Supplemental Digital Content is available in the text.
Keywords: bias, hospital mortality, intensive care units, models/statistical, pediatrics, sepsis/classification
Abstract
OBJECTIVES:
To evaluate the performance of pragmatic imputation approaches when estimating model coefficients using datasets with varying degrees of data missingness.
DESIGN:
Performance in predicting observed mortality in a registry dataset was evaluated using simulations of two simple logistic regression models with age-specific criteria for abnormal vital signs (mentation, systolic blood pressure, respiratory rate, WBC count, heart rate, and temperature). Starting with a dataset with complete information, increasing degrees of biased missingness of WBC and mentation were introduced, depending on the values of temperature and systolic blood pressure, respectively. Missing data approaches evaluated included analysis of complete cases only, assuming missing data are normal, and multiple imputation by chained equations. Percent bias and root mean square error, in relation to parameter estimates obtained from the original data, were evaluated as performance indicators.
SETTING:
Data were obtained from the Virtual Pediatric Systems, LLC, database (Los Angeles, CA), which provides clinical markers and outcomes in prospectively collected records from 117 PICUs in the United States and Canada.
PATIENTS:
Children admitted to a participating PICU in 2017, for whom all required data were available.
INTERVENTIONS:
None.
MEASUREMENTS AND MAIN RESULTS:
Simulations demonstrated that multiple imputation by chained equations is an effective strategy and that even a naive implementation of multiple imputation by chained equations significantly outperforms traditional approaches: the root mean square error for model coefficients was lower using multiple imputation by chained equations in 90 of 99 of all simulations (91%) compared with discarding cases with missing data and lower in 97 of 99 (98%) compared with models assuming missing values are in the normal range. Assuming missing data to be abnormal was inferior to all other approaches.
CONCLUSIONS:
Analyses of large observational studies are likely to encounter the issue of missing data, which are likely not missing at random. Researchers should always consider multiple imputation by chained equations (or similar imputation approaches) when encountering even only small proportions of missing data in their work.
RESEARCH IN CONTEXT.
Researchers analyzing large clinical datasets will nearly always encounter the problem of missing data. Their approach to managing this missingness can have a significant impact on the potential for bias in their results and the validity of their interpretation.
In this study, we compare the effects of commonly used approaches to managing missing data, using a sample dataset from Virtual Pediatric Systems: complete case analysis, assuming missing values are normal (or abnormal), substituting average values, and multiple imputation by chained equations (MICE).
Our simulations that we evaluated against the known outcomes found that MICE outperformed all the other methods we evaluated, almost irrespectively of the degree of missingness.
Analyzing large datasets of routinely collected data is becoming more commonplace in the development of risk stratification models (1–4), such as the Pediatric Index of Mortality 2 (5), the Pediatric Risk of Mortality score 3 (6), pediatric systemic inflammatory response syndrome (pSIRS) (7), and pediatric quick Sequential [Sepsis-Related] Organ Failure Assessment (qSOFA) (3) models. Electronic medical record (EMR) data quality varies (8), yet these datasets provide a valuable resource, as recently highlighted in the derivation of adjusted sepsis mortality rates from EMR data (9). Unfortunately, clinical datasets typically include incomplete cases (i.e., missing data in some variables), and it is not uncommon to encounter large fractions (e.g., 40% or more) of incomplete observations (10). The preferred option is to apply “multiple imputation” (11–13). Nevertheless, there are simpler alternatives, including “complete case analysis,” in which all cases with missing observations were discarded, or “single imputation,” which depends on operational assumptions, such as presuming that laboratory tests are omitted when clinical suspicion is low.
Multiple imputation is a theory-driven approach (11, 12, 14), which rests on the assumption that data elements are missing according to a probabilistic process, or “missing data mechanism,” which is independent of the unobserved value. This is the case, for example, if observations are unobserved due to purely external mechanisms unrelated to the processes being considered, a situation which is termed “missing completely at random.” A less stringent assumption is that the missing mechanism depends on observable data, for example, if certain elements are likely to be missing according to the age of the subject (assuming of course that age is available on all cases). This type of mechanism falls under the terminology “missing at random.”
Under either of these assumptions, it is feasible to generate sets of values that resemble a random sample from the probability distribution of the unknown value as it relates to the available information, yielding multiple imputed datasets. Following this approach, one applies the complete data estimation algorithm to each of imputed datasets and takes the mean across the resultant estimates. ses for these estimates can be derived from a simple formula combining within sample and between sample variance (14, 15). One popular implementation of this approach is Multiple (or Multivariate) Imputation using Chained Equations (MICE) (16, 17).
Multiple imputation cannot be taken as a panacea (15, 18), as situations may arise where the missing at random condition fails. Observations may not be recorded when a clinical test would 1) yield a predictable or clinically unimportant result and is thus not performed, 2) was not feasible, or 3) was performed, produced a significant result and was acted on immediately, rendering the recording of the result unnecessary. In such cases, these data are not missing at random and the application of multiple imputation may lead to biased results.
The other methods have their limitations as well: complete case analysis is evidently prone to inflation of the variance in estimation due to omission of potentially informative data and can result in bias if the mechanism of missing data are nonrandom and operates selectively in a manner that yields a nonrepresentative set of complete observations (e.g., if data can only be obtained from conscious patients); bias may also result if the operational logic underlying simple imputations (e.g., missing is normal) is faulty (19–23).
The purpose of this study was to evaluate simulations of biased missingness in a dataset of ICU observations to determine how best to overcome this data missingness statistically. The goal was to provide guidance for clinician-scientists around the methodology for performing analyses in the presence of missing clinical data, how clinicians should interpret results with its underlying uncertainty, and to gain a better understanding of the advantages and limitations of using multiple imputation approaches.
MATERIALS AND METHODS
Study Design
The efficacies of approaches designed to contend with missing data were determined by their effect on the estimation of certain parameters of interest. Usually, these are the parameters of a statistical model one would hope to fit if a complete dataset was available. We used a simulation approach to evaluate pragmatic imputations (assuming missing is normal, or assuming missing is abnormal) and complete case analysis, and contrasted their performance with a simple multiple imputation (MICE) approach. We did so using a large complete dataset into which varying degrees of nonrandom missingness were introduced.
This article has been prepared with reference to the guidelines for Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (24) and guidance on the development and reporting of prediction models for the critical care setting (25).
Data Source
The Virtual Pediatric Systems (VPS, LLC, Los Angeles, CA) database contains a prospective observational cohort of consecutive PICU admissions in 117 participating hospitals in the United States and Canada, which uses standardized clinical definitions; VPS ensures data quality control and undergoes extensive quality validation prior to release of the data for analysis (26). VPS does not require completed entries for all fields before a record is accepted into the registry; data identified as “missing” during our analysis do not indicate gaps in VPS data collection/validation processes but rather would not have been recorded by the participating PICU, either for clinical or administrative reasons. The VPS registry is widely used as it provides access to a representative clinical cohort.
Approval was obtained from the University of British Columbia/Children’s & Women’s Health Centre of British Columbia Research Ethics Board (H19-00279) for secondary analysis of data from the VPS database; waiver of written informed consent was granted.
