Abstract
Five hundred specimens (288 genital, 192 dermal, and 20 ocular) were extracted by technologists, and the DNA was assayed by LightCycler PCR (DNA polymerase and thymidine kinase [TK] gene targets) and by conventional tube and shell vial cell culture. One hundred fifty-eight confirmed (by cell culture and TK target PCR) positive and LightCycler-positive specimens were detected during the first 30 PCR cycles. LightCycler PCR-positive results for cycles 31 to 45 (39 of 67 [58.2%]) required confirmation by another PCR target (TK). LightCycler PCR is more sensitive (n = 197; 23.1%) than cell cultures (n = 150) for the routine laboratory detection of herpes simplex virus infections.
For more than 20 years, herpes simplex virus (HSV) has been recognized as an important cause of recurrent infections (genital, dermal, and ocular) of immunologically competent hosts; often, spread of the virus in compromised patients produces life-threatening systemic disease (1, 3, 5, 6, 7).
LightCycler PCR was demonstrated to be a more sensitive (4) and rapid assay than shell vial cell cultures. Importantly, the automated molecular method is a closed system that is completely adaptable for implementation in the routine clinical laboratory with P-2 containment standards.
Genital specimens (n = 288), dermal swab specimens (n = 192), and ocular specimens (n = 20) from patients suspected of having HSV infections were each combined with 2 ml of serum-free medium, and each specimen extract was divided into two equal aliquots. Each of two MRC-5 shell vial cell cultures received 0.2 ml of inoculum from one aliquot. The vials were centrifuged, incubated overnight at 36°C, and stained by the indirect immunofluorescence test as previously described (4).
Nucleic acids were extracted from a 0.2-ml serum-free extract of genital, dermal, and ocular swab specimens by the IsoQuick procedure (Orca Research, Inc., Bothell, Wash.) according to the manufacturer's instructions (2).
LightCycler PCR (Roche Molecular Biochemicals, Indianapolis, Ind.) amplifies and monitors the development of the target nucleic acid by a fluorescence assay after each cycle (denaturation, annealing, and extension). This instrument provided rapid (30 to 40 min) results and automation of PCR by precise air-controlled temperature cycling and provided continuous monitoring of amplicon development by a fluorometer incorporated into the LightCycler (2). Extraction of DNA from specimens, primers for amplification of gene targets (DNA polymerase and thymidine kinase), fluorescence-labeled (using the fluorescence resonance energy transfer principle) probes, composition of master mix, and cycling conditions (times and temperatures), were previously described in detail (2). Melting curve features of the LightCycler software were used to distinguish between the two genotypes of HSV (2).
Overall, 500 specimens (288 genital, 192 dermal, and 20 ocular) were processed for the detection of HSV by LightCycler PCR and by conventional tube and shell vial assay. Two hundred twenty-five (45%) specimens yielded positive results for HSV infection. All were positive by LightCycler PCR (DNA polymerase gene target); seventy-five specimens were positive exclusively by the LightCycler assay. HSV was never detected in cell cultures for which PCR gave negative results (specificity, 100%). Genotype results obtained by LightCycler PCR completely agreed with results of serotyping by monoclonal antibodies in the shell vial assay (HSV type 1, n = 66; HSV type 2, n = 82).
Of the total 225 strains detected by LightCycler PCR, 150 specimens were positive by both (n = 147) or exclusively by conventional tube (n = 2) or shell vial (n = 1) cell cultures. These data can be sorted into two main groups based on the PCR cycle in which HSV DNA was detected by the LightCycler. One hundred fifty-eight specimens were positive for HSV DNA by LightCycler PCR during the first 30 cycles of amplification (range, 9 to 30 cycles). Of these 158 specimens, 135 HSV strains (85.4%) were detected in cell cultures (Table 1). Importantly, of the 23 HSV strains in which specific viral DNA was detected exclusively by LightCycler PCR directed to the DNA polymerase gene, all samples were also positive by LightCycler PCR directed to the thymidine kinase gene target. Therefore, we considered all samples (n = 158) in which HSV was detected by LightCycler PCR and cell cultures (n = 135) or by an alternate PCR assay directed to another gene target of the virus (n = 23) within the first 30 cycles of amplification to be confirmed positive results.
