Abstract
The SARS-CoV-2 virus is continuously evolving, with appearance of new variants characterized by multiple genomic mutations, some of which can affect functional properties, including infectivity, interactions with host immunity, and disease severity. The rapid spread of new SARS-CoV-2 variants has highlighted the urgency to trace the virus evolution, to help limit its diffusion, and to assess effectiveness of containment strategies. We propose here a PCR-based rapid, sensitive and low-cost allelic discrimination assay panel for the identification of SARS-CoV-2 genotypes, useful for detection in different sample types, such as nasopharyngeal swabs and wastewater. The tests carried out demonstrate that this in-house assay, whose results were confirmed by SARS-CoV-2 whole-genome sequencing, can detect variations in up to 10 viral genome positions at once and is specific and highly sensitive for identification of all tested SARS-CoV-2 clades, even in the case of samples very diluted and of poor quality, particularly difficult to analyze.
Keywords: SARS-CoV-2, VOCs, Clade, Genotyping assay, NGS, Wastewater
1. Introduction
In late 2019 a novel coronavirus, named Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), emerged as the responsible for COVID-19 disease. On January 12, 2020, thanks to next-generation sequencing (NGS) approaches, carried out by different research institutes, the first complete genome of SARS-CoV-2 was determined (Kim et al., 2020) and released via the GISAID (https://www.gisaid.org/CoV2020/) and Nextstrain (https://nextstrain.org/ncov) databases.
Being an RNA virus, SARS-CoV-2 constantly changes through mutations, evolves rapidly, and accumulates genetic diversity in relatively short periods, as a consequence, new variants of the virus are expected to occur over time, which can potentially give it new characteristics respect to its ancestral strain (i.e., higher infection capacity) (Korber et al., 2020). Indeed, starting from November 2020, the spread of new SARS-CoV-2 variants able to infect also younger patients or to cause the disease in people without any apparent previous illness, have been detected and associated with higher transmissibility and severity of symptoms (Khan et al., 2021), highlighting a need to monitor the genetic diversity and epidemiology of SARS-CoV-2 throughout the pandemic.
Currently, many different variants of the SARS-CoV-2 virus are circulating globally; however, only a few of them, based on their impact on public health, are considered variants of concern (VOCs). In late December 2020, the 202012/1 variant, also known as B.1.1.7 (or 20I/501Y.V1 or Alpha) on the base of its phylogenetic lineage was the first VOC described in the United Kingdom (UK); subsequently, the B.1.351 lineage (or 20H/501Y.V2 or Beta) was reported in South Africa. A third VOC, P1 (or 20J/501Y.V3 or Gamma), was reported in Brazil in early January 2021. More recently, from April 2021, the B.1.617 (or Delta) was reported in India (Cascella et al., 2021). All these variants seem to spread more easily and quickly than other ones, which is leading to an increasing number of COVID-19 cases with more hospitalizations, and potentially more deaths (Khan et al., 2021). Moreover, cases characterized by co-infection of 2 genetically distinct SARS-CoV-2 lineages in the same patients, which could contribute to the severity of the disease, have been recently reported (Pedro et al., 2021).
Therefore, early diagnosis of SARS-CoV-2 variants in infected patients is essential to control and trace the dynamics of the COVID-19 pandemic. Currently, the principal method used for the rapid detection of viral RNA is the Reverse transcription‑quantitative polymerase chain reaction (RT‑qPCR), based on primers amplification of conserved viral genome regions (Corman et al., 2020). However, this test, widely used in diagnostic laboratories, is able to detect only positivity to the virus but not to rapidly detect specific SARS-CoV-2 variants. It is, therefore, important to identify new testing strategies able to overcome the limitations of standard RT‑qPCR, in order to improve efficiency and efficacy of epidemiological surveillance strategies. The TaqMan-based detection is an alternative method for COVID-19 monitoring, endowed with improved specificity and sensitivity for detecting SARS-CoV-2 RNA (Xiao et al., 2021).
