Abstract
Long non‐coding RNAs (LncRNAs), which are more than 200 nucleotides in length and with limited protein‐coding potential, play vital roles in the pathogenesis, tumorigenesis, and angiogenesis of cancers. Aberrant expression of lncRNAs has been detected in various carcinomas and may be correlated with oncogenesis by affecting related genes expression. Recently, an increasing number of studies have reported on long intergenic non‐protein coding RNA 460 (LINC00460) in human tumor fields. LINC00460 is upregulated in diverse cancer tissues and cells. The upregulated expression level of LINC00460 is correlated with larger tumor size, tumor node metastasis (TNM) stage, lymph node metastasis, and shorter overall survival. The regulatory mechanism of LINC00460 was complex and diverse. LINC00460 could act as a competitive endogenous RNA (ceRNA), directly bind with proteins or regulate multiple pathways, which affected tumor progression. Moreover, LINC00460 was also identified to increase drug resistance, and therefore, weaken the effectiveness of tumor treatment. It has become increasingly important to investigate the roles of LINC00460 in various cancers by different mechanisms. Therefore, a more comprehensive understanding of LINC00460 is crucial to expound on the cellular function and molecular mechanism of human cancers. In this review, we refer to studies concerning LINC00460 and provide the basis for the evaluation of LINC00460 as a predicted biomarker or potential therapeutic target in malignancies, and also provide ideas for the future research of lncRNAs similar to LINC00460.
Keywords: cancer, LINC00460, long non‐coding RNA, mechanism, oncogene
In recent years, the regulatory role of long noncoding RNAs (lncRNAs) in human tumors has been gradually recognized. In this review, we respectively showed how lncRNA LINC00460 affected progression of human tumors in different types by multiple mechanisms. A large number of previous researches have demonstrated that LINC00460 as an oncogene plays important roles in tumor growth, development, and metastasis. In view of the important role of LINC00460 in tumorigenesis and development as well as its significance as a biomarker, we summarized the specific role and molecular mechanism of LINC00460 in various tumors, convenient for readers integrally learning about the effect of LINC00460 on diverse cancers. We believe this review is valuable for all the researchers who are interested in human cancers and noncoding RNAs.
INTRODUCTION
Cancer is a predominant reason for death worldwide, and the quantities of cancer cases and deaths rapidly grow as populations grow. 1 Eliminating cancer requires not only improved therapies, but also improved methods to detect cancers at an early stage. Molecular technology plays a vital role in cancer detection and prognosis and has gradually received greater attention. 2 Therefore, we are eager to find a new therapeutic biomarker to improve cancer treatment and prognosis.
An increasing number of studies related to long non‐coding RNAs (lncRNAs) have been reported because of their high specificity and easy detection in tissues, serum, plasma, urine, and saliva. 3 LncRNAs were a novel class of functional RNAs that were longer than 200 nucleotides, characterized by low levels of sequence conservation and expression. 4 LncRNAs regulate various biological functions at different levels, including the epigenetic, transcriptional, and post‐transcriptional levels. 5 Aberrant expressions of lncRNAs in some cancers is associated with tumorigenesis, metastasis, and angiogenesis. 6 , 7 Therefore, lncRNAs could be potential novel biomarkers for cancer diagnosis and therapy. For instance, lncRNA‐differentiation antagonizing non‐protein coding RNA (DANCR) could act as a competing endogenous RNA (ceRNA) that interacts with messenger RNA (mRNA) sequences to restrain the microRNA (miRNA) suppressive effect on target genes and is involved in various cancers. 8 Additionally, lncRNA HOXA‐cluster antisense RNA2 (AS2) has been demonstrated to modulate cellular function and regulate gene expression by complicated regulatory mechanisms. 9
Recently, RNA sequencing data from several cancers, such as lung adenocarcinoma (LC), head and neck squamous carcinoma (HNSCC), breast cancer, bladder cancer, lung squamous cell carcinoma, and renal cell carcinoma, from The Cancer Genome Atlas (TCGA) data portal and identified differentially expressed RNAs were used to construct the lncRNA‐related ceRNA network. 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 After analysis the lncRNA‐related ceRNA network, a common valuable lncRNA correlated with pathogenesis and tumorigenesis was confirmed, namely, long intergenic non‐protein coding RNA 460 (LINC00460). 10 LINC00460 (NR_034119.1, NCBI: https://www.ncbi.nlm.nih.gov) expression was elevated in various cancer types through querying the TCGA database and data analysis of Ensembl database (http://asia.ensembl.org) revealed that LINC00460 has seven transcripts and is located in chr13q33.2 region. 18 The previous studies indicated that the transcripts associated with cancers are mostly LINC00460‐202 and LINC00460‐203. 18 , 19 , 20 LINC00460‐202 transcribed as an 857 nt transcript and has three exons; it has been proved highly expressed in lung cancer. 19 LINC00460‐203 transcribed as a 915 nt transcript and has two exons; it has been demonstrated upregulated in HNSCC 18 and gastric cancer (GC). 20 LINC00460 has been demonstrated to be upregulated in many types of cancers and functions as an important oncogene, the exploration of molecular mechanism of LINC00460 is of great concern and the expression of LINC00460 is involved in the development of various tumors, including HNSCC, colorectal cancer (CRC), 21 lung cancer, 22 papillary thyroid cancer (PTC), 23 hepatocellular carcinoma (HCC), 24 glioma, 25 renal cell carcinoma, 16 meningioma,26 and others. LINC00460 plays a significant role in the pathogenesis and tumorigenesis of diverse cancers and is associated with distant lymph mode metastasis, epithelial‐mesenchymal transition (EMT), and poor overall survival. 26 , 27 Therefore, LINC00460 is one of the most critical regulatory RNAs in human cancers, and it might be a potential and valuable target for therapeutics. In this review, we emphatically discuss the mechanisms and effects of LINC00460 in human cancers (Table 1).
TABLE 1.
