Abstract
Microfluidic devices have been widely applied to trapping and isolation of cells and clusters for controllable intercellular environments and high-throughput analysis, triggering numerous advances in disease diagnosis and single-cell analysis. Passive hydrodynamic cell trapping is one of the simple and effective methods that has been gaining attention in recent years. Our aim here is to review the existing passive microfluidic trapping approaches, including microposts, microfiltration, microwells, and trapping chambers, with emphasis on design principles and performance. We summarize the remarkable advances that hydrodynamic trapping methods offer, as well as the existing challenges and prospects for development. Finally, we hope that an improved understanding of hydrodynamic trapping approaches can lead to sophisticated and useful platforms to advance medical and biological research.
I. INTRODUCTION
Microfluidic isolation and trapping of single-cells and cell clusters shows great potential in advancing medical and biological research.1–3 This is in part due to enabling investigations in cell-to-cell interactions and signaling,4,5 studies of cell heterogeneity in tumors,6 development of immunotherapies,7 investigations of drug responses,8,9 and regulation of stem cell differentiation.10 In conventional cell studies, cells are studied in larger populations in which measurements can only reflect average values summed over multiple cell responses.11 This approach, however, can lead to misinterpretations since it obscures important information about the cells and their statistical nature.11 In addition, cell growth is subject to multiple spatial and temporal cues including cytokine and protein gradients and cell–matrix or cell–cell interactions;12 therefore, better control over the cell environment is desirable when conducting in vitro experiments to achieve long cell viability.
Microfluidic trapping devices immobilize single cells and cell aggregates in a controllable and independent environment, which offers a way to conduct analysis at a single-cell level.11 These microfluidic trapping devices allow integration of cell culture with analytical devices13 as well as the ability to probe biochemical processes that govern cell behavior,14 which permits genetic,15,16 physiological,17,18 and biochemical cell studies19,20 on a single-cell to organ scale. Microfluidic trapping devices also offer a potential for 3D cell culture to better mimic the in vivo microenvironment, allowing researchers to study physical effects on cell function and behavior3,21 and with the scale-down of research protocols, results can be obtained in only a few seconds instead of hours or days.22
Microfluidic strategies for trapping cells can be generally categorized as active contactless cell trapping and contact cell immobilization by surface treatment or hydrodynamics.2,11 The former comprises optical,23 dielectrophoretic,24 acoustic,25 and magnetic26 trapping approaches, while the latter consists of chemically driven cell attachment to surfaces and passive hydrodynamic trapping.11 The approaches such as contact-free active immobilization and affinity-based cell attachment provide high purity,27 accuracy,11 and the potential of high-throughput,28 but are complex and costly. In addition, for tumor cells with down-regulated or lost epithelial markers, such as EpCAM, affinity-based cell attachment methods lose efficacy.29 Therefore, passive hydrodynamic trapping methods have been promoted in recent years.
Micropost arrays, microfiltration, microwell, and chamber trapping are the four main categories of hydrodynamic trapping. Compared with active microfluidic methods, hydrodynamic strategies are much simpler and offer higher throughput and lower cost.30 These passive techniques rely on inherent hydrodynamic forces and channel geometries and other physical obstacles in the channels.31 These strategies may also allow for high cell viability, mature chip fabrication crafts, easy operation, and integration with other downstream analysis. However, deficiencies in the accuracy and flexibility still remain.30 Less complexity in operation and fabrication process need to be the tendency during development.32
Publications on passive hydrodynamic cell trapping have increased from 2 to around 80 papers per year in the past two decades (Fig. 1), suggesting the burgeoning research field and market prospects. A number of review articles33–35 on cell trapping have already been published covering both passive and active approaches, but not specifically focusing on hydrodynamic trapping. Yet, a growing number of trapping devices rely on or are combined with passive hydrodynamic methods; thus, an in-depth review on hydrodynamic cell trapping is necessary for future chip design considerations and cell analysis applications. Narayanamurthy et al.32 presented an extensive review of hydrodynamic trapping of single cells in 2017, where they summarized major works from 2005 to 2016, mainly focusing on single-cell analysis rather than other cellular applications such as cell spheroid formation, co-culturing, and/or real-time monitoring. Other reviews on cell trapping were published some years ago, namely, Kim et al.35 in 2008, Nilsson et al.33 in 2009, and Wu et al.34 in 2017. Since then, many modified designs of trapping structures have been developed and introduced, motivating the need for a new comprehensive review of hydrodynamic trapping. Other reviews also discuss notable works on particle separation and entrapment with microfluidics partly including some hydrodynamic methods.36–38 To add to these discussions, we focus our review on passive trapping techniques.
FIG. 1.
Search the paper topic about “passive*” or “hydrodynamic*” and “microfluidic*” and “capture*” or “trap*” in databases of Web of Science Core Collection, KCI, MEDLINE, Russian Science Citation Index, and SciELO.
Herein, we review microfluidic cell trapping approaches involving only passive forces and physical obstacles including microposts, microfiltration, microwells, and trapping chambers (Fig. 2). We also further categorize trapping techniques to better illustrate the differences of structure design and how hydrodynamics is modified to enhance trapping efficiency. A growing number of trapping devices rely on or are combined with passive hydrodynamic methods; thus, an in-depth assessment on current hydrodynamic cell trapping devices will aid future chip design considerations and cell analysis applications. This review will describe the principle, structure, and performance of each method and summarize the key factors influencing trapping efficiency.
FIG. 2.
Schematics of hydrodynamic cell trapping methods.
II. MICROPOST ARRAY TRAPPING
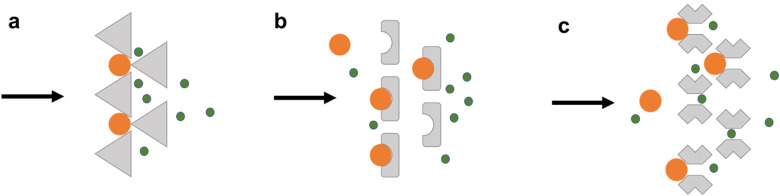
Micropost array-based trapping is a horizontal trapping method32 that can be achieved by simply blocking cells larger than the gap between adjacent posts from flowing through the channel (Fig. 3). In this approach, which was first reported by Lee et al. in 2006,39 micropost spacing must be adapted to the size of the cells to be captured.40 Typically, designing the gap size as 20%–25% of the target cell or cluster diameter is used to immobilize cells even with deformation, while avoiding clogging.41 Shape, dimensions, and layout modifications of microposts have been reported to improve capture efficiency, reduce clogging, and reduce shear stress.42 Polygonal, U-shaped, and other irregularly shaped microposts have all been reported (Fig. 3). From single cells to clusters, from whole blood samples to lysed samples, these all can be captured by microposts of certain shape and dimensions.
FIG. 3.
Schematics of micropost arrays based on (a) polygonal, (b) U-shaped, and (c) butterfly-shaped micropost geometries. Target cells are immobilized in trapping structures, while nontarget cells pass through.
A. Polygonal micropost array
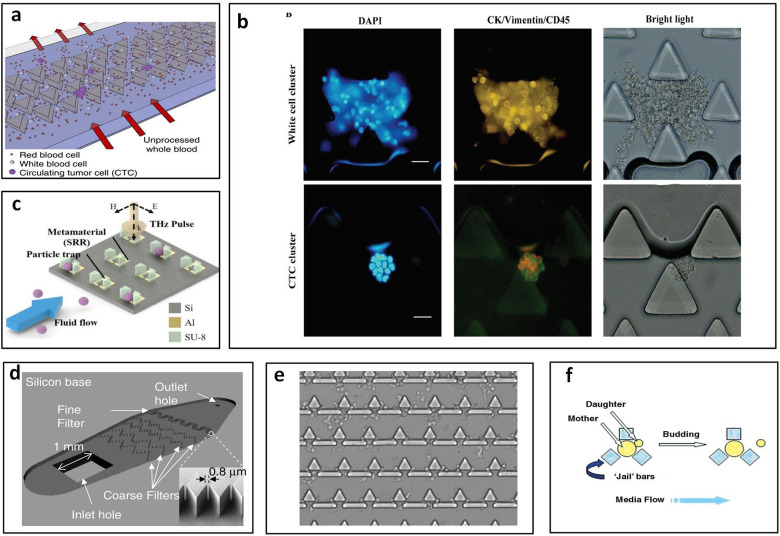
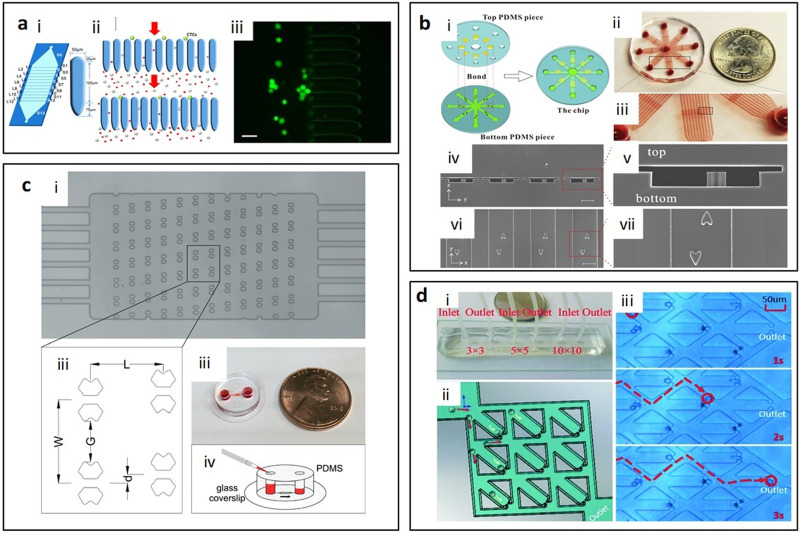
Polygonal micropost arrays, including triangular and diamond shaped microposts, are generally combined in two or more posts as one trap unit. Triangular post arrays usually are used to trap cell clusters, since there is strong cell–cell adhesion in clusters they can be held at vertex angles of triangular posts.43 Sarioglu et al.43 reported a triangular post array for label-free capture circulating tumor cell clusters from whole blood with 99% capture efficiency of at least four-cell clusters. The vertex angles of the triangular posts bifurcate the laminar flow into two 12 × 100 μm2 opening that can hold the circulating tumor cell (CTC) clusters with strong cell–cell adhesion at the bifurcation, while single blood and tumor cells pass through the openings. The parallel array structures increase the total inlet flow rate up to 2.5 ml/h [Fig. 4(a)].43 Gao et al.44 applied triangular pillars with 50 μm and 20 μm gaps [Fig. 4(b)] to block the CTCs and white blood cell clusters and achieved above 94% capture efficiency and 90% recovery rate at 15 ml/h.44 Shih et al.45 developed metamaterial traps with double trapezoids to trap 20 μm microparticles [Fig. 4(c)]. The distance between two trapezoids was designed to be 5 μm. The changes in the THz transmission response of the metamaterial were used to examine the type and quantity of the trapped microparticles.45
FIG. 4.
Polygonal micropost cell trapping arrays. (a) Schematic of triangular microposts in which CTC clusters are captured from unprocessed whole blood while single cells pass through. Reproduced with permission from Sarioglu et al., Nat. Methods 12, 685–691 (2015). Copyright 2015 Springer Nature. (b) Representative fluorescent images of white cell cluster (upper) and CTC cluster (lower) trapped in triangular pillar array. Scale bars, 20 μm. Reproduced with permission from Gao et al., Oncotarget 8, 12917–12928 (2016). Copyright 2016 Authors, licensed under a CC BY 3.0. (c) Schematic of the proposed microfluidic metamaterial sensor (MMS) operating in the THz spectral region. Reproduced with permission from Shih et al., J. Appl. Phys. 121, 023102 (2017). Copyright 2017 AIP Publishing LLC. (d) Schematic of the filter-based microfluidic device with SEM image showing the diamond-shaped pillars. Reproduced with permission from Zhu et al., Biomed. Microdevices 9, 745–750 (2007). Copyright 2007 Springer Nature. (e) White blood cells trapped after whole blood is flowed into the sample inlet. Reproduced with permission from Kuan et al., Sci. Rep. 8, 15345 (2018). Copyright 2018 Authors, licensed under a CC BY 4.0. (f) “Jail” traps for yeast during typical asymmetric budding; daughter cells are then washed away with media flow. Reproduced with permission from Ryley and Pereira-Smith, Yeast 23, 1065–1073 (2006). Copyright 2006 John Wiley and Sons.
