Graphical abstract
Keywords: Phytohormones; Embryogenesis; Desiccation tolerance; Seed development, Seedling establishment
Highlights
-
•
Functional ABA biosynthesis genes show specific roles for ABA accumulation at different stages of seed development and seedling establishment.
-
•
De novo ABA biosynthesis during embryogenesis is required for late seed development, maturation, and induction of primary dormancy.
-
•
ABA plays multiple roles with the key LAFL hub to regulate various downstream signaling genes in seed and seedling development.
-
•
Key ABA signaling genes ABI3, ABI4, and ABI5 play important multiple functions with various cofactors during seed development such as de-greening, desiccation tolerance, maturation, dormancy, and seed vigor.
-
•
The crosstalk between ABA and other phytohormones are complicated and important for seed development and seedling establishment.
Abstract
Background
Seed is vital for plant survival and dispersion, however, its development and germination are influenced by various internal and external factors. Abscisic acid (ABA) is one of the most important phytohormones that influence seed development and germination. Until now, impressive progresses in ABA metabolism and signaling pathways during seed development and germination have been achieved. At the molecular level, ABA biosynthesis, degradation, and signaling genes were identified to play important roles in seed development and germination. Additionally, the crosstalk between ABA and other hormones such as gibberellins (GA), ethylene (ET), Brassinolide (BR), and auxin also play critical roles. Although these studies explored some actions and mechanisms by which ABA-related factors regulate seed morphogenesis, dormancy, and germination, the complete network of ABA in seed traits is still unclear.
Aim of review
Presently, seed faces challenges in survival and viability. Due to the vital positive roles in dormancy induction and maintenance, as well as a vibrant negative role in the seed germination of ABA, there is a need to understand the mechanisms of various ABA regulators that are involved in seed dormancy and germination with the updated knowledge and draw a better network for the underlying mechanisms of the ABA, which would advance the understanding and artificial modification of the seed vigor and longevity regulation.
Key scientific concept of review
Here, we review functions and mechanisms of ABA in different seed development stages and seed germination, discuss the current progresses especially on the crosstalk between ABA and other hormones and signaling molecules, address novel points and key challenges (e.g., exploring more regulators, more cofactors involved in the crosstalk between ABA and other phytohormones, and visualization of active ABA in the plant), and outline future perspectives for ABA regulating seed associated traits.
Introduction
The plant development starts with the seed, followed by the seedling, the vegetative phase, and end with the reproductive phase [1], [2], [3]. Seed production is important for the reproduction and diffusion of many plant species that contain a fully developed embryo and allows the embryo to stay alive during seed maturation and seedling establishment for next-generation initiation [4]. There are two important phases of seed development which include zygotic embryogenesis, seed maturation. Seed maturation occurs as a result of complex, overlapping developmental processes that start from the end of embryogenesis and end when seeds become physiologically independent of the parent plant. It includes a phase of seed storage reserve deposition and the less characterized phase of maturation drying. Furthermore, during maturation, seeds acquire a range of physiological traits i.e. dormancy, vigorous and homogenous germination, after these processes, a viable seedling is established in the field for the life cycle [1], [5]. Seed dormancy and germination are critical phases in the higher plant life cycle and important traits for crop yield, however, both of them are influenced by developmental and environmental signals [6].
Seed dormancy is a key characteristic to prevent viable seed germinating during the harsh and tough growing season [4], [7]. Low seed dormancy level or non-dormant seed increases the risk of seed death and directs the seed to germinate under unfavorable growth conditions, while a high seed dormancy level stops or reduces the seed germination under favorable growth conditions which ultimately reduces the length of the growing season or crop yield [6], [8], [9]. Thus, proper seed dormancy is an important component of plant fitness and provides adaptation to a wide variety of environmental conditions. Further, it is a genetic character influenced by inherence as well as environmental factors. Along with seed later development and maturation, seed dormancy starts to build and reaches a higher level in dry mature seeds known as primary dormancy [10]. In contrast, the induction of dormancy in non-dormant seed due to unfavorable environmental conditions for germination such as light and temperature is known as secondary dormancy [7].
Germination is important that occurs in the lifecycle of all higher plants and has the potential to influence the evolution of traits expressed throughout the life of plants [8]. Seed germination starts when the dormant seed uptakes water and accomplishes when a part of the embryo such as a radicle comes out from the seed coat. The emergence of a radicle by rupturing the seed coat is known as the completion of the germination, however, this procedure depends on the absorption of water by the embryo and activation of a series of physiological events [11]. Germination requires specific environmental conditions, the seed sensitivity to the environment changes continuously as a function to adapt to ambient conditions. Therefore, seed germination depends on endogenous hormonal as well as environmental signals such as temperature, water, and light that allow a dormant seed to germinate successfully [12].
