In PNAS, Xu et al. (1) report that a member of the Argonaute family of proteins, Argonaute 1 (AGO1), is an essential actor in the gene control of flowering. In Arabidopsis thaliana, flowering is inhibited when the floral repressor gene FLOWERING LOCUS C (FLC) is expressed. Therefore, repression of FLC expression is necessary to trigger flowering. Repressors of FLC expression include evolutionary conserved RNA binding proteins such as FCA, FPA, and FLK as well as factors that regulate messenger RNA (mRNA) processing and chromatin structure. A gene can be defined as a stretch of DNA sequence able to be transcribed, rendering either an RNA with a function itself or an mRNA that is translated by the ribosomes into a protein as a final product. Transcription of genes begins at sequences known as promoters, usually located at one of the ends of a gene known as the 5′ end. Promoters are usually not transcribed, but they dictate the beginning of transcription by the enzyme RNA polymerase II at a nucleotide known as the +1 or transcription start site. At the opposite end of the gene, known as its 3′ end, a complex multimolecular assembly determines the end of the RNA transcript. Even though genes are made of double-stranded DNA, only one of the strands serves as a template for the RNA; the other DNA strand is generally not transcribed, but, in some cases, it may be transcribed from a promoter located at the 3′ end of the gene so the RNA polymerase II travels in the opposite direction, giving rise to an antisense RNA. The FLC gene follows this model, with FLC mRNA being produced from the 5′ promoter, and a set of antisense long noncoding RNAs called COOLAIR being produced from the 3′ promoter.
But how does COOLAIR operate in the control of FLC expression and, in consequence, on flowering? To understand this sophisticated gene regulation mechanism that is at the base of our human feelings of joy at the sight of blooming plants, we need to add two new molecular ingredients. First, DNA is not naked in the nucleus but is associated with histones and other proteins forming chromatin. Histones can be modified by nuclear enzymes that add or remove chemical groups to or from specific amino acids of the histone sequence. These are the so-called histone marks, some of which, known as permissive marks, facilitate the passage of the transcribing RNA polymerase II, while others, called repressive marks, impede it by creating a more compact chromatin structure. The second ingredient is a regulated process known as cleavage/polyadenylation that, by cutting at certain phosphodiester bonds, which link RNA nucleotides in a row, can make RNAs shorter than expected.
Having all the ingredients in the bowl, let’s see how they work. In nonflowering conditions, COOLAIR is fully expressed, and the predominant histone mark along the FLC gene is the monomethylated lysine residue 4 of histone H3 (H3K4me1). This is a permissive mark that allows for productive transcription of the FLC mRNA and subsequent repression of flowering. Base pairing of COOLAIR with complementary DNA sequences of FLC forms an RNA–DNA hybrid known as an R-loop. This is instrumental in deposition of the repressive histone mark, as the RNA-binding FCA protein, together with other cleavage/polyadenylation factors, causes cleavage of the nascent COOLAIR RNA. This resolves the R-loop, leading to demethylation of the H3K4me1 and accumulation of the transcriptionally repressive mark H3K27me3. In other words, by making COOLAIR shorter, FCA promotes FLC repression and flowering, through the replacement of permissive chromatin marks by repressive ones (2).
Xu et al. (1) find that AGO1 associates with COOLAIR, promoting its shortening by cleavage/polyadenylation, R-loop resolution, and chromatin silencing.
