Abstract
Mutations in the CBL gene were first identified in adults with various myeloid malignancies. Some patients with juvenile myelomonocytic leukemia (JMML) were also noted to harbor mutations in CBL, but were found to have generally less aggressive disease courses compared to patients with other forms of Ras pathway-mutant JMML. Importantly, and in contrast to most reports in adults, the majority of CBL mutations in JMML patients are germline with acquired uniparental disomy occurring in affected marrow cells. Here, we systematically studied a large cohort of 33 JMML patients with CBL mutations and found that this disease is highly diverse in presentation and overall outcome. Moreover, we discovered somatically acquired CBL mutations in 15% of pediatric patients who presented with more aggressive disease. Neither clinical features nor methylation profiling were able to distinguish patients with somatic CBL mutations from those with germline CBL mutations, highlighting the need for germline testing. Overall, we demonstrate that disease courses are quite heterogeneous even among patients with germline CBL mutations. Prospective clinical trials are warranted to find ideal treatment strategies for this diverse cohort of patients.
Introduction
Juvenile myelomonocytic leukemia (JMML) is a rare and aggressive disease of young children that presents with features of both myelodysplasia and myeloproliferation. To date, hematopoietic stem cell transplantation (HSCT) is the only curative therapy and results in long-term survival of about 50-60% of patients.1,2 The mutational landscape of JMML has been well described in several large studies.3-6 Nearly all patients have initiating mutations in genes that activate the Ras pathway, most commonly, PTPN11, NRAS, KRAS, NF1 or CBL. Secondary mutations are found in approximately 30% of patients at diagnosis and are associated with inferior survival.
CBL mutations were first described in JMML patients in 2009 by our group and others7-10 and account for approximately 15% of all JMML cases.3,11,12 Germline tissue including cord blood and buccal samples were heterozygous for mutations, while leukemia cells were homozygous, indicating that loss of heterozygosity was required for leukemogenesis. The majority of these children presented with syndromic features including facial dysmorphia, growth retardation, cryptorchidism and autoimmune phenomena, specifically vasculitis – a condition that is now called “CBLsyndrome”. Patients with CBL-syndrome are at risk of developing JMML. Surprisingly, several of these patients with confirmed JMML experienced spontaneous resolution of their leukemia. Based on this observation and in stark contrast to other forms of JMML, CBL-syndrome JMML has been thought to be associated with relatively good prognosis. Treatment has evolved accordingly for these patients, who undergo a period of close observation of blood counts and assessment of splenomegaly, with HSCT utilized only after more conventional therapies have failed.13
In overlapping adult myelodysplastic syndromes/myeloproliferative neoplasms, somatically acquired CBL mutations occur in about 10-15% of cases and are associated with a poor prognosis. Several studies in chronic myelomonocytic leukemia (CMML), a disease of adults with many similarities to JMML, have shown that CBL mutations are associated with inferior survival.14-16
In our experience, we have observed that the spectrum of clinical courses of patients with CBL-mutated JMML is wide, ranging from spontaneous resolution to aggressive disease with transformation to acute myeloid leukemia. This observation prompted us to perform a systematic study of clinical presentations and molecular features to identify potential factors that can identify patients who may be observed and those who need immediate therapy.
Methods
Patients
We collected samples and clinical data from 33 patients diagnosed with JMML according to international diagnostic criteria17 who were found to have a clonal CBL mutation. The patients were diagnosed between 2006 and 2019 in 27 different centers across the USA and Canada. Treatment decisions were at the discretion of the treating physicians and were independent of this study. Written consent and specimens were obtained from all patients at the time of routine clinical assessments. The study design was reviewed and approved by the institutional review board of University of California San Francisco, in accordance with the Declaration of Helsinki.
Material and studies
We received bone marrow and/or peripheral blood from all patients, collected at the time of initial diagnosis or shortly thereafter. Mononuclear cells were isolated using a Ficoll method and DNA was extracted using the Gentra Puregene kit (Qiagen, Germany). For identification of genomic mutations, DNA was extracted from bone marrow or peripheral blood mononuclear cells. DNA from peripheral blood was used for DNA methylation analysis. The germline status of CBL mutations was determined using material from either a buccal swab, cord blood or after sorting CD3+ T cells from peripheral blood. A somatic mutation was defined as a mutation that was identified in leukemia tissue (peripheral blood or bone marrow), but had a minimum variant allele fraction (VAF) of <5% in any germline tissue (epithelial cells from buccal swab, cord blood, or T cells).
Next-generation sequencing of juvenile myelomonocytic leukemia-associated mutations
Genomic DNA samples were sequenced using a custom amplicon- based sequencing approach (Paragon Genomics, Hayward, CA, USA) targeting 26 genes that are known to be recurrently mutated in JMML18 (Online Supplementary Table S1 and Online Supplementary Methods). A VAF of 0.05 (=5%) at diagnosis was required for reporting.
Targeted MethylSeq library preparation, sequencing and hierarchical clustering
Genomic DNA (300 ng) was bisulfite converted and 100 ng of converted single-strand DNA were used as the input for a custom 3000 CpG loci targeted MethylSeq assay (Tecan). Single-index library pools were sequenced on the Illumina NovaSeq with paired-end 150 base-pair (bp) mode averaging 20x106 reads per library preparation. MethylSeq hierarchical clustering and classification were performed according to the international consensus definition12 as described in the Online Supplementary Methods.