TABLE 1.
Detection of herpes simplex virus DNA according to cycle number by LightCycler PCR and cell culture assays
| Cycle no. | No. positive by:
|
||
|---|---|---|---|
| LightCycler PCR
|
Cell culture | ||
| DNA polymerase gene | Thymidine kinase gene | ||
| 0–30 | 158 | 23a | 135 |
| 31 | 8 | 5 | 1 |
| 32 | 10 | 7 | 4 |
| 33 | 6 | 6 | 2 |
| 34 | 4 | 2 | 2 |
| 35 | 6 | 3 | 2 |
| 36 | 10 | 7 | 2 |
| 37 | 4 | 2 | 1 |
| 38 | 10 | 5 | 1 |
| 39 | 3 | 1 | 0 |
| 40 | 2 | 1 | 0 |
| 41 | 4 | 0 | 0 |
| 42–45 | 0 | 0 | 0 |
| Subtotal for cycles 31–45 | 67 | 39 | 15 |
| Total | 225 | 62 | 150 |
For cycles 1 to 30, the LightCycler PCR thymidine kinase gene assay was performed only on discrepant (LightCycler PCR-DNA polymerase-positive, cell culture-negative) samples.
Of the initial total of 75 discrepant samples, in which HSV was detected exclusively by the LightCycler PCR directed to the DNA polymerase gene, 67 (89%) of the HSV DNA-positive specimens were amplified from cycles 31 through 41 (Table 1). Thirty-nine of these 67 samples (58.2%) were also HSV DNA positive by the alternative LightCycler PCR directed to the thymidine kinase gene target.
LightCycler PCR is ideally suited for implementation in the routine P-2 biosafety level clinical laboratory due to characteristics of the instrument, such as the physical (closed reaction vessel) and chemical (uracyl-DNA glycosylase) controls for amplicon containment, rapid cycling format (45 cycles in 30 to 40 min), capability for product genotyping (melting curves), and potential for nucleic acid quantitation.
Carryover amplicon contamination should not be a concern, since the target DNA extract is added to the reaction after the cuvettes are seated in the carousel. Thus, the only risk of cuvette breakage occurs prior to the addition of the specimen. Secondly, uracyl-DNA glycosylase is present in the reaction mix. Finally, PCR “real-time” detection occurs in closed vessels. Upon completion of the analysis, cuvettes are removed and discarded into disposable bags in a safety cabinet equipped with a UV light. Collectively, in two large validation studies of LightCycler PCR, we processed 700 specimens (448 genital, 230 dermal, and 22 ocular) for the laboratory diagnosis of HSV infections. Importantly, of the 285 LightCycler confirmed-positive specimens (203 genital [45.3%], 77 dermal [33.5%], and 5 ocular [22.7%]), HSV was detected in only 217 shell vial cell culture specimens (76%) (2).
