SUMMARY
The persistence of anti-viral immunity is essential for protection and exhibits profound heterogeneity across individuals. Here, we elucidate the factors that shape maintenance and function of anti-viral T cell immunity in the body by comprehensive profiling of virus-specific T cells across blood, lymphoid organs, and mucosal tissues of organ donors. We use flow cytometry, T cell receptor sequencing, single-cell transcriptomics, and cytokine analysis to profile virus-specific CD8+ T cells recognizing the ubiquitous pathogens influenza and cytomegalovirus. Our results reveal that virus specificity determines overall magnitude, tissue distribution, differentiation, and clonal repertoire of virus-specific T cells. Age and sex influence T cell differentiation and dissemination in tissues, while T cell tissue residence and functionality are highly correlated with the site. Together, our results demonstrate how the covariates of virus, tissue, age, and sex impact the anti-viral immune response, which is important for targeting, monitoring, and predicting immune responses to existing and emerging viruses.
In brief
Through comprehensive cellular and molecular analysis of virus-specific T cells in circulation and across multiple lymphoid and mucosal tissues, Poon et al. elucidate how maintenance and function of the human anti-viral immune response against ubiquitous viruses influenza and cytomegalovirus are shaped by virus, tissue, age, and sex.
Graphical Abstract
INTRODUCTION
The human immune response to virus infection is remarkably heterogeneous. As starkly revealed in the global coronavirus disease 2019 (COVID-19) pandemic where over 170 million people became infected with the novel respiratory virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), some people experienced mild or no symptoms, while others succumbed to severe disease. There is a well-established association of COVID-19 severity with age and sex; children are largely spared from disease, older adults are more vulnerable with higher mortality, and women fare better than men (Wu and McGoogan, 2020). The factors that drive the magnitude, quality, and protective efficacy of the human immune response to this and other viruses remain unknown. Moreover, the age- and sex-related influences on anti-viral immunity are incompletely understood and important for promoting protective immunity to current and future pathogens.
T lymphocytes are essential for anti-viral immunity, and tissue localization is critical for T cell function and maintenance. During a virus infection, dendritic cells at the tissue site of infection migrate to the tissue draining lymph node (LN), where they present viral antigens to activate naive, virus-specific T cells to proliferate and differentiate to tissue-homing effector T cells for coordinating viral clearance through multiple mechanisms. After resolution of infection, expanded populations of virus-specific T cells die by contraction or persist as memory subsets, including central memory (TCM) and effector memory (TEM) cells, which migrate through lymphoid and non-lymphoid/peripheral tissues, respectively, or become terminal effector cells (TEMRAs) (Masopust et al., 2001; Sallusto et al., 1999, 2004). Memory T cells can also persist as non-circulating, tissue-resident subsets (TRM cells) in multiple non-lymphoid and lymphoid sites; TRM cells are distinguished from circulating memory T cells based on their specific retention in tissues, expression of canonical markers CD69 and CD103, and a distinct transcriptional profile (for reviews, see Mackay and Kallies, 2017; Masopust and Soerens, 2019; Szabo et al., 2019b). Importantly, both CD4+ and CD8+ TRM cells can mediate optimal protective immunity to viruses in mice (Paik and Farber, 2021a), including influenza and SARS-CoV in the lung (Paik and Farber, 2021b; Teijaro et al., 2011; Wu et al., 2014; Zhao et al., 2016) and lymphocytic choriomeningitis virus (LCMV) in mucosal sites (Schenkel et al., 2014). These studies demonstrate the importance of tissue-localized T cells in maintaining and directing anti-viral protective immunity.
Much of our knowledge of human anti-viral immunity, including innate responses, cytokines, antibodies, and T cells, is limited to blood (for a review, see Pulendran and Davis, 2020). However, blood contains a small fraction of the total immune cells throughout the body and lacks tissue-resident immune cells, including TRM (Farber, 2021; Poon and Farber, 2020; Weisberg et al., 2021b). The generation of memory T cell responses to infection and vaccination can be followed in peripheral blood (Akondy et al., 2017; Dan et al., 2021; Graham et al., 2020; Hammarlund et al., 2003; Thom et al., 2021), though studies of infection or vaccination sites have revealed distinct dynamics, functions, and immune cell compositions compared with blood (Guvenel et al., 2020; Patel et al., 2018; Szabo et al., 2021; Poon et al., 2021). Defining the full heterogeneity of virus-specific T cell responses therefore requires comprehensive profiling across multiple sites.
We have established a human tissue resource to obtain blood and multiple lymphoid and mucosal tissues from individual organ donors of all ages, enabling investigations of immune cells and responses across tissues over life (for reviews, see Poon and Farber, 2020; Weisberg et al., 2021b). Our prior studies using these tissues reveal that T cell subset composition, phenotype, function, and transcriptome profiles are specific to the tissue and exhibit site-specific variations with age (Kumar et al., 2018). We identified TRM cells (expressing specific phenotypes and transcriptional profiles) as the predominant T cell subset in mucosal, exocrine, and barrier sites throughout the body across age (Kumar et al., 2017, 2018; Thome et al., 2014). Analysis of T cells specific for cytomegalovirus (CMV), a systemic persistent virus, revealed broad distribution of CMV-specific T cells across sites that varied between individuals (Gordon et al., 2017), although the bases for these variations were not clear.
Here, we present a comprehensive cellular and molecular analysis of virus-specific T cells in blood and tissues from individual donors of all ages, revealing the role of virus, site, age, and sex in shaping anti-viral T cell immunity. We examine CD8+ T cells specific for prevalent but distinct viruses—influenza A (flu) virus as a ubiquitous acute virus confined to the respiratory tract, compared with CMV—in blood, primary and secondary lymphoid organs, and mucosal sites from 58 organ donors aged 1–78 years. We show that virus specificity and tissue localization are the key factors in shaping anti-viral T cell immune responses in the human body, with age and sex influencing T cell subset differentiation. Importantly, virus specificity is correlated with T cell tissue tropism and the clonal repertoire, while tissue localization is associated with differential maintenance of T cell subsets and functional responses. Our findings elucidate how different factors contribute to the heterogeneity of the human anti-viral immune response, which is important for designing targeted approaches to protect against new and emerging viruses.
RESULTS
Heterogeneous maintenance of virus-specific CD8+ T cells across human tissue sites
We analyzed virus-specific T cells across blood and multiple tissue sites of individual organ donors directly ex vivo by examining their frequency, distribution, phenotype, T cell receptor (TCR) expression, and functional profile (see schematic in Figure 1A). Mononuclear cells were isolated from blood, bone marrow (BM), spleen, lung, lung draining lymph node (LLN), iliac lymph node (ILN), mesenteric lymph node (MLN), and jejunum obtained from 58 donors, aged 1–78 y (Table S1), as previously described (Carpenter et al., 2018; Gordon et al., 2017; Granot et al., 2017; Kumar et al., 2017; Sathaliyawala et al., 2013; Thome et al., 2014). For detection of T cells specific for the ubiquitous viruses, flu and CMV, we used fluorescently labeled multimer reagents consisting of MHC class I receptor proteins (HLA-A1, -A2, -A24, -B7) complexed with immunodominant peptide epitopes encountered during natural infection as previously determined (flu: MP, NP, PB1; CMV: IE-1, pp50, pp65; Alam and Sant, 2011; Assarsson et al., 2008; Malik et al., 2017), compared with staining with negative control HLA-multimers containing irrelevant peptides (Figure S1). Only CMV-seropositive individuals exhibited significant frequencies of CMV-specific T cells across sites (Gordon et al., 2017), while flu-specific T cells were identified in nearly all donors.
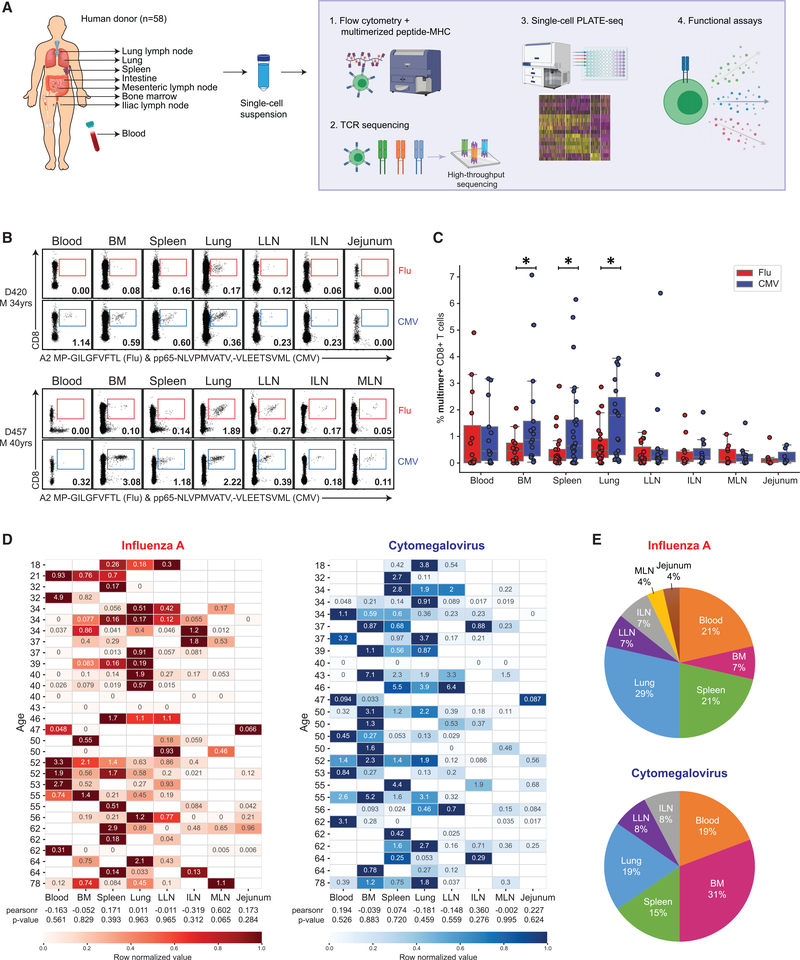
Figure 1. Differential maintenance of flu- and CMV-specific CD8+ T cells across diverse tissue sites.
(A) Schematic diagram illustrating human tissues obtained and experimental workflow for this study.
(B) Distribution of influenza A (flu)-specific (red) and cytomegalovirus (CMV)-specific (blue) CD8+ T cells in different human tissues of two representative donors (D420, top; D457, bottom) based on staining with multimer reagents containing viral epitopes shown in representative flow cytometry plots (see Figure S1 for gating strategy). Numbers indicate frequency of multimer+ cells within total CD8+ T cells.
(C) Frequencies of flu-multimer+ (red) and CMV-multimer+ (blue) CD8+ T cells from 7–27 donors for each tissue site.
(D) Heatmaps showing frequency of flu-specific (left) and CMV-specific (right) CD8+ T cells in blood and tissues sites for individual donors from which data from two or more tissues were obtained. Donors are arranged by increasing age, and color intensity of each cell is based on row normalization of (minimum to maximum [min-max] scaled) values (white cells indicate no sample). Pearson correlation analysis of age and multimer frequency is indicated by “pearsonr” correlation coefficients, and p values are listed underneath each heatmap.
(E) Pie charts showing percentage of donors for which the indicated tissue contains the greatest frequency of flu-multimer+ (top) or CMV-multimer+ (bottom) cells among the tissue sites studied for that donor.
Statistical significance for comparison of means was calculated by unpaired t test and indicated by *p ≤ 0.05. BM, bone marrow; ILN, iliac lymph node; LLN, lung draining lymph node; M, male; MLN, mesenteric lymph node.
Based on multimer staining, we found differences in the frequency and tissue distribution of flu- compared to CMV-specific T cells, which also varied between individuals. Both flu- and CMV-specific T cells were distributed in multiple sites, particularly in blood, BM, spleen, lungs, and LN, with very low-to-negligible frequencies in intestines (Figures 1B and 1C). In the BM, spleen, and lung, there was a significantly greater frequency of CMV-compared with flu-specific CD8+ T cells, which also varied between donors (Figure 1C). This finding may relate to CMV being a persistent virus which has been detected in these sites in seropositive individuals (Gordon et al., 2017). Heatmaps with color intensities based on row-normalized frequencies of virus-specific T cells revealed that certain sites contained higher frequencies of flu- or CMV-specific CD8+ T cells within each individual, which varied between individuals, but not with age (Figure 1D). From this analysis, a substantial frequency of donors (29%) was found to have the highest frequency of flu-specific T cells in the lung relative to other sites, while for CMV-specific T cells, a substantial frequency (31%) of donors had highest frequencies in the BM compared with other sites (Figure 1E). These results indicate that the frequency and tissue distribution of virus-specific T cells is shaped in part by the tissue tropism of the virus.
