Significance
Multiple studies have implicated dozens of risk loci that may be associated with Alzheimer’s disease (AD), but common mechanisms underlying how they may contribute to disease onset or progression remain elusive. This study identifies cell-specific roles for Drosophila orthologs of AD risk genes in lipid droplet formation that, when disrupted, lead to neurodegeneration. Our work reinforces a critical role for the sequestration of peroxidated lipids in glia, and places Apolipoprotein E ε4 (APOE4) with other AD risk factors in the transfer process of lipids from neurons to glia to form lipid droplets.
Keywords: peroxidated lipid transfer, Alzheimer’s disease, GWAS, Drosophila, lipid droplet
Abstract
A growing list of Alzheimer’s disease (AD) genetic risk factors is being identified, but the contribution of each variant to disease mechanism remains largely unknown. We have previously shown that elevated levels of reactive oxygen species (ROS) induces lipid synthesis in neurons leading to the sequestration of peroxidated lipids in glial lipid droplets (LD), delaying neurotoxicity. This neuron-to-glia lipid transport is APOD/E-dependent. To identify proteins that modulate these neuroprotective effects, we tested the role of AD risk genes in ROS-induced LD formation and demonstrate that several genes impact neuroprotective LD formation, including homologs of human ABCA1, ABCA7, VLDLR, VPS26, VPS35, AP2A, PICALM, and CD2AP. Our data also show that ROS enhances Aβ42 phenotypes in flies and mice. Finally, a peptide agonist of ABCA1 restores glial LD formation in a humanized APOE4 fly model, highlighting a potentially therapeutic avenue to prevent ROS-induced neurotoxicity. This study places many AD genetic risk factors in a ROS-induced neuron-to-glia lipid transfer pathway with a critical role in protecting against neurotoxicity.
Alzheimer’s disease (AD) affects ∼2% of the United States population and defines ∼70% of dementia cases (1). AD is pathologically defined by the aberrant accumulation of amyloid-β (Aβ) peptides into extracellular plaques and hyperphosphorylated tau into neurofibrillary tangles. Aβ has been a major focus of how AD is initiated and has been a target of therapeutic approaches (2), but many strategies aimed at reducing Aβ accumulation have failed to mitigate disease progression (3). There is evidence that Aβ-plaques exist in some individuals without consequence to cognition, supporting a hypothesis that multiple insults combine to induce disease (4).
Much of the research focus in recent years has centered on the identification of genetic risk factors of AD. Genome-wide association studies (GWAS) have identified over 40 risk variants associated with AD (5 –10). Some of the variants from these studies are in or near genes that encode proteins involved in lipid regulation (e.g., TREM2, ABCA7) and clathrin-mediated endocytosis (e.g., BIN1, CD2AP, AP2A2, PICALM, and RIN3). How these genes affect the demise of neurons is not yet clear (11). The highest genetic risk factor for AD, Apolipoprotein E ɛ4 (APOE4), is present in ∼40 to 60% of AD patients, and is strongly associated with earlier disease onset (11, 12) and impaired fatty acid metabolism (13). Homozygous carriers for APOE4 are 8 to 12 times more likely to develop AD than noncarriers (14), while individuals carrying the ɛ2 allele of APOE (APOE2) have reduced risk of developing AD (15). We hypothesize that APOE modulates AD risk by mediating lipid transfer between neurons and glia and that the reduced lipid transport capacity of APOE4 (16, 17, 18) limits this transport.
Other insults, in addition to genetic variants, may modulate severity and onset of disease, including oxidative stress caused by accumulation of reactive oxygen species (ROS). ROS can damage proteins, lipids, and nucleic acids (19, 20). When properly regulated, ROS can provide beneficial effects to the cell (21, 22), but proves damaging upon elevated levels of ROS, as with age or when proper control mechanisms become depleted (20). Numerous studies using postmortem tissue from individuals with preclinical AD, mild cognitive impairment, and AD document ROS elevation, including accumulation of peroxidated lipids (19, 23 –27). Whether ROS is a cause or consequence of disease remains an open question but it is evident that ROS production is exacerbated by Aβ42-mediated neurotoxicity (23, 28) and persistent neuroinflammation (29, 30). One hypothesis that ties these studies together is that a vicious cycle between ROS and Aβ production exists, thereby enhancing the speed and severity of disease progression. Hence, it is important that we understand the interactions between genetic variation and oxidative stress in order to reveal the complex etiology of AD.
The complexity of AD pathogenesis and progression is further illustrated by the observation that many AD risk genes are expressed in glia in addition to neurons, suggesting that disruptions of these genes may impact multiple cell types in the brain. There is increasing evidence for an important role of dysregulation of glial lipid metabolism in AD (5, 31, 32). Interestingly, Alois Alzheimer described “adipose saccules” in glial cells of AD patients over a century ago (33, 34), but the link between neurodegeneration and lipid droplet (LD) accumulation in glia has only recently been documented by us and others (35 –37). LD formation has also recently been documented in aged mouse microglia and is associated with defects in microglial phagocytosis as well as increased ROS and proinflammatory cytokine production (38). Recent evidence is quickly mounting that lipids are inextricably linked with pathogenic mechanisms in AD and other neurodegenerative diseases (39 –41).
Insights in the process of LD formation in the nervous system were gained using a fly model of ROS-induced photoreceptor neurodegeneration. In this model, neuron:glia interactions can be readily probed due to the stereotypic morphology of the fly retina in which photoreceptor neurons are surrounded by pigment glia ( SI Appendix, Supplementary Information and Fig. S1A ), with Drosophila glia having homolgous functions to vertebrate glia (42). We show that elevated levels of ROS in neurons induces the formation of glial LDs by transferring peroxidated lipids produced in neurons to glia in a process mediated by the apolipoprotein Glial Lazarillo (GLaz; homolog to human APOD) ( SI Appendix, Fig. S1B ). Similarly, the transfer of lipids from cultured vertebrate neurons that are stressed and physically separated from glia has also been documented to be dependent on APOE (16, 18). Glial LDs are neuroprotective when ROS levels are elevated (16, 35) ( SI Appendix, Supplementary Information ). Defective mitochondria produce ROS, which activates the JNK and SREBP transcription factors that drive lipid synthesis. These lipids become peroxidated in the presence of ROS and are subsequently exported to pigment glia, where they are sequestered in LDs ( SI Appendix, Fig. S1B ).
While ROS induction causes the eventual demise of photoreceptor neurons, activation of neuronal lipogenesis, in the absence of ROS, induces LD formation but not neurodegeneration (36). Hence, lipid peroxidation, but not lipid production itself, causes photoreceptor neurotoxicity. The production and transfer of lipids from neurons to glia is a highly dose-sensitive process, as even single copy loss of critical glial LD formation genes causes a significant reduction in glial LD formation (16, 36). Subtle alterations in expression of genes that affect glial LD formation may lead to progressive loss of the neuroprotective effects associated with glial LD formation. LD loss, due to loss of one copy of GLaz, is fully rescued by expression of human APOE2 or APOE3. However, expression of human APOE4 cannot restore glial LD formation and promotes neurodegeneration, suggesting that APOE4 is a loss-of-function (LOF) allele for glial LD formation (16). Interestingly, a pharmacological agonist of ABCA1 has been shown to restore APOE4 lipidation and ameliorate Aβ42/tau pathologies in a mouse model of human APOE expression (43, 44), but its role in LD formation has not been explored.
Given the enrichment of AD risk-associated genes in lipid handling and endocytosis from human GWAS ( SI Appendix, Supplementary Information ), we tested whether these genes are involved in glial LD formation and neuroprotection from ROS. To test this hypothesis, we targeted candidate orthologs of AD risk genes via RNAi using photoreceptor- or pigment glia-specific expression drivers to determine the effect of loss of these genes on glial LD formation and neurodegeneration ( SI Appendix, Fig. S1C ). We demonstrate that the fly homologs of AD risk genes (ABCA1, ABCA7, LRP1, VPS26, VPS35, PICALM, CD2AP, and AP2A2) play a role in the formation of glial LDs, providing a mechanistic link by which AD risk genes may affect neuronal demise. Additionally, we show that ROS synergizes with Aβ42 to accelerate neuronal death in flies and amyloid plaque formation in mice. Finally, we show that an ABCA1 agonist peptide, previously shown to enhance the lipid-binding properties of APOE4, restores glial LD formation in a humanized fly model of APOE4. Together, these data place AD risk genes in a functional pathway that connects ROS with LD formation and Aβ toxicity.
Results
ABCA Transporters Are Required in Neurons for Glial LD Formation.
We have demonstrated that the apolipoprotein encoding gene, GLaz, is required for the transfer of peroxidated lipids from neurons to glia, but the proteins that are required for lipid transfer across neuronal membranes to apolipoproteins remain unknown. Among the AD risk genes are the genes encoding the adenosine triphosphate-binding cassette transporter A1 (ABCA1) and A7 (ABCA7), lipid floppases that transfer lipids from the inner leaflet to the outer leaflet of the cell membrane making them available for export to APOE acceptor particles (45 –47). Missense variants in both ABCA1 and ABCA7 have been associated with increased risk of developing AD (48 –52) and ABCA7 is known to facilitate clearance of Aβ (53).
