Abstract
The 3′ poly(A) structure improves translation of a eukaryotic mRNA by 50-fold in vivo. This enhancement has been suggested to be due to an interaction of the poly(A) binding protein, Pab1p, with eukaryotic translation initiation factor 4G (eIF4G). However, we find that mutation of eIF4G eliminating its interaction with Pab1p does not diminish the preference for poly(A)+ mRNA in vivo, indicating another role for poly(A). We show that either the absence of Fun12p (eIF5B), or a defect in eIF5, proteins involved in 60S ribosomal subunit joining, specifically reduces the translation of poly(A)+ mRNA, suggesting that poly(A) may have a role in promoting the joining step. Deletion of two nonessential putative RNA helicases (genes SKI2 and SLH1) makes poly(A) dispensable for translation. However, in the absence of Fun12p, eliminating Ski2p and Slh1p shows little enhancement of expression of non-poly(A) mRNA. This suggests that Ski2p and Slh1p block translation of non-poly(A) mRNA by an effect on Fun12p, possibly by affecting 60S subunit joining.
The 5′ cap (7-methyl-GpppG…) and 3′ poly(A) structures of eukaryotic mRNAs are both critically important for translation, for mRNA transport from the nucleus, and for mRNA stability in both the nucleus and the cytoplasm. The in vivo requirement for translation for the 3′ poly(A) structure, estimated by electroporation of mRNAs into living cells, is about 50-fold, while that for the 5′ cap is about 20-fold (11, 12).
The effects of the 5′ cap on translation are mediated by the cap-binding protein, eukaryotic translation initiation factor 4E (eIF4E) (Cdc33p), which associates with eIF4G in the eIF4F complex to promote binding of the mRNA to the 40S ribosomal subunit (through eIF3). A 43S preinitiation complex consisting of the 40S subunit, eIF3, and eIF2-GTP-Met-tRNA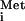 is thought to bind to an mRNA mediated by interaction between eIF3 and eIF4G. This complex scans from the cap at the 5′ end of the mRNA to the first AUG. There, with the help of eIF5 (Tif5p) and eIF5B (Fun12p), 60S subunit joining occurs and translation begins (27; reviewed in references 9 and 37). 60S subunit joining requires both eIF5 and eIF5B. While eIF5 promotes GTP hydrolysis by eIF2, enabling release of the initiation factors from the 40S subunit, eIF5B has its own GTP binding activity and hydrolyzes GTP in a ribosome-dependent reaction (27).
is thought to bind to an mRNA mediated by interaction between eIF3 and eIF4G. This complex scans from the cap at the 5′ end of the mRNA to the first AUG. There, with the help of eIF5 (Tif5p) and eIF5B (Fun12p), 60S subunit joining occurs and translation begins (27; reviewed in references 9 and 37). 60S subunit joining requires both eIF5 and eIF5B. While eIF5 promotes GTP hydrolysis by eIF2, enabling release of the initiation factors from the 40S subunit, eIF5B has its own GTP binding activity and hydrolyzes GTP in a ribosome-dependent reaction (27).
The role of the 3′ poly(A) structure in mRNA translation is not yet completely clear. The poly(A) binding protein (Pab1p) is believed to mediate many of the effects of the 3′ poly(A) structure (reviewed in reference 34). Because the poly(A) tail apparently has roles in nuclear processes as well as cytoplasmic events (22), dissecting these mechanisms is difficult. The PAB1 gene is essential, indicating that at least one of the functions of Pab1p is critical to the cell.
It has recently been observed that eIF4G and Pab1p bind to each other in the presence of poly(A) (38). This binding would be expected to circularize the mRNA, since eIF4G is attached to the 5′ cap by its association with eIF4E and to the 3′ poly(A) by its binding to Pab1p. It has been suggested that this is how the poly(A) tail functions to promote translation of mRNAs. However, mutations of eIF4G that eliminate the binding to Pab1p do not affect the growth rate of cells (40), suggesting Pab1p may activate translation by a different mechanism in vivo.
Biochemical evidence suggested that the role of poly(A) was to promote joining of 60S subunits to the 40S subunit waiting at the initiator AUG (23), but these data showed only a twofold effect and doubts have been raised about this conclusion. A pab1ts mutant accumulates free 60S subunits at the nonpermissive temperature (32), just the result expected if 60S joining is the defective step. However, the relationship of the action of the joining factors Fun12p and Tif5p to the poly(A) structure has not been examined.
Although there is a strict requirement for the 3′ poly(A) structure for translation, both in vivo and in vitro, recent work indicates that this requirement is one imposed by the inhibition of translation of non-poly(A) mRNAs by two homologous nonessential RNA helicases, Ski2p and Slh1p (20, 21, 35, 41). Mutations in SKI2 and SLH1, or other nonessential genes involved in this activity (SKI3, SKI7, and SKI8; see also reference 3), derepress translation of non-poly(A) mRNAs introduced by electroporation, the naturally poly(A)− mRNAs produced by the L-A and L-BC double-stranded RNA (dsRNA) viruses of yeast or the poly(A)− mRNAs produced from an RNA polymerase I promoter. Further, ski2Δ slh1Δ double mutants treat poly(A)+ and poly(A)− mRNAs the same, translating them at the same rate and for the same duration (35). Ski2p, Ski3p, and Ski8p are found in a cytoplasmic complex (4), but their precise role is unclear. As in yeast, there are two human Ski2p homologues (8, 19, 24, 30).
Here we sought to relate the effects of Ski2p and Slh1p in blocking the translation of non-poly(A) mRNA to the translation-promoting effects of Fun12p and Pab1p. Our results support a role of the poly(A) structure in 60S ribosomal subunit joining promoted by Fun12p (eIF5B) and Tif5p (eIF5) but argue against a role for the Pab1p-eIF4G interaction in mediating the poly(A) requirement for translation. We find that the derepression of translation of non-poly(A) mRNA by the ski2Δ slh1Δ double mutation is abrogated by deficiency of Fun12p, suggesting a model in which Ski2p and Slh1p inhibit Fun12p action on mRNAs lacking a 3′ poly(A) structure.
