Summary
The type VII secretion system (T7SS) of Mycobacterium tuberculosis secretes three substrate classes: Esx, Esp, and PE/PPE proteins, that play important roles in bacterial physiology and host interaction. Five subtypes of T7SS, namely ESX-1 to ESX-5, are present in M. tb. ESX-4 is the progenitor of T7SS but its function is not understood. We investigated the ESX-4 system in Mycobacterium marinum. We show that ESX-4 of M. marinum does not secrete its cognate substrates, EsxT and EsxU, under the conditions tested. Paradoxically, the deletion of eccC4, an essential component of ESX-4, resulted in elevated secretion of protein substrates of ESX-1 and ESX-5. Consequently, the ΔeccC4 mutant was more efficient in inducing actin cytoskeleton rearrangement, which led to enhanced phagocytosis by macrophages. Our results reveal an intimate crosstalk between the progenitor of T7SS and its more recent duplication and expansion, and provide new insight into the evolution of T7SS in mycobacteria.
Subject areas: Molecular biology, Microbiology
Graphical abstract
Highlights
-
•
ESX-4 of M. marinum does not constitute an active secretion system
-
•
Deletion of eccC4 of ESX-4 increases secretion of ESX-1 and ESX-5 substrates
-
•
ΔeccC4 is more efficient in inducing cytoskeletal rearrangement of macrophages
-
•
Deletion of eccC4 does not affect the virulence of M. marinum in zebrafish
Molecular biology; Microbiology
Introduction
Tuberculosis, caused by Mycobacterium tuberculosis (M. tb), is a leading infectious disease with 10 million active cases and 1.5 million deaths annually. M. tb and the closely related pathogen Mycobacterium marinum (M. marinum) use a dedicated protein secretion system, called type VII secretion system (T7SS), to deliver protein substrates into the extracellular milieu(Abdallah et al., 2007). The structural components and substrates of T7SS are typically encoded in linked gene clusters. To date, five genomic loci encoding T7SS have been identified in M. tb, named esx-1 to esx-5 (Abdallah et al., 2007; Bitter et al., 2009; Gey Van Pittius et al., 2001). M. marinum contains four esx loci, esx-1, esx-3, esx-4, and esx-5 (Newton-Foot et al., 2016), and a partially duplicated region of esx-1 containing pe35/pe68_1/esxB_1/esxA_1 (Damen et al., 2020).
The Esx proteins, which belong to the WxG100 family that contain a WxG motif and are ∼100 amino acids long, are substrates of the T7SS (Pallen, 2002). EsxA (ESAT-6), the first Esx protein that was discovered, is a substrate of the ESX-1 of M. tb (Hsu et al., 2003; Sorensen et al., 1995; Stanley et al., 2003). The secretion of EsxA is facilitated by its chaperone and binding partner EsxB (CFP-10), which contains a conserved T7SS secretion signal (YxxxD/E) and forms a 1:1 heterodimer with EsxA in the cytosol (Renshaw et al., 2002). However, a recent study showed that EsxA and EsxB were differentially secreted in MTBVAC, a phoP and fadD26 double deletion mutant of M. tb, suggesting that the co-secretion of EsxA and EsxB could be decoupled under certain conditions (Aguilo et al., 2017). In wild type (WT) M. tb, the folded EsxA and EsxB heterodimer is exported by the secretion apparatus encoded by genes in the esx-1 locus (de Jonge et al., 2007). Studies in M. tb and M. marinum have identified additional substrates of ESX-1, including EspJ (Champion et al., 2014), EspB (Gao et al., 2004; McLaughlin et al., 2007; Solomonson et al., 2015; Xu et al., 2007), EspK (Champion et al., 2014; Sani et al., 2010), EspE (Sani et al., 2010), and EspF(Sani et al., 2010), which are encoded in the extended region of the esx-1 locus, as well as EspA and EspC (Fortune et al., 2005; Lou et al., 2017; Millington et al., 2011; Xu et al., 2007). EspA and EspC are encoded by the espACD operon, which is distantly located to the esx-1 locus. Interestingly, the flanking regions of the espACD operon in the genomes of M. tb and M. marinum are different (Orgeur and Brosch, 2018). In M. marinum, EsxA_1 and EsxB_1 are also substrates of ESX-1 (Damen et al., 2020).
The ESX-5 system is the other well studied T7SS, mostly in M. marinum. EsxN and EsxM are cognate substrates of the ESX-5. In addition, the PE/PPE family proteins are also substrates of ESX-5(Bottai et al., 2012). These proteins are named after the proline-glutamate (PE) and proline-proline-glutamate (PPE) motifs at their N-terminal homology domain, respectively (Cole et al., 1998). The C-terminal domain varies extensively in sequence and length. Although nonpathogenic mycobacteria only contain a few pe/ppe genes, these gene families are greatly expanded in the genome of M. tb (169 pe/ppe genes) and M. marinum (281 pe/ppe genes) (Gey van Pittius et al., 2006). The majority of PE/PPE is secreted by the ESX-5 system (Abdallah et al., 2009).
Phylogenetic analysis suggests that the ESX-4 system is the ancestor of the other four ESX secretion systems in mycobacteria (Gey Van Pittius et al., 2001). More recent studies have further characterized the origin and distribution of the ESX systems (Dumas et al., 2016; Newton-Foot et al., 2016). ESX-4 is present in all sequenced mycobacterial genomes, but its function remains largely unknown. Esx-4 has only been studied in two rapidly growing mycobacterial species, M. smegmatis and M. abscessus. In M. smegmatis, ESX-4 was shown to be required for the conjugal transfer of DNA in the recipient strain (Gray et al., 2016). M. smegmatis ESX-1 is also intimately involved in a special type of DNA conjugation (distributive conjugal transfer), but this function of ESX-1 does not overlap with that of ESX-4 (Coros et al., 2008). A similar horizontal DNA transfer system was recently found in M. canettii, a clade of tubercle bacilli closely related to M. tb, with the difference that it does not involve the ESX-1 system (Madacki et al., 2021). In M. abscessus, ESX-4 was required for the intracellular survival of the bacteria inside amoebae and macrophages (Laencina et al., 2018). However, the esx-4 locus of M. abscessus harbors eccE4, which is absent in most other esx-4 loci (Laencina et al., 2018). Because M. smegmatis contains only ESX-1, ESX-3, and ESX-4, with ESX-2 and ESX-5 absent, and M. abscessus contains only ESX-3 and ESX-4, lacking ESX-1, ESX-2, and ESX-5, the functions of ESX-4 discovered in these organisms may not be shared by slowly growing mycobacterial pathogens including M. tb and M. marinum (Gey Van Pittius et al., 2001; Newton-Foot et al., 2016).
In this study, we performed a detailed study on the function of ESX-4 in M. marinum, which shares several key virulence factors with M. tb and causes disease in fish and amphibians (Tobin and Ramakrishnan, 2008). We show that although the ESX-4 system of M. marinum does not actively secrete its cognate Esx substrates, EsxU and EsxT, under the conditions we tested, i.e., deletion of eccC4, an essential component of ESX-4 apparatus, resulted in elevated secretion of ESX-1 and ESX-5 substrates. This result reveals an intimate crosstalk between the progenitor of T7SSs, ESX-4, and its more recent duplication and expansion, ESX-1 and ESX-5. Our study has provided new insights into the evolution and function acquisition of T7SS in mycobacteria.
Results
Construction of M. marinum ΔeccC4 mutant
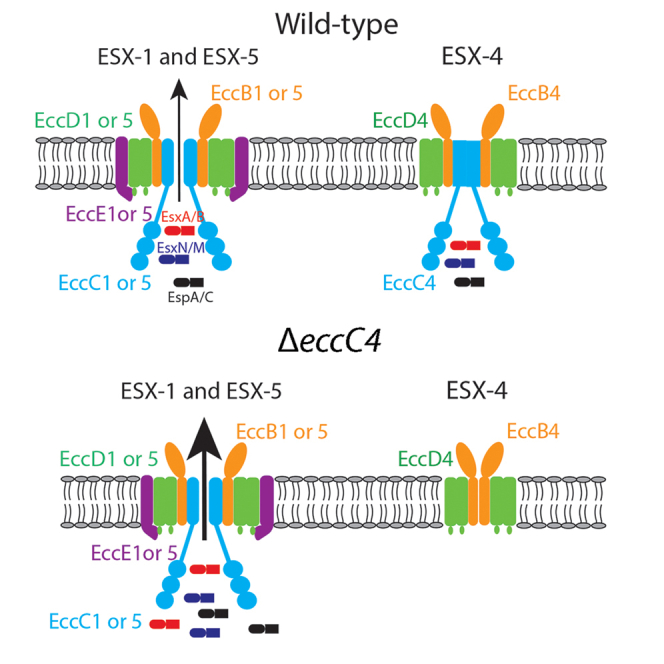
Recent structures of the ESX-5 and ESX-3 secretion apparatuses determined by electron microscopy analysis revealed that the EccB, EccC, EccD, and EccE proteins in each system assemble into oligomeric channels, which provides the pathway for the export of cognate substrates. The substrate secretion is driven by ATP hydrolysis mediated by the ATPase domains of EccC located at the cytosolic face of the inner membrane (Beckham et al., 2017; Famelis et al., 2019; Poweleit et al., 2019). Because EccC provides the energy and is also an integral component of the secretion channel, we constructed an eccC4 deletion strain of M. marinum to study the role of ESX-4 using the specialized phage transducing system (Bardarov et al., 2002).
