Summary
The alarming rise in the emergence of antimicrobial resistance in human, animal and plant pathogens is challenging global health and food production. Traditional strategies used for antibiotic discovery persistently result in the re‐isolation of known compounds, calling for the need to develop more rational strategies to identify new antibiotics. Additionally, anti‐infective therapy approaches targeting bacterial signalling pathways related to virulence is emerging as an alternative to the use of antibiotics. In this perspective article, we critically analyse approaches aimed at revitalizing the identification of new antibiotics and to advance antivirulence therapies. The development of high‐throughput in vivo, in vitro and in silico platforms, together with the progress in chemical synthesis, analytical chemistry and structural biology, are reviving a research area that is of tremendous relevance for global health.
The rise in the emergence of antimicrobial resistance in pathogenic bacteria urges the development of novel approaches to combat pathogenic bacteria. Here, we analyze major strategies aimed at the identification of novel natural product‐based antibiotics from microbial sources and to advance anti‐infective therapy strategies targeting signal transduction pathways.
Introduction
We are facing difficult times economically, ecologically and health‐wise. Fortunately, we live in a scientific age that has shown its potential with the development of several COVID‐19 vaccines in record time, with numerous additional vaccines in different stages of development or applying for a licensing (Brüssow, 2021), reflecting the capacity of our society to overcome a major health crisis.
One of the greatest challenges of our time is the increasing emergence antimicrobial resistance (AMR). AMR microbes are currently causing 2.8 million infections and more than 35 000 deaths per year only in the United States (Centers for Disease Control and Prevention, 2019), as well as 700 000 annual deaths worldwide (Miethke et al., 2021). This global health problem does not discriminate between economic and social levels and affects low‐, middle‐ and high‐income countries (World Health Organization, 2015). Dramatically, if prompt actions are not taken, current estimates indicate that 10 million people will annually die in 2050 as a result of AMR infections (Dadgostar, 2019), surpassing cancer as the leading cause of mortality at a global scale (O’Neil, 2016). AMR will lead to longer hospital stays, increased second‐line treatments and treatment failure. It was estimated that treatment of AMR infections can incur a cost of up to USD 50 000 per patient and episode, and that USD 20 billion per year are necessary to palliate AMR in the United States (Naylor et al., 2018; Dadgostar, 2019). AMR not only impacts on animal and human health but also on agricultural production. Crop production must increase by more than 60% with respect to the current production to feed an expected population of 10 billion people in 2050 (Bhatta et al., 2021). Nevertheless, according to the Food and Agriculture Organization of the United Nations (FAO), plant pathogens are responsible for losses of up to 40% of the annual global crop production, corresponding to a value of USD 290 billion. Traditionally, fighting plant pathogens has been achieved by the use of chemical pesticides that have resulted in the emergence of resistances that in turn hamper the effective treatment of these phytopathogens (Sundin and Wang, 2018).
Thus, urgent actions need to be taken to effectively control human, animal and plant AMR pathogens. Scientifically, this aspect is reflected in the fact that (i) ~ 16% of academic projects are focused on drug discovery (Bem et al., 2015) and (ii) ~ 70% of the projects in the preclinical antibacterial pipeline are aimed at the identification of antibiotics that interact with novel targets (Theuretzbacher et al., 2020). Unfortunately, these research lines are primarily conducted by academia and are generally underfunded. Consequently, future strategies will require an increased cooperation between academic and industrial sectors in order to improve the success rates of novel antibiotic discovery (Miethke et al., 2021). This will require to (re‐) attract pharmaceutical companies to invest into antibiotic research; aspect that will involve governments and policymakers across the globe to quickly legislate means that lessens the enormous capital risks of antibiotic research and development funding.
Although there is a desperate need for the discovery and production of novel antibiotics, the concept of anti‐infective therapy is an attractive alternative to antibiotics (Cegelski et al., 2008; Bjarnsholt et al., 2013; Allen et al., 2014). Anti‐infective therapy is based on targeting molecular mechanisms that lead to disease but that do not interfere with bacterial growth. In other words, this strategy aims at disarming rather than killing bacteria. Bacteria contain a wide range of signal transduction systems such as transcriptional regulators, two‐component systems, chemosensory pathways, phosphoenolpyruvate:sugar phosphotransferase systems, adenylate cyclases and cAMP phosphodiesterases, diguanylate cyclases and c‐di‐GMP phosphodiesterases, extracytoplasmic function sigma factors and Ser/Thr/Tyr protein kinases and phosphatases (Galperin, 2018). These systems sense different types of signals and generate different responses such as transcriptional regulation, control of second messenger levels or chemotaxis. Through the concerted action of these systems, bacteria are able to optimally adapt to their present environment or move chemotactically towards more favourable niches. In the context of anti‐infective therapy, the interference with such signal transduction pathways has the promise to become an efficient strategy to fight pathogenic bacteria (Calvert et al., 2018; Miethke et al., 2021).
