Summary
Faecal Microbiota Transplantation (FMT) is considered as a promising technology to fight against obesity. Wild boar has leanermuscle and less fat in comparison to the domestic pig, which were thought to be related with microbiota. To investigate the function and mechanism of the wild boar microbiota on obesity, we first analysed the wild boar microbiota composition via 16S rDNA sequencing, which showed that Firmicutes and Proteobacteria were the dominant bacteria. Then, we established a high‐fat diet (HFD)‐induced obesity model, and transfer low and high concentrations of wild boar faecal suspension in mice for 9 weeks. The results showed that FMT prevented HFD‐induced obesity and lipid metabolism disorders, and altered the jejunal microbiota composition especially increasing the abundance of the Lactobacillus and Romboutsia, which were negatively correlated with obesity‐related indicators. Moreover, we found that the anti‐obesity effect of wild boar faecal suspension was associated with jejunal N6‐methyladenosine (m6A) levels. Overall, these results suggest that FMT has a mitigating effect on HFD‐induced obesity, which may be due to the impressive effects of FMT on the microbial composition and structure of the jejunum. These changes further alter intestinal lipid metabolism and m6A levels to achieve resistance to obesity.
Our results suggest that the fecal bacteria of wild boar have a mitigating effect on HFD‐induced obesity, which may be due to the impressive effects of wild boar fecal microbiota transplantation on the microbial composition and structure of the jejunum. These changes further alter intestinal lipid metabolism and m6A levels to achieve resistance to obesity.
Introduction
The global obesity problem is persistently prominent and accompanied by increasing morbidity and complications. Solving the problem of obesity is critical for the well‐being of humans and animals. It is well established that intestinal lipid absorption and adipose tissue lipid accumulation contribute largely to obesity and varieties of metabolism disorders (Longo et al., 2019; Dávalos‐Salas et al., 2020). Numerous studies have shown that gut microbiota‐host interactions carry out an invaluable role in obesity (Ley et al., 2005). In particular, the relationship between jejunal microbiota and obesity was gaining more and more attention in recent years. For example, studies have shown that variations of jejunal microbiota respond more directly and dramatically to diet‐induced obesity, which affects lipid absorption by controlling the intestinal epithelial processes during digestion and absorption (Martinez‐Guryn et al., 2018; Chang and Martinez‐Guryn, 2019). Since jejunum is the main site of nutrient and lipid absorption, compared with hindgut microbes, jejunal microbes are more efficiently and straightforwardly involved in regulating the absorption and metabolism of these substances (Chow and Hollander, 1979). Hence, regulating intestinal fat absorption by modulating jejunal microbes may be an alternative strategy for treatment obesity in the future.
The key mechanisms regulating intestinal lipid absorption include: lipid uptake, resynthesis of triglyceride as well as chylomicron packaging and transport (Hussain, 2014). Transmembrane proteins such as CD36, FATPs and FABPs are thought to be engaged in lipid uptake (Glatz and Luiken, 2001, 2018; Lobo et al., 2007). DGAT1, DGAT2 and Mogat2 play critical roles in triglyceride resynthesis (Yen et al., 2008; Nelson et al., 2014). MTP and ApoB48 are key factors in chylomicron packaging and transport (Hussain et al., 1996; Wang and Tran, 1999). Modulating the expression of any of these components may affect intestinal lipid absorption.
Faecal microbiota transplantation (FMT), known for the ability to metastasize the host phenotype, is often used in the treatment of obesity and inflammatory bowel diseases (IBD) (Smits et al., 2013). For instance, after receiving lean donor mice or human faecal bacteria, obese recipients ultimately exhibit a reduced acquisition of an obese phenotype (Vrieze et al., 2012; Kulecka et al., 2016). FMT from healthy donors successfully cures patients with gastrointestinal Clostridium difficile infection (Weingarden and Vaughn, 2017). Despite the growing understanding of FMT as an alternative treatment, studies to date have been limited on the response of jejunal microbiota to FMT. In addition, wild boars are known to be leaner and less fat compared with domestic pigs, while these differences may originate from the composition of microbiota (Gérard, 2016; Yang et al., 2016; Bergamaschi et al., 2020). Therefore, it will be interesting to explore whether their faecal bacteria can alleviate obesity by regulating the lipid absorption and metabolism in small intestines.
In this study, we found that the faecal bacteria obtained from a wild boar were associated with lipid metabolism. To further evaluate its effects on lipids metabolism, we established a HFD‐induced obesity model and treated with faecal bacteria from wild boars. The aim of the study is to investigate the effect of wild boar FMT on HFD‐induced obesity and to explore potential probiotics in wild boar FMT which could assist in alleviating obesity. These findings will open a new avenue for developing therapeutic strategies to solve the problem of obesity.
Results
The composition of wild boar faecal microbiota and its anti‐obesity effects on HFD‐fed mice
The wild boar faeces were analysed by 16s rDNA sequencing. As shown in Fig. S1A‐B, Firmicutes and Proteobacteria were the predominant order at the phylum level, while Enterobacteriaceae, Oscillospiraceae and Lachnospiraceae were the main bacteria at the family level. The functional prediction of bacteria suggested that wild boar faecal microbiota had the highest abundant microbial genes of metabolism which encompass energy metabolism and lipid metabolism (Fig. S1C and D).
To investigate the function of FMT on energy metabolism and lipid metabolism, mice were fed a HFD and orally administered different concentrations of wild boar faecal suspension for 9 weeks. As shown in the morphological observations of mice in Fig. 1A, FMT alleviated the obesity induced by HFD in mice. Consistently, the body weight and relative weight gain in the HFD group increased (P < 0.01) from the second week, while both concentrations of FMT (P < 0.01) inhibited HFD‐induced weight gain from the third week (Fig. 1B and C). However, no differences (P > 0.05) were observed in food intake and energy intake among the HFD group, HFD+LF group and HFD+HF group (Fig. 1D and E). Similarly, FMT alleviated HFD‐induced obesity as evidenced by a reduction in organ weights including epididymal fat, subcutaneous fat and liver. Interestingly, the weight of epididymal fat and subcutaneous fat in HFD+HF group was less (P < 0.05) than the HFD+LF group (Fig. 1F). Histological analysis of epididymal fat exhibited a decrease (P < 0.05) in the number of adipocytes per unit area in mice given FMT compared to the HFD group, no differences (P > 0.05) were observed between the HFD+LF and HFD+HF groups (Fig. 1G and H). The adipocyte size distribution frequency in each group indicated that the proportion of small‐sized adipocytes in the epididymal fat increased and the percentage of medium and large adipocytes decreased after gavage of wild boar faecal suspension (Fig. 1I). The above results suggested that FMT could functionally alleviate HFD‐induced obesity.
Fig. 1.
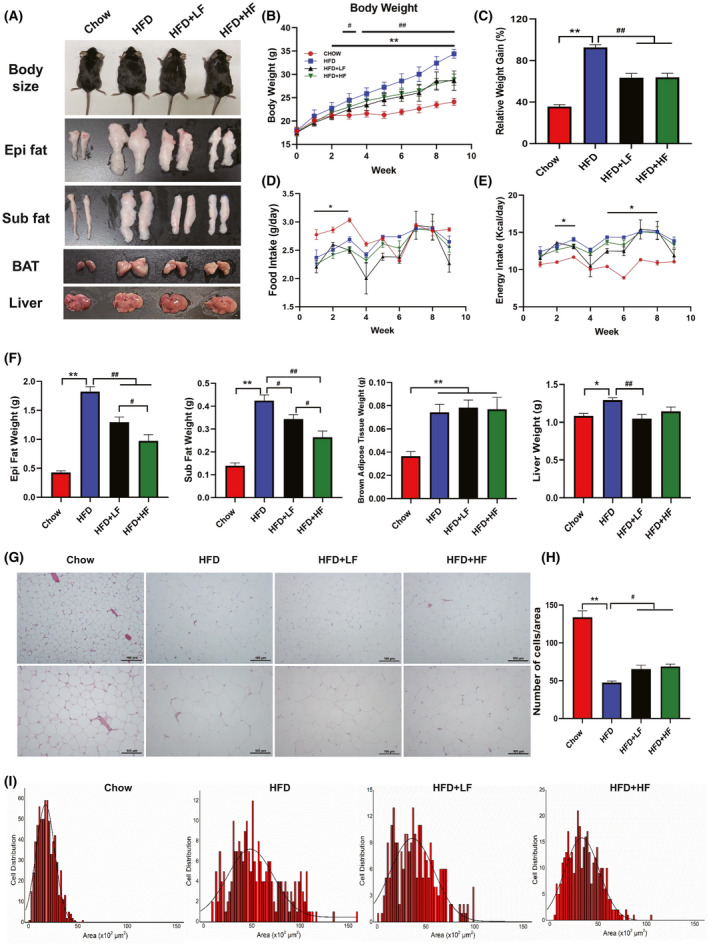
FMT attenuated high‐fat diet‐induced obesity.
