Abstract
Ribosome RNA (rRNA) accounts for more than 80% of the cell's total RNA, while the physiological functions of rRNA modifications are poorly understood. Mutations of 18S rRNA m6A methyltransferase METTL5 cause intellectual disability, microcephaly, and facial dysmorphisms in patients, however, little is known about the underlying mechanisms. In this study, we identified METTL5 protein complex and revealed that METTL5 mainly interacts with RNA binding proteins and ribosome proteins. Functionally, we found that Mettl5 knockout in mESCs leads to the abnormal craniofacial and nervous development. Moreover, using Mettl5 knockout mouse model, we further demonstrated that Mettl5 knockout mice exhibit intellectual disability, recapitulating the human phenotype. Mechanistically, we found that Mettl5 maintains brain function and intelligence by regulating the myelination process. Our study uncovered the causal correlation between mis-regulated 18S rRNA m6A modification and neural function defects, supporting the important physiological functions of rRNA modifications in human diseases.
Keywords: 18S rRNA, Intellectual disability, METTL5, N6-methyladenosine (m6A), Neural development
Introduction
RNA modifications play essential functions in posttranscriptional gene expression regulation. Modifications on mRNAs, for example, m6A mRNA modification, can regulate mRNA splicing, export, transport and translation and therefore regulates diverse biological processes.1 In addition, modifications on tRNAs regulate tRNA stability and function, playing essential functions in regulation of global mRNA translation.2 Mis-regulations of mRNA or tRNA modifications often result in developmental diseases and cancers.3, 4, 5, 6, 7
Ribosome RNA (rRNA) is highly abundant, accounting for more than 80% of the cell's total RNA.8 rRNA is the RNA component of the mRNA translation machinery ribosomes and is essential for protein synthesis. Similar to mRNAs or tRNAs, rRNAs also contain different kinds of modifications. These modified residues are conservatively located in functionally important regions of the ribosomes,9 suggesting that rRNA modifications play important roles in regulating ribosome function. Indeed, rRNA modification machinery and changes in the extent of rRNA modifications connect with development, genetic diseases and cancers.10 However, physiological functions of rRNA modifications are still poorly understood.
Recent studies suggested rRNA modification is essential for neural development and brain function, such as spatial cognition,11 corpus callosum development12 and migration of neurons.13 In addition, it has been reported that the 28S rRNA m6A methyltransferase ZCCHC4 plays important function in translation and tumorigenesis.14 Mutations of the 18S rRNA m6A methyltransferase METTL5 was reported in patients with intellectual disability, microcephaly, and facial dysmorphisms,15,16 however, little is known about the underlying molecular mechanisms.
In this study, we verified Mettl5 is an 18S rRNA m6A methyltransferase and interacts with ribosome proteins and RNA binding proteins by m6A MeRIP-Seq and proteomic analysis, and then we combined in vitro mESCs differentiation models and in vivo Mettl5 knockout mouse model uncovered the critical function of Mettl5 mediated 18S rRNA m6A modification in regulation of neural development and brain function. Moreover, our data revealed that knockout of Mettl5 results in hypomyelination of the neurons, impairing learning and memory disability in mice.
Materials and methods
Generation of the Mettl5 knockout mouse allele
Mettl5+/− mice with C57BL/6N background were generated by using CRISPR-Cas9 systems. The exon 2, exon 3 and exon 4 of Mettl5 were deleted in the knockout Mettl5 allele. Mice were genotyped for the targeted allele by PCR using tail DNA. All experiments were performed in compliance with the relevant laws and institutional guidelines, and were approved by the Laboratory Animal Center of Sun Yat-sen University.