Although inhalation of droplets is the major transmission route of the virus (Lipsitch et al., 2003), and therefore identification of viral RNA in nasopharyngeal swabs is the method of election to monitor COVID-19 propagation, monitoring the virus in wastewaters is also a useful source of epidemiological data (Lodder and de Roda Husman, 2020). Indeed, methods that allows the identification of SARS-CoV-2 RNA in wastewater offer the opportunity to trace circulation of the virus in a community, whose geographical localization and size can be predetermined by the wastewater sampling method selected, or to make comparisons between different geographic areas and over time to assess, for example, viral genome evolution and diffusion kinetics (Kitajima et al., 2020). In this case, however, the testing power is limited by the high dilution of viral load in wastewaters, that can make it difficult to detect its presence with current methods, unless effective enrichment steps are applied to lower the detection limits (Ikner et al., 2012).
Based on these premises, we describe here a rapid and sensitive SARS-CoV-2 genotyping assay panel, that allows discrimination between wild-type (Wuhan strain) and the most relevant SARS-CoV-2 strains, for their early detection in clinical samples, such as RNA extracted from nasopharyngeal swabs and as well as to wastewater samples.
2. Materials and methods
2.1. Nasopharyngeal sample collection
SARS-CoV-2 RNA samples extracted from nasopharyngeal swabs of anonymous subjects affected by COVID‑19 infection, positively confirmed by RT‑qPCR amplification of SARS-CoV-2, were kindly provided from different Campania Region Hospitals included in the SCIRE (SARS-CoV-2 Italian Research Enterprise) collaborative study (Lai et al., 2021).
2.2. cDNA synthesis
Complementary DNA (cDNA) was obtained via reverse transcription with the SensiFAST cDNA Synthesis Kit (Bioline) starting from 2 ul of RNA and according to the following protocol: 25°C for 10 minute, 42°C for 50 minute, 85°C for 5 minute, 4°C hold.
2.3. Quantitative polymerase chain reaction (qPCR)
SARS-CoV-2 RNA was monitored using qPCR of N gene (Primer Forward: GGGGAACTTCTCCTGCTAGAAT; Primer Revers: CAGACATTTTGCTCTCAAGCTG); before used, they were validated against positive (synthetic SARS-CoV-2 RNA Twist Bioscience, MT007544.1 Control 1) and negative (water) control. The Synthetic SARS-CoV-2 RNA was used to generate a calibration standard curve to quantify SARS-CoV-2 RNA amplified and define the optimal cDNA concentration to use in the assays. qPCR was carried out on the LightCycler 480 System (Roche-Life Science) according to standard procedures. Each reaction was performed in a final volume of 10 µl containing 0.7 pmol/µl of each primer, 5 µl of SensiFAST SYBR Lo-ROX Kit (Bioline) and 1,5 µl of cDNA with a continuous detection at 0.11°C/sec increment for 10 minute to verify the presence of a single amplification product. The results were analysed using the LightCycler 480 Software and exported into Microsoft Excel file for further analysis.
2.4. Genotyping assays for variants screening
The target sequences were submitted to Custom TaqMan Assay Design Tool (Thermo Fisher Scientific) to design appropriate sequence-specific forward and reverse primers and TaqMan probes. Each assay includes 2 probes, a VIC dye labeled probe to detect the reference sequence and a FAM dye labeled probe to detect the alternative sequence. TaqMan assay ID numbers and sequences are listed in Supp. Table 1. Genotyping assays were performed following manufacturer's instruction using 1,5 μl of cDNA mixed with 2X TaqMan Genotyping Master Mix and custom 20X TaqMan probes (Thermo Fisher Scientific) in 10 μl reaction volume. All samples were screened on the LightCycler 480 System (Roche-Life Science) according to standard procedures.
2.5. Wastewater sample collection
Raw sewage samples (wastewater) were collected between December 2020 and April 2021 from different WWTPs (Waste Water Treatment Plant) located in the provinces of Naples and Salerno (Campania Region). Composite samples, representing 24-hour period were collected and stored at 4°C until use.
2.6. Wastewater concentration and RNA extraction
On the arrival, samples underwent a 30 minutes treatment at 56°C to increase the safety of the protocol and then concentrated with polyethylene glycol (PEG) precipitation. 45 ml of each wastewater were centrifuged at 4°C for 30 minute at 4500g to pellet wastewater solids; then clarified wastewater was mixed with 10% PEG and 2.25% NaCl and centrifuged at 4°C for 2 h at 12000g (La Rosa et al., 2021). The pellet was finally resuspended in the appropriate volume of PBS and entirely used for RNA extraction with the Total RNA purification kit (Norgen, biotel corp.), following manufacturer instructions and eluting 50 ul. SARS-CoV-2 RNA presence was evaluated using qPCR of N gene, obtaining a Ct values ranging from 33-38.5.
