Abstract
OBJECTIVE
Sporadic brain arteriovenous malformation (BAVM) is a tangled vascular lesion characterized by direct artery-to-vein connections that can cause life-threatening intracerebral hemorrhage (ICH). Recently, somatic mutations in KRAS have been reported in sporadic BAVM, and mutations in other mitogen-activated protein kinase (MAPK) signaling pathway genes have been identified in other vascular malformations. The objectives of this study were to systematically evaluate somatic mutations in MAPK pathway genes in patients with sporadic BAVM lesions and to evaluate the association of somatic mutations with phenotypes of sporadic BAVM severity.
METHODS
The authors performed whole-exome sequencing on paired lesion and blood DNA samples from 14 patients with sporadic BAVM, and 295 genes in the MAPK signaling pathway were evaluated to identify genes with somatic mutations in multiple patients with BAVM. Digital droplet polymerase chain reaction was used to validate KRAS G12V and G12D mutations and to assay an additional 56 BAVM samples.
RESULTS
The authors identified a total of 24 candidate BAVM-associated somatic variants in 11 MAPK pathway genes. The previously identified KRAS G12V and G12D mutations were the only recurrent mutations. Overall, somatic KRAS G12V was present in 14.5% of BAVM lesions and G12D was present in 31.9%. The authors did not detect a significant association between the presence or allelic burden of KRAS mutation and three BAVM phenotypes: lesion size (maximum diameter), age at diagnosis, and age at ICH.
CONCLUSIONS
The authors confirmed the high prevalence of somatic KRAS mutations in sporadic BAVM lesions and identified several candidate somatic variants in other MAPK pathway genes. These somatic variants may contribute to understanding of the etiology of sporadic BAVM and the clinical characteristics of patients with this condition.
Keywords: arteriovenous malformation, cerebrovascular malformation, somatic mutation, MAPK pathway, genotype-phenotype correlation, intracerebral hemorrhage, vascular disorders
BRAIN arteriovenous malformation (BAVM), which results in blood shunting directly from feeding arteries to draining veins, is a rare cerebrovascular lesion that occurs in approximately 0.01% of the population.1,2 Patients with BAVM experience many neurological outcomes, including headache, seizure, deficit, and life-threatening intracerebral hemorrhage (ICH).3–5 However, the mechanisms underlying BAVM development remain largely unknown, and there is currently no effective medical therapy for intervention in BAVM development or prevention of rupture. Interventional treatment options, such as microsurgical resection, embolization, and radio-surgery, are not always feasible and may lead to complications or disability.6–9 A better understanding of BAVM development and the risk factors of ICH is critical for developing new treatments and improving outcomes.
The pathogenesis of sporadic BAVM has long been suspected to involve somatic mutations on the basis of observations of many other vascular anomalies, which feature somatic mutations either alone or in combination with germline mutations.10–15 Recently, somatic activating mutations in KRAS, a gene in the mitogen-activated protein kinase (MAPK) signaling pathway, were reported in a majority of sporadic BAVM lesions,16 and this finding was corroborated by other studies.17–21 Given that somatic mutations in several other MAPK pathway genes have been reported in other vascular anomalies,12,22–24 we hypothesized that somatic mutations in multiple MAPK pathway genes may contribute to the etiology or clinical phenotype of BAVM.
We performed whole-exome sequencing (WES) on paired BAVM tissue and blood DNA samples obtained from 14 patients with sporadic BAVM, and we evaluated 295 genes in the MAPK signaling pathway to identify those genes with somatic mutations in BAVM lesions. The detection of the two KRAS mutations (G12V and G12D) that were recently reported in patients with BAVM16–21 was used to design a modified pipeline to detect and prioritize other candidate somatic variants in the MAPK pathway. Finally, we evaluated the prevalence and associations of somatic KRAS mutations with three clinical phenotypes of BAVM: lesion size (maximum diameter), age at diagnosis, and age at ICH.
Methods
Study Cohort
All patients were prospectively enrolled in the University of California, San Francisco (UCSF) BAVM Project between 2002 and 2017 and had no family history of BAVM, hereditary hemorrhagic telangiectasia, or other known genetic syndromes. Diagnosis of BAVM was confirmed with catheter angiography and/or pathology evaluation. Informed consent for genetic studies was obtained, and the study was approved by the Committee on Human Research at UCSF.
For the discovery cohort, we analyzed data from 14 White patients with a diagnosis of sporadic BAVM and both BAVM tissue that had been frozen in optimal cutting temperature (OCT) compound and blood available for analysis. All patients who underwent microsurgical resection of BAVM had not received prior interventional treatment, except 1 patient whose first treatment was embolization. The patients were evenly split in terms of sex and between those with ruptured and unruptured BAVMs. All patients were White, the most common race/ethnicity in the UCSF BAVM cohort, to minimize confounding due to genetic differences.
