Abstract
A molecular epidemiological study of Mycobacterium simiae strains isolated from AIDS patients in Guadeloupe was performed by the random amplification of polymorphic DNA (RAPD) and pulsed-field gel electrophoresis (PFGE) of DraI- or XbaI-digested bacterial DNAs. A comparison of RAPD profiles suggested a similarity of banding patterns within a group of patients (two clusters of two and three patients), but the available epidemiological and clinical information did not support this finding. PFGE, on the other hand, showed that all the patients were contaminated with individual isolates. Combined numerical analysis performed by compiling the PFGE patterns obtained after XbaI and DraI digestions of bacterial DNAs suggested the occurrence of polyclonal infection in three of nine patients. Our results do not support a common source of M. simiae infection in Guadeloupe.
Although Mycobacterium simiae infections associated with human disease have been reported (1, 7, 8, 16, 21), only rare cases of disseminated disease due to M. simiae have been described; one case involved a patient with multiple sclerosis (12), and the other cases concerned patients with AIDS (5, 6, 9, 19). The natural habitat and the mechanism of transmission of M. simiae to animals and humans are still not clear. M. simiae has been recovered from hospital water supplies (5, 10, 25) as well as from sphagnum vegetation of Madagascar (13). In the Caribbean island of Guadeloupe, a sudden increase in M. simiae isolation from patients over a 6-year period (1992 to 1997) was observed. In this period, there were 22 confirmed M. simiae isolates from 9 patients, compared to a single M. simiae strain isolated during the previous 6-year period of 1986 to 1991. This increase was obviously linked to the AIDS epidemic, as eight of the nine patients were coinfected with the human immunodeficiency virus, with CD4 cell counts below 50/mm3. A similar increase in M. simiae isolation has also been reported by other investigators in the United States (21). As studies correlating the clinical and epidemiological data of patients with molecular typing of M. simiae isolates have not been published, a molecular epidemiological study of M. simiae isolated in Guadeloupe was performed to elucidate whether the recent increase in M. simiae infections could be traced to a common source of infection.
Bacterial strains.
Mycobacteria were isolated from pathological samples at the Pasteur Institute of Guadeloupe and identified using routine bacteriological procedures (2) and PCR-restriction fragment length polymorphism analysis of the hsp65 gene (4, 17). A total of 22 strains corresponding to the nine patients were collected and grown as fresh Löwenstein-Jensen slants at 37°C. The clinical and epidemiological data on the patients are summarized in Table 1.
TABLE 1.
Clinical and epidemiological data on patients with M. simiae strains
| Patient isolate | Origin of specimen | Mo and yr of isolation | Age (yr) | Sexa | Hospital | HIV statusb | Patient status | Remarks |
|---|---|---|---|---|---|---|---|---|
| A1 | Sputum | Sept. 1995 | 75 | F | 1 | − | Alive | Polyclonal infection (A1 to A3 form a cluster separate from the A4 to A6 cluster) |
| A2 | Gastric washing | Oct. 1995 | ||||||
| A3 | Gastric washing | Dec. 1995 | ||||||
| A4 | Gastric washing | Sept. 1996 | ||||||
| A5 | Sputum | Jan. 1997 | ||||||
| A6 | Gastric washing | Jan. 1997 | ||||||
| B | Bone marrow | Jan. 1996 | 33 | M | 2 | + | Dead | |
| C1 | Gastric washing | Jan. 1996 | 47 | M | 2 | + | Dead | Identical isolates |
| C2 | Bronchial aspirate | Feb. 1996 | ||||||
| D | Blood | Feb. 1992 | 40 | F | 2 | + | Dead | |
| E1 | Gastric washing | July 1994 | 34 | M | 2 | + | Dead | Polyclonal infection (E1 is distinct from the clustered isolates E2 to E5) |
| E2 | Blood | July 1994 | ||||||
| E3 | Blood | Jan. 1995 | ||||||
| E4 | Blood | Jan. 1995 | ||||||
| E5 | Blood | Mar. 1995 | ||||||
| F1 | Gastric washing | Feb. 1993 | 47 | F | 1 | + | Dead | Identical isolates |
| F2 | Bronchial aspirate | Jan. 1994 | ||||||
| G1 | Blood | Sept. 1995 | 35 | M | 2 | + | Dead | Polyclonal infection |
| G2 | Blood | Sept. 1995 | ||||||
| H | Blood | Mar. 1996 | 47 | M | 2 | + | Dead | |
| I1 | Bronchial aspirate | Dec. 1997 | 71 | F | 3 | Unknown | Alive | Identical isolates |
| I2 | Gastric washing | Dec. 1997 |
F, female; M, male.