2.7. Preamplification step
Preamplification was performed for wastewater samples as described by Baak-Pablo et al., 2010. Briefly, to 5 μl of cDNA (out of 20 μl), 5 μl of a mix (final concentration 0.2X) containing all TaqMan probes and 10 μl of preamplification master mix (Applied Biosystems) were added and amplified on thermal cycler machine according to the following program: 10 minute at 95°C, 50 cycles of 15 second at 95°C and 4 minute at 60°C, 10 minute at 99°C, hold at 4°C. PCR amplicons were purified with 2.5X AMPure XP magnetic beads (Beckman Coulter) and 1.5 μl of the eluted product was used for TaqMan assay screening, as described above.
2.8. SARS-CoV-2 whole genome sequencing
SARS-CoV-2 whole genome sequencing was applied to both swab and wastewater samples. Indexed libraries were prepared from 12 µl of RNA swabs and 38 µl of wastewater RNA using a targeted amplicon-based NGS panel protocol by Paragon Genomics according to manufacturer's instruction. Libraries were pooled in equimolar amounts. The pooled samples were sequenced on Illumina NextSeq-500 or Miseq System in a 2 × 150 paired-end reads format at a final loading concentration of 1,5 pmol. The obtained reads were aligned on SARS-CoV-2 genome (primary assembly MN908947.3). Mutations were identified using Freebayes v.1.0.2 (Garrison and Marth, 2012). Clade assignment was performed using COVIDEX (Cacciabue et al., 2020) and Nextclade (https://clades.nextstrain.org/). The swab sequences are available in GISAID EpiCoV Database (Supp. Table 2). The Raw sequencing data wastewater samples have been deposited in the EBI ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) with accession number E-MTAB-11261.
3. Results
3.1. Selection and design of genotyping assay
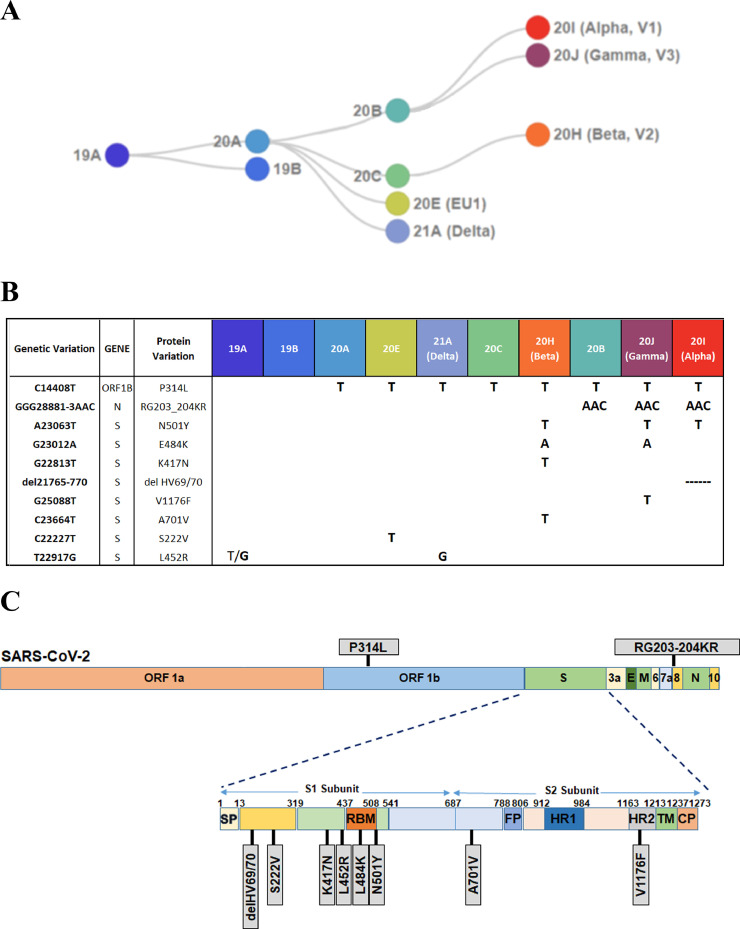
In this study, we have designed, developed, and validated a method for the identification of the main viral strains (Fig. 1 A), starting from RNA extracted from 2 types of matrices, nasopharyngeal swabs and wastewater. The method is based on the use of 10 different TaqMan genotyping assays, each capable of discriminating between WT (Wild-Type) and mutant genotypes (Fig. 1B), selected between the mutations characterizing the main virus lineages circulating and classified as VOCs. The genetic variations tested include point mutations, triplet substitutions and triplet deletions. Among the 10 mutations analyzed, most of them fall in the subunit 1 (S1) of the S gene while 2 are present in subunit 2 (S2); the remaining 2 fall into the N gene and in the ORF1b (Fig. 1C).