For the replication cohort, we analyzed an additional 56 patients with sporadic BAVM, of whom 8 had undergone embolization and 1 had undergone radiosurgery prior to microsurgical resection. The replication cohort included patients with BAVM with full clinical data, and sufficient OCT-frozen tissue available for analysis. Thus, the replication cohort included patients with multiple ethnic backgrounds and is more representative of the patients included in the overall UCSF BAVM Project cohort.
Of 70 patients included in the entire cohort, 43 had ICH. Demographic and phenotypic data for all patients are shown in Supplemental Table 1.
DNA Extraction
Blood genomic DNA was previously isolated and banked with standard DNA extraction techniques. For BAVM tissue, samples were snap-frozen in liquid nitrogen after resection and banked at −80°C in OCT compound. Frozen tissue was cryosectioned in bulk, and genomic DNA was isolated with the DNeasy Blood & Tissue Kit (QIAGEN).
WES Library Preparation and Sequencing
We used Nextera Rapid Capture Exome kits (Illumina) for exome enrichment and library preparation, with 250 ng DNA per sample. Paired-end sequencing (2 × 76 bp) was performed on the Illumina HiSeq 2500 platform at the Broad Institute, Cambridge, Massachusetts.
Somatic Variant Calling and Subsequent Filtering
Sequencing reads were mapped to the human reference genome assembly (hg19) with the Burrows-Wheeler Alignment tool,25 then preprocessed according to the Genome Analysis Toolkit (GATK) best practice workflows for somatic short-variant discovery.26,27 Mutect2 in GATK version 3.6 was used for calling somatic single-nucleotide variants (SNVs) with default parameters. We devised a customized workflow to prioritize candidate somatic variants associated with BAVM, with filters applied sequentially (Supplemental Fig. 1 and Supplemental Table 2). Because the TLOD (theta logarithm of the odds) values (i.e., initial LOD threshold for calling tumor variant) of both KRAS mutations (G12V and G12D) were greater than 4.2, we used TLOD ≥ 4.2 as the first filter. Finally, SnpEff28 and PolyPhen-229 (HumDiv-trained) were used for variant annotation and functional effect prediction, respectively. Variants predicted to be “probably damaging” were retained.
Evaluation of Mutational Background
We applied MutSigCV30 and MUFFINN31 to identify significantly mutated genes harboring mutations associated with BAVM. Because MutSigCV requires both silent and nonsilent variants to evaluate the background mutation rate, we used the 12,272 SNVs that passed the first four filters of the analysis (Supplemental Fig. 1). Oncotator32 was used to generate the input mutation file required by MutSigCV. For MUFFINN, we used the 3624 SNVs that passed all filters of the analysis (Supplemental Fig. 1), network algorithm NDsum, and functional network HumanNet.
Evaluation of Sequencing Coverage
We used ExomeCQA33 to assess the coverage distributions of the sequencing reads in the study cohort, and we used bedtools to calculate the coverage files required. Cohort Coverage Sparseness was used to assess the coverage of all exons across the genome, and Unevenness was used to evaluate the coverage of a given exon across all samples.
Digital Droplet Polymerase Chain Reaction
For validation and replication, we assayed KRAS G12V and G12D by using commercially available digital droplet polymerase chain reaction (ddPCR) assays according to the manufacturer’s instructions (Bio-Rad Unique Assay ID dHsaCP2000005 for mutant G12V, dHsaCP2000001 for mutant G12D, dHsaCP2000006 for wild-type G12V, and dHsaCP2000002 for wild-type G12D). The QX100 ddPCR system (Bio-Rad) was used for droplet generation and data acquisition. A total of 40 ng DNA per reaction was used for a maximum sensitivity of 0.025%. We included a positive control (somatic mutation positive), negative control (wild type only), and nontemplate control on all plates. Data analysis included determination of allelic burden with Bio-Rad QuantaSoft Analysis Pro software. All assays were performed in duplicate or triplicate. Furthermore, we required a minimum of 250 signal-positive droplets per patient and 100 signal-positive droplets per well.
Statistical Analysis
We included samples in the statistical analysis if they passed quality control for both ddPCR assays for G12D and G12V. We calculated summary statistics for patient and BAVM characteristics as mean ± standard deviation (SD) for numeric variables or as count and percentage for categorical variables. For our primary analysis, we tested whether the percentages of G12D or G12V alleles were associated with three outcomes: 1) age at tissue collection; 2) BAVM lesion size (maximum lesion diameter in millimeters); and 3) age at first ICH. For outcomes 1 and 2, we used linear regression models, adjusted for sex and prior ICH, and report exponentiated regression coefficients, or proportional increase (PI), with 95% confidence intervals (CIs). For outcome 3, we used a Cox proportional hazards model, adjusted for sex and BAVM size, and report hazard ratio (HR) and 95% CI; patients without ICH at age of tissue collection were censored. Secondary analyses evaluated individually G12D and G12V allelic burden as a percentage, and we also dichotomized patients according to the presence or absence of either somatic mutation (allelic burden threshold of 0.5%). Data analysis was conducted with Stata version 15.1 (StataCorp LLC).
