Abstract
Most plant roots have multiple cortex layers that make up the bulk of the organ and play key roles in physiology, such as flood tolerance and symbiosis. However, little is known about the formation of cortical layers outside of the highly reduced anatomy of Arabidopsis. Here we use single-cell RNAseq to rapidly generate a cell resolution map of the maize root, revealing an alternative configuration of the tissue formative transcription factor SHORT-ROOT (SHR) adjacent to an expanded cortex. We show that maize SHR protein is hypermobile, moving at least eight cell layers into the cortex. Higher-order SHR mutants in both maize and Setaria have reduced numbers of cortical layers, showing that the SHR pathway controls expansion of cortical tissue to elaborate anatomical complexity.
One sentence summary:
Single-cell RNA-seq maps the maize root transcriptome uncovering a mechanism that regulates cortex layer number.
Roots are radially symmetrical organs composed of three fundamental tissue types, the epidermis on the outside, the ground tissue at the middle, and a core of vascular elements plus pericycle that lie in a central cylinder known as the stele (1). The ground tissue is further divided into two different cell types, the endodermis and cortex, which are arranged as concentric layers around the stele. Variations in ground tissue patterning, particularly the number of cortex cell layers, are common across species and represent one of the defining features giving rise to interspecies root morphological diversity (1). This diversity allows plants to cope with biotic and abiotic stress and adapt to challenging environments. For example, maize cortex plays a role in drought and flood tolerance and hosts colonization of beneficial mycorrhizal associations that reduce stress and improve nutrient uptake (2-5). Therefore, an ongoing question in root biology is how tissue patterning is adjusted to produce divergent root morphologies, and what alterations in the genetic networks control developmental differences among species.
A current limitation is that our knowledge of radial patterning mechanisms in roots comes largely from the study of Arabidopsis, which possesses a simple root anatomy. In Arabidopsis, only two ground tissue layers develop in primary development -- one endodermal and one cortical -- that originate from an asymmetric cell division at or near the initials or stem cells (6). This division is controlled by the SHORT-ROOT (SHR)/SCARECROW (SCR) genetic pathway (7, 8). Mutants in either transcription factor develop a monolayered ground tissue. In addition, SHR mutants lack an endodermis, giving SHR a role in both tissue formation and cell identity (9).
SHR functions as a mobile signal whose protein travels from the stele, where the gene is transcribed, into the surrounding endodermis, where it induces the expression of the downstream transcription factor SCR (8). The pathway then triggers the division that generates the cortex and endodermis layers (9). In maize, the SHR-SCR pathway has been implicated with a role in leaf anatomy (10, 11). Movement of SHR from the stele further out into the cortex could potentially cause extra cell divisions, giving rise to multiple cortex layers (12). Supporting this idea, the SHR-SCR pathway was recently implicated in cortical cell division during nodule formation in the dicot Medicago (13). However, the role of SHR in the expansion of cortical layer number in maize, a monocot, is not known.
Dye penetrance labeling for rapid tissue profiling
We sought to produce a high-resolution spatial and temporal map of gene expression in a complex root that could provide clues to the genetic networks controlling morphological diversity in patterning. Therefore, we generated cell type-specific gene expression profiles using high-throughput single cell RNAseq (scRNAseq) to profile maize roots. Maize is a valuable model for comparative studies because its roots develop multilayered cortical tissues (8-9 cortex cell layers) within the root meristem, and it is amenable to protoplast generation, an essential step in plant scRNAseq (14). However, a challenge of scRNAseq studies in species for which genomic resources are limited is the correct identification of cell types. The use of homologs of Arabidopsis markers obtained by high-throughput cell sorting did not provide a clear identification of morphologically homologous cell types in maize. This is likely because gene orthology is not always apparent and localization over such broad evolutionary distance is not well conserved.
