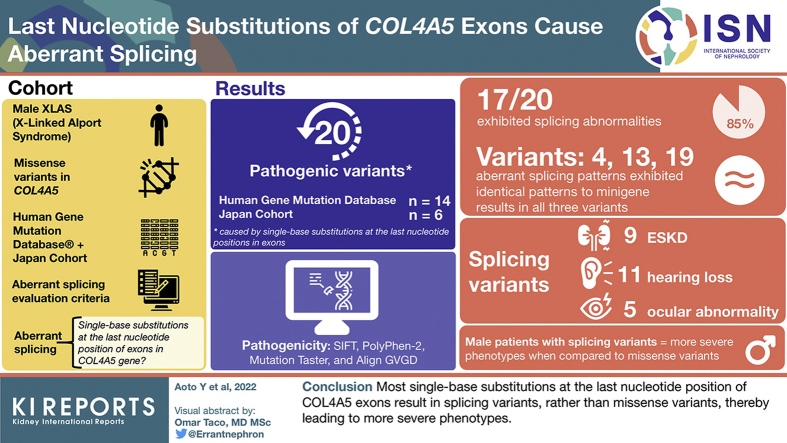
Abstract
Introduction
COL4A5 is a causative gene of X-linked Alport syndrome (XLAS). Male patients with XLAS with nonsense variants have the most severe phenotypes of early onset end-stage kidney disease (ESKD); those with splicing variants have middle phenotypes and those with missense variants have the mildest phenotypes. Therefore, genotyping for male patients with XLAS can be used to predict kidney prognosis. Single-base substitutions at the last nucleotide position in each exon are known to affect splicing patterns and could be splicing variants. Nevertheless, in XLAS, these variants are generally considered to be missense variants, without conducting a transcript analysis, which underestimates some patients as having mild phenotypes. This study aimed to investigate whether single-base substitutions at the last nucleotide position of COL4A5 exons cause aberrant splicing.
Methods
In total, 20 variants were found in the Human Gene Mutation Database (n = 14) and our cohort (n = 6). We performed functional splicing assays using a hybrid minigene analysis and in vivo transcript analyses of patients’ samples when available. Then, we investigated genotype–phenotype correlations for patients with splicing variants detected in this study by comparing data from our previous studies.
Results
Among the 20 variants, 17 (85%) caused aberrant splicing. Male patients with splicing variants had more severe phenotypes when compared with those with missense variants. Findings from the in vivo analyses for 3 variants were identical to those from the minigene assay.
Conclusion
Our study revealed that most single-base substitutions at the last nucleotide position of COL4A5 exons result in splicing variants, rather than missense variants, thereby leading to more severe phenotypes.
Keywords: COL4A5, genotype–phenotype correlation, last nucleotide position, missense variants, single-base substitutions, splicing
Graphical abstract
COL4A5 (NM: 000495.4) encodes type IV collagen α5 chain and is a causative gene of XLAS. XLAS is a hereditary kidney disease that causes heterogeneous renal manifestations, from hematuria alone to ESKD accompanied by sensorineural deafness and ocular abnormalities.1,2 Male patients with XLAS exhibit proteinuria and hematuria in the early stage of childhood and develop ESKD at a median age of 25 to 35 years.3,4 In male patients with XLAS, genotype–phenotype correlation is evident; patients with nonsense variants have the most severe phenotypes of early onset ESKD, whereas patients with splicing variants have moderate phenotypes, and patients with missense variants have mild phenotypes.3, 4, 5, 6 Patients with nonsense variants, splicing variants, and missense variants developed ESKD at a median age of 18, 28, and 40 years, respectively, in our cohort.4 In contrast, the genotype–phenotype correlation does not exist in female patients with XLAS.7,8 Renin–angiotensin–aldosterone system inhibitor treatment can improve kidney prognosis, especially when patients have missense variants, for which this treatment has been found to work more effectively.4,9,10 Therefore, genotyping for male cases is important in the prediction of kidney prognosis.
Type IV collagen α3, α4, and α5 chains form the triple-helix structure in the glomerular basement membrane. Type IV collagen α5 chain consists of the N-terminal domain, collagenous domain, and C-terminal noncollagenous (NC1) domain. The collagenous domain consisted of the nonhelical region (NC2) and triple-helical region.11 In the triple-helical region, the amino acid sequence strictly repeats glycine (Gly) on every third position (Gly-Xaa-Yaa)n, and Gly in every third position contributes to the stability of the triple-helical structure.12,13 In contrary, Gly missense variants in the triple-helical region have pathogenicity owing to disruption of the normal folding of the triple-helical structure14 and reduce protein secretion.15, 16, 17 For this reason, most Gly missense variants in the COL4A5 triple-helical region are caused by Gly substitutions regardless of the position of the single-base substitutions, and male patients with XLAS with these pathogenic variants have mild phenotypes.18,19 Nevertheless, single-base substitutions, especially located at the last nucleotide position of each exon, sometimes affect splicing patterns.20, 21, 22, 23 In fact, in the COL4A5 gene, several single-base substitutions at the last nucleotide position in each exon have been reported and interpreted as missense variants in the Human Gene Mutation Database (Cardiff, United Kingdom). Nevertheless, these variants may be correlated with splicing variants, not missense variants, and the kidney prognosis of these variants may be more severe than the expected conventional missense variants.
The aim of this current study is to investigate the possibility that single-base substitutions at the last nucleotide position of exons in COL4A5 gene cause aberrant splicing. To predict the kidney prognosis, we need to determine which variants lead to missense variants and which variants lead to splicing abnormalities.
Methods
Editorial Policies and Ethical Considerations
The study was conducted in accordance with the Declaration of Helsinki and the ethical guidelines issued by the Ministry of Health, Labor, and Welfare of Japan (2017). Moreover, it was approved by the Institutional Research Committee of Kobe University Graduate School of Medicine. Informed consent was obtained from all participants included in the study, which was in accordance with the guidelines for the patients’ benefit. Therefore, patients could refuse to participate in this study.