In this perspective article, we focus on current and future approaches aimed at identifying novel natural product‐based antibiotics from microbial sources as well as in the development of anti‐infective therapy strategies that target microbial signal transduction pathways relevant for virulence.
The need to diversify microbial isolation sources to improve antibiotic discovery rates
Historically, Actinobacteria have been the main source of antimicrobial compounds and around two thirds of all clinically used antibiotics are produced by members of this phylum (van Bergeijk et al., 2020). Remarkably, the analysis of the new chemical entities identified over the last 40 years revealed that natural product‐based compounds continue to be the main source of antibacterial compounds (Newman and Cragg, 2020). However, the probability of identifying new antibiotics from Actinobacteria using classical methods (i.e. bioactivity‐guided isolation through the screening of environmental isolates) is very low and has been estimated at less than one in a million (van Bergeijk et al., 2020), which is urging the scientific community to advance new experimental and in silico alternatives to improve discovery rates. In this regard, there is much controversy as to whether efforts to identify new natural product‐derived antibiotics should continue to be focused on the screening of soil Actinobacteria or whether the phylogenetic range should be broadened and new or poorly characterized taxa included (Lewis, 2020; Gavriilidou et al. 2021). In this context, the advances in the development of genome mining methods for the prediction of natural products biosynthetic genes from genome sequence data are starting to provide some answers to these questions. Indeed, the success of improved genome mining approaches has increased the discovery rate of novel bioactive secondary molecules (Blin et al., 2021; Medema et al., 2021). The enormous potential of these in silico strategies is reflected in a recent survey of ~ 170 000 bacterial genomes and ~ 10 000 bacterial metagenome assembled genomes which has been deposited in BioRxiv (Gavriilidou et al. 2021). Major conclusions from this work are: (i) only 3% of the bacterial biosynthetic potential for natural product synthesis has been explored experimentally and (ii) although actinobacterial strains showed the highest natural product biosynthetic diversity, less studied taxa also exhibit strong potential for the synthesis of new antibiotics. Consequently, efforts should focus on exploring new environments and ecological niches as alternative isolation sources of new antibiotic producers such as the human microbiome (Donia et al., 2014), sponge microbiota (Wilson et al., 2014) or rhizospheric soils (Crits‐Christoph et al., 2018; Carrión et al., 2019), as well as on expanding the taxonomic range of potential antibiotics producing bacteria. For instance, the enormous potential of this experimental diversification has been exemplified by the reconstruction of bacterial genomes from grassland soil metagenomes (Crits‐Christoph et al., 2018). This seminal study allowed the identification of several under‐investigated bacterial phyla like Gemmatimonadetes, Verrucomicobia, Acidobacteria and Rokubacteria as prolific sources of secondary metabolites of the non‐ribosomal peptide, polyketide and terpene classes, among others. Outstandingly, most of the natural product biosynthetic gene clusters from these organisms were novel and some of the identified bacteria dedicated up to 14% of their genomes to the production of secondary metabolites (Crits‐Christoph et al., 2018), which highlights the relevance of taxa alternative to the Actinobacteria phylum as sources of novel antibiotics.
Whereas genome mining methods lay the groundwork for the identification of novel antibiotics, advanced analytical chemistry is required to confirm the identity of the desired molecule, an aspect that is hampered when the secondary metabolites are produced at low titres. To improve the efficiency of antibiotic discovery, great progress is being made in the development of algorithms that allow to establish associations between biosynthetic gene clusters and the structure of corresponding bioactive molecule, as well as to predict the function of the molecule of interest from available sequence information (Medema et al., 2021). The advance of these and additional technologies will help prioritize among candidate antibiotic biosynthetic clusters and to rapidly improve performance of antibiotic discovery in the short‐term future.
Microfluidics approaches to enhance bacterial cultivability aimed at antibiotic discovery
Although genome mining approaches permit access to untapped secondary metabolites, one of the main constraints encountered in the field of antibiotic discovery at present is the fact that many potential source bacteria are recalcitrant to cultivation. Indeed, current data indicate that < 1% of environmental bacteria are cultivable under standard growth conditions (Lloyd et al., 2018), which hinders the identification of novel antibiotics. In order to improve bacterial cultivability, current strategies are mainly focused on developing high‐throughput culture methods (Lagier et al., 2018). In addition, alternative approaches permit bacterial growth in their natural environment using, for example, the isolation chip (iChip) technology, which is based on the use of semi‐permeable membranes that permit free diffusion of specific nutrients and growing factors from the environment into the bacterial culture (Lewis, 2020; Miethke et al., 2021). In the context of searching for specific culture conditions for the growth of uncultured antibiotic‐producing bacteria, extraordinary progress has been made with the development of high‐throughput microfluidics approaches (Hengoju et al., 2020; Matilla, 2021). Within this expanding research field, microfluidics permits partitioning complex bacterial communities into droplets containing a single cell. These droplets function as micro‐reactors where physiological and biochemical parameters can be monitored and optimized. Remarkably, such systems have successfully identified culture conditions for novel antibiotics producers (Hengoju et al., 2020; Matilla, 2021). For instance, droplet‐based systems mimicking environmental conditions in situ have enabled the cultivation of a higher microbial diversity as compared with conventional culture techniques, including prolific antibiotic producers (Mahler et al., 2021). Importantly, droplet microfluidics coupled to mass spectrometry has been effectively used for the detection and monitoring of secondary metabolites in bacteria (Hengoju et al., 2020) and the rapidly improving performance of analytical chemistry techniques is expected to provide a major boost to microfluidics‐based approaches for antibiotic discovery.