Fig. 1.
SARS-CoV-2 strains and specific mutations. (A) Phylogenetic tree reporting the major SARS-CoV-2 circulating clades (adapted from https://clades.nextstrain.org/). (B) Table summarizing 10 different genetic variations screened by TaqMan assays; for each variation, the corresponding gene position, protein variation and clade and VOC are reported. (C) Schematic representation of SARS-CoV-2 genome, containing 11 ORFs (open reading frame) that codify for 27 different proteins. Grey boxes evidence amino acid changes caused by variations. Spike protein, containing the higher number of mutations, is enlarged to highlight the affected subunit (S1, S2) of the protein.
3.2. SARS-CoV-2 genotyping assay performance
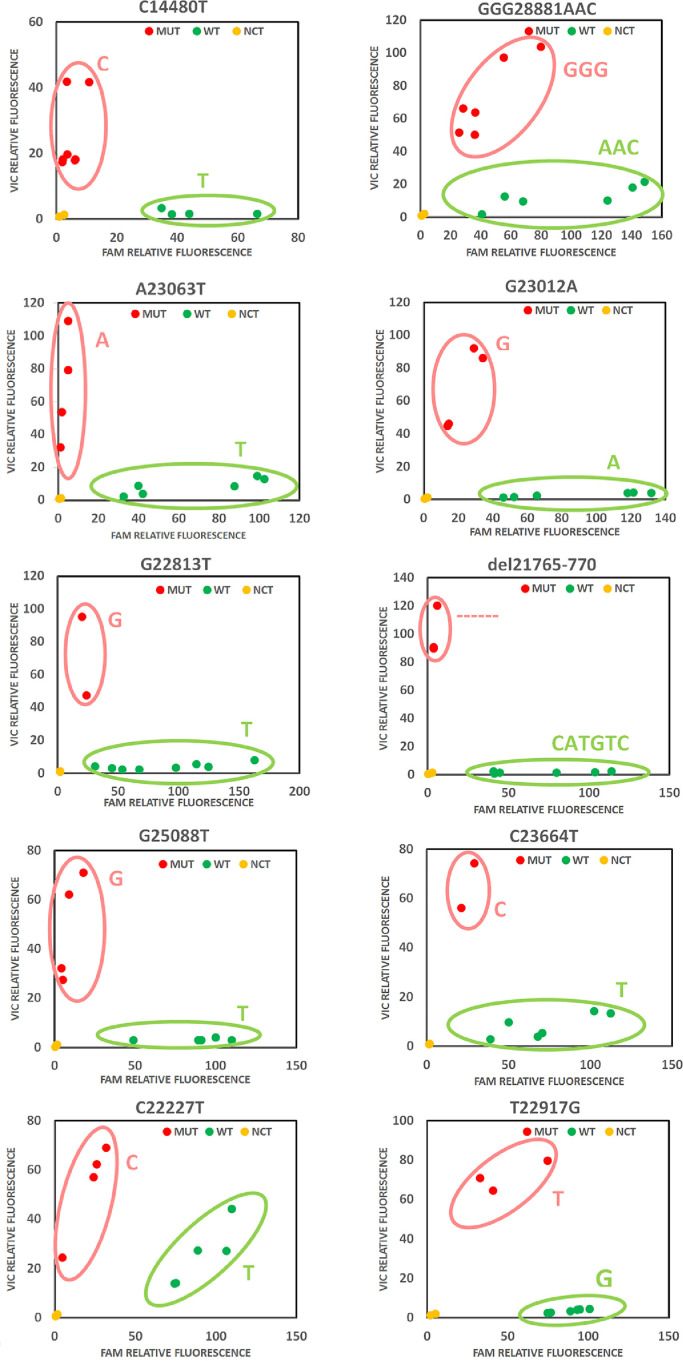
RNAs obtained from swabs samples of COVID-19-patients, previously characterized by SARS-COV-2 whole-genome sequencing (WGS) (Lai et al., 2021) (see Supp. Table 2 for data availability on GISAID database), were used to test the performance of the designed genotyping assay panel. RNAs were tested with the 10 assays (Fig. 1B). The allelic discrimination plots showed for all assays 2 distinct clusters, formed based on the signal intensity ratio of the 2 probes used (VIC and FAM), representing the 2 different genotypes (Fig. 2 and Supp. Table 3). All probes reported achieved a success rate of 100%, correctly discriminating between the 2 alternative sequences with no conflicting results with the sequencing data.