Statistical analyses
Spontaneous resolution of disease was defined as normalization of monocyte counts in the peripheral blood and reduction of spleen to normal size without any JMML-directed therapy. Persistent disease was defined as not achieving complete remission according to the International Working Group definition19 with or without therapy excluding HSCT. Overall survival was defined as time from initial diagnosis to death from any cause and was estimated using the Kaplan-Meier method. The median follow- up time was estimated using the reverse Kaplan-Meier method. Differences in clinical features between patients with germline CBL mutations and those with somatic-only CBL mutations were tested for significance using the Fisher exact test for categorical variables and the Mann-Whitney U test for continuous variables. The level of statistical significance for all tests was a Pvalue less than 0.05. All calculations were performed using Microsoft Excel (version 16.16.3), GraphPad Prism software (version 8.0) and R (version 3.4.1).
Results
Patients’ characteristics
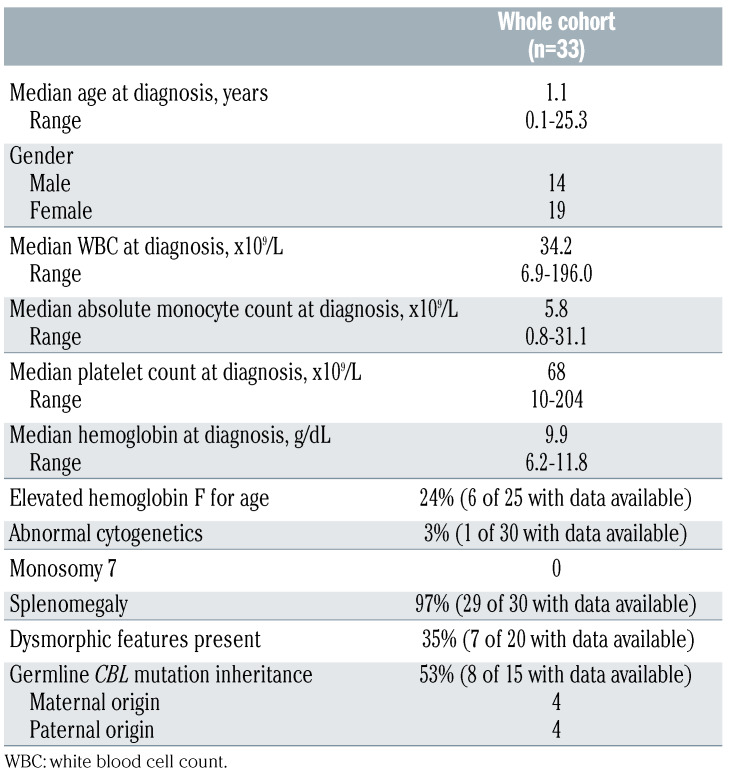
The cohort consisted of 33 patients with a median age at diagnosis of 1.1 years (range, 1 month - 25 years). The median follow-up in this study was 3.5 years (range, 0.2-12.1 years). The patients’ characteristics at diagnosis are shown in Table 1. Additional clinical information for each patient can be found in Online Supplementary Table S2.
Mutational spectrum of CBL-mutated juvenile myelomonocytic leukemia and occurrence of secondary mutations
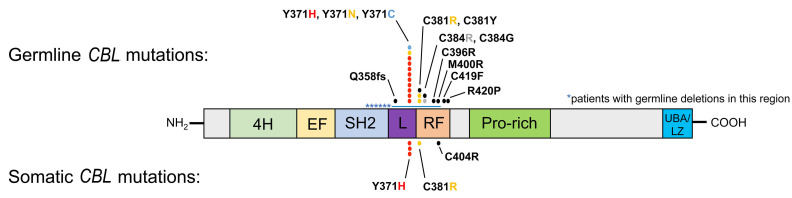
Clonal CBL mutations were confirmed for every patient in the cohort and were exclusively located in the linker and ring finger domains, as previously reported7,16,20 (Figure 1 and Online Supplementary Table S3). Twenty-eight patients had germline alterations of CBL, including six patients with deletions affecting these domains. All but two of these patients with germline CBL mutations showed a higher VAF of the same mutation in their tumor as a result of heterozygosity (all VAF are shown in Online Supplementary Table S3). Of the two patients without heterozygosity, one had a deletion and the other a splice site mutation, compatible with previous reports in other patients.21 Interestingly, we identified five patients (15%) in our cohort with somatic- only CBL mutations. The somatic-only mutations were located at the same hotspot positions as those in the germline population and were homozygous in four patients and heterozygous in one patient. We then conducted further experiments in these five patients with somatic-only CBL mutations. To confirm that these mutations were in fact somatic in nature, buccal swabs were tested in four patients and a cord blood sample in the remaining patient. No exonic or intronic alterations of CBL or any other Ras pathway genes were found in the germline samples of these five patients by targeted deep sequencing. In one of the patients with a somatic-only CBL mutation, we discovered a heterozygous RUNX1 p.R166Q germline mutation, which has been reported in familial platelet disorder with propensity to myeloid malignancy syndrome.
