Abstract
Aberrant activation of the non-receptor kinase c-Abl is implicated in the development of pathogenic hallmarks of Parkinson’s disease, such as α-synuclein aggregation and progressive neuronal loss. c-Abl-mediated phosphorylation and inhibition of parkin ligase function lead to accumulation of parkin interacting substrate (PARIS) that mediates α-synuclein pathology-initiated dopaminergic neurodegeneration.
Here we show that, in addition to PARIS accumulation, c-Abl phosphorylation of PARIS is required for PARIS-induced cytotoxicity. c-Abl-mediated phosphorylation of PARIS at Y137 (within the Krüppel-associated box domain) drives its association with KAP1 and the repression of genes with diverse functions in pathways such as chromatin remodelling and p53-dependent cell death. One phosphorylation-dependent PARIS target, MDM4 (a p53 inhibitor that associates with MDM2; also known as MDMX), is transcriptionally repressed in a histone deacetylase-dependent manner via PARIS binding to insulin response sequence motifs within the MDM4 promoter. Virally induced PARIS transgenic mice develop c-Abl activity-dependent Parkinson’s disease features such as motor deficits, dopaminergic neuron loss and neuroinflammation. PARIS expression in the midbrain resulted in c-Abl activation, PARIS phosphorylation, MDM4 repression and p53 activation, all of which are blocked by the c-Abl inhibitor nilotinib. Importantly, we also observed aberrant c-Abl activation and PARIS phosphorylation along with PARIS accumulation in the midbrain of adult parkin knockout mice, implicating c-Abl in recessive Parkinson’s disease. Inhibition of c-Abl or PARIS phosphorylation by nilotinib or Y137F-PARIS expression in adult parkin knockout mice blocked MDM4 repression and p53 activation, preventing motor deficits and dopaminergic neurodegeneration. Finally, we found correlative increases in PARIS phosphorylation, MDM4 repression and p53 activation in post-mortem Parkinson’s disease brains, pointing to clinical relevance of the c-Abl-PARIS-MDM4-p53 pathway.
Taken together, our results describe a novel mechanism of epigenetic regulation of dopaminergic degeneration downstream of pathological c-Abl activation in Parkinson’s disease. Since c-Abl activation has been shown in sporadic Parkinson’s disease, PARIS phosphorylation might serve as both a useful biomarker and a potential therapeutic target to regulate neuronal loss in Parkinson’s disease.
Keywords: c-Abl, PARIS, MDM4, Parkinson’s disease, p53
See Giesert (doi:10.1093/brain/awab412) for a scientific commentary on this article.
Kim et al. demonstrate that c-Abl phosphorylation of PARIS Y137 contributes to epigenetic repression of MDM4, p53 activation and neurotoxicity. Since c-Abl is implicated in Parkinson’s disease pathogenesis, PARIS Y137 phosphorylation by c-Abl could serve as a new therapeutic target and/or biomarker for Parkinson’s disease.
See Giesert (doi:10.1093/brain/awab412) for a scientific commentary on this article.
Introduction
Parkinson’s disease is mainly defined by progressive loss of dopaminergic neurons, which eventually advances to neurodegeneration in nondopaminergic cells in other brain regions.1 Dopamine, oxidative or mitochondrial stresses have been attributed to neuronal death in Parkinson’s disease pathogenesis.2,3 Moreover, α-synuclein aggregation and its propagation to various brain regions contribute to the progression of diverse motor and non-motor symptoms associated with Parkinson’s disease.4,5 Although oxidative stress and α-synuclein pathology appear to broadly stimulate many signalling pathways, it has been suggested that c-Abl, a stress sensor and non-receptor tyrosine kinase, is a major mediator of dopaminergic neuron loss in response to these Parkinson’s disease-relevant stimuli.6-9 Pathogenic mechanisms of Parkinson’s disease, including dopaminergic neuron loss on 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) intoxication or α-synuclein preformed fibril injection, are largely prevented by genetic or pharmacological inhibition of aberrant c-Abl activation, suggesting a critical role of c-Abl in Parkinson’s disease pathogenesis.6–11 Activated c-Abl can phosphorylate a diverse set of protein substrates.12,13 In relation to Parkinson’s disease pathogenesis, c-Abl-dependent phosphorylation of α-synuclein has been shown to accelerate its aggregation.6 Moreover, parkin, a recessive Parkinson’s disease gene encoding an E3 ubiquitin ligase, is a phospho-substrate of c-Abl.2,3 Activated c-Abl on MPTP or α-synuclein pathology results in parkin phosphorylation and dysfunction.3,7 Among several known parkin substrates, accumulation of parkin interacting substrate [PARIS, also known as zinc finger protein 746 (ZNF746)] seems to play a major role in dopaminergic neurodegeneration downstream of c-Abl-induced parkin inactivation, since PARIS knockout prevented α-synuclein preformed fibril -initiated Parkinson’s disease pathologies.7 The fact that parkin and α-synuclein are c-Abl substrates and that c-Abl inhibition can substantially prevent Parkinson’s disease pathology in mouse models suggest that additional c-Abl substrates might play important roles in Parkinson’s disease pathology.
PARIS also known as ZNF746 is a Krüppel-associated box (KRAB) zinc finger protein, and its expression is regulated by parkin-mediated ubiquitination.14 PINK1-mediated phosphorylation of PARIS facilitates its ubiquitination by parkin and subsequent proteasomal degradation.15 In this regard, PARIS accumulation has been observed in mouse brains with parkin or PINK1 ablation- and α-synucleinopathy- (α-synuclein transgenic mice and α-synuclein preformed fibril injection model) mediated parkin inactivation.7,14,15 In addition, the increased PARIS expression levels observed in post-mortem human brains of sporadic Parkinson’s disease suggest its clinical relevance in Parkinson’s disease pathogenesis.14 A few target genes that are repressed by PARIS have been reported. The master regulator peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1α, encoded by PPARGC1A) is transcriptionally repressed by PARIS accumulation via its binding to insulin response sequence (IRS) motifs in the PPARGC1A promoter.14 Downregulation of PGC-1α by PARIS leads to dysregulated mitochondrial biogenesis and antioxidant defence. In addition, PARIS has been shown to inhibit transketolase transcription, thereby affecting glucose metabolism.16 In a more recent study, PARIS overexpression in C2C12 cells upregulated the expression of cellular senescence markers via p53 induction.17 Although the structure of PARIS implies its functions in KRAB domain-mediated histone deacetylase (HDAC) or repressor complex recruitment, epigenetic targets of PARIS remain to be discovered.
Apoptotic neuronal death has been induced in several animal models of Parkinson’s disease including MPTP, 6-OHDA, rotenone toxin injection models and α-synucleinopathy models.8,18 Activation of p53 is a key event in the triggering of apoptotic programmed cell death, and p53 activity is tightly regulated by several mechanisms.19 p53 itself can be post-translationally modified to regulate its own activation.20 Further, the MDM2 E3 ligase can polyubiquitinate p53, thereby labelling p53 for proteasomal degradation.21 Interestingly, MDM2 can be phosphorylated and inactivated by the stress sensor c-Abl, thereby affecting p53 activation.21 Moreover, MDM4 (in association with MDM2) can potentiate p53 inhibition. In addition to the regulation of p53 stability, MDM4 can inhibit p53 transcriptional activity.22,23 In α-synucleinopathy mouse models, activation of both c-Abl and p53 has been observed.6,8 Elucidating the molecular pathways that activate p53 in response to c-Abl activation will provide insight into potential therapeutic interventions to prevent apoptotic neuron loss in Parkinson’s disease.
In this study, we discovered that PARIS is phosphorylated at Y137 by activated c-Abl. Microarray analysis revealed that gene clusters in various biological pathways, such as chromatin modification, the cell cycle process and epigenetic regulation of gene expression, are regulated in a phosphorylation-dependent manner. In particular, PARIS phosphorylation leads to p53 activation and apoptotic cell death by negatively regulating MDM4 transcription. Moreover, the c-Abl-PARIS-MDM4-p53 signalling pathway was examined in both adeno-associated virus (AAV)-PARIS injected mice and adult conditional parkin knockout mice. c-Abl activation seems to play critical role in dopaminergic neurodegeneration in ventral midbrain stereotactically injected AAV-PARIS mice because treatment with the c-Abl inhibitor nilotinib largely abrogated PARIS-induced neuropathology and molecular alterations. Since we observed enhanced PARIS phosphorylation and MDM4 repression in Parkinson’s disease post-mortem brains, aberrant c-Abl phosphorylation of PARIS might be an important therapeutic target for the treatment of Parkinson’s disease.
Materials and methods
Chemicals and antibodies
STI-571 (imatinib; Sigma, Cat No SML1027), ATP (Invitrogen, Cat No PV3227), 3-aminobenzamide (3-AB; Sigma, Cat No A0788), Z-VAD-FMK (Sigma, Cat No C2105), Trichostatin A (TSA; Sigma, Cat No T8552), and nilotinib (Cayman Chemical, Cat No 641571-10-0) were used in this study. The following primary antibodies were used: rabbit GFP antibody (Cell Signaling Technology; Cat No 2956; 1:5000), mouse GFP antibody (Santa Cruz Biotechnology, Cat No sc-9996, 1:500), mouse FLAG antibody (Sigma; Cat No F1804; 1:50 for immunoprecipitation), rabbit PARIS (ZNF746) antibody (Proteintech; Cat No 24543-1-AP; 1:5000), rabbit antibody to phosphorylated c-Abl (Cell Signaling Technology; Cat No 2868; 1:5000), mouse c-Abl antibody (Sigma; Cat No A5844; 1:5000), rabbit MDM4 antibody (Proteintech; Cat No 17914-1-AP; 1:5000), mouse antibody to phosphorylated p53 (Cell Signaling Technology; Cat No 9284; 1:5000), mouse p53 antibody (Santa Cruz Biotechnology; Cat No sc-126; 1:5000), mouse PGC-1α antibody (Calbiochem, Cat No ST1202), rabbit NRF1 antibody (Abcam, Cat No ab34682), rabbit tyrosine hydroxylase (TH) antibody (Novus Biologicals; Cat No NB300-109; 1:2000), mouse TH antibody (ImmunoStar; Cat No 22941; 1:2000), mouse parkin antibody (Cell Signaling Technology; Cat No 4211; 1:5000), rabbit acetyl-histone H3 antibody (Merck Millipore; Cat No 06-599; 1:20), rabbit Histone H3 antibody (Cell Signaling Technology; Cat No 4620; 1:20), horseradish peroxidase (HRP)-conjugated mouse FLAG antibody (Sigma; Cat No 8592; 1:5000), HRP-conjugated mouse HA antibody (Cell Signaling Technology; Cat No 2999; 1:5000), and HRP-conjugated β-actin mouse antibody (Sigma; Cat No A3854; 1:10 000). The following secondary antibodies were used: HRP-conjugated goat antibody to mouse IgG (Genetex; Cat No GTX-213111-01, 1:5000), HRP-conjugated goat antibody to rabbit IgG (Genetex; Cat No GTX-213110-01; 1:5000), biotin-conjugated goat antibody to rabbit IgG (Vector Laboratories, Cat No BA-1000, 1:1000), Alexa Fluor 568 donkey anti-rabbit IgG (Invitrogen, Cat No A10042), Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen; Cat No A11008), Alexa Fluor 568 donkey anti-mouse IgG (Invitrogen; Cat No A10037), Alexa Fluor 488 donkey anti-mouse IgG (Invitrogen; Cat No A21202), Alexa Fluor 405-conjugated goat antibody to rabbit IgG (ab175652, Abcam), and Alexa Fluor 405-conjugated goat antibody to mouse IgG (ab175660, Abcam).
Cell culture and transfection
Human neuroblastoma SH-SY5Y cells (ATCC) were grown in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO Cat No 11995073) containing 10% foetal bovine serum (FBS; vol/vol; GIBCO Cat No 26140079) and penicillin-streptomycin antibiotic solution (100 U/ml; Sigma, Cat No P4333). SH-SY5Y cells were grown at 37°C in a humidified atmosphere consisting of 5% CO2/95% air. X-tremeGENETM HP transfection reagent (Roche, Cat No 6366546001) was used for transient transfection of SH-SY5Y cells following the manufacturer’s instructions. For mouse dopaminergic neuron culture, neural precursor cells were isolated from ventral midbrains dissected from mouse embryos [embryoinc Day (E)10–12], as previously described.24 Isolated neural precursor cells were plated onto glass-coated dishes with poly-l-ornithine (30 μg/ml in distilled, deionized water overnight) and fibronectin (2 μg/ml in deionized water overnight) and then expanded in N2 medium supplemented with b27 (Gibco), insulin (5 μg/ml; Sigma), basic fibroblast growth factor (bFGF; 20 ng/ml; R&D Systems) and epithelial growth factor (EGF; 20 ng/ml; R&D Systems). During neural precursor cell expansion (3 days), the old media were replaced with fresh media daily. Expanded neural precursor cells were transferred to newly coated plates after cell dissociation by gentle pipetting in Hanks’ Balanced Salt Solution buffer. Neural precursor cells were then transfected using Lipofectamine® 3000 reagent (ThermoFisher Scientific, Cat No L3000015) and differentiated into dopaminergic neurons by culturing in N2 medium supplemented with b27 (ThermoFisher Scientific, Cat No 17504044), insulin (Sigma, Cat No I9278) and ascorbic acid (0.2 mM; Sigma) for 7 days.