We successfully deleted the open reading frame (ORF) of eccC4 in M. marinum strains BAA-535 and 1218R (Figure S1). The ΔeccC4 mutant of M. marinum BAA-535 was used for all subsequent experiments.
ESX-4 of M. marinum does not constitute an active secretion system
The ΔeccC4 mutant grew equally well as the WT strain in 7H9 or Sauton media. To determine whether the deletion of eccC4 affects the substrate secretion of ESX-4, we grew the ΔeccC4 and WT strains in Sauton media. The culture supernatant and cell lysate were prepared and analyzed by Western blot using antisera against EsxU and EsxT, which are the predicted cognate substrates of ESX-4. Surprisingly, neither EsxU nor EsxT were detected in the culture supernatant of WT and ΔeccC4 (Figure 1A). As controls, GroEL2, an intracellular protein, was detected in the cell lysate but not in the culture supernatant, whereas antigen 85A and 85B, both secreted proteins, were detected in the culture supernatant and cell lysate.
Figure 1.
EsxT and EsxU were not secreted in M. marinum
(A) Western blot analysis of EsxT and EsxU secretion. The cell lysates (P) and culture supernatants (S) of WT, ΔeccC4, and the complemented strain (COM) were prepared and analyzed by Western blot using antisera raised against each indicated protein. Data are representative of three biologically independent experiments.
(B) Western blot analysis of the EsxT-EsxU fusion protein. P: cell lysates, S: culture supernatant. Data are representative of three biologically independent experiments.
Because EsxA and EsxB of the ESX-1 system were secreted as a 1:1 heterodimer, and to further test the EsxT and EsxU secretion, we introduced a 9-amino acid linker to generate an EsxT-EsxU fusion protein. A FLAG tag was also added to the N-terminus of EsxT. The DNA encoding this fusion protein was cloned into plasmid vector pME, and the resulting construct was transformed into WT and ΔeccC4. Western blot analysis showed that the EsxT-EsxU fusion protein was successfully expressed in both WT and ΔeccC4 cells and was detected in the cell lysate. However, the fusion protein was not detected in the culture supernatant of either strain (Figure 1B).
Taken together, our results suggest that the ESX-4 system of M. marinum does not mediate the secretion of its cognate substrates, EsxT and EsxU, under the experimental conditions we tested.
Deletion of eccC4 affects substrate secretion of ESX-1 and ESX-5
To gain a better understanding on the role of ESX-4 in protein secretion, we performed a proteomic experiment using the Tandem Mass Tag system coupled with LC-MS/MS analysis to identify and quantify proteins in the supernatant of WT and ΔeccC4 cultures. Three independent experiments were performed and the results are shown (Figure 2A). We found that 10 proteins were consistently at higher levels (>1.5-fold, p < 0.05) in the culture supernatant of the ΔeccC4 mutant than the WT strain in each of the three independent experiments. Remarkably, of these 10 proteins, EsxA, EspJ, EspC are the substrates of ESX-1, and PE35, which is encoded in the esx-1 locus, is required for EsxA and EsxB secretion (Brodin et al., 2006; Chen et al., 2013). Other five proteins (EsxN, PE-PGRS62, PE-PGRS20, and PPE10) are substrates of ESX-5 (Figure 2B).
Figure 2.
Higher levels of ESX-1 and ESX-5 substates were detected in the culture supernatant of ΔeccC4
(A) Quantitative proteomic analysis of the culture supernatant of ΔeccC4 and WT. Three independent experiments were performed and the results (ΔeccC4/WT) are shown in volcano plots.
(B) The 10 proteins that were consistently at higher levels (≥1.5 fold, p < 0.05) in the culture supernatant of ΔeccC4 than WT in each of the three independent experiments. Substrates of ESX-1 and ESX-5 are indicated.
(C) Western blot analysis of the C-terminal His-tagged EspJ in the culture supernatant of ΔeccC4 and WT. Data are representative of four independent experiments.
(D) Relative abundance of His-tagged EspJ in the culture supernatant of ΔeccC4 and WT. Data are from four biologically independent experiments and values are plotted as mean ± SD. ∗, p < 0.05, Mann–Whitney U test.
EsxA_1 and EsxB_1, which are substrates of ESX-1 (Damen et al., 2020), also appeared at higher levels in the supernatant of ΔeccC4 cultures than in WT cultures. The level of EsxB_1 was >1.5-fold in the mutant supernatant in two of the three independent experiments (2.1-fold, 1.4-fold, and 2.1-fold) compared to WT. The level of EsxA_1 in the mutant supernatant was 2.5-fold, 1.4-fold, and 1.2-fold of that in the WT in three independent experiments.
To confirm the proteomic data, we cloned the espJ gene containing a sequence encoding a C-terminal His-tag into the expression vector pMV261. The resulting construct was transformed into the ΔeccC4 and WT strains and the secretion of the recombinant EspJ protein was examined. Consistently, the level of recombinant EspJ in the culture supernatant was 2.3-fold higher in ΔeccC4 than WT (Figures 2C and 2D).
Taken together, these results indicate that the deletion of eccC4 of ESX-4 resulted in elevated secretion of the ESX-1 and ESX-5 substrates.
ΔeccC4 exhibits enhanced phagocytosis by macrophages
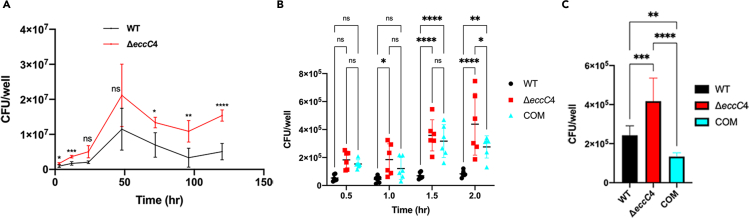
To characterize the function of ESX-4, we next performed macrophage infection experiments with the ΔeccC4 and WT strains. For this experiment, J774 macrophage cells were infected with ΔeccC4 and WT at a multiplicity of infection (MOI) of 0.5, and after a 3-h incubation at 32°C to allow phagocytosis to occur, the cells were washed and incubated with fresh media containing 1 mg/mL gentamicin to kill extracellular bacilli. The intracellular bacteria were enumerated at different time points over a period of 120 h. We found that after the 3-h incubation, the intracellular counts of ΔeccC4 were two times higher than that of WT, which resulted in overall higher intracellular numbers for ΔeccC4 at later time points (Figure 3A). This suggested that ΔeccC4 was more efficient at entering J774 macrophages. To examine this more closely, we repeated the infection experiments with the same MOI but counted the intracellular bacteria at much earlier time points and shorter intervals. Indeed, there is a general trend that the intracellular number of ΔeccC4 was higher than that of WT or the complemented strain during the 2 h infection experiments (Figure 3B).
Figure 3.
ΔeccC4 exhibited enhanced phagocytosis by macrophages
(A) J774 macrophages were infected with ΔeccC4 and WT (MOI = 0.5) for 3 h. The cells were then washed and incubated. At different time points, cells were lysed and intracellular bacteria were enumerated. Data (mean ± SD) are from three biologically independent experiments, each with two technical replicates.
(B) J774 macrophages were infected with ΔeccC4 and WT (MOI = 0.5) at the indicated time points and then the intracellular bacteria were enumerated. Data are from three biologically independent experiments, each with two technical replicates.
(C) BMDMs were infected with ΔeccC4 and WT (MOI = 0.5) for 2 h and intracellular bacteria were enumerated. Data are from three biologically independent experiments, each with two technical replicates. For (A) and (B), two-way ANOVA was performed for statistical analysis. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001; ns, not significant. For (C), one-way ANOVA was performed. ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001; ns, not significant.
To test if the above phenotype could be observed in primary macrophages, we performed the same experiment with bone marrow derived macrophages (BMDMs) isolated from mice. The results showed that ΔeccC4 was more efficient at invading the primary macrophages than WT or the complemented strain (Figure 3C).
