Abstract
The increasing world population and living standards urgently necessitate the transition towards a sustainable food system. One solution is microbial protein, i.e. using microbial biomass as alternative protein source for human nutrition, particularly based on renewable electron and carbon sources that do not require arable land. Upcoming green electrification and carbon capture initiatives enable this, yielding new routes to H2, CO2 and CO2‐derived compounds like methane, methanol, formic‐ and acetic acid. Aerobic hydrogenotrophs, methylotrophs, acetotrophs and microalgae are the usual suspects for nutritious and palatable biomass production on these compounds. Interestingly, these compounds are largely un(der)explored for purple non‐sulfur bacteria, even though these microbes may be suitable for growing aerobically and phototrophically on these substrates. Currently, selecting the best strains, metabolisms and cultivation conditions for nutritious and palatable microbial food mainly starts from empirical growth experiments, and mostly does not stretch beyond bulk protein. We propose a more target‐driven and efficient approach starting from the genome‐embedded potential to tuning towards, for instance, essential amino‐ and fatty acids, vitamins, taste,... Genome‐scale metabolic models combined with flux balance analysis will facilitate this, narrowing down experimental variations and enabling to get the most out of the ‘best’ combinations of strain and electron and carbon sources.
Renewable H2‐ and CO2‐derived compounds as novel resource framework for microbial biomass production
Food production and the planetary boundaries are already beyond the limits of sustainability (Steffen et al., 2015), yet even more pressure on Earth’s carrying capacity is expected with the global population projected to reach 9.2–12.3 billion people by the turn of the century and the rise in living standards (Gerland et al., 2014). Structural changes of the agricultural‐based food chain are required to increase food production and simultaneously reduce the environmental footprint (Horton, 2017). An alternative route to conventional food is the production of microorganisms as a source of human food also known as microbial protein or single‐cell protein (Pikaar et al., 2018). The cultivation of microorganisms for food has many environmental benefits compared to agricultural crop production such as a reduction in arable land expansion, greenhouse gas emissions, nitrogen pollution and water use (Matassa et al., 2016; Pikaar et al., 2018).
Despite the environmental advantages of microbial protein as a food ingredient, studied and industrial production ways are mostly based on agricultural products or fossil fuels as a source of electron donors and/or carbon sources such as molasses, sucrose, starch, methane from natural gas, n‐alkanes and methanol (Nasseri et al., 2011). To truly revolutionize sustainable food production, uncoupling from agriculture or non‐renewable fossil fuels is needed, and hence another resource usage framework. Strong potential lies in the ‘green’ electrification of the chemical industry. This entails electricity production from photovoltaics or wind turbines during off‐peak hours followed by water or carbon dioxide (CO2) reduction with the renewable electron (Martens et al., 2017). First, hydrogen gas (H2) is generated through water electrolysis as a fuel for heat, energy or transportation in the so‐called hydrogen economy (Marbán and Valdés‐Solís, 2007). Secondly, CO2 is reduced into simple C1 or C2 building blocks such as methane (CH4), methanol (CH3OH), formic acid (HCOOH) and acetic acid (CH3COOH) as starting point for (bio)chemical synthesis a.k.a carbon capture and utilization (Martens et al., 2017; Satanowski and Bar‐Even, 2020). Renewable H2 or CO2‐derived compounds can then be used as an electron donor and/or carbon source for more sustainable microbial protein production (Pikaar et al., 2018; Linder, 2019).
The more traditional R&D targets for growing microbial food in a largely land‐ and fossil‐free manner are (aerobic) hydrogen oxidizing bacteria, methylotrophs, acetotrophs or (phototrophic) microalgae respectively cultivated on H2, C1 compounds, CH3COOH and CO2 (Linder, 2019; Fig. 1). Other potentially appealing microbes for microbial production are purple non‐sulphur bacteria (PNSB; Alloul et al., 2021b). They are typically explored photoheterotrophically for resource recovery and environmental technology (Alloul et al., 2018; Cerruti et al., 2020). Even though PNSB are extremely metabolic versatile, research studying their growth for aerobic or phototrophic hydrogen or methylotrophy is rather limited (Fig. 1).
Fig. 1.