Fig. 2.
SARS-CoV-2 variants detection with genotyping assays. Allelic discrimination plots representing the two different genotypes (green dots for WT, red dots for Mutant, yellow dots for non-template control) screened by 10 TaqMan genotyping assay. For each panel, the tested genomic position is reported above.
Moreover, in order to test the efficiency of every TaqMan genotyping assay, we performed serial dilutions of the swab samples cDNA. Data are reported as scatter plots (Supp. Fig. 1A, 2A, 3A and Supp. Table 4). The different dilutions were also subjected to standard qPCR with primers targeting SARS-CoV-2 N gene in order to obtain absolute quantification of viral copies. Synthetic SARS-CoV-2 RNA of the initial isolated Wuhan strain, bringing no mutations, has been used as a standard for the absolute quantification. As reported in Table 1 efficiency values (Ex) were measured for each probe using the Ct slope method. In this case, for all samples, Ct values derived from FAM or VIC-labeled TaqMan probe amplification of 4-5 dilution points were considered for the linear regression graph generation. Data are reported as FAM or VIC threshold cycles versus Log10 copies/ul (Suppl. Fig 1B-C, 2B-C, 3B-C). Each probe was able to distinguish the variant type even at a low limit of detection (LOD), which is up to high Ct values ranging from 35−37. The amplification efficiency of all tested probes was calculated from the slope of the graph using the following equation:
Table 1.
SARS-CoV-2 genotyping assay panel performance.
| TaqMan assay | Reporter dye detected |
|||
|---|---|---|---|---|
| FAM |
VIC |
|||
| Slope | Efficiency | Slope | Efficiency | |
| C14480T | -3,33 | 2,00 | -3,50 | 1,93 |
| GGG28881AAC | -3,34 | 1,99 | -3,41 | 1,97 |
| A23063T | -3,36 | 1,99 | -3,42 | 1,96 |
| G23012T | -3,39 | 1,97 | -3,38 | 1,98 |
| G22813T | -3,43 | 1,96 | -3,35 | 1,99 |
| del21765-770 | -3,37 | 1,98 | -3,35 | 1,99 |
| G25088T | -3,35 | 1,99 | -3,34 | 1,99 |
| C23664T | -3,34 | 1,99 | -3,35 | 1,99 |
| C22227T | -3,35 | 1,99 | -3,35 | 1,99 |
| T22917G | -3,38 | 1,98 | -3,37 | 1,98 |
Assay performances were evaluated by Ct slope method using serial dilutions of cDNA. In the table are listed the slope and PCR efficiency measured for each genotyping assay in the two florescence channels (FAM and VIC), see also Supp. Fig 1, 2, 3.
As can be noticed, in all cases we found an amplification efficiency ranging from 93 to 99%, with an average of 97%. These results suggest that the developed assays present high sensitivity and reproducibility in SARS-CoV-2 variants detection.
