Abstract
Streptococcus oligofermentans is an early colonizer of the oral microbiome with documented bactericidal activity against the oral pathogen Streptococcus mutans. S. oligofermentans has been observed to possess the typical comABCDE competence regulon found within most oral streptococci, however the competence stimulating peptide (CSP) responsible for QS activation and the regulatory role of the competence regulon have yet to be explored. Herein, we have both confirmed the identity of the S. oligofermentans CSP and utilized a wide range of phenotypic assays to characterize its regulatory role in competence, biofilm formation, and hydrogen peroxide formation. To determine the importance of each amino acid residue in CSP:ComD binding, we performed systematic replacement of amino acid residues within the S. oligofermentans CSP and developed a luciferase-based reporter system to assess the ability of these mutated analogs to modulate the competence regulon. Additionally, we performed CD analysis on mutated CSP analogs to determine the correlation between peptide secondary structure and QS activation. To further explore S. oligofermentans’ potential as a biotherapeutic against S. mutans infection, lead QS activators and inhibitors were used in interspecies competition assays to assess the effect of QS modulation on interactions between these two species. Lastly, we have documented a lack of S. oligofermentans induced cytotoxicity, highlighting the potential of this native flora as a biotherapeutic with minimal health risks.
Keywords: Quorum sensing, streptococcus, peptide, biotherapeutic, antibiotic resistance
TOC Figure:
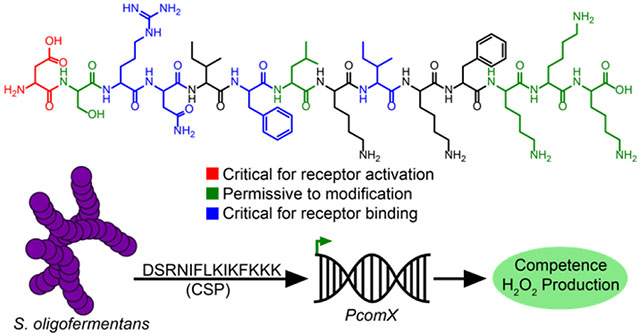
Introduction
The human oral cavity is colonized by a diverse array of microorganisms, including at least six different distinct groups of streptococci.1–3 Many of these oral streptococci act as opportunistic pathogens and several species, including Streptococcus mutans, are primary causative agents of dental carries, a condition that in the US alone results in over $100 billion dollars a year in medical costs.4–7 As streptococci are among the earliest colonizers of the oral microenvironment, they play an important role in the development and final composition of the oral microbiome.1, 8–9 In many cases, streptococci, such as S. mutans, produce bacteriocins or other chemical compounds, as well as complex biofilms, all of which can impede the growth of other members of the oral microbiome, aiding in their colonization of the microenvironment.10–11 Of particular interest is the ability of S. mutans to outcompete other oral streptococci through the production of high amounts of lactic acid, acidifying the surrounding microenvironment and resulting in the killing of competing bacteria.10–12
A relatively newly described member of the oral microbiome is Streptococcus oligofermentans, a member of the mitis group of streptococci that was first isolated from caries-free patients in China.13–14 Compared to other oral streptococci, S. oligofermentans is relatively non-pathogenic, having only been observed in low incidence among patients with bacterial endocarditis.15 Despite its lack of bacterial pathogenesis, S. oligofermentans plays an important role within the oral microbiome, making it of particular interest to study. Specifically, there has been shown to be a decrease in the amount of S. mutans in the presence of S. oligofermentans within the oral microbiome.5, 16–19 Tong and coworkers elucidated that S. oligofermentans possesses lactic acid oxidase (lox) genes that convert lactic acid produced by S. mutans into hydrogen peroxide (H2O2), contributing to the hydrogen peroxide-mediated killing of S. mutans.5 This relatively unique quality suggests that it may be possible to utilize S. oligofermentans as a biotherapeutic against S. mutans, preventing the formation of dental caries. However, the expression of this desired quality within S. oligofermentans is relatively understudied, and there is little information of how S. oligofermentans regulates other phenotypes that have been recognized to play an important role in the colonization of the oral microbiome by other streptococci.
It has been well established that many oral streptococci use a process known as quorum sensing (QS) to regulate the expression of important phenotypes including biofilm formation, bacterial competence, and bacteriocin production to aid in colonization of the oral microbiome.20–25 QS is a method of communication used by both gram-positive and gram-negative bacteria to coordinate the expression of group behavior phenotypes when the bacterial cell density reaches a threshold concentration.20–23, 25 Gram-negative bacteria typically use small molecule N-acyl-homoserine lactones (AHLs), whereas gram-positive bacteria utilize autoinducing peptides (AIPs) as their primary signaling molecule to active QS.25–27 In streptococci the main QS circuitry, termed the competence, or ComABCDE, regulon, is comprised of five different components, and is centered around an AIP known as the competence stimulating peptide (CSP).28–29 Typically, the CSP pro-peptide (ComC) is processed and secreted by an ABC transporter (ComAB) as the mature CSP signal.28 Once a minimal threshold level of the CSP signal is reached in the extracellular environment, the CSP binds and activates the transmembrane histidine-kinase receptor (ComD).28 This binding event triggers phosphorylation of a response regulator (ComE), resulting in further expression of QS genes (comABCDE) and the expression of the alternative sigma factor ComX, the master regulator of competence associated genes and phenotypes (Figure 1).28–29
Figure 1.
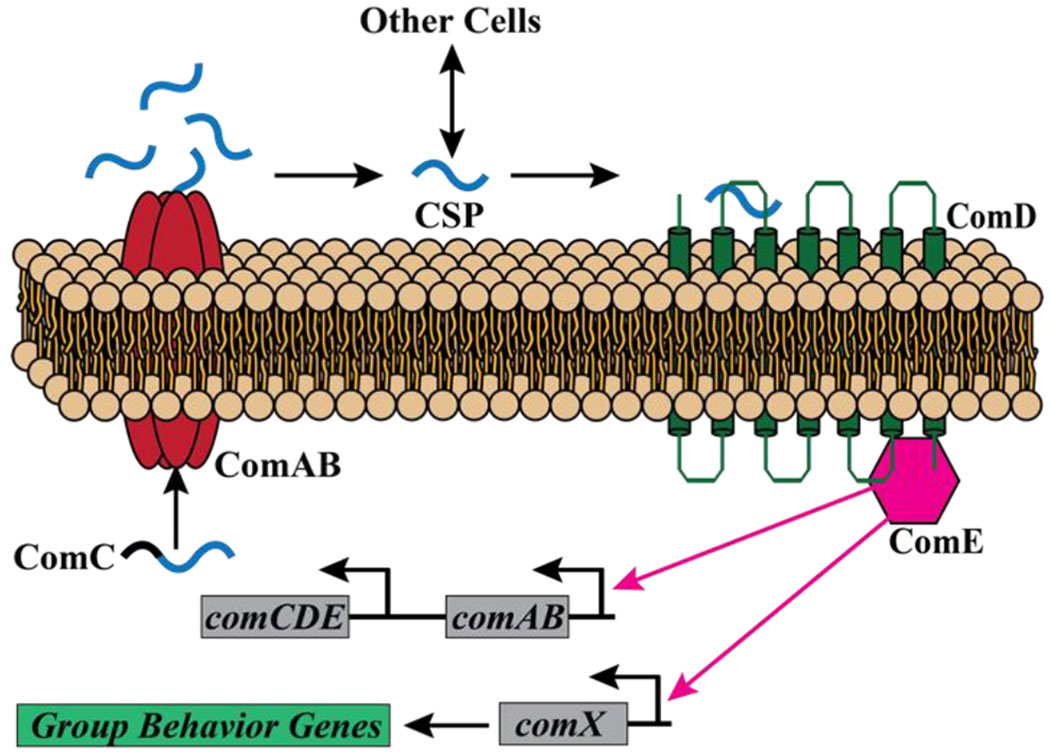
The general streptococcal quorum sensing pathway. ComC is processed and transported to the extracellular environment as the CSP by ComAB. Upon reaching a minimal threshold concentration the CSP binds to its cognate histidine kinase receptor ComD. Activation of ComD leads to phosphorylation of ComE, a response regulator, resulting in further activation of the comABCDE operon and expression of comX, the master regulator of group behavior genes.