As another approach for detection of flu- and CMV-specific T cells independent of HLA type, we stimulated mononuclear T cells from multiple sites with peptide megapools (MPs) that allow simultaneous presentation of a large number of virus-specific epitopes predicted using the Immune Epitope Database and Analysis Resource (IEDB) and bioinformatic approaches (see STAR Methods) (Carrasco Pro et al., 2015; Vita et al., 2019; Weiskopf et al., 2015, 2020). T cell reactivity to each virus was assessed by production of multiple cytokines and cytolytic mediators by intracellular staining (Figure S2A). Consistent with our results from multimer analysis, we were able to detect flu- and CMV-reactive T cells in the BM, spleen, lungs, and LLN of multiple donors, with variable responses between donors (Figures S2B and S2C). Notably, flu-responsive T cells in the lung and LLN exhibited features of multifunctional memory T cells (Figures S2B and S2C), and highly elevated frequencies of CMV-reactive T cells were detected in the BM of some donors (Figure S2C). Together, these results indicate that flu- and CMV-reactive T cells are dispersed across multiple sites, although flu-specific memory T cells are more biased to the lung and associated LN.
Role of tissue localization, virus specificity, and age in subset differentiation of virus-specific CD8+ T cells
To understand the factors controlling maintenance of anti-viral T cells, we investigated the subset differentiation and phenotype of flu- and CMV-specific CD8+ T cells across tissues and age (18–78 y). We previously determined that tissue sites contain distinct compositions of memory T cell subsets as defined by cell surface expression of CCR7 chemokine receptor and CD45RA: TCM (CD45RA−CCR7+); TEMRA (CD45RA+CCR7−); and TEM (CD45RA−CCR7−), which is further delineated into circulating CD69− TEM and non-circulating CD69+ CD103+ TRM cells (Figure S1A) (Sathaliyawala et al., 2013; Thome et al., 2014). The subset composition of virus-specific T cells in each site is shown relative to the profile of polyclonal T cells (Figure 2A) and as compiled frequencies for both flu- and CMV-specific T cells (Figure 2B; see Table S3 for individualized data for each donor). Overall, the subset composition of flu- or CMV-specific T cells within a site was distinct from the subset composition of polyclonal T cells in that site and often differed between the two virus specificities (Figure 2A).
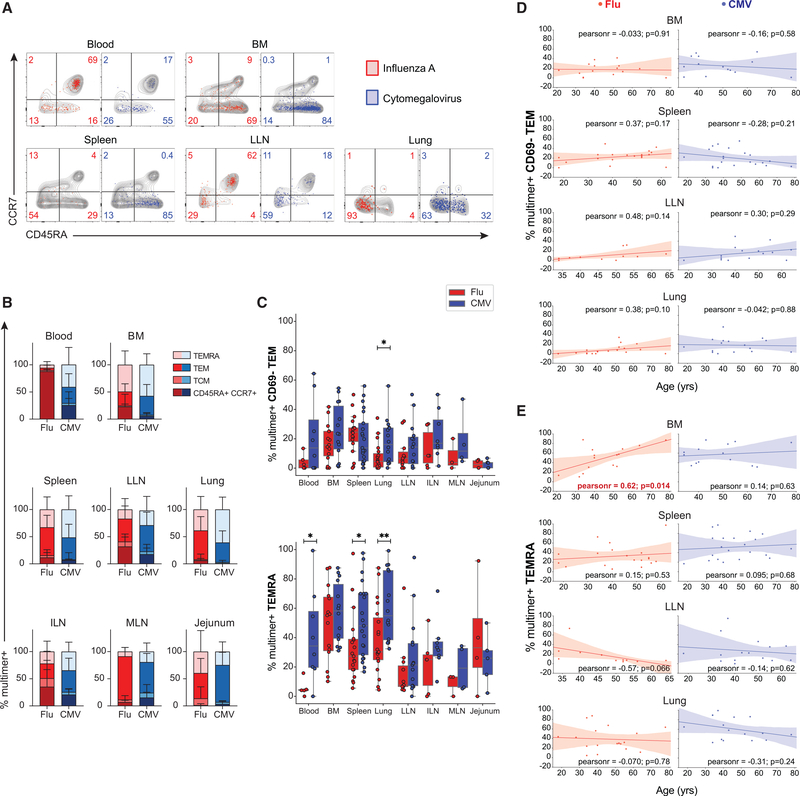
Figure 2. Antigen-specific CD8+ T cell subset differentiation and phenotype are based on virus specificity independent of age.
(A) T cell subset distribution of flu-specific (red) and CMV-specific(blue) CD8+ T cells based on CD45RA and CCR7 expression (CD45RA+/CCR7+; CD45RA−/CCR7+, TCM; CD45RA−/CCR7−, TEM; CD45RA+/CCR7−, TEMRA) shown in representative flow cytometry plots. Gray contour plots depict CD45RA and CCR7 expression of total CD8+ T cells within the same sample. Numbers indicate frequency of multimer+ virus-specific cells within each subset.
(B) Subset distribution of flu-specific (shades of red) and CMV-specific (shades of blue) CD8+ T cells compiled from 3–21 donors in blood and indicated tissue sites.
(C) Frequencies of flu-multimer+ (red) and CMV-multimer+ (blue) CD8+ T cells maintained as CD69− TEM cells (top) and TEMRAs (bottom) from 3–21 donors for each tissue site.
(D and E) Frequencies of flu-specific (red, left) and CMV-specific (blue, right) CD8+CD69− TEM cells (D) and CD8+ TEMRAs (E) in indicated tissue sites as a function of age for each individual donor. The line of best fit was determined by Pearson correlation; “pearsonr” refers to correlation coefficient, p values are indicated for each comparison, and red font indicates significant correlation for BM.
Statistical significance for comparison of means was calculated by unpaired t test and indicated by **p ≤ 0.01; *p ≤ 0.05. Subset frequencies shown from donors with ≥10 multimer+ T cells.
The majority of both flu- and CMV-specific T cells were maintained as CD45RA− CCR7− TEM cells and CD45RA+CCR7− TEMRAs, although the relative distribution of these subsets varied between sites and virus specificity. There were also flu-specific CD45RA+CCR7+ T cells in the blood and LNs of some donors, a phenotype usually associated with naive T cells but also shown to be expressed by long-lived memory T cells in circulation (Akondy et al., 2017). The majority of flu-specific T cells were TEM cells in spleen, lungs, multiple LNs, and jejunum, while flu-specific TEMRAs were present mostly in BM (Figure 2B). CMV-specific T cells were maintained predominantly as TEMRAs in the BM, spleen, and lung and as TEM cells in LNs (Figure 2B). Between the different virus specificities, there were significantly higher frequencies of CMV-specific compared with flu-specific circulating CD69− TEM subset in the lung (Figure 2C). Additionally, higher frequencies of CMV-specific compared with flu-specific TEMRAs were detected in multiple sites (blood, spleen, and lung) (Figure 2C). The frequency of flu- or CMV-specific T cells that are CD69− TEM or TEMRAs in each site did not vary significantly with age, with the exception of flu-specific TEMRAs in BM, which increased significantly with age (Figures 2D and 2E). Similar frequencies of circulating CD69− TEM and TEMRAs were observed for donors where detection of virus-specific T cells involved use of multimers for HLA-A2 or multiple HLA alleles (Figures S3A and S3B). Because CD69− TEM and TEMRAs are circulating subsets, with TEMRAs found at high frequencies in blood-rich sites (Miron et al., 2021;Thome et al., 2014), these results are consistent with CMV-reactive T cells being more circulating compared with flu-specific T cells.
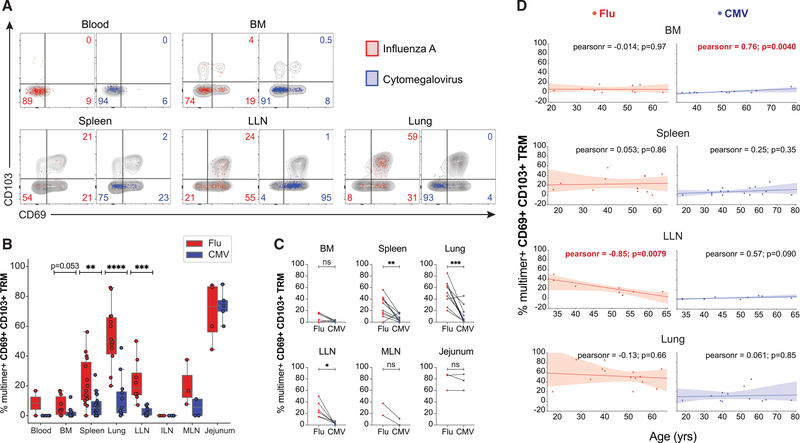
We examined the extent to which flu- and CMV-specific TEM cells exhibit phenotypes of tissue residence based on the expression of tissue residency markers CD69 and CD103, which distinguish human TRM from circulating TEM cells across tissue sites (Kumar et al., 2017). Virus-specific TRM cells were detected only in tissues and not blood (Figures 3A and 3B). Importantly, a higher frequency of flu-specific T cells was maintained as TRM in spleen, lung, and LLN compared with low-to-negligible frequencies of CMV-specific TRM in these and other sites (Figure 3B); this difference was observed across donors where detection of virus-specific T cells involved use of multimers for HLA-A2 or multiple HLA alleles (Figure S3C). The frequency of flu-specific TRM cells was highest in the lung (Figure 3B), consistent with mouse studies (Teijaro et al., 2011; Turner et al., 2014). Paired analysis for individual donors revealed that flu-specific TRM cells are maintained at higher frequencies than CMV-specific TRM cells for each site, with the exception of the jejunum, where the low frequencies of virus-specific T cells detected were all TRM cells (Figure 3C), consistent with TRM predominance in that site (Kumar et al., 2017; Weisberg et al., 2019). With age, the frequency of virus-specific TRM cells remained constant over six decades of life, except for a marked decrease in frequency of flu-specific TRM cells in the LLN and a slight accumulation of CMV-specific TRM cells in the BM (Figure 3D). These results suggest that long-term maintenance of tissue residency of virus-specific CD8+ T cells is shaped primarily by the nature of the infection independent of age.
Figure 3. Establishment of tissue residency correlates with virus specificity and site.
(A) Expression of tissue residency markers CD69 and CD103 by flu-specific (red) and CMV-specific (blue) CD8+ T cells in indicated tissue sites shown in representative flow cytometry plots with gray contour plots depicting total CD8+ T cells within the same sample; numbers indicate frequency of multimer+ virus-specific cells.
(B) Frequencies of flu-multimer+ (red) and CMV-multimer+ (blue) CD69+CD103+CD8+ TRM cells gated on TEM cells from 2–18 donors for each site. Statistical significance for comparison of means was calculated by unpaired t test and indicated by ****p ≤ 0.0001; ***p ≤ 0.001; **p ≤ 0.01.
(C) Paired frequencies within individual donors of flu-multimer+ (red) and CMV-multimer+ (blue) CD69+ CD103+ CD8+ TRM cells from 2–10 donors for each site. Statistical significance between flu- and CMV-multimer+ TRM cells within each tissue site was determined by paired t test and indicated by ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; ns, not significant.
(D) Frequencies of flu-specific (red, left) and CMV-specific (blue, right) TRM cells in indicated tissue sites as a function of age for each individual donor. The line of best fit, Pearson coefficient, and p value are indicated for each comparison; red font indicates significant correlation for LLN. TRM cell frequencies shown from donors with ≥5 multimer+ TEM cells.