Analyzed Variants
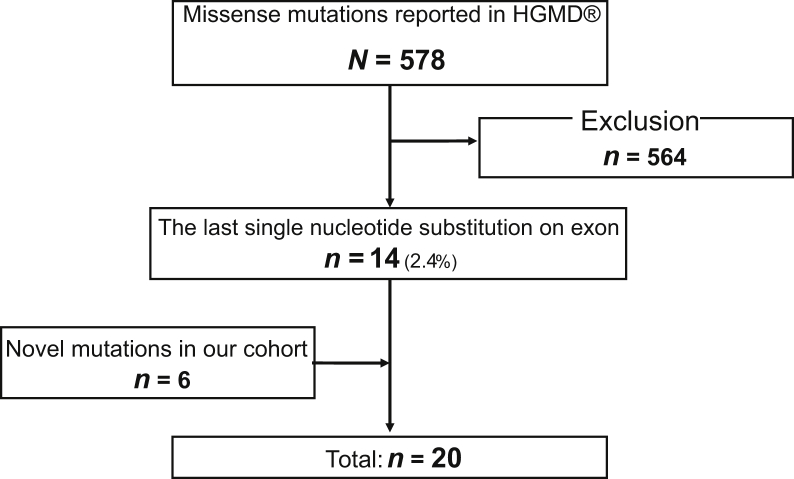
We identified all reported missense variants in COL4A5 from Human Gene Mutation Database (professional release 2021.1). Among 578 missense variants, 14 (2.4%) pathogenic variants caused by single-base substitutions at the last nucleotide positions in exons were included in this study (Figure 1 and Table 1).5,24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36 In addition, 6 novel variants in our cohort were included. Finally, 20 variants were included in this study.
Figure 1.
Flow diagram of variant selection. In total, 578 missense variants in COL4A5 were identified from HGMD and 14 missense variants caused by single-base substitutions at the last nucleotide position in each exon were included. There were 6 novel variants that were included in our cohort. Finally, 20 variants were included in this study. HGMD, Human Gene Mutation Database.
Table 1.
In vitro (minigene) and in vivo results and patient characteristics
| No. | Exon | Variant |
In vitro |
In vivo |
Pathogenicity | Sex | Age, yr | ESKD, yr | Deafness | Ocular abnormality | ID | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide | Amino acid | Transcription | cDNA | |||||||||
| 1 | 15 | c.891A>T | Arg297Ser | Normal | NA | Unknown | Female | 54 | (−) | (−) | (−) | A917 |
| 2 | 19 | c.1165G>A | Gly389Arg | Exon 19 deletion | ND | Splicing | Female | 16 | (+) | (+) | (−) | Weber et al.33 |
| 3 | 21 | c.1423G>A | Gly475Ser | 36 bp deletion | ND | Splicing | Male | ND | 27 | (+) | (+) | Bekheirnia et al.5 |
| Male | ND | (+) | (−) | (−) | ||||||||
| 4 | 25 | c.1948G>T | Gly650Cys | Exon 25 skipping | Exon 25 skipping | Splicing | Male | 31 | 11 | (+) | (−) | A375 |
| Female | 34 | (−) | (−) | (−) | ||||||||
| Female | 58 | 30 | (−) | (−) | ||||||||
| Female | 46 | 27 | (−) | (−) | ||||||||
| Female | 67 | 35 | (+) | (−) | ||||||||
| Female | ND | 68 | (+) | (−) | ||||||||
| 5 | c.1948G>A | Gly650Ser | Exon 25 skipping | ND | Splicing | ND | Wang et al.30 | |||||
| 6 | 28 | c.2244G>T | Lys748Asn | Exon 28 skipping | ND | Splicing | ND | Hertz et al.27 | ||||
| 7 | 30 | c.2509G>A | Gly837Ser | Exon 30 skipping | ND | Splicing | Male | ND | Zhang et al.36 | |||
| 8 | 31 | c.2677G>C | Gly893Arg | 18 bp deletion | ND | Splicing | Male | 44 | 37 | (+) | (+) | Mohammad et al.28 |
| Male | 43 | 28 | (+) | (+) | ||||||||
| Female | 16 | (−) | (−) | (−) | ||||||||
| c.2677G>C (+c.384+1G>A) | Gly893Arg (+c.384+1G>A) | Female | 20 | (−) | (−) | (−) | ||||||
| 9 | c.2677G>A | Gly893Ser | 18 bp deletion | ND | Splicing | ND | Hanson et al.29 | |||||
| 10 | 32 | c.2767G>C | Gly923Arg | Exon 32 skipping | NA | Splicing | Female | 2 | (−) | (−) | (−) | A268 Abe et al.32 |
| Female | 34 | (−) | (−) | (−) | ||||||||
| Female | 8 | (−) | (−) | (−) | A906 | |||||||
| Female | 31 | (−) | ND | ND | ||||||||
| 11 | 35 | c.3106G>A | Gly1036Arg | Normal | NA | Missense | Female | 11 yr | (−) | (−) | (−) | A604 |
| Female | Adult | (−) | ND | ND | ||||||||
| 12 | 37 | c.3373G>A | Gly1125Arg | Exon 37 skipping | ND | Splicing | Female | 30 | (−) | ND | ND | Bullich et al.34 |
| Female | 31 | (−) | ||||||||||
| Female | 12 | (−) | ||||||||||
| 13 | 41 | c.3790G>A | Gly1264Arg | Exon 41 skipping | Exon 41 skipping | Splicing | Male | 53 | 26 yr | (+) | (−) | A21/A399 |
| Female | 49 | (−) | (−) | (−) | ||||||||
| Male | 21 | (−) | ND | ND | ||||||||
| 14 | 42 | c.3924G>C | Gln1308His | Exon 42 skipping | ND | Splicing | Male | ND | 13 yr | (+) | (+) | Bekheirnia et al.5 |
| 15 | 43 | c.3997G>A | Gly1333Ser | Exon 43 skipping | ND | Splicing | Male | ND | Plant et al.25 | |||
| 16 | 44 | c.4069G>C | Gly1357Arg | Exon 44 skipping | NA | Splicing | Female | 38 | (−) | (−) | (−) | A685 |
| 17 | c.4069G>A | Gly1357Ser | Exon 44 skipping | ND | Splicing | Male | ND | 22 | ND | ND | Plant et al.25 | |
| 18 | 46 | c.4297G>A | Gly1433Ser | Normal | NA | Missense | Male | 21 | (−) | (−) | (−) | A771 |
| 19 | 48 | c.4688G>A | Arg1563Glu | Exon 48 skipping | Exon 48 skipping | Splicing | Male | 16 | (−) | (+) | (−) | A582 |
| Female | 48 | 16 | (−) | (−) | ||||||||
| Female | 25 | (−) | (−) | (−) | A910 | |||||||
| Male | 21 | (−) | (+) | (−) | Zhou et al.24 | |||||||
| Female | 44 | (−) | (−) | (−) | ||||||||
| Male | 36 | 29 | (+) | (−) | ||||||||
| Female | ND | (−) | ND | ND | ||||||||
| Female | 53 | (−) | (+) | ND | ||||||||
| Female | ND | (−) | ND | ND | ||||||||
| Male | Dead | 27 | (+) | (+) | ||||||||
| Female | 11 | (−) | (−) | (+) | Han et al.35 | |||||||