The native CSP sequence of S. oligofermentans was previously deduced from genomic analysis, but its identity was not confirmed through isolation from bacterial supernatants. Moreover, the proposed native 14-amino acid S. oligofermentans CSP has been shown to upregulate bacterial competence in a dose-dependent manner through exogenous dosing of the synthetic peptide.14 However, the role of the competence regulon in S. oligofermentans ability to modulate its microenvironment and the expression of other bacterial phenotypes has yet to be explored. In this study we set out to confirm the identity of the S. oligofermentans CSP by isolating it from S. oligofermentans supernatants, characterize the regulatory role of the competence regulon within S. oligofermentans, and to gain a deeper understanding of the molecular mechanisms that drive CSP:ComD binding and subsequent activation of the QS circuitry. To this end, we employed sequencing, protein extraction, and high-resolution mass spectrometry to elucidate the structure of the S. oligofermentans CSP. Following confirmation of the CSP sequence, we utilized several phenotypic assays to determine what role the S. oligofermentans competence regulon plays in the phenotypic expression of bacterial competence, biofilm formation, hydrogen peroxide production, and virulence factor production. To assess the structure activity relationships (SARs) behind CSP:ComD interactions, we performed a full alanine screen of the S. oligofermentans CSP and developed a luciferase based reporter gene assay to measure the degree of QS activation that results from CSP:ComD binding and subsequent ComD activation for all CSP analogs. We then performed structural analysis of all alanine screen analogs using circular dichroism (CD) spectroscopy to further correlate CSP structure and function. Finally, we designed and synthesized a library of second generation truncated and double mutant CSP derivatives based on trends observed in our initial findings. Our analysis revealed that in S. oligofermentans the expression of several key bacterial phenotypes is controlled by the competence regulon. Additionally, we have both identified several residues within the S. oligofermentans CSP that are critical for ComD binding and activation and developed the first CSP derived inhibitors of the S. oligofermentans competence regulon.
RESULTS AND DISCUSSION
Prediction and isolation of the S. oligofermentans CSP from cell-free supernatants
Tong and coworkers have previously predicted the S. oligofermentans CSP based on genomic data, synthesized the predicted 14-amino acid CSP with the sequence DSRNIFLKIKFKKK, and exhibited that the synthetic peptide is capable of inducing competence in S. oligofermantans.14 To ensure that our tested S. oligofermentans strain contained the same CSP sequence, we performed PCR amplification and sequencing of the comC gene as previously described,14 obtaining a predicted sequence that corresponded to that of the previously reported 14-amino acid peptide (Figure 2). Several studies have reported that ComC will sometimes undergo additional processing beyond cleavage at the double glycine repeat to afford a mature CSP signal, either through cleavage of the exported CSP by an extracellular SepM protease,6 or through further processing by the ComAB transporter.30 To ensure that this predicted CSP sequence correlated to that of the mature CSP afforded by processing of ComC by the ComAB transporter, we attempted to isolate the processed CSP from bacterial supernatants. Following precipitation of the excreted crude peptide mixture from cell-free supernatants of S. oligofermentans, we fractionated the total peptides/proteins using reverse-phase high-performance liquid chromatography (RP-HPLC) and observed masses similar to that of the predicted CSP (Figure 3). Further purification of this fraction allowed us to obtain a purified peptide fraction (>97%) with a mass corresponding to the expected mass for the predicted 14-mer CSP, suggesting that this is indeed the mature peptide sequence.
Figure 2.
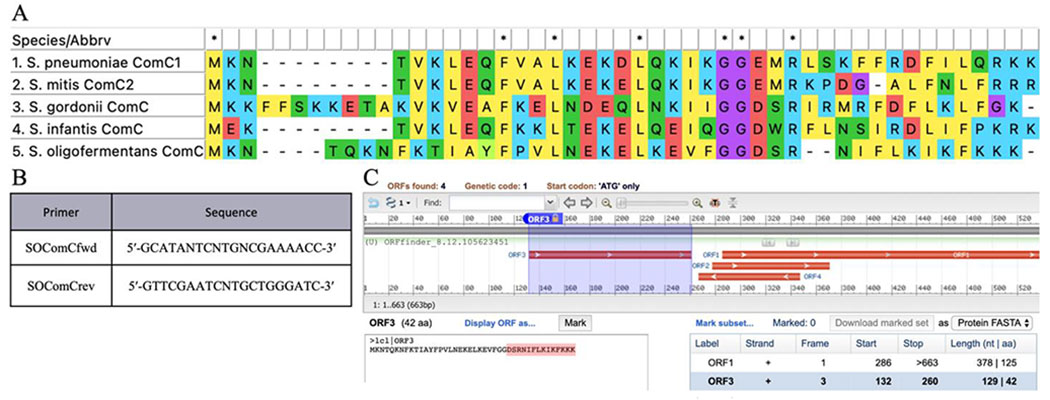
(A) ClustalW alignment of the comC gene of several mitis group streptococci, ComC is typically cleaved following the double glycine repeat to afford a mature CSP signal with a negatively charged N-terminal residue. (B) Primers and probes used for amplification of comC. (C) Identification of the S. oligofermentans CSP following comC sequencing.
Figure 3.
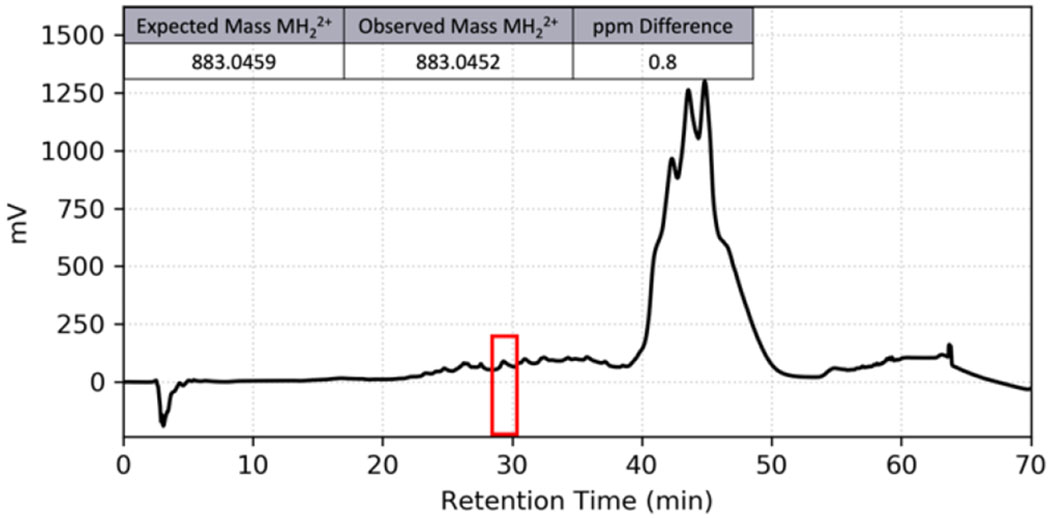
Isolation and detection of the S. oligofermentans CSP from crude cell-free supernatants. RP-HPLC chromatogram of total proteins isolated from the supernatant sample and High Resolution ESI-TOF MS of the fraction collected from 28 to 30 min (red). See the Supporting Information for full experimental details.
Comparison of isolated and synthetic CSP
To further confirm the identity of the S. oligofermentans CSP, we synthesized the predicted CSP sequence using Fmoc solid-phase peptide synthesis (see the Supporting Information for full experimental details). We then compared the purified synthetic peptide to the purified peptide isolated from cell-free supernatants. HPLC analysis of both individual peptides revealed a dominant peak with the same retention time, and combined fractions of these peptides resulted in a single peak that possessed the same retention time as the individual fractions (Figure 4). In addition, ESI-TOF MS analysis of the isolated and synthetic peptides exhibited corresponding exact masses within 5 ppm error of the expected mass of the 14-mer CSP (Figure 4). Lastly, we performed MS/MS analysis of the isolated CSP. The MS/MS results validated the connectivity of the CSP sequence (Figure S-7). Together, these results confirmed that the CSP sequence for S. oligofermentans is the 14-amino acid peptide, DSRNIFLKIKFKKK.
Figure 4.
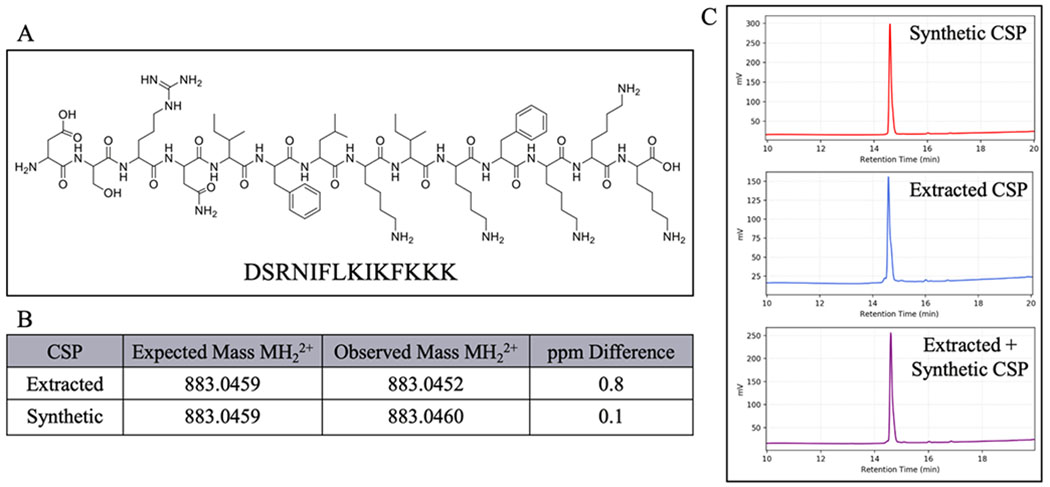
Comparison of purified synthetic and isolated S. oligofermentans CSP. (A) Proposed structure of the 14 amino acid S. oligofermentans CSP. (B) Comparison of masses of synthetic and extracted peptides by ESI-TOF MS (C) Comparison of analytical RP-HPLC chromatograms of purified natural, synthetic, and natural and synthetic CSP.
