Abstract
T-cell prolymphocytic leukemia (T-PLL) is mostly characterized by aberrant expansion of small- to medium-sized prolymphocytes with a mature post-thymic phenotype, high aggressiveness of the disease and poor prognosis. However, T-PLL is more heterogeneous with a wide range of clinical, morphological, and molecular features, which occasionally impedes the diagnosis. We hypothesized that T-PLL consists of phenotypic and/or genotypic subgroups that may explain the heterogeneity of the disease. Multi-dimensional immuno-phenotyping and gene expression profiling did not reveal clear T-PLL subgroups, and no clear T-cell receptor a or b CDR3 skewing was observed between different T-PLL cases. We revealed that the expression of microRNA (miRNA) is aberrant and often heterogeneous in T-PLL. We identified 35 miRNA that were aberrantly expressed in T-PLL with miR-200c/141 as the most differentially expressed cluster. High miR- 200c/141 and miR-181a/181b expression was significantly correlated with increased white blood cell counts and poor survival. Furthermore, we found that overexpression of miR-200c/141 correlated with downregulation of their targets ZEB2 and TGFbR3 and aberrant TGFb1- induced phosphorylated SMAD2 (p-SMAD2) and p-SMAD3, indicating that the TGFb pathway is affected in T-PLL. Our results thus highlight the potential role for aberrantly expressed oncogenic miRNA in T-PLL and pave the way for new therapeutic targets in this disease.
Introduction
T-cell prolymphocytic leukemia (T-PLL) is a relatively rare disease accounting for approximately 2% of mature lymphocytic leukemia patients older than 30 years.1 T-PLL is characterized by an uncontrolled expansion of malignant, mature, postthymic T lymphocytes in the bone marrow (BM), lymph nodes, liver and spleen, resulting in hepato-splenomegaly, lymphadenopathy, skin lesions and a high leukocyte count. Whole-transcriptome analysis of T-PLL cells revealed that the most significantly deregulated genes are involved in T-cell receptor signaling, cytokine signaling and p53-controlled apoptosis.2 In addition, frequent recurrent mutations in DNA damage repair/tumor suppressor genes (e.g., ATM, TP53, MSH3, MSH6, SAMHD1, PARP10, HERC1, HERC2), and oncogenes (JAK1 and JAK3) have been described in T-PLL.3,4 The majority of T-PLL cases exhibit a complex karyotype.3 Many distinct chromosomal abnormalities, including common chromosomal translocations involving chromosome 8, 11 and 14, deletions in 11q, 8p and 7q34q36 and gains of 8q and 8p11p12 have been reported.2,5 However, most T-PLL cases have abnormalities of chromosome 14, of which inv(14)(q11q32) and t(14:14)(q11;q32) are found most frequently.5 Chromosome 14 abnormalities bring the T-cell leukemia/lymphoma 1A (TCL1A, TCL1) oncogene on chromosome 14q32 in close proximity to the T-cell receptor-a/d*************(TRA/TRD) regulatory region on chromosome 14q11, or more rarely to T-cell receptor-b (TRB) regulatory elements located on 7q35, resulting in overexpression of TCL1.6 TCL1 is an adapter protein of 14 kDa that functions in kinase complexes and enhances T-cell receptor-mediated pro-survival signaling.7 The gene encoding mature T-cell proliferation-1 (MTCP1), is highly homologous to TCL1 and is involved in the less frequent translocations t(X;14)(q28;q11) and t(X;7)(q28;q35).5 The total incidence of translocations involving TCL1 and MTCP1 is around 90%.5 The high incidence of TCL1 and MTCP1 translocations strongly suggest that this family of proteins plays a key role in the pathogenesis of T-PLL. In full agreement, different TCL1 and MTCP1 transgenic mouse models demonstrate a role for TCL1 and MTCP1 in the initiation of malignant transformation to overt leukemia similar to T-PLL,8-10 albeit with a long latency of 12-20 months. These data indicate that secondary oncogenic events are required for full oncogenic transformation of the mature T cells. Identification of co-operating leukemia genes in the Em-TCL1 transgenic mouse with a Sleeping Beauty transposon- mediated mutagenesis screen revealed increased nuclear factor (NF)-kB signaling as a collaborating event in TCL1-mediated leukemogenesis.11 The role for small noncoding RNA in the pathogenesis of T-PLL has not been investigated yet.
MicroRNA (miRNA) are an abundant class of small noncoding RNA of 19-24 nucleotides, which guide the RNAinduced silencing complex (RISC) to reverse and partially complementary sequences in the 3’-untranslated regions (3’-UTR) of mRNA and control gene expression through translational inhibition and mRNA destabilization.12 Aberrant expression, biogenesis and activities of miRNA are hallmarks of human cancer, including malignant hematological disorders.13-17 Numerous miRNA have been identified as diagnostic biomarkers for human leukemia.16 Some miRNA are potent oncogenic drivers of leukemogenesis, while others have critical tumor-suppressing activities. 18-22
Here, we characterized a cohort of 31 T-PLL cases by cytomorphology, immuno-phenotyping and immunogenetic analysis, as well as mRNA and miRNA profiling to identify novel mechanisms that contribute to leukemic transformation. We revealed that the expression of miRNA is aberrant, though often heterogeneous, in T-PLL. Additionally, we present the first evidence that aberrant expression of miR-200c/141 affects TGFb-controlled mechanisms in T-PLL that may contribute to its pathogenesis.
Methods
Patient cohort
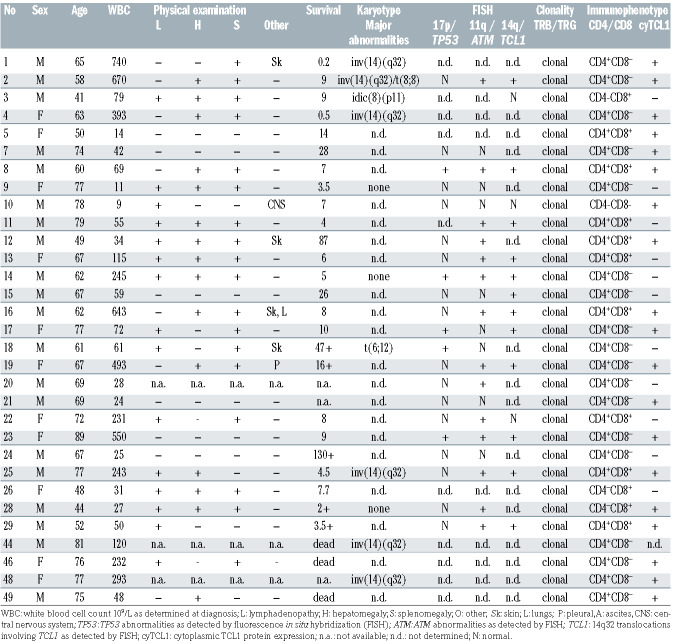
The patient cohort consisted of 31 well-defined T-PLL cases diagnosed between 1985 and 2011. Sampling was done at the moment of diagnosis. Diagnosis of T-PLL is based on a combination of clinical features, morphology, cytogenetics and immunophenotype. The cohort consisted of males (62%) and females (38%) with a median age at diagnosis of 66 years (range, 41-89 years) (Table 1). In our cohort, 52% of the T-PLL patients were CD4+ (n=14), 33% were CD4+ CD8+ (n=9), 11% were CD8+ (n=3), 4% were CD4–CD8– (n=1) and one was not determined. All patients were characterized for known aberrations in genes such as ATM, TP53, TCL1 and IGH. Use of T-PLL patient samples was approved by the Erasmus MC Medical Ethics Committee (MEC- 2015-617). A cohort of 31 T-PLL cases was characterized morphologically, cytogenetically and immunophenotypically. We could isolate high quality RNA from 23 T-PLL patients for gene expression profiling. The average RIN value of the T-PLL samples was 8.7 with a standard deviation of 0.9. Of these, the RNA of 21 T-PLL cases was sufficient for miRNA expression profiling. All RNA samples resulted in high-quality cDNA libraries. Four additional T-PLL samples (T-PLL44, 46, 48 and 49) were used for validation experiments and Western Blotting.