Determined by serology.
RAPD analysis.
Bacterial DNA was prepared from fresh culture using cetyltrimethylammonium bromide (Merck, Darmstadt, Germany) as reported previously (22). DNA was precipitated with isopropanol, pelleted by centrifugation, washed with 70% alcohol, dried, and finally recovered in TE (10 mM Tris, 1 mM EDTA [pH 8]) and adjusted to give a concentration of 1 μg/ml. Amplification was performed in a 25-μl volume containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 0.001% gelatin, 100 μM concentrations of each of the four deoxyribonucleoside triphosphates (dATP, dGTP, dTTP, and dCTP), 5 pmol of primer (10-mer RAPD primers A01 to A20, reference RAF020; Bioprobe Systems, Montreuil-sous-Bois, France), 50 ng of template DNA, and 0.5 U of Thermus aquaticus DNA polymerase (Gibco-BRL Life Technologies, Cergy-Pontoise, France). The amplification mixture was overlaid with 50 μl of mineral oil and was subjected to 45 cycles of amplification (Perkin-Elmer Corp., Norwalk, Conn.) as follows. Samples were incubated at 94°C for 1 min to denature the DNA, 60°C for 1 min to anneal the primers, and 72°C for 1 min to extend the annealed primers. Each amplification experiment included a negative control sample without DNA. The amplification product was analyzed by electrophoresis on a 2% agarose gel (Gibco-BRL Life Technologies) and a DNA molecular weight marker VI (Boehringer Mannheim, Mannheim, Germany). Gels were stained with ethidium bromide and photographed on a UV transilluminator. A total of 20 primers were initially screened for the ability to produce discriminatory polymorphism and reproducible results. As slight variations in banding patterns were noted even when the same DNA controls were analyzed simultaneously, isolates were routinely assayed in duplicate.
PFGE analysis.
Bacteria were grown in 5 ml of Middlebrook 7H9 complete broth. Cultures were inoculated into 40 ml of fresh medium to an optical density at 650 nm of 0.08 and incubated at 37°C to an optical density at 650 nm of 0.3. Plugs were prepared as previously described (11), and bacterial DNA was digested with 30 U of DraI or 60 U of XbaI (Gibco-BRL Life Technologies) at 37°C for 2 h. After digestion, the plugs were loaded into a 1% (wt/vol) agarose gel (Gibco BRL). Large restriction fragments were separated using the contour-clamped homogeneous electric field DRIII pulsed-field gel electrophoresis (PFGE) apparatus (Bio-Rad, Richmond, Calif.) for 24 h at 14°C and 6 V/cm with a switch time of 1 to 40 s for DraI and for 20 h at 14°C and 6 V/cm with a switch time of 1 to 30 s for XbaI. The interpretation of the PFGE patterns was done according to Tenover guidelines before computer-assisted interpretation of the results (18). As visual analysis of PFGE profiles may not be sufficient for comparing highly banded patterns, computer-assisted analysis of patterns was performed using the Taxotron software (Institut Pasteur, Paris, France). The unweighted pair group method using arithmetical averages (14) was used for comparing the patterns directly by using the Taxotron software and to generate the dendrograms. Visual control of the gels was always performed to check for similarities or identities of the patterns. External reference markers (used every five lanes) included the lambda PFG marker for DraI PFGE and low-range PFG markers for XbaI (BioLabs, Beverly, Mass.), which allowed comparison of patterns within a 3 to 4% error tolerance. Identical strains were strains harboring the same number of bands at the same positions or differing by no more than two bands. Combined numerical analysis of DraI and XbaI PFGE patterns was performed as reported previously (15).