| Male | 26 | (−) | (+) | (−) | Gross et al.26 | |||||||
| Male | ND | Average 34.5 |
ND | ND | Pont-Kingdon et al.28 | |||||||
| Male | ||||||||||||
| Male | ||||||||||||
| Male | ||||||||||||
| Male | ||||||||||||
| 20 | 50 | c.4976G>A | Ser1659Asn | Exon 50 skipping | ND | Splicing | ND | Wang et al.30 | ||||
bp, base pair; ESKD, end-stage kidney disease; ID, identification; NA, not available; ND, no data; No., number.
In Vitro Splicing Assay
Minigene Analysis
Genomic DNA of all wild-type (WT) samples and 8 patient samples (numbers [Nos.] 1, 4, 10, 11, 13, 16, 18, 19) in our cohort were extracted from whole blood using QuickGene DNA Whole Blood Kit S (Kurabo Industrial Ltd., Osaka, Japan) or QuickGene-Auto S DNA Blood Kit (Kurabo Industrial Ltd.), in accordance with the manufacturer’s instructions. Fragment primers were designed to contain 1 exon and >100-base pair upstream and downstream flanking introns to this exon (Supplementary Table S1). Nevertheless, when the intron was small, primers were sometimes designed to contain <100-base pair flanking introns. Because the length of intron 15 was short, the fragment for the exon 15 variant was designed containing introns 14 to 16 in COL4A5. When the minigene results revealed a normal splicing pattern, we re-examined them and prepared a new fragment, which contained both exons with the variant and its downstream exons. To create hybrid minigene constructs, we used the H492 vector that we modified previously, which is based on the pcDNA 3.0 mammalian expression vector and contains a multicloning site (Invitrogen, Carlsbad, CA) (Supplementary Figure S1).37
Infusion cloning for 8 variants (Nos. 1, 4, 10, 11, 13, 16, 18, 19) and all WT samples was reacted using Infusion HD Cloning Kit (Takara Bio Inc., Tokyo, Japan) according to the manufacturer’s instructions. For the reported 12 variants in Human Gene Mutation Database (Nos. 2, 3, 5–9, 12, 14, 15, 17, 20) that were not observed in our cohort, variations were introduced by site-directed mutagenesis using Prime STAR Mutagenesis Basal Kit (Takara Bio Inc.), following the manufacturer’s instructions.
Plasmid DNA was confirmed by sequencing using YH303 and YH304 primers (Supplementary Table S1) and transfected into HeLa and HEK293T cells using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. Total RNA was extracted from these cells 24 hours after transfection using the RNeasy Plus Mini Kit (QIAGEN GmbH, Hilden, Germany) following the manufacturer’s instructions. Total RNA (1 μg) was reverse-transcribed using RNA to cDNA EcoDry Premix (Double Primed) (Takara Bio Inc.) following the manufacturer’s instructions. Polymerase chain reaction of reverse-transcribed cDNA was performed using YH307 and YH308 primers. Polymerase chain reaction products were analyzed by electrophoresis on a 1.5% agarose gel using an φ×174-Hae III digest marker, and direct sequencing was performed.
Aberrant Splicing Evaluation Criteria
Abnormal splicing patterns were determined according to the following criteria. Results of electrophoresis reveal the following: (i) WT with only normal splicing and the variant with only aberrant splicing; (ii) WT with only normal splicing and the variant with both normal and aberrant splicing; (iii) WT with both normal and aberrant splicing and the variant with only aberrant splicing; and (iv) normal and aberrant splicing in each WT and variant, with a higher proportion of aberrant splicing in the mutant than in WT.
In vivo Splicing Analysis
When patients’ samples were available, the total mRNA was isolated from peripheral leukocytes using RiboPure-Blood (Thermo Fisher Scientific) and then reverse-transcribed into cDNA using RNA to cDNA EcoDry Premix (Double Primed) (Takara Bio Inc.) following the manufacturer’s instructions. Polymerase chain reaction and direct sequencing were performed using relevant primer pairs (Supplementary Table S1).
In silico Analysis
The pathogenicity of each missense variant was evaluated using SIFT, PolyPhen-2, Mutation Taster, and Align GVGD. We predicted aberrant splicing in each variant using Human Splicing Finder Professional (https://hsf.genomnis.com/home), SD-SCORE,23 and EX-SKIP (https://ex-skip.img.cas.cz). As for the splice site score in both original and variant sequences, MaxEntScan scores were obtained from Alamut Visual (Sophia Genetics Company, Boston, MA). In both original and variant sequences, the numbers of exonic splicing enhancers and exonic splicing silencers were obtained from EX-SKIP.
