Abstract
Conditions have been optimized for the use of a multiplex PCR for the detection of vancomycin-resistant enterococci in nosocomial surveillance specimens. Seven primer sets targeting the genes vanA, vanB, vanC1, vanC2/C3 Enterococcus faecalis-specific, Enterococcus faecium-specific, and rrs (16S rRNA) were used in one reaction tube. The PCR method developed in the present study is simple and reliable for the rapid characterization of vancomycin-resistant enterococci.
The first report in Japan of a vancomycin-resistant enterococcal clinical isolate was in 1996 (5). Since then, vancomycin-resistant enterococci (VRE) have emerged as important nosocomial pathogens (6). In May 1998, a patient colonized with vanA-containing Enterococcus faecium was transferred from an outside hospital to the Okayama University Hospital (Okayama, Japan). Since this was the first case of VRE in our hospital, it was urgent to establish a surveillance system for VRE. Although references available in the literature provided helpful methods to screen for VRE, we found that some modifications were needed to improve detection and characterization. The multiplex PCR assay, which allows simultaneous detection of glycopeptide-resistance genotypes (vanA, vanB, vanC1, and vanC2/C3) and genes encoding d-alanine–d-alanine ligases specific for E. faecalis (ddlE. faecalis) and E. faecium (ddlE. faecium), was developed by Dutka-Malen et al. in 1995 (3). A rapid alkaline method to prepare the template DNA was used, but simpler methods to prepare the template DNA have since been described (1, 2, 4, 7, 13). Initially, we used heat treatment (94°C, 10 min) to prepare the template DNA (13) and added six primer sets for the multiplex PCR assay (3). But we observed only a faint band for the vanA and ddlE. faecalis targets, nonspecific bands for the vanB and ddlE. faecalis targets, or no band for the ddlE. faecium target (an erratum for the ddlE. faecium primer has been published [3]). Some investigators reported that the use of four primer sets (for vanA, vanB, vanC1, and vanC2/C3) clearly reduced the sensitivity of the PCR assay (4, 11). The objective of this study was to optimize the multiplex PCR assay to allow the detection of four glycopeptide resistance genotypes (vanA, vanB, vanC1, and vanC2/C3) and the identification to the species level of four clinically relevant species of enterococci (E. faecalis, E. faecium, E. gallinarum, and E. casseliflavus). This assay also includes 16S rRNA (rrs) as an internal PCR control to improve reliability (1–3, 11, 13), the importance of which has previously been shown (7, 9, 10). Below, we describe an optimized, simple, and reliable multiplex PCR assay containing seven primer sets for the rapid characterization of VRE.
Cell suspensions of presumptive VRE colonies from selective medium (bile esculin azide agar containing 6 μg of vancomycin per ml) after 15 to 48 h of incubation at 37°C were prepared to a density equivalent to a McFarland standard of 3 in 50 μl of 7.5% Chelex 100. Since the colonies on VRE selective medium were usually small after a 24-h, and even a 48-h, incubation, cell suspensions were more easily prepared by using a swab rather than a wire loop. After incubation for 24 h on selective medium, if the colonies were small, we inoculated a presumptive VRE colony in 500 μl of brain heart infusion broth containing 6 μg of vancomycin per ml and incubated it at 37°C overnight. If we observed different colony morphologies for presumptive VRE colonies with a black halo on VRE selective