Mortality Risk Score Models
To illustrate the effect of various approaches and their performance, we performed a simulation experiment. Specifically, we used the coefficients of two logistic regression models generated by creating simple mortality risk score models for children with sepsis. First, we created a model using pSIRS criteria (7), which though not designed as a mortality risk score is increasingly used as such, and which used age-specific vital signs criteria from Goldstein et al (7) for abnormal heart rate (HR), abnormal respiratory rate (RR), abnormal temperature, and abnormal WBC (leukocyte) count. Second, we created a model using pediatric qSOFA scores (3), which used age-specific vital signs criteria from Goldstein et al (7) for abnormal RR and abnormal systolic blood pressure (SBP), while applying the corrected thresholds from their letter to the editor (27), and a Glasgow Coma Scale (GCS) score less than or equal to 11 for abnormal mental status (28).
Patient Population
The analysis used data from all children (age < 18 yr) admitted to 117 PICUs between January 1, 2017, and December 31, 2017. Each child was admitted to a PICU one or more times, yielding a total of 123,035 unique patient episodes, for each of which the VPS database reports a number of different clinical observations. Note, this cohort was further reduced to create “complete populations” for simulation analysis.
Creating a Complete Dataset
Data were analyzed using R statistical software (R Foundation for Statistical Computing, Vienna, Austria) (29). Data science and visualization packages, including “tidyverse” (Version 1.3.0), “mice” (Version 3.13.0), “pROC” (Version 1.17.0.1), “ROCR” (Version 1.0.11), and “viridis” (Version 0.6.0) were used for statistical analysis and experimentation. We have provided the R code files, which were used to build the models and generate results (https://github.com/part-cw/PCCM_2021_ImputationModel). Statistical properties including bias, variance, and the coverage rate of different estimation procedures necessitate an underlying ground truth knowledge of the model coefficients. Two “complete populations” were defined: 48,485 subjects for whom a pSIRS score could be calculated with complete data and 32,293 subjects for whom a qSOFA score could be calculated with complete data. We then fit the corresponding logistic regression models to both complete populations and the model performance (fit) was evaluated using the area under the receiver operating characteristic curve (AUC), computed using five-fold cross-validation in 150-run experiments.
Observation of Missingness Patterns to Motivate Simulation Strategy
The amount of missing data and the mechanism by which missingness occurs should inform the choice of estimation method. Thus, the proportion of missing values for GCS, used to determine abnormal mentation, and WBC were evaluated as a function of the reporting PICU size and the percentage of observations in the dataset submitted by each PICU that contain values outside the normal range. Both variables were chosen as they exhibited a high degree of missingness, namely 59% for WBC and 72% for GCS, and because their missingness was somewhat informative of other variables (Table 1). While it is likely these data were not missing at random, our retrospective analysis cannot distinguish the nature of these missingness patterns; that is, we cannot determine whether these data are missing at random (e.g., data lost in transcription or the laboratory system unavailable) or not missing at random (e.g., because the test was not ordered for clinical reasons or is not uploaded to the VPS system). These variables are frequently included in PICU prognostic scores and are, hence, useful examples for this simulation experiment.
Table 1.
Overview of Study Population With Demographics and Risk Factors, Split by Outcome
| Patient Characteristic | Total (n = 118,826) | Died (n = 2,600) | Survived (n = 116,226) |
|---|---|---|---|
| Age category | |||
| 0 d to < 1 wk | 1,862 (2%) | 136 (5%) | 1,726 (1%) |
| 1 wk to < 1 mo | 2,898 (2%) | 101 (4%) | 2,797 (2%) |
| 1 mo to < 2 yr | 39,545 (33%) | 893 (34%) | 38,652 (33%) |
| 2 to < 6 yr | 23,781 (20%) | 428 (16%) | 23,353 (20%) |
| 6 to < 13 yr | 26,036 (22%) | 538 (21%) | 25,498 (22%) |
| 13 to < 18 yr | 24,704 (21%) | 504 (19%) | 24,200 (21%) |
| Gender | |||
| Female | 53,073 (45%) | 1,123 (43%) | 51,950 (45%) |
| Male | 65,742 (55%) | 1,477 (57%) | 64,265 (55%) |
| Length of stay (d) | 4.2 (± 11.1) | 13.1 (± 27.3) | 4.0 (± 10.3) |
| Pediatric Index of Mortality 2 (5) probability of death | 2.4 (± 7.9) | 31.1 (± 35.2) | 1.7 (± 4.2) |
| Pediatric Risk of Mortality score 3 (6) probability of death | 2.1 (± 9.5) | 40.3 (± 39.0) | 1.3 (± 5.0) |
| Temperature | |||
| Abnormal | 26,201 (22%) | 1,642 (63%) | 24,559 (21%) |
| Normal | 90,733 (76%) | 850 (33%) | 89,883 (77%) |
| Missing | 1,892 (2%) | 108 (4%) | 1,784 (2%) |
| Heart rate | |||
| Abnormal | 57,392 (48%) | 1,801 (69%) | 55,591 (48%) |
| Normal | 61,245 (52%) | 792 (30%) | 60,453 (52%) |
| Missing | 189 (0%) | 7 (0%) | 182 (0%) |
| Respiratory rate | |||
| Abnormal | 108,830 (92%) | 2,194 (84%) | 106,636 (92%) |
| Normal | 9,930 (8%) | 396 (15%) | 9,534 (8%) |
| Missing | 66 (0%) | 10 (0%) | 56 (0%) |
| Systolic blood pressure | |||
| Abnormal | 26,821 (23%) | 1,567 (60%) | 25,254 (22%) |
| Normal | 91,348 (77%) | 1,016 (39%) | 90,332 (78%) |
| Missing | 657 (1%) | 17 (1%) | 640 (1%) |
| Mentation | |||
| Abnormal | 7,351 (6%) | 934 (36%) | 6,417 (6%) |
| Normal | 25,437 (21%) | 132 (5%) | 25,305 (22%) |
| Missing | 86,038 (72%) | 1,534 (59%) | 84,504 (73%) |
| WBC count | |||
| Abnormal | 20,838 (18%) | 1,244 (48%) | 19,594 (17%) |
| Normal | 28,233 (24%) | 799 (31%) | 27,434 (24%) |
| Missing | 69,755 (59%) | 557 (21%) | 69,198 (60%) |
Data are presented as mean (± sd) for continuous variables or n (%) for categorical variables.
Inducing Missingness
We artificially induced missingness into both complete populations to create five datasets for evaluating imputation approaches with different proportions of missing data (10%, 30%, 50%, 70%, and 90% of subjects with missing values) under biased sampling schemes intended to mimic the missingness patterns observed in the original dataset. Specifically, we elected to simulate two simple missingness scenarios—one, where the probability for WBC presence depended on abnormal temperature, and one, where the probability of GCS presence depended on abnormal SBP. The “50% missing” datasets were the primary focus of our evaluation.
Under the first biased sampling scheme, a subject’s probability of selection was governed by the sampling weight assigned to their binary “abnormal temperature” status. This simulation mimics the missing mechanism observed in the complete dataset by featuring a weighted sampling scheme (without replacement) based on temperature—a variable distinguishing subjects with recorded/missing WBC in the complete dataset (Table 1). WBC missingness was artificially induced in the 48,485 patient episodes in which all four pSIRS components were recorded. For example, if sampling weight equaled 0.2 and missingness was 50%, given a fixed number of remaining observations, then a subject with a temperature in the normal range was four times more likely to be selected to have their WBC missing, than a subject with a temperature in the abnormal range. Weights ranged from 0.05 to 0.95 and the sampling schemes were repeated 150 times under each combination of missing proportion and sampling weight. We employed this approach for the model including WBC; that is, for the pSIRS model.