Genotype–Phenotype Correlation
We conducted a genotype–phenotype correlation analysis of splicing variants in this study compared with modified data from our previous report of missense variants, intronic splicing variants, and nonsense variants.4 Given that Ile194Val is not considered pathogenic because of its high allele frequency among healthy Japanese people (2.1%) and because an in vitro analysis revealed that Glu633Lys causes aberrant splicing, we decided not to classify these variants as missense and recognized the latter variant as splicing in the modified data from our previous report.
Statistical Analyses
JMP software version 14 (SAS Institute Inc., Raleigh, NC) was used for data analysis. Continuous and categorical data were compared using Pearson’s χ2 test and Fisher’s exact test. Cumulative event rates were calculated using the Kaplan–Meier method. Two-tailed P <0.05 were considered statistically significant.
Results
Existence of Aberrant Splicing Based on Single-Base Substitutions at the Last Nucleotide Position in Exons
In 20 variants, 17 (85%) exhibited splicing abnormalities that were detected by minigene analysis (Supplementary Figure S2 and Table 1). All 17 variants fulfilled our aberrant splicing evaluation criteria compared with the WT results. In addition, in in vivo analysis, aberrant splicing patterns exhibited identical patterns to minigene results in all 3 variants (nos. 4, 13, 19) (Supplementary Figure S3). Therefore, we concluded that the pathogenicity of these variants was caused by aberrant splicing.
In contrast, 3 of 20 variants were concluded as not causing aberrant splicing. Of these 3 variants, 2 (nos. 11, 18) were Gly missense variants in the triple-helical regions. In silico analysis revealed pathogenic activities for these variants (Table 2). Therefore, we concluded that these 2 variants were Gly missense variants. The other variant (no. 1) was a non-Gly missense variant (p.[Arg297Ser]) in the collagenous domain, and in silico analysis of SIFT and Mutation Taster evaluated this variant as pathogenic. Nevertheless, the mechanism of pathogenicity in this variant could not be determined.
Table 2.
Results of in silico analysis
|
In vitro |
No. | Exon | Variant |
In silico (missense) |
In silico |
In silico (splicing) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Splicing | Nucleotide | Amino acid | SIFT | PolyPhen-2 | Mutation taster | Align GVGD | MaxEntScan |
ESS/ESE (count) |
HSF | EX-SKIP | SD-SCORE | ||||
| WT | Mut | WT | Mut | ||||||||||||
| Aberrant | 2 | 19 | c.1165G>A | Gly389Arg | D (score: 0) | Pro (score: 1.000) | DC (prob: 1) | C25 (GV: 00.00–GD: 55.27) | 8.76 | 4.44 | 10/23 | 10/23 | Aberrant | Normal | Aberrant |
| 3 | 21 | c.1423G>A | Gly475Ser | D (score: 0.01) | Pro (score: 1.000) | DC (prob: 1) | C65 (GV: 60.00–GD: 97.30) | 5.13 | −3.12 | 2/62 | 2/61 | Aberrant | Normal | Aberrant | |
| 4 | 25 | c.1948G>T | Gly650Cys | D (score: 0) | Pro (score: 1.000) | DC (prob: 1) | C65 (GV: 0.00–GD: 158.23) | 10.86 | 8.31 | 0/25 | 0/25 | Aberrant | Normal | Aberrant | |
| 5 | c.1948G>A | Gly650Ser | D (score: 0) | Pro (score: 1.000) | DC (prob: 1) | C55 (GV: 0.00–GD: 55.27) | 10.86 | 10.08 | 0/25 | 0/25 | Aberrant | Normal | Normal | ||
| 6 | 28 | c.2244G>T | Lys748Asn | D (score: 0.04) | Pro (score: 0.999) | DC (prob: 1) | C0 (GV: 109.22–GD: 46.95) | 3.93 | 0 | 13/16 | 13/16 | Aberrant | Normal | Aberrant | |
| 7 | 30 | c.2509G>A | Gly837Ser | D (score: 0) | Pro (score: 0.993) | DC (prob: 1) | C55 (GV: 0.00–GD: 55.27) | 9.45 | 5.2 | 16/13 | 16/11 | Aberrant | Aberrant | Aberrant | |
| 8 | 31 | c.2677G>C | Gly893Arg | D (score: 0) | Pro (score: 1.000) | DC (prob: 1) | C65 (GV: 0.00–GD: 128.13) | 8.55 | 4.34 | 4/30 | 4/30 | Aberrant | Normal | Aberrant | |
| 9 | c.2677G>A | Gly893Ser | D (score: 0) | Pro (score: 1.000) | DC (prob: 1) | C55 (GV: 0.00–GD: 55.27) | 8.55 | 3.86 | 4/30 | 4/30 | Aberrant | Normal | Aberrant | ||
| 10 | 32 | c.2767G>C | Gly923Arg | D (score: 0) | Pro (score: 1.000) | DC (prob: 1) | C65 (GV: 0.00–GD: 125.13) | 8.49 | 2.1 | 8/18 | 8/17 | Aberrant | Aberrant | Aberrant | |
| 12 | 37 | c.3373G>A | Gly1125Arg | D (score: 0) | Pro (score: 1.000) | DC (prob: 1) | C65 (GV: 0.00–GD: 125.13) | 8.59 | 1.89 | 10/29 | 10/29 | Aberrant | Normal | Aberrant | |
| 13 | 41 | c.3790G>A | Gly1264Arg | Tole (score: 0.15) | Pro (score: 1.000) | DC (prob: 1) | C0 (GV: 170.55–GD: 2.75) | 9.8 | 7.44 | 19/19 | 19/19 | Aberrant | Normal | Aberrant | |