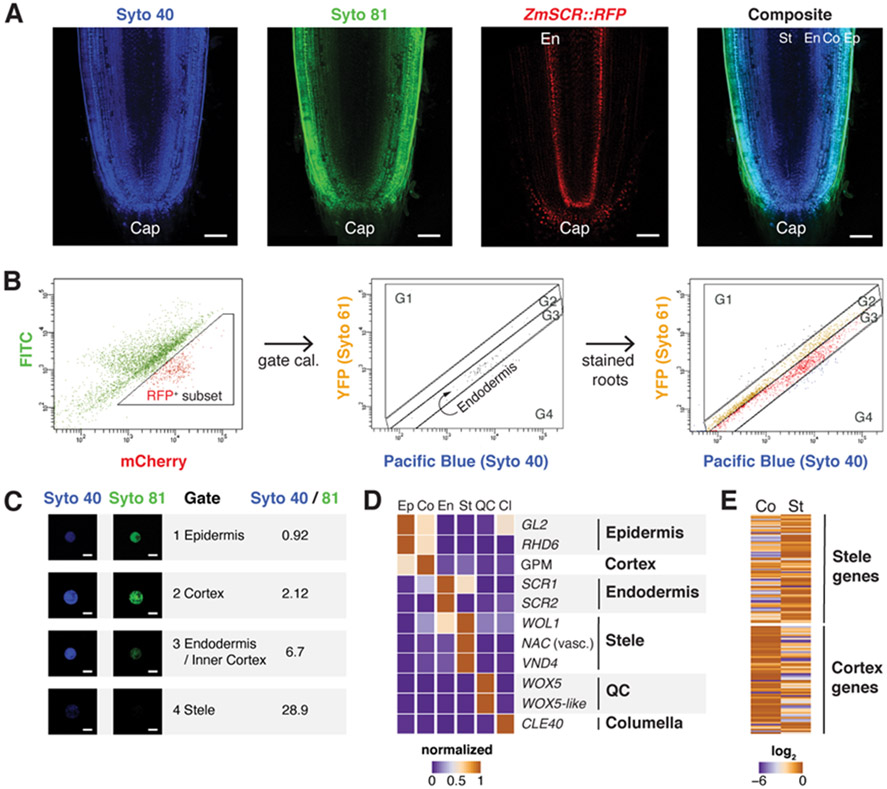
To overcome this problem, we first took advantage of the concentric arrangement of tissues in roots to develop a technique to fluorescently mark cell layers by dye penetrance labeling (DPL). In brief, a highly penetrant dye (Syto 40 - blue) stains the entire root with low but detectable staining in stele, whereas a weakly penetrant dye (Syto 81 - green) stains the outer tissue layers strongly, with a gradual drop in signal intensity towards the inner tissues (Fig.1A). This dual labeling was reproducible across roots and batches and had a negligible effect on transcription (Fig. S1). The approach allowed us to enrich for different concentric tissue layers using blue/green ratios in Fluorescence Activated Cell Sorting (FACS). We calibrated dye ratio to radial position by using DPL on a line expressing a fluorescent protein driven by the SCR promoter (ZmSCR1::RFP; Fig. 1B), which expresses in the endodermis. RFP positive cells were used to calibrate a reference dye ratio for this middle layer, allowing demarcation of inner and outer tissues (Fig. 1C, Fig. S2A). We dissected seminal root tips (5 mm and, in one subset, 5 to 8mm the from tip) and then rapidly enzymatically digested their cell walls, sorting cells belonging to different tissue layers using their specific dye ratio. We also generated a set of whole meristem protoplasts vs intact root controls to filter out any effects of cell wall digestion. Digested and undigested controls clustered closely together and replicate samples yielded consistent profiles (Fig. S2B-D). In addition, we obtained expression profiles of the root cap by dissection and quiescent center (QC), using FACS on a stable QC marker line, ZmWOX5::tagRFPt (Fig.S2E). To validate the entire dataset, we compared the six tissue expression profiles to known markers and to a previous study that used mechanical separation of inner vs outer layers (15), finding 80% agreement or higher (Fig. 1D,E). We also used a panel of conserved and well-characterized markers to validate ICI expression profiles, showing close agreement (Fig. S3, Table S2B). In this manner, we developed a set of at least 170 markers for each radially arranged tissue (Table S1).
Fig. 1. Dye Penetrance Labeling (DPL) and tissue transcriptome analysis in maize.
