Abstract
Purpose
The aim of Copenhagen Hospital Biobank-Cardiovascular Disease Cohort (CHB-CVDC) is to establish a cohort that can accelerate our understanding of CVD initiation and progression by jointly studying genetics, diagnoses, treatments and risk factors.
Participants
The CHB-CVDC is a large genomic cohort of patients with CVD. CHB-CVDC currently includes 96 308 patients. The cohort is part of CHB initiated in 2009 in the Capital Region of Denmark. CHB is continuously growing with ~40 000 samples/year. Patients in CHB were included in CHB-CVDC if they were above 18 years of age and assigned at least one cardiovascular diagnosis. Additionally, up-to 110 000 blood donors can be analysed jointly with CHB-CVDC. Linkage with the Danish National Health Registries, Electronic Patient Records, and Clinical Quality Databases allow up-to 41 years of medical history. All individuals are genotyped using the Infinium Global Screening Array from Illumina and imputed using a reference panel consisting of whole-genome sequence data from 8429 Danes along with 7146 samples from North-Western Europe. Currently, 39 539 of the patients are deceased.
Findings to date
Here, we demonstrate the utility of the cohort by showing concordant effects between known variants and selected CVDs, that is, >93% concordance for coronary artery disease, atrial fibrillation, heart failure and cholesterol measurements and 85% concordance for hypertension. Furthermore, we evaluated multiple study designs and the validity of using Danish blood donors as part of CHB-CVDC. Lastly, CHB-CVDC has already made major contributions to studies of sick sinus syndrome and the role of phytosterols in development of atherosclerosis.
Future plans
In addition to genetics, electronic patient records, national socioeconomic and health registries extensively characterise each patient in CHB-CVDC and provides a promising framework for improved understanding of risk and protective variants. We aim to include other measurable biomarkers for example, proteins in CHB-CVDC making it a platform for multiomics cardiovascular studies.
Keywords: genetics, cardiology, epidemiology
Strengths and limitations of this study.
Genetic data on 96 308 patients allows for extensive studies of both common and rare cardiovascular diseases.
Extensive information of medical history allows for stratification on a high level of granularity, which can be combined with sociodemographic and other environmental factors.
There is a potential bias caused by the selection of healthy blood donors from the Danish Blood Donor Study as controls in case–control studies.
Introduction
Cardiovascular diseases (CVDs) include diseases of the heart and the major blood vessels and range from asymptomatic disease entities to myocardial infarction, heart failure (HF), sudden cardiac death and stroke. Modifiable risk factors, such as obesity, smoking, hypertension and dyslipidaemia, are shared across several CVDs. Genetic risk factors are also shared across several CVDs, for example, the earliest and most robust genetic marker for coronary artery disease (CAD), the chromosome 9p21 locus, is also associated with stroke, aneurysms and myocardial infarction.1–4 Genetic studies of CAD started with linkage studies that identified monogenic causes of CAD and small candidate gene studies with dubious findings. The field has since evolved with increasing cohorts and large, international consortia such as CARDIoGRAM and CAD C4D Genetics Consortium being established.5–8 These initiatives combined with individual efforts across multiple ancestries have led to the discovery of 171 genome-wide significant CAD risk variants,9 12 HF variants10 and at least 138 atrial fibrillation (AF) loci,11 which have been replicated in independent populations. It is expected that there are many more risk variants to be discovered. Recently, biobanks such as the UK Biobank, Japan Biobank, FinnGen and the Trøndelag Health Study (The HUNT Study)12–15 have contributed to the ethnic diversity within genetic research and increased the number of both participants and phenotypes for large-scale genetic studies including studies within CVDs. Not only have numbers increased, but the detailed information captured on the individual enables a deeper phenotyping of the biobank participants with great potential to pave the way for precision cardiology. Improving our understanding of the complex interactions between genetics, lifestyle factors and individual CVDs necessitates large cohorts, such as Copenhagen Hospital Biobank (CHB)-Cardiovascular Disease Cohort (CVDC), enabling stratification of individuals into more refined and detailed phenotypes.
Here, we present the CHB-CVDC, a potent resource for genetic research within CVDs. The CHB-CVDC is part of the CHB, initiated in 2009 and funded by the Department of Clinical Immunology, Copenhagen University Hospital, Denmark. The CHB takes advantage of the pre-existing blood banking system that in addition to treatment obligations (eg, blood transfusions) aims to support medical research. The data foundation of CHB is a collection of leftover EDTA blood samples from patients hospitalised in the Capital Region of Denmark, who were subject to blood typing or red blood cell antibody screening. Details about data collection, quality assessment and storage can be found in Sørensen et al.16 Presently, CHB contains samples from more than 450 000 individuals.16 Each patient is identified by a Central Person Registry (CPR) number, which facilitates linkage to nationwide registries and electronic patient records. Nationwide registries and electronic patient records are updated continuously. Patients in CHB are over 18 years old and only included once.16
The aim of the CHB-CVDC is to provide a platform for studying genetic, environmental, medical and other factors to further our knowledge of initiation, progression and manifestation of CVDs. CHB-CVDC facilitate studies of individual diseases as well as the shared risk factors, pathophysiology and disease trajectories. The ability to link CHB samples to individual data available from local hospital databases, the Danish National Health Registries, Electronic Patient Records and the Clinical Quality Databases in Denmark facilitates fine-grained stratification of patients into subpopulations. This information can be used to develop for example, risk prediction models for CAD and other CVDs.
