15 and Pf1 subunits of the Rpd3S/Sin3S corepressor complex. J. Mol. Biol. 2012;422:519–531. doi: 10.1016/j.jmb.2012.06.013. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jelinic P., Pellegrino J., David G. A novel mammalian complex containing Sin3B mitigates histone acetylation and RNA polymerase II progression within transcribed loci. Mol. Cell. Biol. 2011;31:54–62. doi: 10.1128/mcb.00840-10. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yochum G.S., Ayer D.E. Pf1, a Novel PHD zinc finger protein that links the TLE corepressor to the mSin3A-histone deacetylase complex. Mol. Cell. Biol. 2001;21:4110–4118. doi: 10.1128/mcb.21.13.4110-4118.2001. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cousineau I., Belmaaza A. EMSY overexpression disrupts the BRCA2/RAD51 pathway in the DNA-damage response: implications for chromosomal instability/recombination syndromes as checkpoint diseases. Mol. Genet. Genom. 2011;285:325–340. doi: 10.1007/s00438-011-0612-5. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai B., Hu Z., Huang H., Zhu G., Xiao Z., Wan W., Zhang P., Jia W., Zhang L. Overexpressed KDM5B is associated with the progression of glioma and promotes glioma cell growth via downregulating p21. Biochem Bioph Res Co. 2014;454:221–227. doi: 10.1016/j.bbrc.2014.10.078. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 17.Limame R., de Beeck K.O., Laere S.V., Croes L., Wilde A.D., Dirix L., Camp G.V., Peeters M., Wever O.D., Lardon F., Pauwels P. Expression profiling of migrated and invaded breast cancer cells predicts early metastatic relapse and reveals Krüppel-like factor 9 as a potential suppressor of invasive growth in breast cancer. Oncoscience. 2014;1:69–81. doi: 10.18632/oncoscience.10. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai X.-.Y., Li S., Wang M., Li X., Yang Y., Xu Z., Li B., Li Y., Xia K., Chen H., Wu H. Krüppel-like factor 9 down-regulates matrix metalloproteinase 9 transcription and suppresses human breast cancer invasion. Cancer Lett. 2018;412:224–235. doi: 10.1016/j.canlet.2017.10.027. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 19.Ying M., Tilghman J., Wei Y., Guerrero-Cazares H., Quinones-Hinojosa A., Ji H., Laterra J. Kruppel-like Factor-9 (KLF9) Inhibits Glioblastoma Stemness through Global Transcription Repression and Integrin α6 Inhibition*. J. Biol. Chem. 2014;289:32742–32756. doi: 10.1074/jbc.m114.588988. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klahan S., Huang W.-.C., Chang C.-.M., Wong H.S.-C., Huang C.-.C., Wu M.-.S., Lin Y.-.C., Lu H.-.F., Hou M.-.F., Chang W.-.C. Gene expression profiling combined with functional analysis identify integrin beta1 (ITGB1) as a potential prognosis biomarker in triple negative breast cancer. Pharmacol. Res. 2016;104:31–37. doi: 10.1016/j.phrs.2015.12.004. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 21.Yin H.-.L., Wu C.-.C., Lin C.-.H., Chai C.-.Y., Hou M.-.F., Chang S.-.J., Tsai H.-.P., Hung W.-.C., Pan M.-.R., Luo C.-.W. β1 integrin as a prognostic and predictive marker in triple-negative breast cancer. Int. J. Mol. Sci. 2016;17:1432. doi: 10.3390/ijms17091432. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porter J.C., Hogg N. Integrins take partners: cross-talk between integrins and other membrane receptors. Trends Cell Biol. 1998;8:390–396. doi: 10.1016/s0962-8924(98)01344-0. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 23.Gilcrease M.Z. Integrin signaling in epithelial cells. Cancer Lett. 2007;247:1–25. doi: 10.1016/j.canlet.2006.03.031. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 24.Simirskii V.N., Wang Y., Duncan M.K. Conditional deletion of beta1-integrin from the developing lens leads to loss of the lens epithelial phenotype. Dev. Biol. 2007;306:658–668. doi: 10.1016/j.ydbio.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belkin A.M., Stepp M.A. Integrins as receptors for laminins. Microsc. Res. Techniq. 2000;51:280–301. doi: 10.1002/1097-0029(20001101)51. https://doi.org/3<280::aid−jemt7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 26.Brubaker K., Cowley S.M., Huang K., Loo L., Yochum G.S., Ayer D.E., Eisenman R.N., Radhakrishnan I. Solution structure of the interacting domains of the Mad–Sin3 complex implications for recruitment of a chromatin-modifying complex. Cell. 2000;103:655–665. doi: 10.1016/s0092-8674(00)00168-9. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 27.Justus C.R., Leffler N., Ruiz-Echevarria M., Yang L.V. In vitro cell migration and invasion assays. J. Vis. Exp. Jove. 2014 doi: 10.3791/51046. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 30.Kwon Y.-.J., Leibovitch B.A., Bansal N., Pereira L., Chung C.