(A) Representative images of a deeply penetrating dye (Syto 40), a superficially penetrating dye (Syto 81), the ZmSCR::tagRFPt marking endodermis, and a composite image of Syto 40 and Syto 81 staining, showing position of the endodermal (Ed) layers in dashed region. (B) Cell sorting gating strategy, showing the ZmSCR::tagRFPt population (left), backgated onto a YFP vs Pacific Blue plot with RFP positive (second from left), and (third from left) the gated boundaries for endodermal, outside of endodermis (G1,G2), and inside of endodermis (G4). (C) Validation of ratiometric cell sorting strategy by collecting sorted cells from gates and quantifying fluorescence from microscopy images. (D) Validation of sorted cell RNA-seq profiles via analysis of known markers. (E) Global validation comparing sorted cells vs. mechanically dissected stele and cortex tissues, with heat map showing expression in sorted cortex vs. stele gates, categorized by previously determined stele and cortex markers. Scale bars are 100 μm in (A) and 15 μm in (C).
A single-cell map of the maize root
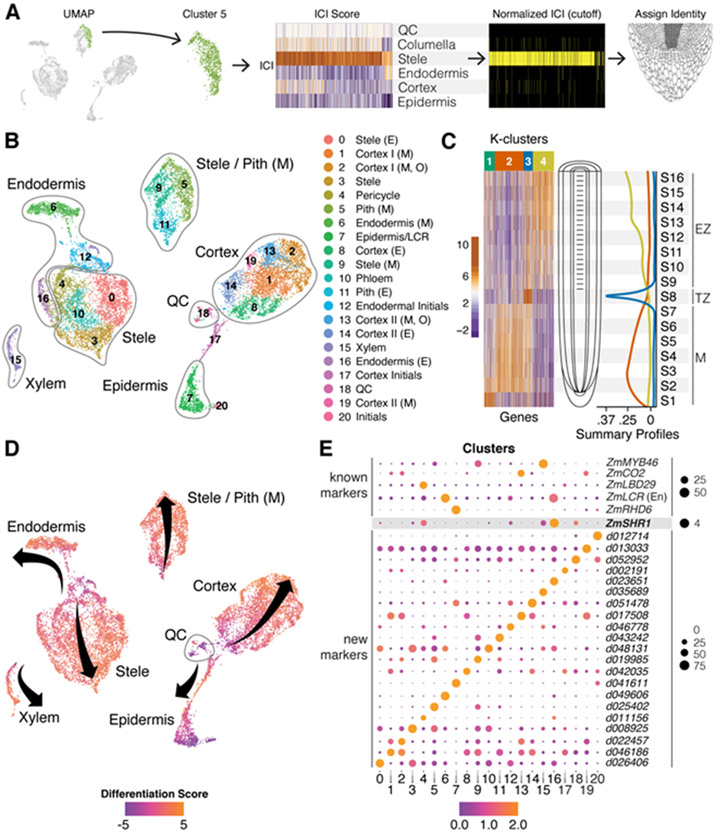
To generate a single cell resolution map of the maize root meristem, we then dissected seminal root tips from 7 day old wild-type B73 maize seedlings and enzymatically digested their cell walls, as above. We then used the cells to prepare single cell cDNA libraries using the 10x Genomics Chromium platform. A total of 14,755 high quality cells were sequenced in three different batches with a mean of 31,105 Unique Molecular Indices/cell and 5,683 detected genes/cell (Fig. S4F). A total of 21 cell clusters were defined and visualized in two dimensions in Seurat using the uniform manifold approximation and projection (UMAP) method (16). To quantify cell identity and classify clusters using the DPL markers, we applied the Index of Cell Identity (ICI) algorithm (17), which generates a cell identity score based on the mean expression of a predefined marker gene set, in this case, from FACS isolated tissues (Fig. 2A & Fig. S5,6). Overall, the technique allowed us to identify all the UMAP clusters, providing a detailed spatial map of transcripts in specific cell types of the maize root (Fig 2B).
Fig. 2. Single-cell RNA-seq spatial and temporal transcriptome maps of the maize meristem.
(A) The ICI method of diagnosing cell of UMAP clusters using known markers and randomization testing (e.g., Cluster 5). (B) Cluster identities as determined by ICI and cell-type specific markers (E,early; M,mature; O,outer; Pith, pith parenchyma). (C) Heat map of highly variant genes along a longitudinal axis of the root meristem. Development patterns show transcripts/markers that peak in the early meristem (M), transition zone (TZ), and elongation zone (EZ). (D) Trajectories of developmental “pseudo-time” in each cell cluster mapped onto the same UMAP depicted in B, where a differentiation score is calculated as a log2 ratio of all EZ/M markers identified in C. Arrow origins represent cells in the meristem near the stem cells for each cell type cluster progressing to more proximal cells. (E) Select known (top) and new markers (bottom) for each cluster. Size of spot represents percent of cells in cluster expressing the marker and color represents their relative expression level in those cells.