Normal T-cell subsets
Peripheral blood from healthy donors were obtained from buffy coats (Sanquin) with approval by the Erasmus MC Medical Ethics Committee (MEC-2016-202). For gene expression profiling, peripheral blood mononuclear cells (PBMC) were first isolated by Ficoll-Paque (density 1.077 g/mL, Pharmacia). CD4+ T cells were labeled with anti-CD3-PerCP-Cy5.5 (cat.#340948) and anti-CD4- PE-Cy7 (cat.#557852) and CD8+ T cells were labeled with anti- CD3-PerCP-Cy5.5 (cat.#340948) and anti-CD8-APC-H7 (cat.#560273)(BD Biosciences). Naïve T cells (CD27+ and CD45RA+), memory (CD27+ and CD45RO+ and effector (CD27– and CD45RA+) T-cell subsets were labeled with, anti-CD45RA-PE (cat.# 555489), anti-CD27-APC (cat.#558664), anti-CD45RO-FITC (cat.#555492) and anti-CD45RA-PE cat.#555489) antibodies (BD Biosciences) (Online Supplementary Table S1). In order to prevent antibody-mediated activation of T cells, all cell populations were kept on ice during antibody staining. Cells were sorted at 4°C and directly in TRIzol (Thermofisher) to prevent cell activation, cell death and/or RNA degradation. Cells fractions were sorted on a FACS Aria II cell sorter (BD Biosciences).
Data availability: i) GSE147930: microarray gene expression data; ii) GSE147931: mRNA sequencing data; iii) GSE147932: small RNA sequencing data.
For a detailed description of the immunophenotyping and cytmorphological analysis, fluorescence in situ hybridization analysis and cytogenetics, T-cell receptor gene rearrangement analysis, Lentiviral miRNA expression vectors and transduction of Jurkat and HeLa cells, gene expression profiling as well as RNA sequencing and data analysis see the Online Supplementary Appendix.
Results
Cytomorphologic, immunophenotypic and immunogenetic analysis of T-cell prolymphocytic leukemia
As T-PLL is a heterogeneous disease,23 we first characterized our T-PLL cohort with respect to clinical, morphological, phenotypical and molecular features (Table 1). Cytomorphologic analysis resulted in three different TPLL subgroups: i) cells with small lymphocytic cell morphology (64%); ii) cells with large blast-like morphology (10%); iii) cells with equivocal morphology (26%) (Online Supplementary Figure S1A). We determined deletions in TP53 (24% of cases tested) and ATM (64% of cases tested) as well as translocations involving TCL1 located on 14q32 (67% of cases tested). However, we did not observe any correlation between cell morphology and different parameters including known aberrations in genes, e.g., ATM, TP53, and TCL1. In order to determine the cell of origin of T-PLL, we performed flowcytometric immunophenotyping of T-PLL samples using the EuroFlow T-CLPD panel24 (www.euroflow.org) taking flow sorted CD4+ naïve (CD4N), CD4+ effector (CD4E), CD4+ memory (CD4M), CD8+ naïve (CD8N), CD8+ effector (CD8E) and CD8+ memory (CD8M) T-cell fractions from healthy individuals as reference. When using only T-PLL cases in a multidimensional APS plot analysis, the majority of T-PLL appeared to cluster according to similarities in their phenotype (Online Supplementary Figure S1B). However, two outliers were noted, T-PLL-10 and T-PLL- 24. Judging by the markers that were found to contribute to PC1 (mainly CD45RA) and PC2 (mainly CD45RA, CD45RO, CD27), the larger T-PLL cluster presumably would have a memory phenotype, while the outliers would resemble the naïve and/or effector T-cell subsets more (Online Supplementary Figure S1B). APS-based analysis of T-PLL cases that were added to fixed APS plots of normal T-cell subsets indeed confirmed that the majority of T-PLL cases did cluster close to or within the CD4+ and CD8+ memory T-cell subsets, whereas some T-PLL were more similar to non-memory subsets (Online Supplementary Figure S1C). As clonal TRA and TRB gene rearrangements with stereotyped CDR3 have proven to be relevant for some T-cell leukemias, e.g., T-large granular lymphocyte leukemia (T-LGL),25,26 we wondered whether this could be relevant for T-PLL as well. First, we asked whether V and J gene usage would define any subgroups of the T-PLL samples. To this end, we performed Sanger sequencing of the clonal TRA and TRB gene rearrangements. In agreement with previously published data,27 sequence analysis of T-PLL samples revealed no clear skewing of V and J gene usage of TRA and TRB genes (Online Supplementary Figure S1D and E). In addition, we did not find stereotyped CDR3 motifs in the productive TRA and TRB gene rearrangements in T-PLL either (data not shown).
Table 1.
Characteristics of the cohort of 31 T-cell prolymphocytic leukemia patients.
In summary, most T-PLL displayed cell morphology and immunophenotypical characteristics reflecting normal memory T cells, with no clear evidence for antigen-driven leukemogenesis.
Figure 1.