ΔeccC4 is more efficient in inducing actin cytoskeletal rearrangement of macrophages
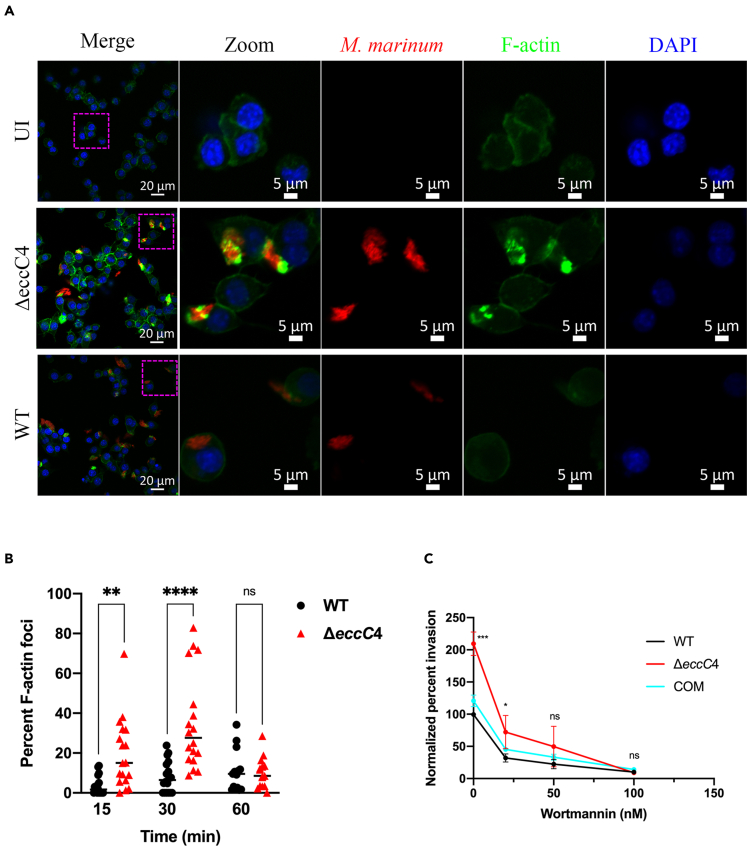
To gain insight into the phagocytosis of ΔeccC4 by macrophages, we performed confocal microscopy analysis. ΔeccC4 and WT were transformed with a plasmid that expresses mCherry and F-actin was labeled with Alexa fluor 488-conjugated phalloidin. At 15 min post infection, ΔeccC4 induced substantially more actin cytoskeletal reorganization and formation of foci than WT (Figure 4A). This phenotype was also observed at 30 min but not at 60 min post infection (Figure 4B), presumably at the latter the phagocytosis process had completed.
Figure 4.
ΔeccC4 was more efficient at inducing actin cytoskeleton rearrangement of macrophages
(A) Confocal microscopy analysis. J774 macrophages were infected with WT or ΔeccC4 carrying a mCherry expressing plasmid for 15 min (MOI = 2). The cells were then fixed, permeabilized, and stained with Alexa fluor 488-conjugated phalloidin. Images are representative of three biologically independent experiments. Highlighted section of the images was zoomed in for better visualization, which shows colocalization of bacteria with the actin foci.
(B) Percentage of actin foci in macrophages infected with WT or ΔeccC4 at the indicated time points post infection. The quantification was done by counting at least 30 cells/field in 6 different fields, and then dividing the number of F-actin foci containing cells by the total number of cells examined. Data are from two independent experiments.
(C) Treatment of macrophages with wortmannin reduced the invasion of M. marinum. J774 macrophages were treated with wortmannin at indicated concentration and then infected with WT, ΔeccC4 or the complemented strain (COM) (MOI = 0.5) for 2 h, and intracellular bacteria were enumerated. Data (mean ± SD) are from three independent experiments. For (B) and (C), two-way ANOVA was performed for statistical analysis. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001; ns, not significant. In (C), only the statistical analysis data between WT and ΔeccC4 was shown. There was no significant difference between WT and COM.
Phosphoinositide 3-kinase (PI3K) plays a critical role in modulating actin cytoskeleton by affecting the cellular levels of phosphatidylinositol 4,5 bisphosphate and phosphatidylinositol (3,4,5)-trisphosphate(Janmey and Lindberg, 2004). To test if the phagocytosis of ΔeccC4 by macrophages is dependent on PI3K, we treated J774 macrophages with wortmannin, a PI3K inhibitor, at different concentrations for 30 min, and then performed the infection experiment with M. marinum for 2 h. Prior treatment of macrophages with wortmannin significantly reduced the invasion of M. marinum and in a concentration dependent manner (Figure 4C). Importantly, the difference in phagocytosis efficiency between ΔeccC4 and WT or the complemented strain was reduced as the concentration of wortmannin increased, suggesting that the enhanced phagocytosis of ΔeccC4 was dependent on PI3K (Figure 4C).
EspJ promotes phagocytosis by macrophages
The enhanced phagocytosis of ΔeccC4 by macrophages could be mediated by one or more proteins that were detected at higher levels in the culture supernatant (Figure 2B). To test this, we expressed and purified recombinant EspJ protein from E. coli (Figure 5A). Fluorescent yellow-green carboxylate-modified latex beads were coated with EspJ or BSA and then used for phagocytosis by J774 macrophages and analyzed by flow cytometry (Figure 5B). Consistent with our hypothesis, we found that coating of latex beads with EspJ significantly enhanced its phagocytosis by macrophages (Figure 5C).
Figure 5.
EspJ promotes phagocytosis by macrophages
(A) SDS-PAGE analysis of the purified EspJ protein. C-terminal His-tagged EspJ was expressed in E. coli and purified.
(B) Fluorescent yellow-green carboxylate-modified latex beads were coated with EspJ or BSA and then used to infect J774 macrophages (MOI = 20) for 2 h and analyzed by flow cytometry.
(C) Quantification of the phagocytosis of beads coated with EspJ or BSA. Results (mean ± SD) are from three independent experiments. One-way ANOVA was performed for statistical analysis. ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗; ns, not significant.
ΔeccC4 is equally virulent as WT in zebrafish
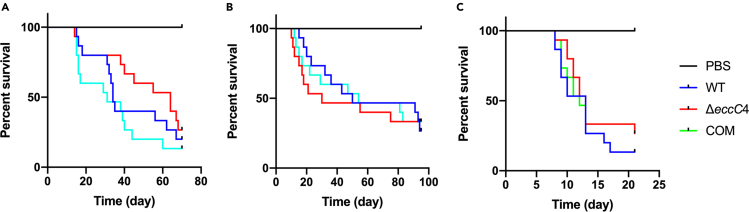
To examine if the enhanced phagocytosis of ΔeccC4 by macrophages observed ex vivo could affect its virulence in vivo, we performed zebrafish infection experiments with ΔeccC4, WT and the complemented strains. Adult zebrafish (N = 15) were infected by intraperitoneal injection with 200 CFU per fish of WT, ΔeccC4, or the complemented strain, and monitored for their survival. Three independent experiments were performed (Figure 6). Log rank analysis found no statistical difference between the survival curves of fish infected with WT, ΔeccC4, or the complemented strain.
Figure 6.
Zebrafish infection with ΔeccC4 and WT of M. marinum
Adult zebrafish (N = 15 per group) were infected by intraperitoneal injection of 200 CFU of WT, ΔeccC4, or the complemented strain and monitored for their survival. The results of three independent experiments (A to C) are shown. There is no statistical difference between the survival curves of fish infected with WT, ΔeccC4 and the complemented strain (log rank analysis).
Discussion
Phylogenetic analysis revealed that the esx-4 genetic locus of M. tb is of ancient origin and is also the only region for which an ortholog could be found in the genomes of other members of high G + C Gram positive bacteria such as Corynebacterium diphtheriae and Streptomyces coelicolor (Gey Van Pittius et al., 2001). It was hypothesized that the esx-4 locus was evolved multiple times in mycobacteria and might take place in the following order: esx-4 → esx-1 → esx-3→ esx-2→ esx-5(Gey Van Pittius et al., 2001). ESX-4-like systems have also been found in plasmids, which appear to be important for the evolution of the ESX systems (Dumas et al., 2016; Newton-Foot et al., 2016). The M. tb complex contains all five ESX systems, and other slowly growing pathogenic mycobacteria such as M. avium and M. marinum contain four ESX systems, whereas rapidly growing mycobacteria such as M. smegmatis contain only three ESX systems (ESX-1, 3, 4) and M. abscessus contains only two (ESX-3 and -4) (Dumas et al., 2016; Newton-Foot et al., 2016). In this study, we performed a detailed characterization of the ESX-4 system in a slowly growing pathogenic mycobacterium, M. marinum. We found that the deletion of eccC4 of esx-4 paradoxically affected the secretion of cognate substrates of ESX-1 and ESX-5. This result reveals an intimate crosstalk between three different ESX systems (ESX-1, ESX-4 and ESX-5) in M. marinum. Izquierdo Lafuente et al. recently expressed a M. tb toxin, CpnT, in M. marinum and found that it is a substrate of ESX-5. Interestingly, the intracellular secretion of CpnT in macrophages requires the combined function of ESX-1 and ESX-4 (Izquierdo Lafuente et al., 2021). Taken together, these findings suggest that different ESX systems may have more functional interplay than previously thought and future studies to elucidate the mechanisms are warranted.