MATERIALS AND METHODS
Strains.
Strains of S. cerevisiae used are shown in Table 1.
TABLE 1.
Strains of S. cerevisiae
| Strain | Genotype | Source or reference |
|---|---|---|
| 3221 | MATahis3 trp1 ura3 L-A-o M-o Gal+ | 35 |
| 4107 | MATahis3 trp1 ura3 slh1::URA3 ski2::HIS3 L-A-o M-o | 35 |
| H117 | MATaura3-52 his1-29 gcn2-101 gcn3-101 ino1 <HIS4-lacZ, URA3> | 15 |
| H272 | MATaura3-52 his1-29 gcn2-101 gcn3-101 ino1 <HIS4-lacZ, URA3> gcd11-508 | 15 |
| H2783 | MATaleu2-3 leu2-112 prt1-kanMX p[PRT1-SalI, URA3] | 28 |
| H1676 | MATaprt1-1 leu2-3 leu2-112 ura3-52 | 28 |
| KAY71 | MATassu2-1 ura3-52 his4 p[URA3] | K. Asano and T. Donahuea |
| KAY73 | MATassu2-1 ura3-52 his4 p[TIF5, URA3] | K. Asano and T. Donahuea |
| YAS43 | MATahis3 leu2 trp1 ura3 | 32 |
| YAS216 | MATα spb2-1 his3 leu2 trp1 ura3 | 32 |
| YAS227 | MATα spb2-1 PAB1::HIS3 ade2 his3 leu2 trp1 ura3 | 32 |
| 4G1 | MATaade2 his3 leu2 trp1 ura3 pep4::HIS3 tif4631::LEU2 tif4632::ura3 p[TIF4632] | This studyb |
| 4G2 | MATaade2 his3 leu2 trp1 ura3 pep4::HIS3 tif4631::LEU2 tif4632::ura3 p[tif4632-233] | This studyb |
| J115 | MATaura2-52 leu2-3 leu2-112 fun12Δ p[FUN12, URA3] | This studyc |
| J116 | MATaura3-52 leu2-3 leu2-112 fun12Δ p[URA3] | This studyc |
| 4697-4A | MATα ura3 trp1 leu2 his3 ski2::HIS3 fun12::LEU2 | This studyd |
| AMS3 | MATα ura3 trp1 leu2 his3 ski2::HIS3 fun12::LEU2 slh1::URA3 | This studye |
KAY71 and KAY73 were a gift of Katsura Asano and Tom Donahue. They were constructed by transforming the ssu2-1 (tif5) strain JRC179-4D with either the empty vector YCplac33 or the TIF5 plasmid pKA235 (1).
Strains 4G1 and 4G2 were constructed in strain YAS1948 (MATa ade2 his3 leu2 trp1 ura3 pep4::HIS3 tif4631::LEU2 tif4632::ura3 p[TIF4632 URA3 CEN]) (generously provided by Alan Sachs). YAS1948 was grown on 5-fluoroorotic acid and transformed with either p[TIF4632, TRP1] to generate 4G1 or transformed with p[tif4632-233, TRP1] to generate 4G2 (as described previously [40]).
Strains were generated by transforming strain J111 (7) with FUN12 plasmid pC479 (6) or empty vector pRS316 (36).
Strain 4697-4A was derived from a cross between 3515 (MATa ski2::HIS3 his3 ura3 trp1 GAL+) and SC224-4B (MATα ura3-52 leu2 fun12::LEU2).
The slh1::URA3 disruption was constructed in 4697-4A (MATα ura3 trp1 leu2 his3 ski2::HIS3 fun12::LEU2) with the disruption plasmid described by Martegani et al. (20), forming strain AMS3. The Ura+ colonies were examined by colony PCR, confirming the absence of the normal gene and the presence of the disruption construct.
Electroporation. (i) Preparation of cells.
A total of 50 ml of cells was grown to an optical density at 600 nm (OD600) of 0.6. Cells were pelleted, resuspended in 4 ml of spheroplast buffer A (50 mM Tris Cl, pH 7.5; 1 mM MgCl2; 30 mM dithiothreitol; 15 mM β-mercaptoethanol; 1 M sorbitol), and 200 μl of a 5-mg/ml solution of zymolase 20T in spheroplast buffer A was added. Incubation at 30°C with gentle swirling was continued until spheroplasts were formed as determined empirically for each strain by diluting the cells 1:100 in sorbitol and in H2O and measuring the decrease in OD600. Spheroplasts were washed gently in 5 ml of spheroplast buffer A and pelleted for 5 min at 1,000 rpm in a SS-34 rotor. Cells were resuspended in 1 ml of spheroplast buffer A, added to 9 ml of YPAD (1% yeast extract, 2% peptone, 2% dextrose, 0.04% adenine sulfate)–1 M sorbitol, and incubated for 90 min at 30°C with gentle swirling to allow the cells to recover from the zymolase treatment. Following recovery, cells were gently washed twice using 5 ml of 1 M sorbitol (spinning at 1,000 rpm for 5 min in an SS-34 rotor). Cells were resuspended in 1 ml of 1 M sorbitol at 2 × 108 cells/ml and used for electroporation. (see references 11 and 12).
(ii) Preparation of mRNAs.
For the measurement of translation in vivo, we used reporter luciferase mRNAs described first by Gallie (12). The T7 promoter-based luciferase constructs were linearized as follows: LUC A0 is linearized with SmaI, and LUC A50 is linearized with DraI. LUC A50 produces an mRNA with a poly(A)50 tail. The capped poly(A)+ (C+A+) and poly(A)− (C+A−) mRNAs were synthesized with Ambion's mMessage mMachine kit. The uncapped mRNAs (C−A+ and C−A−) were synthesized with the MegaScript kit (Ambion). A total of 2 μg of RNA was used per electroporation.
(iii) Electroporation procedure.