A. Morphological observations of mice body size, epi fat, sub fat, BAT and Liver.
B. Body weight.
C. Relative weight gain.
D. Food intake.
E. Energy intake.
F. Organ weight.
G. The representative.
H&E. staining of the epididymal fat, the bars represent 100 μm.
H. Number of cells per area.
I. Distribution of adipocyte size in epididymal fat. Each distribution was obtained from five mice in each group. Data were expressed as means ± SEM (A‐F: n = 8; G–I: n = 5).
* significant difference between Chow and HFD groups.
# significant difference between HFD and HFD+LF or HFD+HF groups, * or #, P < 0.05, ** or ##, P < 0.01.
FMT reduced metabolic disorders in HFD‐fed mice
Obesity is always accompanied by metabolism disorders and is associated with the increased risk of impaired glucose tolerance and development of insulin resistance (Kahn and Hull, 2006; Bouter et al., 2017). Therefore, we further examined the serum lipid profile of each group and performed GTT and ITT to ascertain whether FMT might have favourable metabolic health effects. As shown in Fig. 2A‐E, HFD elevated (P < 0.01) serum glucose, HDL, LDL, TG and TChol level. In turn, supplementation with both low and high concentrations of wild boar faecal suspension suppressed the increases in serum glucose (P < 0.01), serum HDL (P < 0.01), serum TG (P < 0.05), and serum TChol (P < 0.01), while no effects (P > 0.05) were observed on the changes in serum LDL induced by HFD. What is more, compared with mice in the HFD group, mice in the HFD+LF and HFD+HF groups showed a lower blood glucose level following glucose injection and a greater rate of glucose clearance after insulin stimulation (Fig. 2F‐I). Overall, the results suggested that the FMT exerted a positive influence in alleviating HFD‐induced obesity and metabolic disorders.
Fig. 2.
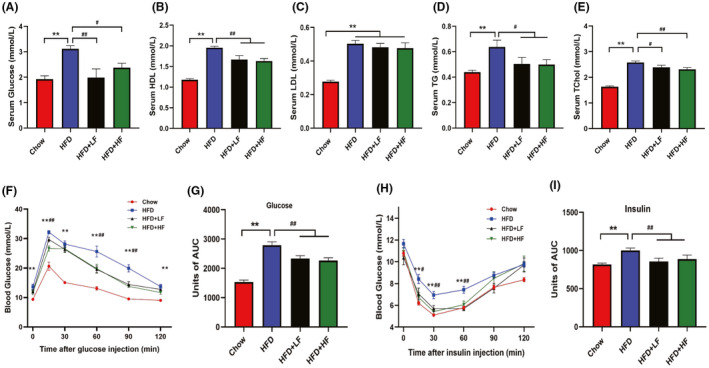
FMT attenuated high‐fat diet‐induced lipid metabolic disorders.
(A–E). Serum glucose level (A) and lipid profiles including HDL (mM) (B), LDL (mM) (C), Triglyceride (TG, mM) (D), Total cholesterol (Tchol, mM) (E).
(F–G) Injection glucose tolerance test (F) and area under the curve (AUC) (G).
(H–I) Insulin tolerance test (H) and area under the curve (AUC) (I). Data were expressed as means ± SEM (n = 8), *significant difference between Chow and HFD groups, #significant difference between HFD and HFD+LF or HFD+HF groups, * or #, P < 0.05, ** or ##, P < 0.01.
FMT attenuated the increase fat absorption in the jejunum induced by HFD
These results indicated that the anti‐obesity effect of FMT was not related to food intake or energy intake of mice (Fig. 1D and E). Intestinal lipid absorption is closely associated with obesity as described in previous studies (Li et al., 2016; Luo et al., 2018), therefore, it is reasonable to assume that FMT may affect intestinal fatty acid absorption in mice to regulate obesity. We next further investigated the effect of FMT on fatty acid absorption in the jejunum. Increased number and size of lipid droplets were observed in the jejunal villi and epithelial cells of HFD‐fed mice, and more lipids were accumulated in both epithelial cells and lamina propria of the mice, while no significant lipid accumulation was observed in the HFD+LF and HFD+HF groups (Fig. 3A and C). Concurrently, Fig. 3B and D showed that HFD reduced the villi crypt ratio and the length of microvilli, causing the loss of microvilli, and FMT effectively prevented this undesirable phenomenon triggered by HFD (P < 0.05). Decreased absorption of lipids in the intestine implies an increase in the excretion of lipids. We further found that compared to the HFD group, HFD+LF and HFD+HF groups showed a trend towards decreased jejunum TG levels and elevated stool TG levels (week 4), the jejunum TG levels were lower in the HFD+HF group than in the HFD+LF group (Fig. 3E and G). The stool TG levels were higher (P < 0.05) in the HFD+HF group mice than in the HFD group at week 8 (Fig. 3H). However, FMT had no effect (P > 0.05) on the jejunum and stool TChol levels of each group (Fig. 3F, I and J). Subsequently, we determined the expression of the major components responsible for fat absorption in the jejunum. Notably, FMT alleviated the HFD‐induced elevation (P < 0.01) of mRNA levels of the fatty acid transport key factors CD36, FABP2, FATP2, FABP4 and FATP4 (Fig. 3K). However, FMT failed to rescue the reduction (P < 0.01) of cholesterol transporter protein NPC1L1 mRNA induced by HFD. The mRNA expression of DGAT2 and ApoB48 involved in triglyceride synthesis and chylomicorn packaging, respectively, were reduced (P < 0.05) in the HFD+LF and HFD+HF groups compared with the HFD group (Fig. 3K). Furthermore, we found that FMT reduced (P < 0.05) the HFD‐induced elevation in FABP2 protein expression (Fig. 3L). Taken together, these results clearly indicated that FMT alleviated HFD‐induced obesity by mitigating increased intestinal lipid absorption.
Fig. 3.
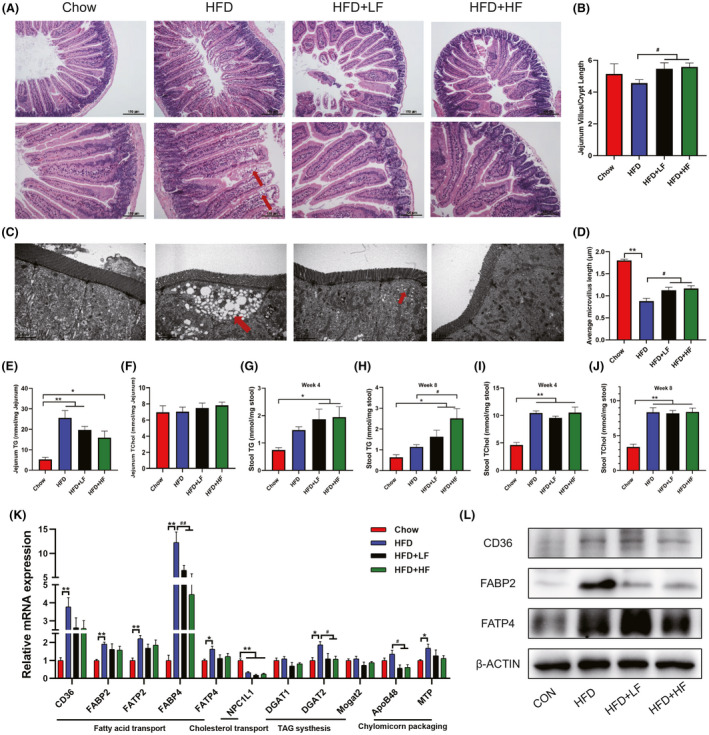
FMT attenuated the increase in jejunum fatty acid absorption induced by high‐fat diet.