The objectives of the present study are to present the features of CHB-CVDC and to evaluate the use of the cohort as a resource for genetic studies by replicating established genetic variants associated with CAD, AF, HF, essential hypertension and cholesterol levels in European populations. Furthermore, we also investigate the potential bias of including the Danish blood donors in the cohort.
Cohort description
Population characteristics
The CHB is a hospital-driven biobank with a broad collection scheme covering a wide range of diseases, designed to facilitate research in health and disease by enabling researchers access to a large resource of well-defined patient samples.16 The CHB-CVDC inclusion criteria contain two components: individuals have (1) to be included in CHB and (2) been assigned at least one of the hospital admission codes presented in table 1. For a more detailed overview of population characteristics, see online supplemental table 1. Only the first assigned CVD was counted. Currently, the CHB-CVDC comprises 96 308 individuals (55% males), and the cohort is increasing as patients are being included on a continuous basis.
Table 1.
Cohort characteristics
| Women | Men | Total | |
| No of patients in CHB-CVDC (%) | 43 479 (45) | 52 829 (55) | 96 308 (100) |
| Year of birth, mean (SD) | 1942.3 (14.8) | 1945.3 (13.0) | |
| Age at first cardiovascular disease, mean (SD) | 63.2 (15.6) | 59.8 (13.6) | |
| Age at inclusion, mean (SD) | 70.5 (14.9) | 67.4 (13.0) | |
| Cardiovascular inclusion ICD-10 codes from the National Patient Registry* | |||
| Hypertension and hypertensive cardiac diseases ICD-10: I10-15 | 16 229 | 13 317 | 29 546 |
| Coronary artery diseases and atherosclerosis ICD-10: I20-25, I70 | 8823 | 15 972 | 24 795 |
| Lipid disorders ICD-10: E78 | 2024 | 2022 | 4046 |
| Cardiac arrhythmia ICD-10: I44-49 | 7177 | 9031 | 16 208 |
| Heart failure, cardiac valve disorders, and myocardial diseases ICD-10: I50, I34-39, I05-09, I40-44 | 3001 | 4263 | 7264 |
| Vascular disorders and aneurysms ICD-10: I71-79 | 1238 | 1883 | 3121 |
| Cerebrovascular diseases and cerebral haemorrhage ICD-10: I60-69 | 3691 | 4710 | 8401 |
| Pulmonary heart diseases and diseases of the pulmonary circulation ICD-10: I26-28 | 751 | 770 | 1521 |
| Vascular kidney disease ICD-10: N17-19 | 545 | 861 | 1406 |
*Patients are stratified by their first assigned cardiovascular diagnosis.
CHB-CVDC, Copenhagen Hospital Biobank-Cardiovascular Disease Cohort; ICD-10, International Statistical Classification of Diseases and Related Health Problems 10th Revision.
bmjopen-2021-049709supp001.pdf (1.2MB, pdf)
Females were older when diagnosed with their first CVD (63.2 years, 95% CI 63.1 to 63.4) compared with males (59.8 years, 95% CI 59.7 to 60.0).
We evaluated the prevalence of comorbidities in the CHB-CVDC using the Charlson Comorbidity Index.17 18 See online supplemental material for a full description. We found that 34 375 individuals (53% males) have at least one comorbidity, and 23 656 individuals (61 % males) have more than five comorbidities (online supplemental table 2). Online supplemental figure 1 gives an overview of the number of comorbidities in different age groups. Consequently, the cohort is well powered to study CVDs in the context of comorbidities. An overview of diagnosis assigned prior to the first CVD is presented in online supplemental table 3. The co-occurrence of CVDs within the cohort is also pronounced, see online supplemental table 4 for an overview.
The most prevalent disease was hypertension, which affected 64 455 CHB-CVDC patients. Cross-referencing with the Danish National Prescription Registry, 63 431 patients had redeemed one or more prescriptions for antihypertensive medication. In addition, 26 581 have been diagnosed with hypercholesterolaemia (online supplemental table 5).
In studies of binary disease traits there is also the opportunity to use blood donors from the Danish Blood Donor Study (DBDS), adding 110 000 individuals to the total cohort population (n=206 308). The DBDS is a large prospective study of blood donors recruited from the blood bank infrastructure across Denmark.19 Sociodemographic details of the Danish blood donors, including sex and age distributions, education, labour market affiliation and level of urbanisation can be found in Burgdorf et al.19 As part of the DBDS, the DBDS Genomic Cohort was later established with the aim to identify genetic predictors important for the healthy donor phenotype.20 Genotyping and imputation of samples in DBDS were performed in the same way as for samples in CHB.20
Follow-up
The 96 308 genotyped individuals currently in the CHB-CVDC are being followed up through Danish national registries, that contain detailed longitudinal information on every contact with the Danish primary and secondary care service. We will on a regular basis update the data collection by extracting information from the many registries to augment the registry-based phenotyping of the cohort. The linked registries are constantly being updated and to date the cohort contains 56 769 patients that are still alive and 39 539 that have died as per information from the Danish Registry of Causes of Death.21 The median time from inclusion (after 2009) to end of follow-up (before 2020) or death is 3.9 years (IQR 1.70–5.73) (online supplemental figure 2). However, because phenotypic data are also included prior to inclusion, the median follow-up time is 41 years. This correspond to the majority of the patients have been followed since the inception of the Danish National Patient Registry in 1977.