Results
WES Data and Read Coverage
Means of 123 million and 119 million reads were generated for the BAVM lesion and blood DNA samples, respectively. Averages of 99.04% and 98.80% of reads were mapped for the lesion and blood DNA samples, respectively. The average (range) sequencing coverage was 108× (88× to 142×) for lesion DNA and 97× (70× to 125×) for blood DNA (Supplemental Table 3). We observed variations in coverage across all samples (Supplemental Table 3) and over the targeted regions within each sample (data not shown).
Identification of Candidate BAVM-Associated Somatic Variants in the MAPK Pathway
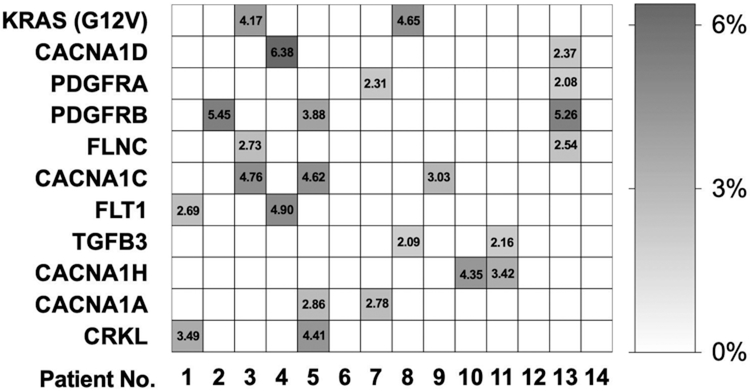
A total of 37,591 somatic variants were identified exome-wide (by using the default parameters of Mutect2). After application of sequential filters (Supplemental Fig. 1), the final number of candidate somatic variants decreased to 3624. We cross-referenced the list of candidate somatic mutations with 295 MAPK pathway genes (included in the Kyoto Encyclopedia of Genes and Genomes database, https://www.genome.jp/kegg/). KRAS was the only MAPK pathway gene with a recurrent mutation (G12V) that was observed in our study (Fig. 1). There were 10 other MAPK pathway genes with candidate somatic variants, each variant unique to 1 patient (Fig. 1). A total of 22 “probably damaging” (PolyPhen2) variants were identified in these 10 MAPK genes (2 or 3 variants per gene) (Fig. 2). Only 1 variant had allelic frequency greater than 6%.
FIG. 1.
Allelic burden (%) of the somatic variants of the patients with sporadic BAVM included in the discovery cohort. The KRAS G12V mutation, the only recurrent somatic mutation in this cohort, is shown on the first line, followed by the 10 MAPK pathway genes harboring candidate somatic mutations in > 1 patient. All results obtained with WES (n = 14).
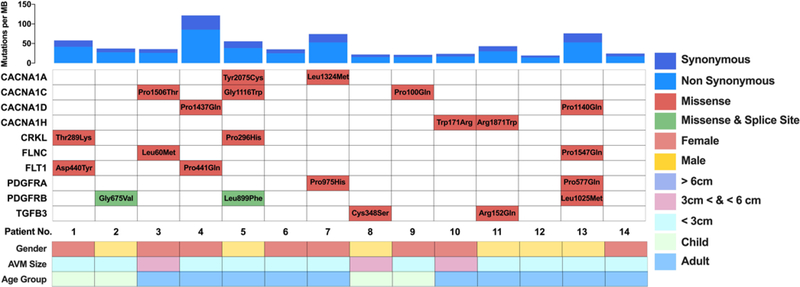
FIG. 2.
Mutational landscape of the MAPK pathway genes with candidate BAVM-associated somatic variants. Upper: Exome-wide somatic mutation rate per megabase (MB) for each patient with sporadic BAVM included in the discovery cohort (n = 14). Lower: Identified candidate somatic variants and phenotypic information (sex, BAVM lesion size, and age group) for each patient.