LINC00460 in human cancers
| Cancer types | Cell lines | Expression | Phenotype | Clinical feature | Clinical value | Role | Related genes and protein | Reference |
|---|---|---|---|---|---|---|---|---|
| Head and neck squamous cell carcinoma | HSC3, Fadu, SAS, WSU‐HN4, WSU‐HN6, WSU‐HN30, SCC‐4, SCC‐9, SCC‐25, CAL27 | Up | Proliferation, invasion, migration, apoptosis, autophagy | Tumor recurrence, lymph node metastasis | Diagnosis, prognosis, therapy | Oncogene | STC2, miR‐206, cleaved‐PARP, Bax, cleaved‐caspase‐3, PRDX1, miR‐612, AKT2, miR‐4443, cyclin D1, p21, miR‐320a | 18 , 28 , 29 , 30 , 31 |
| Esophageal squamous cell carcinoma | EC1, EC9706, KYSE70, TE1, TE13 | Up | Proliferation, cell cycle, migration, invasion, apoptosis | TNM stage, lymph node metastasis, poor prognosis | Diagnosis, prognosis, therapy | Oncogene | CBP, P300, miR‐1224‐5p | 32 , 33 |
| Lung cancer | A549, H1299, H1975, H460, PC9, SPC‐A1, SKLU‐1, Calu‐3, HCC‐78 | Up | Proliferation, migration, invasion, apoptosis | Tumor growth, size, volume, weight | Prognosis, therapy | Oncogene | miR‐769‐5p, EGFR, cleaved caspase‐3, Bcl‐2, Bax, miR‐539, PI3k, Akt, miR‐302c‐5p, FOXA1, miR‐149‐5p | 22 , 34 , 35 , 36 , 37 |
| Papillary thyroid cancer | FTC‐133, 8505C, TPC1, BCPAP | Up | Proliferation, migration, invasion, apoptosis | Tumor size, TNM stage, lymph node metastasis | Diagnosis, therapy | Oncogene | miR‐485‐5p, Raf1, miR‐613, SphK2, miR‐539, MMP‐9, N‐cadherin, vimentin, E‐cadherin | 23 , 38 , 39 |
| Breast cancer | MCF‐7, BT‐474, MDA‐MB‐231, BT‐549 | Up | Cell viability, migration, invasion | Lymph node stage, worse survival outcome | Diagnosis, prognosis | Oncogene | miR‐489‐5p, FGF7, AKT | 40 |
| Gastric cancer | MGC803, BGC823, SGC7901, MKN‐28, MKN‐45, AGS | Up | Proliferation, migration, invasion, apoptosis | Lymph node metastasis, TNM stage | Prognosis, therapy | Oncogene | EZH2, LSD1, CCNG2, miR‐342‐3p, KDM2A, c‐MYC, β‐catenin | 20 , 41 , 42 |
| Hepatocellular carcinoma | HepG2, Hep3B, SNU‐449, THLE‐3 cells, HCCLM3, Huh‐7, LO2 cells | Up | Proliferation, migration, apoptosis | Tumor progression | Diagnosis, therapy | Oncogene | miR‐342‐3p, AGR2, cyclin D1, CDK4, MMP‐3, MMP‐9, Bax, Bcl‐2, miR‐485‐5p, PAK1 | 24 , 43 , 44 |
| Colorectal cancer | HCT116, SW480, HT‐29, LOVO | Up | Proliferation, cell cycle, apoptosis, migration, invasion | Tumor size, advanced tumor stages, lymph node metastasis | Diagnosis, prognosis, therapeutic detection | Oncogene | EZH2, KLF2, CUL4A, miR‐149‐5p, ERG, WWC2, LIMK2, miR‐939‐5p, miR‐433‐3p, ANXA2 | 27 , 45 , 46 , 47 |
| Osteosarcoma | Saos‐2, HOS, U2OS, MG63 | Up | Cell viability, proliferation, migration, apoptosis | Tumor size, distant metastasis | Prognosis, therapy | Oncogene | Cyclin D1, CDK4, CDK6, MMP‐9, FADS1, miR‐1224‐5p | 48 , 49 |
| Nasopharyngeal carcinoma | SUNE‐1, CNE‐1, HNE‐1, CNE‐2, C666‐1, HONE‐1 | Up | Proliferation, migration, invasion, | Poor prognosis | Diagnosis, prognosis, therapy | Oncogene | miR‐30a‐3p, Rap1A, miR‐149‐5p, IL6 | 50 , 51 |
| Ovarian cancer | HO8910, SKOv‐3, A2780, ES‐2 | Up | Proliferation, migration, invasion, apoptosis | Tumor stage, tumor size | Diagnosis, therapy | Oncogene | miR‐338‐3p | 52 |
| Bladder urothelial carcinoma | T24, J82, TCCSUP, UM‐UC‐3 | Up | Proliferation, migration, invasion | Tumor range, metastasis, lymph node | Therapy | Oncogene | miR‐612, FOXK1 | 53 |
| Pancreatic cancer | PC PANC1, SW1990 | Up | Proliferation, migration, invasion | Poor prognosis | Prognosis | Oncogene | miR‐491‐5p | 54 |
| Cervical cancer | HeLa, CaSki | Up | Proliferation | Poor prognosis | Therapy | Oncogene | miR‐503‐5p, miR‐361‐3p, Gli1 | 55 , 56 |
Abbreviations: TNM stage, tumor‐lymph node‐ metastasis stage.
MATERIALS AND METHODS
Search strategy
A systematic literature survey was conducted via electronic searched of PubMed, China National Knowledge Infrastructure (CNKI) and WanFang databases for eligible studies published as of June 1, 2021. The search terms include “long non‐coding RNA”, “lncRNA”, “long intergenic non‐coding RNA”, “LINC00460”, “cancer”, “carcinoma”, “neoplasm”, “tumor”, “prognosis”, “prognostic”, “survival”, “overall survival”, and “OS”. Furthermore, the references in retrieved articles were also manually reviewed for potentially relevant studies.
Inclusive and exclusive criteria
The inclusive criteria were as follows: (1) the expression of LINC00460 was determined; (2) studies on any type of human cancer; (3) for molecular studies; and (4) with full‐text. The exclusive criteria were as follows: (1) letter, review, case report, conference abstract, and meta‐analyses; (2) non‐English papers and non‐human studies; and (3) lack of essential information.
Data extraction and quality control
Two investigators independently extracted data from the eligible studies according to the inclusion and exclusion criteria. The third reviewer verified and then any disagreements were resolved by consensus. The following information was collected: first author name, publication year, cancer type, lncRNA, the detection method of lncRNA LINC00460 expression, biological function, miRNAs, RNA‐binding proteins (RBPs). The process of the study selection is strictly based on the abovementioned eligibility criteria (Figure 1).
FIGURE 1.
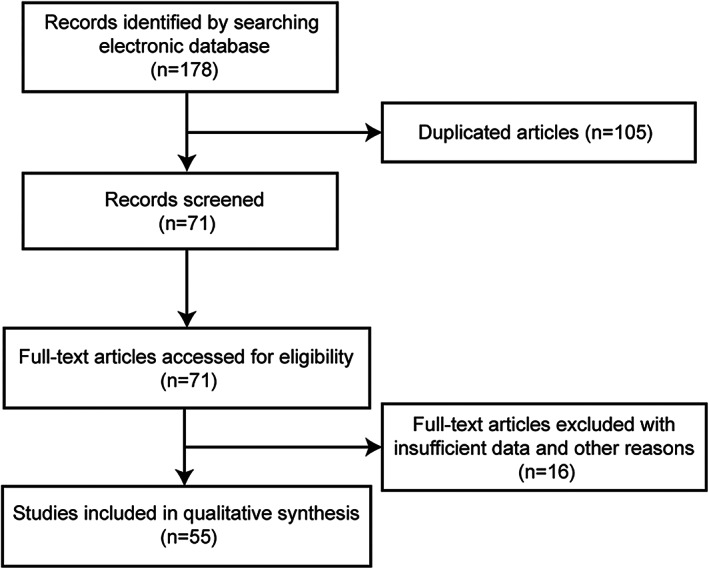
Flow diagram of study selection process
LINC00460 IN VARIOUS CANCERS
Head and neck squamous cell carcinoma
HNSCC is the most common malignant lesion in the head–neck region. 57 The main treatment for the majority of patients is the best supportive care in combination with chemotherapy and targeted therapy. 58 Most recurrences or disease‐specific deaths occur within the first 2 to 3 years, and half of the patients die within 5 years after diagnosis. 59 Therefore, it is necessary to investigate novel valid regulators to detect the disease in the early stage and improve the prognosis of HNSCC patients.
Cao et al. 60 selected a three‐lncRNA panel, including LINC00460, which had the optimal prediction power for HNSCC patients based on analysis. High expression of LINC00460 was detected in HNSCC tissues and was associated with poor survival. 61 In some studies, LINC00460 promoted HNSCC progression by sponging miRNAs and inhibiting their expression. 28 , 29 , 30 , 31 Xue et al. 28 found that LINC00460 silencing affected cell cycle distribution and promoted cell apoptosis and autophagy. The levels of activated apoptosis‐related proteins including PARP, Bax, and caspase‐3 were increased after LINC00460 knockdown. Knockdown of LINC00460 or overexpression of miR‐206 arrested HNSCC cells in the G0/G1 phase of the cell cycle, hindered the cell cycle progression from G1 phase to S phase, and also increased apoptosis. In addition, knockdown LINC00460 or overexpression of miR‐206 downregulated the protein expression of STC2 and blocked the activation of the protein kinase B (AKT) signaling pathway. Therefore, the LINC00460/STC2/miR‐206 axis is critical in influencing the development and progression of HNSCC. 28 Furthermore, Xie et al. 29 found that AKT2 was predicted to be a miR‐612 binding target and the expression of miR‐612 was negatively correlated with AKT2 in HNSCC. LINC00460 could regulate the progression of HNSCC by mediating the miR‐612/AKT2 axials. In addition, to regulate cell proliferation and migration, cyclin D1, p21, E‐cadherin, and N‐cadherin were found to be functional targets of LINC00460. The results indicated that knockdown of LINC00460 decreased the expression of cyclin D1 and increased the expression of p21 to regulate cell proliferation, migration and invasion. 29 Furthermore, Li et al. 30 found that miR‐4443 might act as a tumor suppressor on account of its effect on proliferation, migration, invasion, and apoptosis was also opposite to LINC00460 in HNSCC cells. In addition, some studies have found that LINC00460 also plays a role by binding with RBPs. Jiang et al. 18 found that PRDX1 is an RBP that binds with LINC00460 to affect cell proliferation, migration and EMT in HNSCC. Silencing PRDX1 decreased the expression of LINC00460, whereas PRDX1 overexpression increased the expression of LINC00460. PRDX1 facilitated the transcription of LINC00460 and EMT‐related genes in the nucleus. Knockdown of PRDX1 or LINC00460 remarkably enhanced the level of E‐cadherin and reduced the levels of N‐cadherin, vimentin, ZEB1, and ZEB2 in HNSCC cells 18 (Figure 2).