Diamond micropost trapping also is an efficient and common size-based trapping method. Two neighboring posts combine as one trap site, and the gap between two posts is adjusted based on the particle or cell size. The diamond pillar shape reduces the gap length between any two given pillars and decreases pressure at the filter region to improve trapping efficiency and cell viability.46 Zhu et al.46 optimized the trapping array with diamond shaped pillars in a zigzag layout [Fig. 4(d)] that can prolong the trapping length and increase the capture efficiency of Giardia lamblia (∼12–15 μm long and 5–9 μm wide47), Cryptosporidium parvum cells (∼4–6 μm48), and MCF-7 breast cancer cells (∼20 μm–25 μm49) up to 90.8% ± 5.8%, 89.8% ± 16.6%, and 77.0% ± 9.2%. They designed the zigzag coarse pillar-type filters with a gap of 20 μm to capture MCF-7 breast cancer cells and fine pillar-type filters with 0.8 μm gap size to trap Giardia lamblia and Cryptosporidium parvum cells.46 Zhou et al.50 designed diamond shaped post array with 12 μm wide gap to capture CTCs from blood. They used HeLa cells as the sample in this study. The total trapping efficiency could reach about 70% when the trapping sites were saturated. 97.4% viability was achieved right after capture and 68% of trapped cells showed adherence, as well as the high long-term viability for 11 days.50
In addition to the uniform pillar shape, combination of two or more geometries was used to optimize trapping performance and meet the processing requirement of more complex samples. Kuan et al.51 reported a trapping device for on-chip white blood cell trapping [Fig. 4(e)]. They combined one triangular pillar and two rectangular pillars as one unit with a gap of 2.5 μm, which formed a T-shaped gap to capture white blood cells from whole blood.51 Combining multiple posts as one unit may enable different applications and provide better capture efficiency. Ryley and Pereira-Smith52 described a “jail” design that combines three square posts as one jail-like unit which can capture mother cells (large cells) and let daughter cells (small cells) pass through [Fig. 4(f)].52
B. U-shaped microposts without apertures
U-shaped traps have principally been used for single-cell studies, cell pairing experiments, and tumor spheroid formations.39,53–55 There are two types of design for a U-shaped trap: with apertures and without apertures.39 This geometry has its own trap site unlike polygonal traps that rely on neighboring posts to trap cells. For the traps without apertures, the gap principally is designed on top or bottom of posts allowing fluid to carry cells into traps.
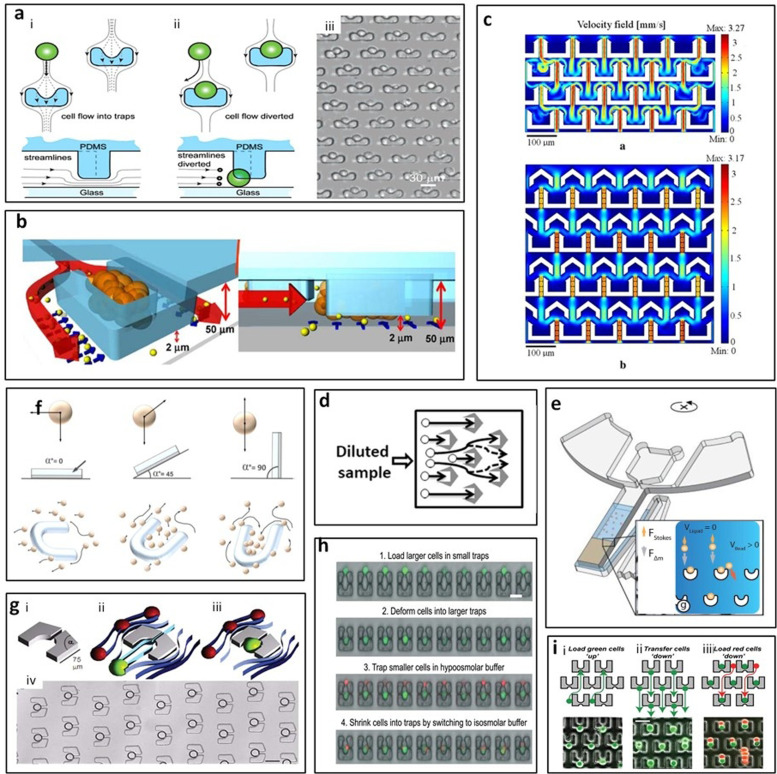
In 2006, Di Carlo et al.56 reported a U-shaped post array biased to trap only single cells and allows for both arrayed culture of individual adherent cells and dynamic control of fluid perfusion [Fig. 5(a)]. Reduced velocities were observed in the trapping region from modeling, with a flow rate of 0.75 ml/min around the trap sites. They achieved consistent trapping of single HeLa cells with about 85% of cells maintained in the trapping site after 24 h.56 Precise positioning of cells and cell-pairs had also been achieved by Zhang et al.,57 demonstrating a “cell printing” technique using hook-shaped traps with high precision. Cells were allowed to extend themselves, therefore, making it possible to quantify biophysical characteristics. Their device achieved high spatial resolution and close to 100% viability with minimal cell damage using a gentle flow rate of <100 μl/min.57 In addition to single cell and cell-pair analysis, tumor spheroid formation and drug discovery can also be performed in a single microfluidic chip. Wu et al.58 reported a U-shaped trap with an inner volume of 35 × 70 × 50 μm3, which can typically contain about ten MCF-7 cells [Fig. 5(b)]. This platform allowed microfluidic self-assembly of spheroids and characterization analysis of spheroid dynamics all in one platform suitable for anticancer drug assays.58
FIG. 5.
U-shaped micropost trapping arrays. (a) (i) and (ii) Schematic of the cell trapping mechanism using flow-through arrayed suspended obstacles. (iii) Phase contrast image of an array of single trapped cells. Scale bar, 30 μm. Reproduced with permission from Di Carlo et al., Anal. Chem. 78, 4925–4930 (2006). Copyright 2006 American Chemical Society. (b) Schematic of one microfluidic spheroid culture trap site. Drug molecules coming with the flow enter through the 2 μm perfusion channel underneath the trap. Reproduced with permission from Wu et al., Biomed. Microdevices 10, 197–202 (2008). Copyright 2008 Springer Nature. (c) The velocity field of the different structures. Reproduced with permission from Chen et al., Microsyst. Technol. 20, 485–491 (2014). Copyright 2014 Springer Nature. (d) Schematic of loading cell sample to the device and physically captured the cells. Reproduced with permission from Chen et al., Biomicrofluidics 11, 054107 (2017). Copyright 2017 AIP Publishing LLC. (e) Beads were loaded into trap under centrifugal force. Reproduced with permission from Burger et al., Lab. Chip 12, 1289–1295 (2012). Copyright 2012 Royal Society of Chemistry. (f) Cell movement in the microfluidic chip in the presence of gravity at different angles (0°, 45°, and 90°). Reproduced with permission from Fu et al., Biofabrication 6, 015009 (2014). Copyright 2014 IOP Publishing, Ltd. (g) U-shaped trap with an aperture was used to trap single droplet. Reproduced with permission from Huebner et al., Lab Chip 9, 692–698 (2009). Copyright 2009 Royal Society of Chemistry. (h) Step-by-step of cell loading and pairing protocol where cell deformation was flow-induced. Reproduced with permission from Dura et al., Lab. Chip 14, 2783–2790 (2014). Copyright 2014 Royal Society of Chemistry. (i) Step-by-step of cell pairing with flow reversal to transfer first cells into larger traps and a second cell type was loaded from the top. Reproduced with permission from Skelley et al., Nat. Methods 6, 147–152 (2009). Copyright 2009 Springer Nature.
Modified U-shaped posts were designed to improve certain performances such as continuous capture and minimizing clogging. Chen et al.59 designed inverted Y-shaped fluidic guides in front of U-shaped traps that caused cells to flow through the space between the structures more easily [Fig. 5(c)], increasing the traps' filling time. Therefore, the capture structures with Y-shaped fluidic guides can continue to capture cells rather than being filling quickly.59 Chen et al.60 modified the trap geometry to a streamlined-shape. This streamlined-shape trap minimizes clogging of cells in capture and release steps [Fig. 5(d)]. Over 85% of cells were captured independent of the input flow speed and over 80% of captured cells were subsequently released.60
While the majority of the micropost trapping systems apply pressure-driven flow to load samples in 2D plane, Burger et al.61 reported a novel centrifugal microfluidic platform using centrifugal force to drive particles trapped in the V-cup [Fig. 5(e)]. This work demonstrated that, depending on the ratio of the active capturing cross section of the cups and the diameter of the beads, the occupancy could be adjusted ranging from single to multi-bead occupancy. When the ratio of beads to cups is ≪1, the capture efficiency was close to 100%.61 In addition to the centrifugal force, gravity also can be applied to trapping devices. Fu et al.62 combined fluid flow and the force of gravity to drive cells into U-shaped traps. The chip was tilted at three angles (0°, 45°, and 90°) and the trapping efficiency of the chip with 90° was highest and four times of the chip with 0° [Fig. 5(f)]. For on-chip downstream spheroid analysis, they applied hydrogel as the substructure, which is non-adhesive to cells to promote the formation of multicellular spheroids.62
C. U-shaped microposts with apertures
To attract more target cells flowing into trapping sites, most designs opened apertures on U-shaped posts to decrease the hydrodynamic resistance and stagnation flow around the traps.63 Kim et al.64 reported a “U-type sieve” model with two apertures that couple the flow field properties within a microchamber. They showed excellent agreement between experimental and computational studies with respect to the cell number per trap and the uniformity of cell distribution within individual microchambers. Compared with previous modeling, they considered both viscous and inertial components to account for both the migration of cells through the microchambers and their deceleration due to confinement in the microsieves.64 Kim et al.65 demonstrated trapping of E. coli cells (∼1 × 3 μm66) in the U-shaped sieve traps. These traps consisted of small openings that are <1 μm in size, creating a flow field that guided cells into the traps. Also, a simple reversal of the flow direction is sufficient to release trapped cells without lysis with a low flow rate (3–5 μl/min).65 Within similar traps, Kukhtevich et al.53 observed that trap arrangement can also affect trap efficiencies. Their semi-circular design resembled that of a “U-type sieve” and was arranged in three rows. Particle displacements with respect to the center of the supply channel affected the trapping probability of a particle for each designated trap in the chamber.53 Kwon et al.67 integrated ZnO nanowires on the microtraps to form mesh-like cage structure to keep E. coli cells anchored to the trap. The apertures in sieves were 3–4 μm wide and 10 μm high. More E. coli cells were trapped and anchored in each unit of nanowire-integrated posts compared to bare posts.67 It may, however, be necessary in some cell studies to retrieve cells once they have been trapped. A way of extracting cells from U-traps is to use hydraulic pressure to selectively open gates of the traps to release the cells inside.68
In addition to cell and particle trapping, artificially generated droplets made up of cell samples or phospholipids are of great interest. Therefore, trapping of these droplets can allow for further investigation of drug screening and lipid–lipid/protein interactions.41,69,70 Huebner et al.41 reported trapping and release of droplets using single aperture U-shape sieves [Fig. 5(g)]. Their results showed that more than 90% droplets trapped by sieves can be released and collected after 45 min.41 Kazayama et al.69 designed U-shaped traps with small back openings for size-selective particle entrapment to investigate artificially generated giant vesicles 10–20 μm in size.69 They designed gap size to be 20%–25% of the target diameter, with the pitch of the traps set at 2 μm larger than the gap to prevent the aggregation of vesicles between the traps. Trapping efficiencies were reported as 64%, 66%, and 70% for their target diameters of 12, 16, and 20 μm, respectively.69
One issue with micropost arrangement is that cells can deform and squeeze through the gaps between posts reducing capture efficiency. Dura et al.71 took advantage of cell deformation and designed U-shaped traps with openings smaller than the cell diameter [Fig. 5(h)]. At high flow rates (100–500 μl/min), cells squeezed into and secured in the traps.71 To protect the viability of cells for long-term, the shear stress and thus the flow rate need to be controlled at a low level (0.2–2 μl/min flow rate),28 which may impact throughput.
D. U-shaped microposts with back traps
Cell–cell interaction studies can also be achieved through U-shaped posts with back traps to pair cells of different types.71,72 Skelley et al.72 reported the post design with front and back traps. After loading the cells to the back traps, cells were then transferred from the back traps to subsequent front traps by reversing flow, and then the second type of cells were loaded into the front traps in order to pair with the first loaded cells [Fig. 5(i)]. They found that cell pairing efficiencies were dependent on cell-loading parameters such as cell concentration, device dimension, loading times, and flow rates. They captured 70%–90% of the cells with an optimal column and row spacing of 20–50 μm.72 In 2015, they adopted the same principles to capture lymphocytes.71 Since lymphocytes are 3× smaller and more deformable than stem cells, they changed the dimensions accordingly to 7–9 μm column spacing, 12–16 μm row spacing, and 8–10 μm cup widths. Additionally, the apertures in between posts and bottom were set as 20%–25% of the total height to prevent cells from squeezing. With the optimized dimensions, 80% cells that entered the array were captured, up to 95% traps were filled and cell pairing efficiencies were between 40% and 85%.71
E. Other micropost geometries
The diversity of micropost shapes includes not only the polygonal and U-shape, but also other irregular shapes like cross, ellipse, and heart. All of them are designed for different applications and trapping targets. Chen et al.73 reported elliptical microposts with 100 μm long slim gaps [Fig. 6(a)], which gradually narrowed from 18 μm to 5 μm to diminish the chances of losing target CTCs. A robust yield of 90% was achieved and it can be used for whole and lyse blood samples with rare CTCs.73 Besides capturing only one kind of single cells, cell pair capturing is also an essential technique for multicellular analysis. Zhu et al.74 designed heart-shaped posts with the gap on top of the posts [Fig. 6(b)] to trap HeLa and MCF-7 cells after successive loading from different directions. The gap at the top of the posts prevents the first cells loss when flowing the second cell type from the opposite direction and allows the secondary cells to touch the posts for capturing. Single cells were captured in 93% traps.74 Weng et al.75 reported a single-cell trapping design using a butterfly-shaped post array [Fig. 6(c)]. A pair of butterfly-shaped post works as one trapping unit with 15 μm aperture between the two posts. The cells were driven by gentle hydrodynamic pressure and the yielding trap occupancy was up to 86.8%. Rat hepatocytes and Brx-142 cells were used as models in this study.75 Mi et al.76 created a device employing the principle of least flow resistance and using hydrostatic force (such that the flow rate was <100 nl/min) for 90% single cell capture. The trapping structures were designed to resemble a resistive electric circuit [Fig. 6(d)]. Cell capture at the first trapping site acts as a plug and, therefore, increases the flow resistance to the site. This structure enables each trap unit to be treated equally and independently.76
FIG. 6.