Phenotypic assays
After confirming the CSP sequence for S. oligofermentans, we set out to determine its regulatory role. Since it has been well established in several species of streptococci that the competence regulon QS pathway is responsible for the regulation of competence, biofilm formation, and virulence factor production, we set out to determine if these processes were under QS control in S. oligofermentans. Additionally, we aimed to determine whether hydrogen peroxide production, the primary means by which S. oligofermentans regulates S. mutans populations within the oral microbiome, was under QS control.
Previous work from Tong and coworkers revealed that bacterial competence was enhanced in response to CSP addition in a dose-dependent manner.14 To observe the effect of exogenous CSP on bacterial competence and confirm these results, we performed transformation assays on S. oligofermentans using four different concentrations of synthetic CSP, 10,000 nM, 1,000 nM, 100 nM, 10 nM, and control with no exogenous CSP. (Figure 5). We observed a dose dependent correlation between CSP concentration and bacterial competence, as the number of transformants was reduced with decreasing CSP concentration, supporting Tong’s earlier observation that competence in S. oligofermentans is under QS control.
Figure 5.

S. oligofermentans competence is dependent upon CSP concentration. Synthetic CSP was added at concentrations of 10,000 nM (A), 1,000 nM (B), 100 nM (C), 10 nM (D), or no exogenous CSP (E). Experiment was repeated three times in triplicate for a total of nine experiments. See Materials and Methods for full experimental details.
As the competence regulon has been determined in other streptococci to regulate biofilm formation, a trait associated with enhanced bacterial pathogenesis, we sought out to determine whether biofilm formation in S. oligofermentans was under QS control. To this end, we performed a crystal violet biofilm quantification assay to assess the formation of bacterial biofilms. Interestingly, our results revealed that under treatment of different exogenous concentrations of CSP (10,000 nM, 1,000 nM, 100 nM, or 10 nM), there was no significant difference in the amounts of biofilms formed compared to untreated wild type S. oligofermentans (Figure 6). As other mitis group streptococci utilize the competence regulon to control biofilm formation, these results cannot entirely rule out the S. oligofermentans QS circuitry playing a role in the expression of this phenotype. Rather, our results indicate that the competence regulon failed to play a significant role in the tested conditions.
Figure 6.
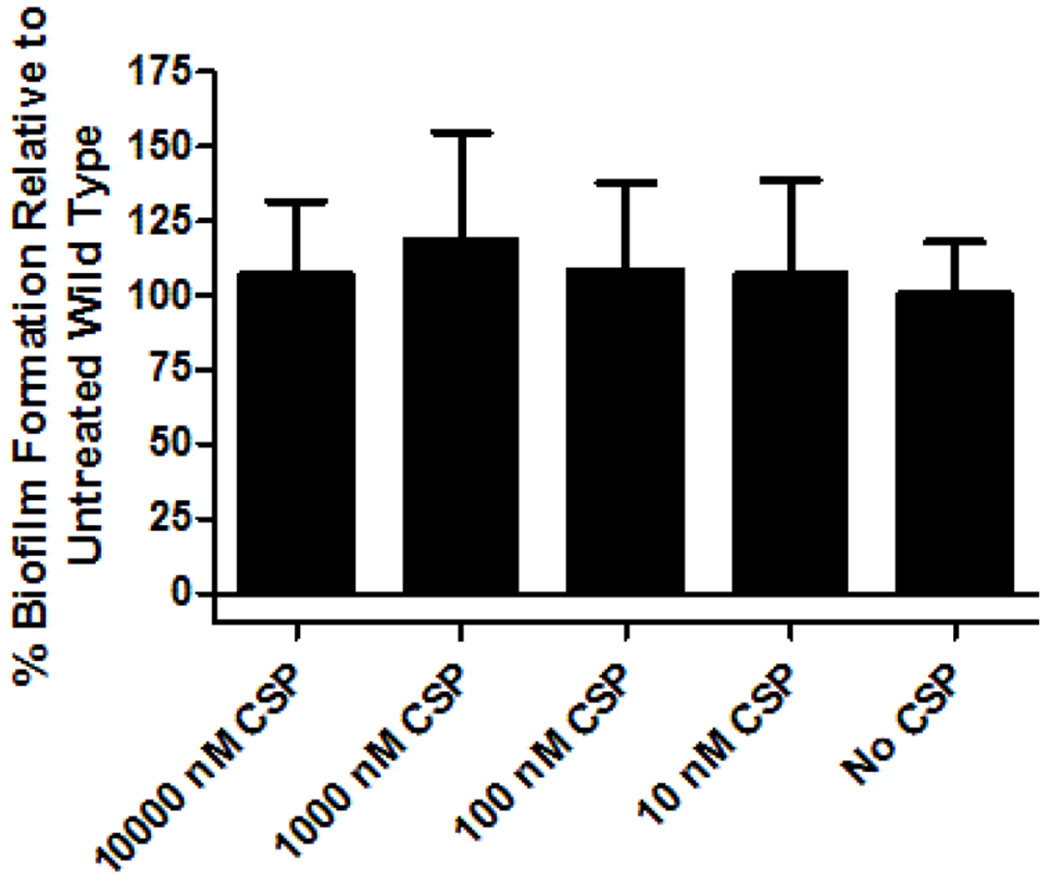
Biofilm formation of S. oligofermentans as a function of CSP concentration. The competence regulon of S. oligofermentans does not appear to have a clear effect on biofilm formation. Experiment was repeated three times in triplicate for a total of nine experiments. See the Supporting Information for full experimental details.
A major aspect of S. oligofermentans that makes it of particular interest is its ability to convert lactic acid to hydrogen peroxide, weaponizing it against other pathogenic oral streptococci including S. mutans. To determine whether this desired capability was under QS control, we assessed the effect of different concentrations of exogenous CSP (10,000 nM, 1,000 nM, 100 nM, or 10 nM) on the ability of S. oligofermentans to produce hydrogen peroxide in THY media containing 10 mM lactic acid, a concentration that Tong and coworkers had previously observed to cause maximal hydrogen peroxide production.5 For CSP concentrations of 100, 1,000, and 10,000 nM, we observed a statistically significant increase in hydrogen peroxide production compared to wild type bacteria untreated with exogenous CSP (Figure 7). These results suggest that the competence regulon is responsible, to some degree, for hydrogen peroxide production in S. oligofermentans, perhaps through downstream upregulation of the lox genes that encode for the lactic acid oxidase that is necessary for hydrogen peroxide production. These results further suggest that hydrogen peroxide production in S. oligofermentans can be modulated through activation of its QS circuitry, potentially providing an approach by which S. oligofermentans can be utilized as a potent biotherapeutic against S. mutans and other pathogenic oral streptococci.
Figure 7.
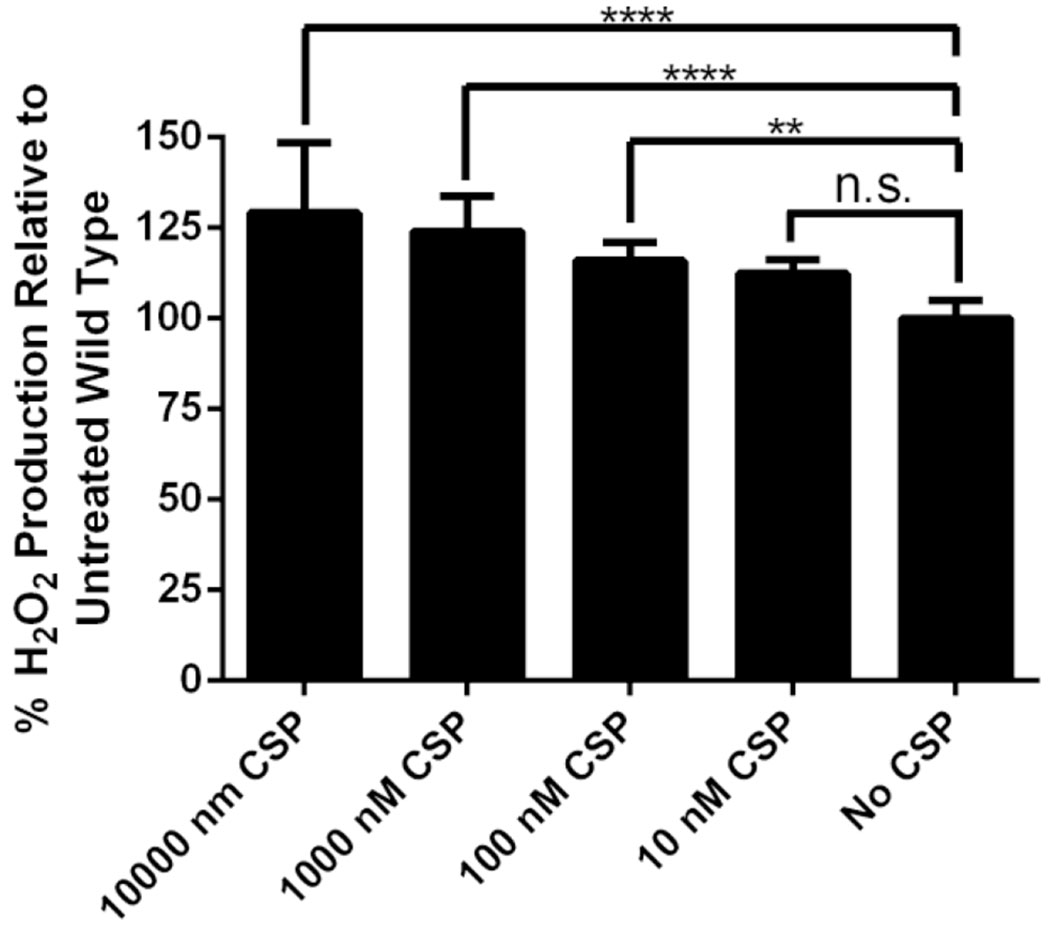
Hydrogen peroxide formation of S. oligofermentans as a function of CSP concentration. Induction of the competence regulon through the addition of exogenous CSP produces a statistically significant increase in the degree of hydrogen peroxide production. Statistical significance was determined using a one-way ANOVA with Bonferroni’s correction; n.s., not significant; **,P≤0.01; ****,P≤0.0001. Experiment was repeated three times in triplicate for a total of nine experiments. See the Supporting Information for full experimental details.