Results and discussion.
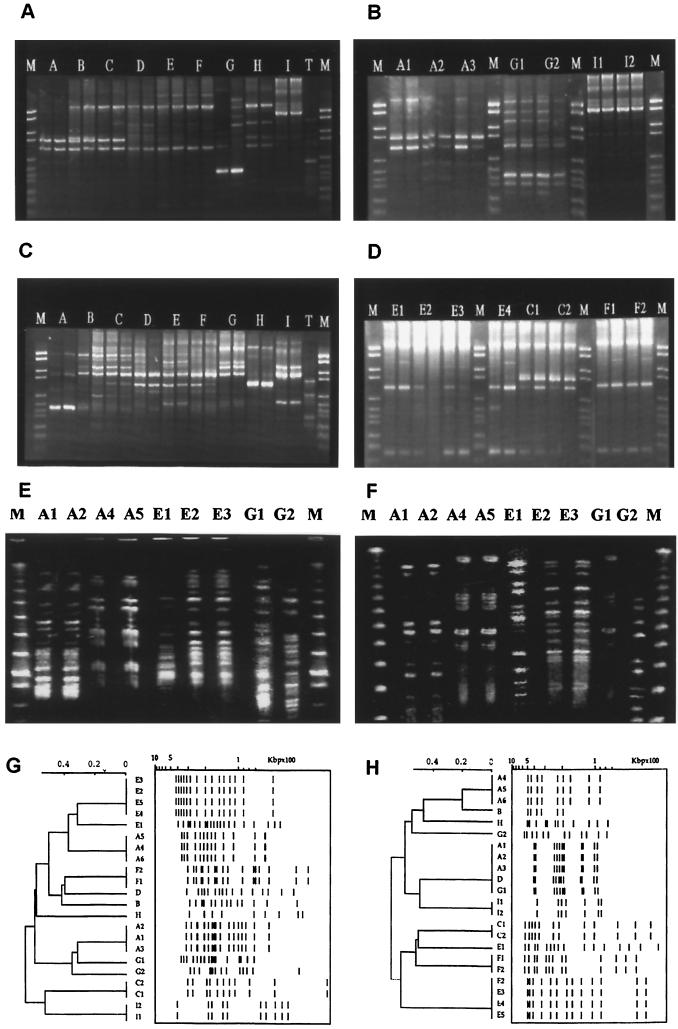
As no species-specific molecular markers are yet available to type M. simiae isolates, we initially attempted to fingerprint the Caribbean M. simiae isolates using the RAPD (24), as it is a rapid typing method based on the use of single primers with arbitrary nucleotide sequences that does not require any prior knowledge of bacterial DNA sequences. However, interactions of primers and their targets during such arbitrary amplification procedures are complex, and amplification profiles for specific oligonucleotide primers are highly dependent on the specific conditions of the reaction. Under our experimental conditions, among the 20 primers assayed we retained 2 primers (primers 4 and 10) (Fig. 1A and C, respectively) which produced discriminatory and reproducible profiles. Results obtained on serial isolates of the patients by primer 4 are illustrated in Fig. 1B and D. Comparison of the RAPD profiles suggested a similarity of banding patterns between patients B and C and among patients D, E, and F, whereas the profiles corresponding to patients A, G, H, and I were unique. Nonetheless, the epidemiological and demographic findings did not support any possible common source of infection between patients B and C or among patients D, E, and F.
FIG. 1.