Overall, missense CBL mutations at residue Y371 were the most common alteration in both groups. One patient with a germline CBL mutation was found to have a secondary somatic CBL mutation with a lower VAF, leading to loss of heterozygosity. No other somatic secondary mutations were found in either cohort using our targeted 26-gene JMML panel at diagnosis.
Comparison of patients with germline or somatic-only CBL mutations
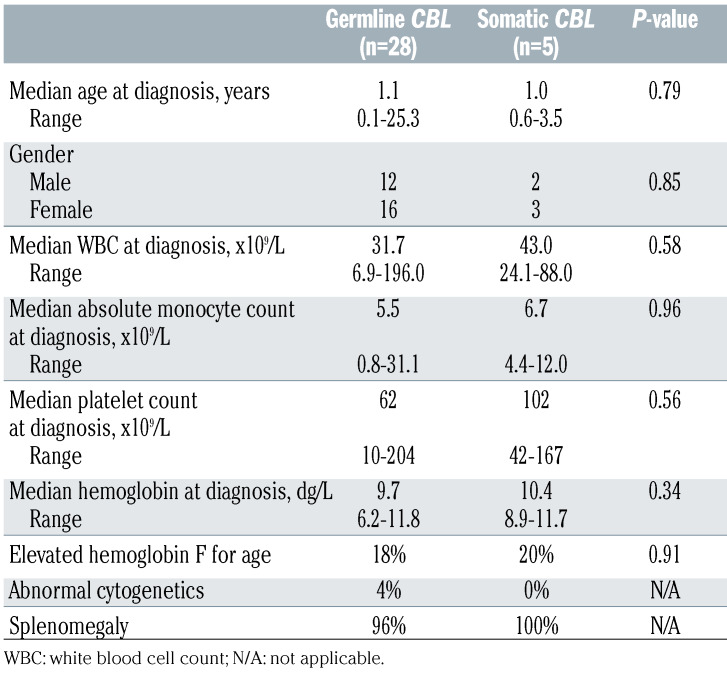
Table 2 describes the clinical presentation of patients with germline CBL mutations compared to those with somaticonly CBL mutations. There were no significant differences between the two cohorts in the patients’ characteristics at diagnosis (Table 2). Of note, patients in both cohorts presented at a median age of 1 year. Clinical signs of dysmorphia could not be used to distinguish between the groups, as 13 of 20 patients with germline CBL mutations had no overtly syndromic features (65%, data not available for 8 patients). None of the patients with somatic CBL mutations presented with obvious signs of dysmorphia. In eight of 15 (53%) patients with germline CBL mutations for whom information was available, the mutation was documented to be inherited from a parent with equal rates of maternal and paternal inheritance.
Outcome and diversity of clinical courses of CBL-mutated juvenile myelomonocytic leukemia
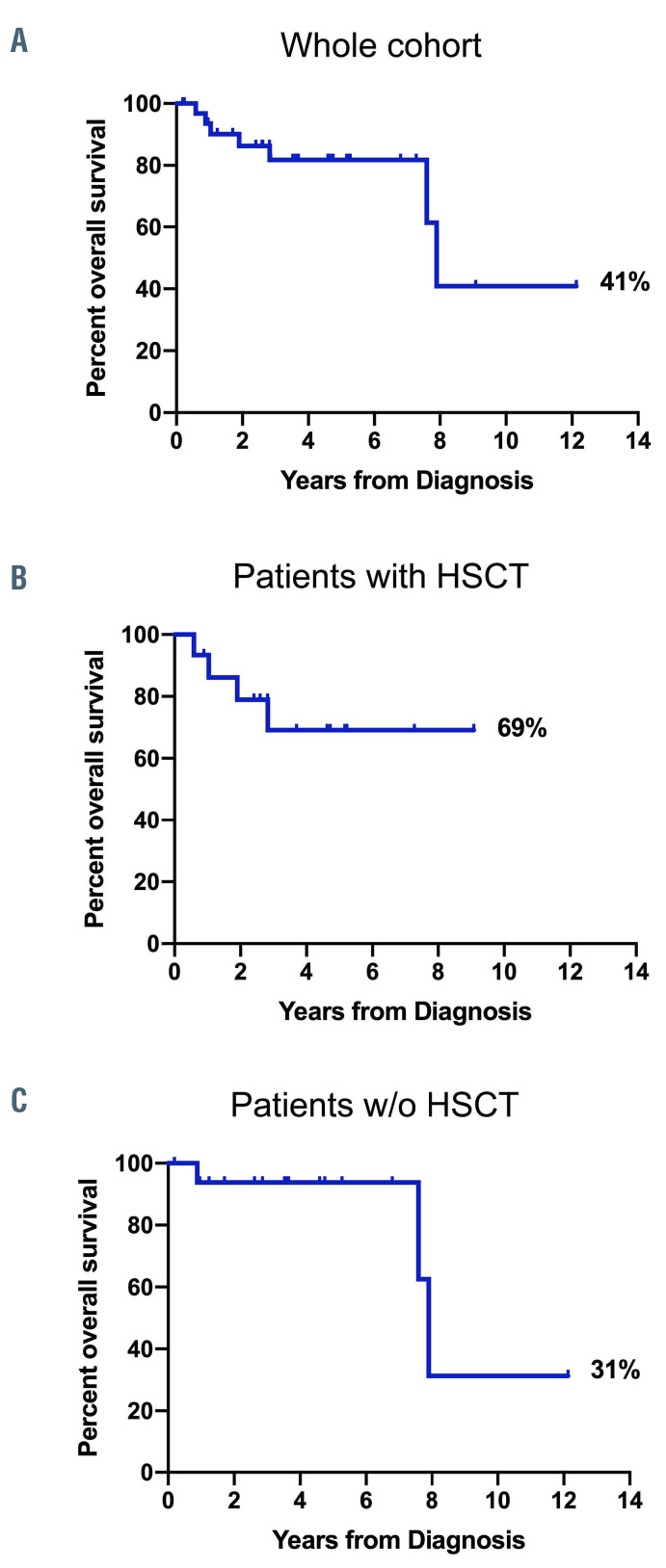
The overall survival rate of the whole cohort after a median follow-up of 3.7 years was 41% (95% confidence interval [95% CI]: 7-75%; median survival: 7.9 years) (Figure 2A). However, 45% of the patients (15 of 33) underwent HSCT at a median time of 0.5 years after initial diagnosis (range, 0.3-2.6 years). Overall survival of patients who underwent HSCT was 69% (95% CI: 36-87%; median survival not reached) compared to 31% (95% CI: 1-74%; median survival: 7.9 years) for patients who did not undergo HSCT (Figure 2B, C). Of note, all three deaths in the non-transplanted cohort were due to other organ failure associated with CBL-syndrome unrelated to JMML. There was no difference in overall survival between the patients with germline CBL mutations and those with somatic CBL mutations (Online Supplementary Figure S1).
Table 1.
Patients’ characteristics at diagnosis.
Figure 1.
Germline and somatic CBL mutations in patients with juvenile myelomonocytic leukemia. Each dot represents one mutation found at the specified codon. The resulting change in amino acid is coded by colors. The stars represent six patients found to have deletions. 4H: four helix bundle; EF: EF-hand like domain; SH2: Src homology 2 domain; L: linker domain; RF: ring finger domain; Pro-rich: proline-rich domain; UBA/LZ: ubiquitin-associated/leucine zipper domain.