Development of S. oligofermentans luciferase reporter strain
In an effort to gain a deeper understanding of the activity of the native CSP and determine the role of each CSP residue in CSP:ComD binding, a luciferase based S. oligofermentans reporter strain was constructed to directly quantify the degree of comX expression upon CSP:ComD binding using an approach similar to that performed by Salvadori and coworkers.31 First, the primer pair SOComXfwd-SOComXrev (Table S-1) was used to amplify the comX promoter region within S. oligofermentans. Both the amplified comX and the plasmid pFW11-luc (SpecR) were then restriction digested using BamHI and NheI. The digested products were then ligated and cloned into Escherichia coli as previously described by Green and coworkers.32 The purified pFW11-luc plasmid containing the S. oligofermentans comX promoter region was then transformed into S. oligofermentans. Incorporation of the reporter plasmid was validated by observing an increase in luminescence following treatment of S. oligofermentans reporter cultures with CSP and 15 μg/mL D-luciferin, compared to a control untreated with CSP (see the Supporting Information for full experimental details).
Design and synthesis of S. oligofermentans CSP alanine screen analogs
To elucidate the SAR for each residue within the 14-amino acid S. oligofermentans CSP, we performed a full alanine screen of the CSP sequence. All alanine screen CSP analogs were constructed using Fmoc solid-phase peptide synthesis on 4-benzyloxbenzyl alcohol (Wang) resin. Following on-resin synthesis of each alanine screen CSP, peptides were cleaved from the solid support, and purified to homogeneity by semipreparative RP-HPLC (see the Supporting Information for full experimental details and peptide characterization).
SAR of the S. oligofermentans CSP
To determine the role of each sidechain residue of the 14-amino acid S. oligofermentans CSP in ComD receptor binding, the activity of synthesized alanine screen peptide analogs was assessed using luciferase reporter gene assays. The S. oligofermentans CSP can be divided into three distinct regions, the N-terminus, the central region, and the C-terminus. Alanine screening of the CSP N-terminus revealed that residues Arg3 and Asp1 play critical roles in receptor binding and activation, respectively. Alanine replacement of residue Asp1 resulted in the conversion of the S. oligofermentans CSP into a ComD inhibitor (Table 1). This observation is consistent with other mitis group streptococci such as S. pneumoniae, as the negatively charged first residue of the CSP has been shown to be critical for receptor activation.33 Interestingly, alanine substitution of residue Arg3 in other mitis group streptococci including S. pneumoniae33 and S. mitis (unpublished data) has resulted in a complete loss of activity. In the case of the S. oligofermentans CSP, while substitution of Arg3 with alanine resulted in a significantly reduced activity compared to the native CSP, the resulting peptide was not completely inactive. Lastly, alanine substitution of Ser2 resulted in a peptide with comparable activity to the native sequence (Table 1). When compared to other mitis group streptococci, these results suggest that the binding site of the S. oligofermentans CSP N-terminus within ComD is more permissive to CSP side chain modification than its homologues among other mitis group streptococci, as Ser2 modification is permissible and Arg3 substitution does not completely abolish CSP activity, a first for the mitis group streptococci. Similar to previous findings for other mitis group streptococci, the first negatively charged N-terminal amino acid plays a critical role in receptor activation, suggesting the presence of a highly conserved motif at this position.
Table 1.
EC50 or IC50 values of alanine screen analogs against the ComD receptora
| Name | EC50 or IC50* (nM)b | 95% CIc |
|---|---|---|
| CSP | 65.7 | 40.4 - 107 |
| CSP-D1A | 957* | 881 - 1040* |
| CSP-S2A | 105 | 72.1 - 153 |
| CSP-R3A | >1000 | >1000 |
| CSP-N4A | -- | -- |
| CSP-I5A | 270 | 159 - 457 |
| CSP-F6A | -- | -- |
| CSP-L7A | 109 | 72.9 - 163 |
| CSP-K8A | 231 | 176 - 304 |
| CSP-I9A | >1000 | >1000 |
| CSP-K10A | 207 | 113 - 381 |
| CSP-F11A | 288 | 188 - 439 |
| CSP-K12A | 115 | 100 - 131 |
| CSP-K13A | 107 | 52 - 219 |
| CSP-K14A | 156 | 123 - 199 |
See the Materials and Methods section for experimental methods. See Supporting Information for detail of reporter strain and plots of agonism or antagonism dose response curves. All assays performed in triplicate.
EC50 or IC50 values determined by testing peptides over a wide range of concentrations.
95% confidence interval.
EC50 not determined due to the analog’s low induction in primary agonism screening assays. See Supporting Information for details.
There is no clear correlation between side chain composition and receptor binding or activation within the core region of the S. oligofermentans CSP. Substitutions to residues Asn4 and Phe6 resulted in peptides that were completely inactive, while substitution to residue Ile9 resulted in a significant loss of activity, suggesting that these residues are critical for receptor binding (Table 1). Alanine substitutions of residues Ile5, Lys8, Lys10, and Phe11 were relatively tolerated resulting in analogs with only a slight decrease in activity compared with the native CSP (3-4.5-fold decrease), whereas substitution of residue Leu7 produced a peptide with activity comparable to that of the native CSP, suggesting that these residues do not play an important role in receptor binding or activation (Table 1). It was previously observed that signal:receptor recognition in S. pneumoniae was mediated by a hydrophobic patch within the CSP core region.33 A similar trend is not observed within the S. oligofermentans CSP as modification of several side chains that facilitate both polar and hydrophobic interactions lead to a loss of activity, whereas the substitution of other side chains produced CSP analogs with activity comparable to the native sequence. Overall, these findings suggest that the receptor binding site for the core region of the S. oligofermentans CSP is likely more promiscuous than that of the N-terminal region.
The C-terminus of the peptide appears to not have a role in either receptor binding or activation as replacement of Lys12, Lys13, or Lys14 with alanine resulted in analogs with comparable activities to the native CSP signal (Table 1). These results indicate that the entire C-terminus of the peptide is dispensable, however the high conservation of a three amino acid charged tail on all mitis group CSPs indicates this region plays an important evolutionary role, likely to increase the overall hydrophilic properties of these peptides to aid in their solubility.
Circular Dichroism analysis of CSP alanine screen analogs
Our SAR analysis of the S. oligofermentans CSP revealed several residues that are critical for receptor binding or activation; however, this analysis was lacking key information as to how these different modifications affected the overall peptide structure. To gain additional structural information and correlate it to bioactivity, we set out to assess the overall structure of the native S. oligofermentans CSP and its alanine screen analogs using Circular Dichroism (CD) spectroscopy. We assessed all CSP analogs in both aqueous (PBS buffer, pH 7.4) (Figure S-6) and membrane mimicking conditions (20% TFE in PBS, pH 7.4) (Figure 8). In aqueous solution the S. oligofermentans CSP was found to be unfolded, but in membrane mimicking conditions this peptide was found to possess a helical conformation. We quantified the percent helicity of the CSP using the mean residue ellipticity at 222 nm as previously reported and found that 18.9% of the CSP sequence is helical (Figure 8). Further evaluation of the alanine analogs of the S. oligofermentans CSP in membrane mimicking conditions revealed that there is no correlation between the degree of helicity and peptide activity, a finding that was atypical for a mitis group streptococcus, as other members of the group have displayed a clear increase in activity in analogs with a higher degree of helicity.33 Alanine substitution of Asn4 produced a peptide that was completely inactive, yet it possessed a degree of helicity higher than the native peptide and the second highest helicity of all tested analogs (Figure 8). Interestingly, replacement of Arg3 was the only alanine substitution that resulted in an analog that did not form an alpha-helix in membrane mimicking conditions, instead exhibiting a spectrum more indicative of a beta-sheet (Figure 8). It has been widely established that the bioactive conformation of a CSP is alpha-helical, and as peptide CSP-R3A exhibited some degree of receptor activation it is likely that the more hydrophobic nature of this peptide forces it to aggregate in solution at higher concentrations, resulting in the beta-sheet observed in this study, while it still likely binds the ComD receptor as an alpha helix.