Distinct flu-specific CD8+ T cell responses in tissues of pediatric versus adult donors
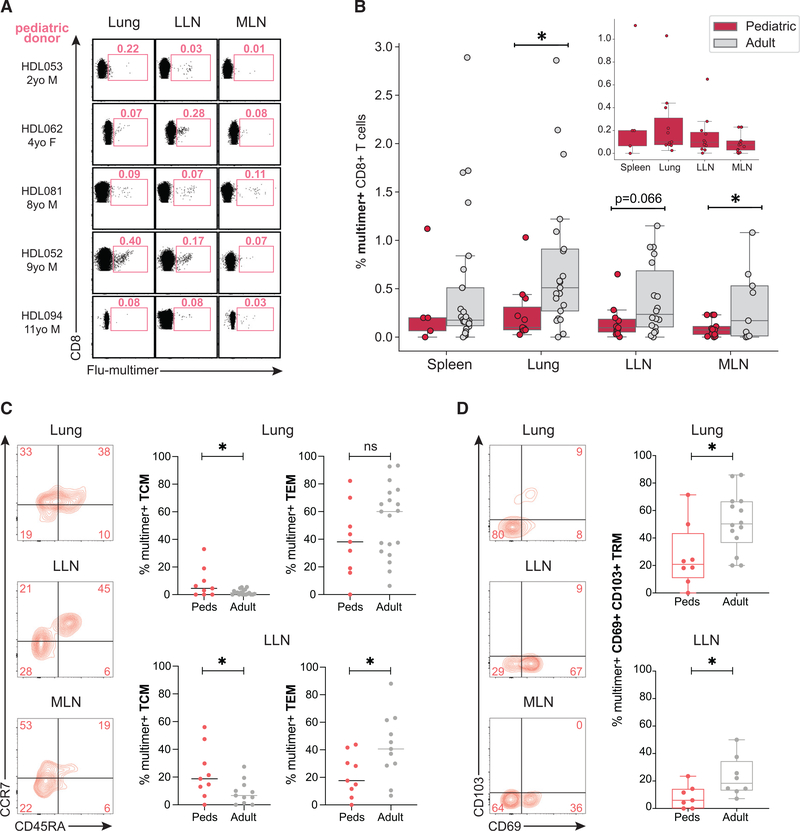
To investigate further the role of age in the differentiation of virus-specific T cells, we analyzed flu-specific T cells in tissues from a cohort of 13 pediatric organ donors, aged 1–11 y (Table S1). We detected flu-specific T cells by multimer staining in the spleen, lung, LLN, and MLN of pediatric donors; the highest frequencies were in the lung and LLN (Figure 4A). However, the frequencies of flu-multimer+ CD8+ T cells in pediatric donor tissues were significantly lower compared with corresponding frequencies in adult donors (Figure 4B). Moreover, the subset distribution and tissue residency also differed between pediatric and adult donors; flu-specific CD8+ T cells in lung and LLN of pediatric donors were maintained at higher frequencies as TCM and lower frequencies as CD69+ CD103+ TRM compared with adult donors (Figures 4C and 4D; see Table S3 for individualized data). These findings demonstrate distinct differences in flu-specific memory T cell generation between children and adults, which could be a result of intrinsic effects of age and/or accumulated exposures to influenza A virus.
Figure 4. Distinct frequencies and subset distribution of flu-specific memory CD8+ T cells between adult and pediatric donors.
(A) Distribution of flu-specific CD8+ T cells in lung, LLN, and MLN of pediatric and adult donors shown in representative flow cytometry plots. Numbers indicate frequency of multimer+ cells within total CD8+ T cell population.
(B) Frequencies of flu-multimer+ CD8+ T cells in pediatric (pink; n = 15) and adult (gray; n = 31) donors from 5–21 donors for each tissue site. Inset shows frequencies of flu-multimer+ cells in pediatric tissues with y axis spanning 0%–1%.
(C) Memory T cell subset distribution of flu-specific CD8+ T cells in indicated tissues of pediatric donors shown in representative flow cytometry plots (left) and compiled in graph (right) showing frequencies of flu-multimer+ CD8+ T cells in pediatric (pink) and adult (gray) donors maintained as TCM (left) and TEM (right) in lung (top) and LLN (bottom) tissue sites. Subset frequencies shown from donors with ≥10 multimer+ T cells.
(D) Expression of tissue residency markers CD69 and CD103 by flu-specific CD8+ T cells in indicated tissues of pediatric donors shown in representative flow cytometry plots (left) and compiled in graph (right) showing frequencies of flu-multimer+ CD69+CD103+CD8+ TRM cells (gated on TEM) in pediatric (pink) and adult (gray) donors in lung (top) and LLN (bottom) tissue sites. TRM frequencies shown from donors with ≥5 multimer+ TEM cells.
Statistical significance for comparison of means was calculated by unpaired t test and indicated by *p ≤ 0.05; ns, not significant.
Sex-related differences in subset delineation of virus-specific T cells
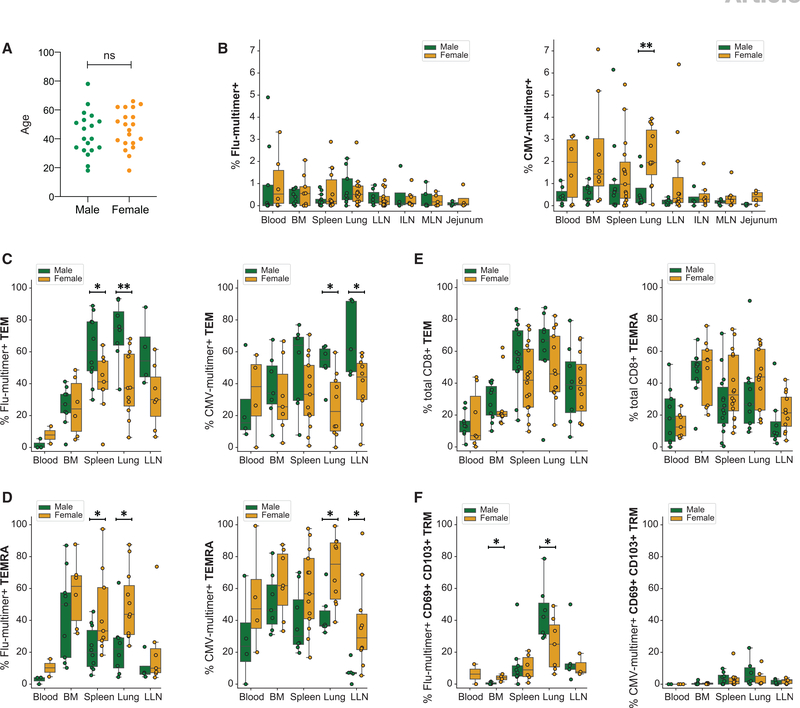
An individual’s sex can also influence the immune responses, as males and females can exhibit distinct T cell responses in peripheral blood (Bongen et al., 2019; Klein and Flanagan, 2016). When stratified by sex, the ages of male and female adult donors in this study were comparable (Figure 5A). However, we found significant differences in subset delineation of virus-specific CD8+ T cells associated with being male or female. Compiled data of CMV-multimer+ T cells across tissue sites show that females maintain a significantly higher frequency of CMV-specific CD8+ T cells in the lung than males do, while such differences were not seen in flu-specific CD8+ T cells between males and females (Figure 5B). Moreover, the subset composition of virus-specific T cells in certain sites differed significantly between males and females; male donors maintained higher frequencies of flu-specific CD8+ TEM cells in the spleen and lung and CMV-specific TEM cells in the lung and LLN compared with females (Figure 5C). Conversely, females exhibited higher frequencies of flu-specific CD8+ TEMRAs in the spleen and lung, and CMV-specific TEMRAs in the lung and LLN compared with male donors (Figure 5D). By contrast, the frequencies of polyclonal TEM and TEMRA subsets among total CD8+ T cells in tissues were comparable between males and females (Figure 5E), indicating that the observed sex-associated differences reflect features of the virus-driven response. Flu-specific TRM cells also differed between sexes; males maintained higher frequencies of lung TRM cells compared with females (Figure 5F). These data suggest that sex influences virus-driven T cell differentiation and maintenance in tissue sites.
Figure 5. Sex-related differences in tissue maintenance of virus-specific CD8+ T cells.
(A) Age range of male and female donors in this study (male: green, n = 19; female: yellow, n = 20).
(B) Frequencies of flu-multimer+ (left) and CMV-multimer+ (right) CD8+ T cells from 3–15 donors for each site stratified by sex.
(C and D) Frequencies of flu-multimer+ (left) and CMV-multimer+ (right) CD8+ TEM cells (C) and CD8+ TEMRAs (D) from 2–13 donors for each site stratified by sex.
(E) Frequencies of total CD8+ TEM cells (top) and TEMRA (bottom) from 7–16 donors for each site stratified by sex.
(F) Frequencies of flu-multimer+ (left) and CMV-multimer+ (right) CD69+ CD103+ CD8+ TRM cells gated on multimer+ cells from 2–10 donors for each site stratified by sex. Subset frequencies shown from donors with ≥10 multimer+ T cells, and TRM frequencies shown from donors with ≥5 multimer+ TEM cells.
Statistical significance for comparison of means was calculated by unpaired t test and indicated by **p ≤ 0.01; *p ≤ 0.05; ns, not significant.
Virus-specific CD8+ T cell clones disperse across sites with distinct expansion and gene usage
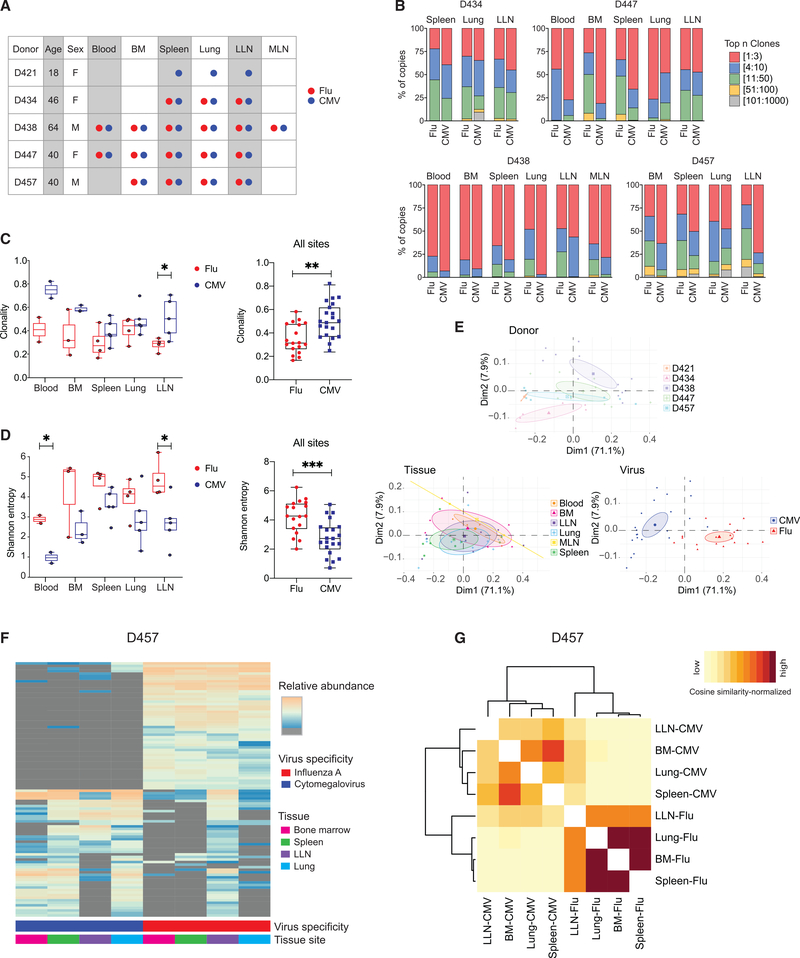
To define the connections between virus-specific T cells in blood and tissues, we analyzed the TCR repertoire of flu- and CMV-specific CD8+ T cells using high-throughput sequencing of the TCRβ-chain gene (TRBV) as described previously (Miron et al., 2021). We sorted multimer+ T cells from five donors across up to six sites per donor (Figure 6A; see Table S2 for cell number, DNA content, and number of clones). In general, the number of CMV-specific CD8+ T cells isolated was much greater compared with flu-specific cells (consistent with the higher frequency overall; Figure 1C), except in some donors where substantial numbers of flu-specific T cells were sorted from the lungs and LLN (Table S2). The number of unique clones obtained for each sample ranged from <100 to >1,000 (Table S2). The majority of the antigen-enriched TCR repertoire (>90%) was composed of 50 clones or fewer (Figure 6B). Notably, CMV-specific CD8+ T cells were more clonally expanded than flu-specific CD8+ T cells within and across specific sites (Figures 6B and 6C). Conversely, flu-specific CD8+ T cells encompassed a more diverse repertoire compared with CMV-specific CD8+ T cells for individual sites (blood and LLN) and across all sites (Figure 6D). These results indicate that repertoire diversity and clonal expansion of virus-specific CD8+ T cells are specific to the virus in a manner consistent across tissue sites and donors.