We observed that all five patients with somatic CBL mutations were refractory to moderately-intense myeloidbased chemotherapy (i.e., 2 g/m2/day cytarabine alone or in combination with 30 mg/m2/day fludarabine).1 Three of these five patients received an allogeneic HSCT, and two are alive and disease-free at the time of last follow-up. One relapsed with frank acute myeloid leukemia following allogeneic HSCT for JMML and died of infectious complications (Vignette 1).
Figure 2.
Kaplan-Meier survival curves for CBL-juvenile myelomonocytic leukemia. (A) Overall survival of the whole cohort (n=33). (B) Overall survival of patients who underwent hematopoietic stem cell transplantation (n=15). (C) Overall survival of patients who did not undergo hematopoietic stem cell transplantation (n=17). One patient who was lost to follow-up was not considered in analyses for (B) or (C). HSCT: hematopoietic stem cell transplantation; w/o: without.
The clinical courses of the 28 patients with germline CBL mutations were heterogeneous. Three patients experienced spontaneous resolution of JMML without any treatment. Two patients underwent splenectomy as their only treatment but both died several years later of CBL-syndromeassociated organ failure (Vignette 2). Twelve patients received allogeneic HSCT following different pretransplant therapies. Nine of the transplanted patients went into remission after the transplant (2 after graft failure [Vignette 5 in the Online Supplementary Material and a separate case previously described by Oshrine.22]). Among the remaining three patients who were transplanted, two died of relapsed JMML and one died of transplant-related complications.
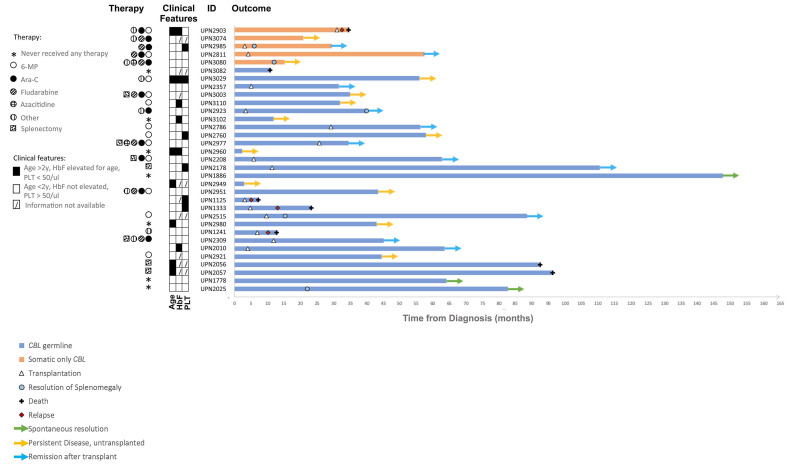
Of the 11 patients who were not transplanted and did not experience spontaneous resolution, nine have persistent disease without having been transplanted to date. The median follow-up of these patients is 3.5 years and their treatments included low-dose chemotherapy (Vignette 3), moderately-intense acute myeloid leukemia-directed chemotherapy, azacitidine, and/or targeted agents. Several patients received no therapy, while others were refractory to multiple lines of therapy. One patient with multiple congenital abnormalities died from complications of multiorgan failure in infancy. One patient was lost to follow up. An overview of all disease courses is shown in Figure 3.