Figure 8.
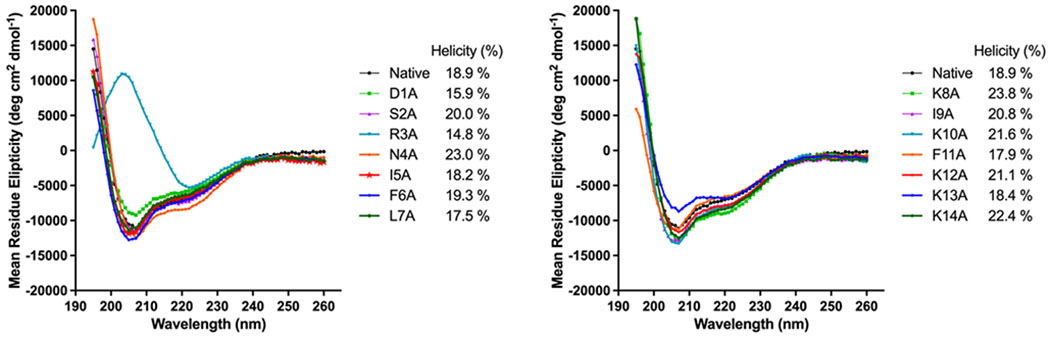
CD spectra of S. oligofermentans alanine screen analogs in membrane mimicking conditions. Most of the alanine screen analogs adopt an alpha helical confirmation in membrane mimicking conditions. However, there does not appear to be a correlation between the degree of alpha helicity and activity.
Second-generation CSP analogs
Our initial analysis of the S. oligofermentans CSP revealed that the N-terminus may be involved in receptor activation, while the C-terminus may be completely dispensable. To further explore these trends in an effort to create enhanced activators and inhibitors of the S. oligofermentans competence regulon, we designed a second-generation library of CSP analogs encompassing sequential truncations of either the three N-terminal residues, or the three C-terminal residues of the native CSP signal. Truncation of one, CSP-des-D1, or two, CSP-des-D1S2, sequential amino acids of the peptide N-terminus produced weak inhibitors of the S. oligofermentans competence regulon (Table 2). Truncation of the entire peptide N-terminus, CSP-des-D1S2R3, produced a peptide that was completely inactive, highlighting the importance of the CSP amino terminus in receptor binding and activation. Moving on to the C-terminus, we determined that truncation of one or two residues produced two peptides with activities comparable to that of the native peptide, CSP-des-K14 and CSP-des-K13K14, whereas truncation of three residues exhibited a more significant decrease in activity. Interestingly, removal of two or three residues from the C-terminus significantly increased the hydrophobic properties of these peptides, as these analogs were found to be harder to dissolve in aqueous solutions, supporting our claim that the purpose of this charged lysine tail is to increase the hydrophilicity of the S. oligofermentans CSP.
Table 2.
EC50 or IC50 values of truncated or double mutation analogs against the ComD receptora
| Name | EC50 or IC50* (nM)b | 95% CIc |
|---|---|---|
| CSP | 65.7 | 40.4 - 107 |
| CSP-des-D1 | >1000* | >1000* |
| CSP-des-D1S2 | >1000* | >1000* |
| CSP-des-D1S2R3 | -- | -- |
| CSP-des-K14 | 179 | 96.4 - 332 |
| CSP-des-K13K14 | 190 | 108 - 333 |
| CSP-des-K12K13K14 | 416 | 165 - 1050 |
| CSP-D1AS2A | 881* | 538 - 1440* |
| CSP-D1AL7A | -- | -- |
| CSP-D1AK13A | >1000* | >1000* |
| CSP-D1AK14A | 537* | 294 - 980* |
| CSP-S2AL7A | -- | -- |
| CSP-S2AK13A | 211 | 161 - 275 |
| CSP-S2AK14A | 201 | 117 - 345 |
| CSP-L7AK13A | -- | -- |
| CSP-L7AK14A | -- | -- |
| CSP-K13AK14A | 166 | 112 - 244 |
See the Materials and Methods section for experimental methods. See Supporting Information for detail of reporter strain and plots of agonism or antagonism dose response curves. All assays performed in triplicate.
EC50 or IC50 values determined by testing peptides over a wide range of concentrations.
95% confidence interval.
EC50 not determined due to the analog’s low induction in primary agonism screening assays. See Supporting Information for details.
As truncation of either the N- or C-terminal regions of the S. oligofermentans CSP was insufficient to produce enhanced QS activators, we hypothesized that double alanine mutation of residues found to be permissive to modification during the initial alanine screening could produce CSP analogs with enhanced receptor binding. Our initial alanine screen revealed that replacement of residues Ser2, Leu7, Lys13, and Lys14 with alanine afforded analogs with activities equivalent to that of the native signal. Double alanine mutant combinations were made with substitutions at two of these positions, with the intent of producing an enhanced activator of the S. oligofermentans competence regulon. Double mutation with residues Ser2, Lys13, and Lys14 all produced CSP analogs with activities similar to that of the native peptide (Table 2). Interestingly, double substitutions incorporating alanine replacement of residue Leu7 displayed a complete loss in agonistic activity, despite the fact that substitution of Leu7 alone was able to activate the competence regulon at a comparable concentration to the native peptide (Table 2).
We previously observed that mutation of the negatively charged residue Asp1 to alanine produced a QS inhibitor, a trend consistent with other mitis group streptococci. In an effort to produce enhanced inhibitors of the competence regulon, double alanine mutant analogs were made with substitutions of Asp1 and the residues Ser2, Leu7, Lys13, or Lys14. Double mutation with Asp1 and Ser2 or Lys13 produced weak QS inhibitors with activities comparable to replacement of Asp1 alone (Table 2). Importantly, double mutation of Asp1 and Lys14 produced the strongest S. oligofermentans QS inhibitor, with a near two-fold improvement in activity compared to replacement of Asp1 alone (Table 2). Dual alanine mutation of positions Asp1 and Leu7 produced a peptide that was unable to interact with the ComD receptor, similar to what was observed for the double mutant activators (Table 2). These results further confirm that while Leu7 alone does not play an important role in ComD binding or activation, mutation of this position likely induces a conformational change that is not tolerable with other mutations of the CSP sequence, leading to a complete loss of ComD binding or activation.
Determining the role of QS modulation in interactions between S. oligofermentans and S. mutans
Previous studies have highlighted how S. mutans strives to achieve dominance of the oral microbiome by either acidifying the microenvironment through the production of lactic acid or using its QS circuitry to coordinate the expression of mutacin, a bacteriocin that can kill off many competing oral microbes.5, 12, 34–35 It has been well documented that S. oligofermentans and S. mutans compete within the microbiome, and that the S. oligofermentans mediated killing of S. mutans is primarily conducted through the conversion of lactic acid into hydrogen peroxide,5 a process that our phenotypic studies have suggested is under control of the competence regulon. As a result of these findings, we hypothesized that by modulating the S. oligofermentans QS circuitry we would be able to tune its response to S. mutans by either enhancing the hydrogen peroxide mediated killing of S. mutans in the presence of a QS activator (native CSP) or decreasing it in the present of a QS inhibitor (CSP D1AK14A). Thus, we tested the effectiveness of QS modulators on S. oligofermentans in the presence of two different S. mutans UA159 strains, one with intact QS circuitry (SMCOM3), and a comC knockout unable to produce mutacin (SMCC3).
In regard to the comC knockout strain SMCC3, we fully expected S. oligofermentans to be able to inhibit the growth of S. mutans due to a lack of mutacin production. Our experiments in several trials showed slight zones of inhibition on S. mutans when in the presence of S. oligofermentans alone, with added lactic acid, or with an added QS activator, or no zone of inhibition when in the presence of a QS inhibitor or added lactic acid and QS activator or QS inhibitor (Figure 9). Furthermore, we predicted that S. oligofermentans would be inhibited by the S. mutans UA159 strain SMCOM3, as the mutacin produced by this strain has been shown to be bacteriocidal against many species of streptococci. Interestingly, in many of the co-culture conditions the growth of neither S. mutans nor S. oligofermentans was inhibited. The only case in which a clear zone of inhibition was observed in several trials was with the pre-treatment of S. oligofermentans with both QS inhibitor and lactic acid, perhaps suggesting that the dual activity of mutacin and an inability to process lactic acid as a biproduct of QS inhibition was sufficient to prevent the growth of S. oligofermentans in the presence of S. mutans (Figure 9). Altogether, we were unable to directly observe conclusive evidence for enhanced S. oligofermentans mediated killing of S. mutans. Nonetheless, since there are many different factors that can influence the interspecies interactions between streptococci, our results cannot entirely rule out QS modulation of S. oligofermentans as an effective approach to counter S. mutans infection. Rather, our results indicate that QS modulation did not have any observable effect in the tested conditions, and that further experiments are required to fully explore the effect of QS on these interactions.