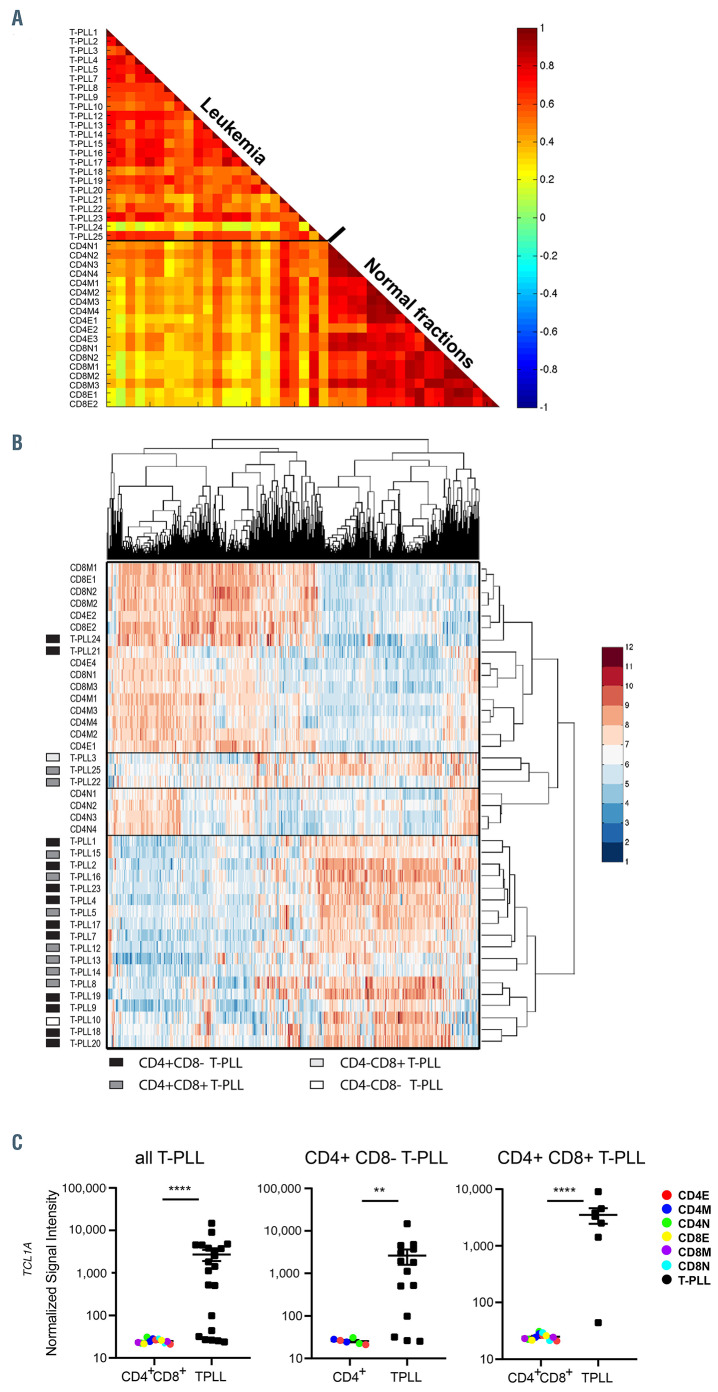
Analysis of gene expression profiles of 23 T-cell prolymphocytic leukemia cases and 18 normal T-cell subsets from healthy controls. (A) A correlation plot showing that T-cell prolymphocytic leukemia (T-PLL) are separated from normal T-cell subsets based on their gene expression profiles. A subset of probesets that showed signal, i.e., for which the median absolute deviation (MAD) from the median exceeds threshold T=0.6 on a log2 scale were selected (total of 3,261 probesets). Color bar represents correlation score. (B) Heatmap of T-PLL cases and normal T-cell subsets after unsupervised clustering (T=0.7; total of 1,904 probesets). Most T-PLL cases tightly cluster, without evident subgroups being detectable. Except for cases 21 and 24, most T-PLL cases cluster, without obvious correlation with immunological subgroups. Color reflects expression level: blue=low, red=high. (C) Expression of the TCL1A oncogene in T-PLL subgroups (all T-PLL, CD4+CD8– T-PLL and CD4+CD8+ T-PLL) compared to indicated normal T-cell fractions. E: effector subset; M: memory subset; N: naïve subset. Significance between the groups was calculated with the Mann-Whitney-U test **P<0.01, ****P<0.0001.
Figure 2.
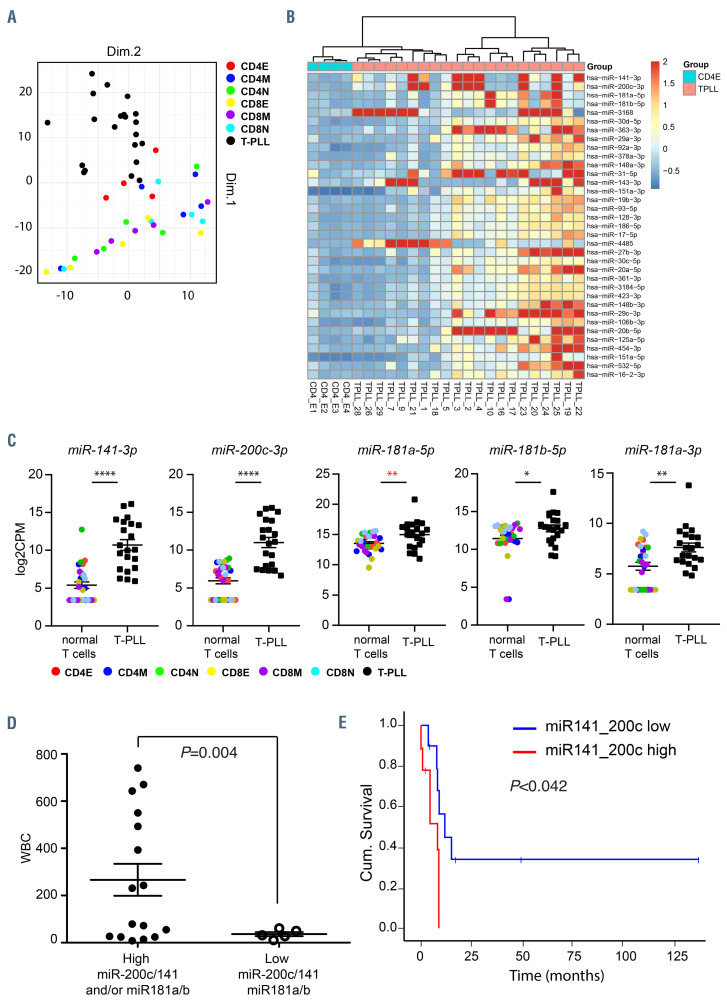
MicroRNA are deregulated in T-cell prolymphocytic leukemia. (A) Principal component analysis (PCA) based on microRNA (miRNA) expression profiles of indicated 21 T-cell prolymphocytic leukemia (T-PLL) cases and T cell fractions. (B) Heatmap of 35 most differential expressed miRNA between CD4+ effector T cells (CD4E) and T-PLL. Heatmap shows the robust normalized z-scores of each miRNA indicated by the color legend and set to maximum 2 (dark red color). The columns show 25 samples, which are hierarchically clustered using their Euclidean distances with complete linkage and Ward’s method. The miRNA are ordered top-down by increased false discovery rate values. (C) Expression of miR-141, miR-200c, miR-181a and miR-181b in T-PLL subgroups (all T-PLL, CD4+CD8– T-PLL and CD4+CD8+ T-PLL) compared to indicated normal T-cell fractions. E: effector subset; M: memory subset; N: naïve subset. Data was tested for normal distribution. Significance between the groups was calculated with the Mann-Whitney-U test (black) or with the Student’s t-test (red) *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. (D) TPLL cases were grouped based on high miR-200c/141 and or miR-181a/b expression (fold-change [FC] >2 compared to normal CD4E, n=16) and low miR-200c/141 and miR-181a/b expression (FC <2, n=5). White blood cell counts (WBC) are displayed. Statistical significance was determined with a Student’s t-test with Welch’s correction. (E) Kaplan Meier plot showing cumulative survival of patients with high vs. low miR-200c/141 expression. Statistical significance was calculated with the Mantel Cox test (log-rank test).
T-cell prolymphocytic leukemia show a relatively homogeneous gene expression profile with TCL1A amongst the highest upregulated genes
We then performed gene expression array analysis of T-PLL samples and normal T-cell subsets. A correlation plot and unsupervised clustering analysis of gene expression showed that most T-PLL samples, with the exception of T-PLL21 and T-PLL24, and to a lesser extent T-PLL3, TPLL22, T-PLL-25, clustered together; at the other end, normal T-cell subsets showed a high correlation to each other (Figure 1A and B).
Next, we focused on CD4+ T-PLL, which is the major group within T-PLL, and compared their gene expression to that of normal CD4+ T-cell subsets. We found 2,282 probe sets differentially expressed (at least 2-fold up- or downregulated) in CD4+ T-PLL compared to normal CD4 subsets. Of the top 100 differentially-expressed probe sets in CD4+ T-PLL, the average expression of most probe sets was upregulated, including the T-PLL characteristic T-cell leukemia driving oncogene TCL1A (Figure 1C; Online Supplementary Table S5). Collectively these data showed that the protein encoding gene expression profile of T-PLL is clearly different from normal T-cell subsets.