Plasmids
Human PARIS was cloned into a pCMV-Tag2A-FLAG vector (FLAG-PARIS, Stratagene). The mutant forms of PARIS (Y121F, Y137F and Y403F) with tyrosine to phenylalanine substitutions at putative phosphorylation sites were generated by mutagenesis of the FLAG-PARIS and pGEX-6P-3-PARIS constructs (QuikChange II Site-Directed Mutagenesis kit; Agilent, Cat No 200522) according to the manufacturer’s instructions. The primer sequences are listed in Supplementary Table 1. The GFP-c-Abl-KA (kinase-active) and pEGFP-C2 constructs have been previously described.3 We purchased plasmids expressing TRIM28 (HA-KAP1, Cat No 10841) and gRNA to c-Abl (pLenti-CRISPR-g-c-Abl, Cat No 77733) from Addgene. CRISPR gRNA to human PARIS was generated by cloning PARIS targeting gRNA sequence (CCTCTTCAGCATGCTCGGAC; cloning oligo 1 = 5′-CACCGCCTCTTCAGCATGCTCGGAC-3′, oligo 2 = 5′-AAACGTCCGAGCATGCTGAAGAGGC-3′) into pLenti-CRISPR-V2 plasmid (Addgene, Cat No 52961). GFP-JAK3 and GFP-Src were cloned into the destination vector pcDNA-DEST53 (Addgene, Cat No 12288-015) from corresponding entry clone library by using gateway cloning. Plasmids expressing β-galactosidase or GFP were used as overexpression controls.
Generation of phosphorylated PARIS (pTyr137) antibodies
Phospho-PARIS-specific rabbit polyclonal antibodies (produced by Young In Frontier) were raised by inoculating the following peptides as antigens: CETLVSLD (phosphorylated) YAISKPEV. Affinity purification of phospho-specific PARIS antibodies from anti-serum was performed using SulfoLinkTM columns (Thermo Fisher Scientific, Cat No 44999) to which target phospho (antigen peptide) or dephopho peptides were immobilized. Phospho-peptide or dephospho-peptide affinity columns were obtained by coupling peptides (1 mg per peptide) to the SulfoLinkTM affinity columns following the manufacturer’s instructions. Phospho-specific PARIS antibodies were purified by serial binding and elution through the phospho-peptide-conjugated column followed by the dephospho-peptide-conjugated column. The concentration of the purified antibody was measured using the BCA kit (ThermoFisher Scientific, Cat No 23227).
RNA extraction and real-time quantitative PCR
For real-time quantitative PCR, total RNA was extracted from cultured SH-SY5Y cells with QIAzol Lysis Reagent (QIAGEN, Cat No 79306) and then treated with DNase I to avoid DNA contamination. Complementary DNA was synthesized from total RNA (1 µg) using the iScriptTM cDNA Synthesis kit (Bio-Rad, Cat No 170-8891). The relative abundance of target mRNA expression was analysed using real-time quantitative PCR (QuantStudioTM 6 Flex Real-Time PCR System, Applied Biosystems) with SYBR Green PCR master mix (Applied Biosystems, Cat No 4309155). The relative mRNA expression levels of target genes were calculated by the ΔΔCt method using GAPDH as an internal loading control. The primer sequences are listed in Supplementary Table 1.
Microarray and Cytoscape analysis
Total RNA was extracted from cultured SH-SY5Y cells with the RNeasy Mini Kit (QIAGEN, Cat No 74104) following the manufacturer’s instructions. The data were summarized and normalized using the robust multi-average method implemented in Affymetrix® Power Tools. We exported the results with gene level RMA analysis and performed differentially expressed gene (DEG) analysis. Genes with >30% alterations in average expression levels and with repeated trend of expression changes (i.e. genes with increase or decrease of transcription compared to control in two experiments) in two independent experiments were used for subsequent DEG analysis. For a DEG set, hierarchical cluster analysis was performed using complete linkage and Euclidean distance as a measure of similarity. Gene enrichment and functional annotation analysis of the selected gene list was performed using Cytoscape ClueGO.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was conducted following the manufacturer’s instructions (Cell Signaling Technology, Cat No 9002). Briefly, SH-SY5Y cells (transfected with mock, FLAG-PARIS wild-type or FLAG-PARIS-Y137F; mock, FLAG-PARIS wild-type with or without TSA) were fixed with 1% formaldehyde at 37°C for 10 min. Glycine quenched samples were lysed in 1 ml of SDS buffer containing protease inhibitors. The lysates were incubated for 10 min on ice and sonicated to shear the DNA. The samples were centrifuged at 9400g at 4°C for 10 min. Precleared supernatant samples were incubated with anti-acetylated histone, anti-histone, anti-FLAG antibodies or rabbit IgG (rIgG)-bound agarose beads followed by several washes. Immunoprecipitated DNA was recovered by phenol-chloroform-ethanol purification after reverse-crosslinking. PCR was performed with input DNA and immunoprecipitated DNA as template using Taq mix (Guangzhou Dongsheng Biotech, Cat No P2012) and the indicated primers. The primer sequences are listed in Supplementary Table 1.
Preparation of proteins and western blot
SH-SY5Y cells were briefly washed with ice-cold PBS, and total protein lysates were prepared in lysis buffer [1% Nonidet P40 and 0.5% sodium deoxycholic acid (NaDOC) in PBS, pH 7.4] supplemented with protease (no. 786-108, G-biosciences)/phosphatase inhibitors (no. 78420, ThermoFisher Scientific). After incubation on ice for 30 min (for complete lysis) with vortexing every 5 min, the lysates were centrifuged at 14 000g for 30 min, and the supernatant was retained for downstream analysis. For protein extraction, the dissected mouse brain tissues were homogenized in lysis buffer (1% Nonidet P40 and 0.5% NaDOC in PBS, pH 7.4, and protease/phosphatase inhibitors) using a Diax 900 homogenizer. The homogenized brain samples were incubated in ice for 30 min (for complete lysis) with vortexing every 5 min, and then centrifuged at 14 000g for 30 min. The supernatants were collected, and protein levels were quantified using the BCA Protein Assay Kit (Pierce) with bovine serum albumin standards. Total protein lysates were subjected to western blot analysis. Lysates were mixed with 2× Laemmli buffer (Bio-Rad) supplemented with β-mercaptoethanol (Bio-Rad) and boiled for 5 min. Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes (0.45 µm; Bio-Rad, Cat No 162-0115) for immunoblotting. Immunoblotting was performed with the indicated antibodies, and the bands were visualized via chemiluminescence (Pierce). Densitometric analysis of the bands was performed using ImageJ software (NIH, rsb.info.nih.gov/ij).
Co-immunoprecipitation
SH-SY5Y cells were washed with ice-cold PBS and harvested with immunoprecipitation buffer (1% Nonidet P40 in PBS, pH 7.4) supplemented with protease/phosphatase inhibitors. The lysate was incubated on ice for 30 min with vortexing every 5 min, followed by three cycles of freezing and thawing for complete lysis. The protein lysates were centrifuged at 14 000g for 30 min. The supernatants were incubated with 50 µl of protein G Sepharose beads (GE Healthcare, Cat No 18-0618-01) and 1 µg of indicated antibodies, and then rotated at 4°C overnight. The next day, the beads were washed three times with ice-cold wash buffer, suspended in 2× Laemmli buffer (Bio-Rad, Cat No 1610737) supplemented with β-mercaptoethanol (Bio-Rad, Cat No 1610710), and boiled for 5 min. The precipitates were separated by SDS-PAGE and subjected to immunoblot analysis with the indicated antibodies.
In vitro kinase assay
GST-PARIS wild-type and phospho-mutant recombinant proteins (2.5 μg) were incubated with 250 ng c-Abl recombinant protein in a kinase buffer (20 mM HEPES; 10 mM MgCl2; 5 mM EGTA; 150 mM NaCl, 20 mM β-glycerol phosphate, 50 μM ATP, pH 7.4), supplemented with or without STI-571 (10 μM), at 30°C for 45 min using a thermomixer (Eppendorf, Cat No 5382000015). Samples were separated by SDS-PAGE and subjected to immunoblot analysis with the indicated antibodies.
Cell viability assay
SH-SY5Y cells were plated in 12-well plates at a density of 0.2 × 106 cells per well. SH-SY5Y cells were transfected with the indicated constructs, and the cells were grown in DMEM containing low serum (2.5% FBS) with or without chemicals at the indicated concentrations. Next, the cells were harvested, washed twice with PBS, resuspended in PBS, mixed with an equal volume of 0.4% trypan blue (wt/vol), and incubated at room temperature for 2 min. Live and dead cells were analysed using a Countess II Automated Cell Counter (Life Technologies). For assessment of apoptotic neuronal death, a terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) assay (Abcam, Cat No ab661110) was used to label DNA fragmentation, according to the manufacturer’s guidelines.
JC-1 assay
Mitochondrial membrane potential was assessed by using the lipophilic cationic JC-1 dye (JC-1 mitochondrial membrane potential assay kit, Cayman Chemical, Cat No 10009172) following the manufacturer’s manual. After SH-SY5Y cells were stained with JC-1 dye for 20 min, and washed twice with assay buffer, JC-1 fluorescence was measured using a SYNERGYneo microplate reader (Bio-Tek) with excitation and emission wavelengths of 535 and 595 nm, respectively, for healthy mitochondria and 485 and 535 nm, respectively, for unhealthy mitochondria.
Immunofluorescence
The cells or floating mouse brain sections fixed with 4% paraformaldehyde (PFA) in PBS were blocked with a blocking solution of 5% normal goat serum (Invitrogen, Cat No 16201) and 0.1% Triton X-100 (Sigma, Cat No X100) in PBS at room temperature for 1 h. The samples were then incubated with combinations of primary antibodies depending on the experiment at 4°C overnight and thereafter washed with PBS containing 0.1% Triton X-100; the samples were then incubated with corresponding fluorescent dye-conjugated secondary antibodies at room temperature for 1 h. After PBS washes, the samples were incubated with DAPI (Sigma, Cat No D9542) solution for 1 min for nuclear counterstaining, and then mounted using Immu-Mount solution (ThermoFisher Scientific, Cat No 9990402). Fluorescent images were obtained using a fluorescence microscope (Axio Imager M2; Zeiss).
Animal experiments
All animal experiments were approved by the Ethical Committee of Sungkyunkwan University and were conducted in accordance with all applicable international guidelines (protocol number: SKKUIACUC2019-04-22-2). Male C57BL/6N mice (3 months old) were purchased from Orient. The floxed parkin mice used have been previously described.14 Animals were maintained in a 12-h dark/light cycle in air-controlled rooms and were provided ad libitum access to food and water. To generate a mouse model in which PARIS is overexpressed in dopaminergic neurons, AAV-Con (1 µl of 6 × 1011 AAV1 vector genomes of CMV promoter per 1 ml PBS; Vector Biolabs, Cat No 7002) or AAV-PARIS (1 µl of 6 × 1011 AAV1 vector genomes of CBA promoter per 1 ml PBS; cloned into pAM-CBA-WPRE-bGHpA plasmid14 and AAV-PARIS produced in the Vector Biolabs) was injected into substantia nigra pars compacta (SNpc; coordinates from bregma, L: 1.2, AP: −3.4, DV: −4.3 mm) of 3-month-old wild-type mice. Conditional adult parkin knockout (parkinfl/fl) mice were established by nigral injection of AAV-GFPCre (1 µl of 2 × 1011 AAV1 vector genomes of CMV promoter per 1 ml PBS; Vector Biolabs, Cat No 7015) to homozygous floxed parkin mice.14 AAV-Y137F-PARIS (1 µl of 6 × 1011 AAV1 vector genomes of CBA promoter per 1 ml PBS; cloned into pAM-CBA-WPRE-bGHpA plasmid14) were produced by the Vector Biolabs and co-injected with AAV-GFPCre into substantia nigra of wild-type and floxed parkin mice to examine the role of PARIS phosphorylation in the parkin knockout background. To test the effect of nilotinib, it was administered [intraperitoneally (i.p.), 50 mg/kg/day] 1 week or [per os (p.o.), nilotinib containing diet, 200 mg nilotinib per 1 kg diet prepared from the Doo Yeol Biotech] 1 month after nigral virus injection for virally induced PARIS overexpression or parkin knockout models, respectively. Daily nilotinib administration began and continued for 2 weeks (50 mg/kg/day, i.p.) or 2 months (200 mg nilotinib per 1 kg diet, p.o.), followed by subsequent behavioural testing and neuropathological analysis for virally induced PARIS overexpression or parkin knockout models, respectively.