Methylation-based clustering of patients’ samples
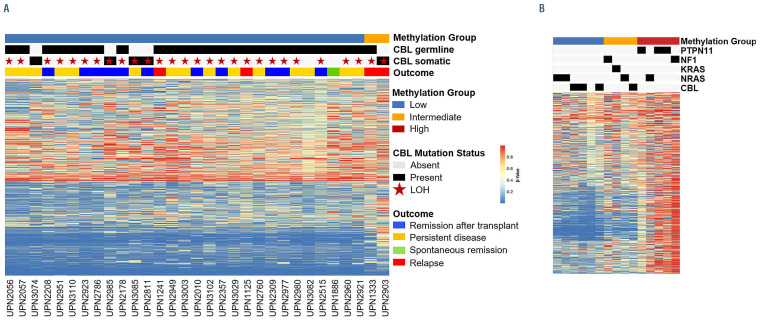
In order to investigate whether DNA methylation profiles are capable of distinguishing patients who will experience spontaneous resolution from those with more aggressive disease, we assessed the methylation of approximately 3,000 CpG loci using peripheral blood of each patient at diagnosis. Patients were designated as having low, intermediate or high methylation based on minimum distance to the centroid of the international consensus definition (Figure 4).1,5,12,23 Thirty-one of the 33 CBL-mutated patients were classified as having low methylation irrespective of whether their mutation was germline or somatic in nature, while the other two patients, one with a germline mutation, the other with a somatic mutation, were classified as having intermediate methylation. Notably, both patients who were classified as having intermediate methylation relapsed after HSCT. Unsupervised hierarchical clustering did not reveal any differences in methylation patterns among patients with germline CBL mutations who experienced spontaneous remission, survived more than 4 years without HSCT or died without receiving treatment.
Table 2.
Comparison of clinical characteristics of patients with germline or somatic-only CBL mutations.
Figure 3.
Swimmer plot showing the clinical course of each patient over time. Each bar represents one patient color-coded based on the presence of germline CBL (blue) or somatic-only CBL (orange). Dates of hematopoietic stem cell transplantation (HSCT), relapse, death or resolution of splenomegaly are depicted by symbols. The current ongoing status of the patient is depicted as a color-coded arrow. Therapeutic agents received by the patient (before HSCT if applicable) are shown on the left side with pattern-coded dots. Clinical features (age, hemoglobin F levels and platelet counts at diagnosis are depicted as filled (if true) or empty (if false) boxes. Dashed boxes are used if data were not available. Information on treatment was missing for patients UPN1125, UPN2949 and UPN 2357; patient UPN2949 was lost to follow-up. HbF: hemoglobin F; PLT: platelet count.
Figure 4.
DNA methylation profiles of patients with CBL-mutated juvenile myelomonocytic leukemia. (A) CBL mutant samples profiled by MethylSeq classified according to the international juvenile myelomonocytic leukemia (JMML) consensus signature. (B) DNA methylation groups low, intermediate, and high were defined using an international cohort of JMML samples described by Schönung et al.12 Each CBL mutant sample was classified into one of the three methylation groups based on minimum distance to the nearest centroid. Heatmaps (A and B) show the b values of 1,386 CpG loci used for methylation classification. LOH: Loss of heterozygosity. Two patients in the low methylation group are not depicted in (A) because their methylation data were generated with an Illumina 450k array.
To demonstrate the wide spectrum of clinical presentations and the variability in treatment decisions we share the following vignettes.
Vignette 1: UPN2903
A previously healthy 12-month-old female infant was noted to have thrombocytopenia with a normal hemoglobin concentration and white blood cell count on a routine full blood count. The patient was asymptomatic and was observed. At 3 years of age, leukoerythroblastosis was noted on her peripheral blood smear, and her fetal hemoglobin concentration was noted to be elevated for age. Her bone marrow was normocellular for age with trilineage hematopoiesis and no dysplasia. Genetic sequencing revealed a CBL p.Y371H mutation at 70% VAF. The patient was noted to have possible facial dysmorphology. A presumptive diagnosis of germline CBL-associated JMML was made, although no germline testing was performed at that time. The patient was observed for nearly 2 years at which point her white cell count increased to 40x109/L. A repeat bone marrow examination was performed, which revealed transformation to acute myeloid leukemia with 20% blasts and a der(16)t(1;16) on cytogenetics resulting in partial deletion of 16q and partial trisomy for 1q. Repeat tumor testing revealed that the CBL mutation had decreased to 52% VAF, and additional NRAS p.G13D (22%) and KRAS p.T58I (3%) mutations were noted. The patient went on to receive various chemotherapeutic regimens, including daunorubicin, cytarabine and etoposide, mitoxantrone and cytarabine, 6-mercaptopurine, hydroxyurea, trametinib and sorafenib, but was never able to achieve remission. The patient was transplanted with 10% bone marrow blasts using busulfan, cyclophosphamide, and melphalan conditioning, but died of infection and graft-versus-host disease shortly after the transplant. Post-mortem, a cord blood sample was analyzed and confirmed the absence of a germline CBL mutation. However, a heterozygous RUNX1 p.R166Q mutation was detected in the cord blood, indicating that the patient had an autosomal dominant familial platelet disorder with propensity to myeloid malignancy syndrome.