A subset of microRNA is differentially expressed in T-cell prolymphocytic leukemia of which miR-141 and miR-200c show prognostic impact
In order to evaluate the role of small non-coding RNA in the T-PLL cohort, we then performed genome-wide miRNA expression profiling by small RNA sequencing of 21 T-PLL cases. Based on miRNA expression, all T-PLL cases clustered together in a principal component analysis (PCA) plot and were most similar to healthy effector CD4 cells (Figure 2A). Therefore, we then compared the expression of individual miRNA in T-PLL with healthy effector CD4 cells. Strikingly, we did not find any miRNA significantly downregulated in T-PLL compared to normal effector CD4 cells (false discovery rate [FDR]<0.05). However, we did identify a set of 35 miRNA that were abundantly expressed and significantly upregulated (FDR<0.05; fold change [log2FC] >1.5) in the T-PLL cohort compared to healthy effector CD4 cells (Figure 2B; Online Supplementary Table S6). These included miR-17-5p, miR-19b-3p, miR- 20a, miR-106b, miR-92a of the oncogenic miR-17~92 clusters and other well-known oncogenic miRNA, including miR-125a, that were also expressed at higher levels in TPLL cases (Online Supplementary Table S6). miRNA of the miR-200c/141 cluster (FDR=0.011; average overexpression FC >55; range, 0.95-1,211) and the miR-181a/b cluster (FDR=0.025; average overexpression FC >20; range, 0.2-451), which were the highest overexpressed miRNA in T-PLL (Figure 2B; Online Supplementary Table S6). Notably, miR-141, miR-200c, miR-181a and miR-181b were significantly overexpressed compared to other normal T-cell fractions (Figure 2C). Subgroups of T-PLL with pronounced overexpression of miR-200c/141 and/or miR-181a/b (T-PLL-1-5,10, 11, 16, 17, 21-25, 28) (range, 10-1,200-fold) significantly correlated with higher white blood cell counts (Figure 2D). Furthermore, after splitting the patients in two groups (>5-fold overexpression compared to effector CD4 cells and <5-fold overexpression compared to effector CD4 cells) and performing a survival analysis, we found that the ten patients with the higher miR-141 and miR-200c levels (T-PLL-1-4, 11, 21, 22, 23, 25 and 28, had a shorter survival compared with the eleven patients with lower levels of miR-141 and miR-200c (T-PLL-5, 9, 10, 16-20, 24, 26, 29) (P=0.042, Mantel-Cox log rank test) (Figure 2E). In contrast, no difference in survival was found based on miR-181 expression (data not shown). A multiple regression analysis with risk factors including age, sex, white blood cell count and immunophenotype did not determine a confounding effect on the survival. Together, these data present the first evidence that miRNA are deregulated but heterogeneously expressed in T-PLL and that miR-141 and miR-200c expression levels may have prognostic value to define a subgroup of T-PLL with relatively poor survival.
The miR-200c/141-ZEB2-TGFb axis is aberrant in T-cell prolymphocytic leukemia
Next, we asked whether aberrant expression of miR- 200c/141 and miR-181a/b results in downregulation of their TargetScan-predicted well-conserved mRNA targets (Online Supplementary Tables S2 to S4) in T-PLL. In order to directly correlate miRNA levels to mRNA expression profiles, we generated transcriptomics data of CD4+ T-PLL samples with high expression levels (>10-fold overexpressed compared to effector CD4 cells) of these miRNA (high miR-200c/141 samples: T-PLL-1, 3, 4, 22, 23, 25; high miR-181a/b samples: T-PLL 3, 4, 10, 16, 17, 23 and 25) (Figure 2B). Effector CD4 T cells from healthy individuals were used as controls, as the miRNA profiles of T-PLL cells appeared most similar to this T-cell subset.
We first analyzed several genes that may be correlated with the upregulation of miR-200c/141 and miR-181a/b. For instance, involvement of enhanced miRNA expression was postulated based on elevated Argonaute-2 (AGO2) expression as a result of chromosome 8 gains in a subset of T-PLL patients.2 However, AGO2 mRNA expression was not significantly increased in our T-PLL cohort (data not shown). The miR-200c/141 cluster is frequently coexpressed with protein tyrosine phosphatase, non-receptor type 6 (PTPN6) through transcriptional read-through and complex 3D chromatin interactions between PTPN6 and miR-200c/141 promoters.28 Accordingly, we found a significant upregulation of PTPN6 in T-PLL samples with enhanced miR-200c/141 expression (FC=3.7; FDR=0.0003; Online Supplementary Figure S2A). Furthermore, MIR200C/141 is known to be transcriptionally upregulated by MYC,41,42 an oncogenic transcription factor that is frequently overexpressed in T-PLL.2,43 In agreement, we noted a significant overexpression of MYC in T-PLL (FC=4.9; FDR=0.0019; Online Supplementary Figure S2B). In normal T-cell development miR-181 controls expression of various phosphatases, such as SHP2, PTPN22, DUSP5, DUSP6 and NRARP, thereby enhancing NOTCH and T-cell receptor signaling.29 In agreement, we found a significant downregulation of DUSP5 mRNA expression in T-PLL cells (FC=5.7; FDR=1.43x10-6; Online Supplementary Figure S2C).
Gene set enrichment analysis (GSEA) showed a significant enrichment of miR-200c/141 and miR-181a/b targets in the downregulated fraction of genes in T-PLL compared to normal effector CD4 T cells (P<0.05) (Figure 3A and B). Ingenuity pathway analysis (IPA) of significantly deregulated genes in T-PLL revealed that the TGFb network is affected in T-PLL, including transforming growth factor b1 (TGFb1) (FC=0.276; P<7.68x10-10), TGFb2 (FC=0.339, FDR=1.85x10-5 and TGFb receptor 3 (TGFbR3) (FC=0.046; FDR=9.66x10-19), which are all significantly downregulated in T-PLL (Online Supplementary Figure S3A), whereas the mRNA expression of TGFbR1 and TGFbR2 was not changed. Our array data showed reduced expression of TGFbR3 compared to all normal T-cell fractions tested, whereas TGFβR1 and TGFbR2 was not changed (Online Supplementary Figure S3B). Furthermore, we noted a reduced TGFbR3 membrane levels in T-PLL with high miR-200c/141 expression compared to normal CD4 T cells (Online Supplementary Figure S3C and D). Notably, despite unchanged TGFbR1 mRNA and protein levels in T-PLL, we did observe reduced TGFbR1 membrane expression on T-PLL compared to normal CD4 T cells (Online Supplementary Figure S3C and E).
Figure 3.