Immunohistochemistry and TH stereology
Animals were intracardially perfused with ice-cold PBS and fixed with 4% PFA. The brain was isolated from the mouse and post-fixed with 4% PFA at 4°C for 1 day, and then embedded in 30% sucrose in PBS at 4°C for 2 days. The cryoprotected brain was processed for sliding microtome (ThermoFisher Scientific, Cat No HM430) sectioning and immunohistochemistry. Coronal sections (35-µm thick) were cut through the brain including the substantia nigra. The fixed floating brain sections were incubated with a rabbit polyclonal anti-TH antibody (1:1000 dilution) followed by sequential incubations with biotinylated goat anti-rabbit IgG and streptavidin-conjugated HRP using the Vectastain ABC kit (Vector Laboratories) according to the manufacturer’s protocol. TH-positive cells were visualized by incubation with 3,3-diaminobenzidine (DAB; Sigma, Cat No D4293) as an HRP substrate. The total number of TH-positive neurons visualized with DAB in the SNpc was determined using the Optical Fractionator probe in the Stereo Investigator software (MicroBrightfield, Williston, VT). All stereological counting was performed blind.
Behaviour tests
Open field test
The open field arena consisted of a rectangular wood box (32 × 32 × 32 cm) divided into 64 (8 × 8) identical regions (4 × 4 cm). The field was subdivided into border, peripheral and central sectors. The central sector included four central squares (2 × 2). The peripheral sector included the 12 squares surrounding the central sector. The remaining squares composed the border sector. The mouse was placed at the centre of the open field and was allowed to explore for 15 min under a dim light. The apparatus was cleaned with diluted 70% ethanol between each trial. A video tracking system (Smart v.3.0 software) was used to record the distance travelled as a measure of locomotor activity. The time spent in the centre and the latency of the first entry into the centre were measured as anxiolytic indicators.
Pole test
Before the pole test, the mice were trained twice. The 9-mm diameter pole is a 23-inch-long metal rod wrapped with bandage gauze. Briefly, the mice were placed on the top of the pole facing head-down. The total latency to reach the bottom of the pole was recorded. The test data are presented as means of the time to reach the base from three trials.
Rotarod test
Before the rotarod test, the mice were trained for 2 days. On Day 3, the mice were placed on an accelerating rotarod cylinder, and the time taken to fall off the cylinder was measured. The speed of the rotarod was slowly increased from 4 to 40 rpm over the course of 5 min. A trial ended if the mouse fell off the rotarod. The data are presented as means of the latency to fall from three trials.
Statistics
Quantified data are presented as mean ± standard error of the mean (SEM). Statistical significance was assessed either using unpaired two-tailed Student’s t-test (two-group) or ANOVA test with Tukey’s HSD post hoc analysis (three or more groups). Differences with a P-value < 0.05 were considered statistically significant. GraphPad Prism software (v.5.03) was used for preparation of all plots and all statistical analyses.
Data availability
Data supporting the findings of this study are included in this paper and Supplementary material. All supporting data are available from the corresponding authors on reasonable request.
Results
c-Abl phosphorylates tyrosine 137 of PARIS, enhancing its interaction with KAP1
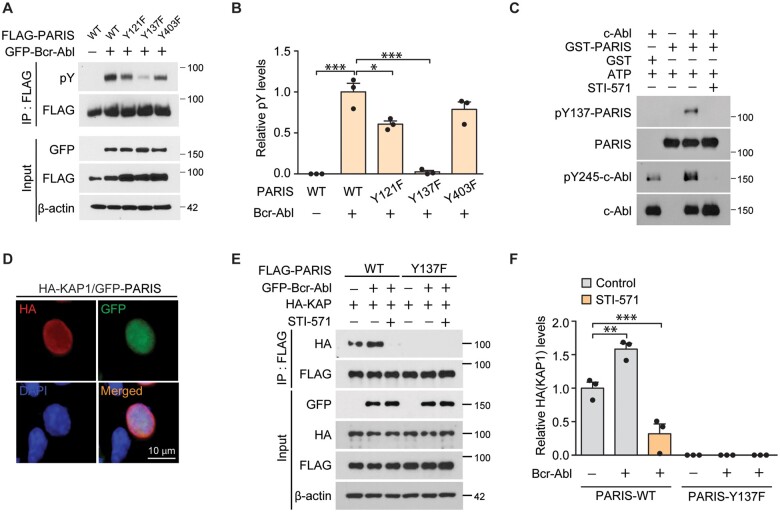
The parkin substrate PARIS is reported to be critical for inducing dopaminergic cell loss downstream of c-Abl activation.7 Moreover, c-Abl is activated under pathological conditions associated with Parkinson’s disease, and activated c-Abl can phosphorylate Parkinson’s disease-associated proteins including parkin and α-synuclein.3,6 To detect any direct interactions between c-Abl and PARIS during dopaminergic cell death in Parkinson’s disease, we first examined whether activated c-Abl is capable of phosphorylating PARIS. Putative tyrosine phosphorylation sites on PARIS (Y121, Y137 and Y403) were predicted with high probability by the in silico phosphorylation site prediction program, NetPhos 2.0. Y121 and Y137 are located within the KRAB domain of PARIS, while Y403 is positioned in between the KRAB and zinc finger domains (Supplementary Fig. 1A). To determine the main sites phosphorylated by c-Abl, SH-SY5Y neuroblastoma cells were transfected with wild-type FLAG-PARIS, or phosphorylation mutants of PARIS with tyrosine to phenylalanine substitutions (Y121F, Y137F and Y403F) together with constitutively active c-Abl (GFP-Bcr-Abl). Co-immunoprecipitation of FLAG-PARIS showed robust tyrosine phosphorylation of wild-type PARIS as monitored using phospho-tyrosine (pY)-specific antibodies (Fig. 1A). We observed marked reduction of tyrosine phosphorylation in the Y137F mutant PARIS as compared to other phosphorylation mutants, indicating that Y137 of PARIS is the main site of c-Abl-mediated phosphorylation (Fig. 1A and B). Amino acid sequence alignment revealed that Y137 within the KRAB domain of PARIS is highly conserved in several mammalian species including mouse, rat, cattle and chimpanzees (Supplementary Fig. 1B). To characterize Y137 phosphorylation of PARIS, we generated rabbit polyclonal antibodies specific for pY137-PARIS. This phospho-specific PARIS antibody recognized robust PARIS phosphorylation in SH-SY5Y cells co-expressing FLAG-PARIS and GFP-Bcr-c-Abl (Supplementary Fig. 1C). However, there was no detectable signal in Y137F-PARIS and GFP-Bcr-c-Abl co-expression samples (Supplementary Fig. 1C), indicating that this antibody is specific to its target. By using this phospho-specific PARIS antibody, we showed relative specificity of PARIS Y137 phosphorylation by Bcr-Abl because co-expression of other tyrosine kinases JAK3 and Src in SH-SY5Y cells failed to induce PARIS Y137 phosphorylation (Supplementary Fig. 1D and E). We also confirmed direct Y137 phosphorylation of recombinant GST-PARIS by c-Abl using in vitro kinase assays (Fig. 1C and Supplementary Fig. 1F and G). Treatment with the c-Abl inhibitor STI-571 abolished c-Abl phosphorylation and PARIS Y137 phosphorylation (Fig. 1C). Mutations of Y121F and Y403F on PARIS had no effect on PARIS Y137 phosphorylation by c-Abl in an in vitro kinase assay (Supplementary Fig. 1F and G).
Figure 1.
Activated c-Abl tyrosine-phosphorylates PARIS. (A) Representative immunoblots of PARIS tyrosine phosphorylation using a phospho-tyrosine specific antibody (pY) on anti-FLAG immunoprecipitation samples from SH-SY5Y cells transiently transfected (48 h) with N-terminal FLAG-tagged wild-type (WT) or mutants with substitutions of phenylalanine for tyrosine residues that are potentially phosphorylated (Y121F, Y137F and Y403F) with or without GFP-Bcr-Abl (constitutively active form). β-Actin serves as an internal loading control. (B) Quantification of relative tyrosine phosphorylation of anti-FLAG immunoprecipitated PARIS wild-type and mutants (n = 3 per group). (C) In vitro kinase assay demonstrating GST-tagged recombinant PARIS (GST-PARIS) phosphorylation at Y137 by recombinant c-Abl kinase assessed by immunoblotting using an anti-pY137-PARIS antibody. STI (10 µM) was used to block c-Abl, and c-Abl activity was monitored with an anti-pY245-c-Abl antibody. Similar results were obtained from two independent experiments. (D) Representative immunofluorescence images depicting the nuclear expression of GFP-PARIS and HA-KAP in SH-SY5Y cells transfected with HA-KAP and GFP-PARIS (48 h) using antibodies against the HA or GFP tags. Scale bar = 10 μm. (E) Immunoblots depicting the association between HA-KAP and FLAG-PARIS in anti-FLAG immunoprecipitation samples from SH-SY5Y cells transfected with the indicated combination of HA-KAP, FLAG-tagged PARIS wild-type or a Y137F phosphorylation-deficient PARIS mutant, and GFP-Bcr-Abl with or without c-Abl inhibitor STI-571 treatment (10 µM). β-Actin serves as an internal loading control. (F) Quantification of HA-KAP levels in the anti-FLAG co-immunoprecipitates from the indicated experimental groups (n = 3 per group). Data are expressed as mean ± SEM. Statistical ANOVA test followed by Tukey’s post hoc analysis or unpaired two-tailed Student’s t-test. *P < 0.05, **P < 0.01 and ***P < 0.001.
Next, we sought to characterize the regulation of PARIS function by Y137 phosphorylation. Potential alterations to the subcellular distribution of PARIS were monitored by immunofluorescence. PARIS overexpression led to nuclear accumulation of PARIS in SH-SY5Y cells, and all phosphorylation-deficient PARIS mutants remained localized to the nucleus (Supplementary Fig. 1H). Since the KRAB domain has been reported to interact with KRAB-associated protein-1 (KAP1, also known as tripartite motif-containing 28), and Y137 is located in the KRAB domain of PARIS, the association between PARIS and KAP1 was examined by immunofluorescence and anti-FLAG co-immunoprecipitation in SH-SY5Y cells transfected with FLAG-PARIS and HA-KAP1. The nuclear co-localization of GFP-tagged PARIS and HA-KAP1 was visualized by their co-labelling in SH-SY5Y cells expressing GFP-PARIS and HA-KAP1 (Fig. 1D). Moreover, wild-type FLAG-PARIS co-immunoprecipitated with HA-KAP1, whereas mutant Y137F PARIS failed to associate with HA-KAP1 (Fig. 1E and F). The association of PARIS and KAP1 was enhanced when GFP-Bcr-Abl was co-expressed (Fig. 1E and F). Together, these results indicate that PARIS is a phospho-substrate of c-Abl and that Y137 phosphorylation of PARIS is required for its efficient association with KAP1.
c-Abl-mediated phosphorylation of PARIS Y137 leads to apoptotic cell death
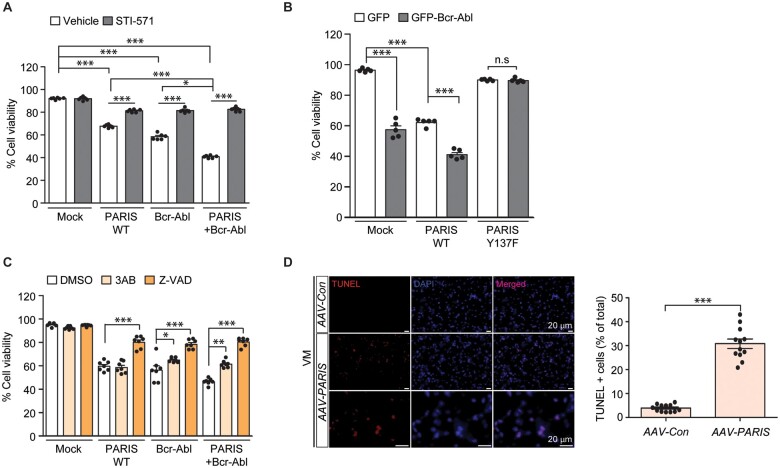
Both c-Abl activation and PARIS accumulation have been implicated in dopaminergic neurodegeneration in Parkinson’s disease.3,14 To determine the role of c-Abl-mediated PARIS phosphorylation in cell death, we evaluated cytotoxicity through a trypan blue exclusion assay in SH-SY5Y cells in response to PARIS and/or c-Abl overexpression. PARIS overexpression in SH-SY5Y cells resulted in ∼40% cytotoxicity, which was enhanced to ∼50% cytotoxicity by co-expression of GFP-Bcr-c-Abl (Fig. 2A). Consistent with the cytotoxic function of c-Abl,6 expression of a constitutively active form of c-Abl alone also led to substantial cytotoxicity in SH-SY5Y cells (Fig. 2A). With GFP-Bcr-Abl expression alone, we observed Y137 phosphorylation of endogenous PARIS, which is completely abolished by STI-571 treatment (Supplementary Fig. 2A and B). The cytotoxicity induced by PARIS and/or c-Abl expression was largely reversed by treatment with the c-Abl inhibitor STI-571 (Fig. 2A). In addition, CRISPR-mediated knockout of c-Abl (Supplementary Fig. 2C and D) also prevented PARIS-induced cytotoxicity in SH-SY5Y cells (Supplementary Fig. 2E). The cytoprotective effects of c-Abl inhibition or genetic ablation in PARIS overexpressing cells indicate the potential involvement of c-Abl activation in response to PARIS accumulation. Indeed, when PARIS is overexpressed, there was overactivation of c-Abl as evidenced by an increase in pY245-c-Abl protein and pY137-PARIS expression (Supplementary Fig. 2F and G). PARIS-induced c-Abl and PARIS phosphorylation was blocked by STI-571 treatment, which is consistent with the cytotoxicity results (Supplementary Fig. 2F and G). The role of PARIS Y137 phosphorylation in cell death was also evaluated in SH-SY5Y cells transiently expressing PARIS wild-type or the Y137F mutant with or without GFP-Bcr-c-Abl. We observed lesser cytotoxicity with PARIS Y137F expression than with PARIS wild-type expression (Fig. 2B). Moreover, co-expression of PARIS Y137F and GFP-Bcr-c-Abl did not produce synergistic cytotoxicity (Fig. 2B). Rather, it appears that PARIS Y137F has a dominant negative effect against GFP-Bcr-c-Abl-induced cytotoxicity, since co-expression of PARIS Y137F and GFP-Bcr-c-Abl resulted in lesser cytotoxicity compared to GFP-Bcr-c-Abl transfection alone (Fig. 2B). Consistent with the requirement of PARIS modification in c-Abl-stimulated cytotoxicity, CRISPR-mediated deletion of PARIS (Supplementary Fig. 2H and I) was largely cytoprotective against GFP-Bcr-Abl induced cell death (Supplementary Fig. 2J).