A second biased sampling scheme was conducted based on the differences in SBP of those subjects with missing or recorded GCS value (Table 1). This simulation features a weighted sampling scheme (without replacement) based on a subject’s SBP. Mentation missingness is artificially induced in the 32,293 patient episodes in which all three qSOFA components were recorded. Again, weights ranged from 0.05 to 0.95 and the sampling schemes were repeated 150 times under each combination of missing proportion and sampling weight. We employed this approach for the model including GCS; that is, for the qSOFA model.
Imputing Missing Data
Next, the following four estimation procedures, or imputation strategies, were applied to the incomplete datasets: 1) MICE, as implemented in the R package “mice” (16), with the five imputed datasets; 2) complete case analysis, in which all episodes with missing observations were discarded (“missing discarded”); 3) all missing observations were assumed to be in the normal range (“missing-as-normal”); and 4) all missing observations were assumed to be outside the normal range (“missing-as-abnormal”). We also investigated using the mode approach to imputation but recognized that a model taking binary predictors (normal or abnormal vital signs) suggests that replacing missing observations with the mode (or even median or mean) value for a given age group will likely be identical to option (3) “missing-as-normal.”
Comparing Imputation Strategies
We compared the four different imputation strategies by comparing the deviation of the imputation models’ coefficients with respect to the true coefficients of the complete population model. Given a sampling weight and missing proportion, the 150 independent estimates of each model’s coefficients were pooled together to obtain the percent bias, root mean square error (RMSE), and coverage rate. As per van Buuren (30), percent bias and RMSE were both calculated based on difference between the average estimate over 150 runs and the true coefficient. For percent bias, we divided the raw bias (difference between the average estimate over 150 runs and the true coefficient) by the true coefficient, taking the absolute value, then multiplying by 100. The RMSE was obtained by taking the square root of the mean of 150 squared differences between the estimates and true parameter value. Finally, the coverage rate is the proportion of CIs (out of 150) that contain the true coefficient value.
To assess the performance differences between groups, we first counted the times MICE resulted in a lower RMSE or percentage bias when compared with missing-as-normal and complete case analysis. Next, we used paired t tests, using a one-tailed test with lower as the alternative hypothesis, for each model’s coefficient for a given sampling weight and missingness, which were summarized as percentages of the total number of coefficients that had a statistically significant p value of less than 0.05.
Fréchet distances between RMSE curves were calculated separately for all pairwise combinations of missing proportions (e.g., comparing MICE at 10% vs MICE at 30%, MICE at 10% vs MICE at 50%, etc.) and summarized in a table as mean, sd, minimum and maximum values for each imputation approach and model coefficient; the Fréchet distance provides a measure of the resemblance between curves, which considers the location and ordering of points on the curves (31).
Finally, the performance of each imputation strategy was evaluated using AUC as a comparison: having induced missingness as described and imputed missing values using the appropriate sampling scheme for pSIRS and qSOFA, AUC values were computed for the logistic regression models built on each of the imputed population datasets, using five-fold cross-validation. The reported AUC values were obtained by taking the mean value of the AUCs computed from 150 runs. These AUCs were compared against the complete population dataset, using Welch’s paired t test, with the significance level set at p equal to 0.01 to correct for multiple comparisons across missingness levels of 10%, 20%, 50%, 70%, and 90%.
RESULTS
The study cohort included 118,826 episodes of ICU admission with sufficiently complete data for analysis; children had a mean age of 6.3 years (sd 5.90 yr) and a 2.19% mortality rate (Table 1); 8,677 (7.3%) of these children had had a previous ICU admission. There were 48,485 subjects for whom a pSIRS score could be calculated with complete data (mortality rate 4.08%), and 32,293 subjects for whom a qSOFA score could be calculated with complete data (mortality rate 3.15%).
Missingness Patterns for Abnormal Mentation (GCS) and Abnormal WBC
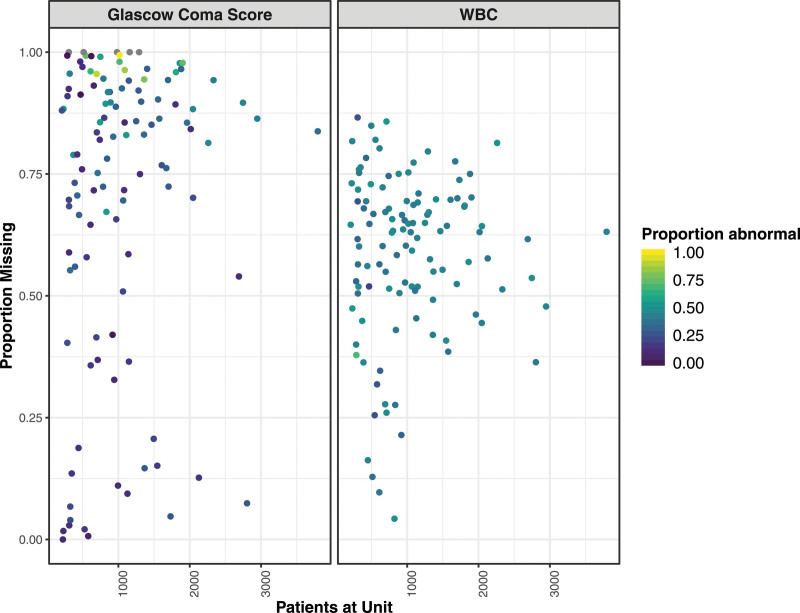
GCS was frequently not reported: in 90 of 117 (76.9%) PICUs, GCS was recorded in less than 50% of episodes, and in 37 of 117 (30.8%) PICUs, GCS was recorded in less than 10% of episodes. Similarly, the proportion of patient episodes without a WBC measurement varies among PICUs, although the range of reporting for WBC (sd = 17%) is narrower than for GCS (sd = 31%) (Fig. 1). There was no evidence of correlation between unit size and the proportions of abnormal GCS (Pearson r = 0.18) or abnormal WBC (r = 0.12); similarly, there was no correlation between unit size and the proportions of missing GCS (r = 0.11) or missing WBC (r = 0.03).
Figure 1.
Proportion of missing values for Glasgow Coma Score, used to determine abnormal mentation, and WBC count by unit size. The proportion of abnormal values for each unit is indicated using the color gradient from purple (all normal) to yellow (all abnormal).
In this dataset, 29% of children with a recorded WBC had an “abnormal temperature,” compared with just 17% of children that were missing a WBC measurement (odds ratio, 2.02; 95% CI, 1.97–2.08; p < 0.001) (Table 2). Similarly, 24% of children with a recorded GCS were listed as having an “abnormal SBP,” compared with just 19% of children with a missing GCS (odds ratio, 1.29; 95% CI, 1.25–1.33; p < 0.001).
Table 2.