Figure 6. CD8+ T cell clonal distribution and diversity are shaped by virus specificity.
(A) Diagram showing donor, tissue, and virus specificity of samples for T cell receptor (TCR) sequencing. flu-multimer+ (red), CMV-multimer+ (blue), and total (gray) CD8+ T cells were sorted from indicated tissue sites of five organ donors.
(B) Clonal abundance plots showing proportion of top n clones per sample for flu- and CMV-specific CD8+ T cells from indicated tissue sites for donors D434 (top left), D447 (top right), D438 (bottom left), and D457 (bottom right).
(C and D) TCR clonality (C; see STAR Methods) and Shannon entropy (D; see STAR Methods) by virus specificity for flu-specific (red) and CMV-specific (blue) CD8+ T cell clones stratified by tissue site (left) and in all tissue sites combined (right).
(E) Principal-component analysis (PCA) of TRBV gene usage based on clone counts per sample, labeled and grouped by donor, tissue, and virus with confidence ellipses plotted around group mean points using the factoextra R package.
(F) Sharing of flu- and CMV-specific CD8+ T cells between indicated sites of donor D457 shown in clone tracking plots with each individual clone denoted by a line shaded by relative abundance within a tissue site.
(G) Cosine similarity between pairwise cell populations of flu- and CMV-specific CD8+ T cell clones in BM, LLN, lung, and spleen of donor D457. Color intensity of each cell is based on the normalization of (min-max scaled) values of sequenced samples.
Statistical significance for comparison of means was calculated by unpaired t test and indicated by ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05.
We further investigated whether usage of TRBV genes was distinct between donors, tissues, and virus specificities. TRBV gene usage for all donors revealed biases in TRBV expression between CMV- and flu-specific T cells; in particular, TRBV19 usage predominated among flu-specific CD8+ T cells across donors and tissues, consistent with findings in blood (Nguyen et al., 2018; Sant et al., 2018) (Figure S4A). Principal-component analysis (PCA) of TRBV gene usage based on donor, tissue, and virus revealed distinct gene usage by donor and virus, but not tissue (Figure 6E). Virus-specific clustering was most apparent, with tight and distinct clustering of TRBV usage for flu- and CMV-specific T cells, showing that the exquisite specificity of human T cells is maintained across sites.
We examined the overlap of virus-specific CD8+ T cell clones across tissue sites to understand how clones are disseminated. Clone-tracking plots for the top 100 clones for each specificity shaded by their relative abundance showed that the most abundant virus-specific CD8+ T cell clones are shared across multiple tissue sites, that certain clones are restricted or enriched in specific sites, and that CMV-specific clones do not exhibit substantial overlap with flu-specific clones (Figures 6F and S4B). The extent of overlap between samples within individual donors was calculated using cosine similarity as done previously (Meng et al., 2017; Miron et al., 2021). Samples containing clones recognizing the same virus had higher cosine similarity and thus greater clonal overlap than samples containing clones for different virus specificities (Figures 6G and S4C). The extent of overlap of CMV-specific clones was higher than for flu-specific T cells for 3/4 donors, and there was greater overlap of clones between BM and spleen in most donors (Figures 6G and S4C). Together, these analyses demonstrate that virus-specific T cell clones disseminate and are maintained in diverse tissue sites, and that clonal expansion and TRBV gene usage are likely driven by the virus-specific response.
Transcriptional and functional signatures of virus-responding T cells segregate by site
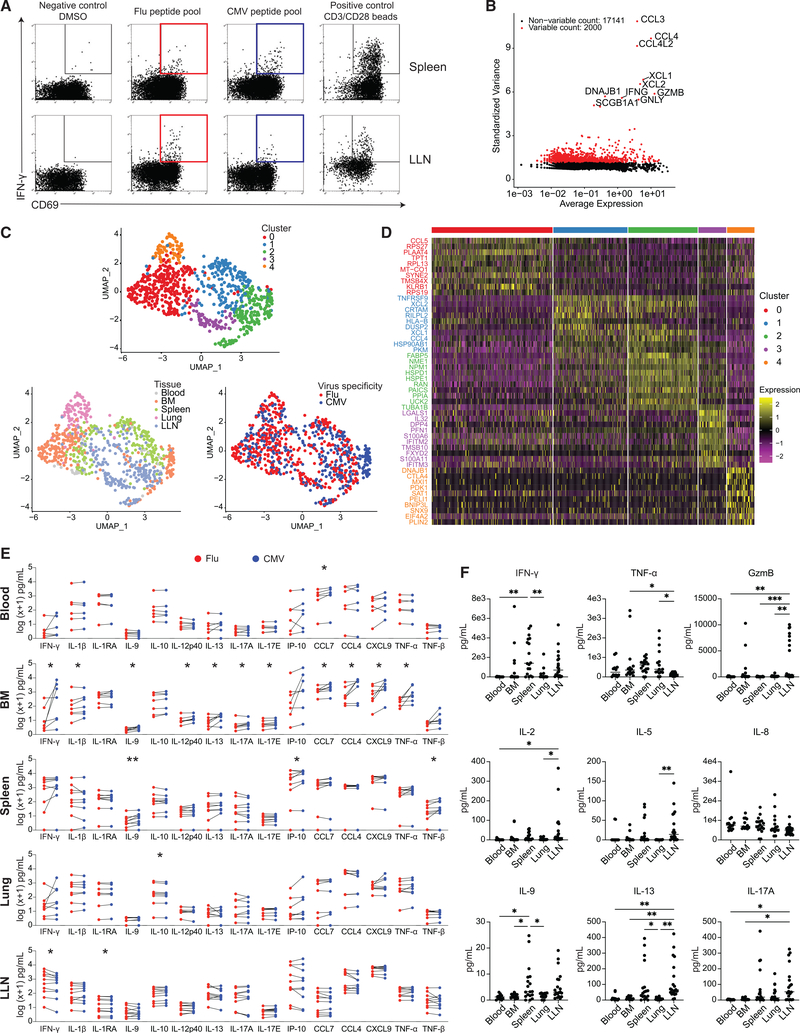
To address how virus specificity and tissue site influence antiviral T cell function, we performed single-cell transcriptome profiling of virus-specific CD8+ T cells from different sites following stimulation with viral antigens. Single-cell suspensions from blood and tissue sites were stimulated in vitro with CMV- or flu-specific peptide MPs, as described above. After 24-h in vitro stimulation with MPs, antigen-responding CD8+ T cells (CD69+ interferon-γ+ [IFN-γ+]) were individually sorted into wells of 96-well plates (Figures 7A, S5A, and S5B), followed by single-cell RNA sequencing (scRNA-seq) using pooled library amplification for transcriptome expression sequencing (PLATE-seq) (see STAR Methods) (Bush et al., 2017; Snyder et al., 2019) and analysis of single-cell transcriptome profiles.
Figure 7. Tissue segregation of the anti-viral T cell response revealed by single-cell transcriptome and functional profiling.
(A) Antigen-responding CD69+IFN-γ+CD8+ T cells in the spleen and LLN of donor D481 following stimulation by DMSO negative control, flu antigen-specific peptide pool, CMV antigen-specific peptide pool, and anti-CD3/CD28 bead positive control, shown in representative flow cytometry plots. Colored gates (red: flu; blue: CMV) indicate populations that were subsequently sorted for sequencing.
(B) Scatterplot showing 2,000 variable genes (red) as determined by calculating average expression and dispersion of each gene. The top 10 most highly variable genes are labeled on the graph.
(C) UMAP embedding of virus-reactive CD8+ T cells colored by cluster as identified via unsupervised hierarchical clustering (see STAR Methods), tissue, or virus specificity.
(D) Heatmap showing top 10 most differentially expressed genes in each cluster. See Table S4 for complete list of differentially expressed genes (padj < 0.1).
(E) Pairwise comparisons of log(x+1) normalized cytokine levels in supernatants from in vitro stimulation of single-cell suspensions from blood (n = 6), BM (n = 7), spleen (n = 9), lung (n = 8), and LLN (n = 10) with flu (red) or CMV (blue) peptide pools. Statistical significance between flu- and CMV-stimulated conditions was calculated by paired t test and indicated by **p ≤ 0.01; *p ≤ 0.05.
(F) Cytokine levels in blood, BM, spleen, lung, and LLN supernatant samples.
Statistical significance was calculated using one-way ANOVA followed by Tukey’s multiple comparisons test indicated by ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05.
The top 10 most highly variable genes expressed by antigen-responding T cells included those encoding cytokines and chemokines (CCL3, CCL4, IFNG, GZMB, GNLY) (Figure 7B), consistent with genes upregulated in activated human CD8+ T cells by scRNA-seq (Szabo et al., 2019a). Unsupervised clustering of 793 cells yielded 5 clusters (see STAR Methods) visualized as Uniform Manifold Approximation and Projection (UMAP) embeddings with individual cells colored by cluster, tissue, or virus specificity (Figure 7C). Some of the clusters corresponded to responses within individual tissues, other clusters represented virus-specific responses from multiple sites, and one sub-cluster corresponded to CMV-specific T cells from the BM (Figures 7C and S6A).
In total, 703 differentially expressed genes (adjusted p ≤ 0.1) were identified among the 5 clusters (Table S4); top differentially expressed genes in each cluster are shown in a heatmap (Figure 7D) and the expression of key genes in UMAP projections (Figure S6B), as described below. Cluster 0, derived from antigen-responding cells from the BM and to a lesser extent the spleen, was enriched for genes associated with T cell function (CCL5, KLRB1, IFITM1, ANXA1, IL7R), transcription factors (STAT1, IRF1), and cell survival (TPT1, XAF1). Cells in cluster 1, from spleen and LLN, expressed genes-encoding chemokines (XCL2, XCL1, CCL4) and transcription factors (EGR2, BATF, NFKB2) and genes associated with cytokine signaling (TNFRSF9 [4-1BB], TNFRSF4 [OX40], IL2RG), tissue residency (CRTAM), immunoregulation (TIGIT), and cell survival (BCL2A1). Cluster 2 was characterized by genes important for anti-viral protection, including genes associated with T cell activation (NME1), as we previously showed (Szabo et al., 2019a), cytokine production (GZMB, IL2RA, IFNG), and lipid metabolism associated with TRM cell survival (FABP5) (Frizzell et al., 2020; Pan et al., 2017). Cluster 3, composed of predominantly LLN cells, included multiple IFN-inducible genes (IFITM2, IFITM3, IFITM1), cytokine genes (IL32, LTB, MIF), and tissue signature genes we previously identified as enriched in T cells in tissues, but not present in blood (Szabo et al., 2019a) (CD226, S100A6, LGALS1). Cluster 4, consisting of antigen-responding T cells from the lung, was enriched for cells expressing the inhibitory receptor CTLA4, which serves as a negative regulator of T cell responses, and the chemokine receptor CXCR4 (Figure 7D; Table S4). Together, these results show differential clustering of gene expression and functional patterns largely correlated to the tissue.
We further analyzed tissue-specific differences in gene expression of antigen-responsive cells by defining gene expression differences by log fold change (logFC) among virus-specific T cells in the BM, lung, and LLN using spleen as a common comparator. We identified a total of 418 genes differentially expressed (adjusted p ≤ 0.1) by antigen-responsive cells in at least one of three comparison pairs, logFC(LLN/spleen) versus logFC(lung/spleen), logFC(BM/spleen) versus logFC(LLN/spleen), and logFC(BM/spleen) versus logFC(lung/spleen) (Figure S5C; Tables S5 and S6), illustrating site-specific influences on antigen-specific T cell responses. In contrast, there were few genes that differed in CMV- compared with flu-reactive T cells; the only significant differences between CMV- and flu-responding T cells were identified in the BM (15 genes, including TNFRSF9 [4-1BB], IFITM1, GZMH) and spleen (1 gene, TNFRSF4 [OX40]) (Figure S5D; Table S7). Together, these findings suggest tissue-mediated regulation of anti-viral T cell function, with CMV-driven influences in the BM.