Figure 2.
PARIS Y137 phosphorylation by c-Abl is required for PARIS-induced cytotoxicity. (A) Trypan blue exclusion cell viability assessment in SH-SY5Y cells transfected with FLAG-PARIS and/or GFP-Bcr-Abl (Bcr-Abl, 48 h) with or without treatment with the c-Abl inhibitor STI-571 (10 µM, 44 h; n = 6 per group). (B) Trypan blue exclusion cell viability assessment in SH-SY5Y cells transfected with FLAG-PARIS or a phospho-deficient Y137F-PARIS mutant with or without GFP-Bcr-Abl (48 h; n = 6 per group). (C) Trypan blue exclusion cell viability assessment in SH-SY5Y cells transfected with FLAG-PARIS and/or GFP-Bcr-Abl (48 h) and treated with the poly (ADP-ribose) polymerase inhibitor 3AB (10 µM, 12 h) or pan-caspase inhibitor Z-VAD (100 µM, 12 h; n = 6 per group). (D) Representative TUNEL assay images of the ventral midbrain from mice that underwent stereotaxic nigral injection of AAV-Con or AAV-PARIS (3 weeks). The coronal brain sections were counterstained with DAPI. Quantification of the percentage of TUNEL-labelled cells in AAV-Con- or AAV-PARIS-injected ventral midbrain regions (right panel, n = 12 sections from three mice per group). Data are expressed as mean ± SEM. Statistical analysis consisted of a one-way ANOVA test followed by Tukey’s post hoc analysis or unpaired two-tailed Student’s t-test. *P < 0.05, **P < 0.01 and ***P < 0.001. n.s. = non-significant. WT = wild-type.
Next, we sought to investigate the major cell death pathways that are regulated by c-Abl-PARIS using pharmacological inhibition of caspase-dependent apoptosis or poly (ADP-ribose) polymerase-1-dependent parthanatos. Interestingly, PARIS-induced cytotoxicity was largely prevented by treatment with the pan-caspase inhibitor Z-VAD, while the poly (ADP-ribose) polymerase inhibitor 3AB had no effect (Fig. 2C), indicating that PARIS toxicity is largely mediated by apoptosis. Similarly, cytotoxicity induced by constitutively active c-Abl was partially blocked by 3AB treatment and was more robustly reversed by Z-VAD treatment. Similar protection by 3AB and Z-VAD was observed in SH-SY5Y cells with PARIS and c-Abl co-expression (Fig. 2C). These results indicate that c-Abl-PARIS-induced cytotoxicity is mainly mediated by apoptosis via caspase activation, although parthanatos is also involved in this process to a lesser extent. Virally induced PARIS overexpression in murine ventral midbrain further confirmed ongoing cell death as shown by an increase in terminal deoxynucleotidyl TUNEL signals (Fig. 2D) that are commonly used as indicators of both apoptotic and parthanatic cell death.
PARIS regulates diverse biological processes through Y137 phosphorylation-dependent gene repression
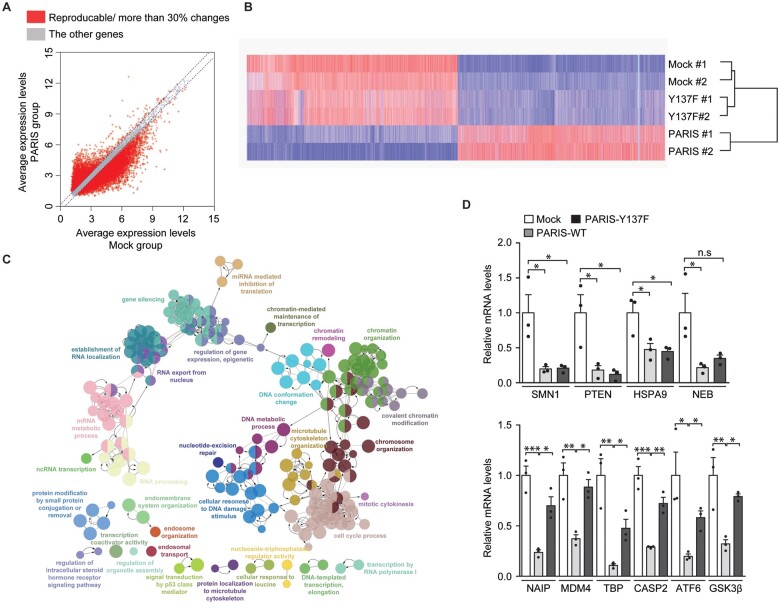
Given that c-Abl phosphorylation of PARIS is critical for apoptotic cell death and PARIS has been shown to function as a transcriptional repressor of a few target genes, we attempted to uncover genes and target biological pathways that may contribute to PARIS-induced cytotoxicity in a phosphorylation-dependent manner. SH-SY5Y cells were transfected with mock, wild-type PARIS, or Y137F PARIS and total RNA was subjected to microarray analysis. We confirmed robust and comparable PARIS wild-type and Y137F expression in SH-SY5Y cells with transfection of the corresponding plasmids, while PARIS Y137 phosphorylation was only induced in PARIS wild-type transfection group (Supplementary Fig. 3A and B). In two independent microarray experiments, average gene expression levels were plotted in a scatter chart depicting the mock versus PARIS wild-type expression groups (Supplementary Table 2 and Fig. 3A). Among a total of 31 128 genes probed, 12 904 DEGs showed >30% reproducible alterations in PARIS group compared to those in the mock transfection group (Fig. 3A). Heat map hierarchical analysis of these DEGs in three experimental groups (including the Y137F PARIS transfection group) revealed that PARIS wild-type expression led to substantial alterations in gene expression, while Y137F PARIS was characterized by expression patterns similar to those of mock transfected cells (Fig. 3B), suggesting that most PARIS-induced changes to gene expression are PARIS Y137 phosphorylation-dependent. When DEGs were further classified into subgroups according to gene regulation patterns, 79% of DEGs were repressed while 21% of DEGs were upregulated by PARIS expression as compared to the mock control (Supplementary Fig. 3C). Most genes downregulated by PARIS expression (96% of total downregulated genes) were Y137 phosphorylation-dependent with >30% reversal of gene expression in the Y137F PARIS group compared to the PARIS wild-type group (Supplementary Fig. 3C). This transcriptomic analysis revealed that the repressive function of PARIS is largely mediated by c-Abl phosphorylation of PARIS Y137.
Figure 3.
Microarray analysis reveals PARIS Y137 phosphorylation-dependent gene regulation. (A) Scatter plots of gene expression in the mock (x-axis) and FLAG-PARIS wild-type (y-axis) transfected SH-SY5Y cells generated by microarray analysis. Genes with >30% reproducible alterations in two independent experiments are shown in red, and only these genes were used for subsequent analyses. (B) Hierarchical cluster analysis of genes from A in SH-SY5Y cells transfected with mock, FLAG-PARIS wild-type, or the Y137F-PARIS mutant (n = 2 per group) using complete linkage and Euclidean distance as a measure of similarity. (C) Functional pathway clustering of the genes that are repressed (reproducible and <30% downregulation) in a PARIS Y137 phosphorylation-dependent manner using the Cytoscape ClueGO plugin. (D) Quantification and validation of select gene transcripts with Y137 phosphorylation-dependent and -independent repression (microarray analysis) by reverse transcription quantitative PCR [RT-qPCR; Y137 phosphorylation-independent genes in microarray: SMN1, PTEN, HSPA9, NEB; Y137 phosphorylation-dependent genes in microarray: NAIP, MDM4, TBP, CASP2, ATF6, GSK3B (GSK3β); n = 3 per group]. Data are expressed as mean ± SEM. Statistical analysis was performed using a one-way ANOVA test followed by Tukey’s post hoc analysis. *P < 0.05, **P < 0.01 and ***P < 0.001. n.s. = non-significant. WT = wild-type.
Next, we performed gene ontology analysis of the DEGs using the ClueGO plugin in Cytoscape. The genes that were regulated by PARIS expression in a phophorylation-independent manner were enriched in diverse biological functions (with 30% enrichment of the selected biological functions): regulation of exo-alpha-sialidase activity (negative) regulation of complement activation, classical pathway, olfactory receptor activity, detection of chemical stimulus involved in sensory perception of smell and negative regulation of chromatin silencing (Supplementary Table 3 and Fig. 3D). There were diverse and large clusters of genes repressed by PARIS in a phosphorylation-dependent manner (Fig. 3C). The biological functions enriched in the list of DEGs (with 20% enrichment of the selected biological functions) include chromatin remodelling and organization, chromatin mediated maintenance of transcription, cell cycle process, cellular response to DNA damage stimulus, regulation of epigenetic gene expression and signal transduction by p53 class mediator (Supplementary Table 3 and Fig. 3C).
For the genes downregulated by PARIS, which account for most DEGs identified by our microarray analysis, we validated a subset of genes (PARIS phosphorylation-dependent and -independent repression) by RT-qPCR. The expression of several genes was confirmed to be downregulated by both PARIS wild-type and PARIS Y137F: survival of motor neuron 1 (SMN1), phosphatase and tensin homologue (PTEN) and heat shock 70 kDa protein 9 (HSPA9) (Fig. 3D). We further validated many genes exhibiting PARIS Y137 phosphorylation-dependent repression: apoptosis inhibitory protein (NAIP), MDM4, TATA box binding protein (TBP), caspase 2 (CASP2), activating transcription factor 6 (ATF6) and glycogen synthase kinase 3 beta (GSK3B) (Fig. 3D).
c-Abl phosphorylation of PARIS regulates epigenetic repression of MDM4 and p53 activation
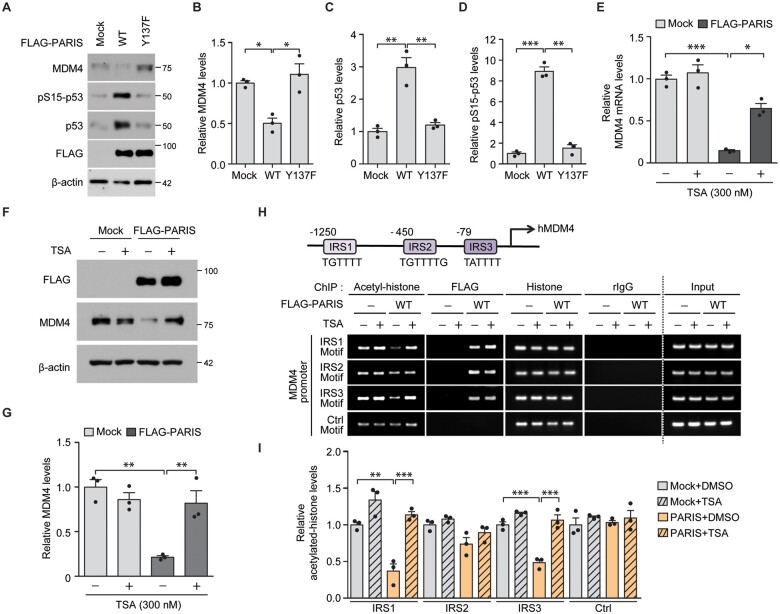
Among the enriched biological functions of genes downregulated by PARIS in a phosphorylation-dependent manner, we hypothesized that the ‘signal transduction by p53 class mediator’ pathway is responsible for cytotoxicity induced by the pathological c-Abl-PARIS interaction. Moreover, p53 pathway activation has been observed in the mouse brains with c-Abl activation or PARIS accumulation.17,25 To elucidate the molecular mechanisms underlying c-Abl-PARIS-induced p53 activation, we focused on the potential PARIS target gene MDM4, a negative regulator of p53 activity, along with MDM2. Interestingly, MDM4 is reported to be repressed in C2C12 cells with PARIS overexpression.17 Consistent with PARIS Y137 phosphorylation-dependent MDM2 mRNA regulation (Fig. 3D), MDM4 protein expression was markedly reduced by PARIS expression in SH-SY5Y cells, whereas Y137F-PARIS expression had no effect on MDM4 protein expression (Fig. 4A and B). PARIS expression led to marked elevation of total p53 and S15-phosphorylated p53 (pS15-p53), which is an indicator of p53 activation (Fig. 4A–D). PARIS-induced MDM4 repression and p53 activation was markedly enhanced when GFP-Bcr-Abl was co-expressed and PARIS Y137 phosphorylation was substantially increased by GFP-Bcr-Abl co-expression (Supplementary Fig. 4A and B). Activation of p53downstream of PARIS expression is PARIS Y137 phosphorylation-dependent since Y137F-PARIS expression (at comparable expression levels to those of wild-type PARIS) failed to activate p53 (Fig. 4A–D). Y137F-PARIS expression normalized MDM4 expression and prevented p53 accumulation even with co-expression of constitutively active GFP-Bcr-Abl (Supplementary Fig. 4A and B). PARIS Y137 phosphorylation-dependent regulation of the MDM4-p53 pathway was further confirmed in dopaminergic neuron cultures transfected with mock, wild-type-PARIS, or Y137F-PARIS (Supplementary Fig. 4C and D). In addition, virally induced PARIS expression in murine ventral midbrain resulted in MDM4 repression and upregulation of p53 and pS15-p53 expression (Supplementary Fig. 4E and F), supporting the existence of the PARIS-MDM4-p53 pathway in the brain.