The most common underlying cause of death is cancer (36%) followed by heart disease (17%) and respiratory disease (10%). However, only 2.6% (n=969) of those that died from a medical reason has undergone autopsy and the exact cause is therefore subject to considerable uncertainty.21 22 Among those who have undergone autopsy a CVD is the most common cause of death (52%). Online supplemental table 6 presents a summary of causes of death for all patients who have died in CHB-CVDC.
Patient and public involvement
Patients and public were not involved in the design of this study.
Genetics
As part of a collaboration between the Copenhagen University Hospital, Rigshospitalet, Denmark and deCODE genetics, Iceland, leftover EDTA blood samples from the patients in CHB-CVDC are sent to deCODE genetics for DNA extraction, genotyping and subsequent imputation. SNP genotyping is performed on the Infinium Global Screening Array from Illumina. Approximately 660 000 common variants are genotyped. A reference panel backbone consisting of whole-genome sequence data from 8429 Danes along with 7146 samples from North-Western Europe from participants in various research projects at deCODE genetics is used for imputation.23–25 The genotyped samples were long-ranged phased using Eagle2 together with 171 298 genotyped samples from North-western Europe. The process used to whole-genome sequence the reference panel backbone, and the subsequent imputation has been extensively described previously.23–25 Genotyping and imputation procedures are identical to the general procedures applied in DBDS and are described in detail previously.20
Databases
Denmark has one of the world’s most comprehensive population registry systems, integrated via the social security number (CPR number) that was established in 1968.26 Nationwide individual-level data that is linkable via the CPR number includes birthdate, place of residence, emigration, immigration, family relationships, education, labour market affiliation, cause and date of death, and much more enabling a deep phenotyping of patients in CHB-CVDC.26–30 The Danish National Patient Registry contains information of all hospitalisations, since 1977. This registry includes date of hospitalisation, diagnoses related to the hospitalisation, hospital examinations and procedures.31 Diagnoses in this registry are classified according to the international classification of diseases, V.8 (1977–1993) and version 10 (1994-).32 Procedural codes are classified using a country-specific coding until 1995, and then according to the Nordic Medico-Statistical Committee Classification of Surgical Procedures.33
Furthermore, electronic patient records that contain clinical notes, laboratory results, images and treatments are available. From the clinical notes we are in the process of extracting smoking, alcohol intake, height, weight and blood pressure measurements using a text mining approach. The electronic health records cover all hospitals in the Capital Region and Region Zealand in the period 2006–2016, and 87% of patients in CHB-CVDC data related to at least one hospital admission.
The laboratory database contains laboratory results from the Departments of Clinical Biochemistry and Clinical Immunology laboratories in Denmark. This database includes results from inpatient, outpatient and emergency encounters. The database includes data dating back to 2008. We are working on making the data from different laboratories comparable.
The Danish National Prescription Registry contains data on all prescription drugs dispensed in Denmark since 1995 and it is mandatory by law for all pharmacies to report to this registry.34 Consequently, it is possible to trace medication use and compliance, assess associations between medications and paraclinical outcomes and through well-established drug-associated phenotypes to investigate diseases that do not require a hospitalisation, such as hypertension and diabetes.34 In combination with the electronic patient records that cover drugs administered during hospital admissions this database is a valuable source for pharmacogenomic studies, from where we can extract information about drug dosage prescribed/administered, dosage changes and adherence over time, adverse reactions and polypharmacy.35
The Danish healthcare system has developed several National Clinical Registries such as the Danish Heart Registry, the AF Database, the Familial Hypercholesterolaemia Database and the Danish Heart Failure Database.36 37 These databases contribute information such as body mass index, smoking, alcohol consumption, diabetes history, previous cardiac surgery, follow-up and outcome data and other related variables.36 37 An extensive overview of registries and databases and their content are given in online supplemental table 7.
Findings to date
To validate CHB-CVDC as a resource for genetic research, we set out to replicate genetic variants associated with CAD, AF, HF, essential hypertension and cholesterol levels identified in prior large meta-analyses of multiple populations.
For each phenotype, we selected a reference study based on the date of publication, the number of individuals in the genetic analysis and the genetic ancestry of the population (preferably European) (table 2). Independent significant genetic variants were retrieved from the reference study and compared with association results from CHB-CVDC. Generalised linear mixed models (Scalable and Accurate Implementation of GEneralized mixed model, SAIGE) and linear mixed models (Bayesian mixed model association method, BOLT-LMM) were applied to obtain association results for the included genetic variants.38 39 For CAD, AF, HF and hypertension, we use cases and controls from the cohort containing both CHB-CVDC and DBDS. Additionally, we compared how the choice of control group impacts the findings, (1) using only patients from CHB-CVDC as cases or controls (2) using only cases from CHB-CVDC and only controls from DBDS. Further details on the methods are available in online supplemental material.
Table 2.