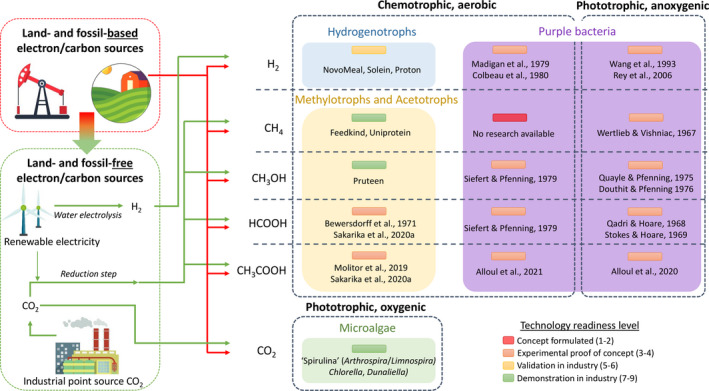
Transition towards electron donors and carbon sources that are not relying on arable land or fossil fuels, enabled by ‘green’ electrification of the chemical industry based on water electrolysis and carbon capture and utilization approaches. These routes can generate several electron donors and/or carbon sources such as hydrogen gas (H2), methane (CH4), methanol (CH3OH), formic acid (HCOOH), acetic acid (CH3COOH) and carbon dioxide (CO2), which are directed to the production of several major classes of chemotrophic and phototrophic microbes and metabolisms. A simplified technology readiness level (TRL), based on the Horizon 2020 work programme definitions, indicates the highest level of maturity of the microbial protein production technology. For commercial products (TRL 5‐9), examples of brand names are indicated, and for others, indicative scientific literature sources are given (TRL 1‐4). Copyright‐free images were sourced from Freepik.com.
Biomass cultivation for microbial protein in general, mainly aims at enhancing the growth rate and (bulk) protein content, rarely targeting nutritious compounds such as essential amino and fatty acids, vitamins and antioxidants or palatability such as taste, odour, texture and appearance. High nutritious biomass quality is, nonetheless, essential to better match the dietary requirements of humans and increase the monetary value of the product. Sensory experience or palatability is also key for the acceptance of food (Lawless, 1991). A study, for instance, showed no clinical side effects from microbial protein intake (12–15% of the daily required nitrogen), yet the taste was critical for the acceptance of food by the participants (Abrahamsson et al., 1971).
Currently, improving the nutritional biomass quality, palatability and growth rate is generally achieved by first selecting or modifying (ethical/legal challenging) the best strain and metabolism followed by optimizing the cultivation conditions (i.e. nutritional quality steering) to get nutrition‐wise the ‘best’ out of the genome. This is typically realized through a non‐targeted and iterative cultivation‐based methodology which is labour‐intensive and empirical. There is a lot of genomic data and techniques available to mine the potential of microbes, yet it is still not commonly applied in the field of microbial protein production. A new approach is, therefore, needed to unlock the full genome‐embedded potential of microbes for nutritious and palatable microbial food and enable more targeted and efficient experimenting.
The usual microbial protein suspects on H2, CO2 and/or C1‐C2 compounds
To date, a variety of (metabolic groups of) microbes has typically been considered for microbial protein production on H2 and C1–C2 compounds with different technology readiness levels Fig. 1. The following section illustrates that strain selection has been briefly touched upon, yet optimizing the cultivation conditions and improving the palatability is often overlooked in literature and, if performed, based on a cultivation‐based approach.
Aerobic hydrogenotrophs, a.k.a. hydrogen oxidizing bacteria, are considered by several companies such as Deep Branch Technology (‘Proton’), NovoNutrients (‘NovoMeal’) and Solar foods (‘Solein’), which all state the importance for the transition towards renewable H2 for protein production (Deep Branch Biotechnology; NovoNutrients; Solar Foods). Unfortunately, none of these companies disclose the used species but Cupriavidus necator has predominantly been studied (Pander et al., 2020). Volova and Barashkov (2010) performed strain selection based on the essential amino acid profile for three species, which included Cupriavidus necator (formerly known as Ralstonia eutropha). However, most studies are focused on the optimization of polyhydroxyalkanoate production by aerobic hydrogenotrophs for valorization as bioplastics instead of maximizing nutritional compounds (Pander et al., 2020).
Production of Methanotrophs with CH4 as electron source, is currently the most mature technology on the market. Companies such as Unibio A/S (‘UniProtein’) and Calysta (‘Feedkind’) are using a strain of Methylococcus capsulatus (Calysta; Unibio Group). A study on strain selection has been performed by D'Mello (1972). They reported that, from six species, M. capsulatus had the highest crude protein content and Methylomonas agile showed to have the highest methionine and cysteine concentration. Literature on nutritional quality steering is more difficult to find and, probably, restricted to undisclosed company reports.