Our results indicate that the ESX-4 of M. marinum does not constitute an active secretion system under the experimental conditions we tested. We did not detect EsxT or EsxU in the culture supernatant of both WT and ΔeccC4 strains. This is in contrast to a recent study of M. abscessus, where the presence of EsxT and EsxU was detected in the culture supernatant of WT, and was reduced in the ΔeccB4 mutant (Laencina et al., 2018). However, the esx-4 locus of M. abscessus contains an eccE4 gene encoding a membrane protein that is absent in M. tb, M. marinum, and M. smegmatis (Laencina et al., 2018). In the structures of ESX-3 and ESX-5 determined by electron microscopy, EccE (EccE3 or EccE5) is a structural component of the secretion apparatus, which together with EccB, EccC, and EccD proteins, assembles into the transmembrane channels (Beckham et al., 2017; Famelis et al., 2019; Poweleit et al., 2019). A copy of eccE gene is also present in the esx-1 and esx-2 loci, although the active transport of substrates has not been demonstrated in ESX-2. It is possible that the absence of eccE4 in the esx-4 locus of M. marinum may compromise its pore-forming capability and consequently cannot transport substrates.
Unexpectedly, although the ESX-4 of M. marinum does not form an active secretion apparatus, the deletion of eccC4 resulted in elevated secretion of substrates ESX-1 and ESX-5 (Figure 2). It is possible that the ATPase domains of EccC4 may interact with substrates of ESX-1 and ESX-5, which act as a ‘sink’ or ‘buffering’ system to allow the coordinated export of cognate substrates by ESX-1 and ESX-5, respectively. This hypothesis is consistent with several observations. Firstly, the substrate specificity of ESX-1, ESX-3, and ESX-5 appears to be mediated by the interactions of the ATPase domains of EccC located at the cytosolic face with its cognate substrates, respectively. For example, the most C-terminal residues, C-terminal to the YxxxD/E secretion signal, of EsxB are required for secretion (Champion et al., 2006). This sequence was also involved in substrate recognition and translocation by binding to the ATPase domains of EccC1 (Rosenberg et al., 2015). Secondly, substrate secretion of the ESX systems appears to be a highly coordinated process because there is a high degree of co-dependency on each other for secretion. For example, the secretion of EspA/EspC by ESX-1 in M. tb is affected by mutations in esxA/esxB and vice versa (Champion et al., 2009; Fortune et al., 2005). It appears that proper functioning of the ESX-1 pathway requires the interaction of multiple substrates with different ATPases before their secretion (Champion et al., 2009). The EccC type ATPase progenitor, EccC4, may retain the ability to bind cognate substrates of ESX-1 and ESX-5. Accordingly, the ΔeccC4 mutant exhibited higher levels of secretion of cognate substrates of ESX-1 and ESX-5, which together likely contribute to the enhanced phagocytosis by macrophages.
Our results showed that disruption of ESX-4 of M. marinum did not affect virulence in zebrafish, suggesting that ESX-4 does not play a major role in virulence in this organism. This is in contrast to the study of M. abscessus, which found that ESX-4 was required for intracellular survival of M. abscessus inside amoebae and macrophages (Laencina et al., 2018). This discrepancy could be explained by the differential evolution and function acquisition of the ESX systems between these two organisms. Of the T7SS in slowly growing mycobacteria including M. tb and M. marinum, ESX-1, ESX-3, and ESX-5 have been functionally characterized. ESX-1 mediates phagosome rupture inside macrophages and the subsequent escape of the bacteria from the phagolysosome, and plays a key role in virulence of M. tb and M. marinum (de Jonge et al., 2007; Hsu et al., 2003; Smith et al., 2008; Tan et al., 2006). ESX-3 and ESX-5 are necessary for iron and fatty acid uptake (Ates et al., 2015; Siegrist et al., 2009). In addition to their roles in nutrient acquisition, ESX-3 and ESX-5 are involved in immune modulation of the host (Abdallah et al., 2008; Tufariello et al., 2016). It appears that although esx-4, the progenitor of the other four esx loci, itself does not play a role in virulence in M. tb and M. marinum, the duplication and expansion (e.g., acquisition of esp and pe/ppe genes) (Abdallah et al., 2008) of this locus during the evolution and host adaptation of these pathogenic organisms have led to more diverse functions of T7SS including roles in nutrient acquisition, virulence, and immune modulation. In contrast, the esx-4 locus might have evolved fewer times in nonpathogenic M. smegmatis and led to different functions, for example, the involvement of ESX-1 in DNA conjugation (Coros et al., 2008). For M. abscessus, a rapidly growing mycobacterium and an opportunistic human pathogen that can cause muco-cutaneous infections, because the esx-4 locus was duplicated only once, the exceptional inclusion of an eccE gene in the esx-4 locus may compensate for the absence of ESX-1 and ESX-5 systems in this organism, and consequently, the ESX-4 plays a more active role in M. abscessus infection of host cells (Laencina et al., 2018).
Limitations of the study
For ESX-4 secretion, we monitored constitutive secretion and cannot exclude that secretion via this system is conditional, and is triggered by some internal or external cues. In addition, we also cannot exclude the possibility that ESX-4 secretes effectors encoded outside the locus and at low levels.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-DDDDK tag Abs | Abcam | Ab1162 |
| Mouse anti- 6 x His tag Abs | Abcam | Ab18184 |
| GroEL2 rabbit antiserum | This paper | N/A |
| Ag85B rabbit antiserum | This paper | N/A |
| Rabbit polyclonal anti-EsxT | This paper | N/A |
| Rabbit polyclonal anti-EsxU | This paper | N/A |
| Ag85A mouse antiserum | This paper | N/A |
| Bacterial and virus strains | ||
| E. coli DH5α | Transgen | Cat# CD201-01 |
| E. coli BL21(DE3) | Transgen | Cat# CD601-02 |
| E. coli NM759 | This paper | N/A |
| Mycobacterium smegmatis mc(2) 155 | ATCC | mc(2) 155 |
| Mycobacterium marinum BAA-535 | ATCC | BAA-535 |
| M. marinum ΔeccC4 | This paper | N/A |
| COM | This paper | N/A |
| M. marinum::mCherry | This paper | N/A |
| M. marinum ΔeccC4::mCherry | This paper | N/A |
| M. marinum::espJ | This paper | N/A |
| M. marinum ΔeccC4::espJ | This paper | N/A |
| M. marinum::esxT-esxU | This paper | N/A |
| M. marinum ΔeccC4::esxT-esxU | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| EspJ | This paper | N/A |
| Alexa fluor 488-conjugated phalloidin | Thermo fisher | A12379 |
| DAPI | Abcam | ab228549 |
| Middlebrook 7H10 | BD difco | Cat# GD-262710 |
| SpeI | NEB | Cat#R3133S |
| XbaI | NEB | Cat#R0145V |
| XhoI | NEB | Cat#R0146V |
| KpnI | NEB | Cat#R3142V |
| PacI | NEB | Cat#R0547V |
| HindIII | NEB | Cat#R3104V |
| NdeI | NEB | Cat#R0111V |
| NheI | NEB | Cat#R3131S |
| MaxPlax Lambda packaging extract | Epicentre | Cat#MP5110 |
| Trizol | Sangon | Cat# B610409 |
| DMEM | Gibco | Cat#11965092 |
| FBS | Gibco | Cat# 10099141C |
| M-CSF | Peprotech | Cat#AF-315-02-100 |
| PrimeScriptTM RT reagent Kit | Takara | Cat#RR037A |
| Tween 80 | Sangon | Cat#A600562 |
| Hygromycin | Sangon | Cat# A600230 |
| kanamycin | Sangon | Cat#A600286 |
| gentamycin | Sangon | Cat# A100304 |
| ampicillin | Sangon | Cat# A610028 |
| Tris | Sangon | Cat# A600194 |
| NaCl | Sangon | Cat# A610476 |
| Triton X-100 | Sangon | Cat# A600198 |
| paraformaldehyde | Sangon | Cat# E672002 |
| urea | Sangon | Cat# A600148 |
| Protease inhibitor cocktail | Sigma | Cat#P8340 |
| IPTG | Sangon | Cat# A600168 |
| Experimental models: Cell lines | ||
| J774A.1 | ATCC | TIB-67 |
| Experimental models: Organisms/strains | ||
| Mouse C57BL/6 | Shanghai SLAC Laboratory Animal Company | N/A |
| Zebrafish AB | EzeRinka | N/A |
| Recombinant DNA | ||
| pJSC284 | This paper | N/A |
| pKOeccC4 | This paper | N/A |
| phLR | This paper | N/A |
| PhLR/pKOeccC4 | This paper | N/A |
| pME | This paper | N/A |
| pME-eccC4 | This paper | N/A |
| pET28a(+) | This paper | N/A |
| pET-SUMO | This paper | N/A |
| pTriEx-4 | This paper | N/A |
| pME-mCherry | This paper | N/A |
| pME-mCherry-FLAG-esxT-esxU | This paper | N/A |
| pMV261 | This paper | N/A |
| Software and algorithms | ||
| Prism 7 | GraphPad | N/A |
| FlowJo VX | BD | N/A |
| Zen black software | Carl Zeiss | N/A |
| Deposited data | ||
| Original western blot images | This paper | https://doi.org/10.17632/jkm8jsfckr.2 |
| Oligonucleotides see Table S1 for this study | ||
Resource availability
Lead contact
Further information and requests for resources and reagents should contact to Dr. Lu Zhang(zhanglu407@fudan.edu.cn).