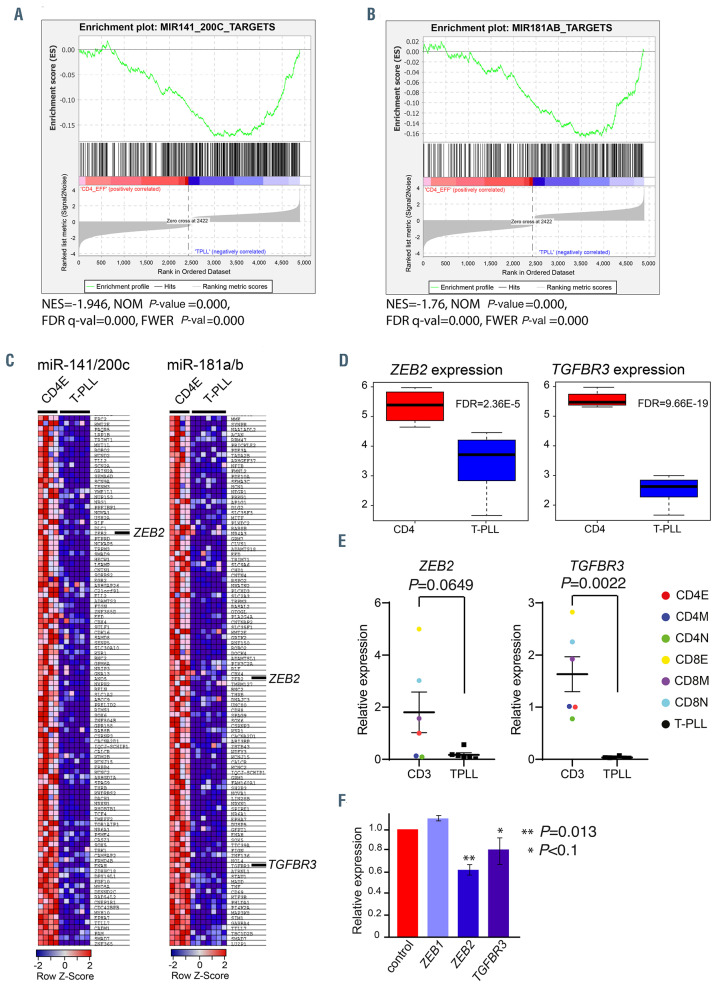
MicroRNA target analysis. (A and B) Gene set enrichment analysis (GSEA) of miR-200c/141 (A) and miR-181a/b (B) predicted targets (www.targetscan.org). In red are shown upregulated genes and in blue downregulated genes. The enrichment is indicated by the green line. NES is the normalized enrichment score. The significance is given by the normalized (NOM) P-value, false discovery rate (FDR) q-val and family-wise error rate (FWER) P-val. (C) Significantly downregulated target genes for the indicated microRNA (miRNA) are shown. (D) Box plots showing the expression of ZEB2 and TGFbR3 in T-cell-prolymphocytic leukemia (T-PLL) compared to effector CD4 T cells. The statistical significance is indicated by the FDR value. (E) Average expression of ZEB2 and TGFbR3 relative to GAPDH and CD4 effector cells is shown. Samples were measured in triplicate. Statistical significance was determined with a Mann-Whitney-U test. E: effector subset; M: memory subset; N: naïve subset. (F) Expression of miR-200c/141 targets in Jurkat cells that are transduced with retroviruses expressing miR-200c/141, relative to the expression in Jurkat cells transduced with empty vector (EV)-control virus (red). Statistical significance was determined with a Student’s t-test.
Figure 4.
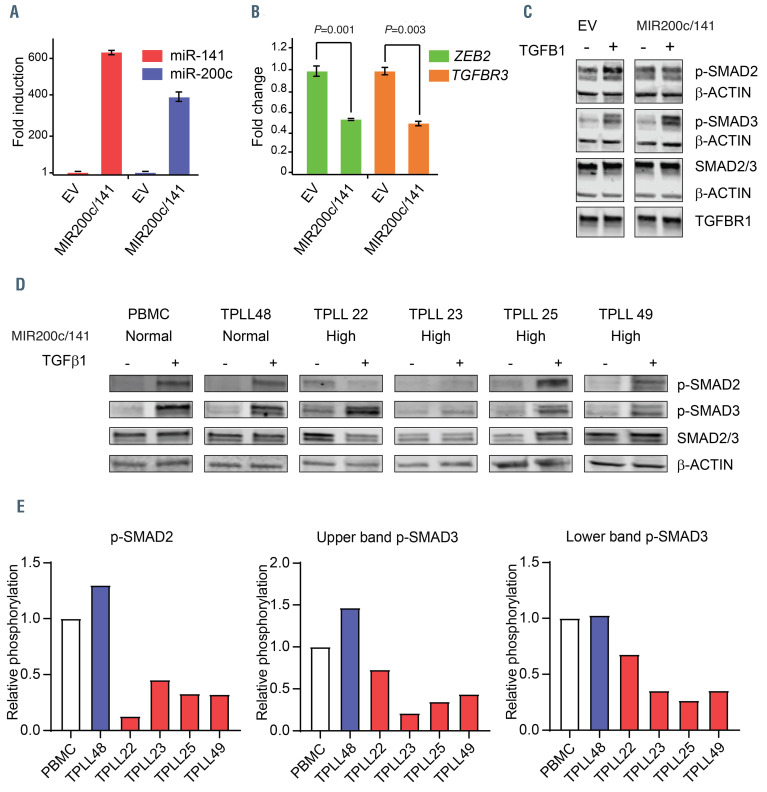
The TGFbR pathway is deregulated in T-cell prolymphocytic leukemia. (A) Overexpression of miR-200c and miR-141 relative to U6 in HeLa-Mir-200c/141 compared to HeLa-empty vector (EV) is plotted. (B) The mRNA expression of ZEB2 and TGFbR3 relative to GAPDH in HeLa-Mir-200c/141 compared to HeLa-EV is plotted. (C) HeLa-Mir-200c/141 and HeLa-EV cells were treated with hu-TGFb1 (10 ng/mL, +) or not stimulated (0 ng/mL, - hu-TGFb1) for 1 hour. Samples of cell lysates were taken and analyzed by western blotting using phospho-specific antibodies against SMAD2 and SMAD3, total SMAD2/3, b-actin and TGFβR1. Data are representative of three independent experiments. (D) Cells were stimulated with hu-TGFb1 (+ hu-TGFb1) or not stimulated (0 ng/mL, - hu-TGFβ1) for 1 hour. Samples of indicated cell lysates were analyzed by western blotting with phospho-specific antibodies against SMAD2 and SMAD3, total SMAD (SMAD2 = upper band, SMAD3 = lower band) and β-actin. (E) Quantification of the data shown in (D). Induction of hu-TGFb1-mediated phosphorylation of indicated phospo-proteins relative to the total SMAD expression level and peripheral blood mononuclear cells (PBMC) values of the different samples are depicted.
A global test on the gene expression profiles of the predicted miR-200c/141 targets in high miR-200c/141 samples and miR-181a/b targets in high miR-181a/b samples revealed that 220 miR-200c/141 targets and 115 miR- 181a/b targets were significantly downregulated in T-PLL compared to effector CD4 cells, including Zinc finger E box-binding homeobox 2 (ZEB2, also known as Smadinteracting protein 1 [SIP1]) and TGFbR3 (Figure 3C and D). We noted that ZEB2 expression is highly variable in normal T-cell fractions with the highest expression in effector CD8 cells and the lowest expression in naïve CD4 T cells (Figure 3E). Furthermore, we confirmed that the expression of ZEB2 in T-PLL is downregulated compared to effector CD4 T cells (Figure 3E). However, TGFbR3 expression is more homogeneously expressed in normal T-cell fractions and significantly downregulated in T-PLL compared to average of all normal T-cell fractions (Figure 3E). In addition, downregulation of ZEB2 and TGFbR3 (>80%) was found in a set of three additional CD4-positive T-PLL samples (T-PLL49, 44, 46) with high miR-200c and miR-141 levels compared to normal PBMC (Online Supplementary Figure S3E). ZEB2 mRNA contains four predicted well-conserved 8-mer sites, one 7-mer-m8 and one 7mer-A1 site for miR-200c-3p, three well-conserved 8-mer sites for miR-141-3p and is a validated target of miR-200c and miR-141.30 ZEB2, has been identified as a SMAD-interacting protein and regulates the transcriptional activities of the TGFb network.31 According to TargetScan, TGFbR3 contains one less conserved miR-200c site in the 3’-UTR. In order to further investigate the functions of miR-200c/141 in regulation of the TGFb network, we transduced Jurkat cells with pLX301-miR-200c/141 (co-expressing EGFP, miR-141 and miR-200c) or empty vector (EV) pLX301 expressing EGFP only. We noted that Jurkat-miR-200c/141 cells showed a 10-fold overexpression of miR-141 and a 5-fold overexpression of miR-200c compared to EV-transduced control Jurkat cells. We also observed a downregulation of both ZEB2 (P=0.013) and TGFbR3 mRNA (P<0.1) in Jurkat-miR-200c/141 cells compared to EV-transduced control Jurkat cells (Figure 3F).
