Abstract
Halomonas venusta, a moderately halophilic, nonfermentative gram-negative rod, is reported for the first time as a human pathogen in a wound that originated from a fish bite.
CASE REPORT
A 55-year-old healthy female was bitten on the right medial ankle by a fish of unknown species while diving in the sea at the Maldive Islands. After a brief painful episode, her ankle and foot became swollen and the wound showed a watery discharge. After returning home, she was seen in the hospital emergency room. The wound showed redness, but neither lymphangitis, lymphadenopathy, fever, nor any other pathology was found. The only laboratory data requested were C-reactive protein (CRP) concentration (21 mg/liter) and a leukocyte count, which was within normal limits. A superficial swab of the wound was taken, and intravenous therapy with imipenem-cilastatin was started. After 1 week the therapy was changed to peroral ciprofloxacin. The wound healed without complications; the CRP concentration was 10 mg/liter 9 days after admission. Antibiotic therapy was discontinued on the 14th day of illness.
The wound swab was inoculated onto blood agar (Columbia blood agar base no. 2 [Becton-Dickinson, Basel, Switzerland] with 5% sheep blood), MacConkey agar (Becton-Dickinson), chocolate agar (GC II agar base with hemoglobin and IsoVitaleX; Becton-Dickinson), and thioglycolate medium without Indicator 135-C (Becton-Dickinson). No anaerobic plates were inoculated since this was a swab of a superficial wound. Microscopically, gram-negative rods and polymorphonuclear leukocytes were seen. Within 48 h at 37°C, a pure culture of mucoid, colorless colonies of a motile gram-negative rod had grown on the media listed above, and there was very slight growth at the top of the thioglycolate medium. The bacterium was oxidase and catalase positive and nonfermentative on triple sugar iron agar slants (Becton-Dickinson). It was inoculated into the API 20 NE (NFT) gallery (bioMérieux, Marcy-l'Etoile, France). The resulting code of 1660757 was interpreted as an “unacceptable profile.” It corresponded to positive reactions for nitrate reduction to nitrite, urea and esculin hydrolysis, and utilization of glucose, N-acetylglucosamine, maltose, gluconate, caprate, malate, citrate, and phenylacetate and to negative reactions for indole production, gelatinase, arginine dihydrolase, and utilization of arabinose, mannose, mannitol, and adipate. In addition, reactions for urease (urea agar base; Becton-Dickinson), esculin hydrolysis (heart infusion agar [Becton-Dickinson] with esculin and iron ammonium citrate), and growth in 6.5% NaCl broth and at 4°C were positive, while starch hydrolysis (on Mueller-Hinton agar [Becton-Dickinson]), reactions for lysine and ornithine decarboxylase (tablets; Rosco Diagnostica, Taastrup, Denmark), and growth on thiosulfate-citrate-bile salts-sucrose (TCBS) agar (Becton-Dickinson) were negative. By National Committee for Clinical Laboratory Standards disk diffusion tests (criteria for members of the family Enterobacteriaceae), the organism was susceptible to ampicillin, amoxicillin-clavulanic acid, piperacillin, cephalothin, cefamandole, cefuroxime, ceftriaxone, ceftazidime, norfloxacin, ciprofloxacin, and sulfamethoxazole-trimethoprim. A zone was noted around a colistin disk (10 μg; Becton-Dickinson) but not around a 150-μg O/129 disk (Oxoid Ltd., Basingstoke, United Kingdom). Cellular fatty acid analysis (MIDI; Microbial ID, Newark, Del.) yielded C18:1ω9c/w12t/ω7c (“Feature 7”; 62.5%), C16:0 (15.8%), C16:1ω7c and C12:03OH (7.2% each), C14:0 (2.9%), and C10:0 (1.9%). This combination of biochemical reactions, antimicrobial susceptibilities, and cellular fatty acids excluded any of the members of the family Vibrionaceae and the nonfermentative gram-negative rods known to cause disease in humans (10). The 16S rRNA of the strain was therefore sequenced.
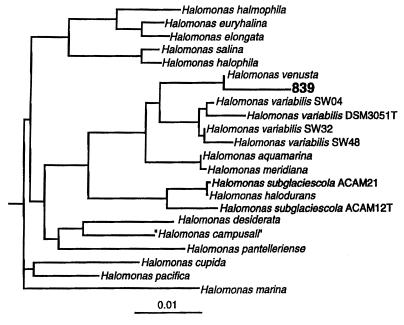
The DNA of the isolate was extracted and sequenced as described previously (6). A search of the current nucleotide databases was done through the network service of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) by use of the BLAST algorithm. The best scores showed a >98% sequence similarity to Halomonas species. By using the phylogenetic analysis program PHYLIP with the sequences of all distinct Halomonas species, a 99% similarity of the sequence of our strain (strain 839) to that of H. venusta was found (Fig. 1).
FIG. 1.
Phylogenetic relationship of Halomonas spp.