For CH3OH, a fully demonstrated technology was developed in the 1970s by Imperial Chemical Industries which used Methylophilus methylotrophuson (‘Pruteen’; Westlake, 1986). Abou‐Zeid and Baghlaf (1983) reported strain selection based on the essential amino acid profile for three species.
Aerobic microbial protein production on HCOOH and CH3COOH is still in the laboratory phase (Bewersdorff and Dostalek, 1971; Molitor et al., 2019; Sakarika et al., 2020a). While for both compounds strain selection has been performed by Sakarika et al. (2020a), no reports have been found, thus far, on nutritional quality steering. For palatability, on the other hand, HCOOH and CH3COOH are probably one of the few compounds that have been studied. A recent publication by Sakarika et al. (2020b) on HCOOH or CH3COOH showed that the type of culture (pure vs. mixed), species and compound affects the sensory properties.
Phototrophic microalgae production on CO2 has been extensively considered and developed for human and animal consumption, albeit more as functional food than as protein source, with numerous companies in Asia and North America (Ritala et al., 2017; Koyande et al., 2019). Screening of commercially available ‘Spirulina’ (genera Arthrospira/Limnospira) and Chlorella revealed a relatively large heterogeneity, and hence optimization potential (Muys et al., 2019). Microalgae are probably the most extensively studied microbes for strain selection and cultivation‐based nutritional quality steering. Most studies focus on improving the essential amino acid profile by species selection (Hempel et al., 2012; Muys et al., 2019) or by changing environmental factors (Ogbonda et al., 2007; Sui et al., 2019b). Essential fatty acids (Hempel et al., 2012), vitamins (Watanabe et al., 2002) and antioxidants (Richmond, 2008; Sui et al., 2019a) have also been explored through species selection and parameter optimization. In terms of palatability, microalgae are also thoroughly explored for flavour, taste and texture (Lafarga, 2019). Many commercial products are available in the form of capsules, powder or integrated in food (e.g. chocolate, crackers, pasta, etc.; (Lafarga, 2019).
Purple bacteria for hydrogenotrophy, methylotrophy and acetotrophy?
PNSB are appealing microbes for nutritious biomass production. Their potential is, first, derived from their highly versatile metabolism (Imhoff, 2006; Alloul et al., 2020), which allows examining a variety of electron and energy sources to steer towards nutritious biomass. Secondly, they have an appealing intrinsic nutritious biomass composition rich in protein with a considerable amount of vitamins (e.g. vitamin B2, B6, C, E, D and folic acid) and carotenoid pigments (e.g. spirilloxanthin, rhodopin, okenone and rhodopinal; Sasaki et al., 1998).
Strain selection and cultivation‐based nutritional quality steering on land‐ and fossil bound electron donors and complex mixtures (e.g. waste streams) have been successful for PNSB. For example, photosynthetic pigments (antioxidant properties) are normally not induced under aerobic dark conditions, yet researchers found that cultivating PNSB at dissolved oxygen concentrations lower than 0.4 mg O2 l−1 triggers the pigment synthesis (Ghosh et al., 1994; Alloul et al., 2021a). Moreover, in previous research, we selected Rhodobacter capsulatus as the most promising PNSB based on growth rate (Alloul et al., 2019). In the same article, we also showed that mixtures of volatile fatty acids improve the growth performance of several pure and mixed PNSB cultures relative to individual volatile fatty acids. In another paper, we observed that fructose as a carbon source enhanced the protein content of several PNSB species compared to growth on volatile fatty acids, alcohols or other sugars (Alloul et al., 2021a).