medium, we analyzed each different colony. After incubation in the broth overnight at 37°C, a 25-μl culture volume was mixed with 25 μl of 15% Chelex 100 (Bio-Rad Laboratories, Hercules, Calif.). If the density of overnight cultures was equivalent to a McFarland standard of less than 2, the cultures were centrifuged and the cell suspension density was increased to make it equivalent to a McFarland standard of 3 in 50 μl of 7.5% Chelex 100. Cell suspensions in 50 μl of 7.5% Chelex 100 were heated for 10 min at 100°C and centrifuged. A 2.5-μl volume of the supernatant was then used for PCR amplification. The seven primer sets shown in Table 1 were added to the reaction mixtures as follows: 5 pmol of the vanA primers; 2.5 pmol each of the vanB, vanC1, vanC2/C3, and rrs primers; 5 pmol of the E. faecalis-specific primers; and 1.25 pmol of the E. faecium-specific primers. The multiplex PCR assay was performed in a total volume of 25 μl containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each deoxynucleotide triphosphate (dATP, dCTP, dGTP, and dTTP), and 0.625 U of Taq DNA polymerase (Takara Shuzo Co., Kusatsu, Japan). DNA amplification was carried out with the following thermal cycling profile: initial denaturation at 94°C for 5 min, 30 cycles of amplification (denaturation at 94°C for 1 min, annealing at 54°C for 1 min, and extension at 72°C for 1 min), and a final extension at 72°C for 10 min in a GeneAmp PCR system 9700 (PE Biosystems, Tokyo, Japan). PCR products were analyzed on a 1.5% SeaKem ME agarose gel (FMC BioProducts, Rockland, Maine) with 0.5× Tris-borate-EDTA buffer. A 100-bp DNA ladder (New England Biolabs, Beverly, Mass.) was used as the molecular size marker. The gels were stained with ethidium bromide and photographed under UV light. Amplification of vanA, vanB, vanC1, vanC2/C3, E. faecalis-specific, E. faecium-specific, and rrs targets produced distinct bands corresponding to their respective molecular sizes that were easily recognizable (Fig. 1). The entire procedure for the multiplex PCR assay (up to 32 samples) can be completed in 4 to 5 h. Each multiplex PCR assay was carried out with a negative control containing all of the reagents without a DNA template.
TABLE 1.
Multiplex PCR primers for detection of VRE
| Primer specificity | Size of PCR product (bp) | Primer pair sequences | Reference |
|---|---|---|---|
| vanA | 1,030 | 5′-CATGAATAGAATAAAAGTTGCAATA-3′ | 2 |
| 5′-CCCCTTTAACGCTAATACGATCAA-3′ | |||
| vanB | 433 | 5′-GTGACAAACCGGAGGCGAGGA-3′ | 2 |
| 5′-CCGCCATCCTCCTGCAAAAAA-3′ | |||
| vanC1 | 822 | 5′-GGTATCAAGGAAACCTC-3′ | 3 |
| 5′-CTTCCGCCATCATAGCT-3′ | |||
| vanC2/C3 | 484 | 5′-CGGGGAAGATGGCAGTAT-3′ | 11 |
| 5′-CGCAGGGACGGTGATTTT-3′ | |||
| E. faecalis | 941 | 5′-ATCAAGTACAGTTAGTCTTTATTAG-3′ | 3 (modified) |
| 5′-ACGATTCAAAGCTAACTGAATCAGT-3′ | |||
| E. faecium | 658 | 5′-TTGAGGCAGACCAGATTGACG-3′ | 1 |
| 5′-TATGACAGCGACTCCGATTCC-3′ | |||
| rrs (16S rRNA) | 320 | 5′-GGATTAGATACCCTGGTAGTCC-3′ | 13 |
| 5′-TCGTTGCGGGACTTAACCCAAC-3′ |
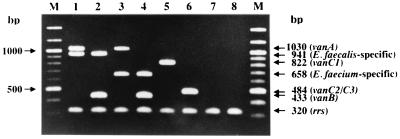
FIG. 1.
Agarose gel electrophoresis of amplified vanA, vanB, vanC1, vanC2, E. faecalis-specific, E. faecium-specific, and rrs genes by the optimized multiplex PCR assay containing seven primer sets. Lanes: M, 100-bp DNA ladder (New England Biolabs); 1, an E. faecalis vanA isolate; 2, an E. faecalis vanB isolate; 3, an E. faecium vanA isolate; 4, an E. faecium vanB isolate; 5, an E. gallinarum vanC1 isolate; 6, an E. casseliflavus vanC2 isolate; 7, a Pediococcus isolate; 8, a Leuconostoc isolate.