Overview of reference studies, number of cases and controls and number of variants investigated
| Phenotype | Reference study (reference) | Cases/controls in reference studies | Replication in current study Cases Controls |
No of variants with concordant direction of effect | Replicated/total | Replicated/power to replicate | |
| Coronary artery disease | Van der Harst 201846 | 122 733/424 528 | 33 746 | 154 311 | 236/241 (98%) | 90/241 (37%) | 90/137 (66%) |
| Atrial fibrillation | Nielsen 201811 | 60 620/970 216 | 30 229 | 157 669 | 137/140 (98%) | 96/140 (69%) | 96/109 (88%) |
| Heart failure | Shah 2020/Arvanitis 202010 49 | 47 309/930 014 10 976/437 573 |
21 443 | 167 068 | 14/15 (93%) | 9/15 (60%) | 9/10 (90%) |
| High density lipoprotein | Global Lipids Genetic Consortium 201350 | 188 577/* | 85 435 | * | 67/68 (99%) | 55/68 (81%) | 55/60 (92%) |
| Low density lipoprotein | Global Lipids Genetic Consortium 201350 | 188 577/* | 81 435 | * | 55/57 (96%) | 35/57 (61%) | 35/41 (85%) |
| Total cholesterol | Global Lipids Genetic Consortium 201350 | 188 577/* | 86 297 | * | 71/72 (99%) | 44/72 (61%) | 44/52 (85%) |
| Triglycerides | Global Lipids Genetic Consortium 201350 | 188 577/* | 83 087 | * | 39/40 (98%) | 29/40 (73%) | 29/32 (91%) |
| Hypertension versus SBP | Evangelou 201840 | 1 006 863/* | 63 431 | 87 752 | 220/258 (85%) | 39/258 (15%) | 39/59 (66%) |
| Hypertension versus DBP | Evangelou 201840 | 1 006 863/* | 63 431 | 87 752 | 260/307 (85%) | 33/307 (11%) | 33/80 (41%) |
If the variant had the same direction of effect and p<0.05 (Bonferroni adjusted), we considered it replicated.
*Case/control setup not applicable. Instead, the total number of samples are listed under cases. A variant was replicated if the effect size of the risk allele had the same direction of effect and a p<0.05 (Bonferroni adjusted). The power to replicate was estimated from the SE and the effect size. The power was set at 80%.
DBP, diastolic blood pressure; SBP, systolic blood pressure.
CAD, AF and HF
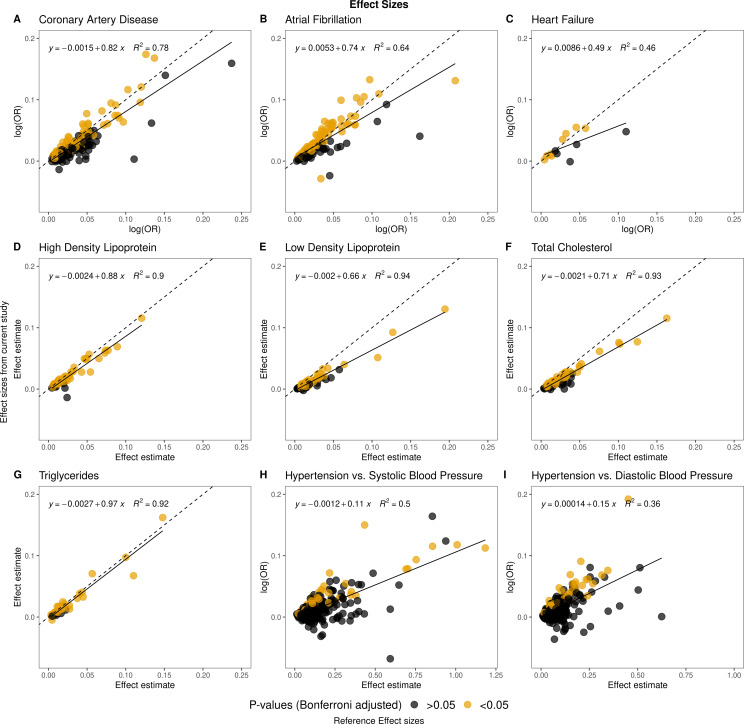
Cases and controls for CAD, AF and HF were defined by International Statistical Classification of Diseases and Related Health Problems 10th/8th Revision (ICD-10/ICD-8) codes (table 2). A positive correlation was found between the observed and published effect sizes for all these phenotypes (figure 1A–C). We observed that we, in general, had smaller effect sizes compared with other studies (see online supplemental table 8). The proportion of replicated variants compared with the number we had power to replicate were 66% for CAD, 88% for AF and 90% for HF (table 2).
Figure 1.
Comparison of effect sizes and reference effect sizes. The effect sizes weighted by risk allele frequency of the reference study (X-axis) are compared with the effect sizes weighted by the risk allele frequency from this study (y-axis). The dotted lines correspond to a correlation of 1 and the dense lines to the observed trendlines.
Cholesterol measurements
A patient’s first measured level of High-Density Lipoprotein (HDL), Low-Density Lipoprotein (LDL), Total Cholesterol (TC) and Triglyceride (TG) were extracted based on NPU codes from the Laboratory Database in Denmark. Detailed information on methods is found in online supplemental material. More than 80 000 corresponding to more than 83% of the CHB-CVDC had at least one cholesterol measurement. We observed that the effect sizes for TG were largely concordant compared with the prior study (figure 1G and online supplemental table 8). For HDL, LDL and TC the effect sizes in this study are lower compared with the effect sizes in the reference study (online supplemental table 8 and figure 1D–F). The proportion of replicated variants compared with the number we had power to replicate exceeds 85% for all the cholesterol analyses. The HDL association analysis where the analysis with most replicated variants (81%).