-.Y., Ariztia E.V., Zelent A., Farias E.F., Waxman S. Targeted interference of SIN3A-TGIF1 function by SID decoy treatment inhibits Wnt signaling and invasion in triple negative breast cancer cells. Oncotarget. 2014;5:88421–88436. doi: 10.18632/oncotarget.11381. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das T.K., Sangodkar J., Negre N., Narla G., Cagan R.L. Sin3a acts through a multi-gene module to regulate invasion in Drosophila and human tumors. Oncogene. 2013;32:3184–3197. doi: 10.1038/onc.2012.326. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis M.J., Liu J., Libby E.F., Lee M., Crawford N.P.S., Hurst D.R. SIN3A and SIN3B differentially regulate breast cancer metastasis. Oncotarget. 2014;5:78713–78725. doi: 10.18632/oncotarget.12805. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ying M., Sang Y., Li Y., Guerrero-Cazares H., Quinones-Hinojosa A., Vescovi A.L., Eberhart C.G., Xia S., Laterra J. Krüppel-like family of transcription factor 9, a differentiation-associated transcription factor, suppresses notch1 signaling and inhibits glioblastoma-initiating stem cells. Stem Cell. 2011;29:20–31. doi: 10.1002/stem.561. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai X., Jiang X., Liu Y., Wang Y., Jiang X., Song G., Qiu H., Zhang Q. Krüppel-like factor 9 upregulates E-cadherin transcription and represses breast cancer invasion and metastasis. Am. J. Cancer Res. 2021;11:3660–3673. [PMC free article] [PubMed] [Google Scholar]
- 35.Jeter C.R., Yang T., Wang J., Chao H., Tang D.G. Concise review: NANOG in cancer stem cells and tumor development: an update and outstanding questions. Stem Cell. 2015;33:2381–2390. doi: 10.1002/stem.2007. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novak D., Hüser L., Elton J.J., Umansky V., Altevogt P., Utikal J. SOX2 in development and cancer biology. Semin. Cancer Biol. 2019;67:74–82. doi: 10.1016/j.semcancer.2019.08.007. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 37.Jhan J.-.R., Andrechek E.R. Triple-negative breast cancer and the potential for targeted therapy. Pharmacogenomics. 2017;18:1595–1609. doi: 10.2217/pgs-2017-0117. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng Y., He C., Wang M., Ma X., Mo F., Yang S., Han J., Wei X. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct. Target Ther. 2019;4:62. doi: 10.1038/s41392-019-0095-0. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toh T.B., Lim J.J., Chow E.K.-H. Epigenetics in cancer stem cells. Mol. Cancer. 2017;16:29. doi: 10.1186/s12943-017-0596-9. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein B.J., Piao L., Xi Y., Rincon-Arano H., Rothbart S.B., Peng D., Wen H., Larson C., Zhang X., Zheng X., Cortazar M.A., Peña P.V., Mangan A., Bentley D.L., Strahl B.D., Groudine M., Li W., Shi X., Kutateladze T.G. The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep. 2014;6:325–335. doi: 10.1016/j.celrep.2013.12.021. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnes V.L., Laity K.A., Pilecki M., Pile L.A. Systematic analysis of SIN3 histone modifying complex components during development. Sci. Rep.-Uk. 2018;8:17048. doi: 10.1038/s41598-018-35093-0. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Ingen H., Lasonder E., Jansen J.F.A., Kaan A.M., Spronk C.A.E.M., Stunnenberg H.G., Vuister G.W. Extension of the binding motif of the Sin3 interacting domain of the mad family proteins †, ‡. Biochem.-Us. 2004;43:46–54. doi: 10.1021/bi0355645. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 43.Pang Y.-.P., Kumar G.A., Zhang J.-.S., Urrutia R. Differential binding of Sin3 interacting repressor domains to the PAH2 domain of Sin3A. FEBS Lett. 2003;548:108–112. doi: 10.1016/s0014-5793(03)00749-x. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 44.Daftary G.S., Zheng Y., Tabbaa Z.M., Schoolmeester J.K., Gada R.P., Grzenda A.L., Mathison A.J., Keeney G.L., Lomberk G.A., Urrutia R. A novel role of the Sp/KLF transcription factor KLF11 in arresting progression of endometriosis. PLoS One. 2013;8:e60165. doi: 10.1371/journal.pone.0060165. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai Q., Liu Y., Zhu P., Kang C., Xu H., Qi B., Wang R., Dong Y., Wu X.Z. SIN3B promotes integrin αV subunit gene transcription and cell migration of hepatocellular carcinoma. J. Mol. Cell Biol. 2018;11:421–432. doi: 10.1093/jmcb/mjy050. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yochum G.S., Ayer D.E. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and transducin-like enhancer of split. Mol. Cell. Biol. 2002;22:7868–7876. doi: 10.1128/mcb.22.22.7868-7876.2002. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pulukuri S.M.K., Rao J.S. Matrix metalloproteinase-1 promotes prostate tumor growth and metastasis. Int. J. Oncol. 2008;32:757–765. [PMC free article] [PubMed] [Google Scholar]