To further investigate the role of tissue localization in shaping functional response to viral antigens, we performed multiplexed quantification of 50 cytokines on supernatant samples collected from viral peptide-mediated stimulations of T cells from multiple sites of 12 donors (Figure S7A). As visualized in heatmaps showing cytokine production normalized to DMSO negative control within each individual donor, cells stimulated with flu MPs or CMV MPs share similar cytokine secretion patterns distinct to the tissue site, with the exception of BM, where CMV-responding T cells exhibit greater levels and cytokine types compared with flu-responding T cells in multiple donors (Figure S7B). Pairwise analysis of cytokine levels showed that CMV-responding BM cells produced significantly greater levels of IFN-γ, interleukin (IL)-1β, IL-9, IL-17A, macrophage inflammatory protein (MIP)-1β, and tumor necrosis factor alpha (TNF-α) compared with flu-responding cells (Figures 7E and S7C). In contrast, there were few differences in cytokine production between flu- and CMV-specific responses in blood, spleen, lung, and LLN (Figures 7E and S7C). Analysis of cytokine production across tissue sites revealed differences in cytokine levels among blood, BM, spleen, lung, and LLN for numerous cytokines (Figures 7F and S7D). Together, the transcriptional and functional results indicate that the recall response of virus-specific T cells upon reactivation is largely shaped by the tissue, with virus-specific functional effects specific to certain sites, such as CMV-specific influences within the BM.
DISCUSSION
Understanding the heterogeneity of the anti-viral immune response in humans requires studying virus-specific T cells in tissue sites where they function and persist. Here, we examined the factors that shape the distribution, subset differentiation, clonal repertoire, and function of anti-viral T cell immune responses to prevalent viruses, flu and CMV. We found that the overall frequency, clonal expansion, tissue distribution, and TCR repertoire of virus-specific CD8+ T cells are determined primarily by the virus type. However, the differentiation and subset delineation of virus-specific T cells were shaped by multiple factors, including tissue site, age, and sex. Moreover, the establishment of virus-specific TRM cells was also a feature of both virus specificity and tissue site. Transcriptional and functional analysis revealed that virus-specific T cell responses are highly correlated to the tissue. Together, our results elucidate the role of multiple factors in shaping the functional maintenance of anti-viral T cell responses throughout the human body, which is important for targeting, monitoring, and predicting immune responses to existing and emerging viruses.
Our tissue resource enables assessment of virus-specific T cells across sites (lungs, intestines, BM, spleen, and LNs) and between different virus types. Here, we focused on two ubiquitous viruses: influenza A as a prevalent acute respiratory virus for which individuals have multiple exposures over their lifetimes, and CMV as a prevalent persisting virus. Our analyses reveal that virus type plays a crucial role in determining the tissue distribution, subset differentiation, and clonal features of virus-specific T cells. Although T cells specific for both virus types could be found in blood and multiple sites (except intestines), flu-specific T cells exhibited biased distribution and functional maintenance in the lungs and LLN consistent with virus tropism, while CMV-specific T cells were present in higher frequencies in multiple sites, particularly in the BM, consistent with CMV establishing persistent infection in hematopoietic cells (Goodrum, 2016). In all donors examined, flu-specific T cells were maintained as TRM cells in lungs (and to a lesser extent in LLN and spleen) consistent with lung TRM establishment to influenza infection in mice (Paik and Farber, 2021a; Teijaro et al., 2011; Wu et al., 2014; Zens et al., 2016). By contrast, CMV-specific T cells were predominantly circulating TEM and TEMRAs, likely because of the systemic nature of CMV infection, and/or the effects of persistent stimulation, as observed in studies in blood (Griffiths et al., 2013; Vescovini et al., 2007). Furthermore, TCR repertoire analysis showed distinct Vβ gene usage and a lack of substantial clonal overlap between CMV- and flu-specific T cells, suggesting that cross-reactivity between virus-specific T cell clones is not extensive. Together, these data demonstrate the critical role that virus plays in long-term maintenance of antiviral immunity.
The acquisition of multiple sites from individual donors allows us to assess the role of tissue localization in shaping the characteristics of virus-specific T cells. As shown here, tissue is a major determinant of the subset composition, phenotype, and function of virus-specific T cells. The tissue environment itself can bias the subset distribution. For example, BM contains more TEMRAs than other sites, while lungs and intestines contain predominantly TRM populations (Kumar et al., 2017; Miron et al., 2018; Thome et al., 2014), thus influencing the phenotype of virus-specific cells in these sites. We also found major associations in the functional response profile of virus-specific T cells with the tissue site. Upon stimulation with virus-specific peptide epitopes, flu- and CMV-specific CD8+ T cells shared transcriptomic profiles consistent with activated human CD8+ T cells (Szabo et al.,2019a), but these profiles segregated primarily based on tissue site, while BM was the only site with differences based on virus specificity. Accordingly, multiplexed quantification of cytokine production revealed a tissue-driven segregation of anti-viral T cell function, with CMV-specific influences within the BM. These results indicate compartmentalized functional maintenance of anti-viral T cell memory that may be uniquely adapted to mediate protection in situ.
Age was highly associated with virus-specific T cell maintenance in specific sites. Although the site-specific subset composition of flu- and CMV-specific T cells did not alter over age (18–78 y) in most sites, flu-specific TRM cells declined in LLN with age, and CMV-specific TRM cells increased slightly with age. We propose that dynamic events occurring in certain sites, such as the replenishment of lung memory T cells from LLN for flu-specific T cells, and the activation and differentiation of CMV-specific T cells in the BM as a result of virus reactivation, results in compartmentalization of age-associated effects. However, when comparing flu-specific T cells in adult versus pediatric tissues, there was a striking difference in the subset composition; there was a significantly higher proportion of TCM and lower frequency of TRM cells in the lungs and LLN of children compared with adults. These results suggest that repeated infections and/or prolonged persistence is necessary for full TRM cell development. The stark differences in anti-viral immune responses in children compared with adults have been identified with antibody and T cell responses to SARS-CoV-2 (Connors et al., 2018; Weisberg et al., 2021a), suggesting intrinsic developmental differences. Dissecting these and other features of pediatric immunity as result of the tissue environment are important for promoting optimal protection at this critical life stage.
Our findings also reveal that biological sex plays a role in shaping maintenance of tissue immunity to viruses. Notably, females have higher frequencies of CMV-specific T cells compared with males, particularly in the lung. Moreover, females have higher frequencies of virus-specific TEMRAs, while males have higher frequencies of TEM cells within lung, spleen, and LLN; males also had higher frequencies of flu-specific lung TRM compared with females. Together, these findings suggest that virus-driven T cell differentiation in females is more skewed to terminal effector generation, a result consistent with mouse studies showing biased generation of short-lived effector cells following infection of female compared with male mice (Yee Mon et al., 2019). Human studies found that female T cells exhibited elevated proliferation, production of pro-inflammatory cytokines, and cytotoxic T cell activity when stimulated compared with male T cells (Abdullah et al., 2012; Hewagama et al., 2009; Sankaran-Walters et al., 2013), which was consistent with TEMRA functional profiles (Buggert et al., 2020; Szabo et al., 2019a). The mechanisms underlying these sex-associated differences will be important to define in future studies for optimizing vaccines and therapies specific to males and females.
In summary, our study provides a comprehensive view of virus-specific T cell memory in humans and the different factors that impact its generation and maintenance. These findings provide a foundation for defining predictive models that can be leveraged to develop individualized vaccines and therapeutic strategies for promoting protective immunity.
Limitations of the study
This study uses human samples from diverse donors to profile multiple aspects of anti-viral immunity. To measure virus-specific T cells responses, we purified T cells from tissue digests using highly optimized protocols; however, lymphocyte isolation from tissues is not quantitative (Steinert et al., 2015), and more precise assessment of frequencies within a site will require methods for detection of rare antigen-specific T cells by imaging. To define further the role of virus, tissue, age, and sex, continued assessment of virus-specific T cells in more donors and across more virus specificities is needed. Although we were able to stratify donors based on age and sex, a larger cohort would allow examination of other factors, such as BMI, comorbidities, race, and ethnicity. As with all human studies, inter-individual variation cannot be fully controlled. Finally, longitudinal assessment of virus-specific T cells was done between individuals and not within individuals, because of limitation of tissue sampling.
STAR*METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents should be directed to and will be fulfilled by lead author Donna L. Farber (df2396@cumc.columbia.edu)
Materials availability
Aliquots of synthesized sets of Influenza A (Flu) and cytomegalovirus (CMV) peptide Megapools (MPs) utilized in this study will be made available upon request and execution of a material transfer agreement (MTA). There may be restrictions to the availability of the peptide reagents due to cost and limited quantity.
Data and code availability
Flow cytomentry and TCR sequencing data reported in this paper have been deposited in the Immunology Database and Analysis Portal (ImmPort) under accession number ImmPort: SDY1885. Single-cell RNA sequencing data reported in this paper has been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GEO: GSE178837.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available form the lead contact upon request
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human tissues were obtained from deceased organ donors at the time of organ acquisition for clinical transplantation through an approved protocol and material transfer agreement with LiveOnNY, the organ procurement organization (OPO) for the New York metropolitan area, as previously described (Carpenter et al., 2018; Dogra et al., 2020; Gordon et al., 2017; Granot et al., 2017; Kumar et al., 2017; Senda et al., 2019; Thome et al., 2014). Human tissues from deceased pediatric donors were obtained at the time or organ acquisition for clinical transplantation through arrangements with multiple OPOs across the US through the Human Atlas of Neonatal Development-Immunity (HANDEL-I) program associated with nPOD (network for Pancreatic Organ Donors with Diabetes) (Pugliese et al., 2014). Donors were free of cancer and seronegative for hepatitis B, C, and HIV. A list of donors from which tissues were used in this study, including donor characteristics and the assays performed with samples from each donor, is presented in Table S1. This study does not qualify as “human subsets” research, as confirmed by the Columbia University IRB, as tissues samples were obtained from brain-dead (deceased) individuals.
METHOD DETAILS
Isolation of single-cell suspensions from tissue samples
Tissue samples were maintained in cold saline, University of Wisconsin (UW) solution, or media and transported to the laboratory within 2–4 hr of organ procurement for adult organs and shipped to the laboratory on ice within 24 hr of organ procurement for pediatric organs. Mononuclear cells were isolated from the blood and BM samples by density centrifugation using Lymphocyte Separation Medium (Corning, cat# 25-072-CI). Spleen, lung, jejunum, and lymph node samples were processed using mechanical and enzymatic digestion, resulting in high yields of live leukocytes, as previously described (Carpenter et al., 2018; Dogra et al., 2020; Gordon et al., 2017; Granot et al., 2017; Kumar et al., 2017; Senda et al., 2019).