Marine bacteria are known to cause wound infections in humans that arise from swimming or walking in salt water or from saltwater contamination of existing lesions (7). Organisms isolated from such infections are halotolerant or halophilic bacteria such as Vibrio spp., Shewanella putrefaciens, and Staphylococcus and Micrococcus spp., as well as normal skin bacteria (9). Fish bites are rare causes of these wounds (3, 8, 9). We describe here a case of human infection with Halomonas venusta, a bacterium thus far not observed in humans, following a fish bite.
H. venusta, a nonpigmented, nonmotile or polar or peritrichously flagellated, nonfermentative, slightly halophilic (optimum, 0.3 to 2.0 M NaCl), gram-negative rod (4) was first described by Baumann et al. (2) in the marine environment of Oahu, Hawaii, as Alcaligenes venustus, but it was later transferred, on the basis of phenotypic characteristics, by some of the same investigators to a new genus, Deleya, as Deleya venusta (1). The reactions given by our isolate are in line with those described for 14 strains of this species (1), even though the investigators had used a different basal medium for substrate utilization. The organism is not covered in the API 20 NE (NFT) database.
Franzmann and Tindall (5) have published a cellular fatty acid profile of Deleya. The major fatty acids of the six known species were C16:1 + C17:0cyc and C18:1 + C19:0cyc, but D. venusta stood out as lacking the cyclopropane acids. The pattern for our strain again conforms to this pattern. Finally, Dobson and Franzmann (4), on the basis of 16S rRNA sequence similarity, placed members of the genus Deleya together with Halomonas and Halovibrio spp. and Paracoccus halodenitrificans into the genus Halomonas. The members of this genus have 4 characteristic nucleotide signatures, in addition to the 15 such signatures associated with the family Halomonadaceae, which includes a cytosine residue at position 486 (Escherichia coli numbering). Our strain possesses these signatures as well (data not shown).
H. venusta should therefore be added to the list of marine organisms capable of causing wound infections in humans. In the absence of an investigation of the fish that cause the bite, we cannot be sure whether the organism originated from the fish or from the ocean itself.
Acknowledgments
We thank Brian Austin for help in the identification of the microorganism described here.
REFERENCES
- 1.Baumann L, Bowditch R D, Baumann P. Description of Deleya gen. nov. created to accomodate the marine species Alcaligenes aestus, A. pacificus, A. cupidus, A. venustus, and Pseudomonas marina. Int J Syst Bacteriol. 1983;33:793–802. [Google Scholar]
- 2.Baumann L, Baumann P, Mandel M, Allen R D. Taxonomy of aerobic marine bacteria. J Bacteriol. 1972;110:402–429. doi: 10.1128/jb.110.1.402-429.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck J D, Spotte S, Gadbaw J J., Jr Bacteriology of the teeth from a great white shark: potential medical implications for shark bite victims. J Clin Microbiol. 1984;20:849–851. doi: 10.1128/jcm.20.5.849-851.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobson S J, Franzmann P D. Unification of the genera Deleya (Baumann et al. 1983), Halomonas (Vreeland et al. 1980), and Halovibrio (Fendrich 1988) and the species Paracoccus halodenitrificans (Robinson and Gibbons 1952) into a single genus, Halomonas, and placement of the genus Zymobacter in the family Halomonadaceae. Int J Syst Bacteriol. 1996;46:550–558. [Google Scholar]
- 5.Franzmann P D, Tindall B J. A chemotaxonomic study of members of the family Halomonadaceae. System Appl Microbiol. 1990;13:142–147. [Google Scholar]
- 6.Goldenberger D, Künzli A, Vogt P, Zbinden R, Altwegg M. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol. 1997;35:2733–2739. doi: 10.1128/jcm.35.11.2733-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoge C W, Watsky D, Peeler R N, Libonati J P, Israel E, Morris J G., Jr Epidemiology and spectrum of Vibrio infections in a Chesapeake Bay community. J Infect Dis. 1989;160:985–993. doi: 10.1093/infdis/160.6.985. [DOI] [PubMed] [Google Scholar]
- 8.Pavia A T, Bryan J A, Maher K L, Hester T R, Jr, Farmer J J., III Vibrio carchariae infection after a shark bite. Ann Intern Med. 1989;111:85–86. doi: 10.7326/0003-4819-111-1-85. [DOI] [PubMed] [Google Scholar]
- 9.Pien F D, Ang K S, Nakashima N T, Evans D G, Grote J A, Hefley M L, Kubota E A. Bacterial flora of marine penetrating injuries. Diagn Microbiol Infect Dis. 1983;1:229–232. doi: 10.1016/0732-8893(83)90022-6. [DOI] [PubMed] [Google Scholar]
- 10.Weyant R S, Moss C W, Weaver R E, Hollis D G, Jordan J G, Cook E C, Daneshvar M I. Identification of unusual pathogenic gram-negative aerobic and facultatively anaerobic bacteria. 2nd ed. Baltimore, Md: The Williams & Wilkins Co.; 1995. [Google Scholar]