KRAS G12V and G12D Mutations
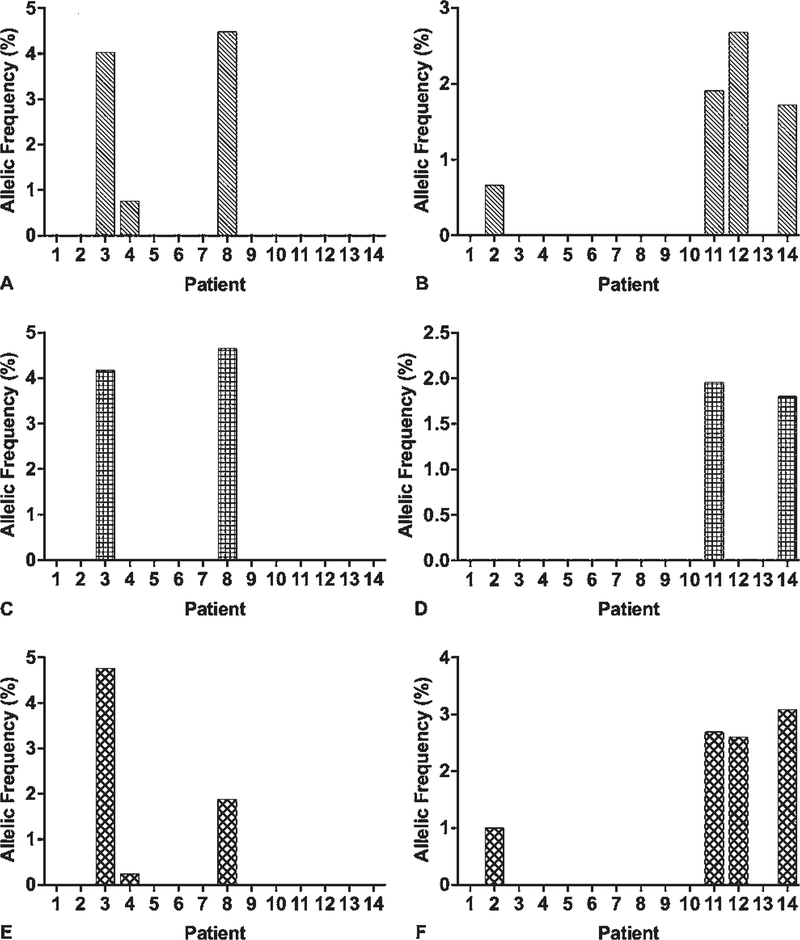
Each of the two previously reported recurrent KRAS mutations16–21 was identified in two different BAVM samples by utilizing Mutect2. The KRAS G12V mutation was among the final group of variants that passed all filters and was identified as a recurrent mutation in our analysis. The KRAS G12D mutation was predicted as “possibly damaging” and was filtered out with functional effect prediction analysis. When we counted the number of sequencing reads that contained these mutations without using any filters, G12V was found in 3 of 14 patients and G12D in 5 patients (Fig. 3A and B); all positive results were subsequently validated with ddPCR (Fig. 3E and F). No samples had positive results for both KRAS mutations. The two KRAS mutations were not detected in any blood samples. No other candidate somatic mutations were identified in KRAS.
FIG. 3.
Identification and validation of KRAS mutations (G12V and G12D) with WES and ddPCR in 14 patients with BAVM. A, C, E: Allelic frequency (%) of KRAS G12V mutation in the 14 patients with BAVM in the discovery cohort. B, D, F: Allelic frequency (%) of KRAS G12D mutation. A and B: Results obtained with WES without filtering. C and D: Results obtained with WES and analyzed with Mutect2. E and F: Results obtained with ddPCR.
Evaluation of Background Mutation Rate
We applied two different approaches, MutSigCV and MUFFINN, to evaluate whether the rate of identified somatic mutations in candidate genes was higher than the expected background mutation rate. Exome-wide, a total of 218 genes were identified as significantly mutated (p < 0.1) with MutSigCV. Six of these were MAPK signaling pathway genes (ATF2, CRKL, DUSP6, KRAS, PDGFB, and PDGFRB). KRAS was the only MAPK pathway gene with recurrent somatic mutation, whereas CRKL and PDGFRB had different mutations in at least 2 patients.
By using MUFFINN, we allocated a score based on the number of candidate mutations and known functional network information to 458 genes. Eleven of these were MAPK pathway genes, including KRAS and all genes listed in Fig. 1. Overall, KRAS, CRKL, and PDGFRB were the only MAPK pathway genes identified as bearing candidate somatic variants associated with BAVM according to both MutSigCV and MUFFINN. Among all MAPK pathway genes, KRAS and PDGFRB were the top two candidate genes containing BAVM-associated variants. MutSigCV analysis identified KRAS as the most significantly mutated gene, followed by PDGFRB; MUFFINN analysis assigned the highest score to PDGFRB, followed by KRAS.
Association of Somatic KRAS Mutations With Clinical Phenotypes of BAVM
By combining the discovery and replication cohorts, we used ddPCR to assay KRAS G12D and G12V in 69 BAVM lesion samples and 32 paired blood samples (Supplemental Table 1). Fifty-six of 69 (81%) samples passed quality control and were included in the statistical analysis. The mean age ± SD at tissue collection was 37.1 ± 17.7 years, and 28 (50%) patients were female. The mean BAVM diameter was 19.2 ± 10.7 mm. Thirty-five (63%) patients had ICH prior to resection at a mean age of 35.7 ± 19.7 years. G12V or G12D somatic mutation was detected in 31 (55%) BAVM DNA samples and in zero blood DNA samples. The somatic mutation allelic burden of KRAS G12D or G12V exceeded 0.5% in 18 (32%) BAVM samples; the highest level detected was 5.0%.