Essential hypertension
Hypertension cases were identified using ICD-10/ICD-8 codes and prescriptions of antihypertensive drugs. This is in contrast to the reference study, where systolic and diastolic blood pressure were measured directly.40 The observed effect sizes and the effect sizes in the reference study were positively correlated, although a smaller proportion of variants had a concordant direction of effect (85%) compared with the phenotypes described above (>90%) (figure 1H, I and table 2). The number of variants we had power to replicate were low (59/258 and 80/307) and the proportion of replicated variants compared with the number of variants we had power to replicate were also low (<67%).
Additional comparisons with the reference studies
All variants had similar allele frequencies as the frequencies in the reference studies (online supplemental figure 3). Overall, association p values were lower in the reference studies, likely due to greater sample sizes (online supplemental figure 4). The variants that showed discordant direction of effect had small effect sizes and large SEs.
We were able to replicate >85% of the variants for six out of nine phenotypes, given the estimated power (power=0.8, α=0.05) (table 2).
Comparison of different study designs
We repeated the association analyses for CAD, AF and HF with only patients from CHB-CVDC as controls and only participants from DBDS as controls, respectively. For all the phenotypes the control group had no effect on the number of variants with concordant direction of effect, but the number of replicated variants decreased when only DBDS was used as controls (online supplemental table 9 and online supplemental figures 5–8). Furthermore, we employed LD Score regression to investigate residual confounding (online supplemental table 10).41 Residual confounding was evaluated through the LD Score regression intercept and the attenuation ratio (the ratio between the intercept and the mean χ2 statistic). An intercept close to one indicates no additional confounding, and a small ratio indicates that the genomic inflation arises from the polygenicity of the phenotype and not residual confounding. Lastly, we also evaluated the genetic correlations to prior studies using LD Score Regression (online supplemental table 11). We used the 1000Genomes EUR v3 LD reference panel for both analyses. Using only DBDS as controls yielded a high level of residual confounding. The genetic correlation analysis indicated the setup using only CHB-CVDC was not significantly different from the main analysis. We could not calculate a genetic correlation for the last study design as the observed heritability was negative.
Other contributions
In a recent study, samples from CHB-CVDC were used together with Icelandic samples and samples from UK biobank, to examine the effects of ABCG5/8 variants on dietary cholesterols and phytosterols and the risk of CAD.42 The authors conclude that phytosterols may be involved in the development of atherosclerosis and clinical trials are needed to investigate whether phytosterols increase risk of CVD.42 43
A genome-wide association study of 6 469 sick sinus syndrome cases and more than a million controls revealed six loci associated to the disease.44 Mendelian randomisation suggests a direct role of AF and lower heart rate in the development of sick sinus syndrome.44
Angioedema is a known adverse drug reaction in individuals treated with antihypertensive ACE inhibitors. A Danish study has found common variants located close to the bradykinin receptor B2 gene to be associated with increased risk of developing angioedema related to treatment with ACE inhibitors.45
Discussion
In this study, we present the features of CHB-CVDC and use the genetic data to validate the registry-based phenotyping in CHB-CVDC. We show a high proportion of variants with a concordant direction of effect between the respective reference studies and the current study. For CAD, AF, HF and cholesterol measurements the proportion exceeded 93%. For the diagnosis of hypertension compared with blood pressure measurements the proportion of variants with the same direction was lower but still 85%.
The proportion of replicated variants exceeded 60% for AF, HF and cholesterol measurements compared with the number we had power to replicate. For CAD and hypertension, this proportion were <67%. The lower proportion could be a consequence of different phenotyping, analysis methods, population differences and the number of cases compared with the number of samples in the reference studies. Furthermore, a high proportion of patients in CHB-CVDC have a hypertension diagnosis, hence it was almost only possible to compare with DBDS that is a cohort of younger and typically healthier individuals (see text below).
It is very likely that having systematic blood pressure measurements available for the present study would have increased the concordance with the studies we set out to replicate. However, we do show that the proxies for blood pressure used that is, diagnosis codes for hypertension and prescription of antihypertensive drugs capture enough signal to replicate some of the strongest signals with comparable effect sizes. We are currently in the process of extracting blood pressure measurement from the unstructured clinical notes using a text mining approach, and they will be available in future studies.
The analyses with DBDS as controls only, show that this setup is decreasing the number of replicated variants and the residual confounding is high. For AF and HF, it seems that DBDS together with patients from CHB-CVDC as controls and CHB-CVDC as controls can be used interchangeably. Nonetheless, careful considerations of study design are necessary and depends on the phenotype under investigation. Here, we have outlined one way of doing so.
For AF, HF and the cholesterol measurements the proportion of replicated variants compared with the number we had power to replicate were >85%. Overall, these results are promising regarding future collaborations with CHB-CVDC as either a discovery or replication cohort.
The comparison of age at their first cardiovascular diagnosis and age at the blood typing test demonstrates that the majority of the patients already had a cardiovascular diagnosis at time of inclusion in the cohort. Such a study design where the event has occurred before the inclusion could lead to a survivor bias, as individuals who died from diseases before the inclusion are not part of the analyses. Nonetheless, as the cohort increases in size over time, this potential issue will disappear.