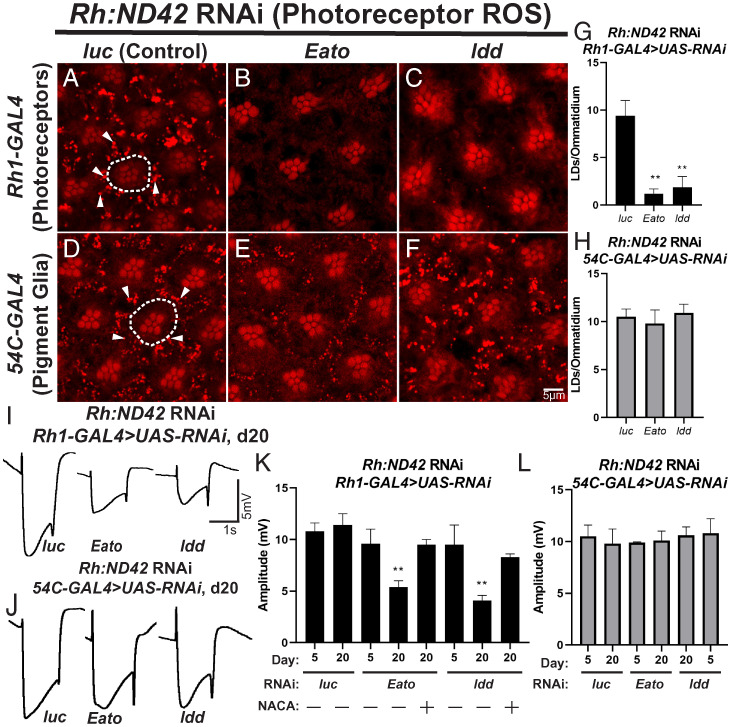
To assess whether ABCA transporters function in glial LD formation in the presence of neuronal ROS we used RNAi-mediated knockdown to reduce ABCA gene expression in glia and neurons. We induced photoreceptor-specific ROS production by constitutive expression of RNAi targeting the mitochondrial complex I gene, ND42 (36). Knockdown of ND42 in photoreceptors induces LD formation in pigment glia, which can be visualized by Nile red staining (Fig. 1 A and D ), but LD phenotypes are not observed in the absence of ROS ( SI Appendix, Fig. S2 A–D ). We then induced expression of RNAi targeting 7 of the 10 fly ABCA genes for which RNAi constructs were available. All selected RNAi constructs efficiently reduced the expression of their respective targets ( SI Appendix, Fig. S3A ) and were expressed in either photoreceptors, using Rh1-GAL4, or pigment glia, using 54C-GAL4. We scored glial LD formation and observed significantly reduced LD formation when Eato and CG34120 (Fig. 1 B, C, and G ), but not CG8908, CG31213, CG1494, or ABCA3 were knocked down in photoreceptors compared to control RNAi (Fig. 1A ). In contrast, when these genes were targeted in glia, no obvious reduction in LD formation was observed (Fig. 1 E, F, and H ). As loss of CG34120 in neurons leads to loss of LDs, we refer to this gene as lipid droplet defective (ldd). Collectively, these data demonstrate that two fly ABCA transporters, Eato and ldd, are required in photoreceptor neurons for glial LD formation. Using gene tree ( SI Appendix, Fig. S4A ) and homology prediction tools ( SI Appendix, Fig. S4B ), we found that the fly genes Eato and ldd are the best orthologs of human ABCA1 and ABCA7 ( SI Appendix, Supplementary Information ).
Fig. 1.
Two ABCA transporters (homologs of human ABCA1 and ABCA7) are required in neurons for glial LD formation. (A–H) LD analysis in fly retina. To induce ROS specifically in photoreceptor neurons, an RNAi against ND42, a mitochondrial complex I subunit, is expressed under the control of the ninaE (Rh) driver. Animals are reared at 29 °C under 12-h light/dark conditions for 24 h after eclosion, prior to isolation of retinas. ROS in photoreceptors induces glial LD formation in control animals (A and D). The photoreceptor rhabdomeres stain positive with Nile red but photoreceptors (dashed lines) do not accumulate LDs. In contrast, pigment glia accumulate LD (arrowheads). Knockdown of Eato and ldd in neurons (B and C), but not in glia (E and F), suppress LD formation, quantified in (G and H, photoreceptor knockdown: black bars, pigment glia knockdown: gray bars), demonstrating a critical role for these genes in neurons for LD formation. Mean ± SEM, one-way ANOVA with Tukey’s post hoc test **P < 0.01 compared to control, n ≥ 10 animals per genotype. (I–L) To assess the functional consequences of LD loss, we performed ERGs at day 5 and day 20. Animals were housed at 29 °C under 12-h light/dark conditions, n ≥ 10 animals per genotype. Representative ERG traces from animals with genotypes indicated (I–J). ERG amplitude quantification (K and L, photoreceptor knockdown: black bars, pigment glia knockdown: gray bars) show that neuronal knockdown of Eato and ldd lead to a severe reduction of ERG amplitude over time, indicative of progressive neurodegeneration, that is rescued by the addition of the antioxidant NACA. Glial knockdown of either Eato or ldd does not affect ERG amplitude. Mean ± SEM, one-way ANOVA with Tukey’s post hoc test *P < 0.05 and **P < 0.01 compared to control, n ≥ 10 animals per genotype.
Loss of LD formation is associated with neurodegeneration (16, 36), and we therefore assessed whether RNAi targeting of Eato and ldd would lead to an age-dependent neurodegeneration. The onset of neurodegeneration in Eato and ldd knockdown flies exposed to neuronal ROS is evidenced by rhabdomere malformation and loss at 20 d posteclosion ( SI Appendix, Fig. S5 A–C ). We also assessed neurodegeneration using the electroretinogram (ERG) assay upon neuronal or glial knockdown of Eato and ldd in the presence of neuronal ROS. ERGs serve as a functional readout of neuronal function and viability (54). ERG amplitudes were quantified in young (5 d posteclosion) and aged (20 d posteclosion) flies. We observed a significant reduction in ERG amplitude with age when Eato and ldd were targeted in neurons, but not when these genes were targeted in pigment glia (Fig. 1 I–L ). We also observed that neurodegeneration is dependent on the presence of ROS, as the addition of the potent antioxidant N-acetylcysteine amide (NACA) rescued the loss of ERG amplitude phenotype observed in the ABCA-targeted backgrounds (Fig. 1K ). Together, these data demonstrate that the ABCA transporters, Eato and ldd, orthologs of the AD risk genes ABCA1 and ABCA7, are required in neurons for glial LD formation.
The APOD Receptor, LRP1, and Retromer Proteins Are Required for Glial LD Formation.
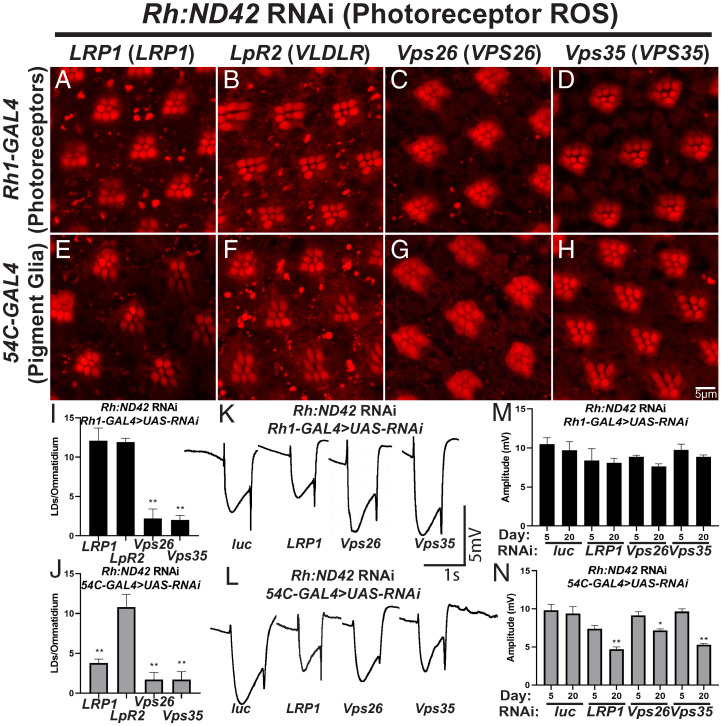
Glial LD formation requires the glial-secreted apolipoprotein, GLaz, but not the neuronally secreted apolipoprotein, NLaz (16). Glial LD formation in a vertebrate neuron:glia cocultures similarly requires the apolipoprotein, APOE (18). As the uptake of lipidated apolipoproteins occurs via endocytosis, we assessed the effects of reduced neuronal or glial expression of genes involved in receptor-mediated endocytosis of apolipoproteins on glial LD formation, beginning with the apolipoprotein receptors, LRP1 and VLDLR (fly LRP1 and LpR2) (55 –57). RNAi targeting these genes was expressed in photoreceptor neurons (Rh1-GAL4) or pigment glia (54C-GAL4) in the presence of neuronal ROS to assess impacts on glial LD formation (knockdown efficiency quantified in SI Appendix, Fig. S3B ; LDs not formed in the absence of neuronal ROS in SI Appendix, Fig. S2 E and F ). Targeting LRP1 in glia, but not neurons, caused a significant reduction in LD formation (Fig. 2 A, E, I, and J ), but neither neuronal nor glial expression of LpR2 RNAi, altered LD formation (Fig. 2 B, F, I, and J ). These data argue that LRP1 is required in glia for LD formation upon neuronal ROS.
Fig. 2.
The APOE receptor, LRP1, and retromer components Vps26 and Vps35 are required for LD formation. (A–J) LD analysis in fly retina. ROS is induced in neurons and RNAi directed against the apolipoprotein receptors (LRP1 and LpR2) or genes critical for retromer function (Vps26 and Vps35) are expressed in neurons (Rh1-GAL4, A–D) and pigment glia (54C-Gal4, E–H). Animals were reared at 29 °C under 12-h light/dark conditions for 24 h prior to isolation of retinas. LRP1 is required in glia (E) but not in neurons (A) to form LD, whereas LpR2 is not required in either cell (B and F). In contrast, the retromer proteins are required in both neurons and glia to form LD (C and D, G and H). Average LD number per ommatidium is quantified (I and J, photoreceptor knockdown: black bars, pigment glia knockdown: gray bars). Mean ± SEM, one-way ANOVA with Tukey’s post hoc test **P < 0.01 compared to control, n ≥ 10 animals for each genotype. (K–N) ERG assays were performed to assess neurodegeneration. Representative traces from animals of genotypes indicated (K and L). Quantification of ERG amplitude (M and N, photoreceptor knockdown: black bars, pigment glia knockdown: gray bars). Glial knockdown of LRP1, VPS26, or VPS35 inhibits LD formation and is associated with an age-dependent neurodegeneration, consistent with a neuroprotective role of glial LD. In contrast, despite LD formation defects when VPS26 or VPS35 were knocked down in neurons, no neurodegeneration or mild neurodegeneration occurs suggesting ROS production or its effects are abrogated. Mean ± SEM, One-way ANOVA with Tukey’s post hoc test *P < 0.05 and **P < 0.01 compared to control, n ≥ 10 animals for each genotype.
We performed ERGs to assess if loss of LRP1 in glia impacts age-dependent photoreceptor loss in animals with elevated levels of ROS in neurons. Compared to control flies, glial but not neuronal knockdown of LRP1 caused reduced ERG amplitudes in an age-dependent manner, indicative of photoreceptor degeneration (Fig. 2 K–N ), which was confirmed by Nile red staining ( SI Appendix, Fig. S5 D and E ). These data suggest that the apolipoprotein receptor, LRP1, promotes glial LD formation and neuroprotection by mediating apolipoprotein endocytosis.