Prior to electroporation, the electroporation cuvettes (0.2-cm electrode gap; Bio-Rad) were kept on ice. Then, 2 μg of each reporter mRNA and 180 μl of yeast spheroplasts were added to a cuvette and pulsed immediately (800 V, 25 F, 1,000 Ω). This results in a pulse that ranges from 20 to 25 ms in duration. Immediately following the electroporation pulse, 1.2 ml of ice-cold YPAD–1 M sorbitol was added to the cuvettes, which were kept on ice.
(iv) Measurement of reporter activity.
The spheroplasts were transferred to ice-cold tubes (Falcon 2059) and placed at room temperature (or at 37°C for the temperature-sensitive mutants) with gentle swirling. Samples were collected at 0, 5, 10, 20, 40, 60, and 120 min. Then, 150 to 200 μl of cells were removed at each time point, the cells were spun down, and the cell pellets were frozen immediately in an ethanol-dry ice bath. To measure luciferase activity, we used the Luciferase Assay Reporter system (Promega). The cell pellets were resuspended in 50 to 100 μl of 1 × Reporter Lysis Buffer (Promega) and vortexed vigorously for 30 s to break open the spheroplasts. Next, 20 μl of the cell lysate was combined with 200 μl of reconstituted Luciferase Assay Substrate (Promega), and the activity was measured immediately in an LKB Wallac 1250 Luminometer.
Introducing the RNAs into the same preparation of spheroplasts results in a 5 to 15% variability in luciferase activity measured per microgram of protein. The light output is proportional to the luciferase concentration with 1.0 light unit corresponding to 165 femtograms of luciferase enzyme (as measured in the Wallac 1250 Luminometer). The activity was normalized to the amount of total protein (measured by Bradford assays).
(v) Kinetics.
Variations in the rate of luciferase expression generally reflect differences in translation rate, while the duration of expression reflects stability of the mRNA (11, 12). The maximum luciferase translation rate (<0.4 U/μg of cell protein/min) is about a 10−6 part of the total protein synthesis in these cells.
Measurement of mRNA turnover.
To measure the effect of ski2Δ slh1Δ on mRNA decay, 200 ml of mutant and wild-type cells was grown to an OD600 of 0.6. Transcription was inhibited by the addition of thiolutin (the kind gift of Edmund Hafner, Pfizer) as described previously (26a), and aliquots of cells were removed at intervals. RNA was extracted using the RNeasy Mini RNA extraction kit (Qiagen) after breaking the cells with glass beads. Extracted RNA was analyzed by the Northern blot method. The membranes were hybridized with either 5′-32P-labeled oligonucleotide probes complementary to regions of the coding sequences of STE2 and URA5 or to an RNA probe complementary to 18S rRNA.
RESULTS
Effect of mutations in eIF3 and eIF2 on translation of electroporated mRNAs.
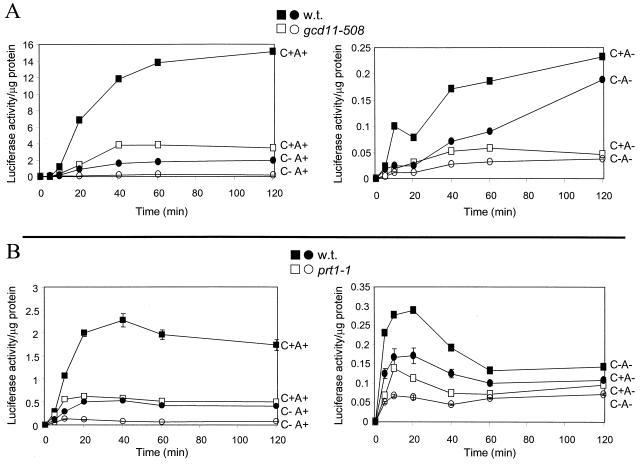
Electroporation faithfully reflected the known requirements of translation in that it requires the 5′ cap and 3′ poly(A) structures. In wild-type cells, the 3′ poly(A) structure increased translation rate by more than 80-fold, while the 5′ cap structure enhanced the translation rate about 8-fold (Fig. 1A). To further assess the fidelity of this method, we tested the effects of prt1-1, with an altered eIF3 subunit (13, 16, 28), and gcd11-508, with an altered eIF2 γ subunit (14). In each case, the translation of electroporated messages was substantially reduced, regardless of the presence of 5′ cap or 3′ poly(A) (Fig. 1). This result is as expected, since eIF2 and eIF3 are necessary for bringing the initiator Met-tRNA and the 40S ribosomal subunit, respectively, into the initiation complex, regardless of the presence or absence of cap or poly(A).
FIG. 1.
(A) Isogenic strains H117 (wild type) and H272 (gcd11-508) were electroporated (1.3 × 108 cells) with 2 μg of RNA. Cells were maintained at 25°C and assayed for luciferase activity at the indicated timepoints as described previously (21). Protein concentrations of all lysates were measured by the Bio-Rad protein assay kit. The results shown are an average of three experiments. The maximum luciferase translation rate (<0.4 U/μg of cell protein/min) is about a 10−6 part of the total protein synthesis in these cells. C+ or C−, mRNA with or without 5′ cap structure; A+ or A−, mRNA with or without 3′ poly(A) structure. (B) Isogenic strains H2783 (wild type) and H1676 (prt1-1) were treated as in panel A except the cells were maintained at 37°C throughout the indicated time course.
Pab1p is required for the poly(A) effect.