m6A MeRIP-Seq (methylated RNA immunoprecipitation and sequencing)
Total RNA was randomly fragmented to size around 100 nucleotides at an appropriate temperature and then subjected to immunoprecipitation (IP) with anti-m6A antibody (Sigma-Aldrich, cat. no ABE572) and protein-A/G magnetic beads (88803, Thermo). The m6A-containing mRNA fragments were captured, washed and eluted by competition with N6-Methyladenosine (M2780, Sigma-Aldrich). The purified RNA fragments from m6A meRIP were used for library construction by MeRIP-qRT-PCR and sequenced (Guangzhou Epibiotek Co., Ltd). The sequence reads mapping (HISAT2), peak calling (exomePeak), peak anno, motif search (HOMER) and gene ontology analysis were performed as previously described.6,17
METTL5 protein complex identification
293T cells transfected with pFLAG-CMV2-hMETTL5 plasmid using Lipofectamine 2000 following the standard protocols. 48 h after transfection, the cells were washed twice with precooled PBS, and then cells were collected and lysed with precooled 500 μl WB/IP lysis buffer. The suspension was centrifuged for 10 min at 13,000 g at 4 °C and the sediment was discarded. The supernatant were incubated with anti-FLAG M2 agarose (A2220, Sigma) at 4 °C for overnight with gentle agitation. After washing, the pulled down proteins were eluted with 1× SDS sample loading buffer. The eluted proteins were separated by gel electrophoresis, gels were stained with Coomassie brilliant blue. The eluted proteins in the gel were identified by Mass Spectrometry. Finally, protein identification software was used to identify the proteins and protein interactions (STRING) in the sample.
mESCs self-renewal and differentiation
mESCs self-renewal and differentiation were performed as previously described.4 To determine self-renewal capacities of the Mettl5 knockout and control mESCs, 105 cells were seeded in 100 mm tissue culture dishes and then the cell numbers were counted at Day 3 and Day 5. For spontaneous differentiation of mESCs, the cell culture dishes were first coated with Poly (2-hydroxyethyl methacrylate) to prevent cell attachment. Then 5 × 106 mESCs were cultured with LIF (−) medium in the Poly 2-pre-coated dishes to induce embryoid body formation for 6 days. To induce the neural lineage differentiation of mESCs, 5 × 105 mESCs were cultured in N2B27 medium for 6 days using a previously established protocol.18
RT-PCR and qRT-PCR
Total RNA was isolated with TRIzol reagent (Sigma) from freshly dissected mouse brains or cultured cells. The cDNAs were prepared from 1 μg total RNA by using PrimeScript™ RT Master Mix (Takara). The quantitative PCR reactions were carried out with STBR Green Premix Pro Taq HS qPCR Kit (Accurate Biology, Changsha, China) on StepOne™ Real-Time PCR System instrument (Thermo).
RNA-seq and gene ontology analysis
RNA sequencing and data analysis was performed by Shenzhen BGI Co., Ltd, China. Briefly, total RNAs from Mettl5−/− mice and their control littermates or cultured cells were extracted by Trizol reagent and purified by using oligo(dT)-attached magnetic beads. Then the cDNA libraries were constructed for high throughput sequencing. The differentially expressed genes were subjected to gene ontology analysis using Toppgene (https://toppgene.cchmc.org).
Morris water maze test
The water maze task was performed as previously described19 with some modifications. It consisted of three phases: 1) 4 days with a visible platform; 2) 4 days with a hidden platform; 3) Probe trials, during which the platform was removed from the maze, lasted 40 s and were performed to assess the retention of previously acquired information. Probe was conducted 48 h later after the last trail of the whole learning process. Mice were tracked by a video camera (Sony) in both trails and probe. Collected data were analyzed by Flyde software (Feidi, Guangzhou, China). Statistical analyses used two-way ANOVA.
Immunofluorescence
Mice brains were dissected and fixed in 4% paraformaldehyde (Biosharp, Hefei, China) for 48h, dehydrated by using graded ethanol, vitrificated by Histo-Clear (Yinxin, Guangzhou, China) and embedded in paraffin. Immunofluorescence of the brain sections (10 μm) was performed using MBP antibody (78896s, CST), fluorescence-labelled secondary antibodies (Biolegend, USA) and DAPI. The fluorescence images were captured with normal fluorescence microscope (Carl Zeiss, Germany) and analyzed with ZEN software.