RAPD (A to D) and PFGE (E to H) profiles of M. simiae isolates. Representative RAPD patterns with the primers 4 (A) and 10 (C) for one isolate per patient and the results obtained with serial isolates from selected patients with primer 4 (B and D) are shown. Panels E and F show representative PFGE patterns obtained with XbaI- and DraI-digested DNAs for patients with polyclonal infections. All of the 22 PFGE profiles obtained are illustrated in dendrograms shown in panels G and H. Samples for RAPD experiments were run in duplicate. T, template DNA control; M, molecular weight marker; A to I, individual patient isolates (A1, A2, etc., represent serial isolates from the same patient). The scale in panels G and H shows the Dice index.
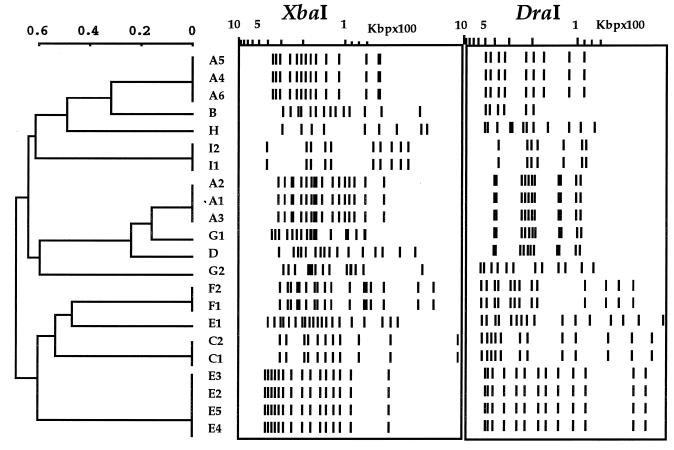
The PFGE results obtained on XbaI- and DraI-digested DNAs are summarized in Fig. 1E to H. Figure 1E and F show representative patterns for patients for whom a polyclonal M. simiae infection was demonstrated, whereas all 22 profiles obtained are illustrated in Fig. 1G and H. The combined numerical analysis of the PFGE results obtained after XbaI and DraI digestions (Fig. 2) showed that all the patients were contaminated with individual isolates and excluded the possibility of a common source of infection. The combined numerical analysis also suggested the occurrence of polyclonal infection in three of the nine patients (patients A, E, and G) (Table 1; Fig. 1 and 2) that was overlooked by the RAPD method. In the case of patient A, identical strains A1 to A3 were isolated during the September to December 1995 period, followed by distinct isolates A4 to A6 a year later (September 1996 to January 1997) (Table 1; Fig. 2 and 3). The polyclonal infection of patient G was also underlined by the susceptibility of isolate G1 to ethambutol and rifabutin, in contrast to the resistance of the isolate G2 to these two drugs.
FIG. 2.
Combined numerical analysis of PFGE results obtained after XbaI and DraI digestions using the unweighted pair group method using arithmetic averages. A to I, individual patient isolates (A1, A2, etc., represent serial isolates from a same patient). The scale shows the Dice index.
The results obtained also show the utility of using two enzymes for PFGE typing. For example, the polyclonal isolate E1 was closely related to isolates E2 to E5 upon XbaI digestion but was easily discriminated upon DraI digestion (Fig. 1 and 2). On the other hand, DraI alone grouped isolates A1 to A3 with D and G1; however, these were easily distinguished from A1 to A3 upon XbaI digestion. We therefore recommend using PFGE on DNAs digested by two distinct enzymes, each being able to generate distinct banding patterns. The final interpretation of clustering of isolates should be preferentially analyzed by combined numerical analysis.