The high-resolution cellular map of the meristem showed multiple cell type subclusters within the stele and cortex, suggesting cellular specialization within that the latter tissue’s multiple cell layers. However, because root cells differentiate as they transition away from the root tip, the possibility remained that some subclusters represented different maturation states of an individual cell type. To distinguish subclusters formed by distinct identity rather than differentiation state, we further generated a set of cell maturation marker transcripts by dissecting 16 longitudinal root slices that together comprised the meristematic, transition, and elongation zones, and subjected the samples to RNAseq analysis (Fig. 2C). Using K-means clustering, we identified three main expression programs: early meristematic (high expression in the meristem and gradual decrease towards the transition zone), transition zone (specific to the mid-maturation point), and post-transition (low expression in the meristematic zone and gradual increase towards the elongation zone). We then generated a cellular differentiation score to label the maturation status of each cell, resolving developmental trajectories of cells in our high-resolution map of cell identities (Fig. 2 D).
High-resolution profiling reveals cortical complexity
In a few cases, the state of cell maturation is indeed the main factor influencing grouping of cells into subclusters. For example, five clusters (8,11,12,14 and 16) had the same identity as adjacent clusters but a different state of maturation. However, the majority of subclusters were composed of cells with a wide range of differentiation states, showing that the grouping in most cases represented distinct cell identities. While two cortex subclusters appeared to be a precursor state of mature cortex (clusters 8 and 14), our analysis confirmed the existence of at least four distinct cortex subtypes (Fig. 2B, clusters 1,2,13,19). Furthermore, using the ROC algorithm in Seurat, which identified 2,436 differentially expressed genes (DEGs) across all clusters, we found 471 transcripts that mark some subset of the four different cortical subtypes (Fig. 2E, Table S3). Thus, we provide quantitative evidence for the sub specialization of cortex that underlies expansion of root complexity, providing strong evidence for cortical cell diversification.
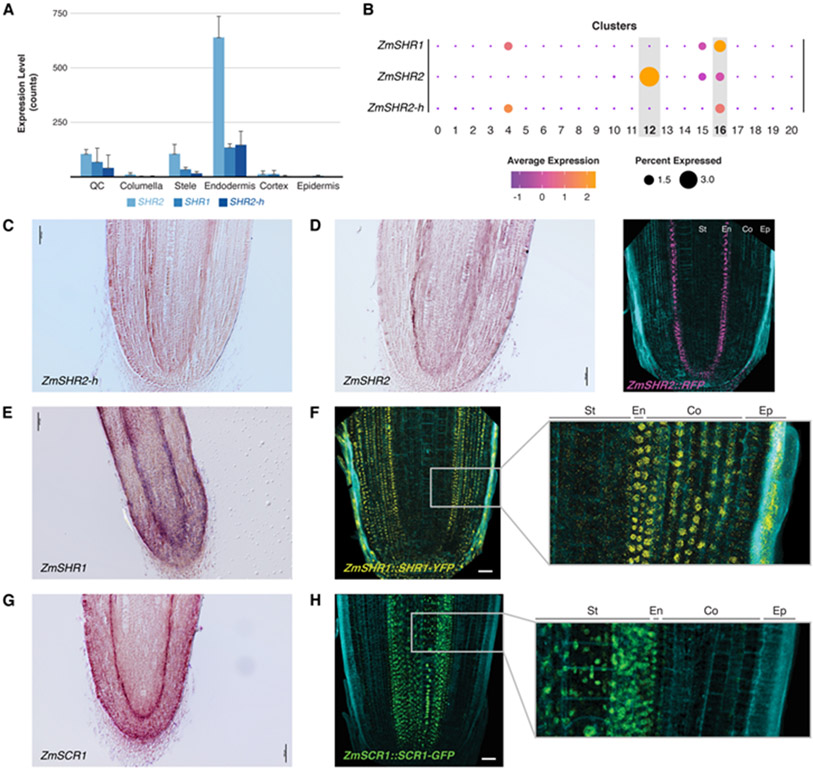
One question that follows is what signaling mechanisms allow maize to form the extra layers that permit cortex sub specialization. We observed that a short list of functional markers with a role in patterning or cell identity in Arabidopsis had conserved localization in homologous tissues in maize (e.g., CO2, cortex; MYB46, xylem; RHD6, epidermis; and LBD29, stele). However, our single-cell data showed that expression of the core patterning gene SHR was specific to the endodermis and not to the stele (Fig. 2E), where the Arabidopsis ortholog is expressed (9). All three maize paralogs of SHR (designated ZmSHR1, ZmSHR2, and ZmSHR2-h) showed the same endodermal enrichment in the profiles generated by DPL and single-cell analysis (Fig. 3A,B). We speculated that the expression of this mobile, division inducing transcription factor adjacent to the cortex could be related to a role in the expansion of that tissue.
