Abstract
The number of studies reporting hormetic responses is rapidly increasing, and quantitative evaluations are needed to improve the understanding of hormetic dose responses. However, there is no standardized methodology to estimate the no-observed-adverse-effect-level (NOAEL) of hormetic dose-response relationships developed using data mined from the published literature. Here, we propose a protocol that can be followed to estimate NOAEL, a process that is illustrated using a specific example. This protocol can be used for maintaining a mutual language (since NOAEL can be defined in different ways), permitting comparisons among different studies, and facilitating cumulative science.
Keywords: Contaminant effect, Exposure-response relationship, Hormesis, Low-dose biological effects, Organismal response, Oxidative stress, Pollution effects, Susceptibility, Tolerance, Toxicological testing
Abbreviations: MAX, maximum stimulatory response; NOAEL, no-observed-adverse-effect-level; PFBA, perfluorobutanoic acid
Graphical abstract
Specification table
| Subject Area |
|
| More specific subject area | Impacts of environmental pollutants |
| Protocol name | Estimation of NOAEL |
| Reagents/tools | n/a |
| Experimental design | Dose response data are mined from published studies, and dose-response relationships are plotted. From the plotted dose-response curves, the no-observed-adverse-effect-level (NOAEL) is estimated for use in meta-data evaluations of hormetic dose response evaluations. |
| Trial registration | n/a |
| Ethics | n/a |
| Value of the Protocol |
|
Description of protocol
Stimulation by low doses of stress appears in the framework of adaptive responses that organisms have evolved to be able to protect themselves against stress and improve their performance under predicted environmental challenges [1,2]. This typically results in a hormetic dose-response relationship, a J-, U-, inverse J-, or inverse U-shaped curve resulting from responses that are opposite between low and high doses/exposures [3]. While hormesis has been neglected and denied the opportunity to mature for a long time, considerable literature published in the recent years has (i) demonstrated the common occurrence of hormesis in numerous organisms exposed to various stresses, (ii) advanced its mechanistic understandings, and (iii) consolidated its quantitative features [4], [5], [6], [7], [8], [9].
An important index of hormetic dose response is the no-observed-adverse-effect-level (NOAEL), which can also be used to estimate the width of the hormetic zone as well as the ratio of NOAEL to the maximum stimulatory response (MAX), serving also as quantitative estimates of biological plasticity [10,11]. However, there is no standardized protocol for estimating NOAEL, yet the methodology used for its estimation may affect the subsequent results [12]. This becomes increasingly important in meta-data studies aiming at mining dose-response data from published literatures to construct dose-response relationships and evaluate them quantitatively. Since a considerable amount of published studies do not present such hormetic dose-response relationships but commonly only the mean responses, it is important to follow a standardized protocol for the estimation of NOAEL, especially in the light of rapidly increasing literature acknowledging hormesis.
Here, we propose a protocol to estimate the NOAEL in hormetic dose-response relationships developed with mined dose-response data. Such a protocol would contribute in harmonizing the methodology, making meaningful comparisons of results of other studies, including the broad Hormesis Database [10,13,14], and facilitating cumulative science.
The protocol is as follow:
-
(1)Dose-response data are extracted from published articles. When data are reported in figures only, data should be extracted using software that have been used widely for meta-data analyses and other scientific purposes, such as ImageJ, GetData Graph Digitizer, Adobe Photoshop, UTHSCSA ImageTool, and LIA32, of which some are non-commercial and freely available to download and use. Note that:
-
•Calibration should be newly done for each figure (and panel for multi-panel figures) from which data are extracted.
-
•Data should be extracted with ideally 1-3 decimals for increased accuracy.
-
•To account for potential measurement error, if the mean value for some of the dose responses is reported in the text, this value should be examined relatively to the value obtained from the data extraction software. The extracted data can be corrected by multiplying the rate of difference between the extracted values and that/those reported by authors. Hence, the text of each paper should be carefully read prior to extracting data.
-
•Similarly, if the figures present data that are % difference from the control response, the control response should be 100%. If the value obtained from the data extraction software is below or above the absolute 100%, the extracted dose-response data of the same response variable should be corrected by multiplying with the rate of difference between the 100% and the extracted value of the control group.