The retromer has been linked to neurodegenerative disease, including AD (58, 59), and serves critical cellular functions in endocytosis and receptor recycling. We hypothesized that the retromer may be important in LRP1 recycling to promote glial LD formation. To test this, we targeted the retromer genes VPS26 and VPS35 via RNAi in our neuronal ROS model (knockdown efficiency quantified in SI Appendix, Fig. S3B ). Vps26 and Vps35 RNAi were expressed in neurons or glia and LD formation and ERG amplitude was assessed. We found that loss of Vps26 and Vps35 in either neurons or glia leads to a significant reduction in glial LDs, suggesting that the retromer is required in both neurons and glia for LD formation (Fig. 2 C, D, and G–J ). We also found no worsening of photoreceptor function over time when Vps26 or Vps35 were knocked down in neurons (Fig. 2 K and M ). In contrast, knockdown of Vps26 and Vps35 in glia caused an age-dependent reduction in amplitude indicative of neurodegeneration (Fig. 2 L and N ). The limited defects of ERG amplitude upon targeting of these genes in neurons suggests that ROS production or the response to ROS production in neurons is blunted or delayed. Western blot analysis to quantify ROS levels provides support for this hypothesis ( SI Appendix, Table S1). The severe loss of ERGs documented in Wang et al. (60) when Vps26 or Vps35 proteins are completely lost in both photoreceptors and glia suggest an additive or synergistic effect between neurons and glia and argues that the retromer is required in both cell types to maintain neuronal health. While further probing of this mechanism is warranted, these data suggest that the neurodegeneration observed when glial retromer function is diminished may be caused by reduced apolipoprotein receptor recycling, thus limiting lipid uptake into glia.
Endocytic AD-Risk Genes Are Required in Glia for Glial LD Formation.
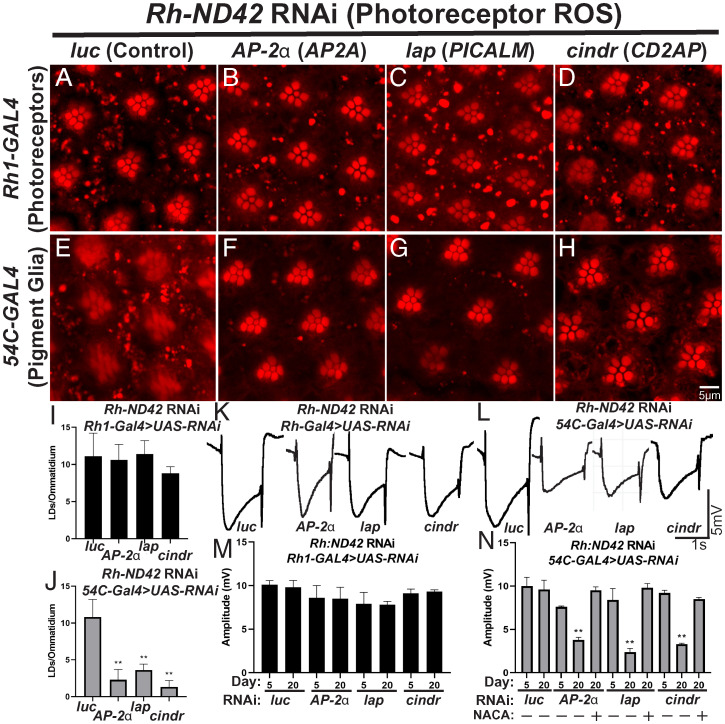
A subset of AD-risk loci map in or near genes involved in endocytosis, including BIN1, CD2AP, PICALM, AP2A2, and RIN3 (61 –63), suggesting that endocytosis may be important for AD pathogenesis. It has been proposed that these genes contribute to AD pathology through their well-characterized function in synaptic transmission in neurons (64 –67). However, there is evidence endocytosis is required for glial LD formation in vertebrate neuron:glia coculture (18). It remains unclear whether endocytic inhibition in neurons, glia, or both cell types causes LD formation inhibition. We therefore set out to examine tissue-specific roles for AD-associated endocytic genes in LD formation. We hypothesized that endocytosis may play a neuroprotective role by aiding the sequestration of toxic, peroxidated lipids from neurons into glial LD.
To examine a role for endocytic AD risk genes in LD formation, we examined LD formation and ERG phenotypes in animals in which homologs of AD-risk genes are targeted, via RNAi, in neurons and glia in the presence of neuronal ROS (knockdown efficiency quantified in SI Appendix, Fig. S3C ; no LD phenotypes are observed in the absence of ROS in SI Appendix, Fig. S2 E and G–I ). We found that knockdown of cindr (CD2AP), Ap-2α (AP2A2), and lap (PICALM) in glia, but not neurons, reduced LD formation (Fig. 3 A–J ), thus implicating these genes as critical components of glial LD formation. In contrast, reduced expression of spri (RIN3) and amph (BIN1) in neurons or glia did not significantly affect LD production ( SI Appendix, Supplementary Information and Fig. S6). Evidence of neurodegeneration is present by day 20 posteclosion in AP-2α, lap, and cindr knockdown flies exposed to neuronal ROS ( SI Appendix, Fig. S5 D and F–H ). We also observed an age-dependent decrease in ERG amplitude upon glial targeting of Ap-2α, lap, and cindr (Fig. 3 K–N ). ERG amplitude deficits were rescued by the addition of the antioxidant NACA (Fig. 3N ), suggesting that ROS and gene dysfunction combine to induce neurodegeneration. Altogether, these data demonstrate that reduced glial endocytosis inhibits the neuroprotective effects of glial LD formation and exacerbates neurodegeneration upon elevated neuronal ROS.
Fig. 3.
AD-associated GWAS genes are required in glia for LD formation upon neuronal ROS induction. (A–J) LD analysis in fly retina. ROS is induced in neurons and RNAi directed against homologs of 5 GWAS genes in photoreceptor neurons (A–D) or glia (E–H). Animals are housed at 29 °C under 12-h light/dark conditions for 24 h prior to isolation of retinas. Expression of RNAi against any genes tested in neurons do not affect the formation of LD in glia significantly (A–D). In contrast, RNAi targeting AP-2a, lap, and cindr in glia reduced LD formation significantly (E–H) as quantified (I, J, photoreceptor knockdown: black bars, pigment glia knockdown: gray bars). Mean ± SEM, one-way ANOVA with Tukey’s post hoc test *P < 0.05 and **P < 0.01 compared to control, n ≥ 10 animals for each genotype. (K–N) ERG assays were performed, as above, to assess neurodegeneration. Animals are housed at 29 °C under 12-h light/dark conditions, n ≥ 10 animals per genotype. Representative traces (K and L) and amplitude quantification (M and N, photoreceptor knockdown: black bars, pigment glia knockdown: gray bars) demonstrate that neuronal knockdown of these genes does not affect ERG amplitude. In contrast, glial knockdown of these genes led to a reduction in LD formation (AP-2a, lap, and cindr) also led to a significant reduction of ERG amplitude in aged animals, showing an age-progressive neurodegeneration, which is rescued by the addition of the antioxidant NACA. Mean ± SEM, one-way ANOVA with Tukey’s post hoc test **P < 0.01 compared to control, n ≥ 10 animals for each genotype.
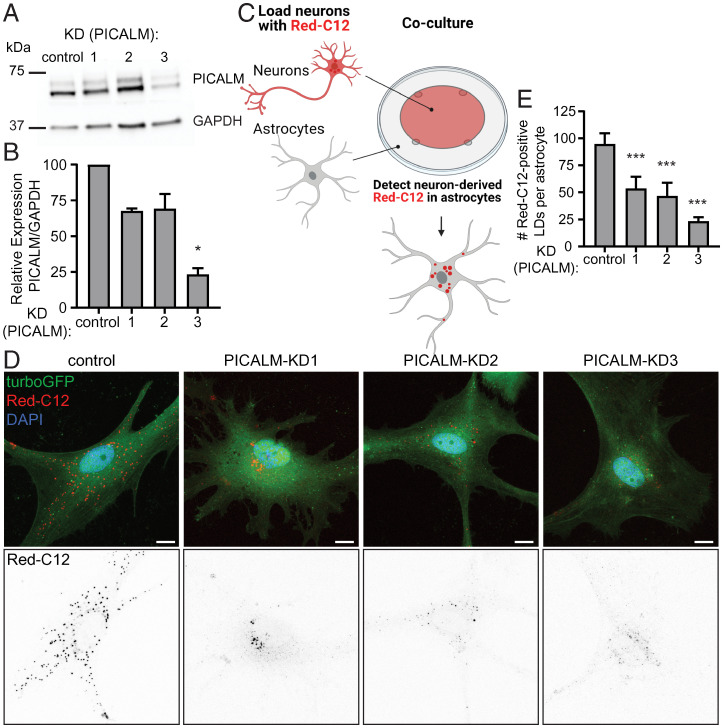
ROS-induced LD formation is conserved in vertebrates (16, 18). We investigated whether knockdown of PICALM, (the ortholog of lap) is required for LD formation in vertebrate glia. To this end, we utilized an established mammalian cell coculture system of rat neurons and astrocytes (68). We utilized lentivirus to deliver three independent short-hairpin RNAs (shRNAs) to reduce PICALM protein, compared to a nontargeting shRNA control, in cultured astrocytes (Fig. 4 A and B ). Independently, we incubated neurons with a fluorescently labeled fatty acid analog Red-C12 overnight and then cocultured the labeled neurons with transduced astrocytes on different coverslips separated by paraffin wax (Fig. 4C ) (18, 68). We found a significant reduction in the transfer of fluorescently labeled fatty acids from neurons to astrocytes when gial PICALM levels are reduced (Fig. 4 D and E ). These data demonstrate that clathrin-mediated endocytosis is critical for the internalization of neuron-derived fatty acids in a mammalian culture system.
Fig. 4.
Lipid transfer between neurons and astrocytes is blunted by knockdown of PICALM. (A) Astrocytes were transduced with lentivirus expressing nontargeting shRNA (control), or three independent PICALM targeting shRNAs (KD1-3). Cell lysates were analyzed by Western blot for PICALM levels and GAPDH as a loading control. (B) Levels of PICALM from transduced astrocytes were quantified and normalized to GAPDH control. Mean ± SEM, Kruskal–Wallis test with Dunn’s posttest *P < 0.05 compared to control, n = 3 from three independent experiments. (C) Schematic of Red-C12 transfer assay. (D) Representative maximum-intensity projections of confocal images of transduced astrocytes following the assay. TurboGFP reporter expression marks transduced cells. (Scale bars, 10 µm.) (E) Quantification of Red-C12+ LDs in astrocytes. Mean ± SEM, one-way ANOVA with Dunnett’s posttest ***P < 0.001 compared to control, n = 6 cells from three independent experiments each.