Micropost trapping arrays with irregularly shaped geometries. (a) Ellipse posts,73 (b) heart-shaped posts,74 (c) butterfly-shaped posts,75 and (d) hook-shaped posts.76 Reproduced with permission from Chen et al., Sci. Rep. 7, 610 (2017). Copyright 2017 Authors, licensed under a CC BY 4.0. Reproduced with permission from Zhu et al., Sens. Actuators B Chem. 283, 685–692 (2019). Copyright 2019 Elsevier. Reproduced with permission from Weng et al., Lab Chip 17, 4077–4088 (2017). Copyright 2017 Royal Society of Chemistry. Reproduced with permission from Mi et al., Lab Chip 16, 4507–4511 (2016). Copyright 2016 Royal Society of Chemistry.
III. CELL TRAPPING BY MICROFILTRATION
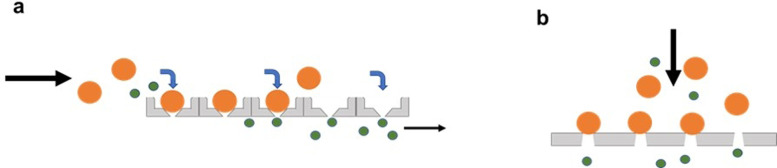
Microfiltration is another size-based hydrodynamic cell trapping method using either horizontal or vertical filters (Fig. 7). To avoid clogging, the crossflow technique is applied in most microfiltration trapping devices.77 Sample solution flows tangentially across the membrane in a crossflow filtration device and the permeable solution flows through the filter. Due to the transmembrane pressure, sample tends to pass the filters but only small cells can flow through the gaps while large cells are captured at trapping sites.78
FIG. 7.
Schematics of microfiltration trapping approaches. Target cells are captured by microfilters in (a) horizontal and (b) vertical arrangements.
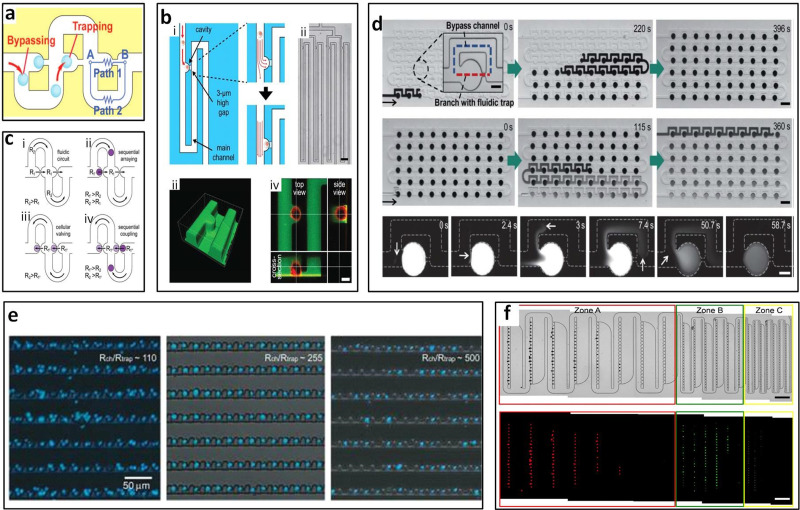
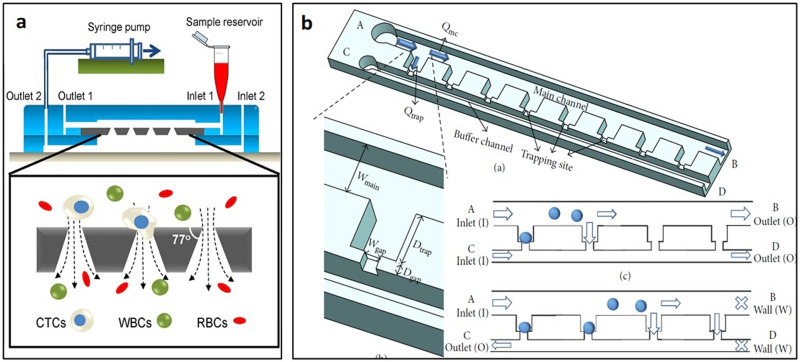
Bypass-channels with sieve-like trapping offer a way to trap cells or clusters sequentially. Since the trapped cells act as plugs, flow resistance at the trapping site increases and subsequent cells flow along the channel bypassing the filled trap.79 Tan and Takeuchi were the first to report the sequential trapping of single cells and to recognize that the flow rate through the trapping site should be higher than the volume flow rate at the main channel, meaning Qtrap/Qmc should be >1 for successful trapping of cells. Sieve-like trapping sites along with the sinuous channel were designed to trap beads in their study [Fig. 8(a)].79 According to the Qtrap/Qmc > 1 principle, modified structure, shape, and size depend on different applications and can be roughly categorized based on the horizontal and vertical trapping designs.
FIG. 8.
Microfiltration with horizontal arrangement. Bypass-channel design (a) allowed for one-bead-to-one-trap79 and (b) single-cell capture.28 Reproduced with permission from Tan and Takeuchi, Proc. Natl. Acad. Sci. U.S.A. 104, 1146–1151 (2007). Copyright 2007 National Academy of Sciences. Reproduced with permission from Kobel et al., Lab Chip 10, 857–863 (2010). Copyright 2010 Royal Society of Chemistry. (c) Cell-pairing also could be achieved by the bypass-channel. Reproduced with permission from Frimat et al., Lab Chip 11, 231–237 (2011). Copyright 2011 Royal Society of Chemistry. (d) Bypass-channel was able to generate static droplet arrays with gradient concentration. Reproduced with permission from Sun et al., Lab Chip 11, 3949–3952 (2011). Copyright 2011 Royal Society of Chemistry. (e) Crossflow microfiltration was able to control number of cells per trap by setting resistance of main channel and trap. Reproduced with permission from Chung et al., Lab. Chip 11, 3949–3952 (2011). Copyright 2011 American Chemical Society. (f) Mixture of 15 μm, 6 μm, and 3 μm particles was sorted by traps in different sizes. Reproduced with permission from Kim et al., Lab Chip 14, 2480–2490 (2014). Copyright 2014 Royal Society of Chemistry.
A. Horizontal arrangement
Horizontal trapping arrangement, which involves flowing the sample along the filter surface with smaller cells passing through the filter pores horizontally, is a common strategy in microfiltration designs. Optimizing geometry of the trapping channel to meet a proper range of flow rate and resistance between the main channel and trap sites is the main method to improve device performance. So far, bypass-channel trapping and crossflow trapping are the two main categories of horizontal arrangement.
B. Bypass-channel trapping
Developed based on Tan and Takeuchi's bypass-channel bead trapping work, Kobel et al.28 reported single cell trapping with bypass-channels, also employing the concept of Qtrap/Qmc > 1. However, with cells, additional requirements including viability and stability of the trapped cells also must be considered. Kobel et al.28 designed trapping structures consisting of a 3 μm high and 12 μm wide horizontal gap on the bottom, which was used to capture non-adherent EG7 cells and achieved a trapping efficiency up to 97% [Fig. 8(b)]. They also identified optimal perfusion rates at 100 nl/min that allowed both a high cell viability (>90%) and a minimization of cell loss (<23%). In addition, automated separation of daughter cells upon division can also be performed in the same device.28 Frimat et al.80 created cellular valving for single-cell pairing [Fig. 8(c)]. Using the same original Qtrap/Qmc > 1 principle but with two trapping sites interfaced with an aperture, they reported a high arraying efficiency of ∼99%.80 After ensuring that Qtrap/Qmc at each trap are in the proper range for trapping single cells, Khalili et al.81 later optimized a single cell trapping model and found the flow resistance ratio of the main channel and the trapping site higher than 3.5 could be used for trapping cells. The trapping length also can be optimized for single cell trapping. For trapping 5 μm yeast cells, 5 μm trapping length is the most suitable since the shorter sites could be blocked by cells while the longer sites would trap multiple cells.81
Besides cell trapping, the bypass-channel also has other novel applications. Bithi and Vanapalli82 applied the bypass-channel to form droplets and then cultured the cells in droplets. After dispensing the cell sample and oil orderly into the oil-prefilled microfluidic device, each bypass-channel loop can park a droplet of 30 nl cell sample and achieve 85%–100% cell capture efficiency. Droplet formation on chip offers benefits of single cell resolution drug assays with a small number of tumor cells or their clusters present in the small sample.82 Sun et al.83 not only applied the bypass-channel to form droplets but also achieved concentration gradients of droplets by inducing diluting plugs of water [Fig. 8(d)]. After sample droplets formed in each trap, the diluting plug, which is water moved into channel, the concentration gradients in the array are produced. This work demonstrated a throughput of nearly 60 different concentrations (in 30 nl drops) in about 10 min using only 2 ml of the sample volume, which is a powerful platform for drug screening.83 Combining the cell traps with electrodes has interested many researchers. By installing electrodes with traps, Zhou et al.84 were able to dynamically monitor single cell lysis by measuring the impedance of trapped cells. In this device, each trap site consists of two sensing electrodes. Depending on flow direction, cells will be captured at the trap facing upstream, while the trap facing downstream is empty.84
Novel modifications of the bypass-channel were also reported. Chung et al.85 achieved >80% efficiency with trapping units that splits incoming flow into two paths (paths A and B) that have different flow resistances, following the Qtrap/Qmc > 1 principle. This method of diverting main flow into trap and bypass channels can ensure single-cell capture. Path A flows through the center of the trapping site and Path B flows through the side channels of the trapping site.85 Similarly, Guan et al.86 used the flow rate ratio principle and then a streamline-based approach to optimize the channel design and thus maximize the capture efficiency. They found that the longer bypass length created streamlines that direct target particles into trap sites and prevented them from flowing to the bypass channels.86 The principle has also been demonstrated with other particles such as protein microcrystals for x-ray diffraction studies.87 Zhou et al.88 developed a valving technique that traps and releases cells upon activation of microfluidic valves. Trapping gaps were designed to be smaller than cells (3–5 μm gaps) immobilizing them in place. Their device demonstrated the entrapment of mouse embryonic stem cells and nuclei with a diameter range of 8–12 μm.88 He et al.89 developed a hydrodynamic shuttling technique able to capture three individual cells of different cell types also by utilizing the bypass channel method.89
C. Crossflow microfiltration
The crossflow microfiltration is also based on the trapping principle Qtrap/Qmc > 1. But compared with bypass-channel trapping, crossflow trapping can offer higher trap concentration. Also, by reversing the flow polarity, full microarray releasing can be accomplished for both microbeads and cells. Released microparticles can be transported to outlet locations and retrieved for subsequent analyses and/or experiments. As a result, microbeads and cells could be processed on-chip to benefit from the advantages associated with microscale systems.90
By optimizing the fluidic resistance of the crossflow channel with respect to the resistance of the loop channel, Chung et al.91 determined the proper range of the resistance ratio of the main channel and trap sites (Rmc/Rtrap). The resistance ratio of ∼110 led to a higher trap occupancy of multiple cells and a higher ratio of ∼255, resulting in the highest single-cell occupancy [Fig. 8(e)]. The optimized trapping mechanism with 25 μm wide trap and 1.8 μm high gap on the top of each trap was designed, which enabled 95% of single-cell occupancy and 94% viability after 24 h. The cells of 8–20 μm diameter at even high-density (up to 5 × 106 cell ml−1) can be used in this device so that this platform can be used for diverse applications such as fundamental studies of stochastic behavior, diagnosis of patient samples, and/or drug screening in cancer and stem cell biology.91 Nourmohammadzadeh et al.92 optimized the Qtrap/Qmc to be 2.8 for trapping and forming ∼300 μm pancreatic islet clusters and 99% ± 2% of the traps are occupied with 95% ± 1% of the occupied traps containing a single particle.92
The height of gap is critical in determining the trapping efficiency.93 Lee's group93 reported that the larger gap height would result in cells squeezing during filtration and the smaller gap height would prohibit both targeted and untargeted cells passing. Therefore, they designed a filtering array to trap single leukemia cells and white blood cells from peripheral blood. The width of traps was set to 10 μm and the height of gap was determined to be 3.3 μm to ensure that WBCs and leukemia cells were trapped while RBCs flow through. Under the flow rate of 0.2 ml/h, the single-cell capturing efficiency was about 80%.93 Similar structure also can be used to analyze embryoid body heterogeneity.94
The trap geometries also impact trapping efficiency. Lawrenz et al.95 evaluated triangular, square, conical, and elliptical traps. Overall, the triangular trap array was the most consistent in filling large amounts of traps with single microparticles where triangular arrays achieved >95% of filled traps having single particles, while the conical, square, and elliptical traps achieved 93%, 78%, and 25%.95 Lu and colleagues96 used a slightly different approach where their method did not depend on resistance changes after traps were occupied, but rather they used guided flow to capture and orient embryos into the traps. Their device consisted of a serpentine channel for bulk flow and resistance channels to guide flow into trap sites resulting in about 90% of traps occupied.96 Hydrodynamic entrapment of non-spherical shaped objects such as a sperm cell that has an oval shaped head (with a length of 9 μm, width of 4.5 μm, and a thickness of 0.5 μm97) was possible with the use of crossflow channels.98 The method used two main channels connected by smaller side channels of 1–2 μm widths, where sperm cells may flow and be squeezed through after a pressure gradient due to the differences in main channel flow rates.98
Particle sorting can also be achieved by crossflow microfiltration. Three different trapping zones connected sequentially in a sinuous channel was reported by Kim et al. for sorting and trapping of cell mixtures [Fig. 8(f)].99 The 15 μm, 6 μm, and 3 μm diameter microspheres were sorted and trapped sequentially with 100%, 98%, and 85% efficiency, respectively.99 The trap size was 16 μm, 8 μm, and 4 μm and the passing gap size was 8 μm, 4 μm, and 2 μm, respectively, for three kinds of microspheres. Therefore, hydrodynamic methods can sort subjects with a resolution as low as 3 μm, with a decreasing efficiency for a smaller particle size due to Brownian diffusive effects.
