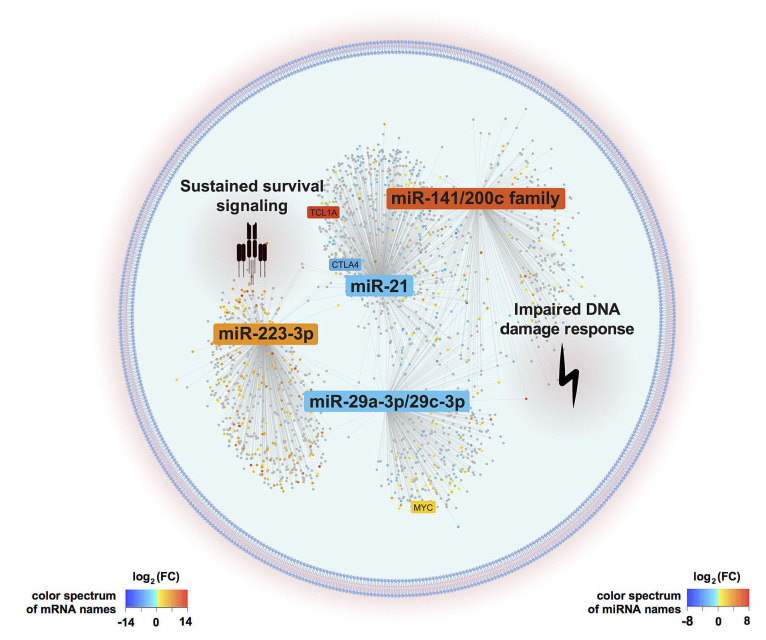
Figure 6.
Graphical summary of postulated microRNA/mRNA -based deregulated networks in T-cell prolymphocytic leukemia. T-cell activity shaped microRNA expression signatures (miR-omes)/ transcriptome networks are displayed as identified by our combinatorial approach of small-RNA and transcriptome sequencing analyses in 41 clinically well characterized T-cell prolymphocytic leukemia (T-PLL) cases. MiR differentially expressed in T-PLL and mRNA associated with these miR (P<0.05) are presented. The background color of miR and mRNA (dots) indicates the fold changes of differential expression as compared to age-matched healthy donor-derived CD3+ pan-T cells (blue=downregulation, red=upregulation). MiR-223-3p, the miR-21 family, the miR-29 family, and the miR-200c/141 family emerged as hallmarks of the T-PLL miR-ome, as they were (i) significantly deregulated among T-PLL cases, (ii) presented putative targets involved in oncogenic pathways of TPLL’s pathobiology, (iii) showed associations with prognostic parameters, and (iv) were already described in the leukemogenesis of other B- and T-cell malignancies. Deregulations of these four miR-families were associated with cooperative effects on DNA damage response pathways as well as on pro-proliferative and cell survival signaling (as revealed by gene set enrichment analysis based on correlated mRNA). Genes previously described as hallmarks of T-PLL (e.g., TCL1A, CTLA4, and MYC) were found within the network of the identified miR-associated mRNA.