-
•Changing the designated controls is so important in meta-analytic evaluations that can even flip synergisms to antagonisms [15]. For the purpose of this protocol, the control in each dose-response relationship is typically the experimental condition with null addition of an exogenously-applied pollutant or other stress-inducing agent. For example, if the effect of microplastics on the swimming ability of zebrafish is studied, the control group is the one with a zero concentration of applied microplastics.
-
•No matter which image analysis software is used, the same one should be used throughout the entire process to decrease potential measurement error.
-
•
-
(2)
All the dose-response data that will be used for developing dose-response relationships should be unified into % of control response (% difference of each treatment from the control). The response to a treatment is calculated as where μχ is the arithmetic mean of a group exposed to a dose/concentration level of χ and μc is the arithmetic mean of the control group (commonly a zero dose; negative control). It should be noted that the low-dose, stimulatory response barely exceeds two fold the control response [13]. Therefore, if multi-fold higher stimulatory responses are found, care should be exercised to cross-check whether the data extraction and calculations up to this point are correct.
-
(3)
A dose-response relationship is built using the % dose-response data. In this protocol, the protocol is illustrated using MS Excel, which is commonly available to any researcher, but other software can also be used. In MS Excel, this is done by selecting the two columns containing the dose (x axis) and response (y axis) data and plotting the data using scatter plots (explained in the following example). Attention should be paid to the dose/concentration units of each treatment, and that the nominal or actual doses/concentrations are used instead of log-transformed ones.
An application of the protocol is illustrated using data reported in a recent study investigating the effect of perfluorobutanoic acid (PFBA), an organofluorine pollutant, on the mean weight gain of beet armyworm Spodoptera exigua (Hübner) larvae reared on soybean leaves [16], and Adobe Photoshop CS4 Extended v.11 (Adobe Systems Incorporated, CA, USA).
-
(1)
The relevant figure in the article is picked, zooming in as much as needed such that the entire figure is visible.
-
(2)
A screenshot is taken (e.g. for many Windows systems the button “prt sc” or “print screen” can be used), and the figure is pasted in an image editing software (e.g. Paint) to be saved as image file.
-
(3)
The image is opened with an image analysis software (Adobe Photoshop here). Following calibration (Fig. 1), the data of 1 and 2 days post treatment are extracted (Fig. 2); note that it is recommended to zoom in as much as needed to increase the accuracy for smaller means in figures with considerable arithmetic differences between data sets.
-
(4)
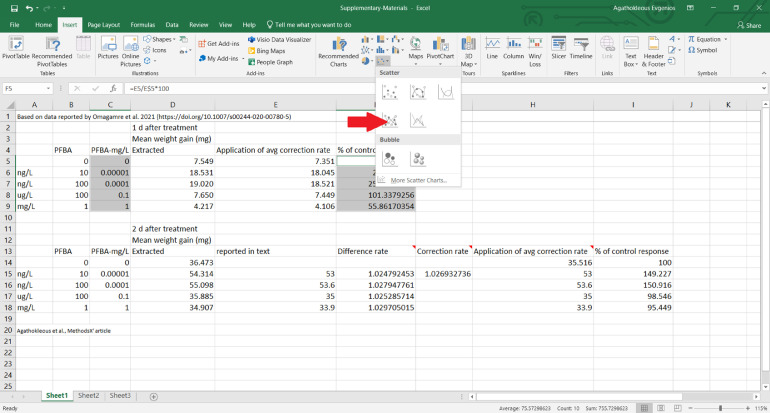
An examination of the text revealed that the values of the mean weight gain were reported for the PFBA treatments (10 ng/L, 100 ng/L, 100 μg/L, and 1 mg/L). Therefore, all the extracted data from this figure are corrected based on the rate of difference between the extracted values and values reported by authors in the text, and the % of control response values are calculated (see MS Excel file in Supplementary Materials).
-
(5)
A dose-response relationship based on the actual data is plotted (Fig. 3). In this study, the concentrations of PFBA were in different units, thus all the concentrations were converted into mg/L. When the data are given as the means of treatment groups presented in tables or in the text, the scatter plot is created following the same procedure, using the reported values without needing to extract the data with an image analysis software.