Fig. 3. SHR and SCR expression in maize endodermis and differences between transcriptional and translational reporters.
(A) Expression of the three ZmSHR paralogs (SHR1,2,2-h) from sorted cells. Error bars are standard deviation. (B) Dot plots representing single cell analysis showing expression of ZmSHR paralogs in endodermis and, at a low level, stele. (C,D) ZmSHR2h in-situ hybridization showing endodermal expression. (D) ZmSHR2 in-situ hybridization and transcriptional reporter. (E) In-situ hybridization of ZmSHR1. (F) Translational reporter for ZmSHR1 with inset showing protein movement into cortex layers. (G) In-situ hybridization of ZmSCR1. (H) Translational reporter SCR1-GFP reporter showing movement into the stele. Scale bars are 50 μm. Cyan in confocal images is autofluorescence at Ex/405.
Mobile SHR regulates cortical complexity in maize and Setaria
To confirm SHR transcript localization, we performed in-situ hybridization on all three SHR mRNAs, confirming their localization in the endodermis (Fig. 3C-E, Fig. S7A-K). A 1.4 kb upstream and downstream transcriptional reporter for ZmSHR2 also showed signal in the root endodermis in agreement with our dye sorted and single cell profiles (Fig. 3D). No signal was found in the stele, confirming that SHR2 transcript localizes to different cell types in maize compared to Arabidopsis. Consistently, the ZmSHR endodermal domain in the root is reminiscent of ZmSHR expression in the shoot bundle sheath (18, 19), which is in an analogous position to root endodermis. In addition, expression of SHR orthologs were shown to localize to the endodermis in date palm (20).
Given evidence that rice SHR proteins are hypermobile when expressed heterologously in Arabidopsis (8), we assessed whether maize SHR protein could move from the endodermis, where it is expressed, into the adjacent cortex. For this, we made a natively expressed protein reporter of ZmSHR1 fused to YFP (ZmSHR1::SHR1-YFP). Indeed, compared to endodermal localization of the ZmSHR1 transcript, the maize SHR1 protein reporter was present in the cortex (Fig. 3F). Moreover, SHR1-YFP signal was not restricted to the adjacent tissue layer as in Arabidopsis but was observed in all cortex layers, suggesting that the endogenous maize SHR1 protein moves through at least 8 cortex layers (Fig. 3F, Fig. S7D). In addition, the ZmSHR1 protein also appeared to be hypermobile when expressed in the endodermis of Arabidopsis (Fig. S8). Whether additional SHR paralogs are mobile is an open question.
SHR’s role in promoting division in Arabidopsis works through direct interaction with SCR (21). Therefore, we also generated maize SCR reporter lines to determine colocalization with SHR . Both in-situ localization and a promoter RFP reporter revealed a signal in the root endodermis, as shown previously (22) and as conserved with localization in Arabidopsis (Fig. 3G, Fig. S7E-H). In addition, we observed low level SCR1 expression in the cortex in the scRNAseq data and in high-sensitivity confocal imaging (e.g., Fig. S7F). However, natively expressed ZmSCR-GFP protein fusions showed a signal in the stele, suggesting that SCR protein in maize moves from cell-to-cell in the opposite direction to SHR (Fig. 3H, Fig. S7G). Our results show that SHR and SCR colocalize in the endodermis and possibly in the cortex. A SCR translational reporter in a second monocot, Setaria viridis (Setaria), showed the same localization in the stele, further corroborating the divergent localization of SCR protein in monocots (Fig. S9A,B).