Once cells are trapped on-chip, cell analyses can be performed for a wide range of aspects, such as apoptosis,100 kinetic cellular response,91 cell growth, coupling, and beating.101 Valero et al.100 reported a microfilter chip to immobilize and real-time monitoring of single cells' apoptotic process. U-shaped trapping sites were designed with 10 μm diameter and 3 μm narrow openings to trap HL60 cells. With this trapping device, apoptosis could be studied in real-time. Espulgar et al.101 demonstrated an integrated microfluidic chip to trap cells and analyze their coupling and decoupling dynamics, which is the first to be observed in cardiomyocyte cell microfluidics. They designed a chip combining filtration, trapping, and monitoring. The cells flowed through the trapping sites with 10, 20, and 50 μm gap tangentially based on the sedimentation effect. After capture, 70%–80% of the trapped cells grew.101
D. Vertical arrangement
Loading the sample on the top of the filter tangentially and letting smaller cells pass through the filter pores vertically can be categorized as vertical microfiltration trapping. Similar to horizontal trapping, vertical trapping also mainly applies the crossflow technique to mitigate the clogging issue while increasing throughput. Different gap shapes and device structures were adapted to achieve the capture of CTCs and cell line cells.
Tang et al.29 developed a crossflow vertical microfiltration with conical-shaped holes to capture CTCs from diluted blood [Fig. 9(a)]. The capture efficiency is up to 95% and the WBC clearance efficiency is 96% when using the flow rate of 0.2 ml/min with filters of 6.5 μm hole diameter.29 Also using vertical microfiltration with conical-shaped holes, Hosokawa et al.102 added the vacuum chamber below the microfiltration to accelerate loading speed and 90% of microcavities loaded cells.102 Hosokawa et al.103 then developed a microcavity array with 2 μm hole and successfully detected 97% of 10–100 lung carcinoma NCI-H358 cells from 1 ml whole blood. In addition, breast, gastric, and colon tumor cell lines that include EpCAM-negative tumor cells, which cannot be isolated by conventional immunomagnetic separation, were successfully recovered on the microcavity array with high efficiency (more than 80%). On average, approximately 98% of recovered cells were viable.103 Their microcavity array system was then used to detect CTC from non-small cell lung cancer patient’s blood. CTCs were detected in 20 of 21 small cell lung cancer (SCLC) patients using the MCA system vs 12 of 21 patients using the CellSearch system.104 Deng et al.105 developed a U-shaped filter between the main channel and the buffer channel [Fig. 9(b)]. The cell suspension flowed along with the filter and the transmembrane pressure resulted in the cells capturing in the trapping sites. The gap size in this study was 5 μm and the target trapped cells were A549 cells. With the flow ratio of the trapping site and the main channel higher than 1, the trapping efficiency was higher than 90%.105
FIG. 9.
Microfiltration with vertical arrangement. Principle of single-cell positioning based on the hydrodynamic filter trapping. Filters can be arranged as (a) arrays29 or (b) single row.105 Reproduced with permission from Tang et al., Sci. Rep. 4, 6052 (2014). Copyright 2014 Authors, licensed under a CC BY 4.0. Reproduced with permission from Deng et al., Sci. World J. 2014, 929163. Copyright 2014 Authors, licensed under a CC BY 3.0.
IV. CELL TRAPPING IN MICROWELLS
For cell culture and cluster formation, the microwell system is an ideal platform that can offer control over parameters such as size, shape, and homogeneity.106 In biological studies, disease diagnosis, and drug screening, microwell platforms play an important role in offering high-throughput data (Fig. 10). A rather straightforward solution to encage cells in a massively parallel format are microwell arrays, where cells are seeded and allowed to settle into microwells by gravity that can hold a single cell or cell aggregates in each position.33,106–108 Unlike micropost and microfiltration approaches, vertical microwells can control cells with certain size,109,110 shape,110 and homogeneity110 in a uniform environment and space for high-throughput and parallel cell or DNA analysis.
FIG. 10.
Schematic of microwell arrays. After being loaded into the microfluidic chip, cells settle down into individual microwells vertically.
One of the advantages of such an array of cell traps is that once trapped, each cell is held at a fixed position on the array. Thus, the transient properties of thousands of cells can be individually monitored by capturing images sequentially using a conventional fluorescence microscope.111 The use of microwell-based intercellular experiments can aid in correctly characterizing the cell dynamics of a single or a small number of cells.30 In addition, since microwells form an array, they allow high-throughput and parallel assays, which are useful in analytical research in cell biology and medical diagnostic tests.112 Microwell dimensions, shape, and arrangement can be adapted to different cell sizes, settle times, and cell seeding densities. All of these microwell parameters and sample properties impact capture efficiency. So far, microwell devices can offer a wide range of applications from single cells capture107,108,111 to multiple cells coculture113 due to their adaptable physical parameters.
Height/diameter ratio of a microwell of ∼1114 has been reported to be the optimal to contain single cells107,115,116 and to form spheroids in microwells.117 For example, a 30 μm well diameter with a 30 μm height was suitable for stem cells (∼30.4 μm in diameter) trapping,116 a 20 μm well diameter with a 21 μm height was best for rat basophil leukemia cells (∼17–20 μm in diameter), while a 25 μm-diameter and 27 μm-height were best for fibroblast cells (∼13–20 μm in diameter).114 Proper microwell dimensions need to be considered to trap the precise number of cells with high efficiency.114 For single-cell studies, the isolation of individual cells in separate traps from bulk solutions need to be optimized to achieve high-throughput analysis. In addition to the fixed well size, Wang et al.111 developed a stretchable circular microwell array for single-cell analysis [Fig. 11(a)]. A microwell array was made of polydimethylsiloxane (PDMS) and stretched uniaxially by a plastic tube controlled by a micromanipulator. Cells were added to the tube and allowed to settle into the expanded microwells. Relaxation of the PDMS by removal of the tube resulted in contraction of the microwells around the cells which were trapped. For trapping 14.1 μm Ba/F3 cells, they set the diameter of the hole on the well as 12 μm which achieved 65.3 ± 7.6% trapping efficiency.111
FIG. 11.
Vertical microwell arrangement. (a) Schematic of uniaxially stretching a PDMS microwell array by pressing with a plastic tube. Reproduced with permission from Wang et al., Anal. Bioanal. Chem. 402, 1065–1072 (2012). Copyright 2012 Springer Nature. (b) Cell docking method with multiple sweeping processes. Reproduced with permission from Park et al., Lab Chip 11, 79–86 (2011). Copyright 2011 Royal Society of Chemistry. (c) The graph of local velocity near the base and wall shear stress in grooves. Alignment of cardiomyocyte cells shown with phase contrast images. In 50 μm grooves, cells were in the upstream corner, while in 75 μm grooves, cells were in downstream corner. Reproduced with permission from Manbachi et al., Lab Chip 8, 747–754 (2008). Copyright 2008 Royal Society of Chemistry. (d) Geometry of microwell affects the trapping rate. Streamlines in triangular microwells had strong recirculation for trapping of a single cell. Reproduced with permission from Park et al., Microfluid. Nanofluidics 8, 263–268 (2010). Copyright 2010 Springer Nature. (e) Circular microwell for the formation of uniform-sized cell spheroids. Reproduced with permission from Kim et al., Lab Chip 12, 4135–4142 (2012). Copyright 2012 Royal Society of Chemistry. (f) Concave microwells offered uniform size of spheroids. Reproduced with permission from Choi et al., Biomaterials 31, 4296–4303 (2010). Copyright 2010 Elsevier. (g) Trapping scheme of an L-microwell. Reproduced with permission from Kim et al., Micromech. Microeng. 25, 035005 (2015). Copyright 2015 IOP Publishing, Ltd. (h) A schematic illustration for the workflow of the floating droplet array (FDA). Reproduced with permission from Labanieh et al., Micromachines 6, 1469–1482 (2015). Copyright 2015 Authors, licensed under a CC BY 4.0. (i) Steps for centrifugation-assisted single-cell trapping (CAScT). Reproduced with permission from Huang et al., Anal. Chem. 87, 12169–12176 (2015). Copyright 2015 American Chemical Society. (j) Microwell array system allowed multiple treatments to cells. Reproduced with permission from Khademhosseini et al., Lab Chip 5, 1380–1386 (2005). Copyright 2005 Royal Society of Chemistry.
In addition to the careful design of well dimensions, Park et al.107 used a multiple sweeping process to move individual budding yeast (3–4 μm in diameter) and Saccharomyces cerevisiae cells (5–10 μm in diameter) into traps [Fig. 11(b)].107 Their circular microwell design had an 8 μm diameter and 8 μm height. Larger diameters from 12 μm to 24 μm resulted in lower single-cell resolution. Additionally, cell density affects the trapping efficiency of their device. To obtain >90% efficiency, cell density needs to be higher than 5.0 × 109 cells per ml, and thus it is not suitable for samples with low cell density.107
Another parameter to consider is the well bottom shape. The geometry of vertical well traps can significantly impact flow streamlines.118 Manbachi et al.119 reported that smaller sized rectangular microgrooves showed great recirculation, where the direction of local velocity near the base of the grooves is opposite that of the main channel flow [Fig. 11(c)]. Local shear stress guided cell alignment following the flow direction in the grooves.119 Park et al.118 simulated the recirculation pattern in the equilateral triangle, square, circle, rhombus, and cone microwells from 3D simulation.