Vignette 2: UPN2056
A 6-month-old patient with several congenital anomalies including hypoplastic kidneys, midgut malrotation, webbed neck, and choanal atresia was suspected of having Kabuki or Noonan syndrome. Over time, he displayed significant developmental delay and short stature. At 3 years of age, this patient developed a myeloproliferative disorder with features suggestive of JMML, including monocytosis, anemia, and juvenile xanthogranulomas, and fewer than 5% blasts in his bone marrow. At this time, tumor sequencing revealed a novel homozygous frameshift mutation in exon 7 of CBL. Germline testing revealed the identical heterozygous deletion. Of note, the patient had an identical twin brother who developed a myeloproliferative disorder at nearly the same time with similar congenital anomalies and was found to carry the same mutation. The patient had a splenectomy at age 4 years, but his monocytosis persisted. The patient had multiple medical comorbidities, including chronic renal disease, seizures and hypothyroidism, and died of complications unrelated to JMML.
Vignette 3: UPN2977
A previously healthy 9-month-old female was evaluated for fatigue and splenomegaly. A full blood count revealed a white blood count of 52x109/L, hemoglobin concentration of 9.0 g/dL, and platelet count of 68x109/L. Bone marrow evaluation demonstrated monocytosis with normal cytogenetics. Genetic testing revealed a germline heterozygous CBL p.Y371H mutation with loss of heterozygosity in the bone marrow. She was initially observed, but subsequently failed to thrive and developed worsening splenomegaly and leukocytosis (peak WBC of 121x109/L). She was started on 6-mercaptopurine with a transient improvement in her white blood cell count. She experienced recurrent splenomegaly, leukocytosis, and thrombocytopenia, so went on to receive two cycles of fludarabine/cytarabine and underwent splenectomy. Despite these interventions, she had persistent leukocytosis. Repeat bone marrow evaluation was consistent with persistent JMML by morphology, but demonstrated new cytogenetic changes with a 3q deletion. Given her refractory JMML and cytogenetic evolution, she received four cycles of azacitidine followed by a haplo-identical peripheral blood stem cell transplant from her mother with busulfan, cyclophosphamide, and melphalan conditioning. She had engraftment failure, so received a stem cell boost and then ultimately received 5/6 unrelated umbilical cord blood transplant with fludarabine, cyclophosphamide, and total body irradiation as conditioning. Her post-transplant course was complicated by recurrent bacteremia and cytomegalovirus viremia. She is now in clinical remission more than 1 year after transplantation and is doing well.
Discussion
In this study, we comprehensively analyzed the largest cohort of CBL-mutated JMML patients described to date. The clinical courses and outcomes of the 33 patients analyzed varied dramatically, and a typical “watch and wait” approach was not appropriate for all patients.
Interestingly, we found that five of the 33 patients had somatically acquired CBL mutations. To our knowledge, this is the first time somatic CBL mutations have been described in JMML. All mutations, whether germline or somatic in nature, were located at the known hotspot loci in the CBL gene. No secondary mutations were found in any other JMML-associated genes in any of these patients at diagnosis. Moreover, patients with germline or somatic CBL mutations did not differ in their clinical presentation or laboratory features at diagnosis. Surprisingly, the overall survival rate of patients with germline CBL mutations in our cohort was lower than that in previous reports. One question that remains unanswered is whether transplantation for germline CBL-mutant patients can effectively decrease or prevent the known non-hematologic complications of CBL-syndrome and potentially increase overall survival. There was a trend towards improved overall survival in patients with germline mutations who underwent HSCT but transplant-related mortality remains a concern.
In a prior study, five of six patients with CBL-syndrome JMML who were not transplanted experienced spontaneous resolution of their hematologic diseases.8 In our cohort, we observed spontaneous resolution in only three of the 18 non-transplanted CBL-mutated JMML patients. In particular, none of the patients with somatic-only CBL mutations experienced spontaneous resolution, which is consistent with other JMML subtypes harboring somatic Ras pathway mutations. In adults with chronic myelomonocytic leukemia, somatic CBL mutations are found in about 10-15% of cases and are associated with a higher rate of transformation to acute myeloid leukemia and inferior overall outcomes.14-16 Similarly, while germline PTPN11 mutations are associated with Noonan syndrome and a typically self-limited form of JMML,24 patients with somatic PTPN11 mutations experience aggressive leukemia and rarely, if ever, have spontaneous remission.23,25 In our cohort, all five patients with somaticonly CBL mutations were refractory to medium-intensity myeloid-based chemotherapy and required HSCT for definitive disease control. We thus maintain that patients with somatic-only CBL-mutated JMML should not be observed without therapeutic intervention, but instead should be treated with HSCT. Furthermore, somatic-only testing of JMML samples is insufficient for proper identification of these patients, and dedicated germline testing should also be performed in all patients with CBL mutations.