Materials availability
The antibodies, antiserum, bacterial strains and recombinant DNA generated in this study are listed in the key resources table as “This paper”. All materials generated in this study are available from lead contact upon reasonable request.
Experimental model and subject details
Animals
All animal procedures were approved by the Animal Care Committee at Fudan University. The 6-8 weeks old C57BL/6 female mice were purchased from Shanghai SLAC Laboratory Animal Company and housed under specific pathogen-free conditions in the Animal Center of the School of Life Science of Fudan University. The 3 months wild-type zebrafish were raised and maintained at 28.5°C in a 14-hour light and 10-hour dark cycle under standard husbandry procedures.
Cell line
J774A.1 cells were cultured in DMEM medium supplemented with 10% FBS at 37°C in 5% CO2.
Methods details
Bacterial strains and growth conditions
Mycobacterium marinum BAA-535 were grown in Middlebrook 7H9 broth supplemented with 10% oleic acid-albumin-dextrose-catalase and 0.05% Tween 80 or on Middlebrook 7H10 agar supplemented with 10% oleic acid-albumin-dextrose-catalase at 30 °C. E. coli DH5α and E. coli BL21 (DE3) were grown in Luria-Bertani medium at 37°C.
Construction of the ΔeccC4 mutant of M. marinum
To generate the ΔeccC4 mutant, the open reading frame (ORF) of eccC4 of M. marinum BAA-535 was replaced with a hygromycin resistance gene using the TM4 phage mediated specialized transduction(Bardarov et al., 2002). The primer pairs used to amplify the left and right fragments of eccC4 to form the allelic exchange substrate were eccC4-LF (5ʹ-GGACTAGTCGGCTCAACGACAAGACG-3ʹ), eccC4-LR (5ʹ-CCGCTCGAGTCGGCAGAAATCGGCATC-3ʹ), eccC4-RF (5ʹ-GCTCTAGACAATGCGTGCGGAAAATC-3ʹ) and eccC4-RR (5ʹ-GGGGTACCGTGTCATCGGCTTCGTCG-3ʹ). The resulting PCR products were digested with SpeI/XhoI and XbaI/KpnI restriction enzymes, respectively, and ligated with the 3650 bp fragment of pJSC284 digested with the same enzymes to generate pKOeccC4 which contained a hygromycin resistance gene. After digestion with PacI, pKOeccC4 was ligated with a phasmid phLR digested by PacI. The resulting vector was packaged with the MaxPlax Lambda packaging extract (Epicentre), followed by transduction into E. coli NM759. The resulting phLR/pKOeccC4 phasmid DNA from the transductant was electroporated into M. smegmatis mc2 -155 and plated for mycobacteriophage plaques at the permissive temperature of 30°C. M. marinum BAA-535 was infected with at least 1010 PFU/mL phage lysate which was prepared from a temperature-sensitive phage plaque at 30°C. Hygromycin-resistant colonies were selected after 1-week post-transduction and confirmed by PCR analysis. The primer pair used to PCR confirmation of the ΔeccC4 mutant were P1 (5ʹ-GACCCCGCCAACCCCACC-3ʹ) and P2 (5ʹ-CGGGCAAGACTCGCCACCAT-3ʹ).
To generate the complement strain of ΔeccC4, a DNA fragment containing the intact eccC4 gene and 600 bp upstream of its start codon was amplified by PCR using the forward primer eccC4-CF (5ʹ-GCTCTAGACAGGACGCCGGACGTGA-3ʹ) and the reverse primer eccC4-CR (5ʹ-CCCAAGCTTTCAATGGTGATGGTGATGATGTCCGGGCTCAGTCCAGGC-3ʹ). M. marinum BAA-535 genomic DNA was used as the template. The PCR product was digested with the restriction enzymes XbaI/HindIII, and ligated into pME which contains a kanamycin resistance gene to generate pME-eccC4. The pME-eccC4 plasmid DNA was electroporated into M. marinum ΔeccC4. Transformants were screened on Middlebrook 7H10 agar containing hygromycin (75 μg/ml) and kanamycin (25 μg/ml).
Molecular cloning
For protein expression and purification in E. coli, target genes were amplified from the genomic DNA of M. marinum BAA-535 or M. tb H37Rv using the following primers: esxT (forward: 5ʹ-CGGGATCCGTGGACCCTGTCCTGTCGTACAACT-3ʹ; reverse: 5ʹ-CCCAAGCTTCTAGCGGGCCCATCCGCC-3ʹ), esxU (forward: 5ʹ-CGGGATCCATGAGTAGTCCGGCAGGTGCCA-3ʹ; reverse: 5ʹ-CCCAAGCTTTCACAGGTCCGCGCCGGC-3ʹ), fbpB (forward: 5ʹ-CGAATTCTTCTCCCGGCCGGGGCT-3ʹ; reverse: 5ʹ-ATAGTCGACTCAGCCGGCGCCTAACGAACTCT-3ʹ), groEL2 (forward: 5ʹ-GCGGCTAGCATGGCCAAGACAATTGCGTACGAC-3ʹ; reverse: 5ʹ-GCGAAGCTTTCAGAAATCCATGCCACCCATGTCG-3ʹ). The PCR products were cloned into the pET-28a (+) vector.
The espJ gene was amplified using the primers (forward: 5ʹ-GGAATTCTATGGCTGAGCCTCTGGCCGT-3ʹ; reverse: 5ʹ-CCCAAGCTTTCAGATCGGAGCTGAGACCGAACCC-3ʹ) and was cloned into the pTriEx-4 vector. The fbpA gene was amplified using the primers (forward: 5ʹ-CGAATTCATGGCATTTTCCCGGCCGGGCTTG-3ʹ; reverse: 5ʹ-ATGAGCTCCTAGGCGCCCTGGGGCGCGGG-3ʹ) and the PCR product was cloned into pET-SUMO.
For expression in M. marinum, the DNA fragment expressing the FLAG-EsxT-EsxU fusion protein was constructed by adding a 9-amino acid linker (GGACTAGTACCCCGAGGATCAACAGGA) between esxT and esxU. The esxT gene was amplified from the genomic DNA of M. marinum BAA-535 using the primers (forward: 5ʹ-GTGGACCCTGTCCTGTCGTACAACT-3ʹ; reverse: 5ʹ-TCCTGTTGATCCTCGGGGTACTAGTCCGCGGGCCCATCCGCC-3ʹ). The esxU gene was amplified using the primers (forward: 5ʹ-GGACTAGTACCCCGAGGATCAACAGGAATGAGTAGTCCGGCAGGTGCCA-3ʹ; reverse:5ʹ-TCACAGGTCCGCGCCGGC-3ʹ). Each PCR product was used as a template for overlap PCR amplification using the primers esxT-MF2(5ʹ-GGAATTCCATATGGTGGATTACAAGGATGACGACGATAAGGACCCTGTCCTGTCGTACAACT-3ʹ) and esxU-MR2 (5ʹ-CGGCTAGCTCACAGGTCCGCGCCGGC-3ʹ)(Horton et al., 1990), as such the two ORFs were fused into a single gene. The esxT-MF2 primer encodes a FLAG tag. The fusion cassette was cloned into NdeI and NheI sites of pME-mCherry, which contains a kanamycin resistance gene, forming the expression plasmid pME-mCherry-FLAG-esxT-esxU.
To generate C-terminal tagged EspJ-6xHis for expression in M. marinum, the ORF of espJ was amplified from genomic DNA of M. marinum BAA-535 using the primers (forward: 5ʹ-GGAATTCATGGCTGAGCCTCTGGCCGT-3ʹ; reverse: 5ʹ -CCCAAGCTTTCAATGGTGATGGTGATGATGGATCGGAGCTGAGACCGAACCCGT-3ʹ. The PCR product was cloned into the EcoRI and HindIII sites of pMV261.