We evaluated the associations between allelic burden of somatic KRAS mutation and three phenotypes of BAVM severity: age at resection, lesion size (maximum diameter), and age at first ICH (Table 1). We found no evidence of an association between somatic KRAS mutation burden and age at BAVM resection (PI 0.7 [95% CI –2.9 to 4.3], p = 0.69), BAVM size (PI 1.4 [95% CI 0.7–3.6], p = 0.18), or age at first ICH (HR 0.87 [95% CI 0.64–1.20], p = 0.40). Individual G12V or G12D mutation burden, or presence of each mutation, was also not significantly associated with any BAVM phenotype (Table 1).
TABLE 1.
Association between allelic burden of somatic KRAS mutation and three phenotypes of BAVM severity
| KRAS Mutation | No. of Patients | Age at Tissue Collection* |
BAVM Size* |
Age at First ICH† |
|||
|---|---|---|---|---|---|---|---|
| Coefficient (95% CI) | p Value | Coefficient (95% CI) | p Value | HR (95% CI) | p Value | ||
|
| |||||||
| G12D or G12V | |||||||
|
| |||||||
| Allelic burden % | 56 | 0.7 (−2.9 to 4.3) | 0.69 | 1.4 (−0.7 to 3.6) | 0.18 | 0.87 (0.64–1.20) | 0.40 |
|
| |||||||
| Presence | 56 | 1.3 (−8.9 to 11.4) | 0.80 | 3.2 (−2.8 to 9.2) | 0.29 | 0.77 (0.35–1.72) | 0.53 |
|
| |||||||
| G12D | |||||||
|
| |||||||
| Allelic burden % | 57 | 2.4 (−2.6 to 7.3) | 0.35 | 1.6 (−1.4 to 4.5) | 0.30 | 0.95 (0.67–1.34) | 0.76 |
|
| |||||||
| Presence | 57 | 1.6 (−9.5 to 12.7) | 0.77 | 2.4 (−4.2 to 9.0) | 0.47 | 0.90 (0.39–2.06) | 0.80 |
|
| |||||||
| G12V | |||||||
|
| |||||||
| Allelic burden % | 65 | −0.6 (−5.1 to 4.0) | 0.80 | 0.9 (−1.6 to 3.4) | 0.48 | 0.87 (0.56–1.35) | 0.53 |
|
| |||||||
| Presence | 65 | 0.4 (−17.1 to 17.9) | 0.97 | 3.0 (−6.6 to 12.5) | 0.54 | 0.68 (0.16–2.96) | 0.61 |
Results of linear regression models, adjusted for sex and prior ICH.
Results of Cox proportional hazards model, adjusted for sex and BAVM size. Patients without ICH at age of tissue collection were censored.
Discussion
We used WES to evaluate paired lesion and blood DNA samples from 14 patients with BAVM for somatic mutations in 295 MAPK pathway genes. Two KRAS mutations (G12V and G12D), which were associated with BAVM in recent studies, were also identified in our study cohort and further validated in a larger cohort with ddPCR. By using these two mutations as a standard and applying a pipeline of stringent filters, we identified candidate BAVM-associated somatic mutations in 10 other MAPK pathway genes.
KRAS Mutations in Patients With BAVM
Overall, KRAS mutations (G12V and G12D) have been identified in at least seven BAVM cohorts,16–21 including ours, which is one of the largest cohorts published to date. Nikolaev et al.16 first reported that 60% of patients with sporadic BAVM carried these KRAS mutations, whereas Hong et al.17 reported that 90% of patients had positive results for KRAS mutation. In our study, we found somatic KRAS mutations in 43% of patients with BAVMs. The lower percentage in our study may be due to, in part, the lower sequencing coverage (70×) of our WES data compared with those of other studies (> 1000×17 or 300×16). However, the reported allelic burden of somatic KRAS mutations was similar in all studies (as high as approximately 5%), including ours. An interesting observation across multiple studies is that G12D was found in more patients with BAVM than G12V, but G12V had a greater average allelic burden (2.01%) than G12D (1.14%). This suggests that a lower allelic burden of the KRAS G12D mutation may be sufficient to produce the BAVM phenotype in comparison with that of G12V, and that G12D may have a stronger functional impact on the KRAS protein in the cerebrovascular context. Functional studies of these KRAS mutations, mostly in epithelial cancer cell lines, suggest that KRAS G12V and G12D mutations may have differential effects on proliferation, selection, and downstream signaling.34,35
Across the seven studies on sporadic BAVM that have been conducted to date, KRAS is the only gene that has been consistently found with recurrent somatic mutations.17–21 This implies that KRAS may play an important role in the etiology of BAVM and may be a potential therapeutic target in the majority of patients with sporadic BAVM. KRAS mutations are commonly found in patients with many types of cancer (including colorectal, pancreatic, and lung cancer36) and other angiogenesis-related disorders such as endometriosis.37
We also evaluated the associations between somatic KRAS mutations (allelic burden or presence) and three phenotypes of BAVM severity (age at resection, lesion size, or age at first hemorrhage) that we hypothesized could be affected by somatic mutation burden; however, we did not detect any statistically significant associations. A potential explanation for the lack of association may be confounding by the variable presence of nonlesion cells in the grossly cryosectioned samples. A more precise evaluation of allelic burden in BAVM lesion cells may be necessary to detect an association: for example, the endothelial cells of BAVM lesions may need to be isolated first with laser capture microscopy. The lack of a statistically significant association between somatic KRAS mutation allelic burden and phenotype could be due to low precision because the 95% CIs are wide (Table 1). For example, the 95% CIs for age at tissue collection and presence of a mutation all have spans greater than 20 years.