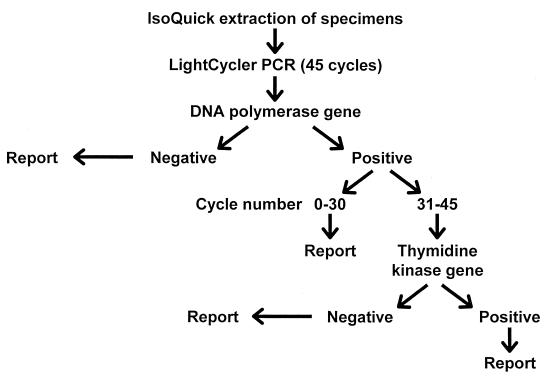
Based on our data, all LightCycler PCR-positive results from amplification cycles 31 to 45 would require confirmation by a second independent PCR assay (Fig. 1). In our study, a confirmatory PCR assay would be required for 67 specimens (225 to 158) analyzed by the initial LightCycler PCR directed to the DNA polymerase gene. The workload implications are that 67 of 225 (29.8%) total LightCycler-positive results would require confirmation by a second LightCycler PCR assay (thymidine kinase target). Ultimately, 39 of these 67 (58.2%) specimens were positive by both LightCycler PCR assays. In total, 158 of 225 specimens (cycles 1 to 30) and 39 of 67 specimens (cycles 31 to 45) were reported as positive for HSV after one or two LightCycler PCR assays (197 of 225) (87.6%). Therefore, 197 confirmed HSV-positive specimens were detected, rather than the 150 detected by cell culture techniques, resulting in an increased sensitivity of 23.1% for LightCycler PCR compared with results for cell culture.
FIG. 1.
Laboratory diagnosis of herpes simplex virus infections.
Direct and indirect costs per procedure, cost of capital, and variable (time for performance is related to the number of specimens processed) and fixed (constant time regardless of the number of samples) effort for performance of the test procedures were all lower for the LightCycler PCR assay than for the shell vial and conventional cell culture procedures. For example, based on our test volumes for processing a specimen from the genital tract, the variable allied health time required for the shell vial cell culture assay was 2.5 times that required for performing the LightCycler PCR assay. Nevertheless, the cumulative direct costs of the two assays differed by only $1.30.
The LightCycler assay for HSV DNA will have a large impact on our laboratory practice since, at the Mayo Clinic, almost three-fourths of all viruses detected in cell cultures during the last five years have been HSV strains. With validation data for LightCycler PCR for the laboratory diagnosis of varicella-zoster virus infection, we plan to extend and implement this technology for the detection of both HSV and varicella-zoster virus from dermal specimens in addition to specimens from genital sources.
REFERENCES
- 1.Brugha R, Keersmakers K, Renton A, Meheus A. Genital herpes infection: a review. Int J Epidemiol. 1997;26:698–709. doi: 10.1093/ije/26.4.698. [DOI] [PubMed] [Google Scholar]
- 2.Espy M J, Uhl J R, Mitchell P S, Thorvilson J N, Svien K A, Wold A D, Smith T F. Diagnosis of herpes simplex virus infections in the clinical laboratory by LightCycler PCR. J Clin Microbiol. 2000;38:795–799. doi: 10.1128/jcm.38.2.795-799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gelven P L, Gruber K K, Swiger F K, Cina S J, Harley R A. Fatal disseminated herpes simplex in pregnancy with maternal and neonatal death. South Med J. 1996;89:732–734. doi: 10.1097/00007611-199607000-00018. [DOI] [PubMed] [Google Scholar]
- 4.Gleaves C A, Wilson D J, Wold A D, Smith T F. Detection and serotyping of herpes simplex virus in MRC-5 cells by use of centrifugation and monoclonal antibodies 16 h postinoculation. J Clin Microbiol. 1985;21:29–32. doi: 10.1128/jcm.21.1.29-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang A H, Graves C R. Herpes simplex hepatitis in pregnancy: a case report and review of the literature. Obstet Gynecol Survey. 1999;54:463–468. doi: 10.1097/00006254-199907000-00026. [DOI] [PubMed] [Google Scholar]
- 6.Kaufman B, Gandhi S A, Louie E, Rizzi R, Illei P. Herpes simplex virus hepatitis: case report and review. Clin Infect Dis. 1997;24:334–338. doi: 10.1093/clinids/24.3.334. [DOI] [PubMed] [Google Scholar]
- 7.Levitz R E. Herpes simplex encephalitis: a review. Heart Lung. 1998;27:209–212. doi: 10.1016/s0147-9563(98)90009-7. [DOI] [PubMed] [Google Scholar]