| 14 | 42 | c.3924G>C | Gln1308His | D (score: 0.02) | Pos (score: 0.560) | DC (prob: 1) | C0 (GV: 75.14–GD: 11.15) | 9.79 | 3.74 | 0/55 | 0/55 | Aberrant | Normal | Aberrant | |
| 15 | 43 | c.3997G>A | Gly1333Ser | D (score: 0) | Pro (score: 1.000) | DC (prob: 1) | C55 (GV: 0.00–GD: 55.27) | 7.51 | 1.24 | 11/14 | 11/14 | Aberrant | Normal | Aberrant | |
| 16 | 44 | c.4069G>C | Gly1357Arg | D (score: 0) | Pro (score: 1.000) | DC (prob: 1) | C65 (GV: 0.00–GD: 125.13) | 10.77 | 7.45 | 18/21 | 18/21 | Aberrant | Normal | Aberrant | |
| 17 | c.4069G>A | Gly1357Ser | D (score: 0) | Pro (score: 1.000) | DC (prob: 1) | C55 (GV: 0.00–GD: 55.27) | 10.77 | 7.61 | 18/21 | 18/21 | Aberrant | Normal | Aberrant | ||
| 19 | 48 | c.4688G>A | Arg1563Glu | D (score: 0) | Pro (score: 1.000) | DC (prob: 1) | C35 (GV: 0.00–GD: 42.81) | 10.48 | 5.2 | 7/12 | 7/12 | Aberrant | Normal | Aberrant | |
| 20 | 50 | c.4976G>A | Ser1659Asn | Tole (score: 0.65) | Pos (score: 0.953) | DC (prob: 1) | C0 (GV: 118.33–GD: 0.00) | 9.65 | 2.69 | 14/19 | 14/19 | Aberrant | Normal | Aberrant | |
| Normal | 1 | 15 | c.891A>T | Arg297Ser | D (score: 0.04) | Pos (score: 0.669) | DC (prob: 1) | C15 (GV: 97.59–GD: 95.93) | 9.35 | 8.46 | 5/31 | 5/29 | Aberrant | Aberrant | Normal |
| 11 | 35 | c.3106G>A | Gly1036Arg | D (score: 0) | Pro (score: 1.000) | DC (prob: 1) | C65 (GV: 0.00–GD: 125.13) | 10.67 | 9.1 | 22/24 | 22/24 | Aberrant | Normal | Aberrant | |
| 18 | 46 | c.4297G>A | Gly1433Ser | D (score: 0) | Pro (score: 1.000) | DC (prob: 1) | C55 (GV: 0.00–GD: 55.27) | 10.77 | 7.61 | 1/33 | 1/34 | Aberrant | Normal | Aberrant | |
D, deleterious; DC, disease casing; ESS, exonic splicing silencer; ESE, exonic splicing enhancer; HSF, human splicing finder; Mut, mutant; No., number; Prob, probably damaging; Pos, possibly damaging; SD-SCORE, SD-SCORE algorithm; Tole, tolerated; WT, wild type.
Genotype–Phenotype Correlation
Among all 20 variants, clinical features of male patients were reported in 7 variants with aberrant splicing and 1 variant with normal splicing (Table 1). Nevertheless, we could not obtain clinical information on the age of ESKD development in 1 male patient with the no. 3 variant5 and 5 male patients with the no. 19 variant.28 Therefore, these patients were excluded from the genotype–phenotype correlation analysis. Finally, we could evaluate 13 male patients with XLAS with aberrant splicing in 6 variants and 1 patient with normal splicing in 1 variant.
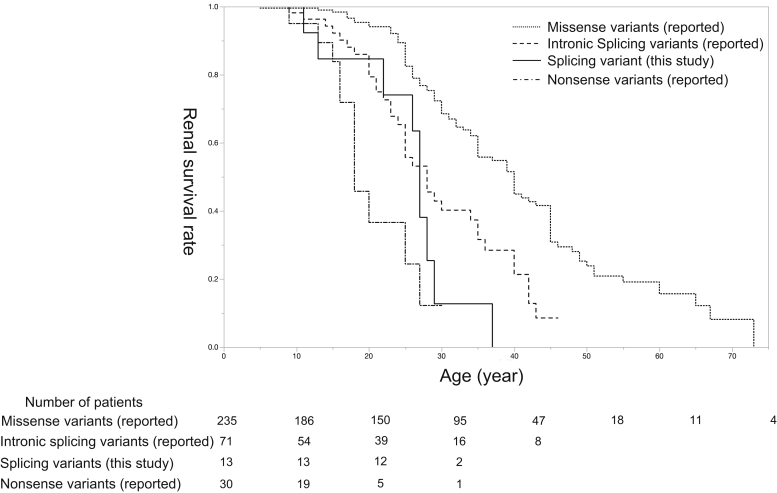
In patients with splicing variants, ESKD, hearing loss, and ocular abnormality were observed in 9, 11, and 5 cases, respectively. Kaplan–Meier analysis revealed that the median age of developing ESKD in these 13 male cases was significantly worse than those with missense variants in our previously reported cohort4 (27 years of age vs. 40 years of age, P < 0.01) (Figure 2). In addition, no significant differences were detected when comparing their data against that from patients with intronic splicing variants (27 years of age vs. 28 years of age, P = 0.72) or nonsense variants (27 years of age vs. 18 years of age, P = 0.09).
Figure 2.
Kidney survival rate in male patients with XLAS. The solid line represents splicing variants detected in this study, and the dotted, dashed, and dot-dashed lines represent missense variants, intronic splicing variants, and nonsense variants reported in our previous study, respectively. The Kaplan–Meier kidney survival analysis results revealed that the median age for developing end-stage kidney disease was significantly lower for patients with splicing variants in our present study compared with those with missense variants in our previous study (27 years of age, 95% CI: 22–29 vs. 40 years of age, 95% CI: 35–45; Wilcoxon: P < 0.01). Nevertheless, there was no significant difference in the median age for developing end-stage kidney disease between patients with splicing variants in this study and those in our previous study with intronic splicing variants (27 years of age, 95% CI: 22–29 vs. 28 years of age, 95% CI: 24–35; P = 0.72) or nonsense variants (27 years of age, 95% CI: 22–29 vs. 18 years of age, 95% CI: 16–27; P = 0.09).
In contrast, the clinical features of only 1 male patient who was found to have only mild phenotypes without aberrant splicing (no. 18) were available: proteinuria was detected at the age of 19 years, and his kidney function was within the normal range at age 21 years.