Figure 9.
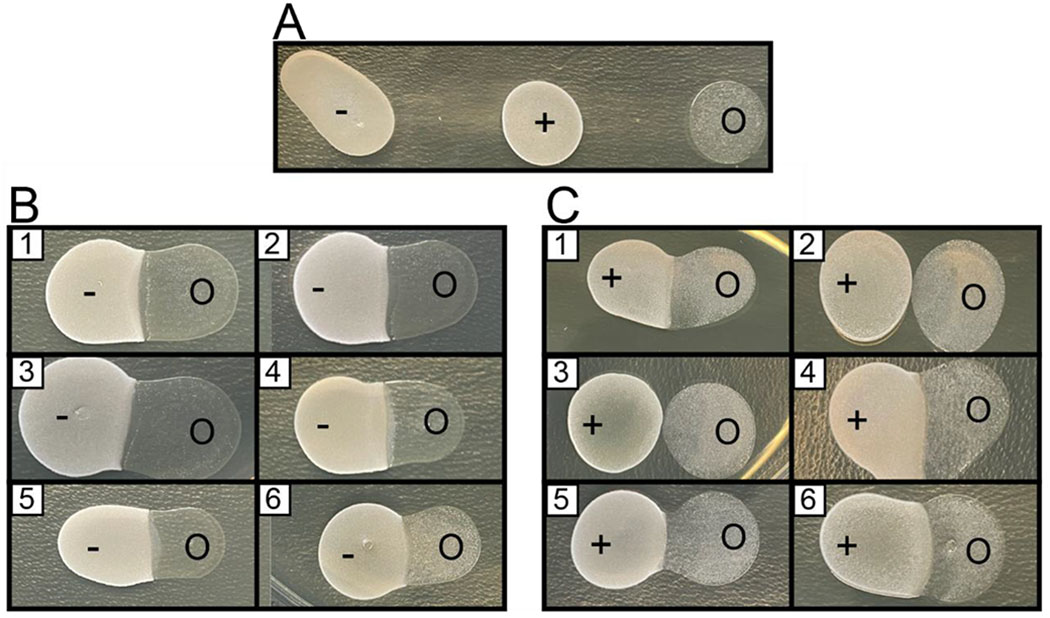
Interspecies competition between S. oligofermentans and S. mutans. A. Individual colonies of S. mutans SMCC3 (UA159, ΔcomC, SpecR) (−), SMCOM3 (UA159, SpecR) (+), and wild type S. oligofermentans (O). B. Competition between SMCC3 (left) and S. oligofermentans (right). C. Competition between SMCOM3 (left) and S. oligofermentans (right). In both B and C, S. oligofermentans was exposed to the following conditions prior to co-inoculation with either S. mutans strain: 1. THY media; 2. THY media + 10 mM lactic acid; 3. THY media + 300 nM native CSP; 4. THY media + 2,500 nM CSP D1AK14A; 5. THY media + 10 mM lactic acid + 300 nM CSP; 6. THY media + 10 mM lactic acid + 2,500 nM CSP D1AK14A. Experiment was repeated three times in triplicate for a total of nine trials. See Materials and Methods for full experimental details.
Assessing the cytotoxicity of S. oligofermentans and CSP analogs
One of the major advantages of S. oligofermentans is its ability to act as a biotherapeutic, outcompeting more pathogenic bacteria within the oral microbiome to promote human health. As many streptococci are known to produce virulence factors associated with cytotoxicity in both human and animal models, we sought to ensure that there was no observable S. oligofermentans or CSP induced cytotoxicity, which would negatively impact the capacity of this proposed approach.35–37 To this end, hemolysis assays against defibrinated rabbit red blood cells (RBCs) conducted in both THY media alone and THY media containing S. oligofermentans were utilized to assess the degree of cytotoxicity attributed to S. oligofermentans or any tested peptides. In THY media alone there was no significant increase in hemolysis in the presence of any of the tested CSP analogs, whereas a significant degree of hemolysis was observed in the .01% Triton X positive control (Figure 10). These results suggest that treatment with CSP alone has no significant cytotoxic effect against the RBCs. Additionally, we observed that there was no significant effect on S. oligofermentans hemolysis upon treatment with the native peptide (CSP), a strong activator (CSP S2A), or a strong inhibitor (CSP D1AK14A) (Figure 10), highlighting both that S. oligofermentans is not likely to cause any significant degree of cytotoxicity, and that any potential cytotoxicity is not the product of a QS-mediated response.
Figure 10.
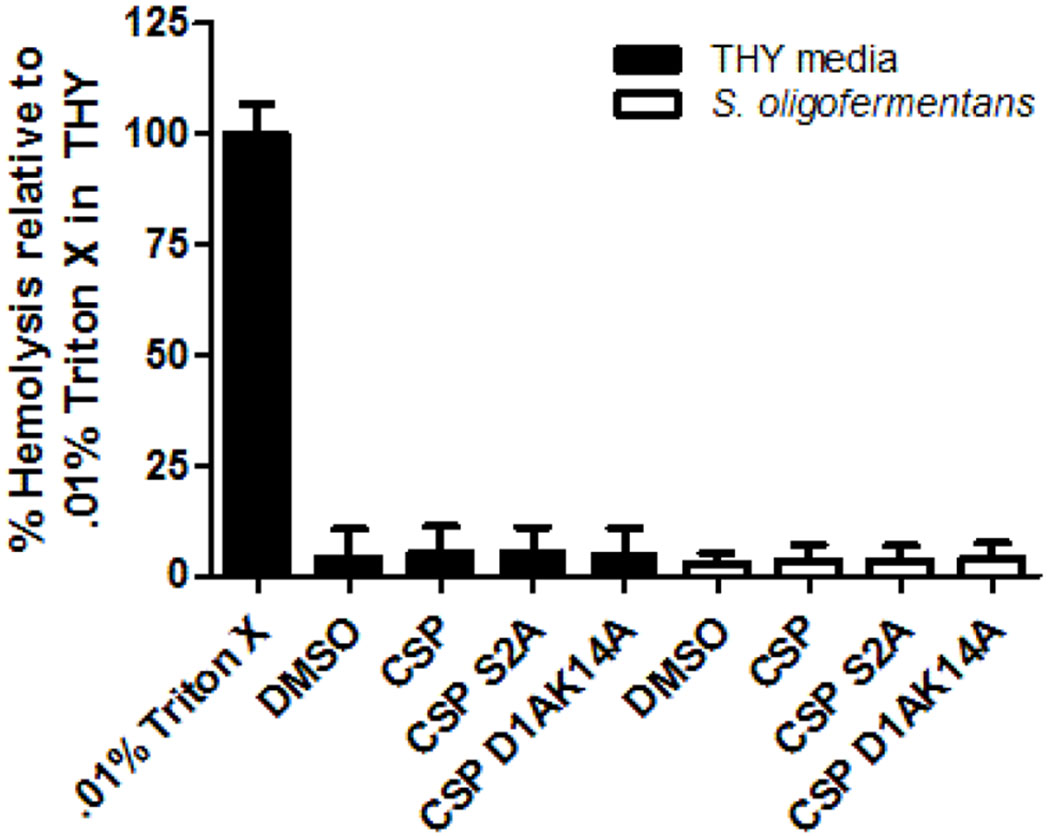
Hemolytic activity of CSP derived QS modulators on defibrinated rabbit red blood cells in the presence of THY media (black) or S. oligofermentans cultures (white). Experiments were performed in triplicate on three separate days. See the Supporting Information for full experimental details.
Summary and Conclusions
It has been well established that streptococcal QS is used to mediate important bacterial phenotypes, and that it plays a critical role in the complex web of inter- and intra-species interactions that occur within the oral microbiome. Herein we have confirmed the identity of the S. oligofermentans CSP as the 14 amino acid peptide DSRNIFLKIFKKK and demonstrated its involvement in the regulation of bacterial competence and hydrogen peroxide production through activation of the competence regulon.