A strong flow recirculation occurs in the triangular microwell thus efficiently catching cells from the main flow [Fig. 11(d)]. Once a cell is captured, cells present in the microwell changes the flow pattern, preventing trapping of other cells.118 Moreover, triangular microwells with dimensions of 50 μm were found to be the most efficient for single cell trapping while providing ample space for cells to grow and spread.118 Computational and experimental study with microbeads also demonstrated an effective trapping of 10–20 μm particles in triangular microwell traps.120
Circular microwell traps were used for spheroid formation after multiple cell trapping was performed in 2012 by Kim et al. [Fig. 11(e)].121 A defined inlet volumetric flow rate (200 μl/h) was used to obtain the proper number of cells settle down in each circular well with 300 μm diameter and 330 μm height. Uniform tumor spheroids with an average diameter of 188.1 μm were formed by trapping a defined number of cells in each well.121 Choi et al.122 found that after loading multiple cells into microwells, concave shaped wells form one uniform spheroid rather than several clusters in one well with a flat bottom [Fig. 11(f)].122 Other studies of tumoroid123 and 3D-cell spheroid124 formation in microwells allowed for in vitro tumor modeling and high-throughput screening. Lee et al.125 developed a deformable truncated L-shaped microwell array for trapping pairs of heterogeneous cells and analyzed cell–cell signaling [Fig. 11(g)]. Each branch of the L-microwell had a dimension of 29 × 18 μm2 and the depth of the entire L-microwell was 30 μm. Truncated L-shape allows to trap only one cell in one branch. Researchers stretched the PDMS substrate to let cells fall into branches and then released the substrate, cells in the L-microwell are slightly squeezed to be held securely in the L-microwells.125
In devices utilizing gravity-based sedimentation, settling time is important to consider since it may be necessary to minimize cellular sedimentation.22 Experiments have provided optimized settling times for different devices varying from 2 to 30 min depending on the well depth, number of wells, and cell density.114,126,127 Longer settling time resulted in higher cell occupancy and consequently prolonged the total experimental time.127 In addition to the gravity-based trapping, buoyancy force can be used to trap low density samples. Due to the low density of droplets, Labanieh et al.128 designed microwells on the top of the chamber and droplets floated into wells. The droplets can be recovered by flipping the device [Fig. 11(h)].128 Centrifugal force was also applied as loading force of trapping devices. Huang et al.129 developed truncated cone shape of the microwells to trap single cells by centrifugation. The efficiency of occupancy could reach approximately 90% within a few seconds when the 24 μm microwells were used [Fig. 11(i)].129
To trap cells selectively and treat cells in multi-phenotype array, Khademhosseini et al.130 presented a system that has been used to trap multiple cell types separately and drive multiple treatments to cell arrays [Fig. 11(j)]. By covering columns of channels to isolate each column of microwells, multiple cell types can be loaded into every channel separately. In addition, delivering of multiple kinds of reagents by serially placing microchannel arrays orthogonally on substrates can realize high-throughput cells analysis.130
Microwell devices have been used in biomedical research such as single-cell analysis,85,108,131–133 in drug screening121 and stem cell differentiation studies.122,126 A cell retainer device was designed for the study of non-adherent primary cells without interfering with cell integrity.134 Single B-lymphocyte analysis was performed in 2007 with microwells at a size of 10 μm diameter and 15 μm depth (less than 10 μm depth resulted in easy dislodgement of cells during wash).133 Jen et al.131 performed high-throughput single-cell chemical lysis in microwell arrays, where they captured HeLa cells in 20 μm and 30 μm diameter wells.131 Recently, Zheng et al.127 used the microwell system where cells were allowed to settle to the bottom overtime to quantify silver nanoparticles in single cells. They achieved 60% single cell occupancy efficiency.127
V. TRAPPING CHAMBER ARRANGEMENT
Microchamber entrapment is the alternative microfluidic trapping strategy. Microchannels can be modified by altering channel dimensions and combining trapping chamber horizontally or vertically; therefore, their resistances and the surface tension can manipulate flow of cells into trapping chambers (Fig. 12).
FIG. 12.
Schematic of trapping chamber. Target cells are squeezed through a constriction microchannel due to cell deformation and then captured in a trapping chamber with larger size, while smaller nontarget cells can flow through.
A. Horizontal arrangement
Cinque et al.135 designed straight channels with arrays of micro-chambers on one side of main channel, which allowed cells flow into the chamber and be captured [Fig. 13(a)]. They found best results with the following ratios of main channel width:neck width:neck length = 5:[1–1.5]:[1.5–2].135 Near 90% trapping efficiency of HeLa cells was achieved by Wang et al.136 in 2015. They created bypass structures in square-wave shaped main channels to test chemotherapeutic drugs on trapped cells.136 Long-term cell culture was achieved using serpentine and tree-pattern microchannels with larger sized trapping chambers for cell growth and proliferation.137 Kamyabi et al.138 designed one microfluidic device for isolating CTC clusters spiked in whole blood, which is equipped with ∼10 000 trap chambers that isolate tumor cell clusters based on their large sizes and dynamic force balance against a pillar obstacle in the trap chamber. At 25 μl/min for blood injection, 100 μl/min as the wash flow rate, and 300 μl/min the release flow rate, a capture efficiency of 66%–87% and a release efficiency of 76%–90% were achieved. Luo et al.139 designed sidewall triangle-chambers for fast cell loading based on gas absorption of degassed PDMS. The length of triangle side is 125 μm and the width of the main channel is 125 μm. All chambers are connected to the main channel with a narrow pass of 20 μm width and 20 μm length [Fig. 13(b)]. After the devices are degassed, buffer solutions with yeast cells can be introduced into the triangle chambers by gas absorption therein. The ratio of cavities with a single cell is 40%. Since the flow velocity in the micro-chambers can be nearly zero, it is easy to change the medium at a flow velocity of 20 μl/h without washing the cells out of the microstructures.139
FIG. 13.
Trapping chamber arrangement. (a) Cells were trapped into side chambers from main channels. Reproduced with permission from Cinque et al., Microelectron. Eng. 88, 1733–1736 (2011). Copyright 2011 Elsevier. (b) After being injected into the main channel, the yeast cell suspension was aspirated into micro-cavities by gas absorption of PDMS surface. Reproduced with permission from Luo et al., Biotechnol. Bioeng. 101, 190–195 (2008). Copyright 2008 John Wiley and Sons. (c) CTCs were captured in cylinder trapping chamber while blood cells could pass through the narrow channels. Reproduced with permission from Ren et al., Anal. Chem. 90, 7526–7534 (2018). Copyright 2008 American Chemical Society. (d) The droplets were captured from channel to cylinder traps due surface. Reproduced with permission from Shi et al., Microfluid. Nanofluidics 15, 467–474 (2013). Copyright 2013 Springer Nature.
B. Vertical arrangement
Ren et al.140 also utilized trapping chambers that can be used to trap CTCs from whole blood. By taking the advantages of cell deformation, CTCs would be squeezed through a constriction microchannel with 8 μm width and height and 100 μm length, and then trapped into a larger sized cylinder chamber with 30 μm width and height and 40 μm length [Fig. 13(c)]. Due to the small size, blood cells can flow pass through the constriction channel without any obstruction. An overall capture ratio >95% can be reached.140 Shi et al.141 integrated the T-junction with trapping chambers to form and trap droplets in one single device. The trapping chambers were added on the top of the channel; since the surface tension within traps was higher than that in the channel, droplets can be captured from channel to traps [Fig. 13(d)]. The following droplets pass through the filled trap along the channel without mixing with the trapped one due to the effect of the surfactant, and then are trapped into the next site.141
VI. SUMMARY AND OUTLOOK
With significant attention being paid to the development of microfluidic systems for trapping of cells, it is important to understand progress made and the existing challenges. The passive hydrodynamic trapping methods offer a simple approach for developing microfluidic systems for enabling cellular assays without the use of external forces.32 As we discuss above, there are many remarkable advantages to microfluidic methods, such as reduction in time, savings in space, conservation of reagents, and need for only small sample volumes (on the order of microliters). Passive microfluidic trapping methods, compared with their active counterparts, are simpler in structure, easier to fabricate, lower in cost, and do not demand skillful operators.142 Only simple trapping geometry and sample flow are required to obtain a high capture efficiency in passive trapping devices, enabling the integration of these devices with upstream sample pretreatment or downstream analysis.
The micropost array is a straightforward and simple method to block target cells from flow. Microfiltration offers a way to trap single cells sequentially79 and delivers the potential for sorting cells and subsequent chemical exposure steps.143 Also, compared with the micropost trapping, once a cell is captured in crossflow microfilters the fluid can bypass this cell, reducing further the pressure and shear stress that cause cell damage.29 In microwells, cells are confined in private and uniform environments enabling for precise control of cell or cluster shape and size.106,118 Last, trapping chambers have been used for long-term cell culturing. With these advantages, high-throughput and rapid analysis are possible in biomedical applications, such as studies of single-cell heterogeneity,6 drug-screening,8,9 stem cell differentiation,10 and cell–cell interactions.4,5
Although the microfluidic cell trapping and analysis are in the early stages, substantial progress has been made. Table I summarizes the key metrics across the four trapping approaches. Most of the passive trapping devices can achieve an efficiency above 80%, with some reporting trapping efficiency with beads and cell clusters close to 100%. Viability above 90% has also been achieved in micropost, microfiltration, and microwell trapping devices, suggesting substantial potential for downstream cell analysis. There are several physical factors that impact chip design and trapping efficiency, such as cell density, deformability, and size, with latter being the most important. Most of the devices are capable of trapping up to tens of thousands of cells and can be integrated (Table I).
TABLE I.
Summary table of the key metrics across the four trapping approaches.
| Microposts | Microfiltration | Microwells | Chambers | |
|---|---|---|---|---|
| Trapping efficiency/occupancy | >70% | >80% | >60% | >60% |
| Flow rate (μl/min) | 0.1–750 | 0.1–200 | N/A or low | 0.3–25 |
| On-chip culture | weeks50,62 | up to 1 week82 | weeks106 | weeks137 |
| On-chip co-culture | Yes55 | Yes80 | Yes113,125 | Yes |
| On-chip cluster formation | Yes58,62 | Yes94 | Yes117,121 | Yes |
| Cell viability | >95% | >90% | ∼100% | >90% |
| Cell size | 1–100 s μm (clusters) | 1–100 s μm (clusters, droplets) | 1–100 s μm (clusters, droplets) | 1–100 s μm (droplets) |
| Capacity | 9–70 000 | 5–10 000 | 1–40 000 | 1–10 000 |
| Fabrication | Soft lithography, ion etching, photolithography | Soft lithography, photolithography, laser drilled PET | Soft lithography, stamping, 3D printing | Soft lithography |
An emerging trend is in the integration of microfluidic trapping devices with either upstream sample preparation (e.g., cell separation, sorting, and enrichment)144–146 or downstream cell analysis or manipulation [e.g., PCR,135 fluorescence in situ hybridization (FISH),147 and drug-screening8,9,58,148]. Table II compares the four cell trapping approaches, focusing on upstream and downstream integration. These integrations offer trapping devices more powerful and comprehensive performance and extend their functionalities to other applications. For example, the integration of trapping devices with droplet microfluidics enables high-throughput screening of single cells or clusters. This integrated platform allows for the encapsulation of single cells and reagents in independent aqueous microdroplets and enables the precise manipulation of these reactors at a very high-throughput.149 To simplify the integration platform, merging the functions of forming droplets and trapping cells into trapping sites150 will be worth advancing. Integration of trapping structures with sensor electrodes could lead to new functionalities as well. Signaling electrodes set on the bottom of trapping sites are able to continuously monitor the changes of single-cell conditions, allowing the studies in the cell dynamic mechanism field. In addition, spectroscopy151 and PCR135 as the downstream analysis systems also worked successfully.
TABLE II.
Comparison of the passive cell trapping methods.
| Microposts | Microfiltration | Microwells | Chambers | |
|---|---|---|---|---|
| Upstream integration | Cell soring | Droplet formation, reagent mixing | Droplet formation, reagent mixing | Droplet formation |
| On-chip downstream analysis | Single-cell analysis,56 drug screening41 cell pairing57,71,72 cluster formation58,62 |
Single-cell analysis,81,100 drug screening82,83 cell pairing80 |
Single-cell analysis,111,118 drug screening,123,124 cell pairing,125 cluster formation117,121 |
Single-cell analysis,140 drug screening136 PCR135 |
| On-chip immunostaining | CK,43 CD45,43 Ki67,43 Vimentin,44 others | ER,91 profilin-1,91 others | CD34,102 CD45,102 Oct4.109 e-eadherin109 others | Oct3/4, CK, others |
| Sensor integration | Impedance sensing electrodes49 | Impedance sensing electrodes84 | … | … |
| On-chip nucleic acid analysis | … | … | Yes108 | Yes135 |
| On-chip protein expression analysis | Yes52 | Yes91,92,94 | Yes109 | Yes |
| On-chip cellular analysis | Yes54,58 | Yes84,92,100 | Yes109,112 | Yes |
| Drug screening | Yes41 | Yes82,83 | Yes123,124 | Yes136 |
| Outstanding issues | Cell deformability, clogging | Cell deformability | Time consuming | … |
While the current hydrodynamic trapping devices offer excellent performance, they are not without limitations. One of the critical challenges is trapping of small objects such as bacteria and exosomes. For example, Brownian diffusion becomes significant and can interfere with the immobilizing of trapped objects.152 Moreover, trapping resolution needs further improvement for more sensitive and accurate capture of these objects. In some current techniques, the passive trapping resolution can be decreased to 3 μm99 (i.e., 3 μm difference in size between target cells and non-target cells). However, investigators usually keep the resolution to about 10 μm to ensure a high capture efficiency.33
Another common challenge of hydrodynamic trapping devices is the limited trapping efficiency for samples with low target cell numbers and deformable cells. One example is trapping of CTCs, which have been receiving broad attention in the microfluidics field due to their strong diagnostic or prognostic potential. However, these cells are extremely rare, with a concentration of only <10 CTCs vs billions of blood cells per 1 ml of blood,153 leading to a sharp decrease in the trapping efficiency. Finding a balance between the target cell density and the trapping efficiency is especially necessary for these samples. Deformability of cells can also negatively impact the trapping efficiency. At high flow rates, cells tend to squeeze through trap apertures resulting in a lower trapping efficiency. Although there are special cases like Ren et al.,140 who captured CTCs by squeezing cells into narrow microchannels, unwanted cell deformity is still detrimental in most of the passive hydrodynamic trapping devices. Cell cluster dissociation also needs to be considered in design of CTC cluster trapping devices, as cell–cell adhesion strength within CTC clusters varies from <28 nN to >524 nN,154 and protecting the intactness of clusters is challenging.