Overview of Study Population Risk Factor Characteristics, Split by Missingness Patterns for Both Leukocytes (WBC Count) and Mentation
| Patient Characteristic | Total (n = 118,826) | Recorded WBC (n = 49,071) | Missing WBC (n = 69,755) | Recorded Mentation (n = 32,788) | Missing Mentation (n = 86,038) |
|---|---|---|---|---|---|
| Age (yr) | 6.3 (± 5.9) | 6.6 (± 6.0) | 6.2 (± 5.8) | 7.5 (± 6.1) | 5.9 (± 5.8) |
| Gender | |||||
| Female | 53,073 (45%) | 21,929 (45%) | 31,144 (45%) | 14,841 (45%) | 38,232 (44%) |
| Male | 65,742 (55%) | 27,132 (55%) | 38,610 (55%) | 17,945 (55%) | 47,797 (56%) |
| Mortality (%) | 2.2 (± 14.6) | 4.2 (± 20.0) | 0.8 (± 8.9) | 3.3 (± 17.7) | 1.8 (± 13.2) |
| Length of stay (d) | 4.2 (± 11.1) | 5.5 (± 12.9) | 3.3 (± 9.5) | 3.3 (± 8.2) | 4.5 (± 12.0) |
| Pediatric Index of Mortality 2 (5) probability of death | 2.4 (± 7.9) | 3.9 (± 11.3) | 1.4 (± 3.9) | 3.2 (± 11.6) | 2.1 (± 5.9) |
| Pediatric Risk of Mortality score 3 (6) probability of death | 2.1 (± 9.5) | 4.2 (± 13.9) | 0.7 (± 3.6) | 3.3 (± 13.8) | 1.7 (± 7.1) |
| Temperature | |||||
| Abnormal | 26,201 (22%) | 14,357 (29%) | 11,844 (17%) | 6,824 (21%) | 19,377 (23%) |
| Normal | 90,733 (76%) | 34,241 (70%) | 56,492 (81%) | 25,541 (78%) | 65,192 (76%) |
| Missing | 1,892 (2%) | 473 (1%) | 1,419 (2%) | 423 (1%) | 1,469 (2%) |
| Heart rate | |||||
| Abnormal | 57,392 (48%) | 24,537 (50%) | 32,855 (47%) | 15,207 (46%) | 42,185 (49%) |
| Normal | 61,245 (52%) | 24,485 (50%) | 36,760 (53%) | 17,561 (54%) | 43,684 (51%) |
| Missing | 189 (0%) | 49 (0%) | 140 (0%) | 20 (0%) | 169 (0%) |
| Respiratory rate | |||||
| Abnormal | 108,830 (92%) | 44,122 (90%) | 64,708 (93%) | 30,073 (92%) | 78,757 (92%) |
| Normal | 9,930 (8%) | 4,916 (10%) | 5,014 (7%) | 2,700 (8%) | 7,230 (8%) |
| Missing | 66 (0%) | 33 (0%) | 33 (0%) | 15 (0%) | 51 (0%) |
| Systolic blood pressure | |||||
| Abnormal | 26,821 (23%) | 14,254 (29%) | 12,567 (18%) | 6,379 (19%) | 20,442 (24%) |
| Normal | 91,348 (77%) | 34,717 (71%) | 56,631 (81%) | 26,330 (80%) | 65,018 (76%) |
| Missing | 657 (1%) | 100 (0%) | 557 (1%) | 79 (0%) | 578 (1%) |
| Mentation | |||||
| Abnormal | 7,351 (6%) | 4,328 (9%) | 3,023 (4%) | 7,351 (22%) | 0 (0%) |
| Normal | 25,437 (21%) | 10,153 (21%) | 15,284 (22%) | 25,437 (78%) | 0 (0%) |
| Missing | 86,038 (72%) | 34,590 (70%) | 51,448 (74%) | 0 (0%) | 86,038 (100%) |
| WBC count | |||||
| Abnormal | 20,838 (18%) | 20,838 (42%) | 0 (0%) | 6,477 (20%) | 14,361 (17%) |
| Normal | 28,233 (24%) | 28,233 (58%) | 0 (0%) | 8,004 (24%) | 20,229 (24%) |
| Missing | 69,755 (59%) | 0 (0%) | 69,755 (100%) | 18,307 (56%) | 51,448 (60%) |
Data are presented as mean (± sd) for continuous variables or n (%) for categorical variables.
Reference Models
The reference coefficients for the pSIRS model, with a moderate population-level AUC of 0.765, were: logit(pSIRS) = 4.107 + (–0.766, abnormal HR) + (–0.679, abnormal WBC) + (0.784, abnormal RR) + (–1.672, abnormal temperature). For the qSOFA model, with an acceptable population-level AUC of 0.880, the reference coefficients were: logit(pqSOFA) = 5.291 + (–3.052, abnormal mentation) + (0.503, abnormal RR) + (–1.621, abnormal SBP).
Effects of pSIRS Model Coefficients if WBC Is Missing in Half of the Episodes
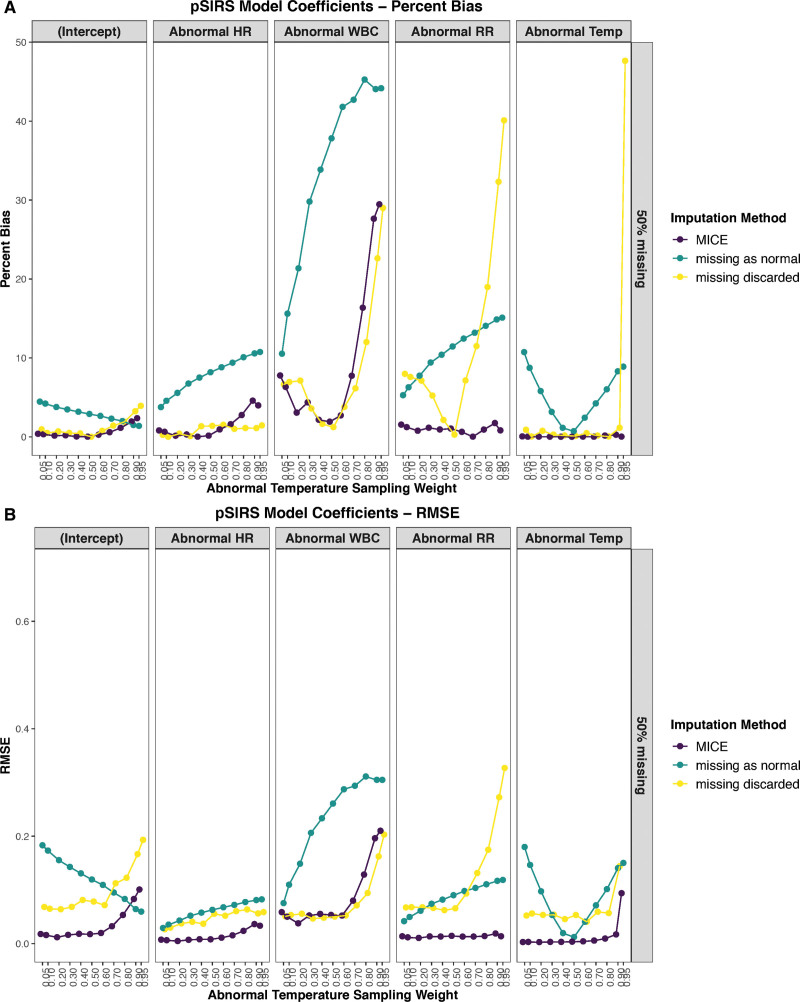
A naive implementation of MICE obtained a lower percent bias (Fig. 2A) in 53 of 55 (96%) simulated conditions and a lower RMSE (Fig. 2B) in 53 of 55 (96%) simulated conditions than assuming missing-as-normal. The MICE algorithm was also preferable to discarding all episodes with missing observations for complete case analysis, as observed by the lower percent bias (Fig. 2A) in 37 of 55 (67%) simulated conditions, and a lower RMSE (Fig. 2B) in 47 of 55 (85%) simulated conditions. Specifically, the RMSE was better for all coefficients, except for abnormal WBC in the missing-as-normal method, and all but the intercept in the complete case analysis method.
Figure 2.