Aβ Synergizes with ROS in Flies and Mice.
Aβ peptides are lipophilic and can bind the apolipoprotein receptor, LRP1, suggesting that altered lipid transfer may also alter amyloidogenesis (69, 70). Moreover, poorly lipidated APOE can aggregate and act as seeds for Aβ plaques (69, 71, 72) and LOF mutations in ABCA1 leads to decreased APOE lipidation and increased amyloidogenesis (73, 74). As ROS-induced glial LD formation is severely affected by AD-associated risk genes and peroxidated lipids accumulate in pre-AD patients (23, 24, 26, 75), we hypothesized that ROS-induced lipid peroxidation may exacerbate Aβ42-induced phenotypes.
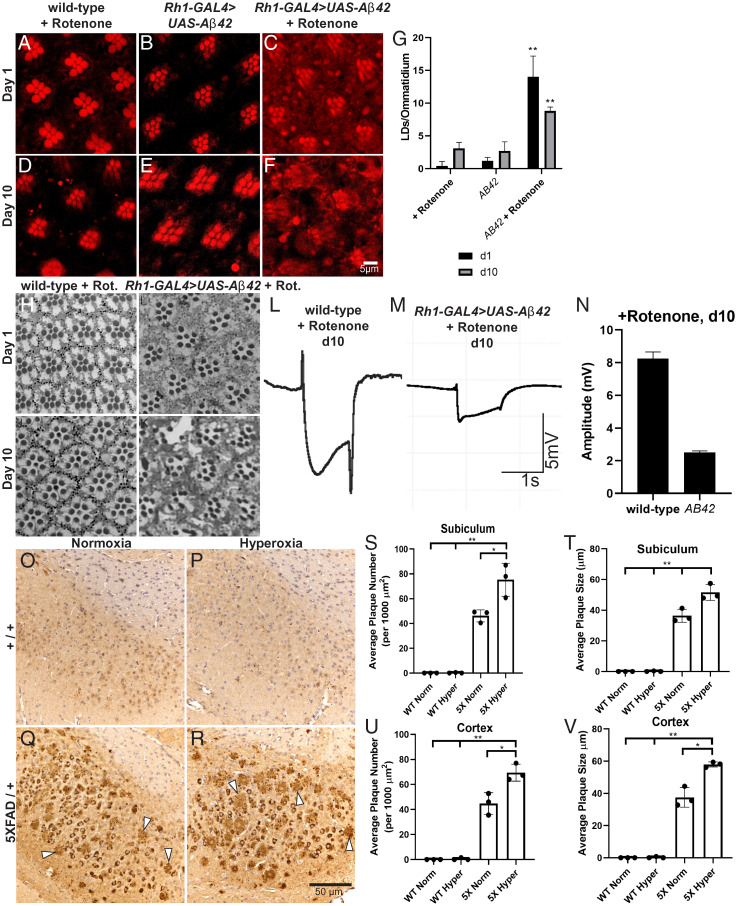
To test this hypothesis, we utilized a well-characterized fly line expressing a secreted form of human Aβ42 that, when expressed in neurons, induces neurodegeneration at >30-d-old flies (76). Wild-type flies and flies expressing Aβ42 in photoreceptor neurons (Rh1-GAL4 > AB42) were exposed to ROS, by feeding flies very low doses of rotenone (a mitochondrial complex I inhibitor) or control food for 10 d (77). This concentration of rotenone induces a minor elevation of ROS (16, 36), but no substantial neurotoxicity and very few LD are observed after 10 d (Fig. 5 A, D, and G ). Animals expressing secreted Aβ42 (using Rh1-GAL4) do not exhibit obvious signs of neuronal death in the fly retina and accumulate very few glial LDs within the same 10-d time frame (Fig. 5 B, E, and G ). In contrast, Aβ42-expressing animals fed 25 µM rotenone exhibit robust glial LD accumulation (Fig. 5 C, F, and G ). The number of LDs observed in these flies decreased between days 1 and 10, indicative of lysis of the LDs, which is associated with cell death as the peroxidated lipids escape from the LD (16, 36). We also observed severe morphological defects of the photoreceptors and glia at day 10 upon rotenone feeding in the Aβ42-expressing flies compared to any other condition tested (Fig. 5 H–K ). The degeneration of the retina in Aβ42 with rotenone flies at day 10 is also associated with a severe loss of ERG amplitude compared to control (Fig. 5 L–N ). These data demonstrate that Aβ42 strongly synergizes with ROS to induce neuronal death.
Fig. 5.
Elevated ROS and the presence of Aβ42 synergize to induce neurodegeneration in flies and mice. (A–G) LD analysis in fly retina. Animals are housed at 29 °C under 12-h light/dark conditions with food changed daily; representative images of ≥10 animals per genotype. Wild-type flies exposed to 25 µM rotenone food for (A) 1 d or (D) 10 d posteclosion were compared with Aβ42-expressing flies at (B) 1 d posteclosion or (E) 10 d posteclosion and with Aβ42-expressing flies exposed to 25 µM rotenone food for (C) 1 d posteclosion and (F) 10 d posteclosion. Note the absence of LD formation with either treatment but the obvious increase in diffuse Nile red staining and the demise of PR by day 10 showing that ROS and Aβ42 synergize to cause the demise of neurons as quantified (G, day 1: black bars, day 10: gray bars). Mean ± SEM, one-way ANOVA with Tukey’s post hoc test **P < 0.01 compared to control, n ≥ 10 animals for each genotype. (H–K) Retinal sections stained with Toluidine blue were obtained from wild-type and Aβ42-expressing flies fed rotenone for 10 d and imaged at 20× magnification. Neurodegeneration is apparent in d10 Aβ42 flies fed rotenone (K). (L–N) Neurodegeneration is also evident by decreased ERG amplitude compared to wild-type, rotenone-fed flies. (O–V) Aβ42 immunohistochemical analysis of 4-mo-old mouse brain sections from wild-type mice reared in normoxic (O) or hyperoxic (P) conditions compared to 5XFAD mice reared in normoxic (Q) or hyperoxic (R) conditions for 3 mo prior to being killed. Arrowheads indicate amyloid plaques, n = 3 animals per genotype and treatment condition. Quantification of average plaque number (S and U) and plaque size (T and V) in the brain regions indicated from mice in (O–R). Plaque size and number is elevated in Aβ-expressing mice exposed to hyperoxia. Mean ± SEM, one-way ANOVA with Tukey’s post hoc test *P < 0.05 and **P < 0.01 compared to control, n = 3 animals for each genotype.
We next tested for an interaction between ROS and amyloid in a vertebrate model using the well-characterized 5XFAD mouse, which expresses mutant human APP and PSEN1 and causes Aβ-plaque formation that has been documented as early as 4 mo of age (78). As rotenone preferentially induces dopaminergic neurotoxicity and is historically used to model PD (79), we instead induced ROS by rearing animals in hyperoxia (80), as evidenced by elevated 4HNE levels by Western blot analysis ( SI Appendix, Fig. S7A ). We assembled cohorts of male and female heterozygous 5XFAD mice and wild-type littermate controls and subjected them to either hyperoxic (55% O2) or normoxic (∼21% O2) conditions for 3 mo beginning at the age of 1 mo (for power analysis, see SI Appendix, Supplementary Information and Table S2). Sagittal brain sections of the mice were stained for Aβ accumulation using established immunohistochemistry techniques (81). We quantified plaque number and size in three regions of the brain: the cortex, subiculum, and hindbrain. In each of these regions, plaque size observed in 5XFAD mice was significantly increased in hyperoxia-treated animals when compared to normoxia-treated animals and plaque number was significantly elevated in the cortex and subiculum of hyperoxia-treated animals (Fig. 5 O–V and SI Appendix, Fig. S7 B and C ), suggesting that elevated ROS exacerbates amyloid plaque formation. Taken together, the data from flies and mice provide evidence for a feed-forward mechanism between ROS and Aβ and suggest that AD phenotypes may be due to interactions between multiple risk factors, including ROS.
A Pharmacological ABCA Agonist Peptide Rescues APOE4-Induced Phenotypes.
Using a humanization strategy in which the fly apolipoprotein, GLaz, is replaced with expression of human APOE, we previously reported that the AD-associated APOE4 allele was much less capable of mediating the transfer of peroxidated lipids from neurons to glia (16). The ABCA1 receptor has previously been shown to drive APOE lipidation (43, 82) and a small ABCA1 agonist peptide, CS6253, restores APOE4 lipidation and ameliorates amyloid plaque and tau phenotypes in mammals (43, 83). Given that neuronally expressed ABCA1 and ABCA7 orthologs in the fly are critical for glial LD formation (Fig. 1), we hypothesized that CS6253-induced elevation of ABCA activity may restore LD formation in APOE4 humanized flies.