Flow cytometry analysis and cell sorting
For flow cytometry analysis, cells were stained in 96-well U-bottom plates protected from light using antibody panels and multimer reagents described in the Key resources table. Briefly, cells were washed with FACS-buffer (PBS with 2% heat-inactivated FBS), then resuspended with multimer reagent staining cocktail for 10 min at room temperature (RT). Cells were then washed again and resuspended in Human TruStain FcX (BioLegend, cat# 422302), followed by surface staining with fluorochrome-conjugated antibodies in FACS-buffer (20 min. at RT), and subsequently fixed in fixation buffer (Tonbo, cat# TNB-1020-L050). For intracellular staining, surface stained cells were fixed for 25 min at RT in fixation buffer (Tonbo, cat# TNB-1020-L050), followed by staining with fluorochrome-conjugated antibodies for 30 min at RT in permeabilization buffer (Tonbo, cat# TNB-1022-L160). Flow cytometry data were acquired on BD LSRII and analyzed using FlowJo V 10.7 software, Prism 8.4.3 software, and custom Python scripts.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies, conventional flow cytometry, and sorting | ||
|
| ||
| Anti-Human CCR7 Alexa Fluor 488 | BioLegend | G043H7; Cat No.: 353205; RRID:AB_10918624 |
| Anti-Human CCR7 APC-Cy7 | BioLegend | G043H7; Cat No.: 353212; RRID:AB_10916390 |
| Anti-Human CD103 BUV395 | BD Biosciences | Ber-act8; Cat No.: 564346; RRID:AB_2738759 |
| Anti-Human CD103 BV421 | BD Biosciences | Ber-act8; Cat No.: 566257; RRID:AB_2739635 |
| Anti-Human CD107a BUV395 | BD Biosciences | H4A3; Cat No.: 565113; RRID:AB_2739073 |
| Anti-Human CD14 PerCP-Cy5.5 | BD Biosciences | M5E2; Cat No.: 550787; RRID:AB_393884 |
| Anti-Human CD19 PerCP-Cy5.5 | Tonbo | HIB19; Cat No.: 65-0199; RRID:AB_2621888 |
| Anti-Human CD3 BUV496 | BD Biosciences | UCHT1; Cat No.: 612940; RRID:AB_2870222 |
| Anti-Human CD3 BV650 | BioLegend | OKT3; Cat No.: 317323; RRID:AB_2563352 |
| Anti-Human CD4 BUV737 | BD Biosciences | SK3; Cat No.: 564305; RRID:AB_2713927 |
| Anti-Human CD4 PE-Cy7 | Tonbo | RPA-T4; Cat No.: 60-0049; RRID:AB_2621831 |
| Anti-Human CD45 Alexa Fluor 700 | BioLegend | HI30; Cat No.: 304023; RRID:AB_493761 |
| Anti-Human CD45 BV711 | BioLegend | HI30; Cat No.: 304050; RRID:AB_2563466 |
| Anti-Human CD45RA Alexa Fluor 700 | BioLegend | HI100; Cat No.: 304120; RRID:AB_493763 |
| Anti-Human CD45RA BV605 | BioLegend | HI100; Cat No.: 304134; RRID:AB_2563814 |
| Anti-Human CD56 PE-Cy5 | BioLegend | 5.1H11; Cat No.: 362515; RRID:AB_2564088 |
| Anti-Human CD69 BV711 | BioLegend | FN50; Cat No.: 310944; RRID:AB_2566466 |
| Anti-Human CD8 BUV496 | BD Biosciences | RPA-T8; Cat No.: 564805; RRID:AB_2869614 |
| Anti-Human CD8 BV510 | BioLegend | SK1; Cat No.: 344732; RRID:AB_2564624 |
| Anti-Human GzmB Alexa Fluor 700 | BD Biosciences | GB11; Cat No.: 560213; RRID:AB_1645453 |
| Anti-Human IFN-γ BB700 | BD Biosciences | B27; Cat No.: 566394; RRID:AB_2744484 |
| Anti-Human Perforin BV510 | BioLegend | dG9; Cat No.: 308120; RRID:AB_2563829 |
| Anti-Human TNF-α PE-Cy7 | BD Biosciences | MAb11; Cat No.: 557647; RRID:AB_396764 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| LIVE/DEAD Fixable Blue Dead Cell Stain | Thermo Fisher Scientific | Cat No.: L23105 |
| Fixable Viability Dye eFluor 780 | Thermo Fisher Scientific | Cat No.: 65-0865-18 |
| Human TruStain FcX | BioLegend | Cat No.: 422302; RRID:AB_2818986 |
| RPMI 1640 | Corning | Cat No.: 10-040-CM |
|
| ||
| Human HLA-multimer reagents | ||
|
| ||
| HLA-A*0101[YSEHPTFTSQY]-CMV pp65 | ProImmune | F266-4A-D (APC) |
| HLA-A*0101[VTEHDTLLY]-CMV pp50 | ProImmune | F739-4A-D (APC) |
| HLA-A*0201[NLVPMVATV]-CMV pp65 | Immudex | WB2132-APC (APC) |
| HLA-A*0201[VLEETSVML]-CMV IE-1 | Immudex | WB2658-APC (APC) |
| HLA-A*2402[QYDPVAALF]-CMV pp65 | ProImmune | F414-4A-D (APC) |
| HLA-A*2402[VYALPLKML]-CMV pp65 | ProImmune | F380-4A-D (APC) |
| HLA-B*0702[TPRVTGGGAM]-CMV pp65 | ProImmune | F045-4A-D (APC) |
| HLA-B*0702[RPHERNGFTVL]-CMV pp65 | ProImmune | F308-4A-D (APC) |
| HLA-A*0101[VSDGGPNLY]-Flu PB1 | ProImmune | F540-4A-D (APC) |
| HLA-A*0101[CTELKLSDY]-Flu NP | Immudex | WA3410-APC (APC) |
| HLA-A*0101[CTELKLSDY]-Flu NP | Immudex | WA3410-PE (PE) |
| HLA-A*0201[GILGFVFTL]-Flu MP | Immudex | WB2161-APC (APC) |
| HLA-A*0201[GILGFVFTL]-Flu MP | Immudex | WB2161-PE (PE) |
| HLA-B*0702[SPIVPSFDM]-Flu NP | Immudex | F1744-4A-D (APC) |
| HLA-B*0702[SPIVPSFDM]-Flu NP | Immudex | F1744-4B-D (PE) |
| HLA-B*0702[QPEWFRNVL]-Flu PB1 | Immudex | F1743-4A-D (APC) |
| HLA-B*0702[QPEWFRNVL]-Flu PB1 | Immudex | F1743-4B-D (PE) |
| HLA-A*0201[ALIAPVHAV]-negative control | Immudex | WB2666-APC (APC) |
| HLA-A*0201[ALIAPVHAV]-negative control | Immudex | WB2666-PE (PE) |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed data | This paper | N/A |
| Flow cytometry and TCR sequencing data | Immunology Database and Analysis Portal | accession number SDY1885 |
| Single-cell RNA sequencing data | NCBI Gene Expression Omnibus | accession number GSE178837 |
|
| ||
| Software and algorithms | ||
|
| ||
| Python version 3.7 | Python (2020) | https://www.python.org/downloads/release/python-370/ |
| Anaconda version 4 | Anaconda (2020) | https://www.anaconda.com/products/individual |
| VDJtools | Shugay et al., 2015 | https://vdjtools-doc.readthedocs.io/en/master/ |
| MiXCR | Bolotin et al., 2015 | https://mixcr.readthedocs.io/en/master/ |
| tcR | Nazarov et al., 2015 | http://imminfo.github.io/tcr/ |
| Seurat | Stuart et al., 2019 | https://github.com/satijalab/seurat/ |
| RStudio version 1.3.1073 | RStudio, Inc. (2020) | https://www.rstudio.com |
| R version 4.0.3 | R Foundation for Statistical Computing (2020) | https://www.R-project.org |
| FlowJo V 10.7 software | Tree Star | https://www.flowjo.com/ |
| Prism 8.4.3 | GraphPad Software, LLC | https://www.graphpad.com/ |
|
| ||
| Other | ||
|
| ||
| EasySep Dead Cell Removal (Annexin V) Kit | STEMCELL Technologies | Cat No.: 17899 |
| EasySep Human CD8+ T Cell Enrichment Kit | STEMCELL Technologies | Cat No.: 19053 |
| Immunocult Human CD3/CD28 T cell activator | STEMCELL Technologies | Cat No.: 10971 |
| IFN-y Secretion Assay – Detection Kit, human (PE) | Miltenyi Biotec | Cat No.: 130-054-202 |
For sorting of multimer+ T cells for TCR sequencing, CD8+ T cells were first enriched from blood, BM, spleen, lung, and lymph nodes using EasySep Human CD8+ T Cell Enrichment Kit (STEMCELL Technologies, cat #19053). Enriched cells were then stained with specific multimers, and subsequently with antibody panels described in the Key resources table. Multimer+ CD8+ T cells were sorted using BD Influx Cell Sorter directly into cell lysis solution (QIAGEN, cat# 158906).
Peptide Megapool design and preparation
The Flu and CMV CD8 MPs were composed of viral-specific epitopes. Those peptides were synthetized as crude material by A&A ltd, San Diego, resuspended in DMSO, and pooled followed by sequentially lyophilization, as previously described. For the CD8 Flu, the MP was developed as previously described (Carrasco Pro et al., 2015; Weiskopf et al., 2015). Epitopes were retrieved by querying the IEDB (http://www.iedb.org) on July 12, 2019 (Vita et al., 2019). The epitopes were extracted using the following query; Organism: Influenza virus (ID:1000204), positive assays only, no B cell assays, no MHC ligand assays, MHC restriction type: MHC class I, host: Homo sapiens. The resulting epitopes were filtered for size (8–11 amino acids) and discovered using one of the following assays: cytotoxicity, ELISPOT, ICS, and multimer/tetramer assay platforms, and response frequency scores > 0. In total, 170 were selected for synthesis. In the case of CD8 CMV, the MP was developed as previously described, based on epitopes of 8–11 residues in length available in the IEDB (http://www.iedb.org) on Jan 14, 2019 using the following query; Organism: human herpesvirus 5 (ID:10359), positive assays only, no B cell assays, no MHC ligand assays, MHC restriction type: MHC class I, host: Homo sapiens. The resultant dataset was further filtered to include only epitopes characterized using cytotoxicity, ELISPOT, ICS, and multimer/tetramer assay platforms, and response frequency scores > 0. In total, 126 epitopes were selected for synthesis.
In vitro T cell stimulations
Mononuclear cells were thawed, and dead cells were removed using the EasySep Dead Cell Removal (Annexin V) Kit (STEMCELL Technologies, cat# 17899). Cells were then plated in 96-well U-bottom plates in RPMI medium containing 10% heat-inactivated human AB serum (Gemini, cat# 507533010) at a concentration of 106 cells/well and left overnight (5% CO2, 37° C). Cells were stimulated by the addition of individual virus-specific MPs (1 μg/mL) for 24 hr. Immunocult Human CD3/CD28 T cell activator (STEMCELL Technologies, cat# 10971) was used as a positive control, and volume of DMSO equal to volume of MP added was used as negative control. Following stimulation, either cell surface staining and PLATE-seq was performed or supernatant was collected for multiplex cytokine secretion assays.
DNA extraction, TRB gene amplification, library preparation, and sequencing
DNA was isolated from cell lysate using the Gentra Puregene Kit (QIAGEN, cat# 159667). The quantity of DNA isolated and sequenced per sample is listed in Table S2. Targeted PCR was used for amplification of TRB sequences from genomic DNA, using a cocktail of forward primers specific for framework region 2 (FR2) sequences of 23 TRBV subgroups (gene families), and 13 TRBJ region reverse primers adapted from the BIOMED2 primer series. Amplicons were purified using the Agencourt AMPure XP beads system (Beckman Coulter, cat# A63881). To generate the sequencing libraries, second-round PCRs were carried out using Nexter-aXT Index Primers S5XX and N7XX. Libraries were sequenced using an Illumina MiSeq in the Human Immunology Core Facility at the University of Pennsylvania. 2×300 bp paired end kits were used for all experiments (Illumina MiSeq Reagent Kit v3, 600 cycle, Illumina, cat# MS102-3003).
TCR read counting and clone mapping
Raw reads are first processed using pRESTO and filtered as previously described (Meng et al., 2017; Miron et al., 2018, 2021). Briefly, sequences are trimmed of poor-quality bases, paired reads are aligned into full length contiguous sequences, short sequences are filtered out, bases with low quality scores are replaced with Ns, and any sequence containing more than 10 such bases is removed from further analysis. Filtered sequences are further processed by MiXCR v3.0.7 (Bolotin et al., 2015) and VDJtools v1.2.1 (Rosenfeld et al., 2017, 2018; Shugay et al., 2015) for clone collapsing using default settings. Clones with only one copy sequences were filtered out.
TCR diversity, clonality, TRBV usage, clonal overlap, and cosine similarity
Clonal abundance plots were generated using the clonal.proportion and vis.top.proportions functions in the tcR package in R (Nazarov et al., 2015). Shannon entropy is a measure of immune diversity, with values ranging from 0 to 1, where 0 indicates no diversity and 1 indicates the most diversity. To calculate Shannon entropy with a given clone denoted x and frequency denoted p(x), . Clonality is a measure of clonal expansion, with values ranging from 0 to 1, where 0 indicates least clonally expanded and 1 indicates most clonally expanded. To calculate clonality, given a clone denoted x, frequency denoted p(x), and total set size of unique clonotypes denoted L, . For analysis of TRBV gene usage, heatmap visualization was generated using CalcSegmentUsage function of VDJtools (Shugay et al., 2015). Principal component analysis was visualized using the factoextra package in R (Kassambara and Mundt, 2020). For clonal overlap analysis, heatmap visualizations were generated using TrackClonotypes function of VDJtools (Shugay et al., 2015). Cosine similarity analysis and heatmap visualization was performed using the cosine.sharing function in the tcR package in R (Nazarov et al., 2015).