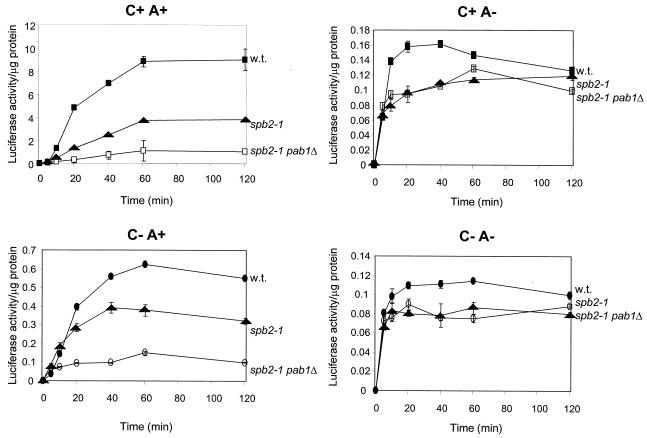
Although the pab1Δ mutation is lethal, it is suppressed by mutation of many of the genes needed for 60S ribosome biogenesis, such as spb2-1, a mutation in ribosomal protein L46 (33). So while it is impossible to compare wild-type and pab1Δ strains, one can compare spb2-1 strains with pab1Δ spb2-1 cells. We found that the pab1Δ mutation selectively lowers translation of poly(A)+ mRNAs, with little effect on poly(A)− mRNAs (Fig. 2). This in vivo result was similar to previously reported in vitro results of Tarun and Sachs (39). This further supports the view that Pab1p is a mediator of the poly(A) effect on translation.
FIG. 2.
Isogenic strains YAS43 (wild type), YAS216 (spb2-1), and YAS227 (spb2-1 pab1Δ) were electroporated with 2 μg of RNA. Cells were maintained at 25°C and assayed for luciferase activity at the times indicated as described in Materials and Methods.
The spb2-1 suppression of pab1Δ does not bypass the requirement for poly(A).
Mutation in any of several genes that reduce the level of free 60S ribosomal subunits makes the PAB1 gene dispensable for growth. It is suggested that the excess of free 40S ribosomal subunits facilitates cap-mediated initiation so that the requirement for the poly(A) (and thus Pab1p) is diminished (39). This hypothesis would predict that deficiency of 60S subunits would by the same mechanism diminish the requirement for the 3′ poly(A) structure. We examined the effect on the poly(A) requirement of the spb2-1 mutation and found that there is a modest general decrease in efficiency of translation of all mRNAs, regardless of the presence of cap or poly(A) structure (Fig. 2). Translation of non-poly(A) mRNA did not increase as predicted in the spb2-1 mutant. In this mutant, poly(A)+ mRNA was still translated 22 times better than was non-poly(A) mRNA.
Pab1p-eIF4G interaction is not necessary for the poly(A) effect.
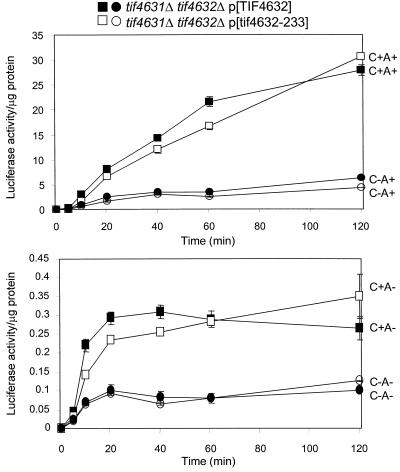
Pab1p interacts with eIF4G in vitro in the presence of poly(A) (38), and this interaction is proposed to be the basis of the requirement for poly(A) for translation (40). This model predicts that a mutation preventing this interaction should reduce or eliminate the requirement for the 3′ poly(A) structure in the presence of competing poly(A)+ mRNAs. We used a strain in which both genes for eIF4G (TIF4631 and TIF4632) are deleted and mutant or wild-type eIF4G is supplied from a plasmid (40). The tif4632-233 mutant eliminates the interaction of eIF4G and Pab1p (40), but we found that this mutation has no effect on the preference for poly(A)+ mRNAs in vivo (Fig. 3). The ratios of translation rate for C+A+ to C+A− mRNA were 46 in the wild type and 47 in the tif4632-233 mutant. The ratios for C−A+ to C−A− were 42 for the wild type and 47 for the mutant. These results suggest that, while Pab1p is a mediator of the poly(A) effect on translation, it must have an action in addition to its interaction with eIF4G.
FIG. 3.
Isogenic strains 4G1 (wild type) and 4G2 (tif4632-233) were electroporated with 2 μg of RNA. Cells were maintained at 25°C and assayed for luciferase activity at the indicated times as described in the text.
3′ poly(A) and the 60S joining step.
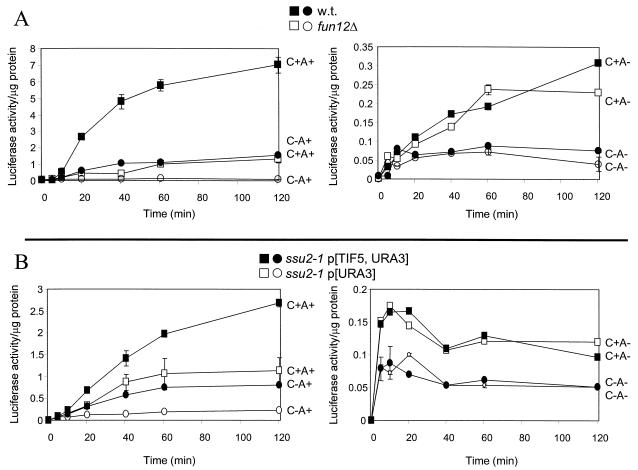
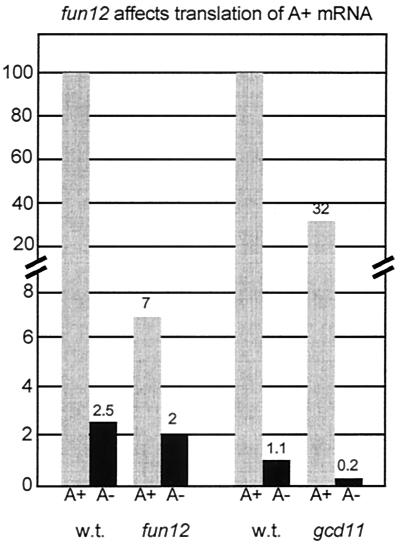
Both eIF5B (Fun12p) and eIF5 (Tif5p) are known to be involved in the 60S ribosomal subunit joining step (5, 27). We found that deletion of FUN12 specifically diminished the translation of poly(A)+ mRNA and had far less effect on non-poly(A) mRNA expression (Fig. 4). The fun12Δ mutation decreased the rate of translation of C+A+ mRNA by >10- fold but reduced the rate of C+A− mRNA by only 1.3-fold. This result suggests that the 3′ poly(A) structure is important for the Fun12p-dependent step, namely, 60S ribosomal subunit joining. In the absence of Fun12p, poly(A) had only a small effect on translation efficiency. The contrast of fun12Δ and gcd11-508 is shown in Fig. 5. While fun12Δ affected primarily poly(A)+ mRNA, gcd11-508 (eIF2) affected translation of poly(A)+ and poly(A)− mRNAs to a comparable extent.