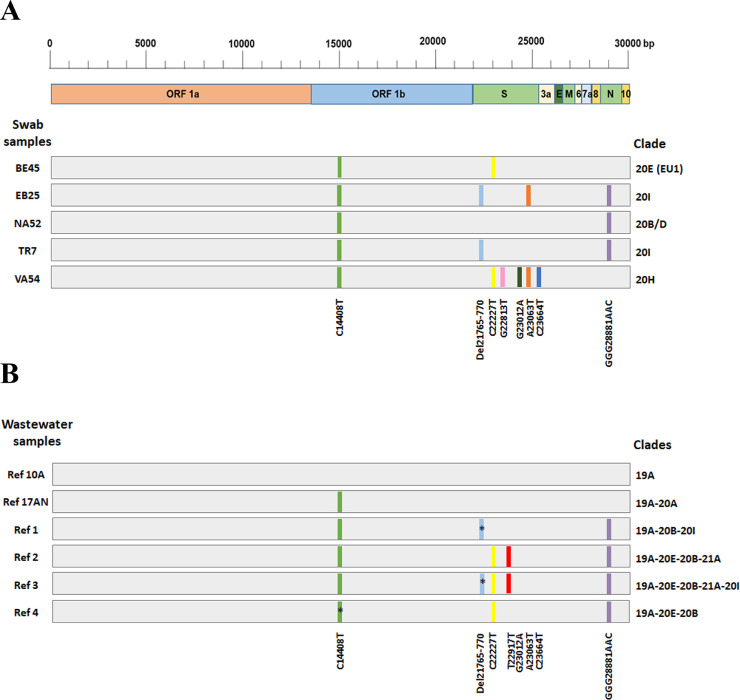
The complete SARS-CoV-2 genotyping panel was tested on 5 swabs to establish the clade assignment ability of our procedure. The results obtained are listed in Supp. Table 6 and summarized in Fig. 3 A. All mutations assed by genotyping assays were confirmed by NGS (see Supp. Table 7). The clade definition was made based on the table reported in Fig. 1B. The panel allowed the correct clade assignment (compare Fig. 3A and Supp. Table 7).
Fig. 3.
Genomic position of SARS-CoV-2 mutations and clade assignment. Schematic representation of the SARS-CoV-2 genome and gene location of the 10 variants (indicated with slash of different colors) tested by TaqMan assay on 5 different swab samples (A) and 6 wastewater samples (B). For each sample (grey bar) the corresponding clade, assigned based on the mutations identified, is reported on the right. Multiple clades in wastewater are due to the presence of several viral strain in the same sample. Black asterisks indicate mutations identified only by the Genotyping assay but not detected by NGS.
3.3. The SARS-CoV-2 genotyping assay panel can also identify co-infections of different virus strains
To assess whether our method was sensitive enough to identify the presence of multiple SARS-CoV-2 strains within the same sample, we analyzed RNAs samples previously tested by NGS that showed mixed viral populations. The selected samples were screened with the specific genotyping assays. The obtained results show that both the WT and variant sequences were efficiently identified (Supp. Fig. 4 and Supp. Table 5). This type of analysis may result very useful for the screening of patients affected by co-infection of different SARS-CoV-2 strains, which could increase the severity of the disease.
3.4. The SARS-CoV-2 genotyping assays panel can identify different virus strains in wastewaters
In order to check if our SARS-CoV-2 genotyping panel was so sensitive to be used also for the screening of low-quality and low-abundant samples, we tested it on wastewater that is generally difficult to analyze. Public health studies show that carrying out tests for the detection of the SARS-CoV-2 in wastewater is an important epidemiological tool for COVID-19 pandemic control. However, this type of sample is generally very difficult to analyze since it contains a mix of very diluted SARS-CoV-2 strains, many contaminants that compromise the sample quality and, above all, the extracted RNA is generally highly degraded. Therefore, in order to be used for NGS or genotyping assays, these samples require different enrichment and purification steps to lower the detection limit of the pathogen virus (see Methods section).