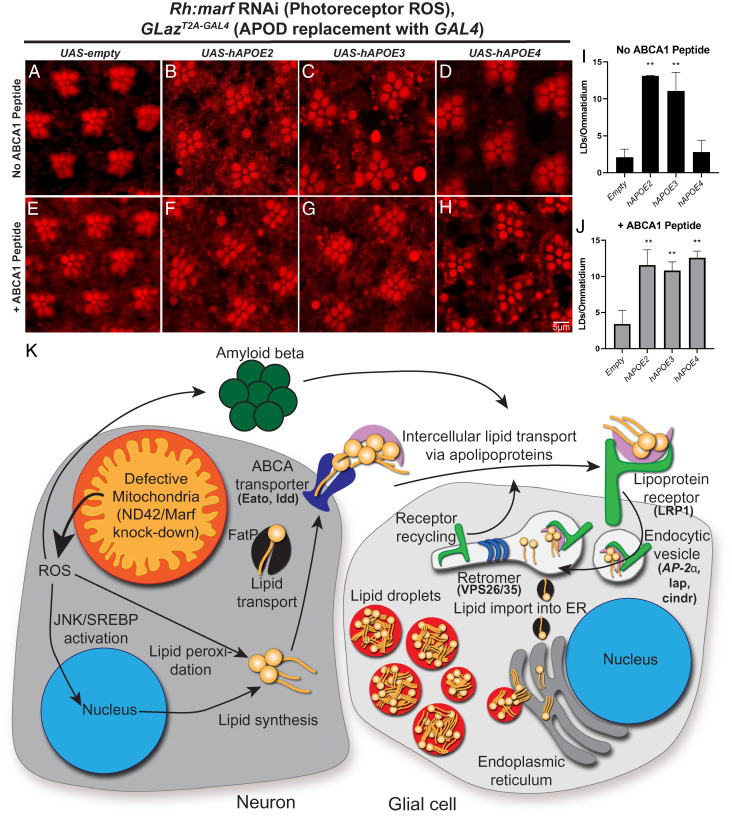
We generated a fly line that expresses a genetically encoded version of CS6253 (43, 44, 83). The peptide sequence was cloned downstream of a secretion signal to enable peptide release from the cell (76). Expression of the peptide was driven by the GLazT2A-GAL4 allele, thus allowing it to be expressed in the same temporal and spatial pattern as GLaz. Neuronal ROS was induced by expressing an RNAi against Marf, the fly homolog of Mitofusin, under control of the ninaE (Rh) promoter, which induces high levels of glial LD accumulation (16). As reported, heterozygous GLazT2A-GAL4 animals have significantly reduced LD production (16) and expression of the peptide does not alter LD formation in this background (Fig. 6 A and E ). Peptide expression in either the APOE2 or APOE3 background does not elevate LD production beyond what is already present without expression of the peptide (Fig. 6 B, C, F, G, I, and J ), suggesting that some maximal amount of LDs have been generated in the presence of APOE2 or APOE3. In contrast, expression of the peptide in the APOE4 background restored LD formation (Fig. 6 D and H–J ). These data suggest that the ABCA1 peptide, CS6253, promotes APOE4 lipidation, thus restoring glial LD formation. Furthermore, neurodegeneration as visualized by loss of photoreceptor rhabdomeres ( SI Appendix, Fig. S8 A and B ) and ERG amplitude deficits ( SI Appendix, Fig. S8 C–F ) is also rescued by this peptide in the APOE4 background. Together, these data implicate a similar mechanism of APOE lipidation in the fly, as has been reported in vertebrate models, and are consistent with the critical role for fly ABCA transporters (ldd and Eato) in LD formation and neuroprotection.
Fig. 6.
An ABCA1 agonist peptide rescues LD formation in the presence of APOE4. (A–J) LD analysis in fly retina. ROS was induced in photoreceptor neurons, as previously reported (17, 40), using an RNAi against marf, the fly ortholog of mitofusin, under the control of ninaE (Rh). Animals are reared at 29 °C under 12-h light/dark conditions for 24 h prior to isolation of retinas; representative images of ≥10 animals per genotype. We utilized a previously characterized allele of Glial Lazarillo (GLaz-T2A:GAL4). LD formation is inhibited in GLaz-T2A-GAL4/+ flies but can be restored by expressing human APOE2 or APOE3, but not APOE4. An ABCA1 agonist peptide was genetically encoded in the fly and expressed in the human APOE variant flies to assess LD formation. Expression of the peptide does not affect LD formation in the presence of APOE2 or APOE3, but fully restores LD formation in the APOE4 expressing flies (E–H) and quantified (I and J, no peptide: black bars, + peptide: gray bars) showing that LD formation is strongly enhanced by the peptide. Mean ± SEM, one-way ANOVA with Tukey’s post hoc test **P < 0.01 compared to control, n = 10 to 15 animals for each genotype. (K) Model of LD accumulation and players identified in this study. We propose a model in which genetic (loss of ABCA, endocytic, or retromer genes) together with ROS sensitize neurons to the presence of amyloid accumulation to induce neurodegeneration. It is likely that this synergy between multiple insults exacerbates neuronal loss in disease. We demonstrated that lipid transfer between neurons and glia requires neuronal ABCA transporters, a glial apolipoprotein receptor, and the retromer, which is required for LRP1 recycling. We propose that endocytosis of lipid particles are processed through lysosomes upon endocytosis. Lysosomes degrade Aβ42 while the lipids are shuttled to the endoplasmic reticulum (ER) to form LD. Hence, this transport of peroxidated lipids and Aβ42 provides a dual protective effect.
Discussion
In this study, we uncovered a pathway in which neurons and glia interact to form LDs upon neuronal ROS (Fig. 6K ), which is neuroprotective. This process requires ABCA transporters (Eato and ldd) and the retromer (VPS26 and VPS35) in neurons together with GLaz (16, 68), glial receptor-mediated endocytic proteins (LRP1, PICALM, CD2AP, and AP2A2), and the retromer (VPS26 and VPS35) in glia. Notably, the genes identified in this study have been implicated in AD and other neurodegenerative diseases (5, 6, 44, 58, 84). Our data implicate a LOF model in which AD risk-associated variants exacerbate disease by limiting lipid transfer and peroxidated lipid sequestration into glial LD. While there is growing evidence that mutations in several AD risk genes are partial LOF mutations (50, 85 –88), we do not rule out that alternative mechanisms, including gain-of-function, are at play in AD, which warrant further exploration. The data presented herein are consistent with the hypothesis that disease is cumulatively and synergistically induced by genetic variants (e.g., LOF variants that disrupt neuron-to-glia lipid transfer) together with other cellular insults (e.g., neuronal ROS).
It has been well documented that ROS levels are elevated with age and in AD (89). Neurons have limited antioxidant capacity and activate various cellular mechanisms in response to ROS (90). Indeed, antioxidant activity is reduced in AD patients and the use of antioxidants as a treatment for AD has been proposed previously, although with mixed outcomes (91 –94), possibly because the treatments were initiated too late in the course of disease or unable to fully penetrate the blood–brain barrier. Understanding how ROS protection is mediated and how these responses go awry may reveal ways to exogenously potentiate the antioxidant response. Regardless of the cause of ROS (e.g., age, environmental stress, or genetic perturbations), oxidative stress may initiate disease in an individual with a previous genetic predisposition to disease, such as APOE4. Other AD risk genes involved in lipid handling and endocytosis may affect the transport of peroxidated lipids from neurons into glia, thus elevating risk for the development of disease. We have shown that the blood–brain barrier crossing antioxidant, NACA strongly suppresses the formation of peroxidated lipids and LD accumulation in flies (16 and this study) and warrants further exploration in disease.
The effects of neuronal ROS served as a platform in which we examined AD-associated risk factors, including ABCA transporters and proteins involved in endocytosis. We found that ABCA transporters in the fly are required in neurons for glial LD formation (Fig. 1). ABCA transporters implicated as risk factors for AD might mediate the export of peroxidated lipids from neurons to glia to protect neurons from the toxic effects of ROS-induced lipid peroxidation in disease. It is noteworthy that loss of either ABCA protein in flies leads to a loss of LD formation and neurodegeneration. This observation may suggest that these proteins function in a dimer or other higher-order complex or that they have different substrates, which are both required for LD formation, such that loss of either protein reduces LD formation. After lipid export from neurons via ABCA proteins, glial LD formation requires several endocytic factors in glia (Fig. 3).
BIN1 is a membrane fission protein that regulates endocytic vesicle size in vertebrates, but it has been implicated in APP processing, as well as tau degradation (61, 95, 96). In flies, the ortholog of BIN1, Amphiphysin (Amph), regulates transverse tubule formation in muscles, which was also shown to be affected in vertebrate mutants (97), but Amph has not been implicated in endocytosis in flies to our knowledge (98, 99). Interestingly, increased expression of BIN1 mediates AD risk by modulating tau pathology (100), which is consistent with our data as we observed no impact on LD formation upon BIN1 loss in our assay ( SI Appendix, Fig. S5). CD2AP is a scaffolding protein that has been implicated in endocytosis and vesicle trafficking as well as APP sorting and processing in flies and vertebrates (101 –103). However, severe LOF alleles of the fly ortholog of CD2AP, cindr, affects synapse maturation as well as synaptic vesicle recycling and release when lost (104). PICALM is a clathrin assembly protein that has been implicated in the import of γ-secretase and APP processing, as well as tau build-up (61, 105). The fly ortholog of PICALM, like-AP180 (lap), acts as a clathrin adaptor to promote clathrin-coated vesicle formation and restrict coated vesicle size as well as the efficacy of synaptic vesicle protein retrieval (106). AP2A2, a member of the AP-2 adaptor protein complex, aids in assembling endocytic components in flies and vertebrates, and is an AD risk factor (62, 107). Finally, RIN3 is a Rab5 guanine nucleotide exchange factor important for recruiting CD2AP and BIN1 to endosomes has been implicated in APP accumulation and tau phosphorylation regulation (63). Based on our data, the fly orthologs of three endocytic genes (CD2AP, AP2A2, and PICALM) play critical functions in glia for LD formation (Fig. 3). Historically, because many endocytic AD-risk genes are known to play a critical role in synaptic transmission, it is thought that their role in AD pathology is related to the function of these genes in neurons. However, single-cell RNA sequencing databases provide evidence for enriched expression of many of the endocytic AD-risk genes in mouse/human glia (108, 109), and our data indicate that fly and vertebrate glia are highly sensitive to partial loss of these genes (Figs. 3 and 4). Hence, even a subtle loss of the encoded proteins in human glia may have different consequences than would a severe loss at synapses in model organisms or cells.
As endocytic vesicles are processed in the cell, the retromer is critical for protein recycling, including cell surface receptors and rhodopsin (rh) (60). We observed reduced LD formation when retromer function was targeted via RNAi in both neurons and glia (Fig. 2). However, neurodegeneration was only observed when the retromer was lost in glia, suggesting different roles for retromer in neurons than in glia. In glia, the receptor, LRP1, is critical for LD formation (present study) and the retromer is required for LRP1 recycling to the membrane for efficient uptake of lipid particles (110), implicating a critical role for retromer-mediated recycling of LRP1 for glial LD formation. In neurons, loss of retromer may lead to progressive neurodegeneration but neurons may be less sensitive to this loss, as knockdown of Vps26 in both glia and neurons causes more severe neuron loss than knockdown of Vps26 in glia alone (73 and this study). It is becoming increasingly evident that the retromer plays critical roles in the maintenance of neurons in AD (58) and its function in glia in the context of AD warrants further exploration.