Activation of silent antibiotic biosynthetic gene clusters
The genetic capacity for the synthesis of secondary metabolites or the possibility to culture the source microorganism outside its natural habitat does not generally guarantee the production of the desired metabolite. This lacking synthesis is primarily due to the intricate regulatory network that controls the expression of secondary metabolite biosynthetic gene clusters, which results in most of them being silent under standard culture conditions. Consequently, these cryptic secondary metabolites represent a hidden source of bioactive compounds with antibiotic properties.
A range of experimental approaches aimed at activating cryptic gene clusters, such as co‐culture methods, heterologous pathway expression, ribosome engineering, promoter exchange, mutation and overexpression of regulatory genes, have proven to be effective in the identification of cryptic antibiotics and are detailed in a number of comprehensive reviews (Rutledge and Challis, 2015; Okada and Seyedsayamdost, 2017; Mao et al., 2018; Kang and Kim, 2021; Scherlach and Hertweck, 2021) (Fig. 1). Additionally, culture‐independent strategies that rely on the chemical synthesis of target compounds based on bioinformatics predictions, named synthetic‐bioinformatic natural products, are also being developed to access silent secondary metabolites (Scherlach and Hertweck, 2021) (Fig. 1). However, we would like to pay here special attention to the intricate regulatory cascades that control secondary metabolites production and the tremendous possibilities derived from the characterization of these regulatory pathways for the activation of cryptic metabolites.
Fig. 1.
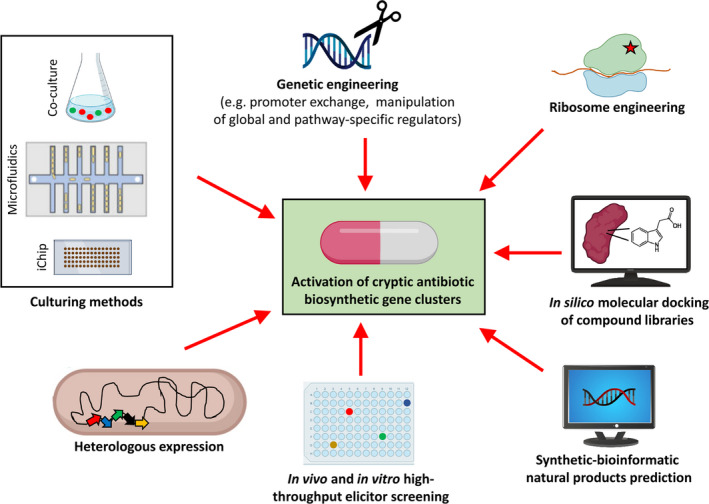
Major in vivo, in vitro and in silico approaches aimed at the activation of cryptic natural product‐based antibiotics.
The biosynthesis of secondary metabolites is energetically costly and the expression of the corresponding biosynthetic gene clusters is tightly regulated by global and pathway‐specific regulators in response to external stimuli (Okada and Seyedsayamdost, 2017; van der Heul et al., 2018; van Bergeijk et al., 2020; Matilla et al., 2021). The identification of these regulators is key to understand the fine‐tuned expression of secondary metabolites, and high‐throughput functional genomics approaches are being developed to uncover complex bacterial regulatory networks (Smith et al., 2021). Many of the regulatory proteins that control antibiotic production sense different signal molecules (Matilla et al., 2021), and the identification of these signals is of major importance to activate cryptic biosynthetic cluster for antibiotic discovery. However, the nature of most of these chemical elicitors is unknown, and this aspect is a major bottleneck in the study of bacterial signal transduction that is of particular relevance in the antibiotic discovery field. To overcome this difficulty, different in vivo and in vitro high‐throughput screening approaches of extensive chemical libraries are being developed (Fig. 1); some of which were shown to be effective in the activation of cryptic antibiotics (Mao et al., 2018; Zhang and Seyedsayamdost, 2020; Scherlach and Hertweck, 2021). Importantly, these screenings can be coupled to mass spectrometry‐based metabolomics to facilitate natural products identification (Zhang and Seyedsayamdost, 2020). As a proof of concept, we have successfully screened in vitro a compound library of ~ 1700 small molecules to identify several phytohormones as ligands of an antibiotic‐specific transcriptional regulator in a plant‐associated bacterium that has resulted in the identification of indole‐3‐acetic acid as a modulator of the production of the antibiotic andrimid (Matilla et al., 2018). Additionally, advances in integrated computational approaches for antibiotic discovery such as in silico docking of virtual compound libraries and molecular dynamics simulations (Lans et al., 2020; Macalino et al., 2020) can be used to identify ligands of key transcriptional regulators and sensor kinases to improve the efficiency in the identification of chemical elicitors of cryptic antibiotics biosynthesis (Fig. 1). Notably, complementary to the in vivo and in vitro approaches described above aimed at activating cryptic biosynthetic clusters, additional high‐throughput proteomics‐based strategies are being implemented for de novo identification of antibiotic targets (Martin et al., 2020). Taken together, the combination of different in vivo, in vitro and in silico high‐throughput screenings will facilitate the discovery of new antibiotic compounds with novel mechanisms of action in the near future.