Figure 4.
PARIS expression leads to p53 activation via PARIS Y137 phosphorylation-dependent epigenetic repression of MDM4. (A) Representative immunoblots examining the expression of MDM4, pS15-p53, p53 and FLAG (PARIS) in SH-SY5Y cells transfected with FLAG-PARIS wild-type or a Y137F mutant (48 h) using the indicated antibodies. β-Actin serves as an internal loading control. (B–D) Relative expression levels of MDM4 (B), p53 (C) and pS15-p53 (D) in the indicated experimental groups from A normalized to the internal loading control (β-actin; n = 3 per group). (E) Quantification of the relative expression of MDM4 mRNA in SH-SY5Y cells transfected (48 h) with mock or FLAG-PARIS and treated with TSA (300 nM, 42 h) determined by RT-qPCR (normalized to internal GAPDH loading control; n = 3 per group). (F) Representative immunoblots of FLAG (PARIS) and MDM4 expression in SH-SY5Y cells transfected (48 h) with mock or FLAG-PARIS and treated with TSA (300 nM, 42 h). (G) Quantification of the relative expression of MDM4 protein in the experimental groups in F normalized to β-actin (n = 3 per group). (H) A schematic diagram depicting the promoter structures of human MDM4 (hMDM4). IRS1, IRS2 and IRS3 motifs are indicated (top). Anti-acetyl-histone and anti-FLAG ChIP analysis of putative IRS (motif 1, 2 and 3) within the MDM4 promoter region in SH-SY5Y cells transfected with mock, or FLAG-PARIS wild-type (48 h, bottom) with or without the HDAC inhibitor TSA (300 nM, 42 h). The non-IRS region within the MDM4 promoter (Ctrl motif) was used as a negative control. Samples immunoprecipitated using either anti-histone antibodies or rabbit IgG were included as experimental controls in ChIP assays. (I) Quantification of relative acetylated histone enrichment on the indicated motifs located within MDM4 promoter determined by PCR amplification of ChIPed DNA in H (n = 3 per group). Data are expressed as mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001 and statistical analysis was performed using an ANOVA test followed by Tukey’s post hoc analysis. WT = wild-type.
Along with identification of MDM4 as a new target of PARIS, we tried to examine potential PARIS phosphorylation-dependent regulation of the PGC-1α-NRF1 pathway that was previously reported to be repressed by PARIS in Parkinson’s disease pathogenesis.14,15 Although PGC-1α failed to pass our cut-off criteria (30% downregulation by PARIS expression compared with mock group) in microarray analysis, PGC-1α transcription appears to be regulated in a PARIS phosphorylation-dependent manner (Supplementary Table 2; log2 value of microarray data for PGC-1α-Mock: PARIS wild-type: PARIS Y137F = 3.59: 3.12: 3.59 versus MDM4-Mock: PARIS wild-type: PARIS Y137F = 6.51: 5.07: 5.48). Although there is only about a 28% decrease of PGC-1α transcript by PARIS wild-type expression, this repression was abolished by Y137F PARIS. Supporting this PGC-1α transcription pattern, there was marked downregulation of PGC-1α protein expression by PARIS expression in SH-SY5Y cells whereas Y137F-PARIS exhibited a diminished capacity to repress PGC-1α expression compared with PARIS wild-type (Supplementary Fig. 4G and H). PGC-1α downstream target NRF1 protein expression was also reduced with PARIS wild-type expression, whereas PARIS Y137F repressed NRF1 expression to a lesser extent (Supplementary Fig. 4G and H). This result indicates that the PGC-1α-NRF1 pathway is in part under regulation by PARIS in a phosphorylation-dependent manner. Interestingly, consistent with the notion that PGC-1α and NRF1 are involved in mitochondrial function and antioxidant defence, the JC-1 assay revealed the impairment of mitochondria membrane potential in SH-SY5Y cells with PARIS wild-type expression (Supplementary Fig. 4I). PARIS Y137F, however, failed to induce mitochondrial damage (Supplementary Fig. 4I). Since the PARIS-MDM4 pathway is novel and identified in an unbiased microarray analysis, we focused on this pathway in the subsequent mechanistic studies.
We discovered that PARIS associates with KAP1 via phosphorylation of Y137 (located within the KRAB domain of PARIS; Fig. 1E and F and Supplementary Fig. 1A). Since KAP1 functions in epigenetic transcriptional repression via recruitment of other repressors including HDACs,26 we examined whether HDACs are involved in MDM4 repression by PARIS overexpression. PARIS-induced repression of MDM4 mRNA and protein expression was largely reversed by the HDAC inhibitor TSA in SH-SY5Y cells (Fig. 4E–G), indicating that histone deacetylation is involved in PARIS-mediated MDM4 repression. Along with rescue of MDM4 repression, the PARIS-induced cytotoxicity was also reversed by TSA treatment in SH-SY5Y cells (Supplementary Fig. 4J). To determine whether PARIS can bind to the MDM4 promoter, ChIP was performed in SH-SY5Y cells expressing PARIS in the presence or absence of TSA. The MDM4 promoter contains three putative IRSs (IRS 1, 2 and 3; Fig. 4H). Anti-FLAG (PARIS) ChIP showed that both FLAG-PARIS bind to all three IRS motifs within the MDM4 promoter in SH-SY5Y cells regardless of HDAC inhibition by TSA (Fig. 4H and I). PARIS wild-type expression led to a marked reduction in the levels of acetylated histones bound to IRS1, and three motifs. This PARIS-induced reduction of histone acetylation on these IRS motifs was completely reversed by the HDAC inhibitor TSA treatment (Fig. 4H and I). We did not detect binding of FLAG-PARIS to a control motif within the MDM4 promoter, and there was no alteration in histone acetylation levels in the control motif. Additionally, we used Y137F-PARIS expression in this ChIP enrichment experiment to determine the potential role of PARIS phosphorylation in MDM4 promoter epigenetic regulation. Similar to PARIS wild-type, Y137F-PARIS retained the ability to bind to the MDM4 promoter (IRS1, 2 and 3 motifs) (Supplementary Fig. 4K and L). However, in contrast to PARIS wild-type, Y137F-PARIS failed to reduce histone acetylation within the IRS1 and 3 motifs of the MDM4 promoter (Supplementary Fig. 4K and L). Together, these results indicate that PARIS-dependent repression of MDM4 expression is mediated by HDAC-dependent epigenetic regulation, and histone deacetylation of IRS motifs in the MDM4 promoter depends on Y137 phosphorylation, which does not affect IRS binding.
c-Abl is required for p53 activation and dopaminergic neuron loss in AAV-PARIS injected mice
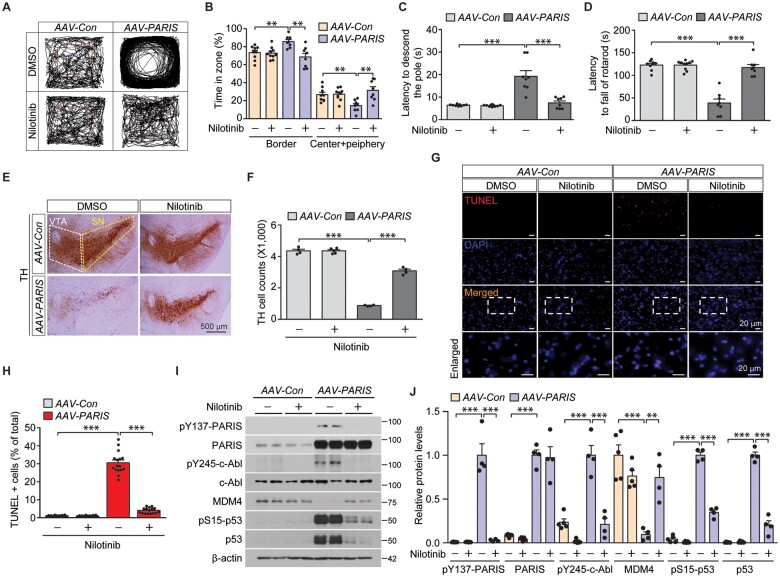
Given the alterations to the c-Abl-PARIS-MDM4 pathway induced by PARIS overexpression in SH-SY5Y cells, we propose that PARIS accumulation can affect c-Abl activity through a feedforward mechanism that then phosphorylates PARIS. To assess whether this is possible in vivo, mice were characterized after intranigral injection of an AAV-PARIS virus or AAV-GFP as a control. Co-immunofluorescence revealed efficient diffusion of nigrally injected AAV-GFP virus to the entire ventral midbrains, transducing multiple cell types including TH-positive dopamine neurons and GFAP-positive astrocytes (Supplementary Fig. 5A–C). Mice with AAV-PARIS nigral injection exhibited elevated anxiety as evidenced by their increased tendency to stay in the border zone in the open-field exploration test and increased latency to enter the centre zone for the first time (Fig. 5A and B and Supplementary Fig. 5A). This enhanced anxiety in mice with viral PARIS expression in the ventral midbrain was abolished by administration of the c-Abl inhibitor nilotinib (Fig. 5A and B and Supplementary Fig. 5D). Moreover, bradykinesia and motor coordination deficits in AAV-PARIS injected mice were also reversed by nilotinib treatment as determined by pole tests and accelerating rotarod tests, respectively (Fig. 5C and D). Virally induced PARIS expression in vivo also induced hyperactivity as shown by increased total distance travelled in an open field test (Supplementary Fig. 5E). This hyperactivity was also blocked by nilotinib treatment (Supplementary Fig. 5E). The results of these behaviour tests indicate that PARIS-induced behaviour and motor deficits are mediated by c-Abl activity.
Figure 5.
Pharmacological inhibition of c-Abl activation restores behaviour/motor deficits and dopaminergic degeneration and prevents MDM4 repression and p53 activation in mice with AAV-PARIS injection. (A) Representative exploratory paths from an open field test of mice that underwent AAV-Con or AAV-PARIS stereotaxic nigral injections (3 weeks) and treatment with the c-Abl inhibitor nilotinib (50 mg/kg/day, i.p. 2 weeks). (B) Anxiety assessment of each experimental mouse group by examining the percentage of exploration time in the border versus the sum of the centre and periphery zones (n = 9 AAV-Con-injected mice, n = 10 AAV-Con-injected mice + nilotinib, and n = 8 AAV-PARIS-injected mice). (C) Pole test for motor function assessment of each experimental mouse group used in B examining the latency to reach the base of vertical pole. (D) Motor coordination of each experimental mouse group used in B determined by the latency to fall in an accelerating rotarod test. (E) Representative TH immunohistochemical staining with Nissl counterstain of substantia nigra from mice that underwent AAV-Con or AAV-PARIS stereotaxic nigral injections (3 weeks) and treatment with the c-Abl inhibitor nilotinib (50 mg/kg/day, i.p. 2 weeks). The substantia nigra and ventral tegmental area regions are indicated by dotted yellow and white lines, respectively. Scale bar = 500 µm. (F) Stereological assessment of TH-positive dopaminergic neurons in the SNpc (injection side) in the indicated mouse groups (n = 4 AAV-Con-injected mice + DMSO, n = 5 AAV-Con-injected mice + nilotinib and n = 4 AAV-PARIS-injected mice). (G) Representative TUNEL assay images of ventral midbrain from mice that underwent stereotaxic nigral injection of AAV-Con or AAV-PARIS (3 weeks) and treatment with the c-Abl inhibitor nilotinib (50 mg/kg/day, i.p. 2 weeks). The coronal brain sections were counterstained with DAPI. (H) Quantification of the percentage of TUNEL-labelled cells in AAV-Con- or AAV-PARIS-injected ventral midbrain regions from mice with or without nilotinib treatment (n = 16 sections from four mice per group). (I) Representative immunoblots examining pY137-PARIS, PARIS, c-Abl, pY245-c-Abl, MDM4, pS15-p53 and p53 expression in the ventral midbrain of AAV-Con- or AAV-PARIS-injected mice with or without nilotinib treatment using the indicated antibodies. (J) Quantification of the relative expression of pY137-PARIS, PARIS, c-Abl, pY245-c-Abl MDM4, pS15-p53 and p53 proteins normalized to β-actin (n = 5 AAV-Con-injected mice and n = 4 AAV-PARIS-injected mice). Data are expressed as mean ± SEM. Statistical analyses was performed using an ANOVA test followed by Tukey’s post hoc analysis or an unpaired two-tailed Student’s t-test. **P < 0.01 and ***P < 0.001. DMSO = dimethyl sulphoxide; WT = wild-type.