Ligand stimulated TGFb type II receptor (TGFbRII) interacts with and phosphorylates TGFbRI, which results in activation of its kinase domain. This allows the TGFbR1 to phosphorylate SMAD2 and SMAD3 transcription factors. TGFbRIII act as a regulator of TGFb signaling and SMAD pathway activation, which is highly contextdependent. 32-34 In order to further investigate the role of miR-200c/141 in the regulation of TGFb-signaling, we transduced HeLa cells with either miR-200c/141 or EGFP only (EV) lentiviruses. HeLa-miR-200c/141 cells had a 397- fold miR-200c and a 624-fold miR-141 overexpression compared to HeLa-EV control cells (Figure 4A). Downregulation of ZEB2 and TGFbR3 mRNA levels in HeLa-miR-200c/141 cells was confirmed by quantitative polymerase chain reaction (qPCR) (Figure 4B). Interestingly, hu-TGFb1 stimulation of HeLa-miR- 200c/141 cells, resulted in increased p-SMAD3 levels, whereas p-SMAD2 was reduced and SMAD2, SMAD3 and TGFbR1 levels remained similar to their expression in control HeLa cells (Figure 4C). Next, we investigated hu- TGFb1-induced SMAD2 and SMAD3 phosphorylation in T-PLL. Notably, the expression of SMAD3 relative to SMAD2 was higher in T-PLL compared to PBMC and HeLa (Figure 4C and D). In addition, we observed a decreased p-SMAD2 and p-SMAD3 induction after hu- TGFb1 stimulation compared to the unstimulated condition in T-PLL with high miR-200c/141 compared to T-PLL with normal levels of miR-200c/141 and healthy PBMC (Figure 4D and E). Together, our results indicate that the miR-200c/141-ZEB2-TGFb- axis is aberrant in T-PLL.
Discussion
In this study, we characterized a cohort of T-PLL patients both phenotypically and molecularly. In full agreement with recently published data,2 we found multiple oncogenic abnormalities in our T-PLL cohort, such as aberrations in ATM, TP53 and high TCL1 overexpression. We did not observe the clinical and morphological heterogeneity of T-PLL due to differences in protein-encoding gene expression profiles as reported in previous publications. 2,35 This discrepancy can be largely explained by the relatively small cohort of T-PLL patients used for this study. Furthermore, we found that T-PLL generally displayed immunological characteristics mostly comparable to memory T cells, without evidence for identical TCR usage pointing towards a common antigen-driven leukemogenesis. Based on miRNA expression, T-PLL cells were most similar to effector T cells.
Our study reports an abnormal expression of miRNA in T-PLL. In total, we identified a set of 35 aberrantly expressed miRNA, that were upregulated compared to normal effector CD4 T cells. Although most T-PLL cases showed a relatively homogenous gene expression profile, T-PLL samples exhibited differential miRNA expression that correlated with blood cell counts and survival. The observation that differentially expressed miRNA are mostly upregulated in T-PLL is striking, since global miRNA depletion is more commonly found in human cancer.14 However, we cannot rule out that some miRNA are actually downregulated but with high degree of variation, such that they are not picked up in this small cohort of T-PLL cases. On the other hand, involvement of enhanced miRNA expression was already postulated based on elevated AGO2 expression as a result of chromosome 8 gains in a subset of T-PLL patients.2 Increased AGO2 expression has been detected in some human cancer types and it is well-established that this results in enhanced miRNA stability and expression levels.36 As we did not find a general AGO2 upregulation in our cohort of T-PLL cases, it remains to be determined why miRNA are upregulated in T-PLL. Some upregulated miRNA in T-PLL concern wellknown oncogenic miRNA, including the oncogenic driver miR-19b, which represses expression of the tumor suppressor gene PTEN37 and miR-125a, which strongly represses the expression of genes involved in apoptosis, such as TP53, PUMA and BAK1.38
Expression of miR-200c/141 is strongly upregulated in a subset of T-PLL. The miR-200c/141 cluster is frequently co-expressed with PTPN6 through transcriptional readthrough and complex 3D chromatin interactions between PTPN6 and miR-200c/141 promoters.28 Accordingly, we found a significant upregulation of PTPN6 in T-PLL samples with enhanced miR-200c/141 expression. It is wellestablished that the expression of miR-200c/141 is strongly induced by oxidative stress.39 As reactive oxygen species (ROS) levels are dramatically increased in T-PLL cells,2 this may therefore largely explain the elevated expression levels of miR-200c/141. Furthermore, MIR200C/141 is known to be transcriptionally upregulated by MYC,40,41 an oncogenic transcription factor that is frequently overexpressed in T-PLL, including our cohort.2,42
In our search for the biological implications of miR- 200c/141 upregulation, we identified a potential role for the miR-200c/141-ZEB2-TGFbR axis in the pathogenesis of T-PLL. The TGFb-signaling pathway regulates critical cellular processes in hematopoietic cells such as proliferation, differentiation, apoptosis and cell migration. Leukemia cells are commonly resistant to TGFb-controlled mechanisms due to mutations and deletion of molecules that are active in the TGFb pathway or due to disruption of the network by oncoproteins.43 We determined miR-200c and miR-141-mediated downregulation of TGFbR3 transcripts in HeLa cells. TGFbR3 is a contextdependent regulator of TGFb signaling and a potent tumor suppressor that is frequently inactivated in various types of cancer.32,34,44 In agreement with the gene expression data, we observed reduced TGFbR3 membrane expression on T-PLL cells. Although the levels of TGFbR1 mRNA and protein were unaffected, we did observe reduced TGFbR1 membrane expression on T-PLL cells. Reduced TGFbR3 expression in HeLa did not affect SMAD2/3 levels, though we noted an aberrant p-SMAD2 and p- SMAD3 level. Furthermore, we found that T-PLL samples with high miR-200c/141 levels have decreased p-SMAD2 and p-SMAD3 levels. Aberrant p-SMAD3 plays critical roles in stemness of tumor cells and progression of malignancies. 45,46 Although this would suggest an important role of p-SMAD proteins in T-PLL leukemogenesis, the oncogenic downstream functions of aberrant TGFbR3 expression and p-SMAD in T-PLL remain to be further investigated.