Why is the finding of this role for AGO1 so relevant? The reason is that Argonaute proteins have been discovered and mainly characterized for their role in posttranscriptional gene silencing (PTGS), a key mechanism discovered as part of the small-RNA revolution. PTGS occurs in the cytoplasm and is mediated by binding of Argonaute proteins to the guide strands of either microRNAs, small interfering RNAs, or PIWI RNAs to be exposed and complementarily interact with target sequences in mRNAs, promoting inhibition of their translation by the ribosomes or their degradation, resulting in a reduction of the abundance of the encoded protein. In the case of mRNA degradation, the Argonaute member involved has an endonucleolytic enzymatic activity known as “slicer” that cleaves the mRNA at the target site of the small RNA (3). Argonaute proteins and PTGS are conserved in all eukaryotes except for the budding yeast Saccharomyces cerevisiae that lacks AGOs. Indeed, both AGOs and PTGS were discovered in plants, and the name of the family originates in the Arabidopsis Argonaute mutant that resembles the tentacles of the cephalopod Argonauta argo (4), whose name is, in turn, inspired by the band of brave sailors (nautae) of the boat Argo who backed Jason in his quest for the Golden Fleece. This is Greek mythology, but there is also a myth in biochemistry stating that, when a new protein is discovered and named and its function is elucidated, that function is frozen as the most important or only possible function of that protein, and any other role discovered later is considered as marginal, artifactual, or directly neglected. AGOs are a good example of the difficulties of breaking the idea of their exclusive canonical roles in the cytoplasm. In fact, over the last decade, there has been compelling evidence for the participation of Argonaute proteins in many nuclear events. These include transcription, alternative mRNA splicing, genome integrity, DNA repair, and long-range chromosomal interactions involving sequences known as insulators and enhancers (5). At the level of transcription, AGOs have been involved in transcriptional gene silencing (TGS) not only in Arabidopsis but also in mammalian cells. Similarly to PTGS, in TGS, nuclear AGOs associate with small RNAs that drive them to target sequences in nascent RNAs, resulting in the local recruitment of histone-modifying or DNA-methylating enzymes that create silencing marks that end up inhibiting transcription of the target gene. AGOs were also reported in transcriptional activation via interaction with RNA polymerase II or to act as coactivators of transcription factors that up-regulate gene expression upon hormone action (6). In alternative splicing, the RNA processing event by which a single gene can generate more than one mature mRNA and protein, AGOs may participate in the creation of intragenic roadblocks to transcriptional elongation through a mechanism similar to TGS, which, in turn, affects cotranscriptional splicing decisions (7). Besides, proteomic studies have also revealed that AGOs associate with a large repertoire of spliceosomal components and splicing factors such as serine-arginine-rich proteins and heterogeneous nuclear ribonucleoproteins, which may regulate splicing, per se, without affecting elongation (8).
Apart from TGS, Argonaute proteins were known to play other nuclear roles in plants. They were found to associate with chromatin preferentially at promoters and 3′ ends of genes, mirroring the pattern of the distribution of RNA polymerase II on active genes (9). Interestingly, AGO’s function at these gene sites seems to be responsive to different stimuli such as plant hormones and biotic or abiotic stresses, which strongly support a physiological role for nuclear AGOs in plants, such as the one now reported by Xu et al. (1) for plant reproduction.
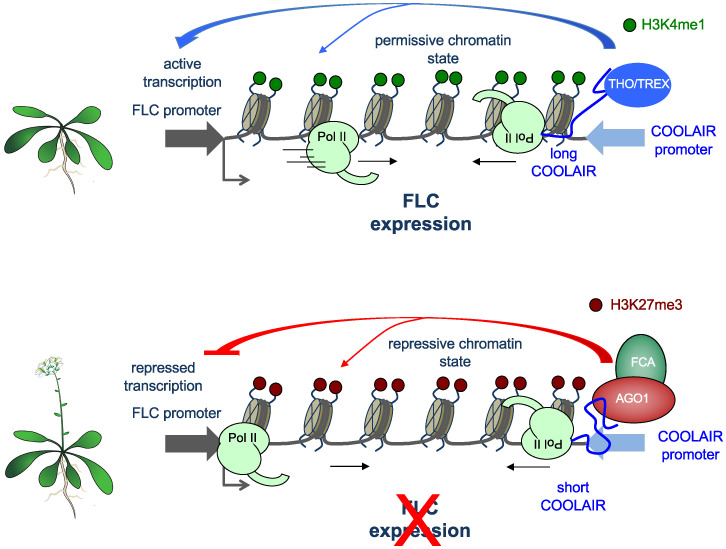