Moreover, the compatibility between upstream, downstream, and trapping technologies is another challenge. For example, to match the flow rate of upstream and trapping sections, the modification of structure designs is required. For downstream analysis, long-term viability of trapped cells needs be considered. Although >90% viability can be maintained for a few hours after capture, long-term viability was seldom reported. One reason is due to the less-than-optimal culture environment in microfluidic devices. Narrow chambers and small cell density limit cell growth and weaken cell-to-cell communications. Further, it is important to keep devices sufficiently compatible where parameters such as channel dimensions, flow rates, or the number of inlets/outlets are easily modified based on the upstream or downstream applications.
Finally, the future directions of this field require creative thinking in the broad context of the microfluidic lab-on-a-chip (LOC) systems. LOC technology is widely seen as the revolutionary future of the current benchtop-based bioanalysis systems. However, most of work to date has focused on maximizing performance of the trapping approach, free from the restrictions posted by integration. As we discussed above, integration needs to consider the interface between the upstream and downstream modules. Adding reagents, removing waste, retrieving cellular/molecular samples, and imaging/detecting outcomes all would pose risks of compromising performance of the trapping device. For example, high throughput requiring high flow speed in the upstream cell sorting module could negatively impact the trapping efficiency, which is better at low speed. Introducing reagents may also change the viscosity of the medium, which, in turn, could alter the performance. Moreover, to preserve the biological properties of the trapped cells, their microenvironment may need to be carefully managed. For example, gas exchange, fluid pressure, and soluble factors must be considered for prolonged on-chip processing and analysis. On the other hand, commercialization of microfluidic devices would ultimately require alternatives to the commonly used PDMS soft lithography, which is not compatible with the mass-production industrial pipelines.
ACKNOWLEDGMENTS
We gratefully acknowledge partial funding support by the University of Illinois Cancer Center and the Loan Hill Department of Bioengineering at the University of Illinois at Chicago.
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created in this study.
REFERENCES
- 1.Weaver W. M. et al. , “Advances in high-throughput single-cell microtechnologies,” Curr. Opin. Biotechnol. 25, 114–123 (2014). 10.1016/j.copbio.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konry T., Sarkar S., Sabhachandani P., and Cohen N., “Innovative tools and technology for analysis of single cells and cell–cell interaction,” Annu. Rev. Biomed. Eng. 18, 259–284 (2016). 10.1146/annurev-bioeng-090215-112735 [DOI] [PubMed] [Google Scholar]
- 3.Charnley M., Textor M., Khademhosseini A., and Lutolf M. P., “Integration column: Microwell arrays for mammalian cell culture,” Integr. Biol. 1, 625 (2009). 10.1039/b918172p [DOI] [PubMed] [Google Scholar]
- 4.Chung S., Sudo R., Vickerman V., Zervantonakis I. K., and Kamm R. D., “Microfluidic platforms for studies of angiogenesis, cell migration, and cell–cell interactions,” Ann. Biomed. Eng. 38, 1164–1177 (2010). 10.1007/s10439-010-9899-3 [DOI] [PubMed] [Google Scholar]
- 5.Zervantonakis I. K., Kothapalli C. R., Chung S., Sudo R., and Kamm R. D., “Microfluidic devices for studying heterotypic cell-cell interactions and tissue specimen cultures under controlled microenvironments,” Biomicrofluidics 5, 013406 (2011). 10.1063/1.3553237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin H. and Marshall D., “Microfluidics for single cell analysis,” Curr. Opin. Biotechnol. 23, 110–119 (2012). 10.1016/j.copbio.2011.11.002 [DOI] [PubMed] [Google Scholar]
- 7.Adriani G. et al. , “Microfluidic models for adoptive cell-mediated cancer immunotherapies,” Drug Discov. Today 21, 1472–1478 (2016). 10.1016/j.drudis.2016.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Y., Wang P.-C., Locascio L. E., and Lee C. S., “Integrated plastic microfluidic devices with ESI-MS for drug screening and residue analysis,” Anal. Chem. 73, 2048–2053 (2001). 10.1021/ac001474j [DOI] [PubMed] [Google Scholar]
- 9.Kim J. et al. , “A programmable microfluidic cell array for combinatorial drug screening,” Lab Chip 12, 1813 (2012). 10.1039/c2lc21202a [DOI] [PubMed] [Google Scholar]
- 10.Geun Chung B. et al. , “Human neural stem cell growth and differentiation in a gradient-generating microfluidic device,” Lab Chip 5, 401–406 (2005). 10.1039/b417651k [DOI] [PubMed] [Google Scholar]
- 11.Johann R. M., “Cell trapping in microfluidic chips,” Anal. Bioanal. Chem. 385, 408–412 (2006). 10.1007/s00216-006-0369-6 [DOI] [PubMed] [Google Scholar]
- 12.El-Ali J., Sorger P. K., and Jensen K. F., “Cells on chips,” Nature 442, 403–411 (2006). 10.1038/nature05063 [DOI] [PubMed] [Google Scholar]
- 13.Dittrich P. S. and Manz A., “Lab-on-a-chip: Microfluidics in drug discovery,” Nat. Rev. Drug Discov. 5, 210–218 (2006). 10.1038/nrd1985 [DOI] [PubMed] [Google Scholar]
- 14.Young E. W. K. and Beebe D. J., “Fundamentals of microfluidic cell culture in controlled microenvironments,” Chem. Soc. Rev. 39, 1036–1048 (2010). 10.1039/b909900j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng Y., Novak R., Shuga J., Smith M. T., and Mathies R. A., “High-performance single cell genetic analysis using microfluidic emulsion generator arrays,” Anal. Chem. 82, 3183–3190 (2010). 10.1021/ac902683t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Easley C. J. et al. , “A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability,” Proc. Natl. Acad. Sci. U.S.A. 103, 19272–19277 (2006). 10.1073/pnas.0604663103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M. B., Srigunapalan S., Wheeler A. R., and Simmons C. A., “A 3D microfluidic platform incorporating methacrylated gelatin hydrogels to study physiological cardiovascular cell–cell interactions,” Lab Chip 13, 2591–2598 (2013). 10.1039/c3lc00051f [DOI] [PubMed] [Google Scholar]
- 18.Ortega-Prieto A. M. et al. , “3D microfluidic liver cultures as a physiological preclinical tool for hepatitis B virus infection,” Nat. Commun. 9, 1–15 (2018). 10.1038/s41467-018-02969-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi J.-W. et al. , “An integrated microfluidic biochemical detection system for protein analysis with magnetic bead-based sampling capabilities,” Lab Chip 2, 27–30 (2002). 10.1039/b107540n [DOI] [PubMed] [Google Scholar]
- 20.Bilitewski U., Genrich M., Kadow S., and Mersal G., “Biochemical analysis with microfluidic systems,” Anal. Bioanal. Chem. 377, 556–569 (2003). 10.1007/s00216-003-2179-4 [DOI] [PubMed] [Google Scholar]
- 21.Sackmann E. K., Fulton A. L., and Beebe D. J., “The present and future role of microfluidics in biomedical research,” Nature 507, 181–189 (2014). 10.1038/nature13118 [DOI] [PubMed] [Google Scholar]
- 22.Whitesides G. M., “The origins and the future of microfluidics,” Nature 442, 368–373 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- 23.Kotnala A., Zheng Y., Fu J., and Cheng W., “Microfluidic-based high-throughput optical trapping of nanoparticles,” Lab Chip 17, 2125–2134 (2017). 10.1039/C7LC00286F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X. et al. , “Microfluidic dielectrophoresis device for trapping, counting and detecting Shewanella oneidensis at the cell level,” Biosens. Bioelectron. 99, 416–423 (2018). 10.1016/j.bios.2017.08.017 [DOI] [PubMed] [Google Scholar]
- 25.Evander M. et al. , “Noninvasive acoustic cell trapping in a microfluidic perfusion system for online bioassays,” Anal. Chem. 79, 2984–2991 (2007). 10.1021/ac061576v [DOI] [PubMed] [Google Scholar]
- 26.Winkleman A. et al. , “A magnetic trap for living cells suspended in a paramagnetic buffer,” Appl. Phys. Lett. 85, 2411–2413 (2004). 10.1063/1.1794372 [DOI] [Google Scholar]
- 27.Park M.-H. et al. , “Enhanced isolation and release of circulating tumor cells using nanoparticle binding and ligand exchange in a microfluidic chip,” J. Am. Chem. Soc. 139, 2741–2749 (2017). 10.1021/jacs.6b12236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobel S., Valero A., Latt J., Renaud P., and Lutolf M., “Optimization of microfluidic single cell trapping for long-term on-chip culture,” Lab Chip 10, 857–863 (2010). 10.1039/b918055a [DOI] [PubMed] [Google Scholar]
- 29.Tang Y. et al. , “Microfluidic device with integrated microfilter of conical-shaped holes for high efficiency and high purity capture of circulating tumor cells,” Sci. Rep. 4, 6052 (2014). 10.1038/srep06052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo T., Fan L., Zhu R., and Sun D., “Microfluidic single-cell manipulation and analysis methods and applications,” Micromachines 10, 104 (2019). 10.3390/mi10020104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhagat A. A. S. et al. , “Microfluidics for cell separation,” Med. Biol. Eng. Comput. 48, 999–1014 (2010). 10.1007/s11517-010-0611-4 [DOI] [PubMed] [Google Scholar]
- 32.Narayanamurthy V., Nagarajan S., Khan A. Y. F., Samsuri F., and Sridhar T. M., “Microfluidic hydrodynamic trapping for single cell analysis: Mechanisms, methods and applications,” Anal. Methods 9, 3751–3772 (2017). 10.1039/C7AY00656J [DOI] [Google Scholar]
- 33.Nilsson J., Evander M., Hammarström B., and Laurell T., “Review of cell and particle trapping in microfluidic systems,” Anal. Chim. Acta 649, 141–157 (2009). 10.1016/j.aca.2009.07.017 [DOI] [PubMed] [Google Scholar]
- 34.Wu J., Chen Q., and Lin J.-M., “Microfluidic technologies in cell isolation and analysis for biomedical applications,” Analyst 142, 421–441 (2017). 10.1039/C6AN01939K [DOI] [PubMed] [Google Scholar]
- 35.Kim S. M., Lee S. H., and Suh K. Y., “Cell research with physically modified microfluidic channels: A review,” Lab Chip 8, 1015–1023 (2008). 10.1039/b800835c [DOI] [PubMed] [Google Scholar]
- 36.Murphy W., Zhang T., B Q., Naler L., Ma S., and Lu C., “Recent advances in the use of microfluidic technologies for single cell analysis,” Analyst 143, 60–80 (2018). 10.1039/C7AN01346A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pratt E. D., Huang C., Hawkins B. G., Gleghorn J. P., and Kirby B. J., “Rare cell capture in microfluidic devices,” Chem. Eng. Sci. 66, 1508–1522 (2011). 10.1016/j.ces.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gross A. et al. , “Technologies for single-cell isolation,” Int. J. Mol. Sci. 16, 16897–16919 (2015). 10.3390/ijms160816897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Carlo D., Aghdam N., and Lee L. P., “Single-cell enzyme concentrations, kinetics, and inhibition analysis using high-density hydrodynamic cell isolation arrays,” Anal. Chem. 78, 4925–4930 (2006). 10.1021/ac060541s [DOI] [PubMed] [Google Scholar]
- 40.Xu X., Li Z., and Nehorai A., “Finite element simulations of hydrodynamic trapping in microfluidic particle-trap array systems,” Biomicrofluidics 7, 054108 (2013). 10.1063/1.4822030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huebner A. et al. , “Static microdroplet arrays: A microfluidic device for droplet trapping, incubation and release for enzymatic and cell-based assays,” Lab Chip 9, 692–698 (2009). 10.1039/B813709A [DOI] [PubMed] [Google Scholar]
- 42.Chen Y. et al. , “Rare cell isolation and analysis in microfluidics,” Lab Chip 14, 626–645 (2014). 10.1039/c3lc90136j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarioglu A. F. et al. , “A microfluidic device for label-free, physical capture of circulating tumor cell clusters,” Nat. Methods 12, 685–691 (2015). 10.1038/nmeth.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao W. et al. , “Analysis of circulating tumor cells from lung cancer patients with multiple biomarkers using high-performance size-based microfluidic chip,” Oncotarget 8, 12917–12928 (2017). 10.18632/oncotarget.14203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shih K. et al. , “Microfluidic metamaterial sensor: Selective trapping and remote sensing of microparticles,” J. Appl. Phys. 121, 023102 (2017). 10.1063/1.4973492 [DOI] [Google Scholar]
- 46.Zhu L. et al. , “Cell loss in integrated microfluidic device,” Biomed. Microdevices 9, 745–750 (2007). 10.1007/s10544-007-9085-z [DOI] [PubMed] [Google Scholar]