Anti‐infective therapy as alternative for the treatment of bacterial infections
The large array of bacterial signal transduction systems illustrates well their importance for bacteria to adapt to their environment and to move to more favourable environmental niches. In the context of fighting pathogenic bacteria, there are a number of examples that illustrate that the interference with signalling processes can be achieved by (i) the application of a key signal molecule that regulates bacterial virulence, (ii) the use of signal antagonists that bind to sensor proteins but do not induce downstream signalling or (iii) the interference with the signalling cascade. An advantage of such strategies is that they typically do not kill bacteria or interfere with growth, preventing the selection of AMR mutants.
The first strategy is best illustrated by Pseudomonas aeruginosa – a human pathogen classified by the World Health Organization as a priority 1 (critical) pathogen for development of novel antibiotics (Tacconelli et al., 2018). It was shown that low concentrations of inorganic phosphate (Pi) induce a lethal phenotype during intestinal colonization (Zaborin et al., 2009). Pi is perceived by the PhoR/B two‐component system (Peng et al., 2017), and transcriptomics studies showed that a modest reduction in the Pi concentration, that is, 1 mM to 0.2 mM, caused increased transcript levels of many virulence genes (Bains et al., 2012). Via animal experimentation it was shown that Pi administration reduces P. aeruginosa infection (Long et al., 2008). In general, surgical interventions cause a significant decrease in the Pi concentration. A mouse model was established in which a surgical intervention was followed by a challenge with P. aeruginosa (Long et al., 2008). It was found that the challenge of the mouse control group resulted in an elevated lethality. However, when mice challenged with P. aeruginosa had previously received oral Pi, the survival rate was significantly higher. The potential of such approach to reduce bacterial virulence is furthermore illustrated by the facts that Pi is cheap and a natural compound that does not cause any undesired side‐effects.
Unfortunately, the knowledge on the identity of signal molecules that determine bacterial virulence is still rather scarce. For example, P. aeruginosa contains 64 sensor kinases of which the large majority were found to be involved in virulence (Francis et al., 2017). However, the signals that control kinase activity have only been determined for a handful of proteins (Francis et al., 2017). Therefore, the precise knowledge of virulence‐related signal molecules will offer elegant strategies to interfere with infectious processes.
Antagonists targeting signal transduction systems in anti‐infective therapy
The canonical topology of sensor proteins like sensor histidine kinases or chemoreceptors consists of an extracytoplasmic ligand‐binding domain (LBD) and a cytosolic domain that is involved in processing the molecular stimulus created by signal binding (Fig. 2). Whereas the cytosolic domains of sensor kinases and chemoreceptors are generally rather conserved in their sequence, there is an enormous diversity among the LBDs. This diversity is reflected in (i) the presence of many different LBD types, such as 80 in chemoreceptors (Ortega et al., 2017) and (ii) by an important sequence divergence with individual LBD families (Gavira et al., 2020). These notions, thus, imply that a specific interference with signal transduction systems can be best achieved by targeting the specific part of sensor proteins, namely the LBD (Fig. 2).
Fig. 2.
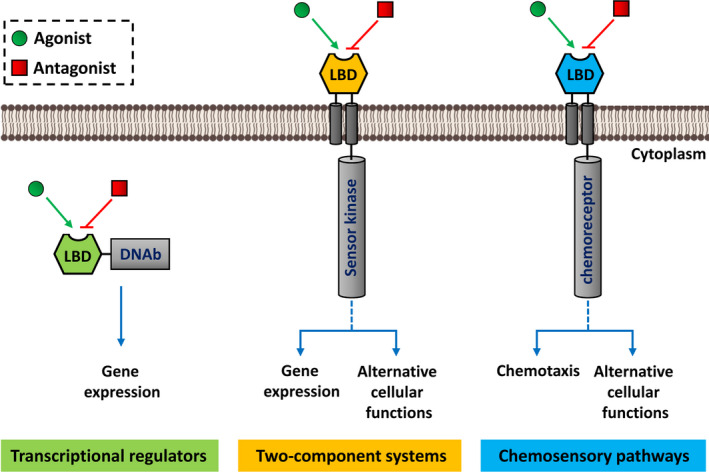
Agonist‐ and antagonist‐mediated modulation of the activity of transcriptional regulators, two component systems and chemosensory pathways. Agonist and antagonists bind to the ligand binding domain (LBD) of sensor proteins inducing or repressing downstream signalling, processes that are represented by triangular and flat arrowheads, respectively. DNAb, DNA‐binding domain.