Effect of varying degrees of abnormal temperature (Temp) sample weighting on the coefficients of the pediatric systemic inflammatory response syndrome (pSIRS) model, when 50% of WBC count values are missing for three approaches: multiple imputation by chained equations (MICE), missing as normal, and complete case analysis (missing discarded). A, The performance using percentage bias of the coefficients, while B shows root mean square error (RMSE) for the coefficients. The model coefficients included the intercept, as well as abnormal heart rate (HR), WBC, respiratory rate (RR), and Temp.
Effects of qSOFA Model Coefficients if Mentation Is Missing in Half of the Episodes
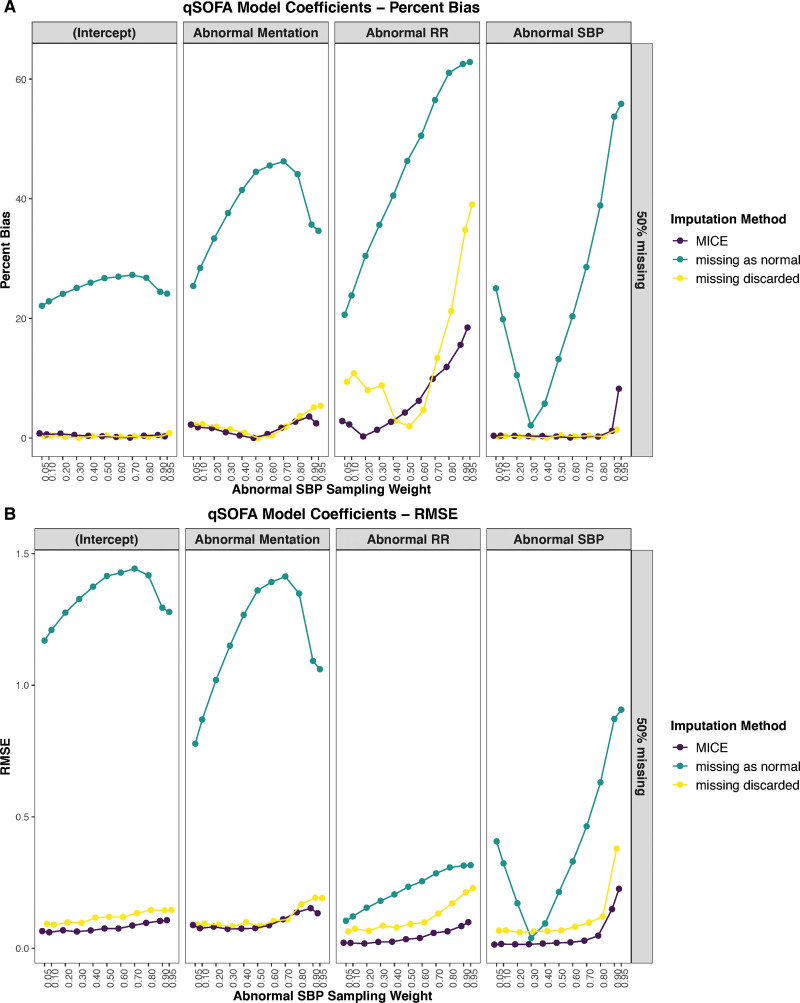
MICE resulted in the lowest RMSE (Fig. 3B) in 43 of 44 (98%) simulated conditions when estimating all four of the model’s coefficients and had a comparable percent-bias (Fig. 3A) to that observed while discarding episodes with missing observations for complete case analysis, in which it was better in 28 of 44 (64%) simulated conditions. The missing-as-normal assumption was inferior irrespective of the metric considered (Fig. 3).
Figure 3.
Effect of varying degrees of abnormal systolic blood pressure (SBP) sample weighting on the coefficients of the quick Sequential [Sepsis-Related] Organ Failure Assessment (qSOFA) model, when 50% of mentation values are missing for three approaches: multiple imputation by chained equations (MICE), missing as normal, and complete case analysis (missing discarded). A, The performance using percentage bias of the coefficients, while B shows root mean square error (RMSE) for the coefficients. The model coefficients included the intercept, as well as abnormal mentation, respiratory rate (RR), and SBP.
Robustness to Degree of Missingness
Compared with complete case analysis, MICE had a statistically significantly difference in RMSE for the pSIRS model coefficients in 43 of 55 (78%) cases and 46 of 55 (84%) for the qSOFA model coefficients. When comparing MICE to missing-as-normal, these values were 52 of 55 (95%) and 31 of 55 (56%), respectively. This comparison of RMSE values is illustrated in a heatmap of p values in Supplementary Figs. A–D (http://links.lww.com/PCC/B899).
Qualitatively, roughly the same patterns emerge irrespective of the proportion of missingness induced and also for both the pSIRS and the qSOFA coefficients (Supplementary Fig. E, http://links.lww.com/PCC/B899). When quantified using Fréchet distances between RMSE curves, MICE proved to be the most robust to the degree of missingness, with the smallest average distance between RMSE curves when examining all four qSOFA coefficients, and four of five pSIRS coefficients (Table 3).
Table 3.
Fréchet Distance Between Root Mean Square Error Curves, Grouped by Model (Pediatric Systemic Inflammatory Response Syndrome or Quick Sequential [Sepsis-Related] Organ Failure Assessment) and Split by Imputation Approach
| Model Coefficient | Multiple Imputation by Chained Equations Curves | Complete Case Analysis Curves | Missing-As-Normal Curves | Missing As Abnormal Curves |
|---|---|---|---|---|
| Pediatric systemic inflammatory response syndrome coefficients | ||||
| (Intercept) | 0.09 (0.04) (0.05–0.15) | 3.33 (4.12) (0.07–8.15) | 0.11 (0.05) (0.05–0.2) | 0.16 (0.07) (0.09–0.3) |
| Abnormal WBC | 0.18 (0.07) (0.10–0.30) | 0.15 (0.07) (0.06–0.29) | 0.19 (0.08) (0.07–0.34) | 0.06 (0.02) (0.02–0.1) |
| Abnormal heart rate | 0.03 (0.01) (0.02–0.05) | 0.11 (0.07) (0.02–0.21) | 0.05 (0.02) (0.02–0.07) | 0.11 (0.04) (0.04–0.17) |
| Abnormal RR | 0.03 (0.01) (0.01–0.05) | 0.29 (0.15) (0.10–0.58) | 0.06 (0.03) (0.02–0.10) | 0.12 (0.05) (0.06–0.21) |
| Abnormal temperature | 0.11 (0.04) (0.01–0.14) | 6.87 (3.46) (0.05–9.69) | 0.12 (0.05) (0.04–0.21) | 0.04 (0.02) (0.02–0.07) |
| Quick Sequential (Sepsis-Related) Organ Failure Assessment coefficients | ||||
| (Intercept) | 0.16 (0.10) (0.04–0.32) | 3.07 (3.81) (0.05–7.57) | 0.73 (0.39) (0.32–1.48) | 0.41 (0.22) (0.13–0.86) |
| Abnormal mentation | 0.17 (0.10) (0.05–0.35) | 0.18 (0.10) (0.07–0.35) | 0.61 (0.28) (0.26–1.17) | 0.15 (0.07) (0.05–0.26) |
| Abnormal systolic blood pressure | 0.34 (0.18) (0.06–0.58) | 3.15 (3.76) (0.11–7.55) | 0.44 (0.16) (0.18–0.66) | 0.44 (0.18) (0.18–0.82) |
| Abnormal RR | 0.09 (0.04) (0.04–0.15) | 0.62 (0.61) (0.08–1.44) | 0.17 (0.07) (0.07–0.30) | 0.79 (0.39) (0.34–1.54) |
RR = respiratory rate. Fréchet distances are presented as mean (sd) (range). The lowest average value in a given row is in boldface font.