Through alanine screening of the S. oligofermentans CSP we were able to gain several insights into the SAR between CSP amino acid side chain moieties and interactions with the ComD receptor. We have demonstrated that the N-terminus of the CSP is critical for receptor binding and activation, and similarly to other mitis group streptococci, it was observed that mutation of the first negatively charged residue to alanine produced a QS inhibitor. Interestingly, we observed that there is no clear trend of activity for mutations to the central region of the CSP, as mutation of both polar or hydrophobic residues produced inactive and active CSP analogs. Moreover, we have demonstrated that the C-terminal region of the CSP signal can be removed without significantly impacting ComD receptor binding or activation. This finding was further confirmed by a retention of peptide activity following truncation of this region. Additionally, through multiple alanine mutation of the CSP we were able to produce the most potent S. oligofermentans QS inhibitor, CSP D1AK14A, which displays a near two-fold greater degree of inhibitory activity compared to CSP D1A.
Comprehensive structural analysis of all alanine screen CSP analogs revealed that, unlike what has been previously observed in other mitis group streptococci, there is no clear trend between alpha helicity and peptide activity for the S. oligofermentans CSP. Additionally, there was no clear trend that would suggest that the CSP may be able to tune the effectiveness of S. oligofermentans against S. mutans in interspecies competition assays, suggesting that future efforts should focus on further probing of these interactions. Hemolysis assays demonstrated that CSPs themselves are relatively non-toxic, and that S. oligofermentans does not produce any virulence factors that could result in undesired toxicity. Overall, this study suggests that S. oligofermentans QS modulators are promising tools for probing interspecies interactions among oral streptococci, or as potential non-toxic additives to help increase the potency of S. oligofermentans as a biotherapeutic against S. mutans infection.
Materials and Methods
General:
Synthesis of peptides was performed through Fmoc solid phase peptide synthesis (SPPS). Peptides were purified to ≥95% as confirmed by analytical reverse phase high-performance liquid chromatography (RP-HPLC). Masses of purified peptides were confirmed with high-resolution ESI-TOFMS. See Tables S-2–S-3 for peptide masses and purities. See supporting information for full experimental details.
Isolation of Crude Peptides from Bacterial Supernatants:
The native S. oligofermentans CSP was extracted from bacterial supernatants using previously described methods with minor modifications.9 See Supporting Information for full experimental details.
Development of S. oligofermentans reporter strain:
The S. oligofermentans luciferase-based reporter strain was constructed using previously described methods with some modifications.31 See Supporting Information for full experimental details.
Luminescence Assay:
Activation Assays.
The ability of synthetic CSP analogs to induce activation of the competence regulon was determined using the S. oligofermentans luciferase reporter strain. First, a single colony of the S. oligofermentans luciferase reporter strain was grown at 37 °C with 5% CO2 in 10 mL THY media (pH 7.3) containing 100 μg/mL spectinomycin for 18 hours. Following incubation, the bacteria were diluted 1:10 in fresh THY media, and incubated for 30 min at 37 °C with 5% CO2. During incubation, clear bottom white 96-well microtiter plates were prepared for activation assays. Initially, activation screening was performed at a high CSP concentration (10,000 nM) for all CSP analogs. For each experimental sample, 2 μL of a 1 mM CSP stock solution in dimethyl sulfoxide (DMSO) was added in triplicate to the clear bottom white 96-well microtiter plate. A total of 2 μL of DMSO was added in triplicate as a negative control, and 2 μL of a 1 mM stock of the native CSP was added in triplicate as a positive control. In the dark, 2 μL of a 15 mg/ml D-luciferin stock was added to each well. Following addition of CSP and D-luciferin, 196 μL of the diluted bacterial culture was added to each well, and the plate was incubated for 30 min at 37 °C. Following incubation, the OD600 and luminescence of each experimental well was measured. Final results were reported as percent activation, which is the ratio between the final blank and OD600 corrected luminescence of experimental values and the positive control. Analogs that exhibited activity > 75% compared to the positive control (see Figures S-2–S-3) were further evaluated using a dose-dependent assay in which peptide stock solutions were diluted with DMSO in serial dilutions (1:2 or 1:3) and assayed as described above. EC50 values, the concentration of the peptide to achieve a half-maximal response, for each activator were calculated using GraphPad Prism. Experiments were performed in triplicate on three separate days.
Inhibition Assays.
Analogs that exhibited a low degree of activation in the initial screening (<50% activation) were evaluated for competitive inhibition of the competence regulon. The ability of synthetic CSP analogs to competitively inhibit comX expression by outcompeting CSP for the receptor binding site was evaluated using the same assay conditions as the activation assays, except inhibition screening introduced the native CSP to each well at a set concentration (300 nM) that was chosen to afford complete activation of the competence regulon, as determined from dose-dependence curves created for the native CSP. As a positive control, 2 μL of CSP and 2 μL of DMSO were added to the same well in triplicate. As a negative control, 4 μL of DMSO was added in triplicate. Then 2 μL of 15 mg/mL D-luciferin was added to each experimental or control well, after which 194 μL of bacteria was added, and the plate was incubated for 30 min at 37 °C. Following incubation, the OD600 and luminescence of each experimental well were measured. Final results were reported as percent activation, which is the ratio between the final blank and OD600 corrected luminescence of experimental values and the positive control. Analogs that exhibited activity < 50% compared to the positive control (see Figures S-4–S-5) were considered competitive inhibitors of the QS circuitry. These peptides were further evaluated in dose-dependent assays where peptide stock solutions were diluted with DMSO in a 1:2 serial dilution and assayed as described above. Inhibitor IC50 values, the concentration of inhibitor required to reduce the QS activation by half, were determined using GraphPad Prism. Experiments were performed in triplicate on three separate days.
Transformation Assay:
A single colony of S. oligofermentans was grown at 37 °C with 5% CO2 in 10 mL THY media (pH 7.3) for 18 hours. Following incubation, S. oligofermentans was diluted 1:25 into fresh THY media. After static incubation at 37 °C with 5% CO2 for 30 min, CSP was added at final concentrations of 10,000 nM, 1,000 nM, 100 nM, or 10 nM, along with 300 ng pDL278 (SpecR). Parallel assays without CSP or without both CSP and plasmid were used as controls to assess the indigenous competence or antibiotic resistance of S. oligofermentans in the tested conditions. After 2 h incubation at 37 °C with 5% CO2, 20 μL of the culture was plated on THY agar containing 400 μg/mL spectinomycin and incubated at 37 °C with 5% CO2 for 48 hours to identify positive transformants. Experiments were performed in triplicate on three separate days.
Biofilm Formation Assay:
Biofilm formation was assessed using previously described methods with minor alterations.38 See Supporting Information for full experimental details.
Hydrogen Peroxide Production Assay:
Hydrogen peroxide production was assessed using previously described methods.39 See Supporting Information for full experimental details.
Interspecies Competition Assays:
A single colony of S. oligofermentans was inoculated into 10 mL THY media (pH 7.3) and grown for 18 h at 37 °C with 5% CO2. Concurrently, single colonies of S. mutans SMCOM3 (UA159, SpecR) and SMCC3 (UA159, ΔcomC, SpecR) were separately inoculated into 10 mL THY media (pH 7.3) containing 100 μg/mL spectinomycin and were grown at 37 °C with 5% CO2 for 18 h. Following growth of the overnight cultures, bacteria were then diluted 1:10 in fresh THY media, and the following experimental conditions were arranged with a total final volume of 500 μL: S. oligofermentans, S. mutans SMCOM3, S. mutans SMCC3, S. oligofermentans + 10 mM lactic acid, S. oligofermentans + 300 nM native CSP, S. oligofermentans + 2,500 nM CSP D1AK14A, S. oligofermentans + 300 nM native CSP + 10 mM lactic acid, S. oligofermentans + 2,500 nM CSP D1AK14A + 10 mM lactic acid. Cultures were then incubated for 2 h at 37 °C with 5% CO2. Immediately following incubation, a 10 μL portion of each S. oligofermentans experimental condition was then spotted adjacently to a 10 μL portion of either S. mutans strain on a THY agar plate. Spotted media was allowed to absorb into the plate for 30 min at RT, followed by incubation at 37 °C with 5% CO2 for 24 h. Following incubation, competition between bacteria was assessed by observing the presence of a zone of inhibition on the spotted cultures. Experiments were performed in triplicate on three separate days.
Hemolysis Assay:
Hemolysis was assessed using previously described methods with minor alterations.40 See Supporting Information for full experimental details.
Circular Dichroism (CD) Spectroscopy:
CD spectra were recorded using an Aviv Biomedical CD spectrometer (model 202-01). All the measurements were performed with a peptide concentration of 100 μM in 1x PBS buffer with 0% or 20% trifluoroethanol (TFE). Measurements were performed using a quartz cuvette (Starna Cells) with a pathlength of 0.1 cm at 25 °C. Samples were scanned once at 3 nm min−1 with a bandwidth of 1 nm and a response time of 20 s over a wavelength range (195 to 260 nm). Single scans were acquired and corrected for their respected solvent concentrations, then converted to mean residue ellipticity (MRE) values using the following equation:
θ is the observed ellipticity in millidegrees, c is the molar peptide concentration, l is the pathlength in centimeters, and n is the number of residues in the peptide sequence. Percent helicity (fH) was calculated for all peptide analogs using the following equation:
is the mean residue ellipticity of the peptide at 222 nm, is the assumed mean residue ellipticity for a peptide with 100% helicity (−44,000 deg cm2 dmol−1), n is the number of residues within the tested peptide sequence, and x is an empirical correction for end effects.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R35GM128651). The S. oligofermentans SS-1917 strain was a generous gift from L. McGee (CDC Streptococcus Lab), and the S. mutans SMCOM3 and SMCC3 strains were a generous gift from D. G. Cvitkovich (University of Toronto). We would like to thank M. J. Tucker (University of Nevada, Reno) for the use of the CD spectrometer.