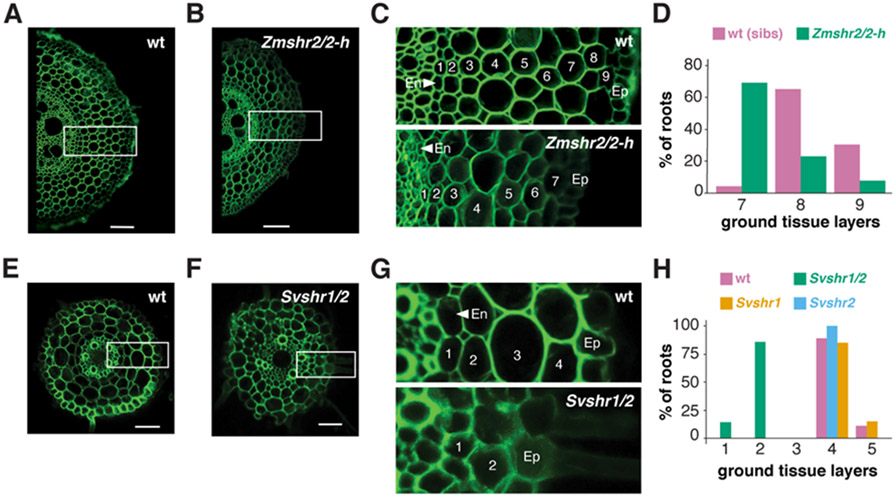
The model that implicates SHR in cortical expansion posits that increased outward movement of the protein could trigger periclinal cell divisions giving rise to extra ground tissue layers (12). To test the model, we targeted the three different maize SHR paralogs to generate loss-of-function mutants in maize using CRISPR-Cas9 (Fig. S10A,B). We recovered mutants in two of the genes (ZmSHR2 and ZmSHR2-h), including the most highly expressed paralog. Single mutants in Zmshr2 or Zmshr2-h had no difference in their root anatomy compared to wild type siblings. However, Zmshr2/2-h double mutants had a significant reduction in the number of cortical layers: with most roots having 7 layers compared to 8 to 9 layers in wild type (Fig 4A-D). Mutants in the single SHR gene in Arabidopsis lack an endodermal layer. However, staining for suberin and morphological analysis showed that Zmshr2/2-h roots still developed an endodermis (Fig. S11). We posit that the remaining functional ZmSHR1 gene in the Zmshr2/2-h background may still enable specification of endodermal identity. Alternatively, ZmSCR1 (and ZmSCR2) may be the primary factors in the specification of the maize endodermis (23). Overall, our results suggest that SHR function in maize is necessary for the full expansion of cortical identity.
Fig. 4. Cortical cell layer analysis in wild type and shr mutants in monocots.
(A,B) Representative maize root cross-sections of wild type in A vs. Zmshr2/2-h double homozygous mutant in B. (C) Enlarged regions from dashed boxes in A and B showing stelar (S), endodermal (Ed), and cortical layers of wild type (top) and Zmshr2/2-h mutant (bottom). (D) Quantification of the cortical cell layers in wild type and heterozygous sibs (n=23) vs. Zmshr2/2-h mutant (n=13, p<0.001, Mann-Whitney Rank Test). (E,F) Representative cross sections of Setaria wild type in E and Svshr1/2 mutants in F. (G) Enlarged regions from dashed boxes in E and F showing stelar (S), endodermal (Ed), and cortical layers of wild type (top) and Svshr1/2 mutant (bottom). (H) Quantification of cortical layers in Setaria wild type (n = 9), Svshr1 single (n = 7), Svshr2 single (n = 6) and double mutants (n =7, p<0.001, Tukey Test after One-Way ANOVA on all genotypes,). Scale bars are 100 μm in (A), (C), and 50 μm in (E), (F). Green is autofluorescence Ex/405, Em/510-535.