Microbial protein production with PNSB on H2‐ and CO2‐derived compounds is largely unexplored and research is mainly limited to phenotypic screenings rather than strain selection or directed towards nutritional compounds. For example, for H2, only two articles were published exploring the fundamentals of chemoautotrophic growth of two Rb. capsulatus strains (Madigan and Gest, 1979; Colbeau et al., 1980). Photoautotrophic growth has also been studied for Rb. sphaeroides, Rhodospirillum rubrum and Rhodopseudomonas palustris, with a dedicated focus on the metabolism, genetic regulation and growth kinetics (Wang et al., 1993; Rey et al., 2006). For CH4, only one article from the 1960s claims that Rps. gelatinosa is able to incorporate CH4 into biomass (Wertlieb and Vishniac, 1967), yet no other research is available whatsoever to support this. CH3OH is better studied with one article from the 1970s performing a kind of strain selection based on phototrophic growth rates of 39 isolates. They showed that a particular strain of Rhodoblastus acidophilus, formerly known as Rhodopseudomonas acidophila, was most suitable for growth on CH3OH (maximum specific growth rate 2.3 d−1; Douthit and Pfennig, 1976). The chemotrophic growth, on the other hand, was not well characterized (Quayle and Pfennig, 1975). For HCOOH, Stokes and Hoare (1969) explored its assimilation on a metabolic level. Growth rates and yields were also studied (Qadri and Hoare, 1968; Siefert and Pfennig, 1979), yet no nutritious compounds nor nutritional quality steering were reported.
Overall, there are still several research opportunities and gaps related to aerobic or phototrophic hydrogen or methylotrophic PNSB production. More research is required for strain selection and cultivation optimization for the production of nutritious and palatable microbial food.
Genome‐informed selection for nutritious biomass
Despite successful accomplishments, research aimed at enhancing the growth performance and nutritional quality of microbial biomass production remains labour‐intensive and is mainly empirical (Fig. 2). Investigations rarely start off with mining the genomic information, even though this is increasingly becoming available. The nec‐plus‐ultra approach to capitalize on the intrinsic biological potential of strains is the utilization of genome‐scale metabolic models (GEM; Oberhardt et al., 2009). These models account for every known metabolic reaction encoded in the genome of an organism and they can be generated from a genome assembly using automated metabolic reconstruction tools (Henry et al., 2010; Machado et al., 2018). GEM can be used to predict the metabolic phenotype of an organism under given growth conditions using flux balance analysis (FBA), a simulation method that calculates the optimal flow of metabolites through the metabolic network and its response to environmental and genetic perturbation. The combination of GEM and FBA allows to predict how the metabolic phenotype responds to different environmental and genetic perturbations and has become a popular computational framework for multiple biotechnological applications (Oberhardt et al., 2009).
Fig. 2.
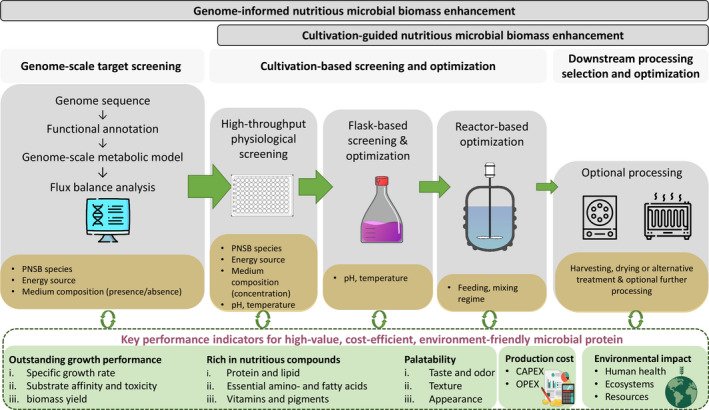
Proposed genome‐scale computational approach for targeted screening and nutritional quality steering. Grey boxes show the necessary steps in the route from computational prediction to downstream processing. Yellow boxes represent the variables of each step in the pipeline. The decreasing height of the green arrows depicts the number of variables (parameter values or options) to select in each step. Each step interacts with the key performance indicator. CAPEX, capital expenditure; OPEX, operational expenditure. This figure has been designed using resources from Freepik.com and flaticon.com.
In the context of nutritious microbial biomass production, this approach enables to mechanistically understand the effect of bioprocess variables such as electron donor and energy source on the relative changes in the fluxes of key pathways, such as those involved in the synthesis of essential amino and fatty acids, vitamins and pigments. This methodology will, therefore, allow narrowing down experimental variations and enable to get most out of the ‘best’ combinations of strain and electron and carbon sources (Fig. 2). Golomysova et al. (2010) for example, have used this approach on the Rb. sphaeroides for H2 production. From several individual and combined carbon sources, they accurately predicted that lactate resulted in the highest H2 productivity. A more extensive screening by Imam et al. (2011) tested the effect of different carbon, nitrogen and growth modes (e.g. aerobic and photoheterotrophic) on the growth rate, hydrogen and polyhydroxybutyrate production. In silico analysis corresponded well with the experimental observations and showed that H2 production was highest for a succinate glutamate mixture.