ZEB1 and ZEB2 are highly related SMAD-interacting transcription factors and important players in the induction of the epithelial–mesenchymal transition, which promotes invasion of tumor cells into the surrounding tissues, drug resistance, cell survival and metastasis.47 ZEB2 binds to SMAD2/3 and to SMAD1/5/8 in ligand-stimulated cells.31,48 ZEB proteins are involved in transcriptional repression by interacting with co-repressor protein CtBP and the NuRD chromatin-remodeling corepressor complex and are transcriptional activators by binding to P300/PCAF.49 ZEB1 and ZEB2 contain multiple miR-200c/141 target sequences in their 3’-UTR.30 Endogenous miRNA levels strongly repress ZEB1 and/or ZEB2 expression in a cell type-dependent fashion. For instance, during EMT miR-200 family members repress translation of both ZEB1 and ZEB2, whereas in CD8+ T cells only ZEB2 levels are downregulated, a process that is presumably controlled by RNA-binding proteins (RBP).30 Overexpression of miR-200 in CD8+ T cells selectively repressed ZEB2 expression, but not ZEB1, and promoted memory T-cell development.30 Interestingly, we found strong ZEB2 downregulation in T-PLL, whereas ZEB1 expression was not affected. The mechanism behind the selective downregulation of ZEB2 in T-PLL cells most-likely involves RBP.30 However, the RBP involved in selective ZEB expression regulation remain to be identified. How downregulation of ZEB2 and the above discussed aberrant p-SMAD3 levels affect target gene expression in T-PLL and how this contributes to oncogenic transformation remains to be unraveled.
We also found a strong upregulation of miR-181a and miR-181b in a subset of T-PLL samples. Upregulation of miR-181 is reported in human cancer, e.g., breast cancer and chronic lymphocytic leukemia (CLL).50-52 Although miR-181 is frequently downregulated in CLL as a whole, enhanced expression of miR-181a is associated with disease progression in trisomy 12 CLL cases.52 However, as this chromosomal aberration is not found in T-PLL,2 miR- 181 is presumably upregulated by a different mechanism. For instance, there is strong evidence that stress-induced STAT3 enhances miR-181 expression in tumor cells,53 a signaling cascade that is commonly activated in T-PLL.2 Enhanced miR-181 expression may have oncogenic transforming functions in T-PLL by affecting multiple pathways. Ectopic expression of miR-181a in normal mouse CD4+ T cells augments the strength and sensitivity of TCR signaling to agonists.54 In normal T-cell development miR-181 controls expression of various phosphatases, such as SHP2, PTPN22, DUSP5, DUSP6 and NRARP, thereby enhancing NOTCH and TCR signaling.29 In agreement, we found a significant downregulation of DUSP5 mRNA expression in T-PLL cells, which may be caused by miR-181-mediated repression. The importance of miR-181 for T-cell leukemogenesis is further demonstrated by an experiment in which genomic deletion of MIR181A1/B1 specifically inhibits NOTCH-induced leukemogenesis in mice through repression of downstream feedback mechanisms of NOTCH and pre-TCR signaling.29 Thus, overexpression of miR-181a1/b1 may contribute to enhanced NOTCH and TCR signaling, thereby enhancing oncogenesis in T-PLL.
In conclusion, we have shown that based on miRNA expression T-PLL is most similar to effector T-cell subsets albeit with some degree of heterogeneity between T-PLL cases. A set of 35 miRNA is aberrantly expressed in T-PLL, with miR-200c/141 being the most upregulated cluster. Aberrant expression of miR-200c/141 affects TGFb-controlled mechanisms that may contribute to the pathogenesis of T-PLL. Additionally, the extent of overexpression of miR-200c and miR-141 appeared to significantly correlate with increased white blood cell counts and poor survival and may thus have prognostic value. Our data highlight a potential role for aberrantly expressed oncogenic miRNA in the pathogenesis of T-PLL, which may pave the way for new therapeutic approaches in this disease.
Supplementary Material
Acknowledgments
We thank Steven Koetzier, Fabiënne van Opstal, Bernard Stikker and Stijn van den Broek for technical assistance during their bachelor and master projects.
Funding Statement
Funding: This work was supported an unrestricted grant from Bayer, Mijdrecht, the Netherlands (to MCKK, KvL, and AWL). CJS and IVZ are supported by Dutch Cancer Society (KWF), grant number: 10948.
References
- 1.Matutes E, Brito-Babapulle V, Swansbury J, et al. Clinical and laboratory features of 78 cases of T-prolymphocytic leukemia. Blood. 1991;78(12):3269-3274. [PubMed] [Google Scholar]
- 2.Schrader A, Crispatzu G, Oberbeck S, et al. Actionable perturbations of damage responses by TCL1/ATM and epigenetic lesions form the basis of T-PLL. Nat Commun. 2018;9(1):697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stengel A, Kern W, Zenger M, et al. Genetic characterization of T-PLL reveals two major biologic subgroups and JAK3 mutations as prognostic marker. Genes Chromosomes Cancer. 2016;55(1):82-94. [DOI] [PubMed] [Google Scholar]
- 4.Johansson P, Klein-Hitpass L, Choidas A, et al. SAMHD1 is recurrently mutated in T-cell prolymphocytic leukemia. Blood Cancer J. 2018;8(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maljaei SH, Brito-Babapulle V, Hiorns LR, Catovsky D. Abnormalities of chromosomes 8, 11, 14, and X in T-prolymphocytic leukemia studied by fluorescence in situ hybridization. Cancer Genet Cytogenet. 1998;103(2):110-116. [DOI] [PubMed] [Google Scholar]
- 6.Virgilio L, Narducci MG, Isobe M, et al. Identification of the TCL1 gene involved in T-cell malignancies. Proc Natl Acad Sci U S A. 1994;91(26):12530-12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herling M, Patel KA, Teitell MA, et al. High TCL1 expression and intact T-cell receptor signaling define a hyperproliferative subset of T-cell prolymphocytic leukemia. Blood. 2008; 111(1):328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gritti C, Dastot H, Soulier J, et al. Transgenic mice for MTCP1 develop T-cell prolymphocytic leukemia. Blood. 1998;92(2):368-373. [PubMed] [Google Scholar]
- 9.Pekarsky Y, Drusco A, Kumchala P, Croce CM, Zanesi N. The long journey of TCL1 transgenic mice: lessons learned in the last 15 years. Gene Expr. 2015;16(3):129-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Virgilio L, Lazzeri C, Bichi R, et al. Deregulated expression of TCL1 causes T cell leukemia in mice. Proc Natl Acad Sci U S A. 1998;95(7):3885-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zanesi N, Balatti V, Riordan J, et al. A Sleeping Beauty screen reveals NF-kB activation in CLL mouse model. Blood. 2013;121(21):4355-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. Metazoan MicroRNAs. Cell. 2018;173(1):20-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111(10):5078-5085. [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834-838. [DOI] [PubMed] [Google Scholar]
- 15.Fernandes Q. MicroRNA: defining a new niche in Leukemia. Blood Rev. 2017; 31(3):129-138. [DOI] [PubMed] [Google Scholar]
- 16.Yeh CH, Moles R, Nicot C. Clinical significance of microRNAs in chronic and acute human leukemia. Mol Cancer. 2016;15(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stavast CJ, Erkeland SJ. The non-canonical aspects of microRNAs: many roads to gene regulation. Cells. 2019;8(11):1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krowiorz K, Ruschmann J, Lai C, et al. MiR- 139-5p is a potent tumor suppressor in adult acute myeloid leukemia. Blood Cancer J. 2016;6(12):e508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alemdehy MF, Haanstra JR, de Looper HW, et al. ICL-induced miR139-3p and miR199a- 3p have opposite roles in hematopoietic cell expansion and leukemic transformation. Blood. 2015;125(25):3937-3948. [DOI] [PubMed] [Google Scholar]