-
(6)
The NOAEL is marked on the dose-response relationship using an image editing software so to be able to estimate its value (Fig. 4).
-
(7)
To estimate the NOAEL, the image file containing the produced dose-response relationship is opened within the image analysis software, and calibration is performed (Fig. 5).
-
(8)
The NOAEL is estimated by measuring the distance from the control (typically a zero concentration/dose) to the line of NOAEL (Fig. 6).
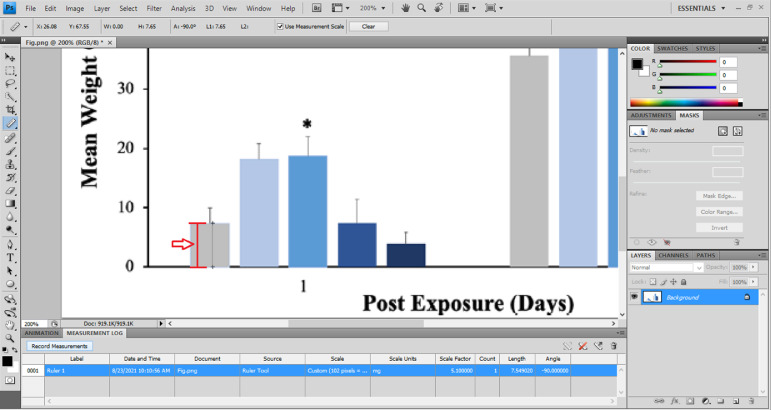
Fig 1.
Calibration for data extraction. Click “Analysis” and then “Ruler tool”. For calibration, follow the path “Analysis” -> “Set Measurement Scale” -> “Custom”. Place the cursor to a point on the y axis where the value is clearly marked, by tick marks or other means (e.g. 0 in this screenshot), press left click, and drag the cursor/ruler up to a different point (e.g. 20 in this screenshot). The pixel length and logical length would be automatically filled. If needed, the logical unit can be manually inputted (e.g. mg in this screenshot). It should be acknowledged that in many published figures the y axis scaling may differ and may not start from zero. In these cases, the difference to zero should be added to or subtracted from the extracted values. Based on data reported in [16]. The red arrow pointing left indicates the selected distance of the ruler tool, from 0 to 20 of y axis. The red vertical line with lower and upper caps is a projection of the selected distance (for presentation purposes). The asterisk indicates statistically significant difference from the control, based on the statistical results in the original paper (no asterisk = no significant difference), just for the user's attention when evaluating the responses qualitatitvely, and has no relevance to this protocol and its calculations.
Fig 2.
Data extraction. Following calibration (Fig. 1), data are extracted. In this screenshot, the control group of mean weight gain one day after the treatment is extracted by placing the ruler tool over the bar of the control and clicking “Record Measurements”. In other words, the cursor is placed onto the x axis (y = 0) where the bar of the control group occurs (the response variable value is null), click is pressed, and the cursor/ruler is dragged up to the top of the bar (maximum value of the response variable). In this case, the value of 7.549 mg was obtained (see column ‘Length’ in the “MEASUREMENT LOG”). If for some reason the ruler tool is not active, click “Analysis” and then “Ruler tool”. Based on data reported in [16]. The same process is repeated for each bar/treatment. The red arrow pointing right indicates the selected distance of the ruler tool. The red vertical line with lower and upper caps is a projection of the selected distance (for presentation purposes). The asterisk indicates statistically significant difference from the control, based on the statistical results in the original paper (no asterisk = no significant difference), just for the user's attention when evaluating the responses qualitatitvely, and has no relevance to this protocol and its calculations.
Fig 3.
Graph making. The data of % of control response are plotted against the treatment doses by selecting “the two columns and then clicking “Insert”, “Scatter”, and then “Scatter with Straight Lines and Markers” (see red arrow pointing right). This file is given as Supplementary Material.
Fig 4.