FIGURE 2.
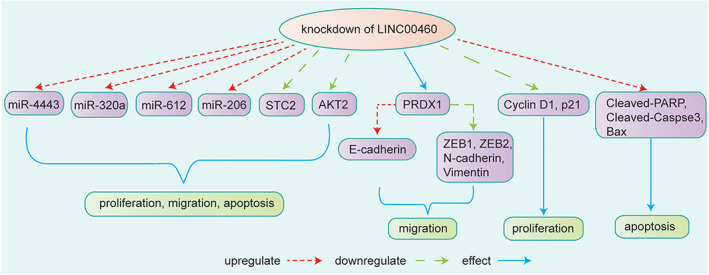
Knockdown of LINC00460 decreased STC2 and promoted cell proliferation, migration, and apoptosis of HNSCC by upregulating miR‐206 or upregulating the expression of miR‐612 by decreasing the expression of AKT or increasing the expression of miR‐4443. 28 , 29 , 30 , 31 LINC00460 affected HNSCC cell proliferation and migration in a PRDX1‐departed manner by regulating the level of E‐cadherin and the levels of N‐cadherin, vimentin, ZEB1, and ZEB2. 18 Knockdown of LINC00460 affected HNSCC cell migration and invasion by increasing the expression of p21 and decreasing the expression of cyclin D1. 29 Knockdown of LINC00460 promoted cell apoptosis by increasing the expression of cleaved‐PARP, Bax, and cleaved caspase‐3 28
Therefore, LINC00460 plays an important role in the development of HNSCC, which is regulated by a variety of mechanisms (specifically sponge miRNAs or directly binds with RBPs). Moreover, the mechanism of LINC00460 involvement in HNSCC needs to be further elucidated, which is of great significance to the application of prognostic and therapeutic target.
Esophageal squamous cell carcinoma
Esophageal cancer (EC) is the eighth most common cancer and the sixth most common cause of cancer‐related death worldwide. There are two main histological types of EC: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). 62 ESCC is one of the most deadly forms of human malignancy, and clinical therapy of ESCC remains limited. 63 Hence, it is indispensable for patients to find a valuable and predictable biomarker to improve the prognosis and overall survival. 64
Liu et al. 65 analyzed the lncRNA expression profile in EC patients’ tissue samples from TCGA and gene expression omnibus (GEO) 66 also identified a novel‐three lncRNA signature including LINC00460 as a predictor of overall survival and disease‐free survival in patients with ESCC. Liang et al. 32 found that LINC00460 was significantly elevated in most of tumor tissues and ESCC cell lines; it was in direct proportion to TNM stage, lymph node metastasis, and poor prognosis. CBP/P300, which were closely related transcriptional coactivators and acetyltransferase enzymes in humans, could directly bind to LINC00460 promoter and active LINC00460 transcribe through histone acetylation. The upregulation of LINC00460 has been demonstrated that promoted ESCC progression through CBP/P300 function. 32 Knockdown of LINC00460 suppressed the metastasis potential and EMT of EC cells by directly binding to miR‐1224‐5p. 33 Therefore, LINC00460 could as a sponge of miRNAs and histone acetylation to regulate the development of ESCC. LINC00460 might be a candidate biomarker for ESCC diagnosis and have tremendous therapeutic value (Figure 3).
FIGURE 3.
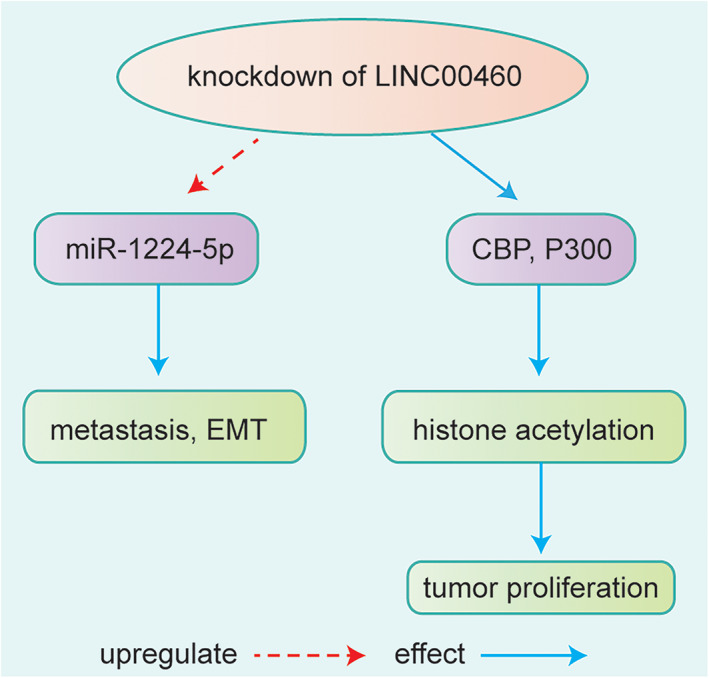
Knockdown of LINC00460 suppressed the metastasis and EMT of esophageal cancer cells by directly binding to miR‐1224‐5p. 33 Knockdown of LINC00460 affected tumor progression and development through CBP/P300 function via influencing histone acetylation 32
Lung cancer
Lung cancer is the leading malignancy incidence rate and mortality rate. 67 It includes two main types: non‐small cell lung cancer (NSCLC) and small cell lung cancer and ~85% of lung cancer consist of NSCLC. 68 Despite advances in health consciousness and systematic treatment, most patients are generally diagnosed at an advanced stage and have a low rate of overall survival. 69
In recent years, research on the molecular mechanism of lung cancer has become a novel method with which to improve diagnosis and prognosis. Li et al. 70 discovered the expression of LINC00460 was remarkably upregulated in NSCLC tissues and was closely related to the TNM stage, lymph node metastasis, and poor prognosis for NSCLC patients. Therefore, it suggested that LINC00460 acts as a valuable target for prognosis and therapy for NSCLC patients.
LINC00460 has emerged as an important regulator of the physiological and pathological processes of NSCLC tumors. Wang et al. 34 indicated that LINC00460 is competitively bound with miR‐539 sites to restrain the suppressive effect on NSCLC cell proliferation. In addition, Ma et al. 35 found that LINC00460 is involved in gefitinib resistance in NSCLC cells by targeting epidermal growth factor receptor (EGFR) through sponging miR‐769‐5p. Zhao et al. 36 discovered that nicotine promoted the development of NSCLC by activating LINC00460 and PI3K/AKT signaling pathway. Specifically, Nicotine affected cell apoptosis mediated by LINC00460 with alterations in Bcl‐2, Bax, and cleaved caspase‐3; moreover, LINC00460 knockdown inhibited the PI3K/AKT signaling pathway, and the effect was also removed by nicotine. 36
NSCLC has a major subtype of LC, and the prognosis of LC also remains poor. The upregulated expression of LINC00460 in LC tissues predicted poor prognosis, and silencing of LINC00460 inhibited cell growth in LC. 71 Ye et al. 22 found that LINC00460 promoted LC cell growth partially by upregulating FOXA1, the special target site for LINC00460 and miR‐302c‐5p, and the effects were partially regulated by the LINC00460/miR‐302c‐5p/FOXA1 axis. LINC00460 also promoted EGFR‐TKI resistance in EGFR‐mutated LC by facilitating the release of inflammatory cytokine interleukin 6 (IL‐6) via function as a decoy for miR‐149‐5p and inducing the EMT process 37 (Figure 4).