There is now a significant body of evidence for the existence of naturally occurring signal antagonists (Busch et al., 2007; Martin‐Mora et al., 2018; Johnson et al., 2021), which are compounds that bind to the same site as the signal molecules but fail to induce downstream signalling (Fig. 2) – a phenomenon likely due to different conformational changes induced by signal and signal antagonist binding (Koh et al., 2016). Antagonists and signal molecules are frequently structurally similar, but the molecular features that determine their agonistic or antagonistic function are currently unclear. However, advances in computational biology have permitted that protein ligands can be identified by high‐throughput in silico molecular docking approaches of compound libraries to three‐dimensional structures of target proteins.
Virulence properties are frequently controlled by quorum‐sensing mechanisms, and the receptors for quorum‐sensing signals are major targets for anti‐infective therapy. A number of in silico docking studies have reported the identification of quorum‐sensing antagonists, as exemplified by (Gnanendra et al., 2013; Nandi, 2016; Vetrivel et al., 2021). This approach is based on the availability of high‐resolution 3D target protein structures, and their absence is a major limitation of this technique. However, the possibility of generating highly precise 3D protein models using the AlphaFold technology has started to revolutionize biology (Jumper et al., 2021; Tunyasuvunakool et al., 2021). These precise models will facilitate the resolution of the crystallographic phase problem using molecular replacement techniques leading to a more rapid resolution of experimental structures or, alternatively, AlphaFold‐created models will be used directly for in silico ligand screening. Such approaches will facilitate the identification of LBD ligands, of which their either agonistic or antagonistic effect on signal transduction needs to be determined experimentally.
A number of studies have reported the identification of inhibitors that target the conserved (cytosolic) part of sensor proteins such as the autokinase domain of sensor kinases (Hirakawa et al., 2020). A major disadvantage of these inhibitors is the necessity to cross the bacterial membrane to reach their targets. As a consequence, most of these compounds are rather hydrophobic that diffuse across the membrane, and in several occasions this hydrophobicity was associated with a reduced specificity of the inhibitor (Hirakawa et al., 2020). In addition, hydrophobic compounds may have the potential to integrate into human membranes that may cause undesired side‐effects. In this context, the specific targeting of extracytoplasmic sensor domains appears to be advantageous.
Concluding remarks and perspectives
Currently, our main weapons to fight pathogenic bacteria are antibiotics that either kill or slow down bacterial growth. However, the rise of AMR infections is alarming, and the identification of new chemicals to combat pathogenic microorganisms is urgently needed. This aspect is reflected in the fact that (i) no novel classes of antibiotics effective against Gram‐negative bacteria have been discovered in more than half a century (Hutchings et al., 2019) and (ii) only ~ 40 antibacterial molecules are currently in clinical trials; a number largely inferior to the approximately 4000 immuno‐oncology agents that are currently under development (Xin Yu et al., 2019). However, advances in multidisciplinary approaches including genomics, metagenomics, proteomics, synthetic biology, high‐throughput culturing methods, bioinformatics and analytical chemistry are contributing to reinvigorate antibiotics research, which will benefit from exploring unconventional ecological niches and under‐investigated microbial taxonomic groups. In addition, anti‐infective therapy is (re‐) emerging as an alternative approach to antibiotics. There is significant promise in targeting sensor domains of signal transduction proteins that are required for full virulence during host infection by a knowledge‐based application of key signal molecules or by signal antagonists. The identification of these molecules will be facilitated by an increasing computing power and precision of molecular docking approaches. Notably, a major bottleneck of such approaches, namely the absence of high‐resolution target structures, will be less important due to the possibilities offered by the AlphaFold technology.
The combined action of antibiotics and antivirulence drugs has been suggested to enable a greater control of pathogenic microorganisms with a reduced occurrence of AMR (Miethke et al., 2021). These strategies are in line with the action plans introduced by the World Health Organization (WHO, 2015) and the European Commission (EC, 2017) to combat the rising of AMR infections at a global scale.
Funding information
This work was supported through grants from the CSIC to MAM (PIE‐202040I003), the Spanish Ministry for Science and Innovation to MAM (PID2019‐103972GA‐I00), the Junta de Andalucía (P18‐FR‐1621) and Spanish Ministry of Economy and Competitiveness (BIO2016‐76779‐P) to TK.
Conflict of interest
The authors declare that there is no conflict of interest.
Acknowledgements
This work was supported through grants from the CSIC to MAM (PIE‐202040I003), the Spanish Ministry for Science and Innovation to MAM (PID2019‐103972GA‐I00), the Junta de Andalucía (P18‐FR‐1621) and Spanish Ministry of Economy and Competitiveness (BIO2016‐76779‐P) to TK.