Single-cell transcriptome profiling of antigen-responsive CD8+ T cells
Single-cell transcriptome profiling was performed on antigen-responsive CD8+ T cells, using pooled library amplification for transcriptome expression (PLATE-seq) (Bush et al., 2017; Snyder et al., 2019). To isolate virus-responding T cells for PLATE-seq, T cells cultured with peptide MP as described above for 24hrs were first stained using the IFN-γ Secretion Assay – Detection Kit, human (Miltenyi Biotec, cat# 130-054-202), which allows isolation of intact IFN-γ-secreting cells using an IFN-γ catch reagent that attaches to CD45 on the surface of leukocytes and subsequent labeling of caught IFN-γ with a fluorochrome-conjugated detection antibody. Then, cells were stained with fluorochrome-conjugated antibodies listed in the Key resources table. Single stained cells were sorted using BD Influx Cell Sorter directly into 96-well plates containing 7.5 μL of lysis buffer [0.2% Triton X-100 (Sigma), SUPERaseIN (1 U/μl) (Thermo Fisher Scientific), 2 mM deoxyribonucleotides (dNTPs) (Thermo Fisher Scientific), and 2 μM reverse transcriptase (RT) primer (Integrated DNA Technologies)]. Library preparation, RNA sequencing was done as described in detail previously (Snyder et al., 2019).
For analysis of PLATE-seq data, for each 96-well plate, empty wells were removed, leaving 981 cells. Several filtering steps were performed for quality control. Non-coding genes and features were removed from the dataset. Cells with less than 200 genes or cells with a mitochondrial percentage greater than 10% were removed. Three outlier cells were removed based on a principal component analysis (PCA) plot, leaving 793 cells in the final dataset. Further data processing was performed using the R package Seurat, designed for single-cell RNA-sequencing analysis (Stuart et al., 2019).
The counts data was log normalized and scaled according to Seurat’s methods. Highly variable features were identified, of which the top 2,000 were plotted and the top ten genes labeled. Dimensionality reduction was performed using the Python implementation Uniform Manifold Approximation and Projection (UMAP), based on these selected 2,000 features and the first 15 components of PCA. A KNN graph based on the Euclidean distance in PCA space was constructed using the FindNeighbors() Seurat function, which then identified five clusters via the Louvain algorithm with a resolution of 0.5. Positive cluster markers were found using differential expression analysis, with a threshold requiring at least 25% of the cluster’s population to express the marker gene. A heatmap was constructed to show the expression levels of the top ten markers for each cluster ranked by FDR value.
Differential expression analyses were performed to identify markers genes that differed between Flu and CMV peptide-stimulated CD8+ T cells for each tissue type. Significant markers with FDR value < 0.1 were identified in the BM Flu versus CMV analysis, as well as one gene in the spleen Flu versus CMV analysis.
Additional differential expression analyses were performed to compare markers between specific tissues, irrespective of virus stimulation. Significant markers with FDR value < 0.1 were identified from spleen versus lung, BM, and LLN CD8+ T cell populations. Log fold change of expression was plotted with pairwise comparisons at each axis, highlighting significant genes in both or either analyses.
Multiplex cytokine secretion assay
Quantification of 50 total human cytokines, chemokines, and growth factors was performed on culture supernatant from in vitro T cell stimulation experiments described above by Eve Technologies Corp. (Calgary, Alberta). Luminex xMAP technology was used for multiplexed quantification of 2 human cytokines in one array (Perforin, Granzyme B) and 48 human cytokines, chemokines, and growth factors in a separate array (sCD40L, EGF, Eotaxin, FGF-2, Flt-3 ligand, Fractalkine, G-CSF, GM-CSF, GROα, IFNα2, IFNγ, IL-1α, IL-1β, IL-1ra, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-17A, IL-17E/IL-25, IL-17F, IL-18, IL-22, IL-27, IP-10, MCP-1, MCP-3, M-CSF, MDC (CCL22), MIG, MIP-1α, MIP-1β, PDGF-AA, PDGF-AB/BB, RANTES, TGFα, TNFα, TNFβ, VEGF-A). The multiplexing analysis was performed using the Luminex 200 system with assay kits sourced by Millipore MILLIPEX (MilliporeSigma, Burlington, Massachusetts, USA) according to the manufacturer’s protocol.
Observed concentrations were calculated with the standard curve based on the fluorescence intensity of the bead population for a specific analyte. For visualization of cytokine/chemokine production by antigen-responding cells in multiple tissues sites within each individual donor, observed concentrations for each analyte were first normalized to observed concentration of DMSO negative control. Normalized concentrations were then scaled across samples for each individual donor on a max absolute scale, with values ranging from −1 to 1 across all analytes, using the MaxAbsScaler features of sklearn.preprocessing function of the Python scikit-learn library (Pedregosa et al., 2011). Heatmap visualizations were generated using the Python data visualization library seaborn (Waskom, 2021). For analysis comparing the production of analytes between cells stimulated with Flu or with CMV MPs across donors for blood, BM, spleen, lung, and LLN, observed concentrations were normalized by log (x+1) followed by statistical analysis by paired t test between Flu and CMV for each analyte in each tissue site.
QUANTIFICATION AND STATISTICAL ANALYSIS
Descriptive statistics of compiled flow cytometry data and statistical testing were performed using Prism (GraphPad). Graphs were generated using the Prism (GraphPad) and the Python matplotlib and seaborn libraries (Hunter, 2007; Waskom, 2021). Differences in means between two sample groups were compared using two-tailed t tests. For comparing paired samples within an individual, we used paired two-tailed t tests. Pearson correlations were used to correlate age and T cell subset frequencies. Multiple group comparisons were done using one-way ANOVA followed by Tukey’s multiple comparisons. P-values below 0.05 were considered as statistically significant. For all figures, **** denotes p value ≤ 0.0001, *** denotes p value ≤ 0.001, ** denotes p value ≤ 0.01, and * denotes p value ≤ 0.05.
Supplementary Material
Highlights.
CD8+ T cells specific for influenza and CMV localize in multiple tissue sites
Persistence of memory CD8+ T cell subsets is shaped by virus tropism and specificity
Regulation of functional responses to viral antigens is primarily tissue mediated
Age and sex influence T cell subset differentiation and tissue residency
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) (grants AI128949, AI06697, AI100119, and HL145547 awarded to D.L.F.; K23AI141686 to T.J.C.). Pediatric sample intake was supported by the Human Atlas for Neonatal Development and Early Life – Immunity (HANDEL-I) program, which was supported by the Helmsley Charitable Trust (to D.L.F. and T.M.B.). This work was supported by NIH contract 75N9301900065 (to A.S. and D.W.). P.D. was supported by a Cancer Research Institute (CRI) Irvington Postdoctoral Fellowship. N.L. was supported by the National Science Foundation Graduate Research Fellowship Program (NSF-GRFP). P.A. Szabo was supported by a Canadian Institutes of Health Research (CIHR) Fellowship. Research reported here was performed in the CCTI Flow Cytometry Core (supported by NIH awards S10RR027050 and S10OD020056), the Sulzberger Columbia Genome Center, the Columbia Single Cell Analysis Core (supported by NIH grant P30-CA013696), and the Perelman School of Medicine Human Immunology Core (supported by NIH grants P30-CA016520 and P30-AI0450080). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We wish to thank the donor families for their generosity and the exceptional efforts of the transplant coordinators and staff of LiveOnNY and nPOD (network for Pancreatic Organ Donors with Diabetes tissue processing core) for making this study possible.
Footnotes
DECLARATION OF INTERESTS
A.S. is a consultant for Gritstone, Flow Pharma, Arcturus, Immunoscape, Cell-Carta, OxfordImmunotech, and Avalia. La Jolla Institute for Immunology (LJI) has filed for patent protection for various aspects of T cell epitope and vaccine design work. E.T.L.P. is an advisor for Roche Diagnostics, Enpicom, The Antibody Society, IEDB, and The American Autoimmune Related Diseases Association. D.L.F. is a consultant for Moderna.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110071.
REFERENCES
- Abdullah M, Chai PS, Chong MY, Tohit ER, Ramasamy R, Pei CP, and Vidyadaran S (2012). Gender effect on in vitro lymphocyte subset levels of healthy individuals. Cell. Immunol. 272, 214–219. [DOI] [PubMed] [Google Scholar]
- Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. (2017). Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S, and Sant AJ (2011). Infection with seasonal influenza virus elicits CD4 T cells specific for genetically conserved epitopes that can be rapidly mobilized for protective immunity to pandemic H1N1 influenza virus. J. Virol. 85, 13310–13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assarsson E, Bui HH, Sidney J, Zhang Q, Glenn J, Oseroff C, Mba-wuike IN, Alexander J, Newman MJ, Grey H, and Sette A (2008). Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J. Virol. 82, 12241–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, and Chudakov DM (2015). MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381. [DOI] [PubMed] [Google Scholar]
- Bongen E, Lucian H, Khatri A, Fragiadakis GK, Bjornson ZB, Nolan GP, Utz PJ, and Khatri P (2019). Sex Differences in the Blood Transcriptome Identify Robust Changes in Immune Cell Proportions with Aging and Influenza Infection. Cell Rep. 29, 1961–1973.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggert M, Vella LA, Nguyen S, Wu VH, Chen Z, Sekine T, Perez-Potti A, Maldini CR, Manne S, Darko S, et al. (2020). The identity of human tissue-emigrant CD8+ T cells. Cell 183, 1946–1961.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush EC, Ray F, Alvarez MJ, Realubit R, Li H, Karan C, Califano A, and Sims PA (2017). PLATE-Seq for genome-wide regulatory network analysis of high-throughput screens. Nat. Commun. 8, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter DJ, Granot T, Matsuoka N, Senda T, Kumar BV, Thome JJC, Gordon CL, Miron M, Weiner J, Connors T, et al. (2018). Human immunology studies using organ donors: Impact of clinical variations on immune parameters in tissues and circulation. Am. J. Transplant. 18, 74–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco Pro S, Sidney J, Paul S, Lindestam Arlehamn C, Weiskopf D, Peters B, and Sette A (2015). Automatic Generation of Validated Specific Epitope Sets. J. Immunol. Res. 2015, 763461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors TJ, Baird JS, Yopes MC, Zens KD, Pethe K, Ravindranath TM, Ho SH, and Farber DL (2018). Developmental Regulation of Effector and Resident Memory T Cell Generation during Pediatric Viral Respiratory Tract Infection. J. Immunol. 201, 432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, Grifoni A, Ramirez SI, Haupt S, Frazier A, et al. (2021). Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 371, eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra P, Rancan C, Ma W, Toth M, Senda T, Carpenter DJ, Kubota M, Matsumoto R, Thapa P, Szabo PA, et al. (2020). Tissue Determinants of Human NK Cell Development, Function, and Residence. Cell 180, 749–763.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber DL (2021). Tissues, not blood, are where immune cells function. Nature 593, 506–509. [DOI] [PubMed] [Google Scholar]
- Frizzell H, Fonseca R, Christo SN, Evrard M, Cruz-Gomez S, Zanluqui NG, von Scheidt B, Freestone D, Park SL, McWilliam HEG, et al. (2020). Organ-specific isoform selection of fatty acid-binding proteins in tissue-resident lymphocytes. Sci. Immunol. 5, eaay9283. [DOI] [PubMed] [Google Scholar]
- Goodrum F (2016). Human Cytomegalovirus Latency: Approaching the Gordian Knot. Annu. Rev. Virol. 3, 333–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CL, Miron M, Thome JJ, Matsuoka N, Weiner J, Rak MA, Igarashi S, Granot T, Lerner H, Goodrum F, and Farber DL (2017). Tissue reservoirs of antiviral T cell immunity in persistent human CMV infection. J. Exp. Med. 214, 651–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham N, Eisenhauer P, Diehl SA, Pierce KK, Whitehead SS, Durbin AP, Kirkpatrick BD, Sette A, Weiskopf D, Boyson JE, and Botten JW (2020). Rapid Induction and Maintenance of Virus-Specific CD8+ TEMRA and CD4+ TEM Cells Following Protective Vaccination Against Dengue Virus Challenge in Humans. Front. Immunol. 11, 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granot T, Senda T, Carpenter DJ, Matsuoka N, Weiner J, Gordon CL, Miron M, Kumar BV, Griesemer A, Ho SH, et al. (2017). Dendritic Cells Display Subset and Tissue-Specific Maturation Dynamics over Human Life. Immunity 46, 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths SJ, Riddell NE, Masters J, Libri V, Henson SM, Wertheimer A, Wallace D, Sims S, Rivino L, Larbi A, et al. (2013). Age-associated increase of low-avidity cytomegalovirus-specific CD8+ T cells that re-express CD45RA. J. Immunol. 190, 5363–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvenel A, Jozwik A, Ascough S, Ung SK, Paterson S, Kalyan M, Gardener Z, Bergstrom E, Kar S, Habibi MS, et al. (2020). Epitope-specific airway-resident CD4+ T cell dynamics during experimental human RSV infection. J. Clin. Invest. 130, 523–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund E, Lewis MW, Hansen SG, Strelow LI, Nelson JA, Sexton GJ, Hanifin JM, and Slifka MK (2003). Duration of antiviral immunity after smallpox vaccination. Nat. Med. 9, 1131–1137. [DOI] [PubMed] [Google Scholar]
- Hewagama A, Patel D, Yarlagadda S, Strickland FM, and Richardson BC (2009). Stronger inflammatory/cytotoxic T-cell response in women identified by microarray analysis. Genes Immun. 10, 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD (2007). Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 9, 90–95. [Google Scholar]
- Kassambara A, and Mundt F (2020). factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package Version. http://www.sthda.com/english/rpkgs/factoextra.