We sought to validate the monocot SHR mutant phenotype with a more severe loss of function by testing its role in Setaria, a close relative of maize. In Setaria, we were able to generate loss of function mutants in the two SHR orthologs using CRISPR-Cas9 (Fig. S12A,B). One Setaria SHR mutant, Svshr2, showed a slight reduction in cortical layers, while a single mutant in the second, Svshr1, showed no phenotype. However, double mutants showed a dramatic reduction in the number of ground tissue layers, resulting in 1-2 layers compared to 4-5 layers in wild type siblings (Fig. 4E-H). These results corroborate the role of SHR in controlling the expansion of cortical layers in two monocots.
The extra cortical divisions mediated by SHR could function through direct interaction with SCR by mediating successive divisions of the cortex-endodermal split near the stem cell niche, where the two proteins overlapped in maize. Alternatively, SHR hypermobility could lead to divisions directly in the cortical layers possibly interacting with low levels of SCR or another protein. At present, we cannot distinguish between these two models.
The results show that SHR has a role in monocots controlling the expansion of cortex, which sets up many traits for environmental acclimation. This illustrates how subtle divergence of a conserved developmental regulator can mediate anatomical complexity that has given rise to specialized functions. Furthermore, the results show that rapid transcriptome mapping using single cell dissection can provide insights into the mechanisms that mediate anatomical diversity. The use of dye labeling to generate a scaffold locational map together with scRNAseq now provides a maize root tissue map that can be used as a reference map in maize and related plants.
Supplementary Material
Table S3. Markers derived from the ROCs in Seurat based on differential expression among the clusters.
Fig. S1. Syto dyes stain consistently and have negligible effect on transcription. Related to Figure 1.
Fig. S2. FACS sorting of RFP positive and dye-stained protoplasts and clustering analyses of tissue transcriptomes. Related to Figure 1.
Figure S3. Corroboration of dye sorting method. Related to Figure 2.
Figure S4. Single cell run statistics. Related to Figure 2.
Fig. S5. ICI analysis using markers from cell sorting experiments to diagnose cluster identity. Related to Figure 2.
Figure S6. Xylem markers from root single-cell RNAseq are xylem enriched in shoot single-cell RNAseq.
Fig. S7. High sensitivity imaging of ZmSCR and ZmSHR reporter lines, in-situ hybridizations, and sense controls. Related to Figure 3.
Fig. S8. ZmSHR1 is hypermobile and capable of generating supernumerary cortical layers in Arabidopsis.
Fig. S9. SvSCR protein is located in the stele in Setaria viridis roots. Related to Fig. 3.
Fig. S10. CRISPR-Cas9 gene editing in ZmSHR genes caused frameshift mutations. Related to Figure 3.
Fig. S11. Endodermal identity analysis by suberin staining in wild type and Zmshr1/3 in maize. Related to Figure 4.
Fig. S12. CRISPR-Cas9 gene editing in SvSHR genes caused frameshift mutations. Related to Figure 4.
Fig. S13. Primer sets used to generate the in-situ probes for the ZmSHR and ZmSCR transcripts.
Table S1. Markers used for ICI representing the tissues profiled by DPL, reporter sorting, or hand dissection.
Table S2. Markers used to refine cluster calls and validate ICI cluster identification.
Acknowledgments
Funding: K.D.B and D.J. are supported by NSF grant IOS-1934388. K.D.B, D.J. and T.R.G. were supported by NSF grant IOS 1445025. K.D.B. is funded by NIH grant R35GM136362. D.J. is funded by NSF IOS-1930101. K.L.G NSF PGRP-23020.
Footnotes
Data materials and availability: All raw and processed/normalized expression data is available through the Gene Expression Omnibus under the SuperSeries accession GSE172302.