(A). The representative H&E staining of the jejunum sections and.
(B) histological scores, the bars represent 100 μm (n = 4).
(C) Ultrastructural changes of lipid droplets and Microvilli under TEM (12 000×).
(D) Average microvillus length (n = 3).
(E–F) Jejunum TG (E) and total cholesterol levels (F) (n = 8).
(G–H) Stool TG levels at week 4 (G) and week 8 (H).
(I–J) Stool TChol levels at week 4 (I) and week 8 (J).
(K) mRNA levels of intestinal fat absorption related genes detected by qPCR.
(L) Proteins levels of CD36, FABP2 and FATP4 by western blot.
Data were expressed as means ± SEM (n = 6 except for those already marked).
*significant difference between Chow and HFD groups, #significant difference between HFD and HFD+LF or HFD+HF groups.
* or #, P < 0.05, ** or ##, P < 0.01.
The effects of FMT on HFD‐induced obesity are associated with jejunum m6A methylation level
As previously reported, m6A levels of liver are affected by HFD, but it has not been reported whether HFD affects intestinal m6A levels (Wu, Li, et al., 2020). Additionally, the gut microbiota has a profound effect on host m6A mRNA modifications, the changes in m6A modification levels are another manifestation of the interaction between commensal bacteria and their hosts (Wang et al., 2019; Jabs et al., 2020). In order to clarify how HFD and wild boar faecal bacteria act at the m6A mRNA modifications, we next examined the levels of m6A modification and the expression of m6A writer and eraser genes in the jejunum of each group. A clear difference in m6A levels of jejunum was observed between the Chow group and HFD group, high concentrations of wild boar faecal suspension prevented the reduction of m6A levels induced by HFD (Fig. 4A and B). As illustrated in Fig. 4C, the mRNA levels of m6A methyltransferases including METTL3, METTL14 and WTAP, the demethylases (FTO and ALKBH5) as well as the m6A binding proteins (YTHDF1 and YTHDF2) showed no difference (P > 0.05) among those groups. Notably, a decreased (P < 0.01) mRNA level of YTHDF3 was detected under HFD treatment. This result suggests that the FMT‐induced changes in the level of jejunum m6A modification may be one of the potential mechanisms for its regulation of lipid metabolism.
Fig. 4.
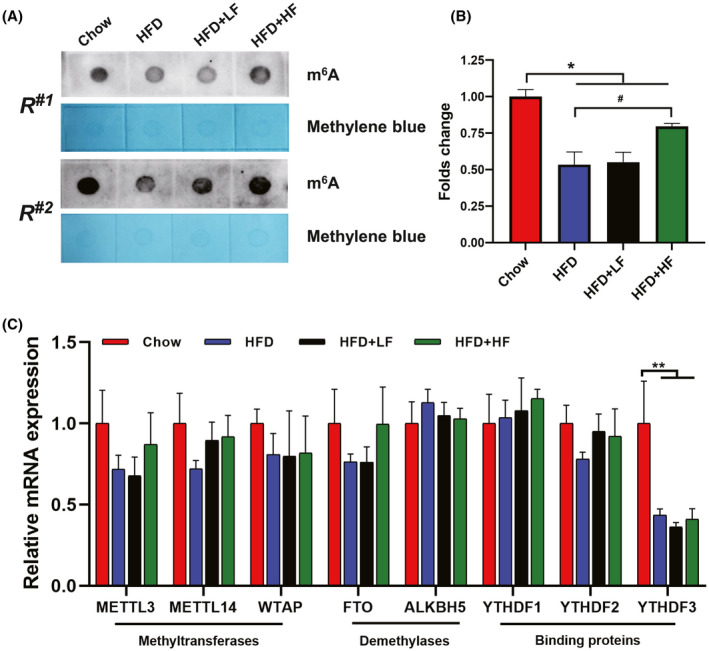
FMT attenuated the decrease in jejunum m6A methylation level induced by high‐fat diet.
(A). m6A dot blot and methylene blue staining.
(B) Densitometry analysis.
(C) mRNA levels of m6A related genes detected by qPCR.
Data were expressed as means ± SEM (n = 6).
*significant difference between Chow and HFD groups.
#significant difference between HFD and HFD+LF or HFD+HF groups.
* or #, P < 0.05, ** or ##, P < 0.01.
FMT regulated the jejunal microbiota in HFD‐fed mice
Numerous studies have shown that intestinal fat absorption is closely related to intestinal microbial composition (Bäckhed et al., 2007; Martinez‐Guryn et al., 2018). Different intestinal microbiota affects fatty acid absorption through distinct mechanisms (Semova et al., 2012). Hence, we would like to further investigate the functions played by jejunal microbiota in regulating jejunal fat absorption. In this study, we found that higher concentrations of faecal suspension (HFD+HF group) led to lower epididymal fat weight, subcutaneous fat weight, jejunum TG levels and higher stool TG levels, m6A levels compared to the HFD+LF group, suggesting that higher concentrations of faecal suspension may serve a superior function in alleviating obesity. Therefore, we further compared the gut microbial composition of the chow, HFD and HFD+HF groups. As shown in Fig. 5A, there was no difference (P > 0.05) in the number of OTUs among the groups. The diversity of jejunal bacteria was higher (P < 0.05) in the HFD group in comparison with the Chow and HFD+HF groups, as indicated by the Shannon index (Fig. 5B). The results of principal coordinates analysis (PCoA) based on weighted unifrac metrics showed that microbiota structure was remarkably different among three groups (R 2 = 0.7494; P = 0.001) (Fig. 5C). The hierarchical clustering tree outcomes on OTU level also indicated a significant separation among the microbiota of the groups (Fig. 5D). Next, the relative abundance of bacteria was further assessed at the phylum, genus and OTU levels respectively (Fig. 5E‐J). Five bacteria with relative abundance above 0.5% in at least one group were demonstrated at the phylum level, namely, Firmicutes, Actinobacteriota, Proteobacteria, Bacteroidota and Desulfobacterota, where the dominant phylum in each group was Firmicutes, which in the HFD+HF group accounted for more than 90% and were more abundant than the Chow (P < 0.001) and HFD (P < 0.05) groups. The relative abundance of Actinobacteriota was second only to Firmicutes in the Chow group and higher (P < 0.01) than HFD and HFD+HF groups (Fig. 5E, H and Fig. S2A). As shown in Fig. 5F and I, the microbiota structure of each group varied at the genus level, and among the 22 bacteria with relative abundance above 1% in either group, 16 bacteria showed differences based on the Kruskal‐Wallis rank sum test. Bifidobacterium was the dominant genus in the Chow group, followed by Lactobacillus. Lactobacillus was the dominant genus in both the HFD and HFD+HF groups, however, compared to the HFD group, the relative abundance of Lactobacillus was higher (P < 0.05) and the proportion of Anaerocolumna was decreased in the HFD+HF group (P = 0.03041) (Fig. S2B). In terms of OTU levels, significant differences were observed in the microbiota structure of each group (Fig. 5G and J). The relative abundance of Bifidobacterium Pseudolongum was the highest in the Chow group, which was higher (P < 0.01) than HFD and HFD+HF groups. Compared with the HFD group, the proportion of Lactobacillus murinus was lower (P < 0.05) in the HFD+HF group, while Lactobacillus johnsonii (P < 0.001), Lactobacillus_reuteri (P < 0.01), Romboutsia (P < 0.05) and Lactobacillus_intestinal (P < 0.05) showed an increase (Fig. S2C). The LDA scores for the abundance of bacteria at the OTU level (LDA > 4) showed that FMT increased the abundance of Lactobacillus and Romboutsia (Fig. 5K). These findings demonstrated that FMT largely altered the structure of jejunal microbiota and provided an idea that FMT might regulate lipid absorption by affecting the abundance of Lactobacillus and Romboutsia in the jejunum rather than mediating the development of jejunal microbiota structure toward that of the Chow group.
Fig. 5.