In conclusion, the results obtained during this investigation do not support a common source of infection for the sudden increase in M. simiae isolation in Guadeloupe, which therefore may be attributed to an increased susceptibility of AIDS patients to opportunistic infections in general. However, one important observation made during this investigation was the fact that none of our highly immunocompromised patients harboring M. simiae infections were simultaneously infected with other atypical mycobacteria or Mycobacterium tuberculosis. Although mixed M. simiae and Mycobacterium avium infections among AIDS patients have been reported (19), we were not able to isolate another mycobacterial species even from patients that were monitored for as long as 16 months and from whom M. simiae was repeatedly isolated (Table 1). Whether this finding underlines a particular immunological background for susceptibility to M. simiae remains an open question.
Concerning the methodology used, we did not find RAPD a satisfactory method for typing M. simiae, as it gave false clustering of isolates that were easily discriminated by using PFGE. Moreover, RAPD was unable to distinguish polyclonal infections of patients A, E, and G. Although it is a rapid method, a number of factors may influence RAPD results, the major variables being DNA quality and concentration, the ratio of primer to bacterial DNA concentration, the concentration of magnesium ions, and the hybridization temperature (3, 20, 23). Moreover, ambiguous polymorphism between distinct isolates may also result from poor discrimination by a primer between alternative priming sites of slightly different nucleotide sequences. Thus, PFGE may be considered appropriate for studying molecular epidemiology of atypical mycobacteria for which no specific insertion sequences are yet described or which do not generate sufficient polymorphism due to a limited number of copies, provided it is performed on DNAs digested by two distinct enzymes, each being able to generate distinct banding patterns.
Acknowledgments
This work was supported through grants from the Délégation Générale au Réseau International des Instituts Pasteur et Instituts Associés, Institut Pasteur, Paris, and Fondation Française Raoul Follereau, Paris, France.
We are grateful to clinicians (F. Cnudde, M.T. Sow, J.M. Gabriel, and M. Strobel) at various hospitals in Guadeloupe for helpful information.
REFERENCES
- 1.Bell R C, Higuchi J H, Donovan W N, Krasnow I, Johanson W G. Mycobacterium simiae: clinical features and follow-up of twenty four patients. Am Rev Respir Dis. 1983;127:35–38. doi: 10.1164/arrd.1983.127.1.35. [DOI] [PubMed] [Google Scholar]
- 2.David H, Lévy-Frébault V, Thorel M F. Méthodes de laboratoire pour mycobactériologie clinique. Paris, France: Commission des Laboratoires de Référence et d'Expertise de l'Institut Pasteur, Institut Pasteur; 1989. [Google Scholar]
- 3.Davin-Regli A, Abed Y, Charrel R N, Bollet C, de Micco P. Variations in DNA concentrations significantly affect the reproducibility of RAPD fingerprint patterns. Res Microbiol. 1995;146:561–568. doi: 10.1016/0923-2508(96)80562-6. [DOI] [PubMed] [Google Scholar]
- 4.Devallois A, Goh K S, Rastogi N. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J Clin Microbiol. 1997;35:2969–2973. doi: 10.1128/jcm.35.11.2969-2973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huminer D, Dux S, Samra Z, Kaufman L, Lavy A, Block C S, Pitlik S D. Mycobacterium simiae infection in Israeli patients with AIDS. Clin Infect Dis. 1993;17:508–509. doi: 10.1093/clinids/17.3.508. [DOI] [PubMed] [Google Scholar]