Consistent with the motor deficits observed in mice with virally induced PARIS expression is the robust and ∼80% or 63% loss of dopaminergic neurons in the SNpc region as determined by unbiased TH or Nissl stereology, respectively (Fig. 5E and F and Supplementary Fig. 5F). Dopamine neuron loss by PARIS was not specific to the SNpc since there was also marked loss of TH-positive neurons in the ventral tegmental area of AAV-PARIS-injected mice (Supplementary Fig. 5G). In addition, we observed loss of TH-stained dopaminergic nerve fibres in the striatum of AAV-PARIS injected mice (Supplementary Fig. 5H and I). Dopaminergic neurodegeneration induced by PARIS expression in both the SNpc and ventral tegmental area was largely reversed by nilotinib treatment (Fig. 5E and F and Supplementary Fig. 5G–I). To determine the presence and extent of progressive and ongoing neurodegeneration by PARIS expression with or without c-Abl inhibition, ventral midbrain sections from virally injected mice (2 weeks post-injection) were subjected to TUNEL labelling. PARIS expression in the ventral midbrain stimulated c-Abl-dependent execution of cell death, as indicated by an increased number of TUNEL-positive cells in mice with AAV-PARIS injection; however, this effect was reversed by nilotinib treatment (to the basal level of AAV-Con-injected mice; Fig. 5G and H).
Molecular pathways downstream of PARIS that are relevant to apoptotic cell death were evaluated by western blot in ventral midbrain tissues from the same experimental mouse groups. Two weeks post-intranigral AAV-PARIS injection, we observed MDM4 downregulation and a substantial increase in the levels of p53 and p53 phosphorylation (Fig. 5I and J), which are consistent with increased ongoing apoptosis (Fig. 5G and H). This alteration to the MDM4-p53 pathway by PARIS expression in vivo requires c-Abl activation, since nilotinib treatment largely blocked this pathway (Fig. 5I and J). Interestingly, the repression of PGC-1α and NRF1 by virally induced PARIS expression was also reversed by nilotinib (Supplementary Fig. 5J and K), suggesting a potential pathological contribution of this pathway downstream of c-Abl-PARIS signalling activation.
Conditional parkin ablation leads to c-Abl-mediated phosphorylation of PARIS, p53 activation and apoptotic neuron loss
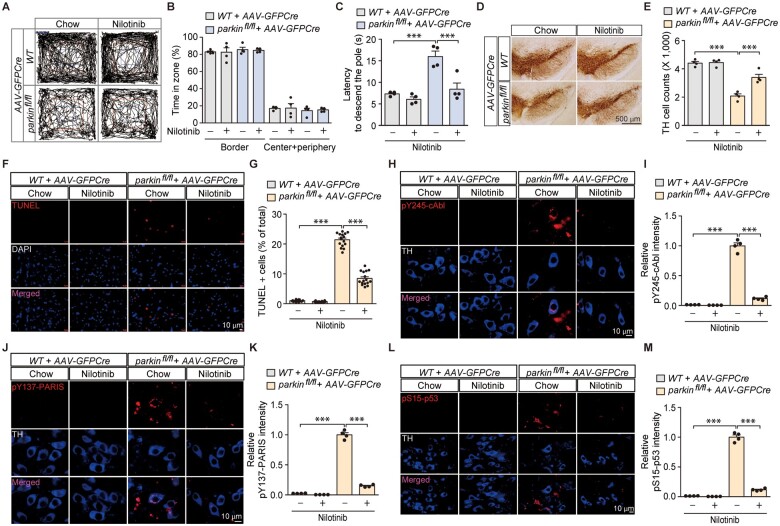
c-Abl activation has been implicated in Parkinson’s disease-associated pathogenesis in α-synucleinopathy and toxin-induced Parkinson’s disease mouse models.2,3,6 Although we showed that accumulation of the parkin substrate PARIS requires c-Abl activity to induce dopaminergic pathology, whether Parkinson’s disease pathologies induced by ablation of the recessive Parkinson’s disease gene parkin involve c-Abl activity have not been evaluated. Conditional ablation of adult parkin in mice has been reported to induce PARIS accumulation and progressive loss of dopaminergic neurons.14 By using this mouse model, we injected AAV-GFPCre into the ventral midbrain of wild-type and floxed parkin (parkinfl/fl) mice, and the role of c-Abl activation was investigated by the c-Abl inhibitor nilotinib administration. Intranigral injection of AAV-GFPCre efficiently transduced the entirety of TH-positive dopaminergic neurons in the substantia nigra of both wild-type and parkinfl/fl mice (Supplementary Fig. 6A). Both parkin knockout and wild-type mice displayed normal exploration (Supplementary Fig. 6B and C) and no difference in the time spent in the centre versus border zones (Supplementary Fig. 6D). However, adult parkin knockout mice took longer to enter the centre zone for the first time after being placed in the open-field arena as compared to controls (Supplementary Fig. 6E), indicating an elevated level of anxiety. In addition, loss of adult parkin in the ventral midbrain of mice induced bradykinesia, since it took significantly longer for the parkin knockout mice to descend to the base in the doparminergic circuit-sensitive pole test (Supplementary Fig. 6F). Parkin knockout mice did not display any difference in motor coordination as compared to controls in the rotarod test (Supplementary Fig. 6G). Although nilotinib treatment had no effect on the time spent in the centre versus border zones in the open-field test (Fig. 6A and B), bradykinesia induced by adult parkin ablation as assessed by the pole test was largely prevented by nilotinib (Fig. 6C).
Figure 6.
Pharmacological inhibition of c-Abl activity in in vivo adult parkin knockout mice prevents motor dysfunction and dopaminergic neurodegeneration with concomitant blocking of PARIS phosphorylation and p53 activation. (A) Representative exploratory paths from an open-field test of 6-month-old wild-type littermate or homozygous floxed parkin mice (parkinfl/fl) nigrally injected with AAV-GFPCre (3 m) and treated with the c-Abl inhibitor nilotinib (200 mg nilotinib per 1 kg diet, p.o. for 2 months) or standard chow diet (chow). (B) Anxiety assessment of each experimental mouse group examining the percentage of exploration time in the border versus the sum of the centre and periphery zones (n = 4 mice per group). (C) Pole test for motor function assessment of each experimental mouse group used in B examining the latency to reach the base of the vertical pole (n = 4 mice per group). (D) Representative TH immunohistochemical staining of substantia nigra from wild-type or homozygous floxed parkin mice (parkinfl/fl) with intranigral injection of AAV-GFPCre with or without nilotinib treatment (200 mg nilotinib per 1 kg diet, p.o. for 2 months). Scale bar = 500 µm. (E) Stereological assessment of TH-positive dopaminergic neurons in the SNpc (injection side) of the indicated mouse groups (n = 4 mice per group). (F) Representative TUNEL assay images of ventral midbrain from wild-type littermate or homozygous floxed parkin mice (parkinfl/fl) that experienced stereotaxic nigral injection of AAV-GFPCre with or without nilotinib treatment (200 mg nilotinib per 1 kg diet, p.o. for 2 months). The coronal brain sections were counterstained with DAPI. Magnified images are shown in the bottom panel. (G) Quantification of the percentage of TUNEL-labelled cells in AAV-GFPCre-injected ventral midbrain regions from wild-type littermate and parkinfl/fl mice with or without nilotinib treatment (n = 16 sections from four mice per group). (H) Representative immunofluorescence images examining the expression of pY245-c-Abl in TH-stained dopamine neurons from the AAV-GFPCre-injected ventral midbrain regions of wild-type littermate and parkinfl/fl mice with or without nilotinib treatment. (I) Quantification of the relative pY245-c-Abl fluorescence signal in the ventral midbrain regions of the indicated experimental groups (n = 4 mice per group). (J) Representative immunofluorescence images examining the expression of pY137-PARIS in TH-stained dopamine neurons from the AAV-GFPCre-injected ventral midbrain regions of wild-type littermate and parkinfl/fl mice with or without nilotinib treatment. (K) Quantification of the relative pY137-PARIS fluorescence signal in the ventral midbrain regions of the indicated experimental groups (n = 4 mice per group). (L) Representative immunofluorescence images examining the expression of pS15-p53 in TH-stained dopamine neurons from the AAV-GFPCre-injected ventral midbrain regions of wild-type littermate and parkinfl/fl mice with or without nilotinib treatment. (M) Quantification of the relative pS15-p53 fluorescence signal in the ventral midbrain regions of the indicated experimental groups (n = 4 mice per group). Data are expressed as mean ± SEM. Statistical analysis was performed using an ANOVA test followed by Tukey’s post hoc analysis or an unpaired two-tailed Student’s t-test. ***P < 0.001. WT = wild-type.
Neuropathological alteration was also assessed in adult parkin knockout mice. Consistent with the previous report,14 adult parkin loss led to ∼50% loss of dopaminergic neurons in the SNpc as determined by anti-TH immunohistochemistry and stereological counting (Supplementary Fig. 6H and I and Fig. 6D and E), which was partially prevented by nilotinib treatment (Fig. 6D and E). We additionally detected an increased number of TUNEL-positive cells in the ventral midbrain of adult parkin knockout mice which was also substantially diminished by nilotinib treatment (Supplementary Figs 6J and K and Fig. 6F and G), indicating ongoing c-Abl mediated cell death under pathological condition of parkin loss. Molecular alterations were also assessed by western blot and immunofluorescence in ventral midbrain tissues from control and adult parkin knockout mice. Along with PARIS accumulation in response to parkin knockout in the ventral midbrain, concomitant c-Abl activation and PARIS Y137 phosphorylation were detected by phospho-specific antibodies for pY245-c-Abl and pY137-PARIS, respectively (Supplementary Fig. 6L and M). Nilotinib substantially blocked the adult parkin ablation-induced c-Abl activation and PARIS phosphorylation in the dopaminergic neurons of adult parkin knockout mice (Fig. 6H–K). Downstream of parkin ablation, MDM4 repression and p53 activation were also observed (Supplementary Fig. 6L and M), which is consistent with ongoing apoptotic cell death as evidenced by an increased percentage of TUNEL-positive cells (Supplementary Fig. 6J and K and Fig. 6F and G). Importantly, nilotinib treatment also strongly blocked marked elevation of pS15-p53 signal in the ventral midbrain of adult parkin knockout mice (Fig. 6L and M). Together, these results from adult parkin knockout mice suggest that c-Abl activation contributes to dysregulation of the PARIS-MDM5-p53 pathway, ultimately leading to progressive apoptotic dopaminergic neuron loss.
PARIS Y137 phosphorylation mediates MDM4 repression, c-Abl and p53 activation and dopaminergic impairment
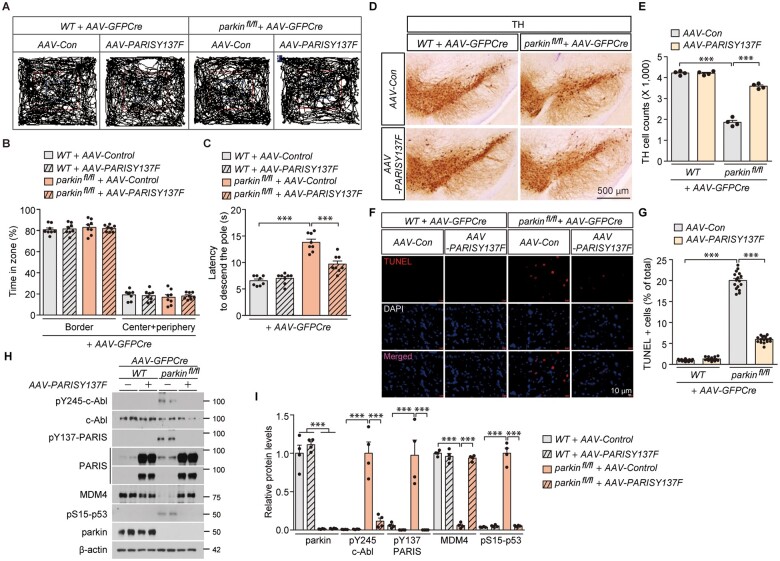
We next sought to investigate potential roles of PARIS Y137 phosphorylation in the pathogenesis of adult parkin knockout mice. We used nigral coinjection of AAV-GFPCre + AAV-Y137F-PARIS in parkin floxed mice since Y137F-PARIS expression functions in a dominant negative fashion in SH-SY5Y cells, potentially antagonizing Y137 phosphorylation of endogenous PARIS and associated downstream pathways (Fig. 2B and Supplementary Fig. 4A and B). Parkin knockout and wild-type mice with or without nigral AAV-PARIS-Y137F injection displayed normal exploration and no difference in the time spent in the centre versus border zones (Fig. 7A and B). Bradykinesia motor deficits in parkin knockout mice were partially reversed by nigral injection of AAV-PARIS-Y137F as assessed by the time to descend to the base in the pole test (Fig. 7C and Supplementary Fig. 6F).