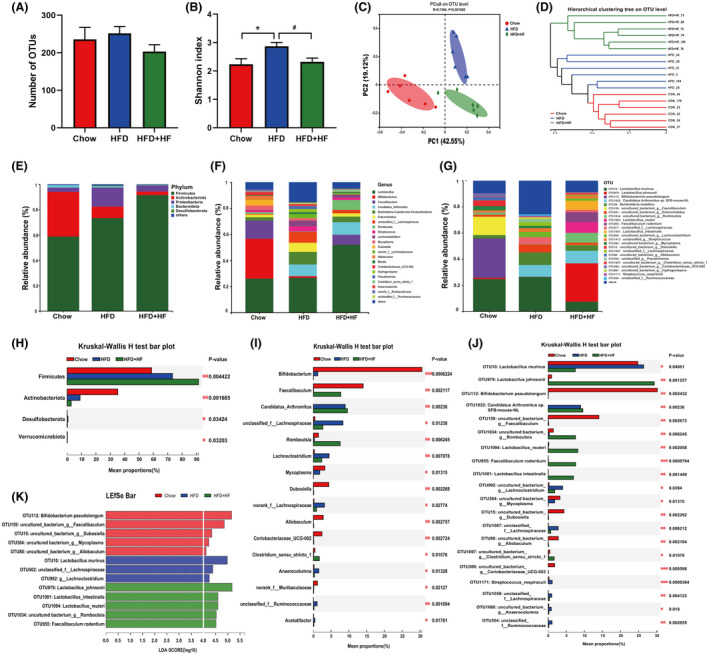
FMT and high‐fat diet altered the overall structure of gut microbiota in the jejunum.
(A). Number of OTUs.
(B) α diversity of gut bacteria based on Shannon index.
(C) PCoA plot of β‐diversity on genus level based on weighted‐unifrac index.
(D) Hierarchical clustering tree on OUT level.
(E–G) Bar plots and at phylum.
(E), genus (F) and OTU level (G).
(H–J) Comparison of dominant phylum (H), genera (I) and OUT (J) in the Chow, HFD and HFD+HF groups.
(K) LEfSe plots based on OUT level (LDA > 4).
Data were expressed as means ± SEM (n = 6).
*significant difference between Chow and HFD groups.
#significant difference between HFD and HFD+HF groups.
* or #, P < 0.05, ** or ##, P < 0.01.
Correlation of jejunal microbiota with HFD‐induced metabolic disorders
Our next objective was to elucidate the correlation between the dominant bacteria in each group and obesity‐induced metabolic disorders, in which Spearman's correlation analysis was employed. As seen in Fig. 6, the four dominant bacteria (Bifidobacterium pseudolongum, g_Faecalibaculum, g_Dubosiella and g_Allobaculum) in the Chow group were significantly negatively associated with obesity indicators (body weight, organ weight and serum lipid profile) and intestinal fat absorption‐related indicators such as TG and Tchol levels of jejunum and stool, FABP2 mRNA expression, while a significant positive correlation was observed with the number of adipocytes and the mRNA expression of NPC1L1. In contrast, g_Lachnoclostridium and f_Lachnospiraceae, the two dominant bacteria in the HFD group, were positively correlated with obesity indicators and showed a significant negative correlation with the number of adipocytes. However, Lactobacillus murinus was not significantly correlated with these indicators, which was consistent with the research of Harley et al. (2013) who reported that Lactobacillus murinus was not accountable for hastening obesity. The correlation between the jejunal microbiota of the HFD+HF group and the metabolic disorders caused by HFD exhibited a similar pattern to that of the Chow group. g_Romboutsia was significantly negatively correlated with the obesity indicators, insulin tolerance and the expression of FABP2, DGAT2 and ApoB48 mRNA. The other four dominant bacteria Lactobacillus johnsonii, Lactobacillus_reuteri, Faecalibaculum rodentium and Lactobacillus intestinalis showed a stronger correlation with intestinal fat absorption related indicators compared with the obesity indicators. Furthermore, the correlation of jejunum m6A methylation level with key jejunum microbiota showed that 4 of the 5 OTUs in Chow group expressed a highly positive correlation with jejunal m6A levels. In contrast, 2 OTUs in HFD group showed a negative correlation with m6A levels. However, HFD+HF group enriched 3 OTUs (Lactobacillus johnsonii, g_Romboutsia and Lactobacillus_reuteri) had a slight positive relationship with m6A levels, and the other 2 OTUs showed a gentle negative correlation (Fig. 6).
Fig. 6.
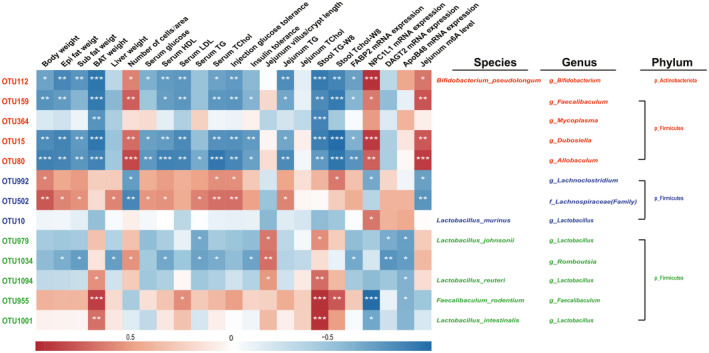
Correlation of jejunum microbiota with HFD‐induced metabolic disorders. The colour of the group is represented by red: Chow group; blue: HFD group; green: HFD+HF group. Data were expressed as means ± SEM (n = 6), *P < 0.05, **P < 0.01 and ***P < 0.001 indicate significant correlation.
Functional predictions of jejunal microbiota by picrust2
Changes in the composition and abundance of the gut microbiota are accompanied by alterations in function. PICRUST 2 was used for metabolic function prediction of the jejunal microbiota in this study. Metabolism and Genetic Information Processing were dramatically different in each group among the 6 pathways at KEGG level 1, while Environmental Information Processing, Cellular Processes and Human Diseases pathways were affected by HFD (Fig. 7A). Since the functions related to lipid metabolism which belong to Metabolism pathway are of interest to us, we further analysed it and noticed 12 pathways in Metabolism at KEGG level 2, of which lipid metabolism, energy metabolism and global and overview maps were the pathways we focused on (Fig. 7B). The findings suggested that lipid metabolism was slightly higher in the HFD+HF group than HFD group, and energy metabolism was the opposite. The global and overview maps were upregulated (P < 0.01) in the HFD group compared to the HFD+HF group. Next, we mined in‐depth for these three pathways and found that contain 32 pathways at KEGG level 3 (data not shown). The heatmap displayed the distribution of the distinct functional pathways among each group with significant variations, with the metabolic functions of jejunal microbiota affiliated to the Chow and HFD+HF groups being more similar in terms of cluster analysis (Fig. 7C).
Fig. 7.
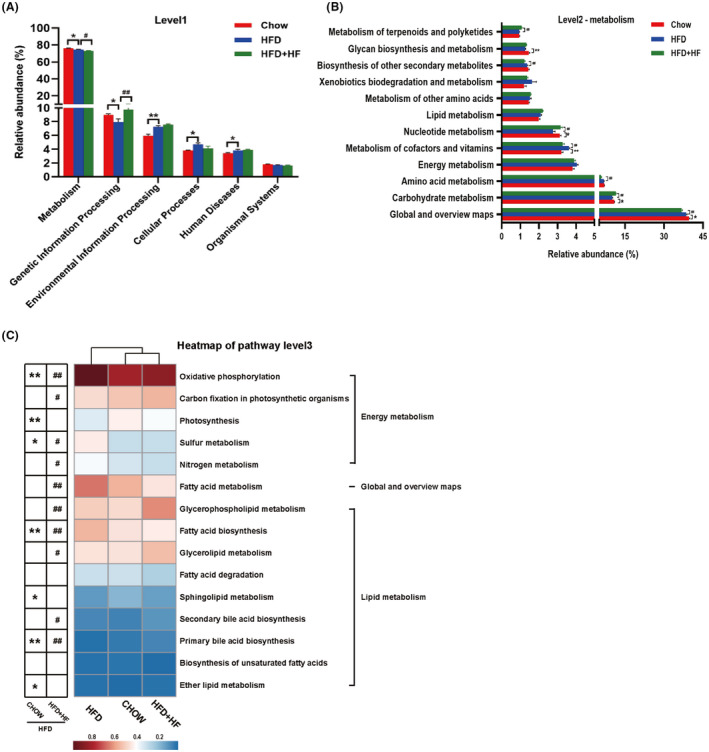
The pathway of different abundances of jejunum microbiota at KEGG levels 1 (A), level 2 (B) and Heatmap of KEGG level 3 (C). Data were expressed as means ± SEM (n = 6), *significant difference between Chow and HFD groups, #significant difference between HFD and HFD+HF groups, * or #, P < 0.05, ** or ##, P < 0.01.