FIG. 4.
Isogenic strains (A) J115 (wild type) and J116 (fun12Δ) or (B) isogenic strains KAY71 (wild type) and KAY73 (ssu2-1/tif5) were electroporated with 2 μg of RNA. Cells were maintained at 25°C and assayed for luciferase activity at the indicated times as described in the text.
FIG. 5.
Comparison of effects of fun12Δ and gcd11-508 mutations on translation rates for poly(A)+ and poly(A)− mRNAs. The comparison is for C+A+ and C+A− mRNAs.
We obtained similar results with a tif5 (ssu2-1) mutation affecting eIF5 (Fig. 4). In this case we saw a decrease of 2.5-fold for C+A+ mRNA. The effect was not as great as for the fun12Δ strain, perhaps because, unlike FUN12, TIF5 is an essential gene and cannot be completely inactivated for the assay.
Ski2p, Fun12p, and the poly(A) requirement.
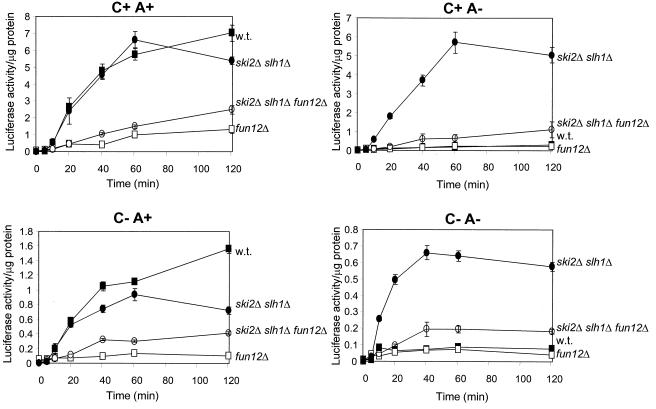
In a FUN12+ strain, a ski2 deletion resulted in a dramatic increase in the efficiency of expression of non-poly(A) mRNA (21). Like Ski2p, Slh1p is a nonessential RNA helicase that represses dsRNA virus copy number (20). In the ski2Δ slh1Δ double mutant poly(A)+ and non-poly(A) mRNAs were translated with equal efficiency for the same duration (35) (Fig. 6). We found that deletion of FUN12 almost completely eliminated this effect (Fig. 6). Most of the increased translation efficiency of a C+A− mRNA seen in a ski2Δ slh1Δ double mutant was lost in the fun12Δ ski2Δ slh1Δ strain. This result is consistent with a model in which Ski2p and Slh1p act through Fun12p.
FIG. 6.
Strains 3221 (wild type), 4107 (ski2Δ slh1Δ), J116 (fun12Δ), and AMS3 (ski2Δ slh1Δ fun12Δ) were electroporated with 2 μg of RNA. Cells were maintained at 25°C and assayed for luciferase activity at the indicated times as described in Materials and Methods. Symbols: ■, wild type; ●, ski2Δ slh1Δ; □, fun12Δ; ○, ski2Δ slh1Δ fun12Δ.
Expression of LUC mRNAs and mRNA turnover.
In addition to translation rates, mRNA turnover can also affect the expression of LUC mRNAs, and both cap and 3′poly(A) affect each process. We used several approaches to distinguish these effects. 32P-labeled LUC mRNAs were electroporated, and their degradation was measured over time by extraction, analysis on gels, and autoradiography. It was found that C+A+, C+A−, C−A+, and C−A− LUC mRNAs all had similar half-lives in the wild type and in ski2 and ski6 mutants (2, 21). Thus, this system did not reflect the known greater instability of mRNAs lacking cap or poly(A), but this result does argue that differences measured in our experiments are due to differences in translation rather than mRNA turnover. Similar experiments comparing ski2Δ slh1Δ double mutants with an isogenic wild-type strain gave similar results (data not shown).
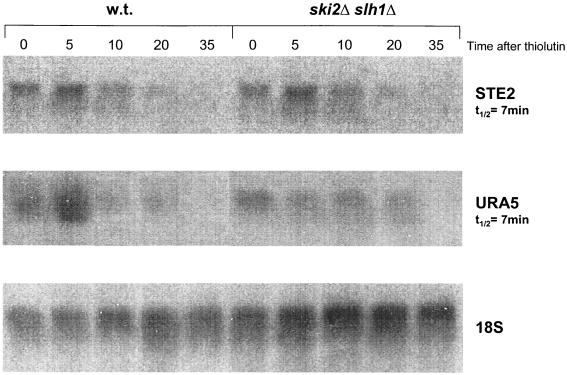
As a second approach to examine the role of mRNA turnover in our results, we examined directly the effect of the ski2Δ slh1Δ double mutation on turnover of endogenous mRNAs. We found no effect on the turnover of URA5 or STE2 mRNAs of the double mutation (Fig. 7), nor was the stability of ACT1 or PGK1 mRNAs affected (35). This confirmed earlier results indicating that unless the major Xrn1p 5′→3′ mRNA degradation system is blocked, the ski2 mutation has no effect on mRNA turnover (18). We further checked that the electroporation procedure does not inactivate the Xrn1p-catalyzed mRNA degradation system (data not shown).
FIG. 7.