Figure 7.
Y137F-PARIS-mediated suppression c-Abl-PARIS pathway rescues MDM4 repression, blocks p53 activation and prevents development of motor deficits and dopamine neuron loss in parkin knockout mice. (A) Representative exploratory paths from an open field test of 6-month-old wild-type littermate or homozygous floxed parkin mice (parkinfl/fl) nigrally injected with AAV-GFPCre (3 months) ± AAV-PARIS-Y137F (3 months, phospho-deficient mutant PARIS). (B) Anxiety assessment of each experimental mouse group examining the percentage of exploration time in the border versus the sum of the centre and periphery zones (n = 8 mice per group). (C) Pole test for motor function assessment of each experimental mouse group used in B examining the latency to reach the base of the vertical pole (n = 8 mice per group). (D) Representative TH immunohistochemical staining of substantia nigra from wild-type littermate or homozygous floxed parkin mice (parkinfl/fl) with intranigral injection of AAV-GFPCre ± AAV-PARIS-Y137F. Scale bar = 500 µm. (E) Stereological assessment of TH-positive dopaminergic neurons in the SNpc (injection side) of the indicated mouse groups (n = 4 mice per group). (F) Representative TUNEL assay images of ventral midbrain from wild-type littermate or homozygous floxed parkin mice (parkinfl/fl) that experienced stereotaxic nigral injection of AAV-GFPCre ± AAV-PARIS-Y137F. The coronal brain sections were counterstained with DAPI. Merged images are shown in the bottom panel. (G) Quantification of the percentage of TUNEL-labelled cells in AAV-GFPCre-injected ventral midbrain regions from wild-type littermate and parkinfl/fl mice ± AAV-PARIS-Y137F (n = 16 sections from four mice per group). (H) Representative immunoblots examining the expression of pY245-c-Abl, c-Abl, pY137-PARIS, PARIS, MDM4, pS15-p53, and parkin in the AAV-GFPCre ± AAV-PARIS-Y137F-injected ventral midbrain regions from wild-type littermate and parkinfl/fl mice using the indicated antibodies. β-Actin serves as an internal loading control. (I) Quantification of the relative expression of pY245-c-Abl, c-Abl, pY137-PARIS, PARIS, MDM4, pS15-p53 and parkin proteins normalized to β-actin in the indicated experimental groups (n = 4 mice per group). Data are expressed as mean ± SEM. Statistical analysis was performed using an ANOVA test followed by Tukey’s post hoc analysis or an unpaired two-tailed Student’s t-test. ***P < 0.001. WT = wild-type.
Neuropathological assessment revealed that adult parkin loss-induced ∼50% loss of dopaminergic neurons in the SNpc was largely prevented by AAV-PARIS-Y137F coinjection (Fig. 7D and E). AAV-PARIS-Y137F injection of wild-type mice had no effect on dopaminergic neuron viability (Fig. 7D and E). Consistent with TH stereological assessment, the increased number of TUNEL-positive cells in the ventral midbrain of adult parkin knockout mice was also substantially diminished by AAV-PARIS-Y137F nigral injection (Fig. 7F and G). Molecular alterations were also assessed by western blot in ventral midbrain tissues from each experimental mouse group. Both c-Abl activation and PARIS Y137 phosphorylation in ventral midbrain tissues from parkin knockout were completely blocked by robust expression of phospho-deficient Y137F-PARIS (Fig. 7H and I). AAV-PARIS-Y137 strongly blocked MDM4 repression and p53 activation in the adult parkin knockout brains (Fig. 7H and I). Together, these in vivo results using phospho-deficient mutant PARIS suggest that PARIS Y137 phosphorylation is critically important in mediating p53 activation, and dopaminergic neurodegeneration in parkin knockout condition.
Clinical relevance of PARIS phosphorylation and MDM4 repression in Parkinson’s disease pathogenesis
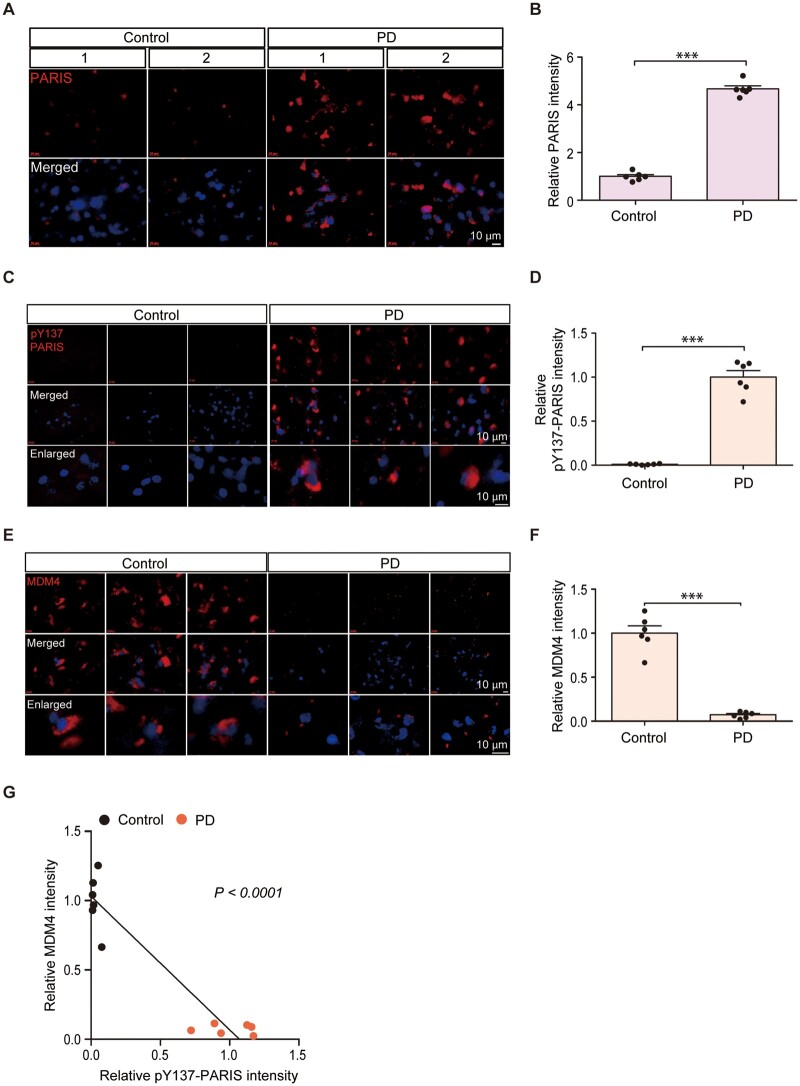
c-Abl overactivation has been noted in post-mortem brain samples from sporadic Parkinson’s disease patients.3 As we found that PARIS is a phospho-substrate of c-Abl, we sought to evaluate the clinical relevance of PARIS Y137 phosphorylation in Parkinson’s disease pathogenesis. Cortex sections from Parkinson’s disease with validated Lewy pathologies and age-matched healthy controls were used to monitor PARIS, pY137-PARIS and MDM4 by immunofluorescence. The expression levels of both PARIS and pY137-PARIS were higher in the Parkinson’s disease cortex sections than in the age-matched controls (Fig. 8A–D). The increase in PARIS Y137 phosphorylation was even more robust as compared to PARIS accumulation in Parkinson’s disease post-mortem brains (Fig. 8A–D). Co-immunofluorescence with the Lewy body marker, pS129-α-synuclein further confirmed high PARIS and pY137-PARIS signals in the Parkinson’s disease patients’ brains with Lewy pathologies (Supplementary Fig. 7A–D). Although PARIS and pY137-PARIS accumulation was observed in both pS129-α-synuclein positive or negative cells in the Parkinson’s disease patients’ brains, there was positive correlation between PARIS/pY137-PARIS and pS129-α-synuclein signals as determined by Pearson correlation analysis (Supplementary Fig. 7A–D). In addition, the expression level of MDM4, a PARIS phosphorylation-dependent target gene, was markedly lower in the Parkinson’s disease sections than in the controls (Fig. 8E and F). Pearson correlation analysis revealed an inverse correlation between PARIS Y137 phosphorylation and MDM4 expression (Fig. 8G). Consistent with high PARIS phosphorylation and low MDM4 expression, we observed high pS15-p53 and total p53 expression in the post-mortem brain sections from patients with Parkinson’s disease as compared to those from age-matched healthy control (Supplementary Fig. 7E–H). These results point to clinical relevance of PARIS Y137 phosphorylation, MDM4 downregulation and p53 activation in Parkinson’s disease pathogenesis.
Figure 8.
Correlative PARIS phosphorylation and MDM4 repression in Parkinson’s disease pathogenesis. (A, C and E) Expression of PARIS, pY137-PARIS and MDM4 in post-mortem cortical sections from Parkinson’s disease patients with cortical Lewy body pathologies and age-matched controls monitored by immunofluorescence using specific antibodies and the nuclear counterstain DAPI. Scale bar = 10 µm. (B, D and F) Quantification of the relative fluorescent intensities of cells from the post-mortem brain tissue sections of Parkinson’s disease patients and age-matched controls (n = 6 control and 6 Parkinson’s disease). (G) Correlation plots and Pearson correlation analysis of pY137-PARIS and MDM4 expression in the post-mortem cortex of Parkinson’s disease patients and age-matched healthy control subjects. Data are expressed as mean ± SEM. ***P < 0.001. Statistical analysis was performed using an unpaired two-tailed Student’s t-test. PD = Parkinson’s disease.
Discussion
c-Abl-mediated PARIS KRAB phosphorylation modulates KAP1 association and epigenetic repression
In this study, we discovered that the parkin interacting substrate PARIS is a phospho-substrate of c-Abl. We also found that c-Abl phosphorylation of Y137 in PARIS (within the KRAB domain) regulates its interaction with KAP1, forming a complex with chromatin remodelling proteins.26 Although PARIS Y137 and its adjacent amino acids are highly conserved in mammals, the phosphorylation-mediated association of KRAB-ZNF with KAP1 has not been previously reported. Crystal structure analysis would shed light on the mechanisms underlying the increased binding of PARIS with KAP1 on Y137 phosphorylation. Moreover, it would be informative to investigate other KRAB-ZNFs that have similar KRAB domains to PARIS to determine whether they are regulated by phosphorylation-dependent KAP1 recruitment.
To our knowledge, the epigenetic regulation of MDM4 transcription and p53 activation has not been previously explored. In this study, we identified PARIS as a transcriptional repressor of MDM4. Interestingly, PARIS can bind to IRS motifs within the MDM4 promoter, regardless of its Y137 phosphorylation status. However, in the context of c-Abl activation and Y137-PARIS phosphorylation, histone acetylation on the MDM4 promoter can be reduced by PARIS, possibly via KAP1 binding and HDAC recruitment. In support of this notion, a phospho-deficient PARIS mutant (Y137F-PARIS) failed to deacetylate histones in the MDM4 promoter and repress its transcription, even though it can bind to the MDM4 promoter. Our discovery of this novel pathway regulating MDM4 transcription (i.e. c-Abl-mediated phosphorylation of PARIS) may contribute to furthering basic and translational research involving aberrant MDM4-p53 activation and neurodegeneration in pathological conditions.
Moreover, it would be informative to further investigate potential epigenetic regulation of the previously reported PARIS target genes and those genes screened from our microarray analysis. Given that the repression of PGC-1α, the previously known PARIS target gene, is partially PARIS phosphorylation dependent and that HDAC regulates PGC-1α transcription,27 future studies regarding potential histone deacetylation on PGC-1α promoter by PARIS would extend our molecular understanding of PARIS-PGC-1α pathway in Parkinson’s disease pathogenesis.
c-Abl activation downstream of parkin loss and PARIS accumulation regulates p53 activation
c-Abl activation is induced by diverse Parkinson’s disease-relevant stresses including mitochondrial stress, DNA damage and α-synucleinopathy.3,6,25 In addition to these challenges more relevant to sporadic Parkinson’s disease cases, we report that c-Abl overactivation can be triggered by loss of parkin, a gene in which mutations account for most early onset recessive familial Parkinson’s disease cases. In line with c-Abl activation following parkin loss, overexpression of the parkin substrate PARIS led to c-Abl hyperactivation in mice with AAV-PARIS nigral injection. Along with c-Abl activation, our virally induced PARIS expression model produced more robust loss of dopamine neurons as compared to that from the previous study,14 presumably due to greater expression levels of PARIS (Fig. 5I and J). It is, however, unclear how PARIS accumulation or parkin ablation leads to c-Abl activation. It can be speculated that PGC-1α-NRF1 repression in response to PARIS accumulation may impair antioxidant defence and subsequently lead to an increase in oxidative stress, which can then trigger c-Abl activation. Supporting this notion and consistent with the previous reports,7,14,15 PARIS expression in both SH-SY5Y cells and mouse brains resulted in a decrease of PGC-1α and NRF1 expression (Supplementary Figs 4G and H and 5J and K). c-Abl activation was consistently observed in these conditions of PGC-1α-NRF1 repression (Supplementary Fig. 2F and G and Fig. 5I and J). Still, further investigation is needed to elucidate the molecular pathways involved in c-Abl activation following parkin knockout and PARIS accumulation. This future study will provide a molecular mechanism which could link PARIS-PGC-1α-NRF1 signalling pathway and c-Abl-PARIS-MDM4 pathway.