Discussion
Various studies reported that the gut microbiota is strongly associated with the development of obesity as an environmental factor involved in the orchestration of fat storage and energy homeostasis (Bäckhed et al., 2004; Chevalier et al., 2015; Zhang et al., 2020). FMT as a gut microbial regulatory strategy with the ability to divert host phenotypes, is considered as a prospering treatment of various human diseases such as obesity (Groot et al., 2017; Zhang et al., 2019). Wild boars are characterized by a very low fat content and high lean mass, which was thought to be related with microbiota (Zhang et al., 2015). The functional predictions of the wild boar faecal microbiota we obtained suggested a substantial involvement in lipid metabolism pathways, which seems to support this phenotype. In this study, we set out to further define the role of the wild boar gut microbiota in HFD‐induced obesity in mice. Multiple indices comprising mouse body size, body weight gain, organ weight as well as distribution of adipocyte size of epididymal fat in mice verified the obesity‐relieving effect of wild boar faecal suspension.
Obesity is responsible for disorders of lipid metabolism and an increased risk of insulin resistance and type 2 diabetes (Kahn et al., 2006; Martinez‐Guryn et al., 2018). Consistent with the previous study (Yin et al., 2018), we found that obesity‐related symptoms in the HFD group, such as serum TG, TChol, VLDL and LDL level were markedly increased. As expected, the wild boar faecal suspension ameliorated the elevated serum levels of TG, TChol and improved insulin sensitivity compared to the HFD mice. These data suggested that FMT exerted a beneficial effect in blocking obesity in mice.
The modulation of intestinal lipid absorption and redistribution in peripheral tissues is the first line‐of‐defence against obesity (Tchernof and Després, 2013; Teng et al., 2020). A variety of factors in the form of dietary cues, intestinal microbiota, as well as the function of absorptive enterocytes may affect intestinal absorption of lipids and contribute to the regulation of obesity (Martinez‐Guryn et al., 2018). Targeted reduction of intestinal lipid absorption is beneficial for controlling obesity, metabolic syndrome, hyperlipidaemia and other diseases (Hussain, 2014; Luo et al., 2018). In this study, we focused on the effect of FMT on intestinal fat absorption. The H&E and TEM results showed that HFD increased intestinal lipid deposition in the intestinal villi and epithelium, while the wild boar faecal suspension alleviated this phenomenon. The notion was further supported by the changed TG levels in the jejunum and stool, suggesting that FMT reduced intestinal lipid absorption upon high‐fat signal. The expression of the key constituents responsible for lipid uptake, triglyceride resynthesis, as well as chylomicron packaging and transport are critical for the occurrence of intestinal fat absorption. Our results indicated that FABP2, but not CD36 and FATP4, was participating in the regulation of lipid uptake by FMT. The key regulators for triglyceride resynthesis DGAT1 and Mogat2 were insensitive to the stimulation of HFD and FMT, but DGAT2 was involved in the process of obesity mitigation by FMT. The component of chylomicron packaging and transport machinery like ApoB48, may be associated with the FMT‐mediated intestinal lipid absorption. Taken together, these results indicated that FMT may alleviate obesity by accelerating lipid excretion and regulating the lipid absorption of intestinal.
It has been reported that FMT induces distinctive shifts in the gut microbiota composition and function (Allegretti et al., 2019). Accumulating evidence pointed to the robust link between gut microbiota and nutrient absorption as well as energy harvesting, and differences in gut microbiota have been demonstrated in lean or obese individuals and mice (Krajmalnik‐Brown et al., 2012; Zong et al., 2020). However, most studies converge on elucidating the role of microbiota in the cecum or colon in obesity. In recent years, emerging investigations showed that microbiota of the jejunum is more sensitive to the effects of fat digestion and absorption, since the jejunum is the main site of fat digestion and absorption (Martinez‐Guryn et al., 2018; Chang and Martinez‐Guryn, 2019). Our data revealed distinct alterations in the diversity and composition of the jejunal microbiota after HFD and FMT treatments. Similar to previous studies reporting that Lachnospiraceae and its genus Lachnoclostridium was associated with obesity (Zhao et al., 2017; Li et al., 2020), our results indicated that microbiota changes induced by HFD shifted towards obesity‐associated microbiota. Interestingly, it was particularly evident that wild boar faecal bacteria increased the abundance of Romboutsia and Lactobacillus such as Lactobacillus_johnsonii and Lactobacillus_reuteri, which have been reported to be involved in regulating lipid metabolism, maintaining intestinal epithelial barrier function and improving diet‐induced obesity and hyperlipidaemia (Li et al., 2019; Russell et al., 2019; Yang et al., 2020). Overall, these results also further supported that FMT could transfer the host phenotype and that wild boar faecal bacteria reshape the microbiota of the mouse jejunum.
‘Microbiota‐gut epigenetics’ is a common manner to regulate the chromatin state of intestinal (Wu, Zhao, et al., 2020), where host RNA modifications in response to microbiota‐host interactions is an area that remains little explored. M6A has been ascertained to be the richest modification existing on eukaryotic mRNA, which is closely associated with obesity (Roundtree et al., 2017; Wang et al., 2020). Interestingly, the gut microbiota has been reported to be involved in regulating m6A levels in the gut, brain, liver and other tissues (Wang et al., 2019; Jabs et al., 2020). We found that wild boar faecal bacteria treatment alleviated the decrease in jejunal m6A levels induced by HFD, and the results of correlation analysis also suggested that the increase in Lactobacilli abundance after faecal bacteria treatment was slightly positively associated with m6A levels. Folate supplied by Lactobacillus or other bacteria could promote RNA methylation by synthesizing the methyl donor S‐adenosylmethionine through the folate and methionine cycle (Wu, Zhao, et al., 2020). We speculated that the increase of Lactobacillus in the intestine caused by wild boar faecal bacteria might play an indispensable role in regulating the jejunal m6A levels, and the significance of jejunal m6A levels may be the combined effect of multiple dominant bacteria. These results implied that variations of intestinal m6A levels respond to the onset of obesity and the mitigating effect of wild boar faecal bacteria on obesity. However, further investigation is needed to explore the precise mechanisms of gut microbiota regulating intestinal lipid absorption and mRNA methylation level.
In summary, our results demonstrated that FMT had a mitigating effect on obesity induced by HFD, which was probably due to the profound effects of FMT on the microbial composition and structure of the jejunum, giving priority to the increase in the relative abundance of Romboutsia and species belonging to the Lactobacillus such as Lactobacillus johnsonii, Lactobacillus_reuteri and Lactobacillus_intestinal. These changes further affect intestinal lipid metabolism and m6A levels, which in turn reduce lipid absorption in the jejunum and promote lipid excretion to ultimately counteract obesity (As shown in the schematic diagram of Fig. 8). However, the hierarchical relationships between gut microbiota and intestinal lipid absorption and host epigenetic modifications as well as their underlying mechanisms remain poorly defined and still need further investigation. It is also a limitation that the study only predicted strains associated with lipid metabolism in obese mice without further screening of key strains for functional validation and mechanism mining.
Fig. 8.
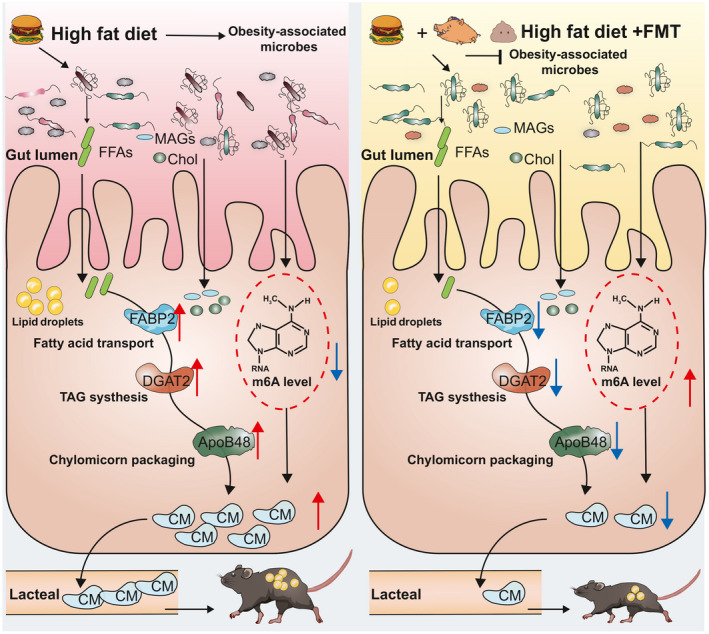
Schematic diagram of the potential mechanisms of the effect of FMT on alleviating HFD‐induced obesity.