FIGURE 4.
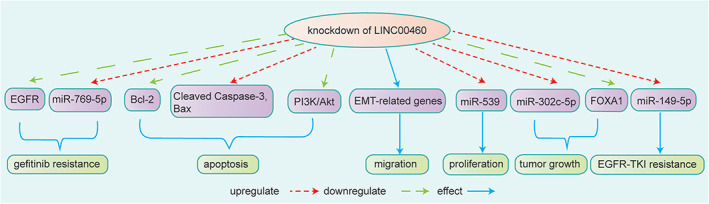
Knockdown of LINC00460 decreased EGFR protein expression by increasing the expression of miR‐769‐5p in NSCLC, which influenced gefitinib resistance. 35 LINC00460 is involved in cell apoptosis by regulating the apoptosis proteins Bcl‐2, Bax, cleaved caspase‐2, and PI3K/AKT. 36 LINC00460 promoted cell migration and invasion through EMT‐related genes. LINC00460 affected cell proliferation via sponging miR‐539. 34 Knockdown of LINC00460 influenced lung adenocarcinoma growth by binding to miR‐302c‐5p and upregulating the expression of FOXA1. 22 LINC00460 also promoted EGFR‐TKI resistance in EGFR‐mutated lung adenocarcinoma by binding to miR‐149‐5p 37
Therefore, LINC00460 acts as a ceRNA sponging miRNAs and inhibited PI3K/AKT signaling pathway to regulate NSCLC progression. LINC00460 might serve as a prognostic indicator and as an effective therapeutic target in the future.
Papillary thyroid cancer
Thyroid tumors are classified as follicle‐derived (thyroid epithelial) neoplasms, other epithelial tumors, non‐epithelial tumors, and secondary tumors. 72 The most common subtype of thyroid cancer is PTC. 73 Despite advances in therapeutic methods, the overall survival of patients with local or distant metastasis is still poor. 74 Further detection should be launched by finding a new sensitive target for PTC and improving the prognosis.
It was found that LINC00460 was upregulated in PTC tissues and was associated with poor prognosis, 38 advanced TNM stage, and lymph node metastasis. 39 Li et al. 38 indicated that LINC00460 promoted cell migration, invasion, and EMT of PTC by sponging miR‐485‐5p to increase the expression of Raf1. Zou et al. 23 also found that LINC00460 knockdown restrained PTC progression by downregulating MMP‐9 expression by sponging miR‐539. In addition, Feng et al. 39 discovered that LINC00460 regulated sphingosine kinase 2 (SphK2) by sponging miR‐613 to partially promote the proliferation, migration, and invasion of PTC cells. Therefore, LINC00460 might be a novel diagnostic and therapeutic target for PTC patients. LINC00460 affected PTC progression mainly by directly binding with miRNAs. Further studies are needed to elucidate the specific mechanisms of LINC000460 in the development of PTC (Figure 5).
FIGURE 5.
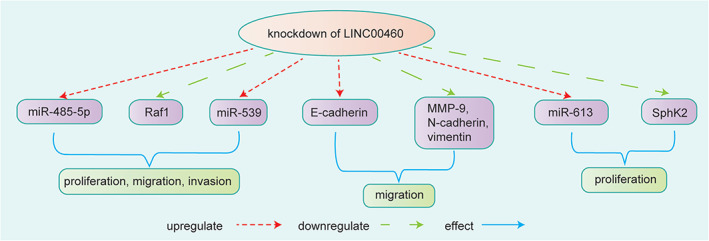
Knockdown of LINC00460 inhibited the proliferation, migration, and invasion of PTC cells by downregulating Raf1 and upregulating miR‐485‐5p. 38 Knockdown of LINC00460 inhibited the migration and invasion of PTC cells by increasing the protein level of E‐cadherin and decreasing the levels of MMP‐9, N‐cadherin, and vimentin. LINC00460 enhanced MMP‐9 protein expression by targeting miR‐539 to facilitate the proliferation and metastasis of PTC. 23 Knockdown of LINC00460 decreased the expression of SphK2 by upregulating miR‐613 to partially affect PTC proliferation 39
Breast cancer
Breast cancer is the second leading cause of cancer death among women after lung cancer. 75 Treatment for breast cancer is based on TNM stage, distant metastasis, patient age and status, and so on. 76 The 5‐year overall survival was connected with early diagnosis; therefore, more valid methods to detect the disease at the early stage and provide potential therapies are needed.
LINC00460 was identified as one of the meaningful lncRNAs from the TCGA database. 12 LINC00460 was overexpressed in breast cancer, and its expression was correlated with distant lymph node metastasis and poor prognosis. 40 Moreover, LINC00460 promoted cell proliferation, migration, and invasion by the miR‐489‐5p/FGF7/AKT axis in breast cancer. 40 A clear understanding of LINC00460 involved in breast cancer is still confusing needed to be further studied, which will accelerate the application of LINC00460 in diagnosis and prognosis (Figure 6).
FIGURE 6.
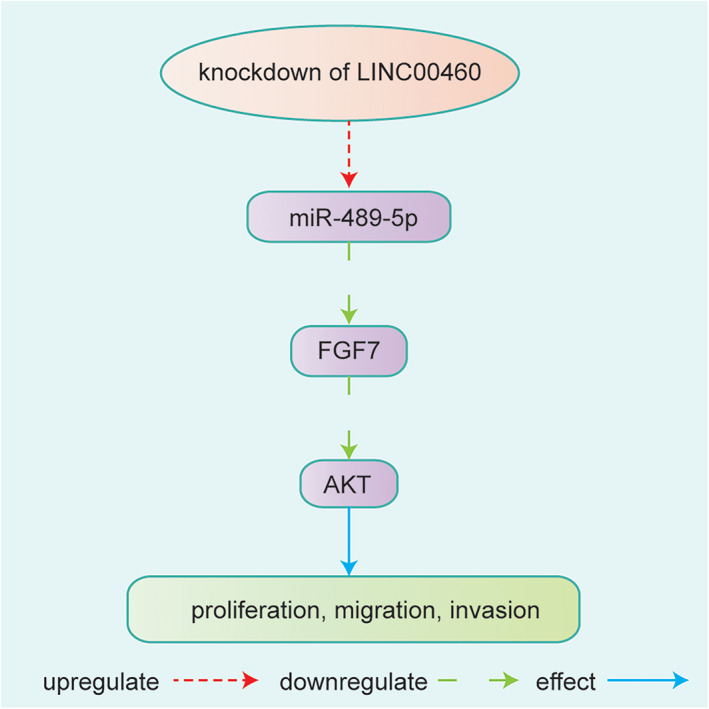
Knockdown of LINC00460 affected breast cancer by inhibiting the expression of miR‐489‐5p, miR‐489‐5p inhibited FGF7‐AKT signaling. LINC00460 affected cell proliferation, migration and invasion through the miR‐489‐5p/FGF7/AKT axis 40
Gastric cancer
GC is the second leading cause of cancer‐related mortality and is correlated with Helicobacter pylori infection, lifestyle factors, and genetics. 77 Despite the advances in current treatment, unfortunately, most patients are diagnosed at an advanced and unresectable stage. 78 Therefore, new investigations and methods must be considered. 79 There are no valid approaches that could be applied to treat advanced GC, so new predictive and prognostic biomarkers are needed to explore for GC patients. 80
Yang et al. 20 found that LINC00460 expression was obviously elevated in GC tissues and that high LINC00460 expression had a poor prognosis in GC patients. The results showed that LINC00460 influenced GC cell proliferation and apoptosis by downregulating CCNG2 via recruiting EZH2 and LSD1. 20 Wang et al. 41 found that LINC00460 upregulated the expression of the cell cycle/cell motility‐associated proteins CCND1, CDK4, vimentin, MMP‐2, and MMP‐9 by competitively binding to miR‐342‐3p; moreover, LINC00460 upregulated KDM2A expression by sponging miR‐342‐3p to partially affect cell proliferation and metastasis in GC. 41 Furthermore, Zhang et al. 42 showed that LINC00460 knockdown reduced c‐Myc and β‐catenin expression to inhibit the Wnt/β‐catenin signaling pathway, which was reported to affect cell proliferation and invasion in several tumors, including GC. 42 Therefore, LINC00460 affected GC progression not only by directly binding with miRNAs or RBPs, but also through the Wnt/β‐catenin signaling pathway. These studies have shown that LINC00460 might be a potential prognostic indicator for GC patients (Figure 7).