Aberrant DNA methylation profiles have shown to be a promising tool for risk stratification in JMML.5,11,23 We therefore assessed the methylome of each patient at diagnosis. Four of the patients had been assessed for methylation in previous methylation studies (UPN1333, UPN2056, UPN2178, UPN2309) that utilized an arraybased approach. The current study assessed methylation status using a sequencing-based approach and all four patients were found to have the same methylation designation as in the previous studies. All but two of the 33 patients clustered with the low-methylation group and both of the patients with intermediate methylation relapsed after HSCT. However, methylation analysis was not able to distinguish between patients who experienced spontaneous remission and those with persistent disease. At this time, methylation analysis does not appear to be a biomarker capable of predicting outcome in patients with CBL-mutated JMML, in contrast to other JMML subtypes.
The optimal treatment for patients who require therapy due to splenomegaly or cytopenias is still unclear. The patients reported in this manuscript were diagnosed between 2006 and 2019. The optimal management of CBL-mutated patients with JMML has evolved over time, and no consensus currently exists given the relatively recent identification of this JMML subtype and the heterogeneity of its presentation. Several experimental studies have hinted at the potential use of targeted therapies in CBL-mutated disease. Mutant CBL is known to engage LYN/PI3K/AKT and JAK2 proteins, and targeted kinase inhibitors of these signaling pathways have been proposed as potential therapeutic strategies.26,27 Preclinical studies have specifically investigated the SRC/LYN inhibitor dasatinib and have shown decreased sensitivity of CBL-mutant JMML cells to granulocyte-macrophage colony-stimulating factor and increased chemotherapy sensitivity.28,29 Another preclinical study using patientderived induced pluripotent stem cells with CBL mutations reported potential efficacy of JAK2 and PI3K/mTOR signaling inhibition and decreased granulocytemacrophage colony-stimulating factor hypersensitivity.30 To date, there have been no prospective clinical trials using these agents in JMML. Further research and international collaboration are warranted to investigate the potential use of targeted therapies in patients with CBLmutated JMML, as a “watch and wait” approach is likely to be effective in only a minority of patients.
Supplementary Material
Acknowledgments
We thank the patients and parents who participated in this study, who made this research possible. We also acknowledge the following physicians who referred patients to our study and without whom this work would not have been possible: Dr. David Becton and Dr. Kimo Stine (Arkansas Children’s Hospital), Dr. Elizabeth Raetz (New York University, Langone), Dr. Paul Shaughnessy (Texas Transplant Institute) and Dr. Alan Ikeda (Children's Specialty Center of Nevada).
Funding Statement
Funding: This work was supported by the National Institutes of Health, National Cancer Institute grants 1U54CA196519 (to MLL, ES); 1U01CA232486 (SKT); 1K08CA184418 (SKT); National Institutes of Health, National Heart, Lung, and Blood Institute grant K08HL135434 (ES); the Leukemia Lymphoma Society (MLL, ES); Cookies for Kids Cancer (MLL); Pediatric Cancer Research Foundation (ES); the V Foundation (ES); the UCSF Catalyst Program (ES); the California Cancer League (ES); the Frank A. Campini Foundation (MLL and ES); the Children’s Hospital of Philadelphia Center for Childhood Cancer Research (SKT) and the German Research Foundation HE 7682/1-1 (AH). Next-generation sequencing was supported by the Center for Advanced Technologies at UCSF and the Computational Biology and Informatics group at the UCSF HDFCCC (supported by NCI grant: 5P30CA082103). MLL is the Benioff Chair of Children’s Health and the Deborah and Arthur Ablin Endowed Chair for Pediatric Molecular Oncology at Benioff Children’s Hospital.
References
- 1.Stieglitz E, Ward AF, Gerbing RB, et al. Phase II/III trial of a pre-transplant farnesyl transferase inhibitor in juvenile myelomonocytic leukemia: a report from the Children's Oncology Group. Pediatr Blood Cancer. 2015;62(4):629-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dvorak CC, Satwani P, Stieglitz E, et al. Disease burden and conditioning regimens in ASCT1221, a randomized phase II trial in children with juvenile myelomonocytic leukemia: a Children's Oncology Group study. Pediatr Blood Cancer. 2018;65(7): e27034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stieglitz E, Taylor-Weiner AN, Chang TY, et al. The genomic landscape of juvenile myelomonocytic leukemia. Nat Genet. 2015;47(11):1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caye A, Strullu M, Guidez F, et al. Juvenile myelomonocytic leukemia displays mutations in components of the RAS pathway and the PRC2 network. Nat Genet. 2015;47(11):1334-1340. [DOI] [PubMed] [Google Scholar]
- 5.Lipka DB, Witte T, Toth R, et al. RAS-pathway mutation patterns define epigenetic subclasses in juvenile myelomonocytic leukemia. Nat Commun. 2017;8(1):2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murakami N, Okuno Y, Yoshida K, et al. Integrated molecular profiling of juvenile myelomonocytic leukemia. Blood. 2018;131(14):1576-1586. [DOI] [PubMed] [Google Scholar]