- 48.Mehner C., Miller E., Nassar A., Bamlet W.R., Radisky E.S., Radisky D.C. Tumor cell expression of MMP3 as a prognostic factor for poor survival in pancreatic, pulmonary, and mammary carcinoma. Gene. Cancer. 2015;6:480–489. doi: 10.18632/genesandcancer.90. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao X., Chen G., Cai H., Wang X., Song K., Liu L., Qiu T., He Y. Aberrantly enhanced melanoma-associated antigen (MAGE)-A3 expression facilitates cervical cancer cell proliferation and metastasis via actuating Wnt signaling pathway. Biomed. Pharmacother. 2020;122 doi: 10.1016/j.biopha.2019.109710. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 50.Chen A., Santana A.L., Doudican N., Roudiani N., Laursen K., Therrien J.-.P., Lee J., Felsen D., Carucci J.A. MAGE-A3 is a prognostic biomarker for poor clinical outcome in cutaneous squamous cell carcinoma with perineural invasion via modulation of cell proliferation. PLoS One. 2020;15 doi: 10.1371/journal.pone.0241551. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Satriyo P.B., Bamodu O.A., Chen J.-.H., Aryandono T., Haryana S.M., Yeh C.-.T., Chao T.-.Y. Cadherin 11 inhibition downregulates β-catenin, deactivates the canonical WNT signalling pathway and suppresses the cancer stem cell-like phenotype of triple negative breast cancer. J. Clin. Med. 2019;8:148. doi: 10.3390/jcm8020148. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen D., Zhou H., Liu G., Zhao Y., Cao G., Liu Q. SPOCK1 promotes the invasion and metastasis of gastric cancer through Slug-induced epithelial-mesenchymal transition. J. Cell. Mol. Med. 2018;22:797–807. doi: 10.1111/jcmm.13357. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh B., Berry J., Shoher A., Ramakrishnan V., Lucci A. COX-2 overexpression increases motility and invasion of breast cancer cells. Int. J. Oncol. 2005 doi: 10.3892/ijo.26.5.1393. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 54.Prasad C.P., Chaurasiya S.K., Guilmain W., Andersson T. WNT5A signaling impairs breast cancer cell migration and invasion via mechanisms independent of the epithelial-mesenchymal transition. J. Exp. Clin. Canc. Res. 2016;35:144. doi: 10.1186/s13046-016-0421-0. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H., Qin J., Jin G., Zou L., Shi J., Han X., Cheng X., Zhang X. Overexpression of Lhx8 inhibits cell proliferation and induces cell cycle arrest in PC12 cell line. Vitro Cell Dev. Biol. 2015;51:329–335. doi: 10.1007/s11626-014-9838-y. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 56.HU Z., XIE L. LHX6 inhibits breast cancer cell proliferation and invasion via repression of the Wnt/β-catenin signaling pathway. Mol. Med. Rep. 2015;12:4634–4639. doi: 10.3892/mmr.2015.3997. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 57.Timbrell S., Aglan H., Cramer A., Foden P., Weaver D., Pachter J., Kilgallon A., Clarke R.B., Farnie G., Bundred N.J. FAK inhibition alone or in combination with adjuvant therapies reduces cancer stem cell activity. Npj Breast Cancer. 2021;7:65. doi: 10.1038/s41523-021-00263-3. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strobl-Mazzulla P.H., Bronner M.E. A PHD12–Snail2 repressive complex epigenetically mediates neural crest epithelial-to-mesenchymal transition. J. Cell Biol. 2012;198:999–1010. doi: 10.1083/jcb.201203098. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodier F., Campisi J. Four faces of cellular senescence. J. Cell Biol. 2011;192:547–556. doi: 10.1083/jcb.201009094. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Graveline R., Marcinkiewicz K., Choi S., Paquet M., Wurst W., Floss T., David G. The chromatin-associated Phf12 protein maintains nucleolar integrity and prevents premature cellular senescence. Mol. Cell. Biol. 2017;37 doi: 10.1128/mcb.00522-16. e00522-16https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grandinetti K.B., Jelinic P., DiMauro T., Pellegrino J., Rodríguez R.F., Finnerty P.M., Ruoff R., Bardeesy N., Logan S.K., David G. Sin3B expression is required for cellular senescence and is up-regulated upon oncogenic stress. Cancer Res. 2009;69:6430–6437. doi: 10.1158/0008-5472.can-09-0537. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang K., Yang J., Yi J. Nucleolar Stress: hallmarks, sensing mechanism and diseases. Cell Stress. 2018;2:125. doi: 10.15698/cst2018.06.139. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeng D., Xiao Y., Zhu J., Peng C., Liang W., Lin H. Knockdown of nucleophosmin 1 suppresses proliferation of triple-negative breast cancer cells through activating CDH1/Skp2/p27kip1 pathway. Cancer Manag. Res. 2018;11:143–156. doi: 10.2147/cmar.s191176. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.