In Silico Analysis
The 5′ splice site scores of all variants were lower than the WT scores, including 3 variants with normal splicing (Table 2). When the 5′ splice site scores by MaxEntScan were compared, the mutant scores were significantly lower in the variants with aberrant splicing than those without aberrant splicing (4.34 ± 0.81 vs. 8.46 ± 0.43, P = 0.03).
The sensitivity/specificity of the HSF, SD-SCORE, and EX-SKIP in our study was 100%/0%, 94.1%/33.3%, and 11.1%/50%, respectively. The numbers of exonic splicing enhancers decreased in 4 variants (nos. 1, 3, 7, 10) and increased in 1 variant (no. 18) (Table 2). Nevertheless, the exonic splicing silencers revealed no change among all 20 variants in this study.
Discussion
In the COL4A5 triple-helical region, the Gly missense variants are known to be pathogenic.14, 15, 16, 17,38 Therefore, these single-base substitutions have been considered to be missense variants. Nevertheless, 15% to 50% of single-base substitutions in exons can cause splicing abnormalities; in particular, canonical sequences in exons at the 5′ splice site have a high potential of disrupting normal splicing.14,20, 21, 22, 23,39 We revealed that 85% (17 of 20) of the single-base substitutions at the last nucleotide position on the exons in COL4A5 led to aberrant splicing (Supplementary Figure S2 and Table 1). In addition, all in vivo transcript analyses for 3 variants (nos. 4, 13, 19) using patient samples exhibited the same splicing pattern with minigene analysis results (Supplementary Figure S3). These results revealed that single-base substitutions at the last nucleotide position of COL4A5 exons are likely to cause aberrant splicing. In our study, in silico tools were found to have high sensitivities but low specificities, so variants with aberrant splicing could not be detected by in silico tools alone. Because male patients with XLAS have a strong genotype–phenotype correlation, transcript analysis is important to reveal whether these variants lead to missense variants or splicing variants.
Male patients with XLAS with nonsense variants have the most severe phenotypes of early onset ESKD; those with splicing variants have middle phenotypes, and those with missense variants have mild phenotypes.3, 4, 5, 6 Our results revealed that those with variants with aberrant splicing exhibited a more severe kidney prognosis than those with previously reported missense variants (Figure 2). Compared with data from our previous reports on intronic splicing and nonsense variants, this study found no significant differences among variants with aberrant splicing. These results support the necessity of investigating splicing patterns to understand kidney prognosis correctly.
In vertebrates, pre-mRNA splicing is caused by U2-dependent spliceosome, which consists of a complex of the following 5 uridine-rich small nuclear ribonucleoproteins (snRNPs): U1, U2, U4, U5, and U6 snRNP proteins and numerous non-snRNP proteins.40 In the first step of spliceosome formation, U1 snRNP recognizes and combines with the exon-intron boundary at the 5′ splice site,41 which includes the last 3 bases of exons and the first 6 nucleotides of introns; its consensus sequence is [(C/A)AG|GT(A/G)AGT].42 Single-base substitutions at the last nucleotide position in each exon lead to a weaker 5′ splice site, which depresses the removal of the upstream intron.43, 44, 45, 46 This mechanism causes changes of specificity or fidelity at the 5′ splice site that suppress the recognition and connection of U1 snRNPs at exon-intron boundaries, and these suppressions reduce upstream 3′ splice site recognition and lead to aberrant splicing.47,48 Nevertheless, this mechanism of aberrant splicing owing to single-base substitutions at the last nucleotide position in exons is still unclear. Actually, in this study, all variants decreased the 5′ splice site scores (Table 2); however, 3 of 20 variants did not have aberrant splicing. In addition, the number of exonic splicing enhancers changes was not characteristic.
This study had several limitations. First, only a small number of variants were included, and our results were mostly achieved through in vitro analysis. Because minigene contains only exons and a portion of flanking introns, in vitro results from the minigene analysis may not always be consistent with the in vivo results. Second, some patients were excluded from the genotype–phenotype correlation analysis owing to a lack of qualifying clinical characteristics. In addition, we excluded 5 male patients with aberrant splicing (no. 19) who had a relatively mild kidney prognosis because we lacked their individual data,28 although this might have distorted the findings from our kidney survival analysis. Third, we did not evaluate synonymous variants resulting from substitution of the last nucleotide position in exons because they have not been considered pathogenic thus far and, therefore, are not registered in the Human Gene Mutation Database. Nevertheless, they may have the potential to cause aberrant splicing. We recognize that single-nucleotide substitutions at the second position from the last nucleotide may also cause aberrant splicing. Both these points warrant further investigation in the future.
In conclusion, we revealed that most single-base substitutions at the last nucleotide of exons did not cause missense variants, but aberrant splicing led to more severe phenotypes. Therefore, when the single-base substitution at the last nucleotide position in each exon in COL4A5 is detected, mRNA analysis should be performed to confirm whether these variants are causing XLAS by missense or splicing variants to accurately predict kidney prognosis.
Disclosure
KI reports receiving grant support from Daiichi Sankyo Co. Ltd.; consulting fees from Kyowa Kirin Co. Ltd. and Boehringer Ingelheim; and lecture fees from Kyowa Kirin Co. Ltd., Chugai Pharmaceutical Co. Ltd., Takeda Pharmaceutical Company, Integrated Development Associates, and Novartis Pharmaceuticals Corporation. KNo reports receiving consulting fees from Kyowa Kirin Co. Ltd. and lecture fees from Kyowa Kirin Co. Ltd., Novartis Pharmaceuticals Corporation, and Chugai Pharmaceutical Co. Ltd. All the other authors declared no competing interests.