Experimental procedures
Animals
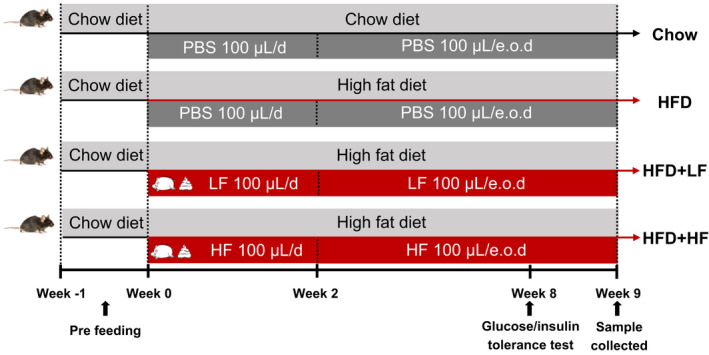
The committee on animal care and use of Zhejiang University (Hangzhou, China) approved all the animal experiments. A total of 48 male C57BL/6N mice aged 4 weeks bought from Shanghai SLAC Laboratory Animal Co. Ltd, were randomly allocated to 4 groups of 12 each, namely, standard chow diet group (Chow group), high‐fat diet group (HFD group), high‐fat diet with gavage of low (HFD+LF group) and high (HFD+HF group) concentrations faecal suspension group respectively. Mice were acclimatized for one week, during which they were fed the standard chow diet and maintained under a 12 h light/dark cycle controlled environment (temperature between 18°C and 22°C; relative humidity between 50 and 60%). For weeks 1–2 of the trial, the Chow and HFD groups were administered 100 µl of phosphate buffered saline (PBS), and the HFD+LF and HFD+HF groups were administered100 µl of different concentrations of faecal suspension by oral gavage daily. After that, each group received PBS or different concentrations of faecal suspension every other day (e.o.d) for 7 weeks (Table 1). The diet ingredient composition and nutrient contents are listed in Table S1. All mice were allowed to feed and drink (distilled water) ad libitum throughout the study. After 9 weeks of the trial, 8 mice in each group were randomly selected for sample collection.
Table 1.
Schematic of experimental procedure.
Faecal microbiota transplantation experiments
Fresh wild boar faecal samples were collected for the preparation of faecal suspension, which were prepared according to Hu et al. (2018) and Hamilton et al. (2012). Briefly, each 10 g of faecal sample was homogenized in 100 ml (1:10 (w/v)) of sterile PBS to prepare a faecal slurry. The slurry was passed through sterile gauze to remove larger particles, and a 70‐µm cell filter was used to remove small particles. The resulting faecal filtrate was centrifuged at 6000 g for 15 min and resuspended in PBS containing 10% sterile glycerol to the original volume or one‐fourth of the original volume to obtain low concentrations (1: 10 (w/v)) and high concentrations (1: 2.5 (w/v)) faecal suspension. All faecal suspensions were stored frozen at – 80°C until used.
Morphology analysis
For H&E staining, the proximal jejunum was fixed in 4% formaldehyde for more than 24 h and embedded in paraffin, followed by slicing and staining with haematoxylin and eosin. Images of the sections were acquired using a Leica DM3000 microscope (LEICA, Wetzlar, Germany) and the villi height and crypt depth were measured using the Image J Software (NIH Image, Bethesda, MD, USA).
Transmission electron microscopy (TEM)
The lipid droplets in the jejunum were observed by TEM as previously described (Luo et al., 2018; Zong, Cao, et al., 2019). The pre‐treated specimens were sectioned in LEICA EM UC7 ultratome (LEICA) and observed in Hitachi H‐7650 (Hitachi, Tokyo, Japan).
Serum glucose level and lipid profiles analysis
The serum glucose level and lipid profiles including high‐density lipoprotein (HDL), low‐density lipoprotein (LDL), triglyceride (TG) and total cholesterol (TChol) were measured with a Hitachi 7600‐020 automatic clinical chemistry analyzer (Hitachi).
Glucose tolerance test (GTT) and insulin tolerance test (ITT)
The GTT and ITT were performed as described by You et al. (2020). At week 8, for GTT, mice were injected intraperitoneally with 400 mg ml‐1 of D‐glucose (2 g kg‐1 body weight; Aladdin, Shanghai, China) after 12 h fasting, and their blood glucose was measured by glucometer (Yuwell, Jiangsu, China) at 0, 15, 30, 60, 90, 120 min post‐injection respectively. Three days after the test of glucose tolerance, ITT was performed with 0.75 U per kg body weight insulin (Aladdin) in mice fasted for 4 h, and blood glucose was monitored as in the GTT.
Measurement of jejunum and faecal TG and TChol levels
Approximately 50 mg of jejunal tissue or stool samples were homogenized in 9 times volume (1: 9 (w/v)) of pre‐cooled ethanol solution and the supernatant was obtained by centrifugation at 12 000 rpm min‐1 for 15 min at 4°C. TG and TChol were performed according to the manufacturer's (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) instructions and measured at 510 nm with a MultiskanSky Microplate Reader (Thermo Fisher Scientific, Waltham, MA, USA).
Quantitative real‐time PCR (qRT‐PCR)
Total RNA of jejunum was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA). The quality of the extracted RNA was assessed using NanoDrop 2000 (Thermo Fisher Scientific). Then 1 µg of RNA was used to reverse transcription reaction with a reverse transcription kit (Invitrogen, Carlsbad, CA, USA). Subsequent qRT‐PCR was performed with gene‐specific primers (Table S2) and SYBR Green dye (Roche, Mannheim, Germany) using a StepOnePlus Real‐Time PCR system (Applied Biosystems, Foster City, USA). The relative mRNA expression of the target gene was analysed by the 2−ΔΔCT method using the β‐actin housekeeping gene as a normalization control.
Western blot analysis
Total protein was extracted from jejunal tissue using Total Protein Extraction kit (KeyGen BioTECH, Nanjing, China). BCA kit (Beyotime, Shanghai, China) was used to determine protein concentration. Proteins were separated on SDS‐PAGE based on size and then transferred to the PVDF membrane for detection with the following antibodies: CD36 (Abcam, #ab133625, Cambridge Science Park, Cambridge, UK), I‐FABP (Proteintech, #21252‐1‐AP, Chicago, IL, USA), Fatp4 (Abcam, #ab200353, Cambridge Science Park, Cambridge, UK), β‐actin (Huabio, #EM21002, Hangzhou, Zhejiang, China), goat anti‐rabbit IgG‐HRP antibody (Huabio, #HA1001, Hangzhou, Zhejiang, China) and goat anti‐mouse IgG‐HRP antibody (Huabio, #HA1001, Hangzhou, Zhejiang, China).
Jejunum contents 16S sequencing and bioinformatics analyses
The jejunal mucosal scrapings were collected and immediately snap‐frozen in liquid nitrogen and stored at −80°C. The QIAamp DNA stool Mini Kit (Qiagen, Hilden, Germany) was used to extract DNA from enteric bacteria in jejunal mucosal scrapings and PCR amplification of the V3‐V4 region (338F: ACTCCTACGGGAGGCAGCA, 806R: GGACTACHVGGGTWTCTAAT) of 16S rDNA. The amplification products were detected by 2% gel electrophoresis and PCR products were recovered by axyprep DNA gel extraction kit (Axygen, San Francisco, CA, USA). The PCR products were detected and quantified by QuantiFluor™‐ST microfluorometer (Promega, Madison, WI, USA) and then Miseq library was constructed for pyrosequencing on the Illumina MiSeq platform (Illumina, Madison, WI, USA). Clean sequences were assigned to the same operational taxonomic units (OTUs) based on a similarity ≥ 97%. Data were analysed on the free online cloud platform (https://cloud.majorbio.com/) of Majorbio (Shanghai, China). Bacterial species were further identified by sequence comparison of OTUs on NCBI. The relative abundance of significant differences in phylum, genus and OTU levels was examined by the Kruskal–Wallis rank sum test. Spearman’s correlation analysis was further analysed to uncover the relationship between jejunal microbiota and intestinal fat absorption.
mRNA extraction and N6‐methyladenosine (m6A) dot blot
Purified mRNA was obtained from total RNA by following the manufacturer's instructions for the GenElute Direct mRNA Miniprep Kit (Sigma, St Louis, MO, USA). The m6A dot blot analysis was performed as described in previous studies (Zong, Zhao, et al., 2019; Zong et al., 2021). In brief, 200 ng mRNA was spotted onto Amersham Hybond‐N+ membrane (GE Healthcare, Marlborough, Middlesex County, USA) and cross‐linked twice with 254 nm, 0.12 J cm‐2 UV before detected with anti‐m6A antibody (Abcam, #ab208577, Cambridge Science Park, Cambridge, UK) and goat anti‐mouse IgG‐HRP antibody (Huabio, #HA1001, Hangzhou, Zhejiang, China). Methylene blue staining was used to indicate the amount of total RNA on the membrane.