Reverse transcription and polymerase chain reaction (RT-PCR) analysis
Mycobacterial cultures (5 mL, OD600 = 1.0) were pelleted and resuspended in 1 mL Trizol. Cells were disrupted by bead beating. The supernatant was then extracted with chloroform: isoamyl alcohol (24:1) and precipitated with isopropanol. Crude RNA samples were treated with gDNA Eraser and reversely transcribed using the PrimeScriptTM RT reagent Kit (Takara) according to the manufacturer’s protocol. The resulting cDNA was used as the template for PCR amplification using primers specific to eccC4 (forward: 5ʹ-CTGTCTGGACTTCGGTGG-3ʹ; reverse: 5ʹ-ATCCTTCAGTGACCCCCT-3ʹ) and sigA (forward: 5ʹ-GCGCCTACCTCAAGCAGAT-3ʹ, reverse: 5ʹ-TGAGGTCCAGAAACGCCATC-3ʹ).
Recombinant proteins purification and antisera preparation
To express the recombinant protein, the pET-28a (+), pet-SUMO or pTriEx-4 constructs were individually transformed into E. coli BL21(DE3) and plated on LB agar containing kanamycin (50μg/mL) or ampicillin (100 μg/mL). Single colonies were randomly selected and grown in LB broth to Log phase and subcultured to 1 L after overnight incubation at 37°C. To induce the expression of protein, E. coli BL21 cultures were grown at 37°C to OD600 = 0.6 and added with 0.5 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) shaking at 220 rpm for 4 h. EspJ, GroEL2, Ag85A and Ag85B proteins were purified from the soluble fraction. The culture was collected by centrifugation at 12,000 rpm for 10 min at 4°C, and resuspended with 1 x PBS. The suspension was sonicated to break the cells. The cell lysates were centrifuged at 12,000 rpm for 20 min at 4°C and the supernatant was collected. The collected supernatant was subjected to Ni-NTA His⋅Bind Resin (Novagen) and purified following the protocol recommended by the manufacturer. The purified proteins were digested using UPL1 at 4°C overnight to remove the SUMO tag. EsxT and EsxU proteins were purified from inclusion body. After IPTG induction, the cultures were collected by centrifuge and resuspended with PBS. The suspension was subjected to sonication to break the cells. The cell lysate was centrifuged at 12,000 rpm for 20 min at 4°C and the pellet collected. The collected pellet was resuspended with 20 mM Tris, pH 8.0, 100 mM NaCl, and 8 M urea, which was then subjected to Ni-NTA His⋅Bind Resin. The urea and imidazole of purified proteins were removed using microfuge tubes with a low molecular weight cut off filter via centrifuge.
Preparation of rabbit anti-sera was conducted following a previous publication (Koul et al., 2000) with minor modification. 500 μg purified recombinant proteins (GroEL2, Ag85B, EsxT or EsxU) and 1 mL of Freund's complete adjuvant mixture was injected into rabbits. Subsequently, three injections of 250 μg each mixed with 1 mL of Freund's incomplete adjuvant were given with an interval of 15 days. Ten days after the final injection, rabbits were bled, and their sera collected. The antibodies against EsxT and EsxU were isolated by passaging the immunized rabbit sera on protein A agarose (Santa Cruz). To prepare the antisera against Ag85A, 10μg purified Ag85A recombinant protein (100 μL) mixed with 100 μL Freund's incomplete adjuvant (Sigma) was injected subcutaneously into C57BL/6 mice. The immunization procedure was repeated two more times (2 weeks apart). Two weeks after the last immunization, mice were sacrificed and their sera collected.
Short-term culture filtrate production and immunodetection
Short-term culture filtrates were prepared following a previous publication (Conrad et al., 2017) with some modifications. M. marinum were grown in Middlebrook 7H9 media with 0.05% Tween-80 to stationary phase. The bacteria pellets were then collected by centrifugation and washed twice with sterilized 1x PBS and transferred to modified Sauton's liquid medium (0.2 g/L KH2PO4, 0.5 g/L MgSO4.7H2O, 2 g/L citric acid, 0.05 g/L ferric ammonium citrate, 60 mL/L glycerol, 4.0 g/L asparagine, pH 7.4) with 0.05% Tween-80, diluted to OD600 = 0.8, and shaking for 48h at 30 °C. Bacteria pellets were harvested by centrifugation at 4000 rpm for 20 min. To prepare whole cell lysates, the bacteria cells were resuspended in 1mL PBS with 1mM PMSF and lysed by sonication. Secreted protein fractions (culture supernatant) were prepared by filtering culture supernatants through a 0.22-μm filter, supplemented with 1 mM PMSF, and concentrated ∼200-fold with an Amicon 3-kDa centrifugal filter (Merck Millipore). The protein concentration in the culture filtrate and bacterial whole cell lysates were quantified using the Bradford assay (Bio-Rad) according to the manufacturer's instruction. A total of 20 μg proteins from culture filtrates or whole cell lysates was separated by SDS-PAGE. Presence of FLAG-EsxT-EsxU, EsxT, EsxU, EspJ-His, Ag85A, Ag85B and GroEL2 were assayed by Western blot, using the following antibodies: Rabbit anti-DDDDK tag Abs (1:5000; Abcam ab1162), rabbit anti-EsxT Abs (1:100), rabbit anti-EsxU Abs (1:100), mouse anti- 6 x His tag Abs (1:4000; Abcam ab18184), rabbit anti-Ag85B or anti-Ag85A sera (1:500) and rabbit anti-GroEL2 serum (1:500).
Tandem mass tagging (TMT) proteomics analysis
The preparation of secreted protein (culture supernatant) was performed as described above. The concentrated protein was dissolved in lysis buffer (8 M urea, 1% Protease Inhibitor Cocktail). The suspension was centrifuged at 12000 g for 10 min at 4°C to remove the debris. The supernatant was collected and the protein concentration was measured according to the manufacturer's instructions of BCA kit. For digestion, 5 mM dithiothreitol was used to reduce the protein solution at 56°C for 30 min. Then, alkylation was performed with 11 mmol/L iodoacetamide at room temperature in darkness for 15 min followed by adding 200 mM TEAB to dilute urea concentration less than 2 M. Subsequently, proteins were digested with trypsin for the first digestion overnight at 37°C (trypsin: protein = 1:50, mass ratio) and successively for a second 4 h digestion (trypsin: protein = 1:100, mass ratio). The peptide was desalted by Strata X C18 SPE column (Phenomenex) and vacuum freeze-dried after trypsin digestion. The dried peptide was reconstituted in 0.5 mol/L TEAB followed by labelling with TMT kit according to the manufacturer's instructions. The TMT labeled peptides were firstly fractionated into 60 fractions and combined into 10 fractions which were dried by vacuum centrifuging by high pH reverse-phase HPLC using Agilent 300Extend C18 column (5 μm particles, 4.6 mm ID, 250 mm length). LC−MS/MS analysis was performed using the EASY-nLC 1000 UPLC system coupled with a Q ExactiveTM Plus (Thermo) mass spectrometer with a nanospray ionization (NSI) source. Maxquant search engine (v.1.5.2.8) was applied to process the resulting MS/MS data. The obtained peptide sequences were searched against the UniProt M. marinum (strain BAA-535) (5418 sequences) database concatenated with a reverse decoy database. The false discovery rate (FDR) was adjusted to <0.01. Volcano plot was generated using Graphpad Prism 7.0.
Macrophage infection assay
To generate bone marrow-derived macrophages (BMDMs), bone marrow was isolated from 6-8 weeks old wild-type C57BL/6 mice. Macrophages were generated by culture of bone marrow cells in medium (DMEM with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 1x β-ME, 10 mM HEPES, and 20 ng/ml recombinant murine M-CSF from Peprotech) for 6 days prior to use in the designated assays.
To determine the intracellular replication of M. marinum, J774A.1 and BMDMs cells were seeded into 24-well plates at a density of 3 × 105 cells per well in DMEM medium supplemented with 10% FBS at 37°C in 5% CO2 overnight. Cells were infected with M. marinum at a multiplicity of infection (MOI) of 0.5 at 32°C for 3 hr in 5% CO2. Cells were then washed three times gently with sterilized 1 × PBS and incubated at 32°C in 5% CO2 for an additional 1 h with fresh medium containing 1 mg/mL gentamycin to kill extracellular bacilli. Cells were again washed twice with PBS and subsequently, incubated at 32°C in 5% CO2 in fresh media with 20 μg/mL gentamycin. At different time points, the infected macrophage monolayers (3-4 wells per strain) were washed 3 times with sterilized 1 × PBS and then lysed with 1 mL of 1% Triton X-100 to release intracellular mycobacteria. The number of intracellular mycobacteria was enumerated by plating appropriate dilutions on Middlebrook 7H10 agar plates containing appropriate antibiotics. Three independent experiments were performed.