As expected, the approach of discarding episodes with missing observations appears to suffer with increasing amounts of missing data. Nevertheless, even with 90% missingness, the performance of the MICE coefficients is likely acceptable for most uses: the coverage rate, whereby the 95% CI of the imputed coefficients included the true value, exceeds 0.95 in 46 of 55 (83.6%) pSIRS coefficient and sampling weight combinations, and 31 of 44 (70.5%) qSOFA coefficient and sampling weight combinations.
Also, see Supplementary Figure E (http://links.lww.com/PCC/B899) for the RMSE values.
Missing As Abnormal
For reference, assuming missing-as-abnormal when estimating all of the model’s coefficients performed much worse in both RMSE (MICE was better in 95% of simulations, missing-as-normal in 82% of simulations, and complete case analysis in 91% of simulations) and percent-bias dimensions (MICE was better in 97% of simulations, missing-as-normal in 79% of simulations, and complete case analysis in 96% of simulations). It was so much worse that we did not include it in any of the plots, as it exceeded the y-axis ranges with which the current data are presented.
Mode Imputation of Missing Values
In our models, the most frequent value was identical to the normal value for each predictor. Thus, the results from mode imputation were identical to the “missing as normal” imputation.
Performance of pSIRS and qSOFA Models in Terms of AUC
For the pSIRS models, in which data had been imputed using MICE, AUCs were in the range 0.764–0.772; these AUCs were actually higher than the population-level AUC of 0.765 for 10% (p < 0.001) and 30% missingness (p < 0.001) and were not different from it for 50%, 70%, and 90% missingness (p > 0.014). Using missing-discarded imputation, AUCs for the pSIRS models were in the range 0.602–0.766; these values were not different to the population-level AUC for 10%, 30%, and 50% missingness (p > 0.040), but performance then deteriorated with a significantly different AUC at 70% (p = 0.004) and 90% missingness (p < 0.001). Using missing-as-normal imputation, AUCs were in the range 0.753–0.765, which were significantly lower than the population-level AUC at all levels of missingness (p < 0.001) (Supplementary Fig. F, http://links.lww.com/PCC/B899).
For the qSOFA models, in which data had been imputed using MICE, AUCs were in the range 0.842–0.884; again, these AUCs were actually higher than the population-level AUC of 0.880 at 10% (p < 0.001) and 30% missingness and not different from it at higher levels of missingness (p > 0.070). Using missing-discarded imputation, AUCs for the qSOFA models were in the range 0.780–0.885; performance deteriorated to a significant degree only at 90% missingness (p = 0.007). Using missing-as-normal imputation, AUCs were in the range 0.734-0.863, which were significantly lower than the population-level AUC at all levels of missingness (p < 0.001) (Supplementary Fig. F, http://links.lww.com/PCC/B899).
DISCUSSION
Analyses of large observational studies will encounter the issue of missing data, which are likely not missing at random. Rooted in convenience, traditional approaches have been either to discard cases with missing observations or to assume that values are in the normal range (20, 23). Increasingly reports of predictive algorithms, big data approaches to medicine, and secondary use of legacy databases have necessarily raised the need to understand data imputation strategies to manage missing data; this is echoed in recent guidance on development and reporting of prediction models for the critical care setting (25).
Key Findings
Simulations performed in this study demonstrate that a simple implementation of MICE resulted in a lower percent bias (in 97% of simulations) and RMSE (in 97% of simulations) than assuming missing-as-normal. In both simulated scenarios—applying pSIRS or qSOFA models—MICE provided the best performance estimates for all model coefficients. When compared with complete case analysis (i.e., discarding cases with missing observations), MICE also resulted in lower RMSE (in 91% of all simulations) and similar, or better, percent-bias (in 66% of all simulations) for all model coefficients. The AUC results for the pSIRS and qSOFA models also suggested that MICE can provide a robust imputation strategy, even for datasets with large proportions of missing data. Missing-as-abnormal always performed worse than any other approach.
MICE was found to be surprisingly robust to large amounts of missing data as similar patterns emerge irrespective of the proportion of missingness induced. This contrasts with the expected degradation in performance that occurs with increasing amounts of missing data when using the method of discarding cases with missing observations. Hence, researchers should consider MICE approaches when encountering missing data in their work—albeit, for small amounts of missingness, doing complete case analysis may also be a reasonable approach, although the threshold for this has not been explicitly evaluated in this study. Caution is advised in adopting a mode, missing-as-normal or missing-as-abnormal assumption, as these were found to be inferior approaches in this study.
Clinical Implications
Given that in most current and past studies, missing data were either discarded or replaced with normal values, the coefficients obtained in such prediction models may be suboptimal; that is, further from the true, as yet unknown, underlying values. This is particularly concerning for studies that reported only small subsets of large datasets due to large overall missingness when combining many different risk factors with smaller degrees of missingness. Many of the risk prediction studies in critical care have used this strategy, which may limit their generalizability (4, 32–34). Furthermore, analyses performed on data from an adult trauma registry also support the case for using multiple imputation: one study found that handling of missing data significantly influenced the assessment of trauma care performance as expressed by the number of unexpected deaths (35); another performed an external validation of a risk model for nosocomial pneumonia after coronary artery bypass graft surgery and found that both missing-as-low-risk and complete case analyses showed unacceptable calibration as per the Hosmer-Lemeshow test, while maintaining reasonable discrimination (36).
Additionally, missingness patterns themselves can be informative; for example, the presence or absence of laboratory values was a potential predictor of inhospital and 30-day mortality in a large ICU database (Medical Information Mart for Intensive Care III database) (37, 38); similarly, an analysis of electronic health record data found that both the presence and timing of a laboratory test order were significantly associated with 3-year survival in outpatients (39). Yet, this approach may limit the transferability of models between different health systems, as seen in the results of a recent sepsis prediction competition (40). This caution may be particularly applicable to machine learning models which, if trained and validated on local clinical data, may not always be generalizable to other clinical contexts (41). Missingness in time-series data remains a problem that may be addressed with forward filling, recurring neural networks (42), or fuzzy-based identification of recoverable or nonrecoverable data (43), to which other approaches are then applied. Identifying the optimal approach to resolve these challenges is an area for future work.
Practical Suggestions for Future Studies
Software development has made it reasonably straightforward to implement the MICE approach; however, like any other complex modeling exercise, it requires careful consideration at each stage of development. The approaches required for imputing values in different data types (normally distributed or skewed continuous data, binary or categorical variables), how to select the variables in the imputation model, and how to decide the appropriate number of imputations all need to be considered (15). The aim is to strike a balance between a sufficiently rich imputation scheme and an unstable model.
Considering the concerns raised by our results, it may be prudent to apply a MICE approach to reexamine the results and conclusions of high-impact studies, which have been used to implement risk stratification tools that are now in clinical use. Similar concerns with imputation methodology, expressed through letters to the editor, prompted the cardiovascular disease risk score (QRISK) (44) to be replaced by a second version (45). Ideally, reanalysis would be performed externally, on a second dataset, but it might be more feasible to call upon the original authors to do so.