REFERENCES
- 1.Esau K, Anatomy of seed plants. (Wiley, New York, ed. 2d, 1977), pp. 550 [Google Scholar]
- 2.Kaldorf M, Kuhn AJ, Schröder WH, Hildebrandt U, Bothe H, Selective Element Deposits in Maize Colonized by a Heavy Metal Tolerance Conferring Arbuscular Mycorrhizal Fungus. Journal of plant physiology 154, 718–728 (1999). [Google Scholar]
- 3.Liu F et al. , Identification and Functional Characterization of a Maize Phosphate Transporter Induced by Mycorrhiza Formation. Plant Cell Physiol 59, 1683–1694 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Zhu J, Brown KM, Lynch JP, Root cortical aerenchyma improves the drought tolerance of maize (Zea mays L.). Plant Cell Environ 33, 740–749 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Gunawardena AH, Pearce DM, Jackson MB, Hawes CR, Evans DE, Characterisation of programmed cell death during aerenchyma formation induced by ethylene or hypoxia in roots of maize (Zea mays L.). Planta 212, 205–214 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Dolan L et al. , Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Benfey PN et al. , Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119, 57–70 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Nakajima K, Sena G, Nawy T, Benfey PN, Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307–311 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Helariutta Y et al. , The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Slewinski TL et al. , Short-root1 plays a role in the development of vascular tissue and kranz anatomy in maize leaves. Mol Plant 7, 1388–1392 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Slewinski TL, Anderson AA, Zhang C, Turgeon R, Scarecrow plays a role in establishing Kranz anatomy in maize leaves. Plant Cell Physiol 53, 2030–2037 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Wu S et al. , A plausible mechanism, based upon Short-Root movement, for regulating the number of cortex cell layers in roots. Proc Natl Acad Sci U S A 111, 16184–16189 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong W et al. , An SHR-SCR module specifies legume cortical cell fate to enable nodulation. Nature 589, 586–590 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Senn A, Pilet P-E, Isolation and some Morphological Properties of Maize Root Protoplasts. Zeitschrift für Pflanzenphysiologie 100, 299–310 (1980). [Google Scholar]
- 15.Opitz N et al. , Extensive tissue-specific transcriptomic plasticity in maize primary roots upon water deficit. J Exp Bot 67, 1095–1107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuart T et al. , Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Efroni I, Ip PL, Nawy T, Mello A, Birnbaum KD, Quantification of cell identity from single-cell gene expression profiles. Genome Biol 16, 9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang YM et al. , Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol 160, 165–177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X et al. , Single-cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Dev Cell 56, 557–568 e556 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao TT et al. , Emergent Protective Organogenesis in Date Palms: A Morpho-Devo-Dynamic Adaptive Strategy during Early Development. Plant Cell 31, 1751–1766 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui H et al. , An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316, 421–425 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Lim J et al. , Molecular analysis of the SCARECROW gene in maize reveals a common basis for radial patterning in diverse meristems. Plant Cell 12, 1307–1318 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes TE, Sedelnikova OV, Wu H, Becraft PW, Langdale JA, Redundant SCARECROW genes pattern distinct cell layers in roots and leaves of maize. Development 146, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. Markers derived from the ROCs in Seurat based on differential expression among the clusters.
Fig. S1. Syto dyes stain consistently and have negligible effect on transcription. Related to Figure 1.
Fig. S2. FACS sorting of RFP positive and dye-stained protoplasts and clustering analyses of tissue transcriptomes. Related to Figure 1.
Figure S3. Corroboration of dye sorting method. Related to Figure 2.
Figure S4. Single cell run statistics. Related to Figure 2.
Fig. S5. ICI analysis using markers from cell sorting experiments to diagnose cluster identity. Related to Figure 2.
Figure S6. Xylem markers from root single-cell RNAseq are xylem enriched in shoot single-cell RNAseq.
Fig. S7. High sensitivity imaging of ZmSCR and ZmSHR reporter lines, in-situ hybridizations, and sense controls. Related to Figure 3.
Fig. S8. ZmSHR1 is hypermobile and capable of generating supernumerary cortical layers in Arabidopsis.
Fig. S9. SvSCR protein is located in the stele in Setaria viridis roots. Related to Fig. 3.
Fig. S10. CRISPR-Cas9 gene editing in ZmSHR genes caused frameshift mutations. Related to Figure 3.
Fig. S11. Endodermal identity analysis by suberin staining in wild type and Zmshr1/3 in maize. Related to Figure 4.
Fig. S12. CRISPR-Cas9 gene editing in SvSHR genes caused frameshift mutations. Related to Figure 4.
Fig. S13. Primer sets used to generate the in-situ probes for the ZmSHR and ZmSCR transcripts.
Table S1. Markers used for ICI representing the tissues profiled by DPL, reporter sorting, or hand dissection.
Table S2. Markers used to refine cluster calls and validate ICI cluster identification.