In addition to the identification of critical LD genes, which overlap with AD risk genes, the data presented here provide a hypothesis for the nonlinear relationship between amyloid burden and clinical severity of disease. Human Aβ42 expression induces neurodegeneration in Drosophila (76), as well as neurological and behavioral phenotypes in mice (78, 111). Notably, production of low levels of ROS or Aβ alone causes a very slow, progressive neurotoxicity. In flies, overexpression of Aβ42 causes neuronal death after ∼30 d (76), but we observed that combining low levels of ROS in Aβ42-expressing flies strongly exacerbated neurodegeneration and elevated ROS enhanced Aβ deposition in 5XFAD mice (Fig. 5). It is noteworthy that Aβ and peroxidated lipids both bind APOE (72, 112), providing a possible mechanism of ROS/Aβ42 synergy. Importantly, APOE4 cannot be adequately lipidated, and lipidation of APOE is required for Aβ42 binding (113 –115). Thus, APOE4 would be unable to properly clear peroxidated lipids and Aβ42, strongly accelerating the demise of neurons. Enhancement of APOE4-mediated lipid clearing could be attained by the ABCA1 agonist CS6253, which restored LD formation in APOE4 flies but did not affect APOE2- or APOE3-mediated LD formation (Fig. 6)
It is noteworthy, given the robust interaction between cellular ROS and amyloid (Fig. 5), that the efficacy in disease modeling in mammals may be enhanced by the addition of ROS, which is currently lacking in the AD field. The addition of ROS in mammalian models comes with various challenges, including the use of toxic drugs (i.e., rotenone) or bulky and expensive equipment (i.e., hyperoxia chambers). Genetic factors that induce ROS may be a more viable option to improve AD models. A study using a mouse model of Leigh syndrome, in which the gene NDUFS4 is knocked out thereby reducing activity of complex I, demonstrated that elevated ROS production induces early lethality (116, 117). These mice have numerous LD in astrocytes and microglia prior to the onset of neuronal loss (36). In hypoxia, these mice live much longer (>170 d) than when reared in normoxic conditions (no animals survived past 75 d) (118). Thus, the addition of ROS by genetic means by, for example removing a single copy of Ndufs4, may prove a viable method to induce ROS in existing AD mammalian models. It is worth noting that the fly ortholog of NDUFAF6 (sicily in flies), another cause of Leigh syndrome, is associated with elevated ROS (16) and was the first mutant in which we observed accumulation of LD in glia (16). Currently, NDUFAF6 has been reported in three GWAS as a risk factor for AD (5, 119, 120), providing further evidence for a link between ROS, LD, and AD.
Although age and mitochondrial dysfunction are obvious potential sources of ROS in AD patients, there may be numerous other conditions that induce ROS production and subsequent lipid peroxidation, LD formation, and neurodegeneration (121). A careful examination of ROS in AD patients and inclusion of ROS in animal models may help begin to provide mechanistic insight into the etiology and progression of AD. We predict that when ROS levels rise, it becomes increasingly difficult for glia to sequester peroxidated lipids into LD, promoting the demise of neurons. Thus, therapeutic approaches aiming to induce glial uptake of lipids to alleviate ROS and clear amyloid should also include approaches to neutralize ROS, such as NACA, to eliminate the long-term consequences of oxidative damage.
Materials and Methods
Information regarding strains, reagents, and tools used in this study can be found in SI Appendix, Supplementary Information and Table S3. Drosophila melanogaster were raised on standard molasses-based laboratory diet at 22 °C under constant light conditions, unless otherwise indicated. UAS-ArgosSS::Peptide transgenic flies were generated using standard methods (122). Experiments using Mus musculus were carried out under the approval of the Animal Care and Use Committee at Baylor College of Medicine.
Whole-mount Nile red and Toluidine blue staining of fly retinas (36, 76), ERG assays (123, 124), as well as RNA extraction, cDNA synthesis, and qRT-PCR were performed as previously described (125). Animal perfusion, sectioning, and immunohistochemistry was performed as in Sillitoe et al. (81). Hippocampal cultures were generated from postnatal day 0 to 1 Sprague-Dawley rats obtained from Charles River Laboratories that arrived at our facility 1 wk prior to birth. These experiments were approved by the Canadian Council of Animal Care at the University of Alberta (AUP#3358). Cultures were prepared as previously described (18, 126). See SI Appendix, Supplementary Materials and Methods for additional details pertaining to these studies.
Quantification and Statistical Analysis.
FIJI (127) was utilized to visualize fly retinal and mouse brain images and all genotypes were blinded prior to quantification. LDs with diameter ≥0.5 µM were manually quantified from fly retinal images. Amyloid plaque number from mouse brain images was manually quantified and amyloid size measurements were taken using the “Measure” tool in FIJI. LabChart 8 (AD Instruments) was used to view and measure the amplitude of ERG traces. Quantification datasets were assembled in Microsoft Excel 365 for computation of Mean ± SEM and one-way ANOVA analysis with post hoc Tukey’s test. For quantification, ≥10 animals per genotype were used unless otherwise indicated. Adjusted P values with a statistical significance cutoff at *P < 0.05 and **P < 0.01. Statistical analysis of knockdown efficiency in rat cells used the Kruskal–Wallis test with Dunn’s posttest using a significance cutoff at *P < 0.05. Analysis of lipid transfer utilized one-way ANOVA with Dunnett’s posttest using a significance cutoff at ***P < 0.001. Post hoc power analyses were performed using GPower 3.1 (128) and effect size (Cohen’s d) was calculated using the formula Cohen’s d = (M 2 − M 1) SD pooled, where SD pooled = √((SD 1 2 + SD 2 2) 2).
Supplementary Material
Acknowledgments
We thank the reviewers for their suggestions to improve the manuscript and the Bloomington Drosophila Stock Center and the Vienna Drosophila Resource Center for providing reagents. We also thank Zhongyuan Zuo, the Bellen Lab EM technician, for assistance with retinal sectioning and imaging. Mouse strains were provided by Huda Zoghbi. Mouse pathological studies were performed by the Cell and Tissue Pathogenesis Core, supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the NIH under Award P50HD103555. Confocal imaging was performed in the Neurovisualization Core of the Intellectual and Developmental Disabilities Research Center, supported by the NIH under Award U54HD083092. This work was supported by grants from the Texas Alzheimer’s Research and Care Consortium under Award 2018-05-11-II (to H.J.B.) and from The National Institute on Aging of the NIH under Award R01 AG073260 (to H.J.B.). M.J.M. was supported by the Medical Genetics Research Fellowship Training Grant from the NIH under Award T32 GM07526-41 and is currently supported by the Brain Disorders & Development Fellowship Training Grant from the NIH under award number T32 NS043124-19. L.D.G. was supported by the Brain Disorders & Development Fellowship Training Grant from the NIH under Award T32 NS043124-18. P.C.M. is supported by a grant from Canadian Institutes of Health Research under Award MFE-164712. M.S.I. is supported by the Canadian Institutes of Health Research under Award 173321, and the Heart & Stroke Foundation of Canada under Award 170722. I.R. is supported by the Alberta Synergies in Alzheimer’s and Related Disorders (SynAD) program funded by the Alzheimer Society of Alberta and Northwest Territories through their Hope for Tomorrow program and the University Hospital Foundation. SynAD operates in partnership with the Neuroscience and Mental Health Institute at the University of Alberta. H.J.B. was supported by the Howard Hughes Medical Institute.
Footnotes
Reviewers: M.F., Oregon Health & Science University Vollum Institute; K.K., University College London; L.P., University College London; and M.W., University of Rochester.
Competing interest statement: J.O.J. is the President and CEO of Artery Therapeutics, Inc. and part owner of a patent of the peptide used in this work (Patent WO2014144708A1).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112095118/-/DCSupplemental.
Data Availability
All data are included in the main text and SI Appendix .