Acknowledgments
This study was supported by Grants-in-Aid for Scientific Research (KAKENHI) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (subject identifications: 16K19642 to TY, 26293203 and 17H04189 to KI, and 19K08726 to KNo); by the Japan Agency for Medical Research and Development (grant number JP19ek0109231h0003 to KI and 19ek0109231s0103 to KNo); and by The Japan Foundation for Pediatric Research (grant no. 19-002 to TH). YA conceived of and directed the study with support from TH, TY, and KNo. YA and KNo wrote the manuscript. AK, SN, SI, EO, RR, NS, and CN established and conducted molecular analysis and interpreted the data. MM established the minigene assay. The manuscript was critically reviewed by all the authors. The authors thank Enago (www.enago.jp) for the English language review of this manuscript. The authors thank all study participants and their families. We are immensely grateful to Mrs. Yoshimi Nozu, Ming Juan Ye, and Yuko Noguchi (Kobe University) for their excellent technical assistance.
Footnotes
Figure S1. Schematic for the hybrid minigene.
Figure S2. Fragment component of minigene and the results of electrophoresis and schematic for transcript analysis from the minigene assay.
Figure S3. Results of electrophoresis and direct sequencing of reverse-transcriptase polymerase chain reaction from patient’s blood.
Table S1. Primer list in this study.
Supplementary Material
Figure S1. Schematic for the hybrid minigene.
Figure S2. Fragment component of minigene and the results of electrophoresis and schematic for transcript analysis from the minigene assay.
Figure S3. Results of electrophoresis and direct sequencing of reverse-transcriptase polymerase chain reaction from patient’s blood.
Table S1. Primer list in this study.
References
- 1.Kashtan C.E. In: GeneReviews®. Adam M.P., Ardinger H.H., Pagon R.A., et al., editors. University of Washington; 1993. Alport syndrome. [Google Scholar]
- 2.Nozu K., Nakanishi K., Abe Y., et al. A review of clinical characteristics and genetic backgrounds in Alport syndrome. Clin Exp Nephrol. 2019;23:158–168. doi: 10.1007/s10157-018-1629-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jais J.P., Knebelmann B., Giatras I., et al. X-linked Alport syndrome: natural history in 195 families and genotype-phenotype correlations in males. J Am Soc Nephrol. 2000;11:649–657. doi: 10.1681/ASN.V114649. [DOI] [PubMed] [Google Scholar]
- 4.Yamamura T., Horinouchi T., Nagano C., et al. Genotype-phenotype correlations influence the response to angiotensin-targeting drugs in Japanese patients with male X-linked Alport syndrome. Kidney Int. 2020;98:1605–1614. doi: 10.1016/j.kint.2020.06.038. [DOI] [PubMed] [Google Scholar]
- 5.Bekheirnia M.R., Reed B., Gregory M.C., et al. Genotype-phenotype correlation in X-linked Alport syndrome. J Am Soc Nephrol. 2010;21:876–883. doi: 10.1681/ASN.2009070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horinouchi T., Nozu K., Yamamura T., et al. Detection of splicing abnormalities and genotype-phenotype correlation in X-linked Alport syndrome. J Am Soc Nephrol. 2018;29:2244–2254. doi: 10.1681/ASN.2018030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamura T., Nozu K., Fu X.J., et al. Natural history and genotype-phenotype correlation in female X-linked Alport syndrome. Kidney Int Rep. 2017;2:850–855. doi: 10.1016/j.ekir.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jais J.P., Knebelmann B., Giatras I., et al. X-linked Alport syndrome: natural history and genotype-phenotype correlations in girls and women belonging to 195 families: a “European Community Alport Syndrome Concerted Action” study. J Am Soc Nephrol. 2003;14:2603–2610. doi: 10.1097/01.asn.0000090034.71205.74. [DOI] [PubMed] [Google Scholar]
- 9.Gross O., Licht C., Anders H.J., et al. Early angiotensin-converting enzyme inhibition in Alport syndrome delays renal failure and improves life expectancy. Kidney Int. 2012;81:494–501. doi: 10.1038/ki.2011.407. [DOI] [PubMed] [Google Scholar]
- 10.Gross O., Tönshoff B., Weber L.T., et al. A multicenter, randomized, placebo-controlled, double-blind phase 3 trial with open-arm comparison indicates safety and efficacy of nephroprotective therapy with ramipril in children with Alport’s syndrome. Kidney Int. 2020;97:1275–1286. doi: 10.1016/j.kint.2019.12.015. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J., Barker D.F., Hostikka S.L., et al. Single base mutation in alpha 5 (IV) collagen chain gene converting a conserved cysteine to serine in Alport syndrome. Genomics. 1991;9:10–18. doi: 10.1016/0888-7543(91)90215-z. [DOI] [PubMed] [Google Scholar]
- 12.Kashtan C.E. Alport syndrome: abnormalities of type IV collagen genes and proteins. Ren Fail. 2000;22:737–749. doi: 10.1081/jdi-100101959. [DOI] [PubMed] [Google Scholar]
- 13.Brodsky B., Persikov A.V. Molecular structure of the collagen triple helix. Adv Protein Chem. 2005;70:301–339. doi: 10.1016/S0065-3233(05)70009-7. [DOI] [PubMed] [Google Scholar]
- 14.Cosgrove D., Liu S. Collagen IV diseases: a focus on the glomerular basement membrane in Alport syndrome. Matrix Biol. 2017;57-58:45–54. doi: 10.1016/j.matbio.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi T., Uchiyama M. Characterization of assembly of recombinant type IV collagen alpha3, alpha4, and alpha5 chains in transfected cell strains. Kidney Int. 2003;64:1986–1996. doi: 10.1046/j.1523-1755.2003.00323.x. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi T., Kakihara T., Uchiyama M. Mutational analysis of type IV collagen alpha5 chain, with respect to heterotrimer formation. Biochem Biophys Res Commun. 2008;366:60–65. doi: 10.1016/j.bbrc.2007.12.037. [DOI] [PubMed] [Google Scholar]