FIGURE 7.
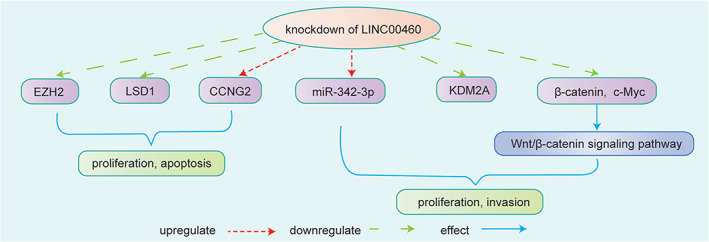
LINC00460 knockdown affected GC cell growth by inhibiting the binding between EZH2 and LSD1, and the inhibitory effect on CCNG2 expression of LINC00460 could be achieved by recruiting EZH2 and LSD1. 20 LINC00460 regulated cell proliferation and invasion by upregulating KDM2A through sponging of miR‐342‐3p. 41 LINC00460 knockdown efficiently inhibited cell proliferation and the Wnt/β‐catenin signaling pathway by reducing c‐Myc and β‐catenin expression 42
Hepatocellular carcinoma
HCC is the most common primary hepatic malignancy in the world, with an increasing worldwide prevalence. 81 Diagnosis of HCC at the earliest possible stage is crucial. Unfortunately, most HCC patients often miss the early diagnosis, leading to poor prognosis and low overall survival. 82 To improve the diagnostic and therapeutic effects, further valid measures should be investigated.
Previous studies indicated that LINC00460 was upregulated in HCC tissues and related to the progression of HCC. 43 , 44 Specifically, knockdown of LINC00460 could suppress the expression of cell cycle‐related proteins cyclin D1/CDK4 and the cell migration‐related proteins MMP‐3/MMP‐9. 43 Moreover, knockdown of LINC00460 inhibited anterior gradient homolog 2 (AGR2) expression by targeting miR‐342‐3p to affect cell proliferation, migration, and apoptosis of HCC. 43 , 44 Furthermore, Tu et al. 24 predicted that the LINC00460/miR‐485‐5p/PAK1 axis is involved in HCC development. The results showed that LINC00460 may be a potential diagnostic marker and valuable therapeutic target for HCC patients (Figure 8).
FIGURE 8.
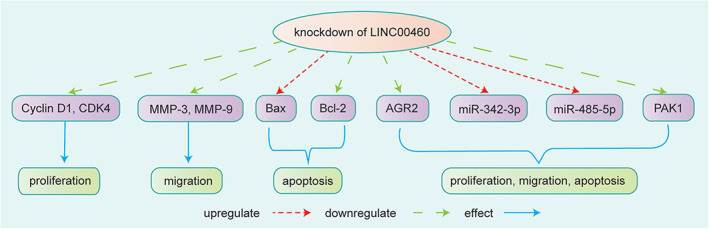
Knockdown of LINC00460 suppressed the expression of the cell proliferation‐related proteins cyclin D1 and CDK4 in HCC, inducing obvious cell cycle arrest. The expression of the cell migration‐related proteins MMP‐3 and MMP‐9 was also greatly suppressed after LINC00460 knockdown. 43 Knockdown of LINC00460 affected cell apoptosis by elevating the expression of Bax and reducing the expression of Bcl‐2. Knockdown of LINC00460 suppressed AGR2 expression by targeting miR‐342‐3p to affect cell proliferation, migration, and apoptosis of HCC. 44 LINC00460 knockdown inhibited PAK1 levels by sponging miR‐485‐5p to affect cell proliferation, migration, and apoptosis 24
Colorectal cancer
CRC is the second‐most in women and third‐most common cancer in men, 83 and CRC ranks second in terms of mortality. 84 The risk of CRC is closely related to genetic and environmental factors. Substantial changes in lifestyle, smoking behavior, alcohol intake, and weight management can improve the risk of CRC. 83 Despite advances in therapeutic methods, patients with CRC present with metastatic disease up to 50% at the time of diagnosis. 85 Lech et al. 86 examined molecular biomarkers correlated with CRC screening, early detection of disease recurrence, and prognostic and predictive factors. Therefore, it is necessary to find valuable biomarkers to better predict diagnosis and improve prognosis.
LINC00460 regulated the cellular processes of CRC by promoting cell proliferation, migration, invasion, and affecting apoptosis or the cell cycle. 21 Lian et al. 45 found that LINC00460 was remarkably upregulated in CRC tissues and was related to larger tumor sizes, advanced tumor node metastasis stages, lymph node metastasis and shorter overall survival in CRC patients. Importantly, LINC00460 promoted CRC cell proliferation by binding to the enhancer of zeste homolog 2 (EZH2) and repressing Krüppellike factor 2 (KLF2); LINC00460 also promoted the expression level of cullin 4A (CUL4A) by competing for miR‐149‐5p to enhance cell proliferation. 45
LINC00460 could also act as a ceRNA and sponge miRNAs in CRC progression. Zhang et al. 46 found that LINC00460 promoted CRC cell metastasis by regulating the miR‐939‐5p/LIMK2 axis. Additionally, knockdown of LINC00460 partially suppressed cell proliferation, migration, invasion, and EMT of CRC by upregulating WWC2 via ERG. 47 Zhang et al. 87 also found that hypomethylated oncogenic LINC00460 could promote CRC metastasis. LINC00460 is also involved in ionizing radiation‐induced radioresistance by mediating EMT processes. 88 In addition, LINC00460 is highly expressed in colon cancer (CC) tissues by targeting miR‐433‐3p and upregulating the expression of ANXA2 to promote cell carcinogenicity 27 (Figure 9).
FIGURE 9.
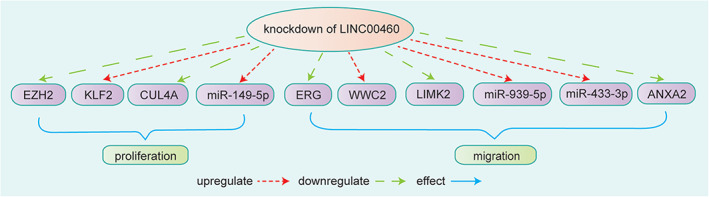
Knockdown of LINC00460 suppressed CRC cell proliferation by upregulating KLF2 via binding to EZH2. Knockdown of LINC00460 inhibited cell proliferation by decreasing the expression of CUL4A and increasing the expression of miR‐149‐5p. 45 Knockdown of LINC00460 suppressed CRC cell metastasis by upregulating WWC2 via ERG. 47 Knockdown of LINC00460 inhibited CRC cell metastasis by decreasing the expression of LIMK2 and miR‐939‐5p sponging. 46 Knockdown of LINC00460 inhibited cell migration by targeting miR‐433‐3p and downregulating the expression of ANXA2 in colon cancer 27
Therefore, LINC00460 could directly bind with miRNAs or RBPs to promote the development of CRC. The studies suggested that LINC00460 could promote CRC progression and might be a diagnostic and prognostic marker for CRC patients. Moreover, LINC00460 might be a potential therapeutic value for CRC radiotherapy.
Other cancers
The expression of LINC00460 was also upregulated in osteosarcoma (OS), nasopharyngeal carcinoma (NPC), ovarian cancer (OC), bladder urothelial carcinoma, pancreatic cancer (PC), and cervical cancer, which might be a valuable prognostic and curative biomarker and potential therapeutic target.