- 47.Adam R. D., “Biology of Giardia lamblia,” Clin. Microbiol. Rev. 14, 447–475 (2001). 10.1128/CMR.14.3.447-475.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leitch G. J. and He Q., “Cryptosporidiosis—An overview,” J. Biomed. Res. 25, 1–16 (2011). 10.1016/S1674-8301(11)60001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arya S. K., Lee K. C., Bin Dah’alan D., Daniel A., and Rahman A. R. A., “Breast tumor cell detection at single cell resolution using an electrochemical impedance technique,” Lab Chip 12, 2362–2368 (2012). 10.1039/c2lc21174b [DOI] [PubMed] [Google Scholar]
- 50.Zhou J. et al. , “The label-free separation and culture of tumor cells in a microfluidic biochip,” Analyst 145(5), 1706–1715 (2020). 10.1039/C9AN02092F [DOI] [PubMed] [Google Scholar]
- 51.Kuan D.-H., Wu C.-C., Su W.-Y., and Huang N.-T., “A microfluidic device for simultaneous extraction of plasma, red blood cells, and on-chip white blood cell trapping,” Sci. Rep. 8, 15345 (2018). 10.1038/s41598-018-33738-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryley J. and Pereira-Smith O. M., “Microfluidics device for single cell gene expression analysis in Saccharomyces cerevisiae,” Yeast 23, 1065–1073 (2006). 10.1002/yea.1412 [DOI] [PubMed] [Google Scholar]
- 53.Kukhtevich I. V., Belousov K. I., Bukatin A. S., Dubina M. V., and Evstrapov A. A., “A microfluidic chip with hydrodynamic traps for in vitro microscopic investigations of single cells,” Tech. Phys. Lett. 41, 255–258 (2015). 10.1134/S1063785015030086 [DOI] [Google Scholar]
- 54.Wlodkowic D., Faley S., Zagnoni M., Wikswo J. P., and Cooper J. M., “Microfluidic single-cell array cytometry for the analysis of tumor apoptosis,” Anal. Chem. 81, 5517–5523 (2009). 10.1021/ac9008463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dura B., Liu Y., and Voldman J., “Deformability-based microfluidic cell pairing and fusion,” Lab Chip 14, 2783–2790 (2014). 10.1039/c4lc00303a [DOI] [PubMed] [Google Scholar]
- 56.Di Carlo D., Wu L. Y., and Lee L. P., “Dynamic single cell culture array,” Lab Chip 6, 1445 (2006). 10.1039/b605937f [DOI] [PubMed] [Google Scholar]
- 57.Zhang K., Chou C.-K., Xia X., Hung M.-C., and Qin L., “Block-cell-printing for live single-cell printing,” Proc. Natl. Acad. Sci. U.S.A. 111, 2948–2953 (2014). 10.1073/pnas.1313661111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu L. Y., Di Carlo D., and Lee L. P., “Microfluidic self-assembly of tumor spheroids for anticancer drug discovery,” Biomed. Microdevices 10, 197–202 (2008). 10.1007/s10544-007-9125-8 [DOI] [PubMed] [Google Scholar]
- 59.Chen J. et al. , “Microfluidic chips for cells capture using 3-D hydrodynamic structure array,” Microsyst. Technol. 20, 485–491 (2014). 10.1007/s00542-013-1933-6 [DOI] [Google Scholar]
- 60.Chen Y., Austin R. H., and Sturm J. C., “On-chip cell labelling and washing by capture and release using microfluidic trap arrays,” Biomicrofluidics 11, 054107 (2017). 10.1063/1.4985771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burger R. et al. , “Array-based capture, distribution, counting and multiplexed assaying of beads on a centrifugal microfluidic platform,” Lab Chip 12, 1289 (2012). 10.1039/c2lc21170j [DOI] [PubMed] [Google Scholar]
- 62.Fu C.-Y. et al. , “A microfluidic chip with a U-shaped microstructure array for multicellular spheroid formation, culturing and analysis,” Biofabrication 6, 015009 (2014). 10.1088/1758-5082/6/1/015009 [DOI] [PubMed] [Google Scholar]
- 63.Kim M.-C. and Klapperich C., “A new method for simulating the motion of individual ellipsoidal bacteria in microfluidic devices,” Lab Chip 10, 2464–2471 (2010). 10.1039/c003627g [DOI] [PubMed] [Google Scholar]
- 64.Kim M.-C., Wang Z., Lam R. H. W., and Thorsen T., “Building a better cell trap: Applying Lagrangian modeling to the design of microfluidic devices for cell biology,” J. Appl. Phys. 103, 044701 (2008). 10.1063/1.2840059 [DOI] [Google Scholar]
- 65.Kim M.-C. et al. , “Programmed trapping of individual bacteria using micrometre-size sieves,” Lab Chip 11, 1089 (2011). 10.1039/c0lc00362j [DOI] [PubMed] [Google Scholar]
- 66.Reshes G., Vanounou S., Fishov I., and Feingold M., “Cell shape dynamics in Escherichia coli,” Biophys. J. 94, 251–264 (2008). 10.1529/biophysj.107.104398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwon D., Kim J., Chung S., and Park I., “A nanowire-integrated microfluidic device for hydrodynamic trapping and anchoring of bacterial cells,” in 2014 IEEE 27th International Conference on Micro Electro Mechanical Systems (MEMS), San Francisco, CA, 2014 (IEEE, 2014), pp. 246–249. 10.1109/MEMSYS.2014.6765621 [DOI] [Google Scholar]
- 68.Kim H. S., Devarenne T. P., and Han A., “A high-throughput microfluidic single-cell screening platform capable of selective cell extraction,” Lab Chip 15, 2467–2475 (2015). 10.1039/C4LC01316F [DOI] [PubMed] [Google Scholar]
- 69.Kazayama Y., Teshima T., Osaki T., Takeuchi S., and Toyota T., “Integrated microfluidic system for size-based selection and trapping of giant vesicles,” Anal. Chem. 88, 1111–1116 1(2016). 10.1021/acs.analchem.5b03772 [DOI] [PubMed] [Google Scholar]
- 70.Paterson D. J., Reboud J., Wilson R., Tassieri M., and Cooper J. M., “Integrating microfluidic generation, handling and analysis of biomimetic giant unilamellar vesicles,” Lab Chip 14, 1806–1810 (2014). 10.1039/C4LC00199K [DOI] [PubMed] [Google Scholar]
- 71.Dura B. et al. , “Profiling lymphocyte interactions at the single-cell level by microfluidic cell pairing,” Nat. Commun. 6, 5940 (2015). 10.1038/ncomms6940 [DOI] [PubMed] [Google Scholar]
- 72.Skelley A. M., Kirak O., Suh H., Jaenisch R., and Voldman J., “Microfluidic control of cell pairing and fusion,” Nat. Methods 6, 147–152 (2009). 10.1038/nmeth.1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen H. et al. , “Highly-sensitive capture of circulating tumor cells using micro-ellipse filters,” Sci. Rep. 7, 1–10 (2017). 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu J. et al. , “Highly efficient microfluidic device for cell trapping and pairing towards cell-cell communication analysis,” Sens. Actuators B: Chem. 283, 685–692 (2019). 10.1016/j.snb.2018.12.078 [DOI] [Google Scholar]
- 75.Weng L. et al. , “A highly-occupied, single-cell trapping microarray for determination of cell membrane permeability,” Lab Chip 17, 4077–4088 (2017). 10.1039/C7LC00883J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mi L. et al. , “A fluidic circuit based, high-efficiency and large-scale single cell trap,” Lab Chip 16, 4507–4511 (2016). 10.1039/C6LC01120A [DOI] [PubMed] [Google Scholar]
- 77.Chen X., Cui D. F., Liu C. C., and Li H., “Microfluidic chip for blood cell separation and collection based on crossflow filtration,” Sens. Actuators B: Chem. 130, 216–221 (2008). 10.1016/j.snb.2007.07.126 [DOI] [Google Scholar]
- 78.Di H., Martin G. J. O., and Dunstan D. E., “A microfluidic system for studying particle deposition during ultrafiltration,” J. Membr. Sci. 532, 68–75 (2017). 10.1016/j.memsci.2017.03.017 [DOI] [Google Scholar]
- 79.Tan W.-H. and Takeuchi S., “A trap-and-release integrated microfluidic system for dynamic microarray applications,” Proc. Natl. Acad. Sci. U.S.A. 104, 1146–1151 (2007). 10.1073/pnas.0606625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frimat J.-P. et al. , “A microfluidic array with cellular valving for single cell co-culture,” Lab Chip 11, 231–237 (2011). 10.1039/C0LC00172D [DOI] [PubMed] [Google Scholar]
- 81.Khalili A., Ahmad A., and R M., “Numerical analysis of hydrodynamic flow in microfluidic biochip for single-cell trapping application,” Int. J. Mol. Sci. 16, 26770–26785 (2015). 10.3390/ijms161125987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bithi S. S. and Vanapalli S. A., “Microfluidic cell isolation technology for drug testing of single tumor cells and their clusters,” Sci. Rep. 7, 41707 (2017). 10.1038/srep41707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun M., Bithi S. S., and Vanapalli S. A., “Microfluidic static droplet arrays with tuneable gradients in material composition,” Lab Chip 11, 3949 (2011). 10.1039/c1lc20709a [DOI] [PubMed] [Google Scholar]
- 84.Zhou Y., Basu S., Laue E. D., and Seshia A. A., “Dynamic monitoring of single cell lysis in an impedance-based microfluidic device,” Biomed. Microdevices 18, 56 (2016). 10.1007/s10544-016-0081-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chung J., Kim Y.-J., and Yoon E., “Highly-efficient single-cell capture in microfluidic array chips using differential hydrodynamic guiding structures,” Appl. Phys. Lett. 98, 123701 (2011). 10.1063/1.3565236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guan A., Shenoy A., Smith R., and Li Z., “Streamline based design guideline for deterministic microfluidic hydrodynamic single cell traps,” Biomicrofluidics 9, 024103 (2015). 10.1063/1.4914469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lyubimov A. Y. et al. , “Capture and x-ray diffraction studies of protein microcrystals in a microfluidic trap array,” Acta Crystallogr. Sect. D: Biol. Crystallogr. 71, 928–940 (2015). 10.1107/S1399004715002308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou Y. et al. , “A microfluidic platform for trapping, releasing and super-resolution imaging of single cells,” Sens. Actuators B: Chem. 232, 680–691 (2016). 10.1016/j.snb.2016.03.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He C.-K., Chen Y.-W., Wang S.-H., and Hsu C.-H., “Hydrodynamic shuttling for deterministic high-efficiency multiple single-cell capture in a microfluidic chip,” Lab Chip 19, 1370–1377 (2019). 10.1039/C9LC00036D [DOI] [PubMed] [Google Scholar]
- 90.Sochol R. D., Dueck M. E., Li S., Lee L. P., and Lin L., “Hydrodynamic resettability for a microfluidic particulate-based arraying system,” Lab Chip 12, 5051 (2012). 10.1039/c2lc40704c [DOI] [PubMed] [Google Scholar]
- 91.Chung K., Rivet C. A., Kemp M. L., and Lu H., “Imaging single-cell signaling dynamics with a deterministic high-density single-cell trap array,” Anal. Chem. 83, 7044–7052 (2011). 10.1021/ac2011153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nourmohammadzadeh M. et al. , “A microfluidic array for real-time live-cell imaging of human and rodent pancreatic islets,” Lab Chip 16, 1466–1472 (2016). 10.1039/C5LC01173F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee D.-H., Li X., Ma N., Digman M. A., and Lee A. P., “Rapid and label-free identification of single leukemia cells from blood in a high-density microfluidic trapping array by fluorescence lifetime imaging microscopy,” Lab Chip 18, 1349–1358 (2018). 10.1039/C7LC01301A [DOI] [PubMed] [Google Scholar]
- 94.Wilson J. L. et al. , “Single-cell analysis of embryoid body heterogeneity using microfluidic trapping array,” Biomed. Microdevices 16, 79–90 (2014). 10.1007/s10544-013-9807-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lawrenz A., Nason F., and Cooper-White J. J., “Geometrical effects in microfluidic-based microarrays for rapid, efficient single-cell capture of mammalian stem cells and plant cells,” Biomicrofluidics 6, 024112 (2012). 10.1063/1.4704521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chung K. et al. , “A microfluidic array for large-scale ordering and orientation of embryos,” Nat. Methods 8, 171–176 (2011). 10.1038/nmeth.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saravia F. et al. , “Differences in boar sperm head shape and dimensions recorded by computer-assisted sperm morphometry are not related to chromatin integrity,” Theriogenology 68, 196–203 (2007). 10.1016/j.theriogenology.2007.04.052 [DOI] [PubMed] [Google Scholar]
- 98.de Wagenaar B. et al. , “Microfluidic single sperm entrapment and analysis,” Lab Chip 15, 1294–1301 (2015). 10.1039/C4LC01425A [DOI] [PubMed] [Google Scholar]
- 99.Kim J., Erath J., Rodriguez A., and Yang C., “A high-efficiency microfluidic device for size-selective trapping and sorting,” Lab Chip 14, 2480–2490 (2014). 10.1039/C4LC00219A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Valero A. et al. , “Apoptotic cell death dynamics of HL60 cells studied using a microfluidic cell trap device,” Lab Chip 5, 49 (2005). 10.1039/b415813j [DOI] [PubMed] [Google Scholar]