- Klein SL, and Flanagan KL (2016). Sex differences in immune responses. Nat. Rev. Immunol. 16, 626–638. [DOI] [PubMed] [Google Scholar]
- Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, Senda T, Sun X, Ho SH, Lerner H, et al. (2017). Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep. 20, 2921–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar BV, Connors TJ, and Farber DL (2018). Human T Cell Development, Localization, and Function throughout Life. Immunity 48, 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay LK, and Kallies A (2017). Transcriptional Regulation of Tissue-Resident Lymphocytes. Trends Immunol. 38, 94–103. [DOI] [PubMed] [Google Scholar]
- Malik A, Adland E, Laker L, Kløverpris H, Fardoos R, Roider J, Sever-insen MC, Chen F, Riddell L, Edwards A, et al. (2017). Immunodominant cytomegalovirus-specific CD8+ T-cell responses in sub-Saharan African populations. PLoS ONE 12, e0189612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D, and Soerens AG (2019). Tissue-Resident T Cells and Other Resident Leukocytes. Annu. Rev. Immunol. 37, 521–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D, Vezys V, Marzo AL, and Lefrançois L (2001). Preferential localization of effector memory cells in nonlymphoid tissue. Science 291, 2413–2417. [DOI] [PubMed] [Google Scholar]
- Meng W, Zhang B, Schwartz GW, Rosenfeld AM, Ren D, Thome JJC, Carpenter DJ, Matsuoka N, Lerner H, Friedman AL, et al. (2017). An atlas of B-cell clonal distribution in the human body. Nat. Bio-technol. 35, 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron M, Kumar BV, Meng W, Granot T, Carpenter DJ, Senda T, Chen D, Rosenfeld AM, Zhang B, Lerner H, et al. (2018). Human Lymph Nodes Maintain TCF-1hi Memory T Cells with High Functional Potential and Clonal Diversity throughout Life. J. Immunol. 201, 2132–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron M, Meng W, Rosenfeld AM, Dvorkin S, Poon MML, Lam N, Kumar BV, Louzoun Y, Luning Prak ET, and Farber DL (2021). Maintenance of the human memory T cell repertoire by subset and tissue site. Genome Med. 13, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarov VI, Pogorelyy MV, Komech EA, Zvyagin IV, Bolotin DA, Shugay M, Chudakov DM, Lebedev YB, and Mamedov IZ (2015). tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinformatics 16, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen THO, Sant S, Bird NL, Grant EJ, Clemens EB, Koutsakos M, Valkenburg SA, Gras S, Lappas M, Jaworowski A, et al. (2018). Perturbed CD8+ T cell immunity across universal influenza epitopes in the elderly. J. Leukoc. Biol. 103, 321–339. [DOI] [PubMed] [Google Scholar]
- Paik DH, and Farber DL (2021a). Anti-viral protective capacity of tissue resident memory T cells. Curr. Opin. Virol. 46, 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik DH, and Farber DL (2021b). Influenza infection fortifies local lymph nodes to promote lung-resident heterosubtypic immunity. J. Exp. Med. 218, e20200218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Tian T, Park CO, Lofftus SY, Mei S, Liu X, Luo C, O’Malley JT, Gehad A, Teague JE, et al. (2017). Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NP, Vukmanovic-Stejic M, Suarez-Farinas M, Chambers ES, Sandhu D, Fuentes-Duculan J, Mabbott NA, Rustin MHA, Krueger J, and Akbar AN (2018). Impact of Zostavax Vaccination on T-Cell Accumulation and Cutaneous Gene Expression in the Skin of Older Humans After Varicella Zoster Virus Antigen-Specific Challenge. J. Infect. Dis. 218 (Suppl_2), S88–S98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. (2011). Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 12, 2825–2830. [Google Scholar]
- Poon MML, and Farber DL (2020). The Whole Body as the System in Systems Immunology. iScience 23, 101509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon MML, Rybinka K, Kato Y, Kubota M, Matsumoto R, Bloom NI, Zhang Z, Hastie KM, Grifoni A, Weiskopf D, et al. (2021). SARS-CoV-2 infection generates tissue-localized immunoloigcal memory in humans. Sci. Immunol. 10.1126/sciimmunol.abl9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugliese A, Yang M, Kusmarteva I, Heiple T, Vendrame F, Wasserfall C, Rowe P, Moraski JM, Ball S, Jebson L, et al. (2014). The Juvenile Diabetes Research Foundation Network for Pancreatic Organ Donors with Diabetes (nPOD) Program: goals, operational model and emerging findings. Pediatr. Diabetes 15, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, and Davis MM (2020). The science and medicine of human immunology. Science 369, eaay4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld AM, Meng W, Luning Prak ET, and Hershberg U (2017). ImmuneDB: a system for the analysis and exploration of high-throughput adaptive immune receptor sequencing data. Bioinformatics 33, 292–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld AM, Meng W, Luning Prak ET, and Hershberg U (2018). ImmuneDB, a Novel Tool for the Analysis, Storage, and Dissemination of Immune Repertoire Sequencing Data. Front. Immunol. 9, 2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Förster R, Lipp M, and Lanzavecchia A (1999). Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Geginat J, and Lanzavecchia A (2004). Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 22, 745–763. [DOI] [PubMed] [Google Scholar]
- Sankaran-Walters S, Macal M, Grishina I, Nagy L, Goulart L, Coolidge K, Li J, Fenton A, Williams T, Miller MK, et al. (2013). Sex differences matter in the gut: effect on mucosal immune activation and inflammation. Biol. Sex Differ. 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant S, Grzelak L, Wang Z, Pizzolla A, Koutsakos M, Crowe J, Loudovaris T, Mannering SI, Westall GP, Wakim LM, et al. (2018). Single-Cell Approach to Influenza-Specific CD8+ T Cell Receptor Repertoires Across Different Age Groups, Tissues, and Following Influenza Virus Infection. Front. Immunol. 9, 1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathaliyawala T, Kubota M, Yudanin N, Turner D, Camp P, Thome JJ, Bickham KL, Lerner H, Goldstein M, Sykes M, et al. (2013). Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity 38, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel JM, Fraser KA, Beura LK, Pauken KE, Vezys V, and Masopust D (2014). T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science 346, 98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senda T, Dogra P, Granot T, Furuhashi K, Snyder ME, Carpenter DJ, Szabo PA, Thapa P, Miron M, and Farber DL (2019). Microanatomical dissection of human intestinal T-cell immunity reveals site-specific changes in gut-associated lymphoid tissues over life. Mucosal Immunol. 12, 378–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugay M, Bagaev DV, Turchaninova MA, Bolotin DA, Britanova OV, Putintseva EV, Pogorelyy MV, Nazarov VI, Zvyagin IV, Kirgizova VI, et al. (2015). VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Comput. Biol. 11, e1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder ME, Finlayson MO, Connors TJ, Dogra P, Senda T, Bush E, Carpenter D, Marboe C, Benvenuto L, Shah L, et al. (2019). Generation and persistence of human tissue-resident memory T cells in lung transplantation. Sci. Immunol. 4, eaav5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert EM, Schenkel JM, Fraser KA, Beura LK, Manlove LS, Igyártó BZ., Southern PJ, and Masopust D (2015). Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell 161, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo PA, Levitin HM, Miron M, Snyder ME, Senda T, Yuan J, Cheng YL, Bush EC, Dogra P, Thapa P, et al. (2019a). Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. 10, 4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo PA, Miron M, and Farber DL (2019b). Location, location, location: Tissue resident memory T cells in mice and humans. Sci. Immunol. 4, eaas9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo PA, Dogra P, Gray JI, Wells SB, Connors TJ, Weisberg SP, Krupska I, Matsumoto R, Poon MML, Idzikowski E, et al. (2021). Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity 54, 797–814.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro JR, Turner D, Pham Q, Wherry EJ, Lefrançois L, and Farber DL (2011). Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187, 5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom R, Tipton T, Strecker T, Hall Y, Akoi Bore J, Maes P, Raymond Koundouno F, Fehling SK, Krähling V, Steeds K, et al. (2021). Longitudinal antibody and T cell responses in Ebola virus disease survivors and contacts: an observational cohort study. Lancet Infect. Dis. 21, 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome JJ, Yudanin N, Ohmura Y, Kubota M, Grinshpun B, Sathaliya-wala T, Kato T, Lerner H, Shen Y, and Farber DL (2014). Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell 159, 814–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DL, Bickham KL, Thome JJ, Kim CY, D’Ovidio F, Wherry EJ, and Farber DL (2014). Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 7, 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vescovini R, Biasini C, Fagnoni FF, Telera AR, Zanlari L, Pedrazzoni M, Bucci L, Monti D, Medici MC, Chezzi C, et al. (2007). Massive load of functional effector CD4+ and CD8+ T cells against cytomegalovirus in very old subjects. J. Immunol. 179, 4283–4291. [DOI] [PubMed] [Google Scholar]
- Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, and Peters B (2019). The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 47 (D1), D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskom ML (2021). seaborn: statistical data visualization. J. Open Source Softw. 6, 3021. [Google Scholar]
- Weisberg SP, Carpenter DJ, Chait M, Dogra P, Gartrell-Corrado RD, Chen AX, Campbell S, Liu W, Saraf P, Snyder ME, et al. (2019). Tissue-Resident Memory T Cells Mediate Immune Homeostasis in the Human Pancreas through the PD-1/PD-L1 Pathway. Cell Rep. 29, 3916–3932.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg SP, Connors TJ, Zhu Y, Baldwin MR, Lin WH, Wontakal S, Szabo PA, Wells SB, Dogra P, Gray J, et al. (2021a). Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat. Immunol. 22, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg SP, Ural BB, and Farber DL (2021b). Tissue-specific immunity for a changing world. Cell 184, 1517–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Cerpas C, Angelo MA, Bangs DJ, Sidney J, Paul S, Peters B, Sanches FP, Silvera CG, Costa PR, et al. (2015). Human CD8+ T-Cell Responses Against the 4 Dengue Virus Serotypes Are Associated With Distinct Patterns of Protein Targets. J. Infect. Dis. 212, 1743–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, van den Akker JPC, Molenkamp R, Koopmans MPG, van Gorp ECM, et al. (2020). Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 5, eabd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, and McGoogan JM (2020). Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA 323, 1239–1242. [DOI] [PubMed] [Google Scholar]
- Wu T, Hu Y, Lee YT, Bouchard KR, Benechet A, Khanna K, and Cauley LS (2014). Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol. 95, 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee Mon KJ, Goldsmith E, Watson NB, Wang J, Smith NL, and Rudd BD (2019). Differential Sensitivity to IL-12 Drives Sex-Specific Differences in the CD8+ T Cell Response to Infection. Immunohorizons 3, 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zens KD, Chen JK, and Farber DL (2016). Vaccine-generated lung tissue-resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight 1,e85832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhao J, Mangalam AK, Channappanavar R, Fett C, Meyerholz DK, Agnihothram S, Baric RS, David CS, and Perlman S (2016). Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity 44, 1379–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Flow cytomentry and TCR sequencing data reported in this paper have been deposited in the Immunology Database and Analysis Portal (ImmPort) under accession number ImmPort: SDY1885. Single-cell RNA sequencing data reported in this paper has been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GEO: GSE178837.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available form the lead contact upon request