First of all, we tested our SARS-CoV-2 genotyping panel using the standard protocol described before, but we were not able to fully analyze the wastewater samples, as many assays generally result in a non-amplification signal, similar to the NCT (data not shown). Subsequently, we tried improving the cDNA production using SARS-CoV-2 genome-specific primers. Despite these enrichment steps, only in few cases, we were able to assign the correct genotype call for all tested probes. Eventually, we were successful by applying a preamplification step before the genotyping tests (Baak-Pablo et al., 2010). In short, we enriched the wastewater samples for sequences surrounding the target variants, performing a PCR reaction containing all genotyping assays in 1 single mix. This step was performed before the genotyping screening test. This procedure has been originally developed to genotype small amounts of cDNA for expression studies or in genotyping traces of DNA isolated from paraffin-embedded tissue (Baak-Pablo et al., 2010), so we speculated that it could be helpful also in our case. 6 wastewater samples collected from November 2020 and April 2021 were tested with the 10 genotyping assays. As reported in Supp. Table 8, applying this protocol, we were able to detect variants at very low limit (Ct values of qPCR of N gene up to 38.5) and obtained a very good call rate, 97% (58/60) of the tested assays. 55 out 58 calls were also confirmed by NGS analyses (Suppl. Table 9). Complete genotyping results of wastewater samples are summarized in Fig. 3B. The first 2 wastewater samples (10 A and 17AN), showed none or just 1 mutation and then can be classified as 19A and 20A clades. The triple substitution GGG28881AAC was identified in 4 samples collected in April 2021, which is characteristic of clades descendants from 20B. 3 samples showed the typical C22227T substitution of 20E (EU1) clade, also defined as the Spanish variant, that was identified in Spain in early summer 2020 and subsequently spread across Europe (Hodcroft et al., 2021). The deletion of 2 amino acids, H69/V70 (del 21765-770) in the Spike protein, is associated with 20I (Alpha, v1) clade, and in the wastewater samples, have been identified in Ref 1 and Ref 3. This deletion was only identified in genotyping assay, but not by NGS (Supp. Table 8-9), showing the greater sensitivity of this method. The H69/V70 deletion has been reported in over 600,000 SARS-CoV-2 genome sequences worldwide and has seen a strong expansion in Europe, Africa, and Asia (Hodcroft et al., 2021). The 2 samples (Ref 1 and Ref 3) lack the A23063T (N501Y) variant, which is also associated with the 20I clade, results confirmed by NGS. Finally, the T22917G variant has been detected in Ref 2 and Ref 3 (Fig. 3). This genotype is characteristic of 21A (Delta) clade, but can be also found in a subpopulation of the original 19A clade (Fig. 1B).
The data obtained show a prevalence in wastewater of 19A and 20B clades in November and December, and the subsequent diffusion of strains 20E, 20I and 21A in April. This result reproduces very well the data of SARS-CoV-2 variants circulating in Italy obtained from patient swab analysis (Lai et al., 2021). Thereafter, we can assert that our genotyping assay panel, with the addition of the preamplification step, is a valuable method to identify SARS-CoV-2 strains in wastewater samples.
4. Conclusions
We developed an efficient and cost-effective SARS-CoV-2 genotyping panel for rapid identification of the most relevant circulating SARS-CoV-2 variants. The proposed method is a flexible solution that allows the screening of a variable number of samples and it can be, where necessary, modulated and implemented according to the emergence of new viral variants. Thanks to the introduction of a preamplification step, all custom probes resulted highly sensitive also for the detection of samples particularly difficult to analyze, such as wastewater, allowing a very low limit of detection.
The application of this method can be routinely applied to both clinical and environmental samples for SARS-Cov-2 epidemiologic investigations, for monitoring strains diffusion in community and geographical areas.
Declaration of competing interests
The authors report no conflicts of interest relevant to this article
Acknowledgments
Acknowledgements
We thanks Prof. Ivan Gentile, Dr. Nicola Schiano Moriello - PO Malattie Infettive and Prof. Giuseppe Portella, Dr. Michele Cennamo, PO Patologia Clinica of “Federico II” University Hospital, Napoli; Dr Angelo Salomone Megna - U.O.C. Malattie Infettive and Dr. Vincenzo Rocco, Dr. Maurizio Fumi - U.O.C. Patologia Clinica of AORN "San Pio" PO G. Rummo, Benevento; Prof. Pasquale Pagliano - U.O.C. Clinica Infettivologica, Prof. Gianluigi Franci - Programma nella Diagnostica Avanzata delle Resistenze Microbiche and Dr. Emilia Vaccaro - U.O.S.D. NAT e Biologia Molecolare of “San Giovanni di Dio e Ruggi d'Aragona” University Hospital, Salerno; Dr Andreina Baj, Dr Angelo Paolo Genoni of Ospedale di Circolo e Fondazione Macchi, Dip. di Medicina e Chirurgia, Università degli Studi dell'Insubria, Varese; Dr. Gregorio Goffredi, Dr. Francesca Marciano - G.O.I. Medicina di Laboratorio e Biologia Molecolare of “Maria Santissima Addolorata” Hospital, Eboli – Salerno for providing nasopharyngeal swab samples.