LINC00460 affected cell viability, cell cycle, apoptosis, migration, invasion, and EMT process by regulating the expression of cyclin D1, CDK4/CDK6 and MMP‐9 in OS. 48 Lian et al. 49 found that LINC00460 facilitated OS cell proliferation, migration, and invasion by regulating FADS1 as a molecular sponge for miR‐1224‐5p in OS. The high expression of LINC00460 was predicted to be associated with poor prognosis in NPC patients, and LINC00460 overexpression facilitated cancer cell migration, invasion, and EMT by targeting miR‐30a‐3p/Rap1A 50 or miR‐149‐5p/IL6 axis. 51 For the OC study, the upregulated expression of LINC00460 was in OC tissues and were associated with tumor stage and tumor size, moreover, LINC00460 promoted OC progression by binding miR‐338‐3p. 52 For bladder urothelial carcinoma, Li et al. 53 indicated that the high expression of LINC00460 was associated with poor survival, promoted cell proliferation and migration, and LINC00460 prompted tumor progression via miR‐612/FOXK1 axis. Moreover, LINC00460 accelerated PC by binding to miR‐491‐5p. 54 For cervical cancer, LINC00460 affected tumor progression by targeting miR‐503‐5p, 55 or by mediating the miR‐361‐3p/Gli1 axis. 56 Therefore, LINC00460 acts as a ceRNA in the pathogenesis and tumorigenesis of cancers base on the above research. This provides the possibility for LINC00460 in the diagnostic and prognostic application (Figure 10).
FIGURE 10.
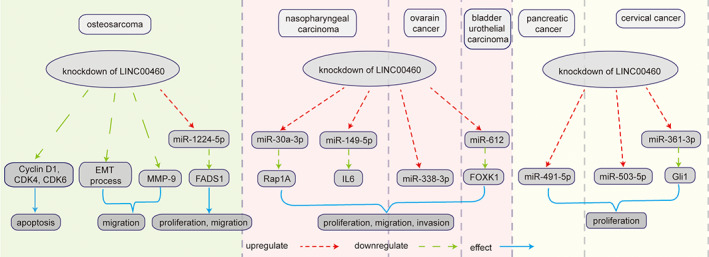
Knockdown of LINC00460 affected OS progression by inducing the cell apoptosis via the decreasing the expression of cyclin D1, CDK4, and CDK6. Knockdown of LINC00460 inhibited cell migration through decreasing the MMP‐9 activity and suppressing the EMT process. 48 Knockdown of LINC00460 decreased the expression of FADS1 and increased miR‐1224‐5p regulating the cell proliferation and migration in OS. 49 Knockdown of LINC00460 suppressed NPC cell proliferation, migration, and invasion by targeting miR‐30a‐3p/Rap1A and miR‐149‐5p/IL6 50 , 51 ; LINC00460 promoted OC progression by binding miR‐338‐3p 52 ; LINC00460 prompted bladder urothelial carcinoma progression via sponging of miR‐612 by elevating FOXK1 expression 53 ; LINC00460 accelerated pancreatic cancer by binding to the suppressor miR‐491‐5p 54 ; knockdown of LINC00460 suppressed proliferation cervical cancer cells by targeting miR‐503‐5p and miR‐361‐3p/Gli1 axis 55 , 56
Summary
More and more evidence suggested that LINC00460 acts as an oncogene in diverse tumors and plays a crucial regulatory role in tumorigenesis. LINC00460 was found to be highly expressed in various tumors tissues and cell lines; moreover, LINC00460 was reported to play vital roles in cellular functions such as proliferation, cell cycle, migration, invasion, autophagy, apoptosis, and others. The regulatory mechanism of LINC00460 was complex and involve many steps, including directly binding with proteins; binding to miRNA to act as a miRNA sponge; or activating signaling pathways, such as PIK3/AKT, Wnt/β‐catenin, etc. The most common mechanism of action of LINC00460 in various tumors is the specific absorption of miRNA as ceRNA. lncRNA‐miRNA‐mRNA regulatory network and key genes are the important steps of tumorigenesis and development, which provides ideas for the exploration of the regulatory mechanisms of LIN00460 in human cancers.
LINC00460 expression was often related to important clinical features such as tumor size, recurrence, TNM stage, lymph node metastasis, or poor prognosis in various cancers, so it might become a potential diagnostic and prognostic biomarker. Furthermore, the research results on LINC00460 provide a preliminary basis for whether it might be a potential target for cancer therapy in the future.
With the development of genomic studies, the role of LINC00460 gradually faded from mystery. However, the exploration of LINC00460 is still in the early stage and further molecular mechanism of LINC00460 should be elucidated.
CONCLUSIONS
With further exploration of the molecular mechanism of lncRNAs, additional knowledge of how lncRNAs affect tumor growth has been reported. LINC00460 is highly expressed in human cancers, and may be a potential biomarker for cancer diagnosis, prognosis, or therapy. As an oncogene, LINC00460 can promote tumor progression and play the role in cell proliferation, migration, invasion, EMT, apoptosis, cell cycle, autophagy, chemoresistance, or radioresistance in various tumors. The mechanisms of the LINC00460 effect are complex. Clarifying the mechanism of LINC00460 is helpful to further study the relationship between lncRNAs and tumor, and is of great value to clarify the pathogenesis of tumors.
CONFLICT OF INTEREST
The authors declare that they have no conflicts interests.
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (82103008), and Shandong Provincial Natural Science Foundation (ZR2020MH192, ZR2020MH188).
Chen X, Song J, Wang X, Sun D, Liu Y, Jiang Y. LncRNA LINC00460: Function and mechanism in human cancer. Thorac Cancer. 2022;13:3–14. 10.1111/1759-7714.14238
Funding information National Natural Science Foundation of China, Grant/Award Number: 82103008; Natural Science Foundation of Shandong Province, Grant/Award Numbers: ZR2020MH188, ZR2020MH192
REFERENCES
- 1. Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends: an update. Cancer Epidemiol Biomarkers Prev. 2016;25(1):16–27. [DOI] [PubMed] [Google Scholar]
- 2. Maruvada P, Fau WW, Wagner PD, Fau WP, Srivastava S, Srivastava S. Biomarkers in molecular medicine: cancer detection and diagnosis. Biotechniques. 2005;38(4S):S9–S15. [DOI] [PubMed] [Google Scholar]
- 3. Chandra Gupta S, Nandan TY. Potential of long non‐coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer. 2017;140(9):1955–67. [DOI] [PubMed] [Google Scholar]
- 4. Zhang Y, Tao Y, Liao Q. Long noncoding RNA: a crosslink in biological regulatory network. Brief Bioinform. 2018;19(5):930–45. [DOI] [PubMed] [Google Scholar]
- 5. Peng WX, Koirala P, Mo YY. LncRNA‐mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kumar MM, Goyal R. LncRNA as a therapeutic target for angiogenesis. Curr Top Med Chem. 2017;17:1750–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thin KZ, Liu X, Feng X, Raveendran S, Tu JC. LncRNA‐DANCR: a valuable cancer related long non‐coding RNA for human cancers. Pathol Res Pract. 2018;214(6):801–5. [DOI] [PubMed] [Google Scholar]
- 9. Wang J, Su Z, Lu S, et al. LncRNA HOXA‐AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–33. [DOI] [PubMed] [Google Scholar]
- 10. Wu X, Sui Z, Zhang H, Wang Y, Yu Z. Integrated analysis of lncRNA‐mediated ceRNA network in lung adenocarcinoma. Front Oncol. 2020;10:554759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang C, Cao W, Wang J, et al. A prognostic long non‐coding RNA‐associated competing endogenous RNA network in head and neck squamous cell carcinoma. PeerJ. 2020;8:e9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang X, Gao C, Feng F, et al. Construction and analysis of competing endogenous RNA networks for breast cancer based on TCGA dataset. Biomed Res Int. 2020;2020:4078596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin YZ, Wu YP, Ke ZB, et al. Bioinformatics analysis of the expression of key long Intergenic non‐protein coding RNA genes in bladder cancer. Med Sci Monit. 2020;26:e920504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu Z, Wang C, Xiang X, Li J, Huang J. Characterization of mRNA expression and endogenous RNA profiles in bladder cancer based on the cancer genome atlas (TCGA) database. Med Sci Monit. 2019;25:3041–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qi L, Zhang T, Yao Y, et al. Identification of lncRNAs associated with lung squamous cell carcinoma prognosis in the competitive endogenous RNA network. PeerJ. 2019;7:e7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang J, Zhang C, He W, Gou X. Construction and comprehensive analysis of dysregulated long non‐coding RNA‐associated competing endogenous RNA network in clear cell renal cell carcinoma. J Cell Biochem. 2019;120(2):2576–93. [DOI] [PubMed] [Google Scholar]