The ski2Δ slh1Δ double mutation does not affect turnover of endogenous STE2 or URA5 mRNAs. Transcription was arrested by the addition of thiolutin (gift of Edmund Hafner, Pfizer, Groton, Conn.), and the RNA was extracted and analyzed by Northern hybridization (26a).
A third criterion for distinguishing the effects on mRNA turnover from those on translation are the kinetics of luciferase synthesis. Differences in expression at the earliest times are most likely due to translation rates and, in cases where the expression is proceeding linearly, rates can be clearly distinguished from yields. Functional half-lives of electroporated C+A+ and C+A− LUC mRNAs varied little between wild-type cells and ski2Δ slh1Δ double mutants (35). Moreover, the half-life in wild-type cells of electroporated C+A+ mRNA (62 min) was only about twice that of C+A− mRNA (36 min), a difference insufficient to account for the >40-fold difference in the expression of these two species (35). Thus, the bulk of the effects we report here are due to changes in translation rates.
Specifically, in the fun12Δ strain, expression from C+A− LUC mRNA has not plateaued before 60 min (Fig. 4A, right), so rates are being measured. The same is true of the expression from C+A− mRNA in the gcd11-508 mutant (Fig. 1A, right). The rate on C+A− mRNA is lowered fivefold by gcd11-508 but is not significantly changed by fun12Δ. This result shows again that translation of C+A− mRNA is affected by the gcd11 mutation but not by the fun12Δ mutation. The mRNA stability is not the process affected. The data for the ssu2-1 (tif5) also shows an effect only on poly(A)+ mRNAs (Fig. 4), but enzyme accumulation plateaus early in the time course, so it remains possible that an effect on translation is obscured by rapid degradation of the non-poly(A) mRNA.
The pab1Δ mutation substantially affects translation of poly(A)+ mRNA (Fig. 2). On C+A− mRNA, expression ceases quickly in both the spb2-1 and spb2-1 pab1Δ mutants, but the double mutant expresses as much luciferase as does the single mutant, and the kinetics are identical, suggesting that there is little effect of pab1Δ on translation of the C+A− mRNA. However, it is possible in this case that an effect on translation is obscured by an opposite effect on mRNA turnover. Likewise, the spb2-1 mutant shows no increase in expression of C+A− mRNA, contrary to the prediction of the 40S abundance model, but an increase in expression might be obscured here by a simultaneous decrease in stability due to the mutant.
DISCUSSION
mRNA electroporation accurately reflects in vivo translation.
The translation of electroporated mRNAs faithfully reflects the in vivo process because it takes place in living viable cells and because it shows the expected requirement for the 5′ cap and 3′ poly(A) structure (12). We further show here that mutations in eIF3 (prt1-1) and eIF2 (gcd11-508) result in the expected diminished translation of all species of electroporated mRNA. We have used this method to dissect the role of the 3′ poly(A) structure in the translation process, relating it to the poly(A) binding protein (Pab1p), the initiation factor eIF4G, the factors eIF5 (Tif5p) and eIF5B (Fun12p) involved in 60S subunit joining, and the RNA helicases Ski2p and Slh1p that regulate the poly(A) requirement for translation.
Interaction of eIF4G and Pab1p does not affect the requirement for 3′ poly(A) for translation.
In the presence of RNA, the poly(A) binding protein (Pab1p) binds to a region of eIF4G included in residues 201 to 317 of eIF4G2 or residues 188 to 300 of eIF4G1 (38, 40). Deletion of this region completely abrogates the binding reaction, as does substitution of residues 233 to 236 of eIF4G2 (RLRK → AVAA) or residues 213 to 216 of eIF4G1 (KLRK → AAAA) (40). This interaction has been proposed to be the mechanism of the poly(A) requirement for efficient translation of mRNAs. Such an interaction is proposed to enhance recruitment of 40S subunits through the eIF4G-eIF3 interaction, as well as promote the recycling of ribosomes that have completed a polypeptide (reviewed in reference 31). However, elimination of the Pab1p-eIF4G interaction by mutation of the interacting domain of eIF4G does not affect the growth rate of yeast cells, suggesting that this model in its simplest form is unlikely. Furthermore, the effect of eliminating this interaction on the in vitro system is seen only with uncapped mRNAs (40). Except for the yeast RNA virus mRNAs, all yeast mRNAs are believed to be capped, and the requirement for a poly(A) for translation is nearly absolute.
One alternative to the simple model described above relies on the fact that if many or most mRNAs lack a poly(A) structure, as in a mutant in poly(A) polymerase, non-poly(A) mRNAs may be found on polysomes (17, 29). Removing the competition with poly(A)+ mRNAs allows non-poly(A) mRNAs to be utilized. If the eIF4G-Pab1p interaction is disrupted and if this is the (or a) mediator of the function of the poly(A), this model would suggest that all messages would be translated well because none would have the poly(A) function. A strong prediction of this model is that in such a strain, translation of a non-poly(A) mRNA would be as efficient as would translation of a poly(A)+ mRNA.
However, we have found that elimination of the Pab1p-eIF4G interaction, by substitution of the interacting region of eIF4G, does not affect the requirement for poly(A) for translation. Even the magnitude of the requirement is unaltered by this change. This indicates that the Pab1p-eIF4G interaction is dispensable for translation and is not the reason why the 3′ poly(A) is essential for translation.
spb2-1 suppression of pab1Δ is not by reducing the requirement for poly(A).
Although pab1Δ is lethal, it is well suppressed by any of a number of mutations producing a deficiency of 60S subunits, including mutations in genes encoding 60S ribosomal subunit proteins and other factors needed for ribosome biogenesis (33). All such mutants have an excess of free 40S subunits, and it has been suggested that these facilitate 40S joining so that the poly(A) structure is no longer as essential for initiation (39). This model predicts that non-poly(A) mRNA should be better translated in the mutant than in the wild-type cells. We tested this hypothesis by examining the effect of spb2-1 on the requirement for poly(A) and found that non-poly(A) mRNA was actually less efficiently translated and that there remained a 22-fold stimulation of translation by the presence of the 3′ poly(A) structure. Similar results have also been reported with mutants in the mak21 gene, which are also involved in 60S subunit biogenesis (10). This indicates that another mechanism must be invoked to explain the viability of spb2-1 pab1Δ strains.