Statistical analysis
Statistical analyses were performed via two-way analysis of variance (ANOVA) or via a one-tailed unpaired Student's t-test. The data are presented as the means ± SEM. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 denote the significance thresholds.
Results and discussion
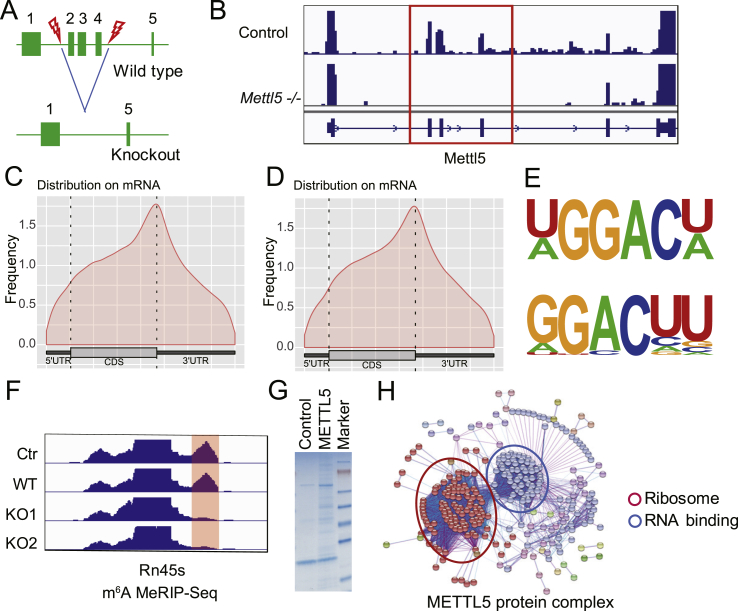
To study the function and molecular mechanism of METTL5 in the pathogenesis of neural diseases, we first knocked out Mettl5 in mouse embryonic stem cells (mESCs). The 2–4 exons of Mettl5 were depleted by CRISPR/Cas9 method (Fig. 1A), which results in frameshift and premature termination of Mettl5 mRNA translation. Then we performed RNA sequencing (RNA-Seq) and confirmed the successful depletion of Mettl5 exons 2–4 in mESCs (Fig. 1B). m6A predominantly targeted the coding regions and 3′ UTR of mRNA transcripts (Fig. 1C, D), which is consistent with the previously reported pattern of m6A peaks.20 Furthermore, a GGAC motif was enriched in m6A peaks on mRNAs in both control and Mettl5 KO mESCs (Fig. 1E). We found no difference on m6A mRNA modification between the control and Mettl5 KO mESCs, suggesting that Mettl5 has little effect on global mRNA m6A patten. However, our m6A methylation immunoprecipitation and sequencing (m6A MeRIP-Seq) showed that Mettl5 knockout reduces the m6A modification on 18S rRNA (Fig. 1F). In addition, proteomic analysis of the METTL5 protein complex revealed that METTL5 mainly interacts with ribosome proteins and RNA binding proteins, including the known interacting protein TRMT112 (Fig. 1G, H and Table S1).16 Overall, our data revealed that Mettl5 interacts with ribosome proteins and functions as an 18S rRNA m6A methyltransferase.
Figure 1.
Mettl5 as an 18S rRNA m6A methyltransferase interacts with ribosome proteins. (A). Schematic diagram of Mettl5 knockout in mESCs. (B). RNA-Seq tracks of Mettl5 mRNA. Deleted Mettl5 exons are indicated with red box. (C). Distribution of m6A targets in control mESCs. (D). Distribution of m6A targets in Mettl5 KO mESCs. (E). Sequence motif identified in m6A MeRIP-seq. The upper is m6A peaks of control mESCs and the bottom is m6A peaks of Mettl5 KO mESCs. (F). m6A MeRIP-Seq tracks of 18s rRNA (Rn18s, chr17:39846354-39848202) within 45s rRNA (Rn45s, chr17:39,840,997-39,850,829). Decreased 18S rRNA peaks are indicated with red highlight. (G). SDS-PAGE of METTL5 protein complex. (H). String view of METTL5 interacting proteins.