- 17. Zhang D, Zeng S, Hu X. Identification of a three‐long noncoding RNA prognostic model involved competitive endogenous RNA in kidney renal clear cell carcinoma. Cancer Cell Int. 2020;20:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang Y, Cao W, Wu K, et al. LncRNA LINC00460 promotes EMT in head and neck squamous cell carcinoma by facilitating peroxiredoxin‐1 into the nucleus. J Exp Clin Cancer Res. 2019;38(1):365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li K, Sun D, Gou Q, et al. Long non‐coding RNA linc00460 promotes epithelial‐mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. [DOI] [PubMed] [Google Scholar]
- 20. Yang J, Lian Y, Yang R, et al. Upregulation of lncRNA LINC00460 facilitates GC progression through epigenetically silencing CCNG2 by EZH2/LSD1 and indicates poor outcomes. Mol Ther Nucleic Acids. 2020;19:1164–75. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Wang X, Mo FM, Bo H, et al. Upregulated expression of long non‐coding RNA, LINC00460, suppresses proliferation of colorectal cancer. J Cancer. 2018;9(16):2834–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ye JJ, Cheng YL, Deng JJ, Tao WP, Wu L. LncRNA LINC00460 promotes tumor growth of human lung adenocarcinoma by targeting miR‐302c‐5p/FOXA1 axis. Gene. 2019;685:76–84. [DOI] [PubMed] [Google Scholar]
- 23. Zou X, Guo ZH, Li Q, Wang PS. Long noncoding RNA LINC00460 modulates MMP‐9 to promote cell proliferation, invasion and apoptosis by targeting miR‐539 in papillary thyroid cancer. Cancer Manag Res. 2020;12:199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tu J, Zhao Z, Xu M, Chen M, Weng Q, Ji J. LINC00460 promotes hepatocellular carcinoma development through sponging miR‐485‐5p to up‐regulate PAK1. Biomed Pharmacother. 2019;118:109213. [DOI] [PubMed] [Google Scholar]
- 25. Feng L, Rao M, Zhou Y, Zhang Y, Zhu Y. Long noncoding RNA 00460 (LINC00460) promotes glioma progression by negatively regulating miR‐320a. J Cell Biochem. 2019;120(6):9556–63. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Xing H, Wang S, Li Q, Ma Y, Sun P. Long noncoding RNA LINC00460 targets miR‐539/MMP‐9 to promote meningioma progression and metastasis. Biomed Pharmacother. 2018;105:677–82. [DOI] [PubMed] [Google Scholar]
- 27. Hong W, Ying H, Lin F, Ding R, Wang W, Zhang M. lncRNA LINC00460 silencing represses EMT in colon cancer through Downregulation of ANXA2 via Upregulating miR‐433‐3p. Mol Ther Nucleic Acids. 2020;19:1209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xue K, Li J, Nan S, Zhao X, Xu C. Downregulation of LINC00460 decreases STC2 and promotes autophagy of head and neck squamous cell carcinoma by up‐regulating microRNA‐206. Life Sci. 2019;231:116459. [DOI] [PubMed] [Google Scholar]
- 29. Xiaoxing Xie GX, Wang Q, Ge Y, Cui X. Long non‐coding RNA LINC00460 promotes head and neck squamous cell carcinoma cell progression by sponging miR‐612 to up‐regulate AKT2. Am J Transl Res. 2019;11(10):6326–40. [PMC free article] [PubMed] [Google Scholar]
- 30. Lin M, Wang ZW, Zhu X. FBXO45 is a potential therapeutic target for cancer therapy. Cell Death Discovery. 2020;6:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang Y, Wang R, Feng L, Ma H, Fang J. LINC00460 promotes cell proliferation, migration, invasion, and epithelial‐Mesenchymal transition of head and neck squamous cell carcinoma via miR‐320a/BGN Axis. Onco Targets Ther. 2021;14:2279–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liang Y, Wu Y, Chen X, et al. A novel long noncoding RNA linc00460 up‐regulated by CBP/P300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci Rep. 2017;37(5):BSR20171019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cui Y, Zhang C, Lian H, et al. LncRNA linc00460 sponges miR‐1224‐5p to promote esophageal cancer metastatic potential and epithelial‐mesenchymal transition. Pathol Res Pract. 2020;216(7):153026. [DOI] [PubMed] [Google Scholar]
- 34. Wang HX, Fau KL, Qin X, Fau QX, Xu J, Fau XJ, et al. LINC00460 promotes proliferation and inhibits apoptosis of non‐small cell lung cancer cells through targeted regulation of miR‐539. Eur Rev Med Pharmacol Sci. 2020;24 (12):6752–8. [DOI] [PubMed] [Google Scholar]
- 35. Ma G, Zhu J, Liu F, Yang Y. Long noncoding RNA LINC00460 promotes the Gefitinib resistance of nonsmall cell lung cancer through epidermal growth factor receptor by sponging miR‐769‐5p. DNA Cell Biol. 2019;38(2):176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao H, Wang Y, Ren X. Nicotine promotes the development of non‐small cell lung cancer through activating LINC00460 and PI3K/Akt signaling. Biosci Rep. 2019;39(6):BSR20182443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nakano Y, Isobe K, Kobayashi H, et al. Clinical importance of long noncoding RNA LINC00460 expression in EGFRmutant lung adenocarcinoma. Int J Oncol. 2020;56(1):243–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li G, Kong Q. LncRNA LINC00460 promotes the papillary thyroid cancer progression by regulating the LINC00460/miR‐485‐5p/Raf1 axis. Biol Res. 2019;52(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Feng L, Yang B, Tang XD. Long noncoding RNA LINC00460 promotes carcinogenesis via sponging miR‐613 in papillary thyroid carcinoma. J Cell Physiol. 2019;234(7):11431–9. [DOI] [PubMed] [Google Scholar]
- 40. Zhu Y, Yang L, Chong QY, et al. Long noncoding RNA Linc00460 promotes breast cancer progression by regulating the miR‐489‐5p/FGF7/AKT axis. Cancer Manag Res. 2019;11:5983–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang F, Liang S, Liu X, Han L, Wang J, Du Q. LINC00460 modulates KDM2A to promote cell proliferation and migration by targeting miR‐342‐3p in gastric cancer. Onco Targets Ther. 2018;11:6383–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang S, Xu J, Wang H, Guo H. Downregulation of long noncoding RNA LINC00460 expression suppresses tumor growth in vitro and in vivo in gastric cancer. Cancer Biomark. 2019;24(4):429–37. [DOI] [PubMed] [Google Scholar]
- 43. Hong H, Sui C, Qian T, Xu X, Zhu X, Fei Q, et al. Long noncoding RNA LINC00460 conduces to tumor growth and metastasis of hepatocellular carcinoma through miR‐342‐3p‐dependent AGR2 up‐regulation. Aging (Albany NY). 2020;12(11):10544–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang J, Li K, Chen J, Hu X, Wang H, Zhu X. Long noncoding RNA LINC00460 promotes hepatocellular carcinoma progression via regulation of miR‐342‐3p/AGR2 Axis. Onco Targets Ther. 2020;13:1979–91. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45. Lian Y, Yan C, Xu H, et al. A novel lncRNA, LINC00460, affects cell proliferation and apoptosis by regulating KLF2 and CUL4A expression in colorectal cancer. Mol Ther Nucleic Acids. 2018;12:684–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y, Liu X, Li Q, Zhang Y. lncRNA LINC00460 promoted colorectal cancer cells metastasis via miR‐939‐5p sponging. Cancer Manag Res. 2019;11:1779–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yuan B, Yang J, Gu H, Ma C. Down‐regulation of LINC00460 represses metastasis of colorectal cancer via WWC2. Dig Dis Sci. 2020;65(2):442–56. [DOI] [PubMed] [Google Scholar]