Although genome‐scale target screening is a powerful tool to predict relative changes in the fluxes of target pathways related to nutritional compounds, its main limitation lies in the fact that it falls short to compute the relative abundance of nutritional compounds in the biomass. In fact, the reconstruction of GEM requires a biomass objective function that describes the biomass composition of the organism in terms of the steady‐state concentration (mmol per gram of dry weight) of its individual components (amino acids, lipids, nucleotides, vitamins, cofactors). An accurately determined biomass objective function is essential to correctly estimate the growth rate and biomass yield on different compounds, yet for the sake of automation, this information is usually extracted from phylogenetically‐close model organisms. Currently, one can only infer how the abundance of a given compound is affected through different perturbations by observing the response at the level of its respective pathway fluxes.
Recently, a variety of methods have been proposed to replace the biomass objective function with objective functions based on gene expression data, but their predictive power seems limited (Machado and Herrgard, 2014). Lachance et al. (2019) propose a computational workflow to generate a species‐specific biomass objective function, but it still requires experimental determination of the relative fraction of the main macromolecules (DNA, RNA, proteins and lipids), and gene expression data to determine the abundance of individual nucleotides and amino acids (Lachance et al., 2019). A new class of models of metabolism and gene expression (ME‐models) extends the traditional GEM by explicitly modelling all the biosynthetic machinery (ribosomes, nucleotides and proteins; O'Brien et al., 2015). ME‐models can fully predict gene and protein expression as well as the biomass composition. However, their reconstruction requires a large knowledgebase and a significant amount of manual curation, making them still unsuitable for integration into a fast‐screening process.
In summary, genome‐driven enhancement of microbial food development shows great potential, particularly based on GEM. The combination of GEM and FBA is the most established and applicable approach as a promising alternative to the typical empirical cultivation‐based methodology. As researchers did successfully for other examples of GEM applications, it is time to embark on showing the efficacy of such approach in selecting the best combinations of strain, metabolism and cultivation conditions. This approach may facilitate the ultimate goal of industrially producing very nutritious and palatable microbial food in a cost‐effective and ecological manner.
Acknowledgements
The authors would like to kindly acknowledge the DOCPRO4 project ‘PurpleTech’ funded by the BOF (Bijzonder onderzoeksfonds; Special research fund) from the University of Antwerp, and the project 'Saraswati 2.0' (821427) funded by the European Union’s Horizon 2020 Research and Innovation programme, for financial support of J.S. and A.A., respectively.
Microbial Biotechnology (2021) 15(1), 6–12
References
- Abou‐Zeid, A.‐Z.‐A. , and Baghlaf, A.O. (1983) Methanol as the carbon source of production of single‐cell proteins (SCP‐s). Zentralblatt für Mikrobiol 138: 451–464. [PubMed] [Google Scholar]
- Abrahamsson, L. , Hambraeus, L. , Hofvander, Y. , and Vahlquist, B. (1971) Single cell protein in clinical testing: a tolerance test in healthy adult subjects comprising biochemical, clinical and dietary evaluation. In Ann Nutr Metab. 13: 186–199. [PubMed] [Google Scholar]
- Alloul, A. , Cerruti, M. , Adamczyk, D. , Weissbrodt, D.G. , and Vlaeminck, S.E. (2020) Control tools to selectively produce purple bacteria for microbial protein in raceway reactors. bioRxiv. 10.1101/2020.1101.1120.912980 [DOI] [PubMed] [Google Scholar]
- Alloul, A. , Ganigué, R. , Spiller, M. , Meerburg, F. , Cagnetta, C. , Rabaey, K. , and Vlaeminck, S.E. (2018) Capture–ferment–upgrade: a three‐step approach for the valorization of sewage organics as commodities. Environ Sci Technol 52: 6729–6742. [DOI] [PubMed] [Google Scholar]