Funding
Work supported by Regione Campania (grants ‘Monitoring the spread and genomic variability of the COVID 19 virus in Campania using NGS technology’, POR Campania FESR2014/2020, CUP:B14I20001980006 and ‘GENOMAeSALUTE’, POR Campania FESR2014/2020, azione 1.5; CUP:B41C17000080007) and DIPMED University of Salerno (grants FARB) to FR, OM and AW; EA is a fellow of Fondazione “U. Veronesi”. CF, IT, JL, VMC are PhD students of the Research Doctorates in 'Molecular and Translational Oncology and Innovative Medical-Surgical Technologies’, University of Catanzaro “Magna Graecia” and Veterinary Sciences of the University of Napoli “Federico II”.
Author contribution
Ylenia D'Agostino: Methodology, Investigation, Writing - Original Draft; Teresa Rocco: Methodology, Investigation; Carlo Ferravante: Formal analysis; Amalia Porta: Methodology, Writing - Review & Editing; Alessandra Tosco: Methodology, Writing - Review & Editing; Valeria Mirici Cappa: Formal analysis; Jessica Lamberti: Investigation; Elena Alexandrova: Investigation; Domenico Memoli: Data Curation; Ilaria Terenzi: Investigation; Concetta Pironti: Investigation; Oriana Motta: Resources, Funding acquisition; Alessandro Weisz: Conceptualization; Project administration, Funding acquisition; Giorgio Giurato: Conceptualization, Data Curation; Francesca Rizzo: Conceptualization, Funding acquisition, Writing - Original Draft;
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.diagmicrobio.2021.115632.
Appendix. Supplementary materials
References
- Baak-Pablo R, Dezentje V, Guchelaar HJ, van der Straaten T. Genotyping of DNA samples isolated from formalin-fixed paraffin-embedded tissues using preamplification. J Mol Diagn. 2010;12:746–749. doi: 10.2353/jmoldx.2010.100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciabue M, Aguilera P, Gismondi MI, Taboga O. Covidex: an ultrafast and accurate tool for virus subtyping. bioRxiv. 2020 doi: 10.1101/2020.08.21.261347. 2020.08.21.261347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M, Rajnik M, Aleem A, Dulebohn SC, Di Napoli R. StatPearls; Treasure IslandFL: 2021. Features, Evaluation, and Treatment of Coronavirus (COVID-19) [PubMed] [Google Scholar]
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:12073907 2012.
- Hodcroft EB, Zuber M, Nadeau S, Vaughan TG, Crawford KHD, Althaus CL, et al. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature. 2021;595:707–712. doi: 10.1038/s41586-021-03677-y. [DOI] [PubMed] [Google Scholar]
- Ikner LA, Gerba CP, Bright KR. Concentration and recovery of viruses from water: a comprehensive review. Food Environ Virol. 2012;4:41–67. doi: 10.1007/s12560-012-9080-2. [DOI] [PubMed] [Google Scholar]
- Khan A, Zia T, Suleman M, Khan T, Ali SS, Abbasi AA, et al. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J Cell Physiol. 2021;236:7045–7057. doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M, Ahmed W, Bibby K, Carducci A, Gerba CP, Hamilton KA, et al. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G, Brandtner D, Mancini P, Veneri C, Bonanno Ferraro G, Bonadonna L, et al. Key SARS-CoV-2 mutations of alpha, gamma, and eta variants detected in urban wastewaters in Italy by long-read amplicon sequencing based on nanopore technology. Water. 2021;13(18):2503. [Google Scholar]
- Lai A, Bergna A, Menzo S, Zehender G, Caucci S, Ghisetti V, et al. Circulating SARS-CoV-2 variants in Italy, October 2020-March 2021. Virol J. 2021;18:168. doi: 10.1186/s12985-021-01638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M, Cohen T, Cooper B, Robins JM, Ma S, James L, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W, de Roda Husman AM. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol Hepatol. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedro N, Silva CN, Magalhaes AC, Cavadas B, Rocha AM, Moreira AC, et al. Dynamics of a dual SARS-CoV-2 lineage co-infection on a prolonged viral shedding COVID-19 case: insights into clinical severity and disease duration. Microorganisms. 2021;9 doi: 10.3390/microorganisms9020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Li Z, Wang X, Wang Y, Wang G, Ren L, et al. Comparison of three TaqMan real-time reverse transcription-PCR assays in detecting SARS-CoV-2. J Virol Methods. 2021;288 doi: 10.1016/j.jviromet.2020.114030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.