Candidate BAVM-Associated Mutations in Other MAPK Pathway Genes
We identified 10 other genes in the MAPK pathway with candidate somatic mutations that were present in multiple patients included in the discovery cohort. Among these, PDGFRA and PDGFRB have differential roles in angiogenesis and vessel stability.38 PDGFRB was one of the three genes (alongside KRAS and CRKL) with candidate somatic mutations that was detected more often than expected. Experimental animal models of BAVM show reduced Pdgfr-β expression,39 and a significant reduction in PDGFRB-positive pericyte coverage has been observed in BAVM vessels and adjacent perivascular space, suggesting a potential mechanism by which damaging somatic mutations in PDGFRB may contribute to BAVM development.40 Recently, activating PDGFRB somatic mutations have also been identified in fusiform (but not saccular) cerebral aneurysms, further supporting a role in cerebrovascular anomaly development.41 CRKL, the third gene with an excess of candidate somatic mutations in BAVM lesions, is a putative proto-oncogene that is upregulated in multiple types of cancers.42 In vitro, amplification of CRKL in EGFR-mutant cells induces drug resistance via activation of ERK and AKT signaling,43 whereas CRKL overexpression promotes cell invasion via upregulation of MMP-9 expression.44
Study Limitations
This study has several limitations. Subfractionation was not used to isolate BAVM lesion cells or endothelial cells, and this may have reduced the amount of mutant DNA molecules in the sample. As discussed above, the relatively low sequencing coverage of our original WES sequencing data set (70× compared with 300× to > 1000× in other studies16,17) reduced sensitivity to detect candidate mutations with allelic representation < 2%. We also observed variation in coverage across samples (Supplemental Table 2 and Supplemental Fig. 2) and uneven coverage of reads over the targeted regions within samples (data not shown), and these further reduced the detection and the accuracy of variant calling.
Genetic Heterogeneity in BAVM
BAVM is a complex lesion with likely genetic heterogeneity. In all previous studies, as well as in ours, somatic KRAS mutations were not found in every patient. We identified several other candidate mutations in MAPK pathway genes; in some cases, somatic mutations in multiple genes were identified in a single patient. Although these mutations remain to be validated, they suggest that a single mutation may be insufficient to induce BAVM formation, and instead multiple hits to genes in the same pathway may be required. These gene hits may come from other mechanisms, e.g., epigenetic inactivation, or BAVM may be related to aberrations in other genes and pathways. This observed genetic heterogeneity ultimately suggests that different therapeutic approaches along with reliable biomarkers may be needed for different patients. In the future, diagnostic sequencing of patient samples (e.g., via targeted sequencing of cell-free DNA or endovascular sampling of lesion cells45) could be used to match patient subgroups with appropriate targeted therapeutic strategies.
Conclusions
Our study suggests that BAVM exhibits somatic mutational heterogeneity, because different combinations of rare somatic mutations in MAPK pathway genes were observed in BAVM lesions (but not in blood DNA). In our study, we replicated the high prevalence of two previously identified KRAS mutations (G12V and G12D), confirming that these two mutations are the most prevalent somatic mutations present in BAVM tissue. We did not find a significant association between somatic KRAS mutation burden and three phenotypes of BAVM severity, but more sensitive detection of mutation burden may be needed. We also identified several new candidate somatic mutations associated with sporadic BAVM, including mutations in PDGFRB and CRKL. Validation and functional studies of these mutations are needed to elucidate the biological processes leading to BAVM.