Strengths and limitations
As a hospital-driven biobank with a comprehensive collection scheme, the CHB covers a wide range of diseases, thereby facilitating large-scale studies with high clinical importance across many patient groups including the patients with CVDs in CHB-CVDC. The collection of routine blood samples from the clinic is a cost-effective biobanking practice, and as the data used for phenotyping already are available through the registries the time and cost of studies using a biobank platform like CHB-CVDC are less than studies using primary data collection.
With genetic data on 96 308 patients, CHB-CVDC comprises a significant increase in the number of cardiovascular patients with genetic data available.10 11 40 46 This is a strength in terms of discovery of new genetic and environmental risk factors and interactions as well as a major strength in the cooperation with other cohorts to increase power of future meta analyses.
The potential to link genetic data to individual-level data from the Danish National Registries is a huge advantage in epidemiological and genetic research. Denmark has one of the world’s oldest registry systems with information of nationwide hospitalisations from the Danish National Patient Registry coupled with the Danish Civil Registration System.26 31 Thus, Denmark is, similar to other Nordic countries, a forerunner of the digitalisation of clinical systems for quality and research purposes. Since these registries are nationwide and the healthcare system is financed by state taxes in Denmark, there is a high population-based coverage. The long-term temporal registration of all hospital admissions, procedures and treatments also enables studies of trajectories of the different diseases and their temporal relations.31
In the last decades, many clinical quality databases relevant for cardiovascular research has been created, for example, Karbase, a nationwide database of vascular surgery from 1993, the Danish Heart Registry with information of all patients with CAD and with a coronary angiography, a percutaneous coronary intervention or a coronary artery bypass graft from 2004.36 47 Hence, we will be able to stratify patients at a high level of detail combined with sociodemographic and other environmental factors.
CHB-CVDC is comparable to other major hospital-driven cohorts where patients are not recruited based on a specific disease. However, as patients were subject to blood typing or red blood cell antibody screening the health condition of the patients are perhaps worse than in other hospital-driven cohorts.
A challenge in designing genome-wide association studies in CHB-CVDC is the selection of controls. There is a potential bias caused by the selection of blood donors from the DBDS as controls. Compared with the patients from CHB-CVDC the DBDS participants are much younger.19 It is inevitable that some of these participants will develop CVDs in the future. Furthermore, the participants from the DBDS are implicitly healthier than the general population and thereby also much healthier than the patients from CHB-CVDC.19 48 This is exemplified by our finding that using only DBDS as controls the number of replicated variants decrease, and the residual confounding is high. Ways to overcome these challenges must be considered in future studies and reflected in the study designs.
Acknowledgments
We would like to thank the patients in CHB-CVDC and the participants in DBDS. We thank deCODE genetics/Amgen and Department of Clinical Immunology, Copenhagen University Hospital for financial support of this study. Furthermore, we thank the staff working in the biobank facilities and in the hospitals whose work make the existence of this cohort sustainable.
Footnotes
Twitter: @Karina_Banasik, @fhq750, @s_brunak, @MetteNyegaard
Collaborators: We encourage scientific collaborations based on data generated in CHB-CVDC and summary statistics from published analyses will be made available upon request. Any collaboration needs first approval by the CHB-CVDC steering committee.
Contributors: IHL, KB, HH and HU conceived and planned the experiments. IHL carried out the analyses. KB, HB, SB, RF-S, HH, ES, LK, CT-P, SRO, CE, MN, HS, UT, DG, OBP, KS and HU contributed to cohort and research design. KB, DW, LT, MAHL, MS, ES, HU and SB established the data infrastructure and data governance design. HB, RF-S, ES, LK, CT-P, OBP, HS, AG, FZ, GBW, AO, GÞ, GM and HU were instrumental to data capture. IHL, KB, ADH, OP, PCH and DW contributed to analyses and interpretation of the results. IHL, KB and HU took the lead in writing the manuscript. HU is the guarantor of this work. All authors provided critical feedback, helped shape the analysis and manuscript, and approved the final version.
Funding: This work was supported by the Novo Nordisk Foundation (NNF) (grant numbers: NNF17OC0027594, NNF14CC0001 and NNF18SA0034956), Innovation Fund Denmark (grant number: 5153-00002B) and Nordforsk, Precision Medicine Heart (grant number: 90580 PM Heart).
Competing interests: The authors affiliated with deCODE genetics/Amgen are employed by the company.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. The study will adhere to the FAIR (http://datafairport.org/: Findable, Accessible, Interoperable and Reusable) concepts. For further access possibilities and contact details please see: https://www.regionh.dk/blodbanken/afdelingen/enheder-paa-rigshospitalet/Sider/biobank.aspx. The data were handled in accordance with 'the Danish Act on Data Protection following the EU Regulation (EU) 2016/ 679, 27 April 2016 pf the European Parliament and of the Council on the protection of natural persons with regard to the processing of personal data and on the free movement of such data, and repealing Directive 95/46/EC'. All data are stored in a private secure cloud of the Danish National Supercomputer for Life Sciences-Computerome.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
CHB is classified as a ‘biobank for future research’. It is part of the Danish National Biobank and has been approved by the Danish Data Protection Agency (general approval number 2012-58-0004, and local number: RH-2007-30−4129/I-suite 00678). Patients included in CHB were informed about their right to refuse the use of their samples for research via the Danish Tissue Utilisation Registry.16 Patients from CHB who have been assigned at least one of the CVD inclusion codes (table 1) are included in CHB-CVDC. Studies under CHB-CVDC, including the use of DBDS as controls, are approved by The National Ethical Committee (1708829, ‘Genetics of CVD’—a genome-wide association study on repository samples from CHB).