- Alloul, A. , Muys, M. , Hertoghs, N. , Kerckhof, F.‐M. , and Vlaeminck, S.E. (2021a) Cocultivating aerobic heterotrophs and purple bacteria for microbial protein in sequential photo‐and chemotrophic reactors. Biores Technol 319: 124192. [DOI] [PubMed] [Google Scholar]
- Alloul, A. , Wille, M. , Lucenti, P. , Bossier, P. , Van Stappen, G. , and Vlaeminck, S.E. (2021b) Purple bacteria as added‐value protein ingredient in shrimp feed: Penaeus vannamei growth performance, and tolerance against Vibrio and ammonia stress. Aquaculture 530: 735788. [Google Scholar]
- Alloul, A. , Wuyts, S. , Lebeer, S. , and Vlaeminck, S.E. (2019) Volatile fatty acids impacting phototrophic growth kinetics of purple bacteria: paving the way for protein production on fermented wastewater. Water Res 152: 138–147. [DOI] [PubMed] [Google Scholar]
- Bewersdorff, M. , and Dostalek, M. (1971) The use of methane for production of bacterial protein. Biotechnol Bioeng 13: 49–62. [DOI] [PubMed] [Google Scholar]
- Calysta (2020). URL http://www.feedkind.com/
- Cerruti, M. , Stevens, B. , Ebrahimi, S. , Alloul, A. , Vlaeminck, S.E. , and Weissbrodt, D.G. (2020) Enrichment and aggregation of purple non‐sulfur bacteria in a mixed‐culture sequencing‐batch photobioreactor for biological nutrient removal from wastewater. Front Bioeng Biotechnol 8: 1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbeau, A. , Kelley, B.C. , and Vignais, P.M. (1980) Hydrogenase activity in Rhodopseudomonas capsulata: relationship with nitrogenase activity. J Bacteriol 144: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deep Branch Biotechnology (2020) [November 2020]. URL https://deepbranchbio.com/
- D'Mello, J.P. (1972) A study of the amino acid composition of methane utilizing bacteria. J Appl Bacteriol 35: 145–148. [DOI] [PubMed] [Google Scholar]
- Douthit, H.A. , and Pfennig, N. (1976) Isolation and growth rates of methanol utilizing Rhodospirillaceae. Arch Microbiol 107: 233–234. [DOI] [PubMed] [Google Scholar]
- Gerland, P. , Raftery, A.E. , Ševčíková, H. , Li, N. , Gu, D. , Spoorenberg, T. , et al. (2014) World population stabilization unlikely this century. Science 346: 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, R. , Hardmeyer, A. , Thoenen, I. , and Bachofen, R. (1994) Optimization of the Sistrom culture medium for large‐scale batch cultivation of Rhodospirillum rubrum under semiaerobic conditions with maximal yield of photosynthetic membranes. Appl Environ Microbiol 60: 1698–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomysova, A. , Gomelsky, M. , and Ivanov, P.S. (2010) Flux balance analysis of photoheterotrophic growth of purple nonsulfur bacteria relevant to biohydrogen production. Int J Hydrogen Energy 35: 12751–12760. [Google Scholar]
- Hempel, N. , Petrick, I. , and Behrendt, F. (2012) Biomass productivity and productivity of fatty acids and amino acids of microalgae strains as key characteristics of suitability for biodiesel production. J Appl Phycol 24: 1407–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, C.S. , DeJongh, M. , Best, A.A. , Frybarger, P.M. , Linsay, B. , and Stevens, R.L. (2010) High‐throughput generation, optimization and analysis of genome‐scale metabolic models. Nat Biotechnol 28: 977–982. [DOI] [PubMed] [Google Scholar]
- Horton, P. (2017) We need radical change in how we produce and consume food. Food Security 9: 1323–1327. [Google Scholar]
- Imam, S. , Yilmaz, S. , Sohmen, U. , Gorzalski, A.S. , Reed, J.L. , Noguera, D.R. , and Donohue, T.J. (2011) iRsp1095: a genome‐scale reconstruction of the Rhodobacter sphaeroides metabolic network. BMC Syst Biol 5: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhoff, J.F. (2006) The phototrophic alpha‐proteobacteria. In The Prokaryotes. Dworkin, M. , Falkow, S. , Rosenberg, E. , Schleifer, K.‐H. , and Stackebrandt, E. (eds). New York: Springer, pp. 41–64. [Google Scholar]
- Koyande, A.K. , Chew, K.W. , Rambabu, K. , Tao, Y. , Chu, D.‐T. , and Show, P.‐L. (2019) Microalgae: a potential alternative to health supplementation for humans. Food Science and Human Wellness 8: 16–24. [Google Scholar]