A role for poly(A) in subunit joining.
An earlier model for the role of poly(A) in translation, based on in vitro data, centered on a role for poly(A) in promoting the 60S subunit joining reaction (23). The description of two factors involved in 60S subunit joining, eIF5 encoded by TIF5 (5) and eIF5B encoded by FUN12 (6, 27), enabled a new test of this model. We find that defects in either of these factors result in decreased translation of poly(A)+ mRNA but little change in the translation of non-poly(A) mRNA. This supports the model in which poly(A) promotes 60S subunit joining by promoting the activity of Fun12p and Tif5p. Our data support a role of Pab1p in mediating the action of poly(A). Thus, Pab1p-poly(A) may influence 60S joining with the 40S subunits at the initiator AUG, thereby increasing the efficiency of translation of poly(A)+ mRNAs.
Role of Ski2p-Slh1p in the action of poly(A).
In the absence of the related RNA helicases Ski2p and Slh1p, there is no difference in either the rate or the duration of translation of poly(A)+ and poly(A)− mRNAs (35). This implies that Ski2p and Slh1p block the translation of non-poly(A) mRNAs. Blocking the Xrn1p-catalyzed 5′→3′ degradation of mRNAs reveals an effect of Ski2p on 3′→5′ mRNA degradation (18). However, the experiments reported here and previously use cells and mRNAs not blocked for the action of the Xrn1p system and, under these conditions, there is no effect of Ski2p on mRNA turnover. We have further shown that mRNA turnover is unaffected by the ski2Δ slh1Δ double mutation (35), and electroporated cells have an active Xrn1 degradation system (A. Searfoss and R. Wickner, unpublished data). These and other considerations (35) indicate that the effects observed here are on translation.
It was previously hypothesized that the Ski proteins act by affecting 60S ribosome biogenesis to produce a requirement for the 3′ poly(A) for subunit joining (21, 25). Indeed, ski6 mutants show both alterations of 60S subunit structure and relative poly(A) independence of translation, supporting this mechanism (2).
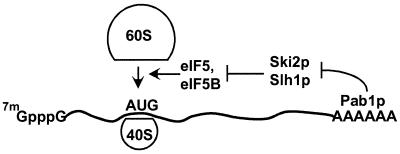
Our finding that most of the increased translation of non-poly(A) mRNA seen in a ski2Δ slh1Δ double mutant is prevented by a further fun12Δ mutation is consistent with a model in which Ski2p and Slh1p block translation by blocking Fun12p action. An heuristic model that summarizes these results is as follows: 3′poly(A)-Pab1p —|Ski2p-Slh1p —|Fun12p-Tif5p → 60S subunit joining. This model qualitatively explains the requirement for poly(A) in wild-type, but not in cells defective for downstream components Ski2p-Slh1p or Fun12p (Fig. 8). It further explains why the ski2Δ slh1Δ strains have elevated translation of poly(A)− mRNAs, whereas the fun12Δ strains have depressed translation specifically of poly(A)+ mRNAs. Finally, it explains why the fun12Δ mutation is epistatic to ski2Δ slh1Δ.
FIG. 8.
Model of the action of poly(A) to promote 60S ribosomal subunit joining. Poly(A)-Pab1p inhibits Ski2p-Slh1p which inhibits Fun12p-Tif5p which promotes 60S ribosomal subunit joining.
While a molecular explanation will await biochemical studies, one could speculate that the schematic interactions shown above reflect actual protein-protein interactions, with Pab1p-poly(A) binding to Ski2p-Slh1p, which in turn bind to Fun12p and/or Tif5p. Recent biochemical evidence suggests the 3′ end of an mRNA can affect subunit joining. Binding of a protein to a 3′UTR site of lipoxygenase mRNA inhibits 60S subunit joining in extracts of erythroid precursor cells (26). Other molecular mechanisms could underlie the functional pathway above; however, our results do implicate the 60S joining step as both a target and a regulated mediator of poly(A)-dependent translation.
ACKNOWLEDGMENTS
We thank Thomas F. Donahue for allowing us to use the ssu2-1 mutant prior to publication, and we are grateful to Alan Hinnebusch, Herman Edskes, and Dan Masison for critical reading of the manuscript.