Clinical enterococcal isolates were used to test the optimized multiplex PCR assay. E. faecalis and E. faecium isolates possessing the vanA or vanB resistance gene were collected from 15 U.S. hospitals in three states over a 6-year period from 1991 through 1996 as part of epidemiological studies (12). E. gallinarum (vanC1), E. casseliflavus (vanC2), Pediococcus, and Leuconostoc isolates were collected during VRE surveillance from June to July 1998 at Okayama University Hospital. A vanA strain (E. faecium ATCC 51559), a vanB strain (E. faecalis ATCC 51299), a vanC1 strain (E. gallinarum ATCC 49573), a vanC2 strain (E. casseliflavus ATCC 25788), and a vancomycin-susceptible E. faecalis strain (ATCC 19433) were used as quality control strains. These isolates and strains were characterized by automated systems and/or conventional biochemical tests, and glycopeptide-resistant genotypes were determined by using PCR assays that contained a single primer set. The optimized multiplex PCR assay was performed on 3 E. faecalis (vanA) isolates, 30 E. faecium (vanA) isolates, 15 E. faecalis (vanB) isolates, 3 E. faecium (vanB) isolates, 6 E. gallinarum (vanC1) isolates, 9 E. casseliflavus (vanC2) isolates, 3 Pediococcus isolates, and 3 Leuconostoc isolates. As shown in Fig. 1, PCR products with a size of ca. 320 bp corresponding to the rrs target (internal control) were observed for all of the isolates (lanes 1 to 8), including a Pediococcus isolate (lane 7) and a Leuconostoc isolate (lane 8). This internal PCR control provided assurance that the clinical specimens were successfully amplified and detected. In addition, one or two intense bands of the expected sizes (Table 1) were generated from VRE isolates. As shown in Fig. 1, two bands (total, three bands) with sizes of ca. 941 and 1,030 bp corresponding to the E. faecalis-specific and vanA targets (lane 1) and two of ca. 433 and 941 bp corresponding to the vanB and E. faecalis-specific targets (lane 2) were generated from an E. faecalis vanA isolate and an E. faecalis vanB isolate, respectively. Two bands (total, three bands) with sizes of ca. 1,030 and 658 bp corresponding to the vanA and E. faecium-specific targets (lane 3) and two of ca. 433 and 658 bp corresponding to the vanB and E. faecium-specific targets (lane 4) were generated from an E. faecium vanA isolate and an E. faecium vanB isolate, respectively. One band (total, two bands) with a size of ca. 822 bp (lane 5) or 484 bp (lane 6) corresponding to the vanC1 (E. gallinarum) or vanC2 (E. casseliflavus) target, respectively, was generated. Of 72 isolates tested, 100% of the expected PCR products with intense bands on the gel were generated.
To optimize the conditions for the multiplex PCR assay, we made changes in the template DNA preparations, primer combinations, primer concentrations, and primer lengths. For preparation of template DNA from VRE cells, several methods have been reported. The method for direct suspension of VRE cells in a PCR mixture (2, 4) is the simplest, but the expected PCR products were not always generated for the multiplex PCR assay. The methods reported in the literature for heat treatment of cell suspensions in saline (13), 5% Chelex 100 (1), lysis buffer containing 7.5% Chelex 100 (7), or the InstaGene Matrix kit (Bio-Rad Laboratories) (14) are much simpler than the rapid alkaline method (3). When the heat treatment of cell suspensions was performed in distilled water or saline for 5 to 15 min at 94 to 100°C, the expected PCR products were not always generated for the multiplex PCR assay. However, when we used the InstaGene Matrix kit, which sequesters cell lysis products, 100% of the expected PCR products were generated. We also tested two methods of heat treatment at 100°C for 5 min (1) or at 95°C for 15 min (7) which use the cation-chelating resin Chelex 100. These methods were as reliable as the InstaGene Matrix method. Based on these studies, we currently heat cell suspensions in 7.5% Chelex 100 for 10 min at 100°C to prepare template DNA from VRE cells.