- 17.Kamura M., Yamamura T., Omachi K., et al. Trimerization and genotype-phenotype correlation of COL4A5 mutants in Alport syndrome. Kidney Int Rep. 2020;5:718–726. doi: 10.1016/j.ekir.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savige J., Storey H., Watson E., et al. Consensus statement on standards and guidelines for the molecular diagnostics of Alport syndrome: refining the ACMG criteria. Eur J Hum Genet. 2021;29:1186–1197. doi: 10.1038/s41431-021-00858-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savige J., Storey H., Il Cheong H., et al. X-linked and autosomal recessive Alport syndrome: pathogenic variant features and further genotype-phenotype correlations. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krawczak M., Reiss J., Cooper D.N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 21.Teraoka S.N., Telatar M., Becker-Catania S., et al. Splicing defects in the Ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am J Hum Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ars E., Serra E., García J., et al. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1 [published correction appears in Hum Mol Genet. 2000;9:659] Hum Mol Genet. 2000;9:237–247. doi: 10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- 23.Sahashi K., Masuda A., Matsuura T., et al. In vitro and in silico analysis reveals an efficient algorithm to predict the splicing consequences of mutations at the 5’ splice sites. Nucleic Acids Res. 2007;35:5995–6003. doi: 10.1093/nar/gkm647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J., Gregory M.C., Hertz J.M., et al. Mutations in the codon for a conserved arginine-1563 in the COL4A5 collagen gene in Alport syndrome. Kidney Int. 1993;43:722–729. doi: 10.1038/ki.1993.103. [DOI] [PubMed] [Google Scholar]
- 25.Plant K.E., Green P.M., Vetrie D., Flinter F.A. Detection of mutations in COL4A5 in patients with Alport syndrome. Hum Mutat. 1999;13:124–132. doi: 10.1002/(SICI)1098-1004(1999)13:2<124::AID-HUMU4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 26.Gross O., Netzer K.O., Lambrecht R., Seibold S., Weber M. Meta-analysis of genotype-phenotype correlation in X-linked Alport syndrome: impact on clinical counselling. Nephrol Dial Transplant. 2002;17:1218–1227. doi: 10.1093/ndt/17.7.1218. [DOI] [PubMed] [Google Scholar]
- 27.Hertz J.M. Alport syndrome. Molecular genetic aspects. Dan Med Bull. 2009;56:105–152. [PubMed] [Google Scholar]
- 28.Pont-Kingdon G., Sumner K., Gedge F., et al. Molecular testing for adult type Alport syndrome. BMC Nephrol. 2009;10:38. doi: 10.1186/1471-2369-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanson H., Storey H., Pagan J., Flinter F. The value of clinical criteria in identifying patients with X-linked Alport syndrome. Clin J Am Soc Nephrol. 2011;6:198–203. doi: 10.2215/CJN.00200110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F., Zhao D., Ding J., et al. Skin biopsy is a practical approach for the clinical diagnosis and molecular genetic analysis of X-linked Alport’s syndrome. J Mol Diagn. 2012;14:586–593. doi: 10.1016/j.jmoldx.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Mohammad M., Nanra R., Colville D., et al. A female with X-linked Alport syndrome and compound heterozygous COL4A5 mutations. Pediatr Nephrol. 2014;29:481–485. doi: 10.1007/s00467-013-2682-6. [DOI] [PubMed] [Google Scholar]
- 32.Abe Y., Iyoda M., Nozu K., et al. A novel mutation in a Japanese family with X-linked Alport syndrome. Intern Med. 2016;55:2843–2847. doi: 10.2169/internalmedicine.55.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber S., Strasser K., Rath S., et al. Identification of 47 novel mutations in patients with Alport syndrome and thin basement membrane nephropathy. Pediatr Nephrol. 2016;31:941–955. doi: 10.1007/s00467-015-3302-4. [DOI] [PubMed] [Google Scholar]
- 34.Bullich G., Domingo-Gallego A., Vargas I., et al. A kidney-disease gene panel allows a comprehensive genetic diagnosis of cystic and glomerular inherited kidney diseases. Kidney Int. 2018;94:363–371. doi: 10.1016/j.kint.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 35.Han K.H., Park J.E., Ki C.S. De novo mutations in COL4A5 identified by whole exome sequencing in 2 girls with Alport syndrome in Korea. Korean J Pediatr. 2019;62:193–197. doi: 10.3345/kjp.2018.06772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X., Zhang Y., Zhang Y., et al. X-linked Alport syndrome: pathogenic variant features and further auditory genotype-phenotype correlations in males. Orphanet J Rare Dis. 2018;13:229. doi: 10.1186/s13023-018-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nozu K., Iijima K., Kawai K., et al. In vivo and in vitro splicing assay of SLC12A1 in an antenatal salt-losing tubulopathy patient with an intronic mutation. Hum Genet. 2009;126:533–538. doi: 10.1007/s00439-009-0697-7. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi T., Uchiyama M. Mutant-type alpha5(IV) collagen in a mild form of Alport syndrome has residual ability to form a heterotrimer. Pediatr Nephrol. 2010;25:1169–1172. doi: 10.1007/s00467-009-1433-1. [DOI] [PubMed] [Google Scholar]
- 39.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wahl M.C., Will C.L., Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Reed R. Mechanisms of fidelity in pre-mRNA splicing. Curr Opin Cell Biol. 2000;12:340–345. doi: 10.1016/s0955-0674(00)00097-1. [DOI] [PubMed] [Google Scholar]
- 42.Zhang M.Q. Statistical features of human exons and their flanking regions. Hum Mol Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 43.Talerico M., Berget S.M. Effect of 5’ splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990;10:6299–6305. doi: 10.1128/mcb.10.12.6299-6305.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grabowski P.J., Nasim F.U., Kuo H.C., Burch R. Combinatorial splicing of exon pairs by two-site binding of U1 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1991;11:5919–5928. doi: 10.1128/mcb.11.12.5919-5928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuo H.C., Nasim F.H., Grabowski P.J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991;251:1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- 46.Cho S., Moon H., Loh T.J., et al. Splicing inhibition of U2AF65 leads to alternative exon skipping. Proc Natl Acad Sci U S A. 2015;112:9926–9931. doi: 10.1073/pnas.1500639112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buratti E., Baralle D. Novel roles of U1 snRNP in alternative splicing regulation. RNA Biol. 2010;7:412–419. doi: 10.4161/rna.7.4.12153. [DOI] [PubMed] [Google Scholar]
- 48.Will C.L., Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.