Microbial Biotechnology (2021) 15(1), 70–78
References
- Allen, R.C. , Popat, R. , Diggle, S.P. , and Brown, S.P. (2014) Targeting virulence: can we make evolution‐proof drugs? Nat Rev Microbiol 12: 300–308. [DOI] [PubMed] [Google Scholar]
- Bains, M. , Fernandez, L. , and Hancock, R.E. (2012) Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa . Appl Environ Microbiol 78: 6762–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bem, A.E. , Velikova, N. , Pellicer, M.T. , van Baarlen, P. , Marina, A. , and Wells, J.M. (2015) Bacterial histidine kinases as novel antibacterial drug targets. ACS Chem Biol 10: 213–224. [DOI] [PubMed] [Google Scholar]
- van Bergeijk, D.A. , Terlouw, B.R. , Medema, M.H. , and van Wezel, G.P. (2020) Ecology and genomics of Actinobacteria: new concepts for natural product discovery. Nat Rev Microbiol 18: 546–558. [DOI] [PubMed] [Google Scholar]
- Bhatta, M. , Sandro, P. , Smith, M.R. , Delaney, O. , Voss‐Fels, K.P. , Gutierrez, L. , and Hickey, L.T. (2021) Need for speed: manipulating plant growth to accelerate breeding cycles. Curr Opin Plant Biol 60: 101986. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt, T. , Ciofu, O. , Molin, S. , Givskov, M. , and Høiby, N. (2013) Applying insights from biofilm biology to drug development ‐ can a new approach be developed? Nat Rev Drug Discov 12: 791–808. [DOI] [PubMed] [Google Scholar]
- Blin, K. , Shaw, S. , Kloosterman, A.M. , Charlop‐Powers, Z. , van Wezel, G.P. , Medema, M.H. , and Weber, T. (2021) antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49: W29–W35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüssow, H. (2021) COVID‐19: vaccination problems. Environ Microbiol 23: 2878–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, A. , Lacal, J. , Martos, A. , Ramos, J.L. , and Krell, T. (2007) Bacterial sensor kinase TodS interacts with agonistic and antagonistic signals. Proc Natl Acad Sci USA 104: 13774–13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert, M.B. , Jumde, V.R. , and Titz, A. (2018) Pathoblockers or antivirulence drugs as a new option for the treatment of bacterial infections. Beilstein J Org Chem 14: 2607–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrión, V.J. , Perez‐Jaramillo, J. , Cordovez, V. , Tracanna, V. , de Hollander, M. , Ruiz‐Buck, D. , et al. (2019) Pathogen‐induced activation of disease‐suppressive functions in the endophytic root microbiome. Science 366: 606–612. [DOI] [PubMed] [Google Scholar]
- Cegelski, L. , Marshall, G.R. , Eldridge, G.R. , and Hultgren, S.J. (2008) The biology and future prospects of antivirulence therapies. Nat Rev Microbiol 6: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2019) Antibiotic resistance threats in the United States. In Department of Health and Human Services. Atlanta: CDC. URL https://stacks.cdc.gov/view/cdc/82532 [Google Scholar]
- Crits‐Christoph, A. , Diamond, S. , Butterfield, C.N. , Thomas, B.C. , and Banfield, J.F. (2018) Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 558: 440–444. [DOI] [PubMed] [Google Scholar]
- Dadgostar, P. (2019) Antimicrobial resistance: implications and costs. Infect Drug Resist 12: 3903–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia, M.S. , Cimermancic, P. , Schulze, C.J. , Wieland Brown, L.C. , Martin, J. , Mitreva, M. , et al. (2014) A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158: 1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Commission (2017) A European One Health Action Plan against Antimicrobial Resistance (AMR). URL https://ec.europa.eu/health/sites/default/files/antimicrobial_resistance/docs/amr_2017_action‐plan.pdf. [Google Scholar]
- Francis, V.I. , Stevenson, E.C. , and Porter, S.L. (2017) Two‐component systems required for virulence in Pseudomonas aeruginosa . FEMS Microbiol Lett 364: fnx104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin, M.Y. (2018) What bacteria want. Environ Microbiol 20: 4221–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavira, J.A. , Gumerov, V.M. , Rico‐Jiménez, M. , Petukh, M. , Upadhyay, A.A. , Ortega, A. , et al. (2020) How bacterial chemoreceptors evolve novel ligand specificities. mBio 11: e03066–e3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavriilidou, A. , Kautsar, S.A. , Zaburannyi, N. , Krug, D. , Müller, R. , Medema, M.H. , and Ziemert, N. (2021) A global survey of specialized metabolic diversity encoded in bacterial genomes. bioRxiv. 10.1101/2021.08.11.455920. [DOI]
- Gnanendra, S. , Mohamed, S. , and Natarajan, J. (2013) Identification of potent inhibitors for Salmonella typhimurium quorum sensing via virtual screening and pharmacophore modeling. Comb Chem High Throughput Screen 16: 826–839. [DOI] [PubMed] [Google Scholar]