- Lachance, J.C. , Lloyd, C.J. , Monk, J.M. , Yang, L. , Sastry, A.V. , Seif, Y. , et al. (2019) BOFdat: Generating biomass objective functions for genome‐scale metabolic models from experimental data. PLoS Comput Biol 15: e1006971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarga, T. (2019) Effect of microalgal biomass incorporation into foods: Nutritional and sensorial attributes of the end products. Algal Res 41: 101566. [Google Scholar]
- Lawless, H. (1991) The sense of smell in food quality and sensory evaluation. J Food Qual 14: 33–60. [Google Scholar]
- Linder, T. (2019) Making the case for edible microorganisms as an integral part of a more sustainable and resilient food production system. Food Security 11: 265–278. [Google Scholar]
- Machado, D. , Andrejev, S. , Tramontano, M. , and Patil, K.R. (2018) Fast automated reconstruction of genome‐scale metabolic models for microbial species and communities. Nucleic Acids Res 46: 7542–7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado, D. , and Herrgard, M. (2014) Systematic evaluation of methods for integration of transcriptomic data into constraint‐based models of metabolism. PLoS Comput Biol 10: e1003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan, M.T. , and Gest, H. (1979) Growth of the photosynthetic bacterium Rhodopseudomonas capsulata chemoautotrophically in darkness with H2 as the energy source. J Bacteriol 137: 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbán, G. , and Valdés‐Solís, T. (2007) Towards the hydrogen economy? Int J Hydrogen Energy 32: 1625–1637. [Google Scholar]
- Martens, J.A. , Bogaerts, A. , De Kimpe, N. , Jacobs, P.A. , Marin, G.B. , Rabaey, K. , et al. (2017) The chemical route to a carbon dioxide neutral world. Chemsuschem 10: 1039–1055. [DOI] [PubMed] [Google Scholar]
- Matassa, S. , Boon, N. , Pikaar, I. , and Verstraete, W. (2016) Microbial protein: future sustainable food supply route with low environmental footprint. Microb Biotechnol 9: 568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor, B. , Mishra, A. , and Angenent, L.T. (2019) Power‐to‐protein: converting renewable electric power and carbon dioxide into single cell protein with a two‐stage bioprocess. Energy Environ Sci 12: 3515–3521. [Google Scholar]
- Muys, M. , Sui, Y.X. , Schwaiger, B. , Lesueur, C. , Vandenheuvel, D. , Vermeir, P. , and Vlaeminck, S.E. (2019) High variability in nutritional value and safety of commercially available Chlorella and Spirulina biomass indicates the need for smart production strategies. Biores Technol 275: 247–257. [DOI] [PubMed] [Google Scholar]
- Nasseri, A.T. , Rasoul‐Amini, S. , Morowvat, M.H. , and Ghasemi, Y. (2011) Single cell protein: production and process. Am J Food Technol 6: 103–116. [Google Scholar]
- NovoNutrients (2020) URL https://www.novonutrients.com/
- Oberhardt, M.A. , Palsson, B.O. , and Papin, J.A. (2009) Applications of genome‐scale metabolic reconstructions. Mol Syst Biol 5: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, E.J. , Monk, J.M. , and Palsson, B.O. (2015) Using genome‐scale models to predict biological capabilities. Cell 161: 971–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbonda, K.H. , Aminigo, R.E. , and Abu, G.O. (2007) Influence of temperature and pH on biomass production and protein biosynthesis in a putative Spirulina sp. Biores Technol 98: 2207–2211. [DOI] [PubMed] [Google Scholar]
- Pander, B. , Mortimer, Z. , Woods, C. , McGregor, C. , Dempster, A. , Thomas, L. , et al. (2020) Hydrogen oxidising bacteria for production of single‐cell protein and other food and feed ingredients. Eng Biol 4: 21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaar, I. , Matassa, S. , Bodirsky, B.L. , Weindl, I. , Humpenöder, F. , Rabaey, K. , et al. (2018) Decoupling livestock from land use through industrial feed production pathways. Environ Sci Technol 52: 7351–7359. [DOI] [PubMed] [Google Scholar]
- Qadri, S.H. , and Hoare, D. (1968) Formic hydrogenlyase and the photoassimilation of formate by a strain of Rhodopseudomonas palustris . J Bacteriol 95: 2344–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quayle, J. , and Pfennig, N. (1975) Utilization of methanol by Rhodospirillaceae. Arch Microbiol 102: 193–198. [DOI] [PubMed] [Google Scholar]