References
- 1. Masdeu J. C., Neuroimaging of diseases causing dementia. Neurol. Clin. 38, 65–94 (2020). [DOI] [PubMed] [Google Scholar]
- 2. Bloom G. S., Amyloid-β and tau: The trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurol. 71, 505–508 (2014). [DOI] [PubMed] [Google Scholar]
- 3. Yiannopoulou K. G., Anastasiou A. I., Zachariou V., Pelidou S. H., Reasons for failed trials of disease-modifying treatments for Alzheimer disease and their contribution in recent research. Biomedicines 7, 97 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Strooper B., Karran E., The cellular phase of Alzheimer’s disease. Cell 164, 603–615 (2016). [DOI] [PubMed] [Google Scholar]
- 5. Kunkle B. W., et al. ; Alzheimer Disease Genetics Consortium (ADGC); European Alzheimer’s Disease Initiative (EADI); Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE); Genetic and Environmental Risk in AD/Defining Genetic, Polygenic and Environmental Risk for Alzheimer’s Disease Consortium (GERAD/PERADES), Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lambert J.-C., et al. ; European Alzheimer’s Disease Initiative (EADI); Genetic and Environmental Risk in Alzheimer’s Disease; Alzheimer’s Disease Genetic Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology, Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–1458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jansen I. E., et al. , Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hao S., Wang R., Zhang Y., Zhan H., Prediction of Alzheimer’s disease-associated genes by integration of GWAS summary data and expression data. Front. Genet. 9, 653 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nazarian A., Yashin A. I., Kulminski A. M., Genome-wide analysis of genetic predisposition to Alzheimer’s disease and related sex disparities. Alzheimers Res. Ther. 11, 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma Y., et al. , Alzheimer’s disease GWAS weighted by multi‐omics and endophenotypes identifies novel risk loci. Alzheimers Dement. 16, e043977 (2020). [Google Scholar]
- 11. Karch C. M., Goate A. M., Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry 77, 43–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ward A., et al. , Prevalence of apolipoprotein E4 genotype and homozygotes (APOE e4/4) among patients diagnosed with Alzheimer’s disease: A systematic review and meta-analysis. Neuroepidemiology 38, 1–17 (2012). [DOI] [PubMed] [Google Scholar]
- 13. Qi G., et al. , APOE4 impairs neuron‐astrocyte coupling of fatty acid metabolism. Alzheimers Dement. 16, e045251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michaelson D. M., APOE ε4: The most prevalent yet understudied risk factor for Alzheimer’s disease. Alzheimers Dement. 10, 861–868 (2014). [DOI] [PubMed] [Google Scholar]
- 15. Li Z., Shue F., Zhao N., Shinohara M., Bu G., APOE2: Protective mechanism and therapeutic implications for Alzheimer’s disease. Mol. Neurodegener. 15, 63–81 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu L., MacKenzie K. R., Putluri N., Maletić-Savatić M., Bellen H. J., The glia-neuron lactate shuttle and elevated ROS promote lipid synthesis in neurons and lipid droplet accumulation in glia via APOE/D. Cell Metab. 26, 719–737.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hatters D. M., Peters-Libeu C. A., Weisgraber K. H., Apolipoprotein E structure: Insights into function. Trends Biochem. Sci. 31, 445–454 (2006). [DOI] [PubMed] [Google Scholar]
- 18. Ioannou M. S., et al. , Neuron-astrocyte metabolic coupling protects against activity-induced fatty acid toxicity. Cell 177, 1522–1535.e14 (2019). [DOI] [PubMed] [Google Scholar]
- 19. Butterfield D. A., Brain lipid peroxidation and Alzheimer disease: Synergy between the Butterfield and Mattson laboratories. Ageing Res. Rev. 64, 101049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grimm A., Eckert A., Brain aging and neurodegeneration: From a mitochondrial point of view. J. Neurochem. 143, 418–431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ristow M., Schmeisser S., Extending life span by increasing oxidative stress. Free Radic. Biol. Med. 51, 327–336 (2011). [DOI] [PubMed] [Google Scholar]
- 22. Thapa A., Carroll N. J., Dietary modulation of oxidative stress in Alzheimer’s disease. Int. J. Mol. Sci. 18, 1583–1596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Butterfield D. A., Boyd-Kimball D., Oxidative stress, amyloid-β peptide, and altered key molecular pathways in the pathogenesis and progression of Alzheimer’s disease. J. Alzheimers Dis. 62, 1345–1367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peña-Bautista C., López-Cuevas R., Cuevas A., Baquero M., Cháfer-Pericás C., Lipid peroxidation biomarkers correlation with medial temporal atrophy in early Alzheimer Disease. Neurochem. Int. 129, 104519 (2019). [DOI] [PubMed] [Google Scholar]
- 25. Zabel M., et al. , Markers of oxidative damage to lipids, nucleic acids and proteins and antioxidant enzymes activities in Alzheimer’s disease brain: A meta-analysis in human pathological specimens. Free Radic. Biol. Med. 115, 351–360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bradley-Whitman M. A., Lovell M. A., Biomarkers of lipid peroxidation in Alzheimer disease (AD): An update. Arch. Toxicol. 89, 1035–1044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moreira P. I., et al. , “The key role of oxidative stress in Alzheimer’s disease” in Oxidative Stress and Neurodegenerative Disorders, Qureshi G. A., Parvez S. H., Eds. (Elsevier, 2007), pp. 267–281. [Google Scholar]
- 28. Cheignon C., et al. , Oxidative stress and the amyloid beta peptide in Alzheimer’s disease. Redox Biol. 14, 450–464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Regen F., Hellmann-Regen J., Costantini E., Reale M., Neuroinflammation and Alzheimer’s disease: Implications for microglial activation. Curr. Alzheimer Res. 14, 1140–1148 (2017). [DOI] [PubMed] [Google Scholar]
- 30. Gray S. C., Kinghorn K. J., Woodling N. S., Shifting equilibriums in Alzheimer’s disease: The complex roles of microglia in neuroinflammation, neuronal survival and neurogenesis. Neural Regen. Res. 15, 1208–1219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walsh J., et al. , Identification and quantification of the basal and inducible Nrf2-dependent proteomes in mouse liver: Biochemical, pharmacological and toxicological implications. J. Proteomics 108, 171–187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Di Paolo G., Kim T.-W., Linking lipids to Alzheimer’s disease: Cholesterol and beyond. Nat. Rev. Neurosci. 12, 284–296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alzheimer A., Uber eine eigenartige Erkrankung der Hirnrinde. Allg. Zeitschrift Rsychiatrie Psych. Medizine 64, 146–148 (1907). [Google Scholar]
- 34. Alzheimer A., Stelzmann R. A., Schnitzlein H. N., Murtagh F. R., An English translation of Alzheimer’s 1907 paper, “Uber eine eigenartige Erkankung der Hirnrinde”. Clin. Anat. 8, 429–431 (1995). [DOI] [PubMed] [Google Scholar]
- 35. Van Den Brink D. M., et al. , Physiological and pathological roles of FATP-mediated lipid droplets in Drosophila and mice retina. PLoS Genet. 14, e1007627 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu L., et al. , Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell 160, 177–190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smolič T., et al. , Astrocytes in stress accumulate lipid droplets. Glia 69, 1540–1562 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marschallinger J., et al. , Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 23, 194–208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin G., Wang L., Marcogliese P. C., Bellen H. J., Sphingolipids in the pathogenesis of Parkinson’s disease and Parkinsonism. Trends Endocrinol. Metab. 30, 106–117 (2019). [DOI] [PubMed] [Google Scholar]
- 40. Chung H. L., et al. , Loss- or gain-of-function mutations in ACOX1 cause axonal loss via different mechanisms. Neuron 106, 589–606.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reed T. T., Lipid peroxidation and neurodegenerative disease. Free Radic. Biol. Med. 51, 1302–1319 (2011). [DOI] [PubMed] [Google Scholar]
- 42. Freeman M. R., Drosophila Central Nervous System Glia (Cold Spring Harbor Laboratory Press, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boehm-Cagan A., et al. , ABCA1 agonist reverses the ApoE4-driven cognitive and brain pathologies. J. Alzheimers Dis. 54, 1219–1233 (2016). [DOI] [PubMed] [Google Scholar]
- 44. Hafiane A., Bielicki J. K., Johansson J. O., Genest J., Novel apo E-derived ABCA1 agonist peptide (CS-6253) promotes reverse cholesterol transport and induces formation of preβ-1 HDL in vitro. PLoS One 10, e0131997 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wahrle S. E., et al. , ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J. Biol. Chem. 279, 40987–40993 (2004). [DOI] [PubMed] [Google Scholar]
- 46. Turton J., Morgan K., “ATP-binding cassette, subfamily A (ABC1), member 7 (ABCA7)” in Genetic Variants in Alzheimer’s Disease, Morgan K., Carrasquillo M., Eds. (Springer, New York, 2013), pp. 135–158. [Google Scholar]
- 47. Tarling E. J., de Aguiar Vallim T. Q., Edwards P. A., Role of ABC transporters in lipid transport and human disease. Trends Endocrinol. Metab. 24, 342–350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen Q., et al. , Influence of four polymorphisms in ABCA1 and PTGS2 genes on risk of Alzheimer’s disease: A meta-analysis. Neurol. Sci. 37, 1209–1220 (2016). [DOI] [PubMed] [Google Scholar]
- 49. Teresa J. C., et al. , Association of genetic variants of ABCA1 with susceptibility to dementia: (SADEM study). Metab. Brain Dis. 35, 915–922 (2020). [DOI] [PubMed] [Google Scholar]
- 50. Fehér Á., et al. , ABCA1 rs2230805 and rs2230806 common gene variants are associated with Alzheimer’s disease. Neurosci. Lett. 664, 79–83 (2018). [DOI] [PubMed] [Google Scholar]
- 51. Van den Bossche T., et al. ; Belgian Neurology Consortium, Phenotypic characteristics of Alzheimer patients carrying an ABCA7 mutation. Neurology 86, 2126–2133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chang Y. T., et al. , ABCA7 polymorphisms correlate with memory impairment and default mode network in patients with APOEε4-associated Alzheimer’s disease. Alzheimers Res. Ther. 11, 103–113 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aikawa T., Holm M. L., Kanekiyo T., ABCA7 and pathogenic pathways of Alzheimer’s disease. Brain Sci. 8, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jaiswal M., Sandoval H., Zhang K., Bayat V., Bellen H. J., Probing mechanisms that underlie human neurodegenerative diseases in Drosophila. Annu. Rev. Genet. 46, 371–396 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lane-Donovan C., Herz J., ApoE, ApoE receptors, and the synapse in Alzheimer’s disease. Trends Endocrinol. Metab. 28, 273–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Herz J., Apolipoprotein E receptors in the nervous system. Curr. Opin. Lipidol. 20, 190–196 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rodríguez-Vázquez M., Vaquero D., Parra-Peralbo E., Mejía-Morales J. E., Culi J., Drosophila lipophorin receptors recruit the lipoprotein LTP to the plasma membrane to mediate lipid uptake. PLoS Genet. 11, e1005356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Muhammad A., et al. , Retromer deficiency observed in Alzheimer’s disease causes hippocampal dysfunction, neurodegeneration, and Abeta accumulation. Proc. Natl. Acad. Sci. U.S.A. 105, 7327–7332 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Berman D. E., Ringe D., Petsko G. A., Small S. A., The use of pharmacological retromer chaperones in Alzheimer’s disease and other endosomal-related disorders. Neurotherapeutics 12, 12–18 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang S., et al. , The retromer complex is required for rhodopsin recycling and its loss leads to photoreceptor degeneration. PLoS Biol. 12, e1001847 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Van Acker Z. P., Bretou M., Annaert W., Endo-lysosomal dysregulations and late-onset Alzheimer’s disease: Impact of genetic risk factors. Mol. Neurodegener. 14, 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nelson P. T., Fardo D. W., Katsumata Y., The MUC6/AP2A2 locus and its relevance to Alzheimer’s disease: A review. J. Neuropathol. Exp. Neurol. 79, 568–584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shen R., et al. , Upregulation of RIN3 induces endosomal dysfunction in Alzheimer’s disease. Transl. Neurodegener. 9, 26–44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gan Q., Watanabe S., Synaptic vesicle endocytosis in different model systems. Front. Cell. Neurosci. 12, 171–197 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Seto E. S., Bellen H. J., Lloyd T. E., When cell biology meets development: Endocytic regulation of signaling pathways. Genes Dev. 16, 1314–1336 (2002). [DOI] [PubMed] [Google Scholar]