REFERENCES
- 1.Asano K, Krishnamoorthy T, Phan L, Pavitt G D, Hinnebusch A G. Conserved bipartite motifs in yeast eIF5 and eIF2Bepsilon, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benard L, Carroll K, Valle R C P, Wickner R B. Ski6p is a homologue of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol Cell Biol. 1998;18:2688–2696. doi: 10.1128/mcb.18.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benard L, Carroll K, Valle R C P, Wickner R B. The Ski7 antiviral protein is an EF1-α homolog that blocks expression of non-poly(A) mRNA in Saccharomyces cerevisiae. J Virol. 1999;73:2893–2900. doi: 10.1128/jvi.73.4.2893-2900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown J T, Bai X, Johnson A W. The yeast antiviral proteins Ski2p, Ski3p and Ski8p exist as a complex in vivo. RNA. 2000;6:449–457. doi: 10.1017/s1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakravarti D, Maitra U. Eukaryotic translation initiation factor 5 from Saccharomyces cerevisiae: cloning, characterization and expression of the gene encoding the 45,346-Da protein. J Biol Chem. 1993;268:10524–10533. [PubMed] [Google Scholar]
-
6.Choi S K, Lee J H, Zoll W L, Merrick W C, Dever T E. Promotion of binding Met-tRNA
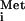 to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar]
to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar] - 7.Choi S K, Olsen D S, Roll-Mecak A, Martung A, Remo K L, Burley S K, Hinnebusch A G, Dever T E. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol Cell Biol. 2000;20:7183–7191. doi: 10.1128/mcb.20.19.7183-7191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dangel A W, Shen L, Mendoza A R, Wu L-C, Yu C Y. Human helicase gene SKI2W in the HLA class III region exhibits striking structural similarities to the yeast antiviral gene SKI2 and to the human gene KIAA0052: emergence of a new gene family. Nucleic Acids Res. 1995;23:2120–2126. doi: 10.1093/nar/23.12.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dever T E. Translation initiation: adept at adapting. Trends Biochem Sci. 1999;24:398–403. doi: 10.1016/s0968-0004(99)01457-7. [DOI] [PubMed] [Google Scholar]
- 10.Edskes H K, Ohtake Y, Wickner R B. Mak21p of Saccharomyces cerevisiae, a homolog of human CAATT-binding protein, is essential for 60S ribosomal subunit biogenesis. J Biol Chem. 1998;273:28912–28920. doi: 10.1074/jbc.273.44.28912. [DOI] [PubMed] [Google Scholar]
- 11.Everett J G, Gallie D R. RNA delivery in Saccharomyces cerevisiae using electroporation. Yeast. 1992;8:1007–1014. doi: 10.1002/yea.320081203. [DOI] [PubMed] [Google Scholar]
- 12.Gallie D R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 13.Hanic-Joyce P J, Singer R A, Johnston G C. Molecular characterization of the yeast PRT1 gene in which mutations affect translation initiation and regulation of cell proliferation. J Biol Chem. 1987;262:2845–2851. [PubMed] [Google Scholar]
- 14.Hannig E M, Cigan A M, Freeman B A, Kinzy T G. GCD11, a negative regulator of GCN4 expression, encodes the γ subunit of eIF-2 in Saccharomyces cerevisiae. Mol Cell Biol. 1992;13:506–520. doi: 10.1128/mcb.13.1.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harashima S, Hinnebusch A G. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:3990–3998. doi: 10.1128/mcb.6.11.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartwell L H, McLaughlin C S. A mutant of yeast apparently defective in the initiation of protein synthesis. Proc Natl Acad Sci USA. 1969;62:468–474. doi: 10.1073/pnas.62.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu C L, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs-Anderson J S, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S-G, Lee I, Kang C, Song K. Identification and characterization of a human cDNA homologous to yeast SKI2. Genomics. 1995;25:660–666. doi: 10.1016/0888-7543(95)80008-a. [DOI] [PubMed] [Google Scholar]
- 20.Martegani E, Vanoni M, Mauri I, Rudoni S, Saliola M, Alberghina L. Identification of gene encoding a putative RNA-helicase, homologous to SKI2, in chromosome VII of Saccharomyces cerevisiae. Yeast. 1997;13:391–397. doi: 10.1002/(SICI)1097-0061(19970330)13:4<391::AID-YEA92>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 21.Masison D C, Blanc A, Ribas J C, Carroll K, Sonenberg N, Wickner R B. Decoying the cap- mRNA degradation system by a dsRNA virus and poly(A)- mRNA surveillance by a yeast antiviral system. Mol Cell Biol. 1995;15:2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrissey J P, Deardorff J A, Hebron C, Sachs A B. Decapping of stabilized, polyadenylated mRNA in yeast pab1 mutants. Yeast. 1999;15:687–702. doi: 10.1002/(SICI)1097-0061(19990615)15:8<687::AID-YEA412>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 23.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translation initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nomura N, Miyajima N, Sazuka T, Tanaka A, Kawarabayashi Y, Sato S, Nagase T, Seki N, Ishikawa K, Tabata S. Prediction of the coding sequences of unidentified genes. II. The coding sequences of 40 new genes (KIAA 0041–KIAA 0080) deduced by analysis of randomly sampled cDNA clones from human immature myeloid cell line KG1. DNA Res. 1994;1:27–35. doi: 10.1093/dnares/1.1.27. [DOI] [PubMed] [Google Scholar]
- 25.Ohtake Y, Wickner R B. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol Cell Biol. 1995;15:2772–2781. doi: 10.1128/mcb.15.5.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostareck D H, Ostareck-Lederer A, Shatsky I N, Hentze M W. Lipoxygenase mRNA silencing in erythroid differentiation: the 3′ UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- 26a.Parker R, Herrick D, Peltz S W, Jacobson A. Measurement of mRNA decay rates in Saccharomyces. In: Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. Vol. 194. San Diego, Calif: Academic Press, Inc.; 1991. pp. 415–423. [Google Scholar]
- 27.Pestova T V, Lomakin I B, Lee J H, Choi S K, Dever T E, Hellen C U T. The ribosomal subunit joining reaction in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 28.Phan L, Zhang X, Asano K, Anderson J, Vornlocher H P, Greenberg J R, Qin J, Hinnebusch A G. Identification of a translation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Proweller A, Butler S. Efficient translation of poly(A)-deficient mRNAs in Saccharomyces cerevisiae. Genes Dev. 1994;8:2629–2640. doi: 10.1101/gad.8.21.2629. [DOI] [PubMed] [Google Scholar]
- 30.Qu X, Yang Z, Zhang S, Shen L, Dangel A W, Hughes J H, Redman K L, Wu L-C, Yu C Y. The human DEVH-box protein Ski2w from the HLA is localized in nucleoli and ribosomes. Nucleic Acids Res. 1998;26:4068–4077. doi: 10.1093/nar/26.17.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sachs A B. Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 447–465. [Google Scholar]
- 32.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 33.Sachs A B, Davis R W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990;247:1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- 34.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 35.Searfoss A M, Wickner R B. 3′ poly(A) is dispensable for translation. Proc Natl Acad Sci USA. 2000;97:9133–9137. doi: 10.1073/pnas.97.16.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonenberg N, Hershey J W B, Mathews M B. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 38.Tarun S Z, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 39.Tarun S Z, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 40.Tarun S Z, Wells S E, Deardorff J A, Sachs A B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Widner W R, Wickner R B. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol Cell Biol. 1993;13:4331–4341. doi: 10.1128/mcb.13.7.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]