Footnotes
Supporting Information
Additional experimental procedures, peptide characterization, primary reporter assay data, dose-response curves for CSP analogs, and supplementary figures. This material is available free of charge via the internet at https://pubs.acs.org/doi/10.1021/acschembio.1c00746.
References
- 1.Abranches J; Zeng L; Kajfasz JK; Palmer SR; Chakraborty B; Wen ZT; Richards VP; Brady LJ; Lemos JA, Biology of Oral Streptococci. Microbiol Spectr 2018, 6 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dewhirst FE; Chen T; Izard J; Paster BJ; Tanner AC; Yu WH; Lakshmanan A; Wade WG, The human oral microbiome. J Bacteriol 2010, 192 (19), 5002–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paster BJ; Olsen I; Aas JA; Dewhirst FE, The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000 2006, 42, 80–7. [DOI] [PubMed] [Google Scholar]
- 4.Forssten SD; Björklund M; Ouwehand AC, Streptococcus mutans, caries and simulation models. Nutrients 2010, 2 (3), 290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tong H; Chen W; Merritt J; Qi F; Shi W; Dong X, Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol Microbiol 2007, 63 (3), 872–80. [DOI] [PubMed] [Google Scholar]
- 6.Bikash CR; Hamry SR; Tal-Gan Y, Structure-Activity Relationships of the Competence Stimulating Peptide in Streptococcus mutans Reveal Motifs Critical for Membrane Protease SepM Recognition and ComD Receptor Activation. ACS Infect Dis 2018, 4 (9), 1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pepperney A; Chikindas ML, Antibacterial Peptides: Opportunities for the Prevention and Treatment of Dental Caries. Probiotics Antimicrob Proteins 2011, 3 (2), 68. [DOI] [PubMed] [Google Scholar]
- 8.Kreth J; Merritt J; Qi F, Bacterial and host interactions of oral streptococci. DNA Cell Biol 2009, 28 (8), 397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrington A; Tal-Gan Y, Identification of Streptococcus gallolyticus subsp. gallolyticus (Biotype I) Competence-Stimulating Peptide Pheromone. J Bacteriol 2018, 200 (14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X; Browngardt CM; Jiang M; Ahn SJ; Burne RA; Nascimento MM, Diversity in Antagonistic Interactions between Commensal Oral Streptococci and Streptococcus mutans. Caries Res 2018, 52 (1-2), 88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemos JA; Burne RA, A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology (Reading) 2008, 154 (Pt 11), 3247–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsui R; Cvitkovitch D, Acid tolerance mechanisms utilized by Streptococcus mutans. Future Microbiol 2010, 5 (3), 403–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong H; Gao X; Dong X, Streptococcus oligofermentans sp. nov., a novel oral isolate from caries-free humans. Int J Syst Evol Microbiol 2003, 53 (Pt 4), 1101–1104. [DOI] [PubMed] [Google Scholar]
- 14.Tong H; Zhu B; Chen W; Qi F; Shi W; Dong X, Establishing a genetic system for ecological studies of Streptococcus oligofermentans. FEMS Microbiol Lett 2006, 264 (2), 213–9. [DOI] [PubMed] [Google Scholar]
- 15.Matta M; Gousseff M; Monsel F; Poyart C; Diebold B; Podglajen I; Mainardi JL, First case of Streptococcus oligofermentans endocarditis determined based on sodA gene sequences after amplification directly from valvular samples. J Clin Microbiol 2009, 47 (3), 855–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tong H; Chen W; Shi W; Qi F; Dong X, SO-LAAO, a novel L-amino acid oxidase that enables Streptococcus oligofermentans to outcompete Streptococcus mutans by generating H2O2 from peptone. J Bacteriol 2008, 190 (13), 4716–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L; Tong H; Dong X, Function of the pyruvate oxidase-lactate oxidase cascade in interspecies competition between Streptococcus oligofermentans and Streptococcus mutans. Appl Environ Microbiol 2012, 78 (7), 2120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao X; de Soet JJ; Tong H; Gao X; He L; van Loveren C; Deng DM, Streptococcus oligofermentans Inhibits Streptococcus mutans in Biofilms at Both Neutral pH and Cariogenic Conditions. PLoS One 2015, 10 (6), e0130962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao X; Yang J; de Soet JJ; Liu H; Gao X; van Loveren C; Deng D, Factors Influencing the Competition between Streptococcus oligofermentans and Streptococcus mutans in Dual-Species Biofilms. Caries Res 2017, 51 (5), 507–514. [DOI] [PubMed] [Google Scholar]
- 20.Bassler BL, How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol 1999, 2 (6), 582–587. [DOI] [PubMed] [Google Scholar]
- 21.Miller MB; Bassler BL, Quorum sensing in bacteria. Annu Rev Microbiol 2001, 55 (1), 165–199. [DOI] [PubMed] [Google Scholar]
- 22.Waters CM; Bassler BL, Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol 2005, 21, 319–346. [DOI] [PubMed] [Google Scholar]
- 23.Ng W-L; Bassler BL, Bacterial quorum-sensing network architectures. Annu Rev Genet 2009, 43, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rutherford ST; Bassler BL, Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2012, 2 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mull RW; Harrington A; Sanchez LA; Tal-Gan Y, Cyclic Peptides that Govern Signal Transduction Pathways: From Prokaryotes to Multi-Cellular Organisms. Curr Top Med Chem 2018, 18 (7), 625–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sturme MH; Kleerebezem M; Nakayama J; Akkermans AD; Vaugha EE; de Vos WM, Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Van Leeuwenhoek 2002, 81 (1-4), 233–43. [DOI] [PubMed] [Google Scholar]
- 27.Patteson JB; Lescallette AR; Li B, Discovery and Biosynthesis of Azabicyclene, a Conserved Nonribosomal Peptide in Pseudomonas aeruginosa. Org Lett 2019, 21 (13), 4955–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cvitkovitch DG; Li YH; Ellen RP, Quorum sensing and biofilm formation in Streptococcal infections. J Clin Invest 2003, 112 (11), 1626–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jimenez JC; Federle MJ, Quorum sensing in group A Streptococcus. Front Cell Infect Microbiol 2014, 4, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrington A; Proutiere A; Mull RW; du Merle L; Dramsi S; Tal-Gan Y, Secretion, Maturation, and Activity of a Quorum Sensing Peptide (GSP) Inducing Bacteriocin Transcription in Streptococcus gallolyticus. mBio 2021, 12 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salvadori G; Junges R; Morrison DA; Petersen FC, Overcoming the Barrier of Low Efficiency during Genetic Transformation of Streptococcus mitis. Front Microbiol 2016, 7, 1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green MR; Sambrook J, The Inoue Method for Preparation and Transformation of Competent. Cold Spring Harb Protoc 2020, 2020 (6), 101196. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y; Koirala B; Sanchez LA; Phillips NR; Hamry SR; Tal-Gan Y, Structure-Activity Relationships of the Competence Stimulating Peptides (CSPs) in Streptococcus pneumoniae Reveal Motifs Critical for Intra-group and Cross-group ComD Receptor Activation. ACS Chem Biol 2017, 12 (4), 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merritt J; Qi F, The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol 2012, 27 (2), 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamiya RU; Taiete T; Gonçalves RB, Mutacins of Streptococcus mutans. Braz J Microbiol 2011, 42 (4), 1248–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadioglu A; Weiser JN; Paton JC; Andrew PW, The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol 2008, 6 (4), 288–301. [DOI] [PubMed] [Google Scholar]
- 37.Fittipaldi N; Segura M; Grenier D; Gottschalk M, Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol 2012, 7 (2), 259–79. [DOI] [PubMed] [Google Scholar]
- 38.McBrayer DN; Cameron CD; Gantman BK; Tal-Gan Y, Rational Design of Potent Activators and Inhibitors of the Enterococcus faecalis Fsr Quorum Sensing Circuit. ACS Chem Biol 2018, 13 (9), 2673–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X; Tong H; Dong X, PerR-regulated manganese ion uptake contributes to oxidative stress defense in an oral streptococcus. Appl Environ Microbiol 2014, 80 (8), 2351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tal-Gan Y; Stacy DM; Foegen MK; Koenig DW; Blackwell HE, Highly Potent Inhibitors of Quorum Sensing in Staphylococcus aureus Revealed Through a Systematic Synthetic Study of the Group-III Autoinducing Peptide. J. Am. Chem. Soc 2013, 135 (21), 7869–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