- 66. Takei K., Haucke V., Clathrin-mediated endocytosis: Membrane factors pull the trigger. Trends Cell Biol. 11, 385–391 (2001). [DOI] [PubMed] [Google Scholar]
- 67. Kaksonen M., Roux A., Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 19, 313–326 (2018). [DOI] [PubMed] [Google Scholar]
- 68. Ioannou M. S., Liu Z., Lippincott-Schwartz J., A neuron-glia co-culture system for studying intercellular lipid transport. Curr. Protoc. Cell Biol. 84, e95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Verghese P. B., et al. , ApoE influences amyloid-β (Aβ) clearance despite minimal apoE/Aβ association in physiological conditions. Proc. Natl. Acad. Sci. U.S.A. 110, E1807–E1816 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ermondi G., et al. , Lipophilicity of amyloid β-peptide 12-28 and 25-35 to unravel their ability to promote hydrophobic and electrostatic interactions. Int. J. Pharm. 495, 179–185 (2015). [DOI] [PubMed] [Google Scholar]
- 71. Sharman M. J., et al. , APOE genotype results in differential effects on the peripheral clearance of amyloid-β42 in APOE knock-in and knock-out mice. J. Alzheimers Dis. 21, 403–409 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lanfranco M. F., Ng C. A., Rebeck G. W., ApoE lipidation as a therapeutic target in Alzheimer’s disease. Int. J. Mol. Sci. 21, 6336 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Koldamova R., Staufenbiel M., Lefterov I., Lack of ABCA1 considerably decreases brain ApoE level and increases amyloid deposition in APP23 mice. J. Biol. Chem. 280, 43224–43235 (2005). [DOI] [PubMed] [Google Scholar]
- 74. Wahrle S. E., et al. , Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J. Clin. Invest. 118, 671–682 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bradley M. A., Xiong-Fister S., Markesbery W. R., Lovell M. A., Elevated 4-hydroxyhexenal in Alzheimer’s disease (AD) progression. Neurobiol. Aging 33, 1034–1044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chouhan A. K., et al. , Uncoupling neuronal death and dysfunction in Drosophila models of neurodegenerative disease. Acta Neuropathol. Commun. 4, 62–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li N., et al. , Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 278, 8516–8525 (2003). [DOI] [PubMed] [Google Scholar]
- 78. Oakley H., et al. , Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sanders L. H., Timothy Greenamyre J., Oxidative damage to macromolecules in human Parkinson disease and the rotenone model. Free Radic. Biol. Med. 62, 111–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ferrari M., et al. , Hypoxia treatment reverses neurodegenerative disease in a mouse model of Leigh syndrome. Proc. Natl. Acad. Sci. U.S.A. 114, E4241–E4250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sillitoe R. V., Stephen D., Lao Z., Joyner A. L., Engrailed homeobox genes determine the organization of Purkinje cell sagittal stripe gene expression in the adult cerebellum. J. Neurosci. 28, 12150–12162 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Krimbou L., et al. , Molecular interactions between apoE and ABCA1: Impact on apoE lipidation. J. Lipid Res. 45, 839–848 (2004). [DOI] [PubMed] [Google Scholar]
- 83. Boehm-Cagan A., et al. , Differential effects of apoE4 and activation of ABCA1 on brain and plasma lipoproteins. PLoS One 11, e0166195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Shinohara M., Tachibana M., Kanekiyo T., Bu G., Role of LRP1 in the pathogenesis of Alzheimer’s disease: Evidence from clinical and preclinical studies. J. Lipid Res. 58, 1267–1281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bertram L., McQueen M. B., Mullin K., Blacker D., Tanzi R. E., Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat. Genet. 39, 17–23 (2007). [DOI] [PubMed] [Google Scholar]
- 86. Vardarajan B. N., et al. , Identification of Alzheimer disease-associated variants in genes that regulate retromer function. Neurobiol. Aging 33, 2231.e15–2231.e30 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Steinberg S., et al. ; DemGene, Loss-of-function variants in ABCA7 confer risk of Alzheimer’s disease. Nat. Genet. 47, 445–447 (2015). [DOI] [PubMed] [Google Scholar]
- 88. Vasquez J. B., Simpson J. F., Harpole R., Estus S., Alzheimer’s disease genetics and ABCA7 splicing. J. Alzheimers Dis. 59, 633–641 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Singh A., Kukreti R., Saso L., Kukreti S., Oxidative stress: A key modulator in neurodegenerative diseases. Molecules 24, 1583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Burnside S. W., Hardingham G. E., Transcriptional regulators of redox balance and other homeostatic processes with the potential to alter neurodegenerative disease trajectory. Biochem. Soc. Trans. 45, 1295–1303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wojtunik-Kulesza K. A., Oniszczuk A., Oniszczuk T., Waksmundzka-Hajnos M., The influence of common free radicals and antioxidants on development of Alzheimer’s disease. Biomed. Pharmacother. 78, 39–49 (2016). [DOI] [PubMed] [Google Scholar]
- 92. Mullan K., et al. , Serum concentrations of vitamin E and carotenoids are altered in Alzheimer’s disease: A case-control study. Alzheimers Dement. (N. Y.) 3, 432–439 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Vina J., LLoret A., Giraldo E., Badia M. C., Alonso M. D., Antioxidant pathways in Alzheimers disease: Possibilities of intervention. Curr. Pharm. Des. 17, 3861–3864 (2011). [DOI] [PubMed] [Google Scholar]
- 94. Frank B., Gupta S., A review of antioxidants and Alzheimer’s disease. Ann. Clin. Psychiatry 17, 269–286 (2005). [DOI] [PubMed] [Google Scholar]
- 95. Ramjaun A. R., Micheva K. D., Bouchelet I., McPherson P. S., Identification and characterization of a nerve terminal-enriched amphiphysin isoform. J. Biol. Chem. 272, 16700–16706 (1997). [DOI] [PubMed] [Google Scholar]
- 96. Takeda T., et al. , Dynamic clustering of dynamin-amphiphysin helices regulates membrane constriction and fission coupled with GTP hydrolysis. eLife 7, e30246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lee E., et al. , Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science 297, 1193–1196 (2002). [DOI] [PubMed] [Google Scholar]
- 98. Zelhof A. C., et al. , Drosophila Amphiphysin is implicated in protein localization and membrane morphogenesis but not in synaptic vesicle endocytosis. Development 128, 5005–5015 (2001). [DOI] [PubMed] [Google Scholar]
- 99. Razzaq A., et al. , Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes Dev. 15, 2967–2979 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chapuis J., et al. , Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol. Psychiatry 18, 1225–1234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ubelmann F., et al. , Bin1 and CD2AP polarise the endocytic generation of beta-amyloid. EMBO Rep. 18, 102–122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Harrison B. J., et al. , The adaptor protein CD2AP is a coordinator of neurotrophin signaling-mediated axon arbor plasticity. J. Neurosci. 36, 4259–4275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Furusawa K., et al. , CD2-associated protein (CD2AP) overexpression accelerates amyloid precursor protein (APP) transfer from early endosomes to the lysosomal degradation pathway. J. Biol. Chem. 294, 10886–10899 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ojelade S. A., et al. , cindr, the Drosophila homolog of the CD2AP Alzheimer’s disease risk gene, is required for synaptic transmission and proteostasis. Cell Rep. 28, 1799–1813.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Baig S., et al. , Distribution and expression of picalm in Alzheimer disease. J. Neuropathol. Exp. Neurol. 69, 1071–1077 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhang B., et al. , Synaptic vesicle size and number are regulated by a clathrin adaptor protein required for endocytosis. Neuron 21, 1465–1475 (1998). [DOI] [PubMed] [Google Scholar]
- 107. González-Gaitán M., Jäckle H., Role of Drosophila α-adaptin in presynaptic vesicle recycling. Cell 88, 767–776 (1997). [DOI] [PubMed] [Google Scholar]
- 108. Zhang Y., et al. , Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89, 37–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zhang Y., et al. , An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Vos D., Kuivenhoven J. A., van de Sluis B., Recycling the LDL receptor to combat atherosclerosis. Aging (Albany NY) 10, 3638–3640 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kobayashi D. T., Chen K. S., Behavioral phenotypes of amyloid-based genetically modified mouse models of Alzheimer’s disease. Genes Brain Behav. 4, 173–196 (2005). [DOI] [PubMed] [Google Scholar]
- 112. Butterfield D. A., Castegna A., Lauderback C. M., Drake J., Evidence that amyloid beta-peptide-induced lipid peroxidation and its sequelae in Alzheimer’s disease brain contribute to neuronal death. Neurobiol. Aging 23, 655–664 (2002). [DOI] [PubMed] [Google Scholar]
- 113. Namba Y., Tomonaga M., Kawasaki H., Otomo E., Ikeda K., Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer’s disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res. 541, 163–166 (1991). [DOI] [PubMed] [Google Scholar]
- 114. Strittmatter W. J., et al. , Apolipoprotein E: High-avidity binding to β-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 90, 1977–1981 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kanekiyo T., Xu H., Bu G., ApoE and Aβ in Alzheimer’s disease: Accidental encounters or partners? Neuron 81, 740–754 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Quintana A., et al. , Fatal breathing dysfunction in a mouse model of Leigh syndrome. J. Clin. Invest. 122, 2359–2368 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Assouline Z., et al. , A constant and similar assembly defect of mitochondrial respiratory chain complex I allows rapid identification of NDUFS4 mutations in patients with Leigh syndrome. Biochim. Biophys. Acta 1822, 1062–1069 (2012). [DOI] [PubMed] [Google Scholar]
- 118. Jain I. H., et al. , Hypoxia as a therapy for mitochondrial disease. Science. 352, 54–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Moreno-Grau S., et al. ; GR@ACE consortium; DEGESCO consortium; Alzheimer’s Disease Neuroimaging Initiative, Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer’s disease and three causality networks: The GR@ACE project. Alzheimers Dement. 15, 1333–1347 (2019). [DOI] [PubMed] [Google Scholar]
- 120. Bellenguez C., et al. , New insights on the genetic etiology of Alzheimer’s and related dementia. medRxiv [Preprint] (2020). 10.1101/2020.10.01.20200659 (Accessed 8 December 2021). [DOI]
- 121. L. D. Goodman , H. J. Bellen , Recent insights into the role of glia and oxidative stress in Alzheimer's disease gained from Drosophila. Curr. Opin. Neurobiol. 72, 32–38 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lee P. T., et al. , A gene-specific T2A-GAL4 library for Drosophila. eLife 7, e35574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Heisenberg M., Separation of receptor and lamina potentials in the electroretinogram of normal and mutant Drosophila . J. Exp. Biol. 55, 85–100 (1971). [DOI] [PubMed] [Google Scholar]
- 124. Verstreken P., et al. , Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron 40, 733–748 (2003). [DOI] [PubMed] [Google Scholar]
- 125. Barish S., et al. , Combinations of DIPs and Dprs control organization of olfactory receptor neuron terminals in Drosophila . PLoS Genet. 14, e1007560 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. G. M. J. Beaudoin, 3rd , et al. , Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat. Protoc. 7, 1741–1754 (2012). [DOI] [PubMed] [Google Scholar]
- 127. Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Faul F., Erdfelder E., Buchner A., Lang A. G., Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav. Res. Methods 41, 1149–1160 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the main text and SI Appendix .