In the present study, we also varied the primer concentrations. We had to use concentrations of the ddlE. faecalis and ddlE. faecium primer sets (reference 3, erratum) that were at least four times as high as the concentrations of the vanB (2), vanC1 (3), and vanC2/C3 primer sets (11) in order to observe intense bands on the gel. We and others (11) have observed that vanA amplification (3) was inhibited by primer combinations. Although a primer set for the E. faecium-specific target published by Cheng et al. (1) inhibited vanA amplification, this primer set was more sensitive than other primer sets. Therefore, we could use a concentration of the E. faecium-specific primer set that was one-half that of the other primer sets (vanB, vanC1, and vanC2/C3) without inhibiting vanA amplification. Reliable results were obtained by using a vanA (2) primer set concentration that was twice that of the other primer sets and an E. faecium-specific primer set (1) concentration that was one-half of the concentration of the other primer sets (vanB, vanC1, and vanC2/C3). Since intense bands were not observed when a primer set for the ddlE. faecalis target (3) was used, we modified the primer set to make it 7 bases longer (each 25 bases). In short, the best result was observed when the primer combinations shown in Table 1 were added to reaction mixtures at 5 pmol of the vanA primers; 2.5 pmol each of the vanB, vanC1, vanC2/C3, rrs primers; 5 pmol of the E. faecalis-specific primers; and 1.25 pmol of the E. faecium-specific primers.
Cell suspensions at a density equivalent to a McFarland standard of 0.5 to 1 or a few colonies from an agar plate have been used by several investigators (1, 2, 4, 7, 13). When we used this concentration, we observed faint bands or no bands for the expected PCR products. When we used thicker cell suspensions at a density equivalent to a McFarland standard of 3 for the multiplex PCR assay containing seven primer sets, 100% of the expected PCR products with intense bands on the gel were generated. Intense bands were not observed when the thermal cycling time was less than 1 min at each temperature. Therefore, we currently use 1 min at each temperature. The best results were obtained when 54°C was used as the annealing temperature.
VRE selective medium (bile esculin azide agar containing 6 μg of vancomycin per ml) has been used for routine screening of nosocomial surveillance specimens for VRE (7, 8, 10). The growth of colonies with a black halo on VRE selective medium must be differentiated from that of gram-positive Pediococcus and Leuconostoc species. The phenotype- and PCR-based schemes reported by Sahm et al. provide efficient methods for detecting and characterizing VRE within 24 h of isolation (10). Gram staining quickly eliminated gram-positive bacilli from further testing. The multiplex PCR assay containing three primer sets for the vanA, vanB, and rrs (16S rRNA) targets was used. The pyrazinamidase and leucine aminopeptidase tests differentiated VRE from Pediococcus and Leuconostoc isolates. However, more thorough follow-up analyses of isolates are often necessary to fully support epidemiologic and infection control efforts in these schemes (10).
The average turnaround time for the PCR method is 48 h from the time of specimen setup, compared to 96 h for the routine phenotypic method (7). Culture on VRE selective medium after an enrichment step is the most sensitive method for detecting VRE (8, 11). On the other hand, a vanA-specific PCR assay for detection of VRE showed sensitivity almost equivalent to that of the culture method after the enrichment step and final results were obtained within 24 to 30 h of specimen setup (8). One limitation of this vanA-specific PCR assay for VRE surveillance was that it did not detect all of the VRE genotypes (8). With the optimized multiplex PCR assay reported here, which allows simultaneous detection of glycopeptide resistance genotypes and identification of clinically relevant enterococci to the species level, we could obtain definitive results within 48 h from the time of specimen setup. If the colonies had grown enough after 15 to 18 h, we could obtain the results within 24 h from the time of specimen setup. PCR methods for detecting and identifying VRE directly from fecal samples (11) are not cost-effective when one is dealing with a large number of specimens and low prevalence. However, it would be very useful when urgent nosocomial surveillance is needed, such as a new VRE case in the hospital. When the method used to prepare template DNA directly from fecal samples (11) is used, followed by the optimized multiplex PCR assay reported in the present study, results can be expected on the day of specimen setup.
In summary, the optimized multiplex PCR assay reported here is an attractive alternative to the currently used methods since it provides simpler and more accurate analysis of the molecular epidemiology of clinical VRE isolates.