Supplementary Material
Acknowledgments
We thank the patients and their families for their participation in our studies, and members of the UCSF BAVM Project and the Center for Cerebrovascular Research for technical assistance and study support. This project was supported in part by the following grants: NIH NS034949 and NS099268 (awarded to H.K.), NS070029 (L.P.), NS027713 and NS112819 (H.S.), AHA 19POST34370066 (S.G.), a Christopher C. Getch fellowship from the Congress of Neurological Surgeons (B.P.W.) and the Michael Ryan Zodda Foundation (H.S.).
Disclosures
Dr. Gao was supported by the China Scholarship Council. Dr. Walcott was supported by a Christopher C. Getch fellowship (Congress of Neurological Surgeons). Mr. Nelson receives salary support from an NIH grant. Dr. Hetts receives support of non–study-related clinical or research effort from Siemens, Stryker, MicroVention, and Cerenovus and owns stock in Filtro Medical. Dr. Kim receives clinical or research support for the study described (includes equipment or material) from NIH.
ABBREVIATIONS
- BAVM
brain arteriovenous malformation
- ddPCR
digital droplet polymerase chain reaction
- GATK
Genome Analysis Toolkit
- ICH
intracerebral hemorrhage
- MAPK
mitogen-activated protein kinase
- OCT
optimal cutting temperature
- PI
proportional increase
- SNV
single-nucleotide variant
- TLOD
theta logarithm of the odds
- UCSF
University of California, San Francisco
- WES
whole-exome sequencing
Footnotes
Supplemental Information
Online-Only Content
Supplemental material is available with the online version of the article.
Supplemental Tables and Figures. https://thejns.org/doi/suppl/10.3171/2020.11.JNS202031.
References
- 1.Berman MF, Sciacca RR, Pile-Spellman J, et al. The epidemiology of brain arteriovenous malformations. Neurosurgery. 2000; 47(2): 389–397. [DOI] [PubMed] [Google Scholar]
- 2.Gabriel RA, Kim H, Sidney S, et al. Ten-year detection rate of brain arteriovenous malformations in a large, multiethnic, defined population. Stroke. 2010; 41(1): 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedlander RM. Clinical practice. Arteriovenous malformations of the brain. N Engl J Med. 2007; 356(26): 2704–2712. [DOI] [PubMed] [Google Scholar]
- 4.Kim H, Al-Shahi Salman R, McCulloch CE, et al. Untreated brain arteriovenous malformation: patient-level meta-analysis of hemorrhage predictors. Neurology. 2014; 83(7): 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim H, Pawlikowska L, Su H, Young WL. Genetics and vascular biology of brain vascular malformations (Chapter 12). In: Grotta JC, Albers GW, Broderick JP, et al. , eds. Stroke: Pathophysiology, Diagnosis, and Management. 6th ed. Churchill Livingstone Elsevier; 2016: 149–162. [Google Scholar]
- 6.Solomon RA, Connolly ES Jr. Arteriovenous malformations of the brain. N Engl J Med. 2017; 377(5): 498. [DOI] [PubMed] [Google Scholar]
- 7.Rutledge WC, Abla AA, Nelson J, et al. Treatment and outcomes of ARUBA-eligible patients with unruptured brain arteriovenous malformations at a single institution. Neurosurg Focus. 2014; 37(3): E8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potts MB, Lau D, Abla AA, et al. Current surgical results with low-grade brain arteriovenous malformations. J Neurosurg. 2015; 122(4): 912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Beijnum J, van der Worp HB, Buis DR, et al. Treatment of brain arteriovenous malformations: a systematic review and meta-analysis. JAMA. 2011; 306(18): 2011–2019. [DOI] [PubMed] [Google Scholar]
- 10.Limaye N, Wouters V, Uebelhoer M, et al. Somatic mutations in angiopoietin receptor gene TEK cause solitary and multiple sporadic venous malformations. Nat Genet. 2009; 41(1): 118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macmurdo CF, Wooderchak-Donahue W, Bayrak-Toydemir P, et al. RASA1 somatic mutation and variable expressivity in capillary malformation/arteriovenous malformation (CM/AVM) syndrome. Am J Med Genet A. 2016; 170(6): 1450–1454. [DOI] [PubMed] [Google Scholar]
- 12.Limaye N, Kangas J, Mendola A, et al. Somatic activating PIK3CA mutations cause venous malformation. Am J Hum Genet. 2015; 97(6): 914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akers AL, Johnson E, Steinberg GK, et al. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum Mol Genet. 2009; 18(5): 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirley MD, Tang H, Gallione CJ, et al. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med. 2013; 368(21): 1971–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soblet J, Limaye N, Uebelhoer M, et al. Variable somatic TIE2 mutations in half of sporadic venous malformations. Mol Syndromol. 2013; 4(4): 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikolaev SI, Vetiska S, Bonilla X, et al. Somatic activating KRAS mutations in arteriovenous malformations of the brain. N Engl J Med. 2018; 378(3): 250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong T, Yan Y, Li J, et al. High prevalence of KRAS/BRAF somatic mutations in brain and spinal cord arteriovenous malformations. Brain. 2019; 142(1): 23–34. [DOI] [PubMed] [Google Scholar]