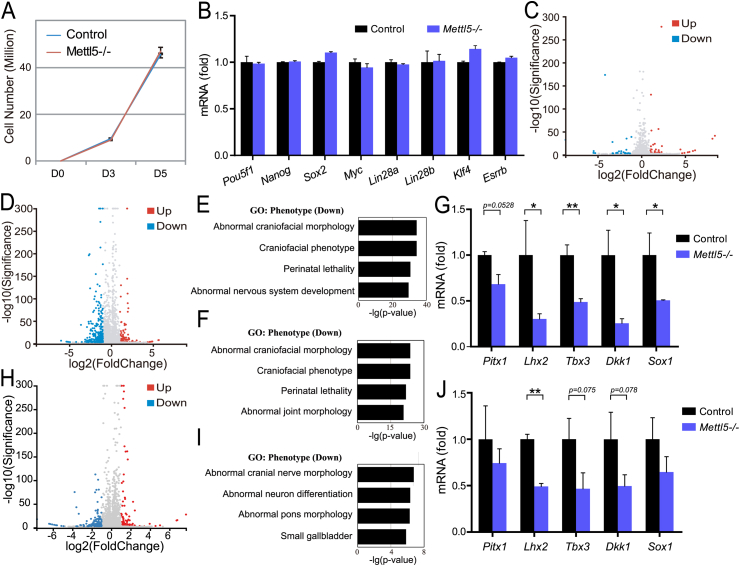
We next determined the function of Mettl5 in regulation of mESCs self-renewal and differentiation. Knockout of Mettl5 does not affect mESC proliferation (Fig. 2A) and has little effect on the expression of mESC markers including Pou5f1 and Nanog (Fig. 2B). Moreover, our RNA-Seq data revealed that Mettl5 knockout has little effect in global mRNA expression in mESCs (Fig. 2C and Table S2), suggesting that Mettl5 is not essential for mESCs self-renewal. Then we induced random differentiation of mESCs using the embryonic body (EB) differentiation model to study the function of Mettl5 in mESC differentiation. RNA-Seq was performed after 6 days of EB differentiation to identify the differentially expressed mRNAs (Fig. 2D and Table S2). Gene ontology revealed that the down-regulated genes in the Mettl5 knockout cells are associated with the abnormal craniofacial and nervous development (Fig. 2E) that is confirmed by gene ontology analysis cross-comparing the differentially expressed genes between control mESCs vs control EB and Mettl5 KO mESCs vs Mettl5 EB (Fig. 2F). The decreased expression of several of the key neural development genes in the Mettl5 knockout mESCs was confirmed by qRT-PCR (Fig. 2G). To further study the role of Mettl5 in neural differentiation, we induced neural lineage differentiation of mESCs using N2B27 induction model. Our results demonstrated that Mettl5 knockout mESCs have impaired neural differentiation capacity, further confirming the critical function of Mettl5 in cranial and neural development (Fig. 2H–J and Table S2). Overall, our data uncovered that mis-regulated Mettl5 results in craniofacial and neural development defects in mESCs, which could explain the intellectual disability and facial dysmorphism phenotypes in the METTL5 mutated human patients.
Figure 2.