- 48. Jiang JJ, Wang FC, Han LP. Long intergenic nonprotein coding RNA 00460 predicts a poor prognosis and promotes tumorigenesis of human osteosarcoma. Mol Med Rep. 2020;21(2):649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lian H, Xie P, Yin N, et al. Linc00460 promotes osteosarcoma progression via miR‐1224‐5p/FADS1 axis. Life Sci. 2019;233:116757. [DOI] [PubMed] [Google Scholar]
- 50. Hu X, Liu W, Jiang X, et al. Long noncoding RNA LINC00460 aggravates invasion and metastasis by targeting miR‐30a‐3p/Rap1A in nasopharyngeal carcinoma. Hum Cell. 2019;32(4):465–76. [DOI] [PubMed] [Google Scholar]
- 51. Kong YG, Cui M, Chen SM, Xu Y, Xu Y, Tao ZZ. LncRNA‐LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR‐149‐5p to up‐regulate IL6. Gene. 2018;639:77–84. [DOI] [PubMed] [Google Scholar]
- 52. Liu X, Wen J, Wang H, Wang Y. Long non‐coding RNA LINC00460 promotes epithelial ovarian cancer progression by regulating microRNA‐338‐3p. Biomed Pharmacother. 2018;108:1022–8. [DOI] [PubMed] [Google Scholar]
- 53. Li J, Huang S, Zhang Y, Zhuo W, Tong B, Cai F. LINC00460 enhances bladder carcinoma cell proliferation and migration by modulating miR‐612/FOXK1 Axis. Pharmacology. 2021;106(1‐2):79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu J, Sun S, Liao W, et al. LINC00460 promotes pancreatic cancer progression by sponging miR‐491‐5p. J Gene Med. 2021;23(6):e3333. [DOI] [PubMed] [Google Scholar]
- 55. Lin L, Xin B, Jiang T, Wang XL, Yang H, Shi TM. Long non‐coding RNA LINC00460 promotes proliferation and inhibits apoptosis of cervical cancer cells by targeting microRNA‐503‐5p. Mol Cell Biochem. 2020;475(1–2):1–13. [DOI] [PubMed] [Google Scholar]
- 56. Li F, Zhu W, Wang Z. Long noncoding RNA LINC00460 promotes the progression of cervical cancer via regulation of the miR‐361‐3p/Gli1 axis. Hum Cell. 2021;34(1):229–37. [DOI] [PubMed] [Google Scholar]
- 57. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32. [DOI] [PubMed] [Google Scholar]
- 58. Specenier P, Vermorken JB. Optimizing treatments for recurrent or metastatic head and neck squamous cell carcinoma. Expert Rev Anticancer Ther. 2018;18(9):901–15. [DOI] [PubMed] [Google Scholar]
- 59. Du E, Mazul AL, Farquhar D, et al. Long‐term survival in head and neck cancer: impact of site, stage, smoking, and human papillomavirus status. Laryngoscope. 2019;129(11):2506–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cao W, Liu JN, Liu Z, et al. A three‐lncRNA signature derived from the atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94–101. [DOI] [PubMed] [Google Scholar]
- 61. Ritu Chaudhary1 XW, Cao Biwei2, De La Iglesia1 J, Masannat1 J, Song1 F, Hernandez‐Prera3 JC, et al. Long noncoding RNA, LINC00460, as a prognostic biomarker in head and neck squamous cell carcinoma (HNSCC). Am J Transl Res. 2020;12(2):684–96. [PMC free article] [PubMed] [Google Scholar]
- 62. Wang Q, Rao Y, Guo X, et al. Oral microbiome in patients with Oesophageal squamous cell carcinoma. Sci Rep. 2019;9(1):19055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Reichenbach ZW, Murray MG, Saxena R, et al. Clinical and translational advances in esophageal squamous cell carcinoma. Adv Cancer Res. 2019;144:95–135. [DOI] [PubMed] [Google Scholar]
- 64. Guan Z, Wang Y, Wang Y, et al. Long non‐coding RNA LOC100133669 promotes cell proliferation in oesophageal squamous cell carcinoma. Cell Prolif. 2020;53(4):e12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu W, Zhang Y, Chen M, Shi L, Xu L, Zou X. A genome‐wide analysis of long noncoding RNA profile identifies differentially expressed lncRNAs associated with esophageal cancer. Cancer Med. 2018;7(8):4181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huang G‐W, Xue Y‐J, Wu Z‐Y, et al. A three‐lncRNA signature predicts overall survival and disease‐free survival in patients with esophageal squamous cell carcinoma. BMC Cancer. 2018;18(1):147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. [DOI] [PubMed] [Google Scholar]
- 68. Alexander M, Wolfe R, Ball D, et al. Lung cancer prognostic index: a risk score to predict overall survival after the diagnosis of non‐small‐cell lung cancer. Br J Cancer. 2017;117(5):744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Villalobos P, Wistuba II. Lung cancer biomarkers. Hematol Oncol Clin North Am. 2017;31(1):13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. YZ Q‐YY. Effects of Linc00460 on cell migration and invasion through regulating epithelial‐mesenchymal transition (EMT) in non‐small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;2018(22):1003–10. [DOI] [PubMed] [Google Scholar]
- 71. Qi G, Kong W, Mou X, Wang S. A new method for excavating feature lncRNA in lung adenocarcinoma based on pathway crosstalk analysis. J Cell Biochem. 2019;120(6):9034–46. [DOI] [PubMed] [Google Scholar]
- 72. Abdullah MI, Junit SM, Ng KL, Jayapalan JJ, Karikalan B, Hashim OH. Papillary thyroid cancer: genetic alterations and molecular biomarker investigations. Int J Med Sci. 2019;16(3):450–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. McLeod DSA, Zhang L, Durante C, Cooper DS. Contemporary debates in adult papillary thyroid cancer management. Endocr Rev. 2019;40(6):1481–99. [DOI] [PubMed] [Google Scholar]
- 74. Lin P, Guo YN, Shi L, et al. Development of a prognostic index based on an immunogenomic landscape analysis of papillary thyroid cancer. Aging (Albany NY). 2019;11(2):480–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64(1):52–62. [DOI] [PubMed] [Google Scholar]
- 76. Maughan KLLM, Ham PS. Treatment of breast cancer. 2011. Am Fam Physician. 2010;81(11):1339–46. [PubMed] [Google Scholar]
- 77. Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J. 2014;55(12):621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39(7):1010428317714626. [DOI] [PubMed] [Google Scholar]
- 79. Machlowska J, Maciejewski R, Sitarz R. The pattern of signatures in gastric cancer prognosis. Int J Mol Sci. 2018;19(6):1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Panarese I, De Vita F, Ronchi A, et al. Predictive biomarkers along gastric cancer pathogenetic pathways. Expert Rev Anticancer Ther. 2017;17(5):417–25. [DOI] [PubMed] [Google Scholar]
- 81. Clark T, Maximin S, Meier J, Pokharel S, Bhargava P. Hepatocellular carcinoma: review of epidemiology, screening, imaging diagnosis, response assessment, and treatment. Curr Probl Diagn Radiol. 2015;44(6):479–86. [DOI] [PubMed] [Google Scholar]
- 82. Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34(2):153–9. [DOI] [PubMed] [Google Scholar]
- 83. Kuipers EJ, Grady WM, Lieberman D, et al. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 85. Xue L, Williamson A, Gaines S, et al. An update on colorectal cancer. Curr Probl Surg. 2018;55(3):76–116. [DOI] [PubMed] [Google Scholar]
- 86. Lech G, Slotwinski R, Slodkowski M, Krasnodebski IW. Colorectal cancer tumour markers and biomarkers: recent therapeutic advances. World J Gastroenterol. 2016;22(5):1745–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang H, Lu Y, Wu J, Feng J. LINC00460 Hypomethylation promotes metastasis in colorectal carcinoma. Front Genet. 2019;10:880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang J, Ding L, Sun G, Ning H, Huang R. Suppression of LINC00460 mediated the sensitization of HCT116 cells to ionizing radiation by inhibiting epithelial‐mesenchymal transition. Toxicol Res (Camb). 2020;9(2):107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]