- 18.Al-Olabi L, Polubothu S, Dowsett K, et al. Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy. J Clin Invest. 2018; 128(4): 1496–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goss JA, Huang AY, Smith E, et al. Somatic mutations in intracranial arteriovenous malformations. PLoS One. 2019; 14(12): e0226852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priemer DS, Vortmeyer AO, Zhang S, et al. Activating KRAS mutations in arteriovenous malformations of the brain: frequency and clinicopathologic correlation. Hum Pathol. 2019; 89: 33–39. [DOI] [PubMed] [Google Scholar]
- 21.Oka M, Kushamae M, Aoki T, et al. KRAS G12D or G12V mutation in human brain arteriovenous malformations. World Neurosurg. 2019; 126: e1365–e1373. [DOI] [PubMed] [Google Scholar]
- 22.Ayturk UM, Couto JA, Hann S, et al. Somatic activating mutations in GNAQ and GNA11 are associated with congenital hemangioma. Am J Hum Genet. 2016; 98(6): 1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couto JA, Vivero MP, Kozakewich HP, et al. A somatic MAP3K3 mutation is associated with verrucous venous malformation. Am J Hum Genet. 2015; 96(3): 480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Couto JA, Huang AY, Konczyk DJ, et al. Somatic MAP2K1 mutations are associated with extracranial arteriovenous malformation. Am J Hum Genet. 2017; 100(3): 546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25(14): 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20(9): 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011; 43(5): 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cingolani P, Platts A, Wang LL, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012; 6(2): 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7(4): 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013; 499(7457): 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho A, Shim JE, Kim E, et al. MUFFINN: cancer gene discovery via network analysis of somatic mutation data. Genome Biol. 2016; 17(1): 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramos AH, Lichtenstein L, Gupta M, et al. Oncotator: cancer variant annotation tool. Hum Mutat. 2015; 36(4): E2423–E2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Shashikant CS, Jensen M, et al. Novel metrics to measure coverage in whole exome sequencing datasets reveal local and global non-uniformity. Sci Rep. 2017; 7(1): 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monticone M, Biollo E, Maffei M, et al. Gene expression deregulation by KRAS G12D and G12V in a BRAF V600E context. Mol Cancer. 2008; 7: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stolze B, Reinhart S, Bulllinger L, et al. Comparative analysis of KRAS codon 12, 13, 18, 61, and 117 mutations using human MCF10A isogenic cell lines. Sci Rep. 2015; 5: 8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braicu C, Buse M, Busuioc C, et al. A comprehensive review on MAPK: a promising therapeutic target in cancer. Cancers (Basel). 2019; 11(10): E1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anglesio MS, Papadopoulos N, Ayhan A, et al. Cancer-associated mutations in endometriosis without cancer. N Engl J Med. 2017; 376(19): 1835–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Ingram DA, Murphy MP, et al. Release of proinflammatory mediators and expression of proinflammatory adhesion molecules by endothelial progenitor cells. Am J Physiol Heart Circ Physiol. 2009; 296(5): H1675–H1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W, Guo Y, Walker EJ, et al. Reduced mural cell coverage and impaired vessel integrity after angiogenic stimulation in the Alk1-deficient brain. Arterioscler Thromb Vasc Biol. 2013; 33(2): 305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winkler EA, Bell RD, Zlokovic BV. Pericyte-specific expression of PDGF beta receptor in mouse models with normal and deficient PDGF beta receptor signaling. Mol Neurodegener. 2010; 5: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karasozen Y, Osbun JW, Parada CA, et al. Somatic PDGFRB activating variants in fusiform cerebral aneurysms. Am J Hum Genet. 2019; 104(5): 968–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song Q, Yi F, Zhang Y, et al. CRKL regulates alternative splicing of cancer-related genes in cervical cancer samples and HeLa cell. BMC Cancer. 2019; 19(1): 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheung HW, Du J, Boehm JS, et al. Amplification of CRKL induces transformation and epidermal growth factor receptor inhibitor resistance in human non-small cell lung cancers. Cancer Discov. 2011; 1(7): 608–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin F, Chengyao X, Qingchang L, et al. CRKL promotes lung cancer cell invasion through ERK-MMP9 pathway. Mol Carcinog. 2015; 54(suppl 1): E35–E44. [DOI] [PubMed] [Google Scholar]
- 45.Sun Z, Lawson DA, Sinclair E, et al. Endovascular biopsy: strategy for analyzing gene expression profiles of individual endothelial cells obtained from human vessels. Biotechnol Rep (Amst). 2015; 7: 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.