References
- 1. Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med 2007;357:443–53. 10.1056/NEJMoa072366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science 2007;316:1488–91. 10.1126/science.1142447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007;316:1491–3. 10.1126/science.1142842 [DOI] [PubMed] [Google Scholar]
- 4. Wellcome Trust Case Control Consortium . Genome-Wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–78. 10.1038/nature05911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Preuss M, König IR, Thompson JR, et al. Design of the coronary artery disease genome-wide replication and meta-analysis (cardiogram) study: a genome-wide association meta-analysis involving more than 22 000 cases and 60 000 controls. Circ Cardiovasc Genet 2010;3:475–83. 10.1161/CIRCGENETICS.109.899443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schunkert H, König IR, Kathiresan S, et al. Large-Scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333–8. 10.1038/ng.784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coronary Artery Disease (C4D) Genetics Consortium . A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet 2011;43:339–44. 10.1038/ng.782 [DOI] [PubMed] [Google Scholar]
- 8. IBC 50K CAD Consortium . Large-Scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet 2011;7:e1002260. 10.1371/journal.pgen.1002260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roberts R, Chang CC, Hadley T. Genetic risk stratification: a paradigm shift in prevention of coronary artery disease. JACC Basic Transl Sci 2021;6:287–304. 10.1016/j.jacbts.2020.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shah S, Henry A, Roselli C, et al. Genome-Wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun 2020;11:163. 10.1038/s41467-019-13690-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nielsen JB, Thorolfsdottir RB, Fritsche LG, et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet 2018;50:1234–9. 10.1038/s41588-018-0171-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–9. 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nagai A, Hirata M, Kamatani Y, et al. Overview of the Biobank Japan project: study design and profile. J Epidemiol 2017;27:S2–8. 10.1016/j.je.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. FinnGen . FinnGen documentation of R5 release, 2021. Available: https://github.com/FINNGEN/finngen-documentation
- 15. Krokstad S, Langhammer A, Hveem K, et al. Cohort profile: the HUNT study, Norway. Int J Epidemiol 2013;42:968–77. 10.1093/ije/dys095 [DOI] [PubMed] [Google Scholar]
- 16. Sørensen E, Christiansen L, Wilkowski B, et al. Data resource profile: the Copenhagen Hospital Biobank (Chb). Int J Epidemiol 2021;50:719–20. 10.1093/ije/dyaa157 [DOI] [PubMed] [Google Scholar]
- 17. Quan H, Sundararajan V, Halfon P, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care 2005;43:1130–9. 10.1097/01.mlr.0000182534.19832.83 [DOI] [PubMed] [Google Scholar]
- 18. Gasparini A. Comorbidity: an R package for computing comorbidity scores. J Open Source Softw 2018;3:648. 10.21105/joss.00648 [DOI] [Google Scholar]
- 19. Burgdorf KS, Simonsen J, Sundby A, et al. Socio-Demographic characteristics of Danish blood donors. PLoS One 2017;12:e0169112. 10.1371/journal.pone.0169112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hansen TF, Banasik K, Erikstrup C, et al. Dbds genomic cohort, a prospective and comprehensive resource for integrative and temporal analysis of genetic, environmental and lifestyle factors affecting health of blood donors. BMJ Open 2019;9:e028401. 10.1136/bmjopen-2018-028401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Helweg-Larsen K. The Danish register of causes of death. Scand J Public Health 2011;39:26–9. 10.1177/1403494811399958 [DOI] [PubMed] [Google Scholar]
- 22. Svendsen MT, Bøggild H, Skals RK, et al. Uncertainty in classification of death from fatal myocardial infarction: a nationwide analysis of regional variation in incidence and diagnostic support. PLoS One 2020;15:e0236322. 10.1371/journal.pone.0236322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kong A, Masson G, Frigge ML, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat Genet 2008;40:1068–75. 10.1038/ng.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gudbjartsson DF, Helgason H, Gudjonsson SA, et al. Large-Scale whole-genome sequencing of the Icelandic population. Nat Genet 2015;47:435–44. 10.1038/ng.3247 [DOI] [PubMed] [Google Scholar]
- 25. Helgadottir A, Thorleifsson G, Gretarsdottir S, et al. Genome-Wide analysis yields new loci associating with aortic valve stenosis. Nat Commun 2018;9:987. 10.1038/s41467-018-03252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schmidt M, Pedersen L, Sørensen HT. The Danish civil registration system as a tool in epidemiology. Eur J Epidemiol 2014;29:541–9. 10.1007/s10654-014-9930-3 [DOI] [PubMed] [Google Scholar]
- 27. Thygesen LC, Daasnes C, Thaulow I, et al. Introduction to Danish (nationwide) registers on health and social issues: structure, access, legislation, and archiving. Scand J Public Health 2011;39:12–16. 10.1177/1403494811399956 [DOI] [PubMed] [Google Scholar]
- 28. Laugesen K, Ludvigsson JF, Schmidt M, et al. Nordic health registry-based research: a review of health care systems and key registries. Clin Epidemiol 2021;13:533–54. 10.2147/CLEP.S314959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jensen VM, Rasmussen AW. Danish education registers. Scand J Public Health 2011;39:91–4. 10.1177/1403494810394715 [DOI] [PubMed] [Google Scholar]
- 30. Petersson F, Baadsgaard M, Thygesen LC. Danish registers on personal labour market affiliation. Scand J Public Health 2011;39:95–8. 10.1177/1403494811408483 [DOI] [PubMed] [Google Scholar]
- 31. Schmidt M, Schmidt SAJ, Sandegaard JL, et al. The Danish national patient registry: a review of content, data quality, and research potential. Clin Epidemiol 2015;7:449–90. 10.2147/CLEP.S91125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. WHO . Classification of diseases (ICD). Available: https://www.who.int/standards/classifications/classification-of-diseases [Accessed 5 Sep 2021].