Mettl5 regulates neural development in vitro. (A). Cell proliferation of Mettl5 KO and control mESC. (B). mRNA expression of stem cell markers in Mettl5 KO and control mESCs. (C). Volcano Plot of RNA-Seq data from Mettl5 KO and control mESCs. (D). Volcano Plot of RNA-Seq data from the Mettl5 KO and control embryonic bodies (differentiated for 6 days). (E). Gene ontology analysis of decreased expression genes in the Day6 Mettl5 KO embryonic bodies. (F). Gene ontology analysis of cross-comparison between control mESCs vs control embryonic bodies and Mettl5 KO mESCs vs Mettl5 embryonic bodies. (G). qRT-PCR analysis of mRNA expression in Mettl5 KO and control embryonic bodies (differentiated for 6 days). (H). Volcano Plot of RNA-Seq data from the Mettl5 KO and control cells (differentiated for 6 days in the N2B27 medium). (I). Gene ontology analysis of decreased expression genes in Mettl5 KO cells (differentiated for 6 days in the N2B27 medium). (J). qRT-PCR analysis of mRNA expression in Mettl5 KO and control cells (differentiated for 6 days in the N2B27 medium).
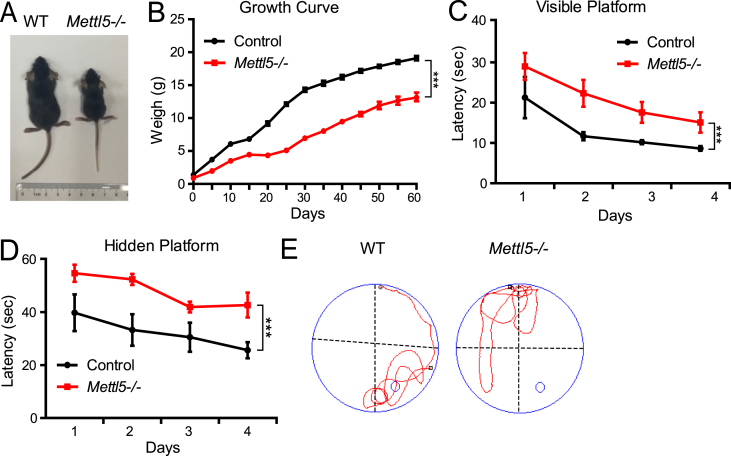
We further established the Mettl5 knockout mouse model to study the in vivo function of Mettl5. The Mettl5 knockout mice are viable but smaller compared to the wild type controls (Fig. 3A, B). Since METTL5 mutated patients and Mettl5 knockout mESCs displayed intellectual disability and neural development defects, we used Morris water maze assays to investigate the spatial learning and memory abilities of the Mettl5 knockout mice.19 Our data revealed that Mettl5 knockout results in impaired learning and memory capacities in the mice (Fig. 3C–E and Table S3), but not the swimming ability (Fig. 3E). Taken together, using knockout mouse model, we demonstrated that Mettl5 knockout mice exhibit intellectual disability, recapitulating the human phenotype. Our in vitro and in vivo data uncovered that Mettl5 is essential for the neural development and intellectual capacity.
Figure 3.
Mettl5 knockout impairs neural development and intelligence in vivo. (A). Representative pictures of Mettl5−/− mice at P14. (B). Growth curve with indicated genotypes (∗∗∗P < 0.001. Control: n = 5, Mettl5.−/−: n = 8). (C). The mean escape latency (±SEM) for mice to reach the visible platform in water maze assay (∗∗∗P < 0.001. Control: n = 6, Mettl5.−/−: n = 6). (D). The mean escape latency (±SEM) for mice to reach the hidden platform (∗∗∗P < 0.001. Control: n = 6, Mettl5.−/−: n = 6). (E). Probe trial was performed 48 h after the last training session by removing the platform.