Acknowledgments
We thank medical technologists at the Clinical Microbiology Laboratory of Okayama University Hospital for their cooperation. We also thank M. J. Zervos for providing VRE strains for the study.
REFERENCES
- 1.Cheng S, McCleskey F K, Gress M J, Petroziello J M, Liu R, Namdari H, Beninga K, Salmen A, DelVecchio V G. A PCR assay for identification of Enterococcus faecium. J Clin Microbiol. 1997;35:1248–1250. doi: 10.1128/jcm.35.5.1248-1250.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark N C, Cooksey R C, Hill B C, Swenson J M, Tenover F C. Characterization of glycopeptide-resistant enterococci from U. S. hospitals. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. . (Erratum, 33:1434.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Free L, Sahm D F. Detection of enterococcal vancomycin resistance by multiplex PCR. In: Persing D H, editor. PCR protocols for emerging infectious diseases. Washington, D.C.: ASM Press; 1996. pp. 150–155. [Google Scholar]
- 5.Fujita N, Yoshimura M, Komori T, Tanimoto K, Ike Y. First report of the isolation of high-level vancomycin-resistant Enterococcus faecium from a patient in Japan. Antimicrob Agents Chemother. 1998;42:2150. doi: 10.1128/aac.42.8.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ike Y, Tanimoto K, Ozawa Y, Nomura T, Fujimoto S, Tomita H. Vancomycin-resistant enterococci in imported chickens in Japan. Lancet. 1999;353:1854. doi: 10.1016/S0140-6736(99)01060-0. [DOI] [PubMed] [Google Scholar]
- 7.Jayaratne P, Rutherford C. Detection of clinically relevant genotypes of vancomycin-resistant enterococci in nosocomial surveillance specimens by PCR. J Clin Microbiol. 1999;37:2090–2092. doi: 10.1128/jcm.37.6.2090-2092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roger M, Faucher M-C, Forest P, St-Antoine P, Coutlée F. Evaluation of a vanA-specific PCR assay for detection of vancomycin-resistant Enterococcus faecium during a hospital outbreak. J Clin Microbiol. 1999;37:3348–3349. doi: 10.1128/jcm.37.10.3348-3349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenstraus M, Wang Z, Chang S-Y, DeBonville D, Spadoro J P. An internal control for routine diagnostic PCR: design, properties, and effect on clinical performance. J Clin Microbiol. 1998;36:191–197. doi: 10.1128/jcm.36.1.191-197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahm D F, Free L, Smith C, Eveland M, Mundy L M. Rapid characterization schemes for surveillance isolates of vancomycin-resistant enterococci. J Clin Microbiol. 1997;35:2026–2030. doi: 10.1128/jcm.35.8.2026-2030.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satake S, Clark N, Rimland D, Nolte F S, Tenover F C. Detection of vancomycin-resistant enterococci in fecal samples by PCR. J Clin Microbiol. 1997;35:2325–2330. doi: 10.1128/jcm.35.9.2325-2330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thal L A, Donabedian S, Robinson-Dunn B, Chow J W, Dembry L, Clewell D B, Alshab D, Zervos M J. Molecular analysis of glycopeptide-resistant Enterococcus faecium isolates collected from Michigan hospitals over a 6-year period. J Clin Microbiol. 1998;36:3303–3308. doi: 10.1128/jcm.36.11.3303-3308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van de Klundert J A M, Vliegenthart J S. PCR detection of genes coding for aminoglycoside-modifying enzymes. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology. Washington, D.C.: American Society for Microbiology; 1993. pp. 547–552. [Google Scholar]
- 14.You I, Kariyama R, Zervos M J, Kumon H, Chow J W. In-vitro activity of arbekacin alone and in combination with vancomycin against gentamicin- and methicillin-resistant Staphylococcus aureus. Diagn Microbiol Infect Dis. 2000;36:37–41. doi: 10.1016/s0732-8893(99)00104-2. [DOI] [PubMed] [Google Scholar]