- Rey, F.E. , Oda, Y. , and Harwood, C.S. (2006) Regulation of uptake hydrogenase and effects of hydrogen utilization on gene expression in Rhodopseudomonas palustris . J Bacteriol 188: 6143–6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond, A. (2008) Handbook of Microalgal Culture: Biotechnology and Applied Phycology. John Wiley & Sons. [Google Scholar]
- Ritala, A. , Häkkinen, S.T. , Toivari, M. , and Wiebe, M.G. (2017) Single cell protein—State‐of‐the‐Art, Industrial Landscape and Patents 2001–2016. Front Microbiol 8: 2001–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakarika, M. , Candry, P. , Depoortere, M. , Ganigué, R. , and Rabaey, K. (2020a) Impact of substrate and growth conditions on microbial protein production and composition. Biores Technol 317: 124021. [DOI] [PubMed] [Google Scholar]
- Sakarika, M. , Sosa, D.A.T. , Depoortere, M. , Rottiers, H. , Ganigué, R. , Dewettinck, K. , and Rabaey, K. (2020b) The type of microorganism and substrate determines the odor fingerprint of dried bacteria targeting microbial protein production. FEMS Microbiol Lett 367: fnaa138. [DOI] [PubMed] [Google Scholar]
- Sasaki, K. , Tanaka, T. , and Nagai, S. (1998) Use of photosynthetic bacteria for the production of SCP and chemicals from organic wastes. In Bioconversion of waste materials to industrial products. Martin, A.M. (ed). Boston, MA: Springer, pp. 247–292. [Google Scholar]
- Satanowski, A. , and Bar‐Even, A. (2020) A one‐carbon path for fixing CO 2: C1 compounds, produced by chemical catalysis and upgraded via microbial fermentation, could become key intermediates in the valorization of CO 2 into commodity chemicals. EMBO Rep 21: e50273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefert, E. , and Pfennig, N. (1979) Chemoautotrophic growth of Rhodopseudomonas species with hydrogen and chemotrophic utilization of methanol and formate. Arch Microbiol 122: 177–182. [Google Scholar]
- Solar Foods (2020) URL https://solarfoods.fi/
- Steffen, W. , Richardson, K. , Rockstrom, J. , Cornell, S.E. , Fetzer, I. , Bennett, E.M. , et al. (2015) Planetary boundaries: Guiding human development on a changing planet. Science 347: 736–746. [DOI] [PubMed] [Google Scholar]
- Stokes, J.E. , and Hoare, D.S. (1969) Reductive pentose cycle and formate assimilation in Rhodopseudomonas palustris . J Bacteriol 100: 890–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui, Y. , Muys, M. , Van de Waal, D.B. , D'Adamo, S. , Vermeir, P. , Fernandes, T.V. , and Vlaeminck, S.E. (2019a) Enhancement of co‐production of nutritional protein and carotenoids in Dunaliella salina using a two‐phase cultivation assisted by nitrogen level and light intensity. Biores Technol 287: 121398. [DOI] [PubMed] [Google Scholar]
- Sui, Y. , Muys, M. , Vermeir, P. , D'Adamo, S. , and Vlaeminck, S.E. (2019b) Light regime and growth phase affect the microalgal production of protein quantity and quality with Dunaliella salina . Biores Technol 275: 145–152. [DOI] [PubMed] [Google Scholar]
- Unibio Group (2020) URL https://www.unibio.dk/
- Volova, T.G. , and Barashkov, V.A. (2010) Characteristics of proteins synthesized by hydrogen‐oxidizing microorganisms. Appl Biochem Microbiol 46: 574–579. [PubMed] [Google Scholar]
- Wang, X. , Modak, H.V. , and Tabita, F.R. (1993) Photolithoautotrophic growth and control of CO2 fixation in Rhodobacter sphaeroides and Rhodospirillum rubrum in the absence of ribulose bisphosphate carboxylase‐oxygenase. J Bacteriol 175: 7109–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, F. , Takenaka, S. , Kittaka‐Katsura, H. , Ebara, S. , and Miyamoto, E. (2002) Characterization and bioavailability of vitamin B12‐compounds from edible algae. J Nutr Sci Vitaminol (Tokyo) 48: 325–331. [DOI] [PubMed] [Google Scholar]
- Wertlieb, D. , and Vishniac, W. (1967) Methane utilization by a strain of Rhodopseudomonas gelatinosa . J Bacteriol 93: 1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake, R. (1986) Large‐scale continuous production of single cell protein. Chem Ing Tech 58: 934–937. [Google Scholar]