Statistical analysis
All outcomes were analysed using one‐way ANOVA test of SPSS version 26.0. (SPSS, Inc., Chicago, IL, USA) and Duncan’s multiple tests were performed to compare the statistical differences. In addition, the GraphPad Prism version 8.0 (GraphPad Prism, San Diego, CA, USA), R version 4.0.5 and Origin 9.0 software (Origin Lab Corporation, Wellesley Hills, MA, USA) were used to visualize the data for graphing. Data are expressed as the mean ± SEM.
Conflict of interest
The authors declare no conflict of interests.
Acknowledgements
This work was funded by Zhejiang Provincial Natural Science Foundation of China (Grant No. LQ21C170002), the National Natural Science Foundation of China (Grant No. 32002185 and 31630075), and China Agriculture Research System of MOF and MARA (CARS‐35). We are sincerely grateful to Hangzhou Zoo (Zhejiang, China) for providing the faeces of wild boars.
Microbial Biotechnology (2021) 15(1), 337–352
Contributor Information
Yizhen Wang, Email: yzwang321@zju.edu.cn.
Xin Zong, Email: zongxin@zju.edu.cn.
Data Availability Statement
The datasets used and/or analysed in this study are available from the corresponding authors on reasonable request. Sequence data supporting the results of this study have been deposited in the NCBI’s Sequence Read Archive (SRA BioProject No. PRJNA745266 and PRJNA747860).
References
- Allegretti, J.R. , Mullish, B.H. , Kelly, C. , and Fischer, M. (2019) The evolution of the use of faecal microbiota transplantation and emerging therapeutic indications. Lancet 394: 420–431. [DOI] [PubMed] [Google Scholar]
- Bäckhed, F. , Ding, H. , Wang, T. , Hooper, L.V. , Koh, G.Y. , Nagy, A. , et al. (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101: 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed, F. , Manchester, J.K. , Semenkovich, C.F. , and Gordon, J.I. (2007) Mechanisms underlying the resistance to diet‐induced obesity in germ‐free mice. Proc Natl Acad Sci USA 104: 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi, M. , Maltecca, C. , Schillebeeckx, C. , McNulty, N.P. , Schwab, C. , Shull, C. , et al. (2020) Heritability and genome‐wide association of swine gut microbiome features with growth and fatness parameters. Sci Rep 10: 10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouter, K.E. , van Raalte, D.H. , Groen, A.K. , and Nieuwdorp, M. (2017) Role of the gut microbiome in the pathogenesis of obesity and obesity‐related metabolic dysfunction. Gastroenterology 152: 1671–1678. [DOI] [PubMed] [Google Scholar]
- Chang, E.B. , and Martinez‐Guryn, K. (2019) Small intestinal microbiota: the neglected stepchild needed for fat digestion and absorption. Gut Microbes 10: 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier, C. , Stojanović, O. , Colin, D.J. , Suarez‐Zamorano, N. , Tarallo, V. , Veyrat‐Durebex, C. , et al. (2015) Gut microbiota orchestrates energy homeostasis during cold. Cell 163: 1360–1374. [DOI] [PubMed] [Google Scholar]
- Chow, S.L. , and Hollander, D. (1979) A dual, concentration‐dependent absorption mechanism of linoleic acid by rat jejunum in vitro. J Lipid Res 20: 349–356. [PubMed] [Google Scholar]
- Dávalos‐Salas, M. , Mariadason, J.M. , Watt, M.J. , and Montgomery, M.K. (2020) Molecular regulators of lipid metabolism in the intestine ‐ Underestimated therapeutic targets for obesity? Biochem Pharmacol 178: 114091. [DOI] [PubMed] [Google Scholar]
- Gérard, P. (2016) Gut microbiota and obesity. Cell Mol Life Sci 73: 147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatz, J.F. , Luiken, J.J. , and Bonen, A. (2001) Involvement of membrane‐associated proteins in the acute regulation of cellular fatty acid uptake. J Mol Neurosci 16: 123–132. discussion 151–7. [DOI] [PubMed] [Google Scholar]
- Glatz, J.F.C. , and Luiken, J. (2018) Dynamic role of the transmembrane glycoprotein CD36 (SR‐B2) in cellular fatty acid uptake and utilization. J Lipid Res 59: 1084–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot, P.D. , Frissen, M.N. , Clercq, N.D. , and Nieuwdorp, M. (2017) Fecal microbiota transplantation in metabolic syndrome: history, present and future. Gut Microbes 8: 253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, M.J. , Weingarden, A.R. , Sadowsky, M.J. , and Khoruts, A. (2012) Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. Am J Gastroenterol 107: 761–767. [DOI] [PubMed] [Google Scholar]
- Harley, I.T. , Giles, D.A. , Pfluger, P.T. , Burgess, S.L. , Walters, S. , Hembree, J. , et al. (2013) Differential colonization with segmented filamentous bacteria and Lactobacillus murinus do not drive divergent development of diet‐induced obesity in C57BL/6 mice. Mol Metab 2: 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J. , Ma, L. , Nie, Y. , Chen, J. , Zheng, W. , Wang, X. , et al. (2018) A microbiota‐derived bacteriocin targets the host to confer diarrhea resistance in early‐weaned piglets. Cell Host Microbe 24: 817–832.e8. [DOI] [PubMed] [Google Scholar]
- Hussain, M.M. (2014) Intestinal lipid absorption and lipoprotein formation. Curr Opin Lipidol 25: 200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, M.M. , Kancha, R.K. , Zhou, Z. , Luchoomun, J. , Zu, H. , and Bakillah, A. (1996) Chylomicron assembly and catabolism: role of apolipoproteins and receptors. Biochim Biophys Acta 1300: 151–170. [DOI] [PubMed] [Google Scholar]
- Jabs, S. , Biton, A. , Bécavin, C. , Nahori, M.‐A. , Ghozlane, A. , Pagliuso, A. , et al. (2020) Impact of the gut microbiota on the m6A epitranscriptome of mouse cecum and liver. Nat Commun 11: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn, S.E. , Hull, R.L. , and Utzschneider, K.M. (2006) Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444: 840–846. [DOI] [PubMed] [Google Scholar]
- Krajmalnik‐Brown, R. , Ilhan, Z.E. , Kang, D.W. , and DiBaise, J.K. (2012) Effects of gut microbes on nutrient absorption and energy regulation. Nutr Clin Pract 27: 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulecka, M. , Paziewska, A. , Zeber‐Lubecka, N. , Ambrozkiewicz, F. , Kopczynski, M. , Kuklinska, U. , et al. (2016) Prolonged transfer of feces from the lean mice modulates gut microbiota in obese mice. Nutr Metab (Lond) 13: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley, R.E. , Bäckhed, F. , Turnbaugh, P. , Lozupone, C.A. , Knight, R.D. , and Gordon, J.I. (2005) Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102: 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Song, J. , Zaytseva, Y.Y. , Liu, Y. , Rychahou, P. , Jiang, K. , et al. (2016) An obligatory role for neurotensin in high‐fat‐diet‐induced obesity. Nature 533: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Qi, C. , Zhu, H. , Yu, R. , Xie, C. , Peng, Y. , et al. (2019) Lactobacillus reuteri improves gut barrier function and affects diurnal variation of the gut microbiota in mice fed a high‐fat diet. Food Funct 10: 4705–4715. [DOI] [PubMed] [Google Scholar]
- Li, X. , Wang, Y. , Xing, Y. , Xing, R. , Liu, Y. , and Xu, Y. (2020) Changes of gut microbiota during silybin‐mediated treatment of high‐fat diet‐induced non‐alcoholic fatty liver disease in mice. Hepatol Res 50: 5–14. [DOI] [PubMed] [Google Scholar]
- Lobo, S. , Wiczer, B.M. , Smith, A.J. , Hall, A.M. , and Bernlohr, D.A. (2007) Fatty acid metabolism in adipocytes: functional analysis of fatty acid transport proteins 1 and 4. J Lipid Res 48: 609–620. [DOI] [PubMed] [Google Scholar]