- 101.Espulgar W. et al. , “Single cell trapping and cell–cell interaction monitoring of cardiomyocytes in a designed microfluidic chip,” Sens. Actuators B: Chem. 207, 43–50 (2015). 10.1016/j.snb.2014.09.068 [DOI] [Google Scholar]
- 102.Hosokawa M. et al. , “High-Density microcavity array for cell detection: Single-cell analysis of hematopoietic stem cells in peripheral blood mononuclear cells,” Anal. Chem. 81, 5308–5313 (2009). 10.1021/ac900535h [DOI] [PubMed] [Google Scholar]
- 103.Hosokawa M. et al. , “Size-Selective microcavity array for rapid and efficient detection of circulating tumor cells,” Anal. Chem. 82, 6629–6635 (2010). 10.1021/ac101222x [DOI] [PubMed] [Google Scholar]
- 104.Hosokawa M. et al. , “Size-based isolation of circulating tumor cells in lung cancer patients using a microcavity array system,” PLoS One 8, e67466 (2013). 10.1371/journal.pone.0067466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Deng B. et al. , “Parameter screening in microfluidics based hydrodynamic single-cell trapping,” Sci. World J. 2014, 929163 (2014). 10.1155/2014/929163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moeller H.-C., Mian M. K., Shrivastava S., Chung B. G., and Khademhosseini A., “A microwell array system for stem cell culture,” Biomaterials 29, 752–763 (2008). 10.1016/j.biomaterials.2007.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Park M. C., Hur J. Y., Cho H. S., Park S.-H., and Suh K. Y., “High-throughput single-cell quantification using simple microwell-based cell docking and programmable time-course live-cell imaging,” Lab Chip 11, 79–86 (2011). 10.1039/C0LC00114G [DOI] [PubMed] [Google Scholar]
- 108.Wood D. K., Weingeist D. M., Bhatia S. N., and Engelward B. P., “Single cell trapping and DNA damage analysis using microwell arrays,” Proc. Natl. Acad. Sci. U.S.A. 107, 10008–10013 (2010). 10.1073/pnas.1004056107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hwang Y.-S. et al. , “Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11,” Proc. Natl. Acad. Sci. U.S.A. 106, 16978–16983 (2009). 10.1073/pnas.0905550106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Karp J. M. et al. , “Controlling size, shape and homogeneity of embryoid bodies using poly(ethylene glycol) microwells,” Lab Chip 7, 786–794 (2007). 10.1039/b705085m [DOI] [PubMed] [Google Scholar]
- 111.Wang Y., Shah P., Phillips C., Sims C. E., and Allbritton N. L., “Trapping cells on a stretchable microwell array for single-cell analysis,” Anal. Bioanal. Chem. 402, 1065–1072 (2012). 10.1007/s00216-011-5535-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim S.-H., Lee G. H., and Park J. Y., “Microwell fabrication methods and applications for cellular studies,” Biomed. Eng. Lett. 3, 131–137 (2013). 10.1007/s13534-013-0105-z [DOI] [Google Scholar]
- 113.Khademhosseini A. et al. , “Co-culture of human embryonic stem cells with murine embryonic fibroblasts on microwell-patterned substrates,” Biomaterials 27, 5968–5977 (2006). 10.1016/j.biomaterials.2006.06.035 [DOI] [PubMed] [Google Scholar]
- 114.Rettig J. R. and Folch A., “Large-scale single-cell trapping and imaging using microwell arrays,” Anal. Chem. 77, 5628–5634 (2005). 10.1021/ac0505977 [DOI] [PubMed] [Google Scholar]
- 115.Randall L., V C., Kalinin Y., Jamal M., Manohar T., and Gracias D. H., “Three-dimensional microwell arrays for cell culture,” Lab Chip 11, 127–131 (2011). 10.1039/C0LC00368A [DOI] [PubMed] [Google Scholar]
- 116.Rosenthal A., Macdonald A., and Voldman J., “Cell patterning chip for controlling the stem cell microenvironment,” Biomaterials 28, 3208–3216 (2007). 10.1016/j.biomaterials.2007.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shinohara M. et al. , “Combination of microwell structures and direct oxygenation enables efficient and size-regulated aggregate formation of an insulin-secreting pancreatic β-cell line,” Biotechnol. Prog. 30, 178–187 (2014). 10.1002/btpr.1837 [DOI] [PubMed] [Google Scholar]
- 118.Park J. Y. et al. , “Single cell trapping in larger microwells capable of supporting cell spreading and proliferation,” Microfluid. Nanofluidics 8, 263–268 (2010). 10.1007/s10404-009-0503-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Manbachi A. et al. , “Microcirculation within grooved substrates regulates cell positioning and cell docking inside microfluidic channels,” Lab Chip 8, 747 (2008). 10.1039/b718212k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tongmanee T. et al. , “Development of a triangular micro well for single cell trapping—Computational study,” in 2015 8th Biomedical Engineering International Conference (BMEiCON) 2015 (IEEE, 2015), pp. 1–4. 10.1109/BMEiCON.2015.7399540 [DOI] [Google Scholar]
- 121.Kim C., Bang J. H., Kim Y. E., Lee S. H., and Kang J. Y., “On-chip anticancer drug test of regular tumor spheroids formed in microwells by a distributive microchannel network,” Lab Chip 12, 4135 (2012). 10.1039/c2lc40570a [DOI] [PubMed] [Google Scholar]
- 122.Choi Y. Y. et al. , “Controlled-size embryoid body formation in concave microwell arrays,” Biomaterials 31, 4296–4303 (2010). 10.1016/j.biomaterials.2010.01.115 [DOI] [PubMed] [Google Scholar]
- 123.Lee S. W. et al. , “A cell-loss-free concave microwell array based size-controlled multi-cellular tumoroid generation for anti-cancer drug screening,” PLoS One 14, e0219834 (2019). 10.1371/journal.pone.0219834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee G. H. et al. , “Networked concave microwell arrays for constructing 3D cell spheroids,” Biofabrication 10, 015001 (2017). 10.1088/1758-5090/aa9876 [DOI] [PubMed] [Google Scholar]
- 125.Lee G.-H. et al. , “Deformable L-shaped microwell array for trapping pairs of heterogeneous cells,” J. Micromech. Microeng. 25, 035005 (2015). 10.1088/0960-1317/25/3/035005 [DOI] [Google Scholar]
- 126.Chin V. I. et al. , “Microfabricated platform for studying stem cell fates,” Biotechnol. Bioeng. 88, 399–415 (2004). 10.1002/bit.20254 [DOI] [PubMed] [Google Scholar]
- 127.Zheng L.-N. et al. , “Determination of silver nanoparticles in single cells by microwell trapping and laser ablation ICP-MS determination,” J. Anal. At. Spectrom. 34, 915–921 (2019). 10.1039/C8JA00438B [DOI] [Google Scholar]
- 128.Labanieh L., Nguyen T. N., Zhao W., and Kang D.-K., “Floating droplet array: An ultrahigh-throughput device for droplet trapping, real-time analysis and recovery,” Micromachines 6, 1469–1482 (2015). 10.3390/mi6101431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang L., Chen Y., Chen Y., and Wu H., “Centrifugation-Assisted single-cell trapping in a truncated cone-shaped microwell array chip for the real-time observation of cellular apoptosis,” Anal. Chem. 87, 12169–12176 (2015). 10.1021/acs.analchem.5b03031 [DOI] [PubMed] [Google Scholar]
- 130.Khademhosseini A. et al. , “Cell docking inside microwells within reversibly sealed microfluidic channels for fabricating multiphenotype cell arrays,” Lab Chip 5, 1380–1386 (2005). 10.1039/b508096g [DOI] [PubMed] [Google Scholar]
- 131.Jen C.-P., Hsiao J.-H., and Maslov N. A., “Single-cell chemical lysis on microfluidic chips with arrays of microwells,” Sensors 12, 347–358 (2011). 10.3390/s120100347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lin C.-H. et al. , “A microfluidic dual-well device for high-throughput single-cell capture and culture,” Lab Chip 15, 2928–2938 (2015). 10.1039/C5LC00541H [DOI] [PubMed] [Google Scholar]
- 133.Tokimitsu Y. et al. , “Single lymphocyte analysis with a microwell array chip,” Cytometry A 71A, 1003–1010 (2007). 10.1002/cyto.a.20478 [DOI] [PubMed] [Google Scholar]
- 134.Deutsch M. et al. , “A novel miniature cell retainer for correlative high-content analysis of individual untethered non-adherent cells,” Lab Chip 6, 995 (2006). 10.1039/b603961h [DOI] [PubMed] [Google Scholar]
- 135.Cinque L., Yamada A., Ghomchi Y., Baigl D., and Chen Y., “Cell trapping, DNA extraction and molecular combing in a microfluidic device for high throughput genetic analysis of human DNA,” Microelectron. Eng. 88, 1733–1736 (2011). 10.1016/j.mee.2010.12.058 [DOI] [Google Scholar]
- 136.Wang Y. et al. , “A microfluidic digital single-cell assay for the evaluation of anticancer drugs,” Anal. Bioanal. Chem. 407, 1139–1148 (2015). 10.1007/s00216-014-8325-3 [DOI] [PubMed] [Google Scholar]
- 137.Chen H., Sun J., Wolvetang E., and Cooper-White J., “High-throughput, deterministic single cell trapping and long-term clonal cell culture in microfluidic devices,” Lab Chip 15, 1072–1083 (2015). 10.1039/C4LC01176G [DOI] [PubMed] [Google Scholar]
- 138.Kamyabi N. et al. , “A microfluidic device for label-free isolation of tumor cell clusters from unprocessed blood samples,” Biomicrofluidics 13, 044111 (2019). 10.1063/1.5111888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Luo C. et al. , “A fast cell loading and high-throughput microfluidic system for long-term cell culture in zero-flow environments,” Biotechnol. Bioeng. 101, 190–195 (2008). 10.1002/bit.21877 [DOI] [PubMed] [Google Scholar]
- 140.Ren X. et al. , “Entrapment of prostate cancer circulating tumor cells with a sequential size-based microfluidic chip,” Anal. Chem. 90, 7526–7534 (2018). 10.1021/acs.analchem.8b01134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shi Y. et al. , “High throughput generation and trapping of individual agarose microgel using microfluidic approach,” Microfluid. Nanofluidics 15, 467–474 (2013). 10.1007/s10404-013-1160-6 [DOI] [Google Scholar]
- 142.Zhou J., Mukherjee P., Gao H., Luan Q., and Papautsky I., “Label-free microfluidic sorting of microparticles,” APL Bioeng. 3, 041504 (2019). 10.1063/1.5120501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Banaeiyan A. A., Ahmadpour D., Adiels C. B., and Goksör M., “Hydrodynamic cell trapping for high throughput single-cell applications,” Micromachines 4, 414–430 (2013). 10.3390/mi4040414 [DOI] [Google Scholar]
- 144.Kulasinghe A., Zhou J., Kenny L., Papautsky I., and Punyadeera C., “Capture of circulating tumour cell clusters using straight microfluidic chips,” Cancers 11, 89 (2019). 10.3390/cancers11010089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhou J. et al. , “Isolation of circulating tumor cells in non-small-cell-lung-cancer patients using a multi-flow microfluidic channel,” Microsyst. Nanoeng. 5, 1–12 (2019). 10.1038/s41378-019-0045-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhou J. et al. , “Isolation of cells from whole blood using shear-induced diffusion,” Sci. Rep. 8, 9411 (2018). 10.1038/s41598-018-27779-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Matsunaga T. et al. , “High-efficiency single-cell entrapment and fluorescence in situ hybridization analysis using a poly(dimethylsiloxane) microfluidic device integrated with a black poly(ethylene terephthalate) micromesh,” Anal. Chem. 80, 5139–5145 (2008). 10.1021/ac800352j [DOI] [PubMed] [Google Scholar]
- 148.Dong L. et al. , “Integrated microfluidic device for drug studies of early C. elegans embryogenesis,” Adv. Sci. 5, 1700751 (2018). 10.1002/advs.201700751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Brouzes E. et al. , “Droplet microfluidic technology for single-cell high-throughput screening,” Proc. Natl. Acad. Sci. U.S.A. 106, 14195–14200 (2009). 10.1073/pnas.0903542106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dewan A., Kim J., McLean R. H., Vanapalli S. A., and Karim M. N., “Growth kinetics of microalgae in microfluidic static droplet arrays,” Biotechnol. Bioeng. 109, 2987–2996 (2012). 10.1002/bit.24568 [DOI] [PubMed] [Google Scholar]
- 151.Liberale C. et al. , “Integrated microfluidic device for single-cell trapping and spectroscopy,” Sci. Rep. 3, 1258 (2013). 10.1038/srep01258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Karimi A., Yazdi S., and Ardekani A. M., “Hydrodynamic mechanisms of cell and particle trapping in microfluidics,” Biomicrofluidics 7, 021501 (2013). 10.1063/1.4799787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Miller M. C., Doyle G. V., and Terstappen L. W. M. M., “Significance of circulating tumor cells detected by the CellSearch system in patients with metastatic breast colorectal and prostate cancer,” J. Oncol. 2010, 1–8 (2010). 10.1155/2010/617421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mutlu B. R. et al. , “In-flow measurement of cell-cell adhesion using oscillatory inertial microfluidics,” Lab Chip 20, 1612–1620 (2020). 10.1039/D0LC00089B [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created in this study.