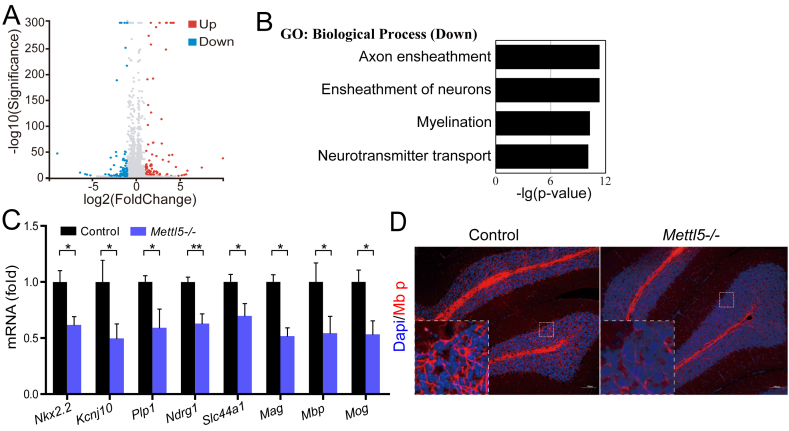
To study the underlying molecular mechanisms of Mettl5 in regulation of intellectual disability pathogenesis, we performed RNA-Seq of the brain to identify its downstream targets (Fig. 4A and Table S3). Our sequencing result revealed that genes down-regulated in the brains of Mettl5 knockout mice are involved in the neuron ensheathment, myelination and neurotransmitter transport processes (Fig. 4B). Neuron ensheathment with myelin protects and insulates the axons and thus facilitates the quick and accurate transmission of electrical impulses.21,22 Myelination is critical for various brain functions including learning capacity and intelligence.24 Our qRT-PCR and immunofluorescent assays confirmed the decreased expression of key neural development genes and myelin genes in the brain of Mettl5 knockout mice (Fig. 4C, D), supporting that Mettl5 maintains brain function and intelligence by regulating the myelination process.
Figure 4.
Mettl5 regulates the myelination process. (A). Scatter plot of mRNA expression by using RNA-Seq data from the Mettl5 KO and control brain tissues at P28. (B). Gene ontology analysis of decreased expression genes in Mettl5 KO brain tissues at P28. (C). qRT-PCR analysis of mRNA expression in the Mettl5 KO and control brain tissues at P28. (∗P < 0.05, ∗∗P < 0.01). (D). Immunostainings of myelin basic protein (Mbp) in cerebellum from mice with indicated genotypes. Scale bar, 100 μm.
Overall, using in vitro mESCs differentiation models, we revealed that knockout of Mettl5 leads to neural differentiation defects in mESCs. We then established the first Mettl5 knockout mouse model and uncovered the critical function of Mettl5 mediated 18S rRNA m6A modification in regulation of neural development and brain function. Moreover, our data revealed that knockout of Mettl5 in mouse results in decreased myelination of the neurons, leading to the impaired learning and memory disability in mice, supporting the role of Mettl5 in regulation of neural function.15,25,26 This study uncovered novel insights into the causal correlation between mis-regulated 18S rRNA m6A modification and neural function defects, supporting the important physiological functions of rRNA modifications in human diseases.
Author contributions
L.W., Z.P., D.C., YZ.J., and S.L. conceived the study, designed the experiments and supervised the project. L.W., Y.L., R.L., P.Y., J.M., M.C., H.H., X.W., G.W., F.L., and S.L. acquired the data. L.W., D.C., YZ.J., S.L. wrote the manuscript.
Conflict of interests
The authors delare no conflict of interest.
Acknowledgements
This work was supported by Excellent Youth Scholars grant from National Natural Science Foundation of China (No. 81922052), Distinguished Young Scholars grant from Natural Science Foundation of Guangdong (No. 2019B151502011), and the Guangzhou People's Livelihood Science and Technology Project (No. 201903010006).
Footnotes
Peer review under responsibility of Chongqing Medical University.
The GEO accession number for RNA sequencing and m6A MeRIP-Seq data is: GSE153140.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2020.07.004.
Contributor Information
Quan Yuan, Email: yuanquan@scu.edu.cn.
Yi-Zhou Jiang, Email: jiangyz@szu.edu.cn.
Shuibin Lin, Email: linshb6@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following are the supplementary data to this article:
Identification of METTL5 protein complex.
RNA-Seq of Mettl5 knockout and control mESCs.
RNA-Seq of Mettl5 knockout and control brain samples.