- 6.Koeck J L, Debord T, Fabre M, Vincent V, Cavallo J D, Le Vagueresse R. Disseminated Mycobacterium simiae infection in a patient with AIDS: clinical features and treatment. Clin Infect Dis. 1996;23:832–833. doi: 10.1093/clinids/23.4.832. [DOI] [PubMed] [Google Scholar]
- 7.Krasnow I, Gross W. Mycobacterium simiae infection in the United States. Am Rev Respir Dis. 1975;111:357–360. doi: 10.1164/arrd.1975.111.3.357. [DOI] [PubMed] [Google Scholar]
- 8.Lavy A, Yoshpe-Purer Y. Isolation of Mycobacterium simiae from clinical specimens in Israel. Tubercle. 1982;63:279–285. doi: 10.1016/s0041-3879(82)80016-0. [DOI] [PubMed] [Google Scholar]
- 9.Lévy-Frébault V, Pangon B, Buré A, Katlama C, Marche C, David H L. Mycobacterium simiae and Mycobacterium avium-M. intracellulare mixed infection in acquired immune deficiency syndrome. J Clin Microbiol. 1987;25:154–157. doi: 10.1128/jcm.25.1.154-157.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthews J H, Warren N G. Mycobacterium simiae. Am Rev Respir Dis. 1983;127:788–789. doi: 10.1164/arrd.1983.127.6.788b. [DOI] [PubMed] [Google Scholar]
- 11.Picardeau M, Varnerot A, Rauzier J, Gicquel B, Vincent V. Mycobacterium xenopi IS1395, a novel insertion sequence expanding the IS256 family. Microbiology. 1996;142:2453–2461. doi: 10.1099/00221287-142-9-2453. [DOI] [PubMed] [Google Scholar]
- 12.Rose H D, Dorff G J, Lauwasser M, Sheth N K. Pulmonary and disseminated Mycobacterium simiae infection in humans. Am Rev Respir Dis. 1982;126:1110–1113. doi: 10.1164/arrd.1982.126.6.1110. [DOI] [PubMed] [Google Scholar]
- 13.Schroder K H, Kazda J, Muller K, Muller H J. Isolation of Mycobacterium simiae from the environment. Zentbl Bakteriol. 1992;277:561–564. doi: 10.1016/s0934-8840(11)80482-2. [DOI] [PubMed] [Google Scholar]
- 14.Sneath P, Sokal R. Numerical taxonomy: the principles and practices of classification. W. H. San Francisco, Calif: Freeman and Co.; 1973. [Google Scholar]
- 15.Sola C, Horgen L, Devallois A, Rastogi N. Combined numerical analysis based on the molecular description of Mycobacterium tuberculosis by four-repetitive sequence-based DNA typing systems. Res Microbiol. 1998;149:349–360. doi: 10.1016/s0923-2508(98)80440-3. [DOI] [PubMed] [Google Scholar]
- 16.Sriyabhaya N, Wonswatana S. Pulmonary infection caused by atypical mycobacteria: a report of 24 cases in Thailand. Rev Infect Dis. 1981;3:1085–1089. doi: 10.1093/clinids/3.5.1085. [DOI] [PubMed] [Google Scholar]
- 17.Telenti A, Marchesi F, Balz M, Bally F, Böttger E C, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres R A, Nord J, Feldman R, La Bombardi V, Barr M. Disseminated mixed Mycobacterium simiae-Mycobacterium avium complex infection in acquired immunodeficiency syndrome. J Infect Dis. 1991;164:432–433. doi: 10.1093/infdis/164.2.432. [DOI] [PubMed] [Google Scholar]
- 20.Tyler K D, Wang G, Tyler S D, Johnson W M. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J Clin Microbiol. 1997;35:339–346. doi: 10.1128/jcm.35.2.339-346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valero G, Peters J, Jorgensen J H, Graybill J R. Clinical isolates of Mycobacterium simiae in San Antonio, Texas. An 11 yr review. Am J Respir Crit Care Med. 1995;152:1555–1557. doi: 10.1164/ajrccm.152.5.7582293. [DOI] [PubMed] [Google Scholar]
- 22.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams J G, Hanafey M K, Rafalski J A, Tingey S V. Genetic analysis using random amplified polymorphic DNA markers. Methods Enzymol. 1993;218:704–740. doi: 10.1016/0076-6879(93)18053-f. [DOI] [PubMed] [Google Scholar]
- 24.Williams J G, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolinsky E. Nontuberculous mycobacteria and associated diseases. Am Rev Respir Dis. 1979;119:107–159. doi: 10.1164/arrd.1979.119.1.107. [DOI] [PubMed] [Google Scholar]