Marking the no-observed-adverse-effect-level (NOAEL) before estimating it. Determine the NOAEL, i.e. the point where the curve crosses the line of 100 % of control response to enter the high-dose zone of adversity, and insert a line crossing the x axis using an image editing software, such as Paint. In cases like the one examined here that there is a large concentration gap between the last concentration occurring within the hormetic zone and the next concentration in the high-dose zone, the curve may not directly cross the line of 100 % of control response, thus creating some uncertainty in deciding where the NOAEL should be marked. However, the NOAEL can be marked on the last point where the curve is clearly on the 100% of control response without entering the below 100 % of control response area. The control group is typically a negative control (zero dose of chemical treatment) in studies evaluating the effects of pollutants on organisms. This protocol is not for risk assessments and deriving safety limits, but for scientific evaluations of the quantitative characteristics of hormesis, and, thus, this is a less important matter.
Fig 5.
Calibration for no-observed-adverse-effect-level (NOAEL) estimation. The dose-response relationship image file is opened within the image analysis software. Click “Analysis” and then “Ruler tool”. For calibration, follow the path “Analysis” -> “Set Measurement Scale” -> “Custom”. Place the cursor to a point on the x axis (e.g. 0 mg/L in this screenshot), click, and drag it up to a different point (e.g. 0.4 mg/L in this screenshot). The pixel length and logical length would be automatically filled. If needed, the logical unit can be manually inputted (e.g. mg/L in this screenshot). Based on data reported in [16].
Fig 6.
Estimation of the no-observed-adverse-effect-level (NOAEL) estimation following calibration (Fig. 5). In this screenshot, the NOAEL is extracted by placing the ruler tool over the distance between 0 mg/L and the vertical line of NOAEL and clicking “Record Measurements”. In this case, the value of 0.159 mg/L was obtained (see column ‘Length’ in the “MEASUREMENT LOG”). If for some reason the ruler tool is not active, click “Analysis” and then “Ruler tool”.
This protocol can be used to estimate the NOAEL in dose-response relationships (with negative control) developed from data reported in published studies to improve the understanding of the quantitative features of hormesis. In cases where some publications might exist only in hard copy, the same protocol can be followed using images of the relevant figures that are scanned with a scanner connected to a computer. To minimize potential unconscious confirmation bias, i.e. the phenomenon where researchers’ research outcomes might be affected by researchers’ expectations and the belief of what might be ‘correct’ [17,18], followers of this protocol are encouraged to consider the application of blind observation and/or the derivation of the NOAEL of the same dose-response relationship by 2-3 reviewers and using the average value. To further facilitate the harmonization of the protocol and reduce the researcher-to-researcher variability, followers of this protocol may consider practicing with the data given in Supplementary Materials and the figures presented herein before attempting to work with new data.
Direct submission or co-submission
Co-submissions are papers that have been submitted alongside an original research paper accepted for publication by another Elsevier journal
Direct Submission
Declaration of Competing Interest
The same authors are also the editors of the journal issue within which this paper is published; however, none of the authors was involved in the peer-review process of the manuscript. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
E.A. acknowledges multi-year support from The Startup Foundation for Introducing Talent of Nanjing University of Information Science & Technology (NUIST), Nanjing, China (No. 003080). Sponsor had no involvement in the study design; the collection, analysis or interpretation of the data; preparation of the manuscript or the decision where to submit the manuscript for publication.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.mex.2021.101568.