- 20.Wojtowicz EE, Lechman ER, Hermans KG, et al. Ectopic miR-125a expression induces long-term repopulating stem cell capacity in mouse and human hematopoietic progenitors. Cell Stem Cell. 2016;19(3):383-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cimmino A, Calin GA, Fabbri M, et al. miR- 15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944-13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stavast CJ, Leenen PJM, Erkeland SJ. The interplay between critical transcription factors and microRNAs in the control of normal and malignant myelopoiesis. Cancer Lett. 2018;427:28-37. [DOI] [PubMed] [Google Scholar]
- 23.Laribi K, Lemaire P, Sandrini J, Baugier de Materre A. Advances in the understanding and management of T-cell prolymphocytic leukemia. Oncotarget. 2017;8(61):104664-104686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Dongen JJ, Lhermitte L, Bottcher S, et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia. 2012;26(9):1908-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrido P, Ruiz-Cabello F, Barcena P, et al. Monoclonal TCR-Vbeta13.1+/CD4+/NKa+/ CD8-/+dim T-LGL lymphocytosis: evidence for an antigen-driven chronic T-cell stimulation origin. Blood. 2007;109(11):4890-4898. [DOI] [PubMed] [Google Scholar]
- 26.Sandberg Y, Kallemeijn MJ, Dik WA, et al. Lack of common TCRA and TCRB clonotypes in CD8(+)/TCRalphabeta(+) T-cell large granular lymphocyte leukemia: a review on the role of antigenic selection in the immunopathogenesis of CD8(+) T-LGL. Blood Cancer J. 2014;4(1):e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotrova M, Novakova M, Oberbeck S, et al. Next-generation amplicon TRB locus sequencing can overcome limitations of flow-cytometric Vbeta expression analysis and confirms clonality in all T-cell prolymphocytic leukemia cases. Cytometry A. 2018; 93(11):1118-1124. [DOI] [PubMed] [Google Scholar]
- 28.Batista L, Bourachot B, Mateescu B, Reyal F, Mechta-Grigoriou F. Regulation of miR- 200c/141 expression by intergenic DNAlooping and transcriptional read-through. Nat Commun. 2016;7:8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fragoso R, Mao T, Wang S, et al. Modulating the strength and threshold of NOTCH oncogenic signals by mir-181a-1/b-1. PLoS Genet. 2012;8(8):e1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan T, Dominguez CX, Amezquita RA, et al. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8(+) T cell fates. J Exp Med. 2018; 215(4):1153-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verschueren K, Remacle JE, Collart C, et al. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5'-CACCT sequences in candidate target genes. J Biol Chem. 1999; 274(29):20489-20498. [DOI] [PubMed] [Google Scholar]
- 32.Vander Ark A, Cao J, Li X. TGF-beta receptors: In and beyond TGF-beta signaling. Cell Signal. 2018;52:112-120. [DOI] [PubMed] [Google Scholar]
- 33.Bilandzic M, Stenvers KL. Betaglycan: a multifunctional accessory. Mol Cell Endocrinol. 2011;339(1-2):180-189. [DOI] [PubMed] [Google Scholar]
- 34.Gatza CE, Oh SY, Blobe GC. Roles for the type III TGF-beta receptor in human cancer. Cell Signal. 2010;22(8):1163-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durig J, Bug S, Klein-Hitpass L, et al. Combined single nucleotide polymorphismbased genomic mapping and global gene expression profiling identifies novel chromosomal imbalances, mechanisms and candidate genes important in the pathogenesis of T-cell prolymphocytic leukemia with inv(14)(q11q32). Leukemia. 2007; 21(10): 2153-2163. [DOI] [PubMed] [Google Scholar]
- 36.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131(6):1097-1108. [DOI] [PubMed] [Google Scholar]
- 37.Han YC, Vidigal JA, Mu P, et al. An allelic series of miR-17 approximately 92-mutant mice uncovers functional specialization and cooperation among members of a microRNA polycistron. Nat Genet. 2015;47(7):766-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaham L, Binder V, Gefen N, Borkhardt A, Izraeli S. MiR-125 in normal and malignant hematopoiesis. Leukemia. 2012;26(9):2011-2018. [DOI] [PubMed] [Google Scholar]
- 39.Mateescu B, Batista L, Cardon M, et al. miR- 141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17(12):1627-1635. [DOI] [PubMed] [Google Scholar]
- 40.Chen P, Guo X, Zhang L, et al. MiR-200c is a cMyc-activated miRNA that promotes nasopharyngeal carcinoma by downregulating PTEN. Oncotarget. 2017;8(3):5206-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28(20):3157-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsi AC, Robirds DH, Luo J, et al. T-cell prolymphocytic leukemia frequently shows cutaneous involvement and is associated with gains of MYC, loss of ATM, and TCL1A rearrangement. Am J Surg Pathol. 2014; 38(11):1468-1483. [DOI] [PubMed] [Google Scholar]
- 43.Dong M, Blobe GC. Role of transforming growth factor-beta in hematologic malignancies. Blood. 2006;107(12):4589-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernabeu C, Lopez-Novoa JM, Quintanilla M. The emerging role of TGF-beta superfamily coreceptors in cancer. Biochim Biophys Acta. 2009;1792(10):954-973. [DOI] [PubMed] [Google Scholar]
- 45.Tarasewicz E, Jeruss JS. Phospho-specific Smad3 signaling: impact on breast oncogenesis. Cell Cycle. 2012;11(13):2443-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naka K, Jomen Y, Ishihara K, et al. Dipeptide species regulate p38MAPK-Smad3 signalling to maintain chronic myelogenous leukaemia stem cells. Nat Commun. 2015;6:8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fardi M, Alivand M, Baradaran B, Farshdousti Hagh M, Solali S. The crucial role of ZEB2: From development to epithelial-tomesenchymal transition and cancer complexity. J Cell Physiol. 2019;234(9):14783-14799. [DOI] [PubMed] [Google Scholar]
- 48.Conidi A, Cazzola S, Beets K, et al. Few Smad proteins and many Smad-interacting proteins yield multiple functions and action modes in TGFbeta/BMP signaling in vivo. Cytokine Growth Factor Rev. 2011;22(5-6):287-300. [DOI] [PubMed] [Google Scholar]
- 49.Scott CL, Omilusik KD. ZEBs: Novel players in immune cell development and function. Trends Immunol. 2019;40(5):431-446. [DOI] [PubMed] [Google Scholar]
- 50.Mansueto G, Forzati F, Ferraro A, et al. Identification of a new pathway for tumor progression: microRNA-181b up-regulation and CBX7 down-regulation by HMGA1 protein. Genes Cancer. 2010;1(3):210-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strotbek M, Schmid S, Sanchez-Gonzalez I, et al. miR-181 elevates Akt signaling by cotargeting PHLPP2 and INPP4B phosphatases in luminal breast cancer. Int J Cancer. 2017; 140(10):2310-2320. [DOI] [PubMed] [Google Scholar]
- 52.Visone R, Rassenti LZ, Veronese A, et al. Karyotype-specific microRNA signature in chronic lymphocytic leukemia. Blood. 2009;114(18):3872-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39(4):493-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li QJ, Chau J, Ebert PJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129(1):147-161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.