Immunofluorescence confocal microscopy
J774A.1 cells (5 x 105) were cultured in 20-mm diameter glass-bottom dishes overnight at 37°C in 5% CO2 and infected with M. marinum expressing mCherry at a MOI of 2. At each time point, infected cells were washed twice with 1x PBS and fixed with 4% paraformaldehyde in 1x PBS for 10 min. Cells were then permeabilized with 0.1% Triton X-100 in 1x PBS for 5 min, blocked with 1% BSA in 1x PBS for 1h, and stained with Alexa fluor 488-conjugated phalloidin (Thermo scientific A12379) and DAPI (Abcam, ab228549) for 20 min following the protocol recommended by the manufacturer. Stained cells were protected with ProLong live antifade reagent diluted with 1x PBS. The fluorescence images were documented using a Zeiss LSM880 confocal microscope and Zen black software.
Flow cytometry-based latex beads phagocytosis assay
A 2.5 μL sample of a stock suspension of fluorescent yellow-green carboxylate-modified latex beads (Sigma L4655) was mixed with 0.2 mL PBS containing 30 μg BSA or EspJ for 2 hr at 4°C, then centrifuged at 15000 rpm for 10 min. The supernatant was removed. Protein coated latex beads were resuspended with equal volume DMEM. J774A.1 cells were cultured at a density of 3 × 105 cells per well in 24-well plates at 37°C in 5% CO2 overnight. J774A.1 cells were incubated with 30 μL latex beads suspension at a MOI of 20 for 2 hr at 37°C in 5% CO2. Cells were detached and washed four to five times with ice-cold PBS, then were fixed with 4% paraformaldehyde and analyzed on a BD FACS Calibur flow cytometer. Flow cytometry data were analyzed using FlowJo software.
Zebrafish infection
Zebrafish infection with M. marinum was performed following a previous publication (Wu et al., 2017). A dosage of 200 CFU bacteria per fish was applied to infect adult zebrafish intraperitoneally followed by monitoring their survivals. PBS was used as the negative control.
Quantification and statistical analysis
Statistical analyses were performed by GraphPad Prism version 7.0 software (GraphPad, San Diego, CA, USA). Two-tailed Student's test or Mann–Whitney U-test was used to determine the statistical significance between two samples (∗p < 0.05). For more than two samples, statistical significance was determined by one-Way ANOVA or two-way ANOVA follow by Bonferroni test with a statistical threshold of p < 0.05. Error bars depict mean SD.
Acknowledgments
This study was supported by grants from the National Key R&D Program of China (No.2018YFD0500900), China’s 13th Five Year Programs for the prevention and cure of great infectious diseases (2018ZX10731301-001-005), and grants from Canadian Institutes of Health Research (CIHR) PJT-156261 and PJT-173353 to JL.
Author contributions
J.L. conceptualized the work. Y.W., Y.T., C.L., J.Z., J.M., J.J. and X.G. performed the experiments. L.Z., J.L. and G.Z., Y.L. supervised the experiments. Y.W. and J.L. analyzed the data and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103585.
Contributor Information
Lu Zhang, Email: zhanglu407@fudan.edu.cn.
Jun Liu, Email: jun.liu@utoronto.ca.
Supplemental information
Data and code availability
Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon reasonable request.
This study did not generate any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abdallah A.M., Savage N.D., van Zon M., Wilson L., Vandenbroucke-Grauls C.M., van der Wel N.N., Ottenhoff T.H., Bitter W. The ESX-5 secretion system of Mycobacterium marinum modulates the macrophage response. J. Immunol. 2008;181:7166–7175. doi: 10.4049/jimmunol.181.10.7166. [DOI] [PubMed] [Google Scholar]
- Abdallah A.M., van Pittius N.C.G., Champion P.A.D., Cox J., Luirink J., Vandenbroucke-Grauls C.M., Appelmelk B.J., Bitter W. Type VII secretion—mycobacteria show the way. Nat. Rev. Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- Abdallah A.M., Verboom T., Weerdenburg E.M., Gey van Pittius N.C., Mahasha P.W., Jimenez C., Parra M., Cadieux N., Brennan M.J., Appelmelk B.J., Bitter W. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol. Microbiol. 2009;73:329–340. doi: 10.1111/j.1365-2958.2009.06783.x. [DOI] [PubMed] [Google Scholar]
- Aguilo N., Gonzalo-Asensio J., Alvarez-Arguedas S., Marinova D., Gomez A.B., Uranga S., Spallek R., Singh M., Audran R., Spertini F., Martin C. Reactogenicity to major tuberculosis antigens absent in BCG is linked to improved protection against Mycobacterium tuberculosis. Nat. Commun. 2017;8:16085. doi: 10.1038/ncomms16085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ates L.S., Ummels R., Commandeur S., van de Weerd R., Sparrius M., Weerdenburg E., Alber M., Kalscheuer R., Piersma S.R., Abdallah A.M., et al. Essential role of the ESX-5 secretion system in outer membrane permeability of pathogenic mycobacteria. PLoS Genet. 2015;11:e1005190. doi: 10.1371/journal.pgen.1005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardarov S., Bardarov S., Pavelka M.S., Sambandamurthy V., Larsen M., Tufariello J., Chan J., Hatfull G., Jacobs W.R. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology (Reading) 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- Beckham K.S., Ciccarelli L., Bunduc C.M., Mertens H.D., Ummels R., Lugmayr W., Mayr J., Rettel M., Savitski M.M., Svergun D.I., et al. Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis. Nat. Microbiol. 2017;2:17047. doi: 10.1038/nmicrobiol.2017.47. [DOI] [PubMed] [Google Scholar]
- Bitter W., Houben E.N., Bottai D., Brodin P., Brown E.J., Cox J.S., Derbyshire K., Fortune S.M., Gao L.-Y., Liu J. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottai D., Di Luca M., Majlessi L., Frigui W., Simeone R., Sayes F., Bitter W., Brennan M.J., Leclerc C., Batoni G. Disruption of the ESX-5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Mol. Microbiol. 2012;83:1195–1209. doi: 10.1111/j.1365-2958.2012.08001.x. [DOI] [PubMed] [Google Scholar]
- Brodin P., Majlessi L., Marsollier L., de Jonge M.I., Bottai D., Demangel C., Hinds J., Neyrolles O., Butcher P.D., Leclerc C., et al. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect. Immun. 2006;74:88–98. doi: 10.1128/IAI.74.1.88-98.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion M.M., Williams E.A., Pinapati R.S., Champion P.A. Correlation of phenotypic profiles using targeted proteomics identifies mycobacterial esx-1 substrates. J. Proteome Res. 2014;13:5151–5164. doi: 10.1021/pr500484w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion P.A., Champion M.M., Manzanillo P., Cox J.S. ESX-1 secreted virulence factors are recognized by multiple cytosolic AAA ATPases in pathogenic mycobacteria. Mol. Microbiol. 2009;73:950–962. doi: 10.1111/j.1365-2958.2009.06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion P.A., Stanley S.A., Champion M.M., Brown E.J., Cox J.S. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science. 2006;313:1632–1636. doi: 10.1126/science.1131167. [DOI] [PubMed] [Google Scholar]
- Chen J.M., Zhang M., Rybniker J., Boy-Rottger S., Dhar N., Pojer F., Cole S.T. Mycobacterium tuberculosis EspB binds phospholipids and mediates EsxA-independent virulence. Mol. Microbiol. 2013;89:1154–1166. doi: 10.1111/mmi.12336. [DOI] [PubMed] [Google Scholar]
- Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., 3rd, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Conrad W.H., Osman M.M., Shanahan J.K., Chu F., Takaki K.K., Cameron J., Hopkinson-Woolley D., Brosch R., Ramakrishnan L. Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc. Natl. Acad. Sci. 2017;114 doi: 10.1073/pnas.1620133114. 201620133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coros A., Callahan B., Battaglioli E., Derbyshire K.M. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol. Microbiol. 2008;69:794–808. doi: 10.1111/j.1365-2958.2008.06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damen M.P.M., Phan T.H., Ummels R., Rubio-Canalejas A., Bitter W., Houben E.N.G. Modification of a PE/PPE substrate pair reroutes an Esx substrate pair from the mycobacterial ESX-1 type VII secretion system to the ESX-5 system. J. Biol. Chem. 2020;295:5960–5969. doi: 10.1074/jbc.RA119.011682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge M.I., Pehau-Arnaudet G., Fretz M.M., Romain F., Bottai D., Brodin P., Honore N., Marchal G., Jiskoot W., England P., et al. ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J. Bacteriol. 2007;189:6028–6034. doi: 10.1128/JB.00469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas E., Christina Boritsch E., Vandenbogaert M., Rodriguez de la Vega R.C., Thiberge J.M., Caro V., Gaillard J.L., Heym B., Girard-Misguich F., Brosch R., Sapriel G. Mycobacterial pan-genome analysis suggests important role of plasmids in the radiation of type VII secretion systems. Genome Biol. Evol. 2016;8:387–402. doi: 10.1093/gbe/evw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famelis N., Rivera-Calzada A., Degliesposti G., Wingender M., Mietrach N., Skehel J.M., Fernandez-Leiro R., Bottcher B., Schlosser A., Llorca O., Geibel S. Architecture of the mycobacterial type VII secretion system. Nature. 2019;576:321–325. doi: 10.1038/s41586-019-1633-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune S.M., Jaeger A., Sarracino D.A., Chase M.R., Sassetti C.M., Sherman D.R., Bloom B.R., Rubin E.J. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc. Natl. Acad. Sci. U S A. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L.Y., Guo S., McLaughlin B., Morisaki H., Engel J.N., Brown E.J. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol. Microbiol. 2004;53:1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x. [DOI] [PubMed] [Google Scholar]