To help in FLC silencing, Xu et al. (1) show that AGO1 associates not only with COOLAIR but also with subunits of RNA polymerase II, splicing factors, and the so-called THO/TREX protein complex, known to link transcription to RNA processing. Paradoxically, while, as mentioned above, AGO1 promotes COOLAIR shortening, R-loop resolution, and removal of the permissive histone mark H3K4me1, the THO/TREX complex has the opposite effect, that is, promotion of long COOLAIR and, in consequence, FLC expression (Fig. 1). In summary, AGO1 and the THO/TREX complex act antagonistically, and the fact that they are able to physically interact suggests that, depending on their relative concentrations, their activities could be mutually neutralized. Xu et al. show that the “slicer” activity of AGO1 is necessary for its function in FLC repression, although it is not completely clear why. On the other hand, it is still uncertain whether AGO1 action on FLC repression requires charging of small RNAs. In fact, there are nuclear functions of Argonaute proteins that are mediated by small RNAs, but there is also increasing evidence that AGO proteins can be recruited to chromatin sites independently of small RNA guidance (5). Such an independence does not preclude that nuclear AGOs act via interaction with nascent transcripts in a sequence-independent manner.
Fig. 1.
Molecular bases of the control of the FLC gene expression by AGO1, COOLAIR RNA processing, and chromatin state. (Top) Expression of FLC, promoted by the THO/TREX complex, prevents flowering. (Bottom) AGO1 and FCA collaborate to promote repression of FLC expression through COOLAIR shortening and the replacement of permissive (H3K4me1) by repressive (H3K27me3) histone marks, which triggers flowering. Pol II, RNA polymerase II.
If there is a word to characterize the multiple cytoplasmic and nuclear roles of Argonaute proteins in different organisms, it is “complexity.” More and more, they appear to be necessary partners of every molecular process involving RNA. Undoubtedly, the FLC/COOLAIR example represents a fascinating and unexpected novelty with fundamental consequences in the living world.
Acknowledgments
Our research is supported by the Consejo Nacional de Investigaciones Científicas y Técnicas of Argentina, the Agencia Nacional de Promoción de la Investigación, el Desarrollo Tecnológico y la Innovación of Argentina, and the Lounsbery Foundation.
Footnotes
The authors declare no competing interest.
See companion article, “Antagonistic cotranscriptional regulation through ARGONAUTE1 and the THO/TREX complex orchestrates FLC transcriptional output,” 10.1073/pnas.2113757118.
References
- 1.Xu C., Fang X., Lu T., Dean C., Antagonistic cotranscriptional regulation through ARGONAUTE1 and the THO/TREX complex orchestrates FLC transcriptional output. Proc. Natl. Acad. Sci. U.S.A. 118, e2113757118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu C., et al. , R-loop resolution promotes co-transcriptional chromatin silencing. Nat. Commun. 12, 1790 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Höck J., Meister G., The Argonaute protein family. Genome Biol. 9, 210 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohmert K., et al. , AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170–180 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nazer E., Gómez Acuña L., Kornblihtt A. R., Seeking the truth behind the myth: Argonaute tales from “nuclearland.” Mol. Cell, in press. [DOI] [PubMed] [Google Scholar]
- 6.Gómez Acuña L. I., et al. , Nuclear role for human Argonaute-1 as an estrogen-dependent transcription coactivator. J. Cell Biol. 219, e201908097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naftelberg S., Schor I. E., Ast G., Kornblihtt A. R., Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 84, 165–198 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Batsché E., Ameyar-Zazoua M., The influence of Argonaute proteins on alternative RNA splicing. Wiley Interdiscip. Rev. RNA 6, 141–156 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Liu C., et al. , Arabidopsis ARGONAUTE 1 binds chromatin to promote gene transcription in response to hormones and stresses. Dev. Cell 44, 348–361.e7 (2018). [DOI] [PubMed] [Google Scholar]