References
- 1.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duechler M., Leszczynska G., Sochacka E., Nawrot B. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell Mol Life Sci. 2016;73(16):3075–3095. doi: 10.1007/s00018-016-2217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu P.J., Shi H., He C. Epitranscriptomic influences on development and disease. Genome Biol. 2017;18(1):197. doi: 10.1186/s13059-017-1336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin S., Liu Q., Lelyveld V.S., Choe J., Szostak J.W., Gregory R.I. Mettl1/Wdr4-Mediated m(7)G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol Cell. 2018;71(2):244–255. doi: 10.1016/j.molcel.2018.06.001. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torres A.G., Batlle E., Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med. 2014;20(6):306–314. doi: 10.1016/j.molmed.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Lin S., Choe J., Du P., Triboulet R., Gregory R.I. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol Ther. 2019;27(4):710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noller H.F. Ribosomal RNA and translation. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 10.Sloan K.E., Warda A.S., Sharma S., Entian K.D., Lafontaine D.L.J., Bohnsack M.T. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017;14(9):1138–1152. doi: 10.1080/15476286.2016.1259781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang T., Chen P., Li W., et al. Cognitive deficits in mice lacking Nsun5, a cytosine-5 RNA methyltransferase, with impairment of oligodendrocyte precursor cells. Glia. 2019;67(4):688–702. doi: 10.1002/glia.23565. [DOI] [PubMed] [Google Scholar]
- 12.Yuan Z., Chen P., Zhang T., Shen B., Chen L. Agenesis and hypomyelination of corpus callosum in mice lacking Nsun5, an RNA methyltransferase. Cells. 2019;8(6):552. doi: 10.3390/cells8060552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen P., Zhang T., Yuan Z., Shen B., Chen L. Expression of the RNA methyltransferase Nsun5 is essential for developing cerebral cortex. Mol Brain. 2019;12(1):74. doi: 10.1186/s13041-019-0496-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma H., Wang X. N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol. 2019;15(1):88–94. doi: 10.1038/s41589-018-0184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richard E.M., Polla D.L., Assir M.Z., et al. Bi-allelic variants in METTL5 cause autosomal-recessive intellectual disability and microcephaly. Am J Hum Genet. 2019;105(4):869–878. doi: 10.1016/j.ajhg.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Tran N., Ernst F.G.M., Hawley B.R., et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47(15):7719–7733. doi: 10.1093/nar/gkz619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8(1):176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 18.Ying Q.L., Stavridis M., Griffiths D., Li M., Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21(2):183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 19.Vorhees C.V., Williams M.T. Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat Protoc. 2006;1(2):848–858. doi: 10.1038/nprot.2006.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ke S., Alemu E.A., Mertens C., et al. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev. 2015;29(19):2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherman D.L., Brophy P.J. Mechanisms of axon ensheathment and myelin growth. Nat Rev Neurosci. 2005;6(9):683–690. doi: 10.1038/nrn1743. [DOI] [PubMed] [Google Scholar]
- 22.Milbreta U., Lin J., Pinese C., et al. Scaffold-mediated sustained, non-viral delivery of miR-219/miR-338 promotes CNS remyelination. Mol Ther. 2019;27(2):411–423. doi: 10.1016/j.ymthe.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenzie I.A., Ohayon D., Li H., et al. Motor skill learning requires active central myelination. Science. 2014;346(6207):318–322. doi: 10.1126/science.1254960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leismann J., Spagnuolo M., Pradhan M., et al. The 18S ribosomal RNA m(6) A methyltransferase Mettl5 is required for normal walking behavior in Drosophila. EMBO Rep. 2020;21(7):e49443. doi: 10.15252/embr.201949443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ignatova V.V., Stolz P., Kaiser S., et al. The rRNA m(6)A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes Dev. 2020;34(9–10):715–729. doi: 10.1101/gad.333369.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of METTL5 protein complex.
RNA-Seq of Mettl5 knockout and control mESCs.
RNA-Seq of Mettl5 knockout and control brain samples.