Development of any prediction model for the critical care domain should include consideration of how to manage missing values in the data used to train and test the model and provide transparent reporting of missing data. A range of approaches to evaluating model performance should be considered before recommending a specific model for clinical use, although as this was not our aim, it was not a primary concern in the present study. Further guidance on these issues is available (25).
MICE cannot be adopted as a universal solution to data missingness, as its uncritical use can still lead to biased results if data are not missing at random (15, 18). Care should be taken to consider potential causes of data missingness on a variable-by-variable basis and whether data are likely to be missing systematically (e.g., it may be known that a missing clinical measurement denotes a healthy patient), which may guide a specific imputation strategy. In the absence of evidence supporting specific assumptions about data missingness patterns, multiple imputation may offer a solution that is relatively robust to misinterpretation of the data missingness mechanism. In addition to the pattern of missingness, the characteristics of the primary data, including possible correlations based on physiologic relationships between variables, may also help inform imputation strategies. For example, forward filling would not be appropriate (or even possible) if patients are only sampled once; for example, in cases in which their worst value is reported for a given variable.
Limitations
The main limitation of the current study is our choice of simulation strategy, particularly the introduction of missingness based on only a single variable. Also, for “knowing” the true underlying values, our simulation population was necessarily one of complete cases, and hence the reference coefficients, against which we benchmarked, may not be representative of the larger North American cohort from which the data were obtained. This approach has the advantage of using recorded true values for outcomes, instead of performing imputation to create reference values, and this may increase the realism of the underlying ground truth.
We also believe that applying a simple missingness pattern, instead of a complex multivariable one, allows for easier interpretation of the results. It is left for future work to demonstrate the same superiority of MICE with more complex missingness mechanisms and patterns; for example, one could introduce missingness in related variables of pH, bicarbonate and arterial carbon dioxide concentration, or explore missingness in related variables, such as oxygen saturation, where the missingness depended on both high HR and low blood pressure, as a surrogate of poor perfusion. Future work should also consider feature engineering approaches, such as adding new features to denote missing data in existing variables when the missingness mechanism can be reliably established. These approaches are not easily generalizable, but missingness may be informative in a given context or institution, as it reflects practice patterns.
We used dichotomous predictors rather than continuous (age-adjusted) variables. This approach can be criticized for causing information loss and depends on the threshold selection for model performance; however, it is much simpler and is commonly encountered in other settings—especially when using manually calculated, rather than computer-derived, risk scores, such as Weiss et al (9) have described. Similarly, we selected a binary outcome (mortality) with low prevalence; continuous outcomes, such as predicting PICU length of stay, may result in better models, but with a less relevant and more difficult to ascertain outcome.
This study was not designed to define or identify a threshold of acceptable missingness and any such limit we might determine from these data would not be generalizable to other datasets. Finally, we have not presented results for last observation carried forward (also known as sample-and-hold or forward filling) (46). Although mode imputation was found to be equivalent to the “missing as normal” approach in this study, these results cannot be extrapolated to scenarios with continuous predictors, and thus mean, median or mode approaches need to be considered in future comparisons of imputation strategies with missing data. That said, these approaches have also been evaluated elsewhere and are generally discouraged (47, 48).
CONCLUSIONS
Analyses of large observational studies will typically encounter the issue of missing data, which are likely not missing at random. Simulations performed in this study demonstrate that MICE is an effective strategy when nonmissing variables in a dataset are predictive of the missing variables and that even a naive implementation of MICE significantly outperforms approaches, such as discarding cases with missing data or assuming missing values are in the normal range. Researchers should consider MICE (or similar approaches) when encountering even small proportions of missing data in their work.
AT THE BEDSIDE.
MICE is a robust approach, which researchers should consider when encountering even small degrees of missingness in large clinical datasets; statistical guidance may be required to implement MICE appropriately.
Suboptimal strategies for dealing with missing data, including complete case analysis in some situations, can lead to errors in model coefficients, which may contribute to misinterpretation of results and invalidate their generalizability.
The concerns identified by our results suggest it may be prudent to apply a MICE approach to reexamine the conclusions of high-impact studies that have been used to support risk stratification tools currently in clinical use.
ACKNOWLEDGMENTS
We wish to thank the Virtual Pediatric Systems team for their support in obtaining the data. Data were provided by Virtual Pediatric Systems, LLC. No endorsement or editorial restriction of the interpretation of these data or opinions of the authors has been implied or stated.
Supplementary Material
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/pccmjournal).
This work was supported, in part, by a 2020 BC Children’s Hospital Research Institute External Salary Recognition Award (to Dr. Görges).
Dr. Steif received funding from the University of British Columbia (UBC). Dr. Brant received support for article research from the British Columbia Children’s Hospital Research Institute. Dr. Sreepada’s institution received funding from Mitacs Postdoctoral Fellowship. Dr. Murthy holds a Research Chair from Health Research Foundation and Innovative Medicines Canada. Dr. Görges’ institution received funding from the Natural Sciences and Engineering Research Council of Canada, the Canadian Institutes of Health Research, the Juvenile Diabetes Research Foundation, and the British Columbia Children’s Hospital Research Institute; he received funding from the Michael Smith Foundation for Health Research; he disclosed he is party to a licensing agreement between UBC and NeuroWave Systems for unrelated work, that he was a director for the Society for Technology in Anesthesia, and a principal investigator and coinvestigator for Canada’s Digital Technology Supercluster projects. Mr. West has disclosed that he does not have any potential conflicts of interest.