- Hengoju, S. , Tovar, M. , Man, D.K.W. , Buchheim, S. , and Rosenbaum, M.A. (2020) Droplet microfluidics for microbial biotechnology. In Advances in Biochemical Engineering/Biotechnology. Berlin, Heidelberg: Springer, pp. 1–29. 10.1007/10_2020_140. [DOI] [PubMed] [Google Scholar]
- van der Heul, H.U. , Bilyk, B.L. , McDowall, K.J. , Seipke, R.F. , and van Wezel, G.P. (2018) Regulation of antibiotic production in Actinobacteria: new perspectives from the post‐genomic era. Nat Prod Rep 35: 575–604. [DOI] [PubMed] [Google Scholar]
- Hirakawa, H. , Kurushima, J. , Hashimoto, Y. , and Tomita, H. (2020) Progress overview of bacterial two‐component regulatory systems as potential targets for antimicrobial chemotherapy. Antibiotics (Basel) 9: E635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings, M.I. , Truman, A.W. , and Wilkinson, B. (2019) Antibiotics: past, present and future. Curr Opin Microbiol 51: 72–80. [DOI] [PubMed] [Google Scholar]
- Johnson, K.S. , Elgamoudi, B.A. , Jen, F.‐ E.‐C. , Day, C.J. , Sweeney, E.G. , Pryce, M.L. , et al. (2021) The dCache chemoreceptor TlpA of Helicobacter pylori binds multiple attractant and antagonistic ligands via distinct sites. mBio 12: e0181921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper, J. , Evans, R. , Pritzel, A. , Green, T. , Figurnov, M. , Ronneberger, O. , et al. (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H.‐S. , and Kim, E.‐S. (2021) Recent advances in heterologous expression of natural product biosynthetic gene clusters in Streptomyces hosts. Curr Opin Biotechnol 69: 118–127. [DOI] [PubMed] [Google Scholar]
- Koh, S. , Hwang, J. , Guchhait, K. , Lee, E.‐G. , Kim, S.‐Y. , Kim, S. , et al. (2016) Molecular insights into toluene sensing in the TodS/TodT signal transduction system. J Biol Chem 291: 8575–8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier, J.‐C. , Dubourg, G. , Million, M. , Cadoret, F. , Bilen, M. , Fenollar, F. , et al. (2018) Culturing the human microbiota and culturomics. Nat Rev Microbiol 16: 540–550. [DOI] [PubMed] [Google Scholar]
- Lans, I. , Anoz‐Carbonell, E. , Palacio‐Rodríguez, K. , Aínsa, J.A. , Medina, M. , and Cossio, P. (2020) In silico discovery and biological validation of ligands of FAD synthase, a promising new antimicrobial target. PLoS Comput Biol 16: e1007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, K. (2020) The science of antibiotic discovery. Cell 181: 29–45. [DOI] [PubMed] [Google Scholar]
- Lloyd, K.G. , Steen, A.D. , Ladau, J. , Yin, J. , and Crosby, L. (2018) Phylogenetically Novel uncultured microbial cells dominate earth microbiomes. mSystems 3: e00055–e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J. , Zaborina, O. , Holbrook, C. , Zaborin, A. , and Alverdy, J. (2008) Depletion of intestinal phosphate after operative injury activates the virulence of P. aeruginosa causing lethal gut‐derived sepsis. Surgery 144: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalino, S.J.Y. , Billones, J.B. , Organo, V.G. , and Carrillo, M.C.O. (2020) In Silico strategies in tuberculosis drug discovery. Molecules 25: E665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler, L. , Niehs, S.P. , Martin, K. , Weber, T. , Scherlach, K. , Hertweck, C. , et al. (2021) Highly parallelized droplet cultivation and prioritization of antibiotic producers from natural microbial communities. eLife 10: e64774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, D. , Okada, B.K. , Wu, Y. , Xu, F. , and Seyedsayamdost, M.R. (2018) Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr Opin Microbiol 45: 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, J.K. , Sheehan, J.P. , Bratton, B.P. , Moore, G.M. , Mateus, A. , Li, S.‐ H.‐J. , et al. (2020) A dual‐mechanism antibiotic kills gram‐negative bacteria and avoids drug resistance. Cell 181: 1518–1532.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Mora, D. , Ortega, A. , Perez‐Maldonado, F.J. , Krell, T. , and Matilla, M.A. (2018) The activity of the C4‐dicarboxylic acid chemoreceptor of Pseudomonas aeruginosa is controlled by chemoattractants and antagonists. Sci Rep 8: 2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. (2021) Facing crises in the 21st century: microfluidics approaches for antibiotic discovery. Microb Biotechnol. 10.1111/1751-7915.13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. , Daddaoua, A. , Chini, A. , Morel, B. , and Krell, T. (2018) An auxin controls bacterial antibiotics production. Nucleic Acids Res 46: 11229–11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. , Velando, F. , Martín‐Mora, D. , Monteagudo‐Cascales, E. , and Krell, T. (2021) A catalogue of signal molecules that interact with sensor kinases, chemoreceptors and transcriptional regulators. FEMS Microbiol Rev. 10.1093/femsre/fuab043 [DOI] [PubMed] [Google Scholar]