- 7.Loh ML, Sakai DS, Flotho C, et al. Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood. 2009;114(9):1859-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niemeyer CM, Kang MW, Shin DH, et al. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat Genet. 2010;42(9):794-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiba N, Kato M, Park MJ, et al. CBL mutations in juvenile myelomonocytic leukemia and pediatric myelodysplastic syndrome. Leukemia. 2010;24(5):1090-1092. [DOI] [PubMed] [Google Scholar]
- 10.Perez B, Mechinaud F, Galambrun C, et al. Germline mutations of the CBL gene define a new genetic syndrome with predisposition to juvenile myelomonocytic leukaemia. J Med Genet. 2010;47(10):686-691. [DOI] [PubMed] [Google Scholar]
- 11.Murakami N, Okuno Y, Yoshida K, et al. Integrated molecular profiling of juvenile myelomonocytic leukemia. Blood. 2018;131(14):1576-1586. [DOI] [PubMed] [Google Scholar]
- 12.Schönung M, Meyer J, Nöllke P, et al. International consensus definition of DNA methylation subgroups in juvenile myelomonocytic leukemia. Clin Cancer Res. 2021;27(1):158-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locatelli F, Niemeyer CM. How I treat juvenile myelomonocytic leukemia. Blood. 2015;125(7):1083-1090. [DOI] [PubMed] [Google Scholar]
- 14.Duchmann M, Yalniz FF, Sanna A, et al. Prognostic role of gene mutations in chronic myelomonocytic leukemia patients treated with hypomethylating agents. EBioMedicine. 2018;31:174-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itzykson R, Kosmider O, Renneville A, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31(19):2428-2436. [DOI] [PubMed] [Google Scholar]
- 16.Jankowska AM, Makishima H, Tiu RV, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118(14): 3932-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan RJ, Cooper T, Kratz CP, Weiss B, Loh ML. Juvenile myelomonocytic leukemia: a report from the 2nd International JMML Symposium. Leuk Res. 2009;33(3):355-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hecht A, Meyer J, Chehab FF, et al. Molecular assessment of pretransplant chemotherapy in the treatment of juvenile myelomonocytic leukemia. Pediatr Blood Cancer. 2019;66(11):e27948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niemeyer CM, Loh ML, Cseh A, et al. Criteria for evaluating response and outcome in clinical trials for children with juvenile myelomonocytic leukemia. Haematologica. 2015;100(1):17-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kales SC, Ryan PE, Nau MM, Lipkowitz S. Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Res. 2010;70(12):4789-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strullu M, Caye A, Cassinat B, et al. In hematopoietic cells with a germline mutation of CBL, loss of heterozygosity is not a signature of juvenile myelo-monocytic leukemia. Leukemia. 2013;27(12):2404-2407. [DOI] [PubMed] [Google Scholar]
- 22.Oshrine B. Primary graft failure but treatment success: a case of reversion to heterozygosity after allogeneic hematopoietic cell rransplantation with autologous hematopoietic recovery in a child with CBLrelated juvenile myelomonocytic leukemia. J Pediatr Hematol Oncol. 2021;43(3):e426-e428. [DOI] [PubMed] [Google Scholar]
- 23.Stieglitz E, Mazor T, Olshen AB, et al. Genome-wide DNA methylation is predictive of outcome in juvenile myelomonocytic leukemia. Nat Commun. 2017;8(1):2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choong K, Freedman MH, Chitayat D, Kelly EN, Taylor G, Zipursky A. Juvenile myelomonocytic leukemia and Noonan syndrome. J Pediatr Hematol Oncol. 1999;21(6):523-527. [PubMed] [Google Scholar]
- 25.Matsuda K, Nakazawa Y, Iwashita C, et al. Myeloid progenitors with PTPN11 and nonRAS pathway gene mutations are refractory to treatment with 6-mercaptopurine in juvenile myelomonocytic leukemia. Leukemia. 2014;28(7):1545-1548. [DOI] [PubMed] [Google Scholar]
- 26.Javadi M, Richmond TD, Huang K, Barber DL. CBL linker region and RING finger mutations lead to enhanced granulocytemacrophage colony-stimulating factor (GMCSF) signaling via elevated levels of JAK2 and LYN. J Biol Chem. 2013;288(27):19459-19470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belizaire R, Koochaki SHJ, Udeshi ND, et al. CBL mutations promote activation of PI3K/AKT signaling via LYN kinase. bioRxiv. 2020. Apr 14. doi: 10.1182/blood.2020006528[preprint, not peerreviewed]. [Google Scholar]
- 28.Bunda S, Kang MW, Sybingco SS, et al. Inhibition of SRC corrects GM-CSF hypersensitivity that underlies juvenile myelomonocytic leukemia. Cancer Res. 2013;73(8):2540-2550. [DOI] [PubMed] [Google Scholar]
- 29.Bunda S, Qin K, Kommaraju K, Heir P, Ohh M. Juvenile myelomonocytic leukaemiaassociated mutation in Cbl promotes resistance to apoptosis via the Lyn- PI3K/AKT pathway. Oncogene. 2015;34(6):789-797. [DOI] [PubMed] [Google Scholar]
- 30.Tasian SK, Casas JA, Posocco D, et al. Mutation-specific signaling profiles and kinase inhibitor sensitivities of juvenile myelomonocytic leukemia revealed by induced pluripotent stem cells. Leukemia. 2019;33(1):181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.