REFERENCES
- 1.Mayhew MB, Petersen BK, Sales AP, et al. Flexible, cluster-based analysis of the electronic medical record of sepsis with composite mixture models. J Biomed Inform. 2018; 78:33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balamuth F, Weiss SL, Neuman MI, et al. Pediatric severe sepsis in U.S. children’s hospitals. Pediatr Crit Care Med. 2014; 15:798–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters C, Murthy S, Brant R, et al. Mortality risk using a pediatric quick sequential (sepsis-related) organ failure assessment varies with vital sign thresholds. Pediatr Crit Care Med. 2018; 19:e394–e402 [DOI] [PubMed] [Google Scholar]
- 4.Görges M, Peters C, Murthy S, et al. External validation of the “quick” pediatric logistic organ dysfunction-2 score using a large North American cohort of critically ill children with suspected infection. Pediatr Crit Care Med. 2018; 19:1114–1119 [DOI] [PubMed] [Google Scholar]
- 5.Slater A, Shann F, Pearson G; Paediatric Index of Mortality (PIM) Study Group. PIM2: A revised version of the Paediatric Index of Mortality. Intensive Care Med. 2003; 29:278–285 [DOI] [PubMed] [Google Scholar]
- 6.Pollack MM, Patel KM, Ruttimann UE. PRISM III: An updated pediatric risk of mortality score. Crit Care Med. 1996; 24:743–752 [DOI] [PubMed] [Google Scholar]
- 7.Goldstein B, Giroir B, Randolph A; International Consensus Conference on Pediatric Sepsis. International pediatric sepsis consensus conference: Definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005; 6:2–8 [DOI] [PubMed] [Google Scholar]
- 8.Ho LV, Ledbetter D, Aczon M, et al. The dependence of machine learning on electronic medical record quality. AMIA Annu Symp Proc. 2017; 2017:883–891 [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss SL, Balamuth F, Chilutti M, et al. Identification of pediatric sepsis for epidemiologic surveillance using electronic clinical data. Pediatr Crit Care Med. 2020; 21:113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendelsohn AB, Dreyer NA, Mattox PW, et al. Characterization of missing data in clinical registry studies. Ther Innov Regul Sci. 2015; 49:146–154 [DOI] [PubMed] [Google Scholar]
- 11.Kenward MG, Carpenter J. Multiple imputation: Current perspectives. Stat Methods Med Res. 2007; 16:199–218 [DOI] [PubMed] [Google Scholar]
- 12.Barnard J, Meng XL. Applications of multiple imputation in medical studies: From AIDS to NHANES. Stat Methods Med Res. 1999; 8:17–36 [DOI] [PubMed] [Google Scholar]
- 13.Moons KG, Donders RA, Stijnen T, et al. Using the outcome for imputation of missing predictor values was preferred. J Clin Epidemiol. 2006; 59:1092–1101 [DOI] [PubMed] [Google Scholar]
- 14.Pedersen AB, Mikkelsen EM, Cronin-Fenton D, et al. Missing data and multiple imputation in clinical epidemiological research. Clin Epidemiol. 2017; 9:157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White IR, Royston P, Wood AM. Multiple imputation using chained equations: Issues and guidance for practice. Stat Med. 2011; 30:377–399 [DOI] [PubMed] [Google Scholar]
- 16.van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate imputation by chained equations in R. J Stat Softw. 2011; 45:1–67 [Google Scholar]
- 17.Yucel RM. State of the multiple imputation software. J Stat Softw. 2011; 45:v45/i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayati Rezvan P, Lee KJ, Simpson JA. The rise of multiple imputation: A review of the reporting and implementation of the method in medical research. BMC Med Res Methodol. 2015; 15:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood AM, White IR, Thompson SG. Are missing outcome data adequately handled? A review of published randomized controlled trials in major medical journals. Clin Trials. 2004; 1:368–376 [DOI] [PubMed] [Google Scholar]
- 20.Bell ML, Fiero M, Horton NJ, et al. Handling missing data in RCTs; a review of the top medical journals. BMC Med Res Methodol. 2014; 14:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan TR, Yelland LN, Lee KJ, et al. Treatment of missing data in follow-up studies of randomised controlled trials: A systematic review of the literature. Clin Trials. 2017; 14:387–395 [DOI] [PubMed] [Google Scholar]
- 22.Rombach I, Gray AM, Jenkinson C, et al. Multiple imputation for patient reported outcome measures in randomised controlled trials: Advantages and disadvantages of imputing at the item, subscale or composite score level. BMC Med Res Methodol. 2018; 18:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Flórez ID, Colunga Lozano LE, et al. A systematic survey on reporting and methods for handling missing participant data for continuous outcomes in randomized controlled trials. J Clin Epidemiol. 2017; 88:57–66 [DOI] [PubMed] [Google Scholar]
- 24.Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. Ann Intern Med. 2015; 162:55–63 [DOI] [PubMed] [Google Scholar]
- 25.Leisman DE, Harhay MO, Lederer DJ, et al. Development and reporting of prediction models: Guidance for authors from editors of respiratory, sleep, and critical care journals. Crit Care Med. 2020; 48:623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Virtual Pediatric Systems LLC. VPS PICU Registry. 2017. Available at: http://www.myvps.org/vps-picu. Accessed September 4, 2020
- 27.Gebara BM. Values for systolic blood pressure. Pediatr Crit Care Med. 2005; 6:500. [DOI] [PubMed] [Google Scholar]
- 28.Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. 2016; 315:801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Team. R: A Language and Environment for Statistical Computing. 2004. Available at: https://www.r-project.org. Accessed September 4, 2020
- 30.van Buuren S. Flexible Imputation of Missing Data. Second Edition. Boca Raton, FL, CRC Press, 2018 [Google Scholar]
- 31.Alt H, Godau M. Computing the Fréchet distance between two polygonal curves. Int J Comput Geom Appl. 1995; 05:75–91 [Google Scholar]
- 32.Mudri M, Williams A, Priestap F, et al. Comparison of drugs used for intubation of pediatric trauma patients. J Pediatr Surg. 2020; 55:926–929 [DOI] [PubMed] [Google Scholar]
- 33.Xiao C, Wang S, Fang F, et al. Epidemiology of pediatric severe sepsis in main PICU centers in Southwest China. Pediatr Crit Care Med. 2019; 20:1118–1125 [DOI] [PubMed] [Google Scholar]
- 34.Hooli S, Colbourn T, Lufesi N, et al. Predicting hospitalised paediatric pneumonia mortality risk: An external validation of RISC and mRISC, and local tool development (RISC-Malawi) from Malawi. PLoS One. 2016; 11:e0168126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Reilly GM, Jolley DJ, Cameron PA, et al. Missing in action: A case study of the application of methods for dealing with missing data to trauma system benchmarking. Acad Emerg Med. 2010; 17:1122–1129 [DOI] [PubMed] [Google Scholar]
- 36.Sanagou M, Wolfe R, Leder K, et al. External validation and updating of a prediction model for nosocomial pneumonia after coronary artery bypass graft surgery. Epidemiol Infect. 2014; 142:540–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharafoddini A, Dubin JA, Maslove DM, et al. A new insight into missing data in intensive care unit patient profiles: Observational study. JMIR Med Inform. 2019; 7:e11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber GM, Adams WG, Bernstam EV, et al. Biases introduced by filtering electronic health records for patients with “complete data.” J Am Med Inform Assoc. 2017; 24:1134–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agniel D, Kohane IS, Weber GM. Biases in electronic health record data due to processes within the healthcare system: Retrospective observational study. BMJ. 2018; 361:k1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyna MA, Josef CS, Jeter R, et al. Early prediction of sepsis from clinical data: The PhysioNet/Computing in Cardiology Challenge 2019. Crit Care Med. 2020; 48:210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Futoma J, Simons M, Panch T, et al. The myth of generalisability in clinical research and machine learning in health care. Lancet Digit Health. 2020; 2:e489–e492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipton ZC, Kale DC, Wetzel R. Modeling missing data in clinical time series with RNNs. Proceedings of Machine Learning for Healthcare 2016. Los Angeles, CA, August 19-20, 2016, pp 253–270 [Google Scholar]
- 43.Cismondi F, Fialho AS, Vieira SM, et al. Missing data in medical databases: Impute, delete or classify? Artif Intell Med. 2013; 58:63–72 [DOI] [PubMed] [Google Scholar]
- 44.Hippisley-Cox J, Coupland C, Vinogradova Y, et al. Derivation and validation of QRISK, a new cardiovascular disease risk score for the United Kingdom: Prospective open cohort study. BMJ. 2007; 335:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hippisley-Cox J, Coupland C, Vinogradova Y, et al. Performance of the QRISK cardiovascular risk prediction algorithm in an independent UK sample of patients from general practice: A validation study. Heart. 2008; 94:34–39 [DOI] [PubMed] [Google Scholar]
- 46.Haukoos JS, Newgard CD. Advanced statistics: Missing data in clinical research–part 1: An introduction and conceptual framework. Acad Emerg Med. 2007; 14:662–668 [DOI] [PubMed] [Google Scholar]
- 47.Li P, Stuart EA, Allison DB. Multiple imputation: A flexible tool for handling missing data’. In: JAMA Guide to Statistics and Method. Livingston EH, Lewis RJ. (Eds). New York, NY, McGraw-Hill Education, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desai M, Montez-Rath ME, Kapphahn K, et al. Missing data strategies for time-varying confounders in comparative effectiveness studies of non-missing time-varying exposures and right-censored outcomes. Stat Med. 2019; 38:3204–3220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.