- Medema, M.H. , de Rond, T. , and Moore, B.S. (2021) Mining genomes to illuminate the specialized chemistry of life. Nat Rev Genet 22: 553–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke, M. , Pieroni, M. , Weber, T. , Brönstrup, M. , Hammann, P. , Halby, L. , et al. (2021) Towards the sustainable discovery and development of new antibiotics. Nat Rev Chem 19: 1–24. [Google Scholar]
- Nandi, S. (2016) Recent advances in ligand and structure based screening of potent quorum sensing inhibitors against antibiotic resistance induced bacterial virulence. Recent Pat Biotechnol 10: 195–216. [DOI] [PubMed] [Google Scholar]
- Naylor, N.R. , Atun, R. , Zhu, N. , Kulasabanathan, K. , Silva, S. , Chatterjee, A. , et al. (2018) Estimating the burden of antimicrobial resistance: a systematic literature review. Antimicrob Resist Infect Control 7: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, D.J. , and Cragg, G.M. (2020) Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod 83: 770–803. [DOI] [PubMed] [Google Scholar]
- O’Neill, J. (2016) Tackling Drug‐resistant Infections Globally: Final Report and Recommendations. The Review on Antimicrobial Resistance. London: HM Government and the Wellcome Trust. URL https://amr‐review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf. [Google Scholar]
- Okada, B.K. , and Seyedsayamdost, M.R. (2017) Antibiotic dialogues: induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol Rev 41: 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega, A. , Zhulin, I.B. , and Krell, T. (2017) Sensory repertoire of bacterial chemoreceptors. Microbiol Mol Biol Rev 81: e00033–e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y.‐C. , Lu, C. , Li, G. , Eichenbaum, Z. , and Lu, C.‐D. (2017) Induction of the pho regulon and polyphosphate synthesis against spermine stress in Pseudomonas aeruginosa . Mol Microbiol 104: 1037–1051. [DOI] [PubMed] [Google Scholar]
- Rutledge, P.J. , and Challis, G.L. (2015) Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol 13: 509–523. [DOI] [PubMed] [Google Scholar]
- Scherlach, K. , and Hertweck, C. (2021) Mining and unearthing hidden biosynthetic potential. Nat Commun 12: 3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L.M. , Jackson, S.A. , Gardner, P.P. , and Fineran, P.C. (2021) SorTn‐seq: a high‐throughput functional genomics approach to discovering regulators of bacterial gene expression. Nat Protoc 16: 4382–4418. [DOI] [PubMed] [Google Scholar]
- Sundin, G.W. , and Wang, N. (2018) Antibiotic resistance in plant‐pathogenic bacteria. Annu Rev Phytopathol 56: 161–180. [DOI] [PubMed] [Google Scholar]
- Tacconelli, E. , Carrara, E. , Savoldi, A. , Harbarth, S. , Mendelson, M. , Monnet, D.L. , et al. (2018) Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic‐resistant bacteria and tuberculosis. Lancet Infect Dis 18: 318–327. [DOI] [PubMed] [Google Scholar]
- Theuretzbacher, U. , Outterson, K. , Engel, A. , and Karlén, A. (2020) The global preclinical antibacterial pipeline. Nat Rev Microbiol 18: 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunyasuvunakool, K. , Adler, J. , Wu, Z. , Green, T. , Zielinski, M. , Žídek, A. , et al. (2021) Highly accurate protein structure prediction for the human proteome. Nature 596: 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetrivel, A. , Natchimuthu, S. , Subramanian, V. , and Murugesan, R. (2021) High‐throughput virtual screening for a new class of antagonist targeting LasR of Pseudomonas aeruginosa . ACS Omega 6: 18314–18324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, M.C. , Mori, T. , Rückert, C. , Uria, A.R. , Helf, M.J. , Takada, K. , et al. (2014) An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506: 58–62. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2015) Global Action Plan on Antimicrobial Resistance. Geneva: World Health Organization. URL https://ahpsr.who.int/publications/i/item/global‐action‐plan‐on‐antimicrobial‐resistance [Google Scholar]
- Xin Yu, J. , Hubbard‐Lucey, V.M. , and Tang, J. (2019) Immuno‐oncology drug development goes global. Nat Rev Drug Discov 18: 899–900. [DOI] [PubMed] [Google Scholar]
- Zaborin, A. , Romanowski, K. , Gerdes, S. , Holbrook, C. , Lepine, F. , Long, J. , et al. (2009) Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci USA 106: 6327–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , and Seyedsayamdost, M.R. (2020) Discovery of a cryptic depsipeptide from streptomyces ghanaensis via MALDI‐MS‐guided high‐throughput elicitor screening. Angew Chem Int Ed 59: 23005–23009. [DOI] [PubMed] [Google Scholar]