- Longo, M. , Zatterale, F. , Naderi, J. , Parrillo, L. , Formisano, P. , Raciti, G.A. , et al. (2019) Adipose tissue dysfunction as determinant of obesity‐associated metabolic complications. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, H. , Jiang, M. , Lian, G. , Liu, Q. , Shi, M. , Li, T.Y. , et al. (2018) AIDA selectively mediates downregulation of fat synthesis enzymes by ERAD to retard intestinal fat absorption and prevent obesity. Cell Metab 27: 843–853.e6. [DOI] [PubMed] [Google Scholar]
- Martinez‐Guryn, K. , Hubert, N. , Frazier, K. , Urlass, S. , Musch, M.W. , Ojeda, P. , et al. (2018) Small Intestine Microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe 23: 458–469.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.W. , Gao, Y. , Yen, M.I. , and Yen, C.L. (2014) Intestine‐specific deletion of acyl‐CoA:monoacylglycerol acyltransferase (MGAT) 2 protects mice from diet‐induced obesity and glucose intolerance. J Biol Chem 289: 17338–17349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree, I.A. , Evans, M.E. , Pan, T. , and He, C. (2017) Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, J.T. , Roesch, L.F.W. , Ördberg, M. , Ilonen, J. , Atkinson, M.A. , Schatz, D.A. , et al. (2019) Genetic risk for autoimmunity is associated with distinct changes in the human gut microbiome. Nat Commun 10: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semova, I. , Carten, J.D. , Stombaugh, J. , Mackey, L.C. , Knight, R. , Farber, S.A. , and Rawls, J.F. (2012) Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12: 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits, L.P. , Bouter, K.E. , de Vos, W.M. , Borody, T.J. , and Nieuwdorp, M. (2013) Therapeutic potential of fecal microbiota transplantation. Gastroenterology 145: 946–953. [DOI] [PubMed] [Google Scholar]
- Tchernof, A. , and Després, J.P. (2013) Pathophysiology of human visceral obesity: an update. Physiol Rev 93: 359–404. [DOI] [PubMed] [Google Scholar]
- Teng, B. , Huang, C. , Cheng, C.L. , Udduttula, A. , Yu, X.F. , Liu, C. , et al. (2020) Newly identified peptide hormone inhibits intestinal fat absorption and improves NAFLD through its receptor GPRC6A. J Hepatol 73: 383–393. [DOI] [PubMed] [Google Scholar]
- Vrieze, A. , Van Nood, E. , Holleman, F. , Salojärvi, J. , Kootte, R.S. , Bartelsman, J.F. , et al. (2012) Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143: 913–6.e7. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Song, C. , Wang, N. , Li, S. , Liu, Q. , Sun, Z. , et al. (2020) NADP modulates RNA m(6)A methylation and adipogenesis via enhancing FTO activity. Nat Chem Biol 16: 1394–1402. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Li, Y. , Chen, W. , Shi, H. , Eren, A.M. , Morozov, A. , et al. (2019) Transcriptome‐wide reprogramming of N(6)‐methyladenosine modification by the mouse microbiome. Cell Res 29: 167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Tran, K. , and Yao, Z. (1999) The activity of microsomal triglyceride transfer protein is essential for accumulation of triglyceride within microsomes in McA‐RH7777 cells. A unified model for the assembly of very low density lipoproteins. J Biol Chem 274: 27793–27800. [DOI] [PubMed] [Google Scholar]
- Weingarden, A.R. , and Vaughn, B.P. (2017) Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes 8: 238–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Li, Y. , Yu, J. , Gan, Z. , Wei, W. , Wang, C. , et al. (2020) Resveratrol attenuates high‐fat diet induced hepatic lipid homeostasis disorder and decreases m(6)A RNA methylation. Front Pharmacol 11: 568006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Zhao, Y. , Wang, X. , Kong, L. , Johnston, L.J. , Lu, L. , and Ma, X. (2020) Dietary nutrients shape gut microbes and intestinal mucosa via epigenetic modifications. Crit Rev Food Sci Nutr 12: 1–15. [DOI] [PubMed] [Google Scholar]
- Yang, G. , Hong, E. , Oh, S. , and Kim, E. (2020) Non‐Viable Lactobacillus johnsonii JNU3402 protects against diet‐induced obesity. Foods 9: 1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Huang, X. , Fang, S. , Xin, W. , Huang, L. , and Chen, C. (2016) Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Sci Rep 6: 27427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, C.L. , Stone, S.J. , Koliwad, S. , Harris, C. , and Farese, R.V. Jr (2008) Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res 49: 2283–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, J. , Li, Y. , Han, H. , Chen, S. , Gao, J. , Liu, G. , et al. (2018) Melatonin reprogramming of gut microbiota improves lipid dysmetabolism in high‐fat diet‐fed mice. J Pineal Res 65: e12524. [DOI] [PubMed] [Google Scholar]
- You, W. , Xu, Z. , Sun, Y.E. , Valencak, T.G. , Wang, Y. , and Shan, T. (2020) GADD45α drives brown adipose tissue formation through upregulating PPARγ in mice. Cell Death Dis 11: 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.H. , Shen, L.Y. , Luo, J. , Wu, Z.H. , Jiang, Y.Z. , Tang, G.Q. , et al. (2015) Analysis of carcass and meat quality traits and nutritional values of hybrid wild boars under different crossing systems. Genet Mol Res 14: 2608–2616. [DOI] [PubMed] [Google Scholar]
- Zhang, X.Y. , Chen, J. , Yi, K. , Peng, L. , Xie, J. , Gou, X. , et al. (2020) Phlorizin ameliorates obesity‐associated endotoxemia and insulin resistance in high‐fat diet‐fed mice by targeting the gut microbiota and intestinal barrier integrity. Gut Microbes 12: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Mocanu, V. , Cai, C. , Dang, J. , Slater, L. , Deehan, E.C. , et al. (2019) Impact of fecal microbiota transplantation on obesity and metabolic syndrome‐A systematic review. Nutrients 11: 2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L. , Zhang, Q. , Ma, W. , Tian, F. , Shen, H. , and Zhou, M. (2017) A combination of quercetin and resveratrol reduces obesity in high‐fat diet‐fed rats by modulation of gut microbiota. Food Funct 8: 4644–4656. [DOI] [PubMed] [Google Scholar]
- Zong, X. , Cao, X. , Wang, H. , Xiao, X. , Wang, Y. , and Lu, Z. (2019) Cathelicidin‐WA facilitated intestinal fatty acid absorption through enhancing PPAR‐γ dependent barrier function. Front Immunol 10: 1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, X. , Fu, J. , Xu, B. , Wang, Y. , and Jin, M. (2020) Interplay between gut microbiota and antimicrobial peptides. Anim Nutr 6: 389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, X. , Xiao, X. , Shen, B. , Jiang, Q. , Wa Ng, H. , Lu, Z. , et al. (2021) The N6‐methyladenosine RNA‐binding protein YTHDF1 modulates the translation of TRAF6 to mediate the intestinal immune response. Nucleic Acids Res 49: 5537–5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, X. , Zhao, J. , Wang, H. , Lu, Z. , Wang, F. , Du, H. , and Wang, Y. (2019) Mettl3 deficiency sustains long‐chain fatty acid absorption through suppressing Traf6‐dependent inflammation response. J Immunol 202: 567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed in this study are available from the corresponding authors on reasonable request. Sequence data supporting the results of this study have been deposited in the NCBI’s Sequence Read Archive (SRA BioProject No. PRJNA745266 and PRJNA747860).