Appendix. Supplementary materials
References
- 1.Berry R., López-Martínez G. A dose of experimental hormesis: When mild stress protects and improves animal performance. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2020;242 doi: 10.1016/j.cbpa.2020.110658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burbano M.S.J., Gilson E. The power of stress: The telo-hormesis hypothesis. Cells. 2021;10:1156. doi: 10.3390/cells10051156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duarte-Sierra A., Tiznado-Hernández M.E., Jha D.K., Janmeja N., J A. Abiotic stress hormesis: an approach to maintain quality, extend storability, and enhance phytochemicals on fresh produce during postharvest. Compr. Rev. Food Sci. Food Saf. 2020;19:3659–3682. doi: 10.1111/1541-4337.12628. [DOI] [PubMed] [Google Scholar]
- 4.Calabrese E.J., Agathokleous E. Hormesis: Transforming disciplines that rely on the dose response. IUBMB Life. 2021 doi: 10.1002/iub.2529. [DOI] [PubMed] [Google Scholar]
- 5.Calabrese E.J., Agathokleous E., Kapoor R., Dhawan G., Kozumbo W.J., Calabrese V. Metformin-enhances resilience via hormesis. Ageing Res. Rev. 2021;71 doi: 10.1016/j.arr.2021.101418. [DOI] [PubMed] [Google Scholar]
- 6.Carvalho M.E.A., Castro P.R.C., Azevedo R.A. Hormesis in plants under Cd exposure: from toxic to beneficial element? J. Hazard. Mater. 2020;384 doi: 10.1016/j.jhazmat.2019.121434. [DOI] [PubMed] [Google Scholar]
- 7.Shahid M., Niazi N.K., Rinklebe J., Bundschuh J., Dumat C., Pinelli E. Trace elements-induced phytohormesis: A critical review and mechanistic interpretation. Crit. Rev. Environ. Sci. Technol. 2020;50:1984–2015. doi: 10.1080/10643389.2019.1689061. [DOI] [Google Scholar]
- 8.Jalal A., de Oliveira Junior J.C., Ribeiro J.S., Fernandes G.C., Mariano G.G., Trindade V.D.R., dos Reis A.R. Hormesis in plants: physiological and biochemical responses. Ecotoxicol. Environ. Saf. 2021;207 doi: 10.1016/j.ecoenv.2020.111225. [DOI] [PubMed] [Google Scholar]
- 9.Erofeeva E.A. Plant hormesis and Shelford’s tolerance law curve. J. For. Res. 2021;32:1789–1802. doi: 10.1007/s11676-021-01312-0. [DOI] [Google Scholar]
- 10.Calabrese E.J., Blain R.B. The hormesis database: The occurrence of hormetic dose responses in the toxicological literature. Regul. Toxicol. Pharmacol. 2011;61:73–81. doi: 10.1016/j.yrtph.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Calabrese E.J., Mattson M.P. How does hormesis impact biology, toxicology, and medicine? Npj Aging Mech. Dis. 2017;3:13. doi: 10.1038/s41514-017-0013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agathokleous E., Saitanis C., Markouizou A. Hormesis shifts the no-observed-adverse-effect level (NOAEL) Dose-Response. 2021;19 doi: 10.1177/15593258211001667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calabrese E.J., Agathokleous E., Kozumbo W.J., Stanek E.J., Leonard D. Estimating the range of the maximum hormetic stimulatory response. Environ. Res. 2019;170:337–343. doi: 10.1016/j.envres.2018.12.020. [DOI] [PubMed] [Google Scholar]
- 14.Agathokleous E., Feng Z., Iavicoli I., Calabrese E.J. The two faces of nanomaterials: a quantification of hormesis in algae and plants. Environ. Int. 2019;131 doi: 10.1016/j.envint.2019.105044. [DOI] [PubMed] [Google Scholar]
- 15.Fong C.R., Chiquillo K.L., Gaynus C.J., Grier S.R., Hà B.A., Ryznar E.R., Smith L.L., Sura S.A., Zweng R.C., Anggoro A.W., Moore T.N., Fong P. Flip it and reverse it: Reasonable changes in designated controls can flip synergisms to antagonisms. Sci. Total Environ. 2021;772 doi: 10.1016/j.scitotenv.2021.145243. [DOI] [PubMed] [Google Scholar]
- 16.Omagamre E.W., Ojo F., Zebelo S.A., Pitula J.S. Influence of perfluorobutanoic acid (PFBA) on the developmental cycle and damage potential of the beet armyworm Spodoptera exigua (Hübner) (Insecta: Lepidoptera: Noctuidae) Arch. Environ. Contam. Toxicol. 2020;79:500–507. doi: 10.1007/s00244-020-00780-5. [DOI] [PubMed] [Google Scholar]
- 17.Kardish M.R., Mueller U.G., Amador-Vargas S., Dietrich E.I., Ma R., Barrett B., Fang C.-C. Blind trust in unblinded observation in Ecology. Evolution, and Behavior, Front. Ecol. Evol. 2015;3:51. doi: 10.3389/fevo.2015.00051. [DOI] [Google Scholar]
- 18.Zvereva E.L., Kozlov M.V. Biases in ecological research: attitudes of scientists and ways of control. Sci. Reports. 2021;11 doi: 10.1038/s41598-020-80677-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.