- 33. Nordisk Medicinal-Statistisk Komité . NOMESCO classification of surgical procedures. Copenhagen: Nordic Medico-Statistical Committee, 2010. [Google Scholar]
- 34. Kildemoes HW, Sørensen HT, Hallas J. The Danish national prescription registry. Scand J Public Health 2011;39:38–41. 10.1177/1403494810394717 [DOI] [PubMed] [Google Scholar]
- 35. Rodríguez CL, Kaas-Hansen BS, Eriksson R. Drug interactions in hospital prescriptions in Denmark: prevalence and associations with adverse outcomes 2021. 10.1101/2021.05.27.21257764 [DOI] [PMC free article] [PubMed]
- 36. Özcan C, Juel K, Flensted Lassen J, et al. The Danish heart registry. Clin Epidemiol 2016;8:503–8. 10.2147/CLEP.S99475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schjødt I, Nakano A, Egstrup K, et al. The Danish heart failure registry. Clin Epidemiol 2016;8:497–502. 10.2147/CLEP.S99504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loh P-R, Tucker G, Bulik-Sullivan BK, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet 2015;47:284–90. 10.1038/ng.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou W, Nielsen JB, Fritsche LG, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet 2018;50:1335–41. 10.1038/s41588-018-0184-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Evangelou E, Warren HR, Mosen-Ansorena D, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet 2018;50:1412–25. 10.1038/s41588-018-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bulik-Sullivan BK, Loh P-R, Finucane HK, et al. Ld score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–5. 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Helgadottir A, Thorleifsson G, Alexandersson KF, et al. Genetic variability in the absorption of dietary sterols affects the risk of coronary artery disease. Eur Heart J 2020;41:2618–28. 10.1093/eurheartj/ehaa531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weingärtner O, Patel SB, Lütjohann D. It's time to personalize and optimize lipid-lowering therapy. Eur Heart J 2020;41:2629–31. 10.1093/eurheartj/ehaa445 [DOI] [PubMed] [Google Scholar]
- 44. Thorolfsdottir RB, Sveinbjornsson G, Aegisdottir HM, et al. Genetic insight into sick sinus syndrome. Eur Heart J 2021;42:1959–71. 10.1093/eurheartj/ehaa1108 [DOI] [PubMed] [Google Scholar]
- 45. Ghouse J, Ahlberg G, Andreasen L, et al. Association of Variants Near the Bradykinin Receptor B2 Gene With Angioedema in Patients Taking ACE Inhibitors. J Am Coll Cardiol 2021;78:696–709. 10.1016/j.jacc.2021.05.054 [DOI] [PubMed] [Google Scholar]
- 46. van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res 2018;122:433–43. 10.1161/CIRCRESAHA.117.312086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Eldrup N, Cerqueira C, de la Motte L, et al. The Danish vascular Registry, Karbase. Clin Epidemiol 2016;8:713–8. 10.2147/CLEP.S99506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rigas AS, Skytthe A, Erikstrup C, et al. The healthy donor effect impacts self-reported physical and mental health - results from the Danish Blood Donor Study (DBDS). Transfus Med 2019;29 Suppl 1:65–9. 10.1111/tme.12478 [DOI] [PubMed] [Google Scholar]
- 49. Arvanitis M, Tampakakis E, Zhang Y, et al. Genome-Wide association and multi-omic analyses reveal ACTN2 as a gene linked to heart failure. Nat Commun 2020;11:1122. 10.1038/s41467-020-14843-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Willer CJ, Schmidt EM, Sengupta S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274–83. 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2021-049709supp001.pdf (1.2MB, pdf)
Data Availability Statement
Data are available on reasonable request. The study will adhere to the FAIR (http://datafairport.org/: Findable, Accessible, Interoperable and Reusable) concepts. For further access possibilities and contact details please see: https://www.regionh.dk/blodbanken/afdelingen/enheder-paa-rigshospitalet/Sider/biobank.aspx. The data were handled in accordance with 'the Danish Act on Data Protection following the EU Regulation (EU) 2016/ 679, 27 April 2016 pf the European Parliament and of the Council on the protection of natural persons with regard to the processing of personal data and on the free movement of such data, and repealing Directive 95/46/EC'. All data are stored in a private secure cloud of the Danish National Supercomputer for Life Sciences-Computerome.