Interestingly, the reciprocal interaction between PARIS and c-Abl is analogous to the previously reported reciprocal interaction between α-synuclein and c-Abl. While c-Abl directly phosphorylates α-synuclein potentiating its aggregation tendency, α-synuclein preformed fibril injection triggers c-Abl overactivation. c-Abl inhibition in α-synuclein preformed fibril injection models was sufficient to mitigate α-synuclein pathologies, indicating a requirement for c-Abl activity in α-synucleinopathy.7 Similarly, pharmacological inhibition of c-Abl activation and PARIS Y137 phosphorylation in both the AAV-PARIS transduced mice and adult parkin knockout mice was sufficient to block motor deficits, dopaminergic neurodegeneration and MDM4-p53 alterations even in the presence of PARIS accumulation. These results indicate that PARIS accumulation alone is not sufficient to induce dopaminergic neurodegeneration, but c-Abl-mediated PARIS phosphorylation is required for neurotoxicity of PARIS.
Although we proposed that PARIS phosphorylation via Y137-mediated MDM4 repression might be a key molecular mechanism of p53 activation in Parkinson’s disease, p53 activity is regulated by diverse molecular pathways downstream of c-Abl activation. hosphorylation of p53 and induction of apoptosis by activated c-Abl during stress have been previously reported.25 Moreover, the p53-inhibiting E3 ligase MDM2 is also phosphorylated by c-Abl.21 c-Abl-mediated phosphorylation of MDM2 is known to block its regulation of p53.21 Thus, it is possible that other c-Abl regulated pathways are also involved in neurodegeneration in our Parkinson’s disease animal models of PARIS overexpression or parkin knockout. However, our cell viability assessment and molecular characterization in SH-SY5Y cells indicated that PARIS Y137 phosphorylation by c-Abl is the main contributor to c-Abl cytotoxicity since Y137F-PARIS co-expression largely blocked constitutively active c-Abl-induced cytotoxicity, MDM4 repression and p53 activation. To further characterize the role of the PARIS-MDM4-p53 pathway (downstream of c-Abl activation) in the induction of dopamine neuron loss, it would be necessary to examine whether p53 activation and dopaminergic neurotoxicity can be rescued in the background of PARIS knockout or MDM4 overexpression.
PARIS phosphorylation in mediating diverse Parkinson’s disease pathologies
Microarray analysis in SH-SY5Y cells expressing PARIS wild-type or the Y137F mutant revealed that most DEGs by PARIS are downregulated in a Y137 phosphorylation-dependent manner. These DEGs are enriched in biological functions such as chromatin modification, cell cycle process and epigenetic regulation of gene expression. We report that MDM4 is repressed by PARIS in models of Parkinson’s disease dopaminergic neurodegeneration. Such transcriptomic regulation by PARIS suggests that PARIS regulates other Parkinson’s disease-related pathologies via Y137 phosphorylation-dependent target genes that are yet to be explored. A recent in vivo study using PARIS knockout mice showed that α-synuclein preformed fibril brain injection failed to induce c-Abl activation, dopaminergic neurodegeneration, and α-synuclein aggregation.7 This study suggests the potential involvement of PARIS target genes that contribute to α-synucleinopathies. Our microarray study, however, has limitations in that the extents of ongoing cell death and c-Abl activity are possibly varied among three different experimental groups. Single cell RNAseq trajectory analysis for SH-SY5Y cells expressing PARIS wild-type or Y137F in the background of constitutively active Bcr-Abl would provide more conclusive information about primary PARIS phosphorylation-dependent target gene and pathways.
It is also important to note the prevalence of c-Abl activation in neurodegenerative diseases. c-Abl activation has been noted in both dopaminergic and nondopaminergic neurons. Consistent with this notion we observed strong PARIS Y137 phosphorylation in Parkinson’s disease post-mortem cortex indicating the presence of c-Abl hyperactivation in this brain region. The post-mortem cortex sections from patients with Parkinson’s disease used in this study have been validated for Lewy body pathologies by the Brain Body Donation Program, which could contribute to c-Abl activation and parkin inactivation. While an initial report failed to see PARIS accumulation in the Parkinson’s disease post-mortem cortex samples using western blot,14 our selection of Lewy pathology-evident post-mortem Parkinson’s disease brain sections might account for our observation of PARIS accumulation in cortex of Parkinson’s disease. Since genes that are regulated in a PARIS phosphorylation-dependent manner are involved in diverse biological processes, it is highly likely that neuronal function and circuits are affected in brain regions where c-Abl activity is high, as PARIS potentially dysregulates genes that are necessary for normal neuron function. Future studies are required to screen additional Parkinson’s disease-relevant disease targets for their function(s) in the regulation of Parkinson’s disease-associated pathologies other than neurodegeneration.
PARIS phosphorylation as a therapeutic target and diagnostic marker for Parkinson’s disease
Dysregulated epigenetic regulation in Parkinson’s disease has been recently explored, and some HDAC inhibitors are considered therapeutically beneficial for Parkinson’s disease treatment. However, there have been no reports of specific genes involved in Parkinson’s disease-associated aberrant epigenetic regulation. In this regard, PARIS could be the first Parkinson’s disease-associated transcriptional repressor to be discovered that guides pathological chromatin alterations in specific gene groups. We showed that PARIS binding to IRS motifs in the MDM4 promoter is not sufficient for the inhibition of MDM4 expression. Y137 phosphorylation is required for KAP1 binding and HDAC-mediated histone deacetylation of MDM4 IRS motifs. Similar mechanisms might play roles in the epigenetic regulation of phosphorylation-dependent PARIS target genes. Studies on the epigenetic status of the promoters of PARIS targets in post-mortem Parkinson’s disease brains would identify the clinical relevance of histone modifications in specific gene promoters. Given that HDAC can be nonspecific for pathology-associated gene promoters, inhibiting c-Abl-PARIS pathways may be more efficient in preventing the epigenetic dysregulation of specific disease-associated target genes.
Besides the translational value of targeting the c-Abl-PARIS pathway in Parkinson’s disease treatment, PARIS Y137 phosphorylation could serve as a new diagnostic marker for Parkinson’s disease. Since c-Abl activation strongly correlates with the progressive development of Parkinson’s disease pathology, monitoring the PARIS-Y137 phosphorylation status using our PARIS phospho-specific antibody may be a useful alternative for estimating c-Abl activation. Since α-synuclein phosphorylation is being extensively investigated as a Parkinson’s disease biomarker in human fluid samples, it would be also informative to determine whether Y137 phosphorylated PARIS can be detected in Parkinson’s disease fluid samples and whether it correlates with disease progression.
Supplementary Material
Acknowledgements
We thank Dr Tae-In Kam for providing helpful suggestions regarding the in vitro kinase assay.
Funding
This research was supported by grants (2017M3C7A1043848 to Y.L.) from the National Research Foundation (NRF) funded by the Ministry of Science, ICT, & Future Planning (MSIP). We are also grateful to the Banner Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona for the provision of human biological materials. The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson’s disease and Related Disorders), the National Institute of Aging (P30 AG19610 Arizona Alzheimer’s Disease Core Centre), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- AAV
adeno-associated virus
- ChIP
chromatin immunoprecipitation
- DEG
differentially expressed gene
- HDAC
histone deacetylase
- KRAB
Krüppel-associated box
- IRS
insulin response sequence
- PARIS
parkin interacting substrate
- SNpc
substantia nigra pars compacta
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labelling
References
- 1. Lang AE, Lozano AM. Parkinson’s disease. First of two parts. N Engl J Med. 1998;339(15):1044–1053. [DOI] [PubMed] [Google Scholar]
- 2. Imam SZ, Zhou Q, Yamamoto A, et al. Novel regulation of parkin function through c-Abl-mediated tyrosine phosphorylation: Implications for Parkinson's disease. J Neurosci. 2011;31(1):157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ko HS, Lee Y, Shin JH, et al. Phosphorylation by the c-Abl protein tyrosine kinase inhibits parkin's ubiquitination and protective function. Proc Natl Acad Sci U S A. 2010;107(38):16691–16696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Braak H, Del Tredici K, Rüb U, de Vos RAI, Jansen Steur ENH, Braak E. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol Aging. 2003;24(2):197–211. [DOI] [PubMed] [Google Scholar]
- 5. Luk KC, Kehm V, Carroll J, et al. Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338(6109):949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brahmachari S, Ge P, Lee SH, et al. Activation of tyrosine kinase c-Abl contributes to alpha-synuclein-induced neurodegeneration. J Clin Invest. 2016;126(8):2970–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brahmachari S, Lee S, Kim S, et al. Parkin interacting substrate zinc finger protein 746 is a pathological mediator in Parkinson's disease. Brain. 2019;142(8):2380–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karim MR, Liao EE, Kim J, et al. alpha-Synucleinopathy associated c-Abl activation causes p53-dependent autophagy impairment. Mol Neurodegener. 2020;15(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karuppagounder SS, Brahmachari S, Lee Y, Dawson VL, Dawson TM, Ko HS. The c-Abl inhibitor, nilotinib, protects dopaminergic neurons in a preclinical animal model of Parkinson’s disease. Sci Rep. 2014;4:4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Imam SZ, Trickler W, Kimura S, et al. Neuroprotective efficacy of a new brain-penetrating C-Abl inhibitor in a murine Parkinson's disease model. PLoS One. 2013;8(5):e65129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee S, Kim S, Park YJ, et al. The c-Abl inhibitor, Radotinib HCl, is neuroprotective in a preclinical Parkinson's disease mouse model. Hum Mol Genet. 2018;27(13):2344–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brahmachari S, Karuppagounder SS, Ge P, et al. c-Abl and Parkinson's disease: Mechanisms and therapeutic potential. J Parkinsons Dis. 2017;7(4):589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang JY. The capable ABL: What is its biological function? Mol Cell Biol. 2014;34(7):1188–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shin JH, Ko HS, Kang H, et al. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell. 2011;144(5):689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee Y, Stevens DA, Kang SU, et al. PINK1 primes parkin-mediated ubiquitination of PARIS in dopaminergic neuronal survival. Cell Rep. 2017;18(4):918–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang H, Jo A, Kim H, et al. PARIS reprograms glucose metabolism by HIF-1alpha induction in dopaminergic neurodegeneration. Biochem Biophys Res Commun. 2018;495(4):2498–2504. [DOI] [PubMed] [Google Scholar]
- 17. Bae JH, Jeong HJ, Kim H, et al. ZNF746/PARIS overexpression induces cellular senescence through FoxO1/p21 axis activation in myoblasts. Cell Death Dis. 2020;11(5):359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Venderova K, Park DS. Programmed cell death in Parkinson's disease. Cold Spring Harb Perspect Med. 2012;2(8):a009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kruse JP, Gu W. SnapShot: p53 posttranslational modifications. Cell. 2008;133(5):930–30.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldberg Z, Vogt Sionov R, Berger M, et al. Tyrosine phosphorylation of Mdm2 by c-Abl: Implications for p53 regulation. EMBO J. 2002;21(14):3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Francoz S, Froment P, Bogaerts S, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci U S A. 2006;103(9):3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mancini F, Di Conza G, Monti O, et al. Puzzling over MDM4-p53 network. Int J Biochem Cell Biol. 2010;42(7):1080–1083. [DOI] [PubMed] [Google Scholar]
- 24. Rhee YH, Kim TH, Jo AY, et al. LIN28A enhances the therapeutic potential of cultured neural stem cells in a Parkinson’s disease model. Brain. 2016;139(Pt 10):2722–2739. [DOI] [PubMed] [Google Scholar]
- 25. Yuan ZM, Huang Y, Ishiko T, Kharbanda S, Weichselbaum R, Kufe D. Regulation of DNA damage-induced apoptosis by the c-Abl tyrosine kinase. Proc Natl Acad Sci U S A. 1997;94(4):1437–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oleksiewicz U, Gładych M, Raman AT, et al. TRIM28 and interacting KRAB-ZNFs control self-renewal of human pluripotent stem cells through epigenetic repression of pro-differentiation genes. Stem Cell Rep. 2017;9(6):2065–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galmozzi A, Mitro N, Ferrari A, et al. Inhibition of class I histone deacetylases unveils a mitochondrial signature and enhances oxidative metabolism in skeletal muscle and adipose tissue. Diabetes. 2013;62(3):732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are included in this paper and Supplementary material. All supporting data are available from the corresponding authors on reasonable request.