- Gey Van Pittius N.C., Gamieldien J., Hide W., Brown G.D., Siezen R.J., Beyers A.D. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-10-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey van Pittius N.C., Sampson S.L., Lee H., Kim Y., van Helden P.D., Warren R.M. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 2006;6:95. doi: 10.1186/1471-2148-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T.A., Clark R.R., Boucher N., Lapierre P., Smith C., Derbyshire K.M. Intercellular communication and conjugation are mediated by ESX secretion systems in mycobacteria. Science. 2016;354:347–350. doi: 10.1126/science.aag0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R.M., Cai Z.L., Ho S.N., Pease L.R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Hsu T., Hingley-Wilson S.M., Chen B., Chen M., Dai A.Z., Morin P.M., Marks C.B., Padiyar J., Goulding C., Gingery M., et al. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. U S A. 2003;100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo Lafuente B., Ummels R., Kuijl C., Bitter W., Speer A. Mycobacterium tuberculosis Toxin CpnT Is an ESX-5 Substrate and Requires Three Type VII Secretion Systems for Intracellular Secretion. mBio. 2021;12 doi: 10.1128/mBio.02983-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey P.A., Lindberg U. Cytoskeletal regulation: rich in lipids. Nat. Rev. Mol. Cel. Biol. 2004;5:658–666. doi: 10.1038/nrm1434. [DOI] [PubMed] [Google Scholar]
- Koul A., Choidas A., Treder M., Tyagi A.K., Drlica K., Singh Y., Ullrich A. Cloning and characterization of secretory tyrosine phosphatases of <em>Mycobacterium tuberculosis</em>. J. Bacteriol. 2000;182:5425–5432. doi: 10.1128/jb.182.19.5425-5432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laencina L., Dubois V., Le Moigne V., Viljoen A., Majlessi L., Pritchard J., Bernut A., Piel L., Roux A.L., Gaillard J.L., et al. Identification of genes required for Mycobacterium abscessus growth in vivo with a prominent role of the ESX-4 locus. Proc. Natl. Acad. Sci. U S A. 2018;115:E1002–E1011. doi: 10.1073/pnas.1713195115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y., Rybniker J., Sala C., Cole S.T. EspC forms a filamentous structure in the cell envelope of Mycobacterium tuberculosis and impacts ESX-1 secretion. Mol. Microbiol. 2017;103:26–38. doi: 10.1111/mmi.13575. [DOI] [PubMed] [Google Scholar]
- Madacki J., Orgeur M., Mas Fiol G., Frigui W., Ma L., Brosch R. ESX-1-Independent horizontal gene transfer by Mycobacterium tuberculosis complex strains. mBio. 2021;12 doi: 10.1128/mBio.00965-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin B., Chon J.S., MacGurn J.A., Carlsson F., Cheng T.L., Cox J.S., Brown E.J. A mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog. 2007;3:e105. doi: 10.1371/journal.ppat.0030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millington K.A., Fortune S.M., Low J., Garces A., Hingley-Wilson S.M., Wickremasinghe M., Kon O.M., Lalvani A. Rv3615c is a highly immunodominant RD1 (Region of Difference 1)-dependent secreted antigen specific for Mycobacterium tuberculosis infection. Proc. Natl. Acad. Sci. U S A. 2011;108:5730–5735. doi: 10.1073/pnas.1015153108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Foot M., Warren R.M., Sampson S.L., van Helden P.D., Gey van Pittius N.C. The plasmid-mediated evolution of the mycobacterial ESX (Type VII) secretion systems. BMC Evol. Biol. 2016;16:62. doi: 10.1186/s12862-016-0631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgeur M., Brosch R. Evolution of virulence in the Mycobacterium tuberculosis complex. Curr. Opin. Microbiol. 2018;41:68–75. doi: 10.1016/j.mib.2017.11.021. [DOI] [PubMed] [Google Scholar]
- Pallen M.J. The ESAT-6/WXG100 superfamily -- and a new Gram-positive secretion system? Trends Microbiol. 2002;10:209–212. doi: 10.1016/s0966-842x(02)02345-4. [DOI] [PubMed] [Google Scholar]
- Poweleit N., Czudnochowski N., Nakagawa R., Trinidad D.D., Murphy K.C., Sassetti C.M., Rosenberg O.S. The structure of the endogenous ESX-3 secretion system. Elife. 2019;8 doi: 10.7554/eLife.52983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw P.S., Panagiotidou P., Whelan A., Gordon S.V., Hewinson R.G., Williamson R.A., Carr M.D. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6∗CFP-10 complex. Implications for pathogenesis and virulence. J. Biol. Chem. 2002;277:21598–21603. doi: 10.1074/jbc.M201625200. [DOI] [PubMed] [Google Scholar]
- Rosenberg O.S., Dovala D., Li X., Connolly L., Bendebury A., Finer-Moore J., Holton J., Cheng Y., Stroud R.M., Cox J.S. Substrates control multimerization and activation of the multi-domain ATPase motor of type VII secretion. Cell. 2015;161:501–512. doi: 10.1016/j.cell.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani M., Houben E.N., Geurtsen J., Pierson J., de Punder K., van Zon M., Wever B., Piersma S.R., Jimenez C.R., Daffe M., et al. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010;6:e1000794. doi: 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist M.S., Unnikrishnan M., McConnell M.J., Borowsky M., Cheng T.Y., Siddiqi N., Fortune S.M., Moody D.B., Rubin E.J. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc. Natl. Acad. Sci. U S A. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J., Manoranjan J., Pan M., Bohsali A., Xu J., Liu J., McDonald K.L., Szyk A., LaRonde-LeBlanc N., Gao L.Y. Evidence for pore formation in host cell membranes by ESX-1-secreted ESAT-6 and its role in Mycobacterium marinum escape from the vacuole. Infect. Immun. 2008;76:5478–5487. doi: 10.1128/IAI.00614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomonson M., Setiaputra D., Makepeace K.A.T., Lameignere E., Petrotchenko E.V., Conrady D.G., Bergeron J.R., Vuckovic M., DiMaio F., Borchers C.H., et al. Structure of EspB from the ESX-1 type VII secretion system and insights into its export mechanism. Structure. 2015;23:571–583. doi: 10.1016/j.str.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Sorensen A.L., Nagai S., Houen G., Andersen P., Andersen A.B. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 1995;63:1710–1717. doi: 10.1128/iai.63.5.1710-1717.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S.A., Raghavan S., Hwang W.W., Cox J.S. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc. Natl. Acad. Sci. U S A. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T., Lee W.L., Alexander D.C., Grinstein S., Liu J. The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell Microbiol. 2006;8:1417–1429. doi: 10.1111/j.1462-5822.2006.00721.x. [DOI] [PubMed] [Google Scholar]
- Tobin D.M., Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol. 2008;10:1027–1039. doi: 10.1111/j.1462-5822.2008.01133.x. [DOI] [PubMed] [Google Scholar]
- Tufariello J.M., Chapman J.R., Kerantzas C.A., Wong K.W., Vilcheze C., Jones C.M., Cole L.E., Tinaztepe E., Thompson V., Fenyo D., et al. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc. Natl. Acad. Sci. U S A. 2016;113:E348–E357. doi: 10.1073/pnas.1523321113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Ru H.W., Xiang Z.H., Jiang J., Wang Y.C., Zhang L., Liu J. WhiB4 regulates the PE/PPE gene family and is essential for virulence of Mycobacterium marinum. Sci. Rep. 2017;7:3007. doi: 10.1038/s41598-017-03020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Laine O., Masciocchi M., Manoranjan J., Smith J., Du S.J., Edwards N., Zhu X., Fenselau C., Gao L.Y. A unique Mycobacterium ESX-1 protein co-secretes with CFP-10/ESAT-6 and is necessary for inhibiting phagosome maturation. Mol. Microbiol. 2007;66:787–800. doi: 10.1111/j.1365-2958.2007.05959.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon reasonable request.
This study did not generate any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.