Environmental factors (temperature, soil, nutrition, light, water, humidity, air, pollutants, etc.) influence seeds dormancy and germination as well as other developmental stages of seed [13]. Seed catch signals from the environment, then endogenous pathways involved by phytohormones, such as calcium ion, reactive oxygen species are activated and seed dormancy/germination and other developmental stages are designed accordingly [14], [15]. Phytohormones play key distinct roles in the plant life cycle, from seed maturation, seed germination to the floral transition and abiotic/biotic stress responses [13], [16]. For example auxin, ABA, ET, and GA have been found that have important roles during plant development and in seed dormancy and germination regulation [17], [18], [19]. Plants maintain the availability and level of hormones in different parts of the plant body at different developmental stages in an intricate and balanced manner [20]. ABA is derived from epoxycarotenoid cleavage and is obtained one of important plant-specific hormone among other, and performs various physiological functions in the plant such as in transpiration, improved resistance from temperature (low and high) during plant development, and in the regulation of dormancy and germination [23], [24], [25]. In dormancy and germination control, ABA is one of the key hormones that play a prominent role [10], [21], [22].
Similarly, it is hypothetical that ABA plays a vital role to maintain the dormant form of seeds in a severe environment [19], [21], [26]. Previously, it has been reported that ABA biosynthesis, signaling, and degradation genes play important functions in induction, stabilization, and release of dormancy. The mutation or over-expression of key ABA-related genes results in germination-associated phenotypes [27], [28], [29], [30]. In this review, we focus and discuss the updated findings related to ABA biosynthesis, signaling and degradation, and its versatile functions associated with seed development and seedling establishment, raise some key questions for the future study of ABA function.
Role of ABA biosynthesis genes in seed development
Maternal ABA plays a significant role in embryo development and seed maturation in tobacco and Arabidopsis [31]. But, ABA is also de novo synthesized in embryo and testa during embryo development, as well as accumulates during seed maturation, facilitates late seed maturation processes, synthesis of storage proteins to prevent seed abortion, induces primary dormancy and allows successful germination as well as a successive seedling enterprise [32]. So, de novo synthesis of active ABA plays a more important role in seed development and later germination.
Active ABA is synthesized through an indirect pathway from xanthophylls (e.g., zeaxanthin, violaxanthin, and neoxanthin) [33], [34]. Three types of genes are responsible for the successive steps of ABA biosynthesis such as ZEAXANTHIN EPOXIDATION (ZEP), OXIDATIVE CLEAVAGE OF 9-CIS-EPOXYCAROTENOIDS (NCED), and ABSCISIC ALDEHYDE OXIDATION (AAO) (Fig. 1) [35].
Fig. 1.
Regulation of seed development and dormancy by ABA biosynthesis through the carotenoid pathway started from β-carotene (C40). The complete ABA synthesis process takess place in plastids and cytoplasm where ZEAXANTHIN EPOXIDASE (VPs, ZEP, ABA1/2) converts zeaxanthin into antheraxanthin and all trans-violaxanthin. ABA4 catalyzes the conversion from all-trans-violoxanthin to the all-trans-neoxanthin. The conversion of xanthoxin from 9′-cis-neoxanthin and 9′-cis-violaxanthin is exerted by VP14 and NCEDs (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE), among which the NCEDs display different subcellular localization of plastid or cytoplasm. The oxidation of abscisic aldehyde by AAO3 (ABSCISIC ALDEHYDE OXIDASE3) is responsible for the conversion from abscisic aldehyde into ABA, which in turn induces and maintains seed dormancy. But, it is yet unknown of the factors responsible for the conversion from all-trans-violoxanthin /all-trans-neoxanthin to 9-cis-violoxanthin/9-cis-neoxanthin.
The ZEP/ABA gene was firstly identified in Arabidopsis thaliana and Nicotiana plumbaginifolia [36]. Their mutants (aba1/aba2) with deficient ABA were impaired in the oxidation of zeaxanthin into antheraxanthin and violaxanthin [37], which is thought as an initial step of ABA biosynthesis (Fig. 1). In rice, a Tos17 viviparous mutant was identified to have viviparous germination due to a defect in the oxidation of zeaxanthin during ABA synthesis [38]. Numerous other ABA auxotrophic mutants (vp2, vp5, vp7, and vp9) identified in maize by genetic screening have defects in zeaxanthin epoxidase activity and block the early steps of carotenoid biosynthesis too [39]. All these evidenced that the oxidation of zeaxanthin is an important and conservative phase in the ABA synthesis of the plant. It is always not very clear for the conversion from all-trans-violoxanthin and the all-trans-neoxanthin to 9-cis-violoxanthin and 9-cis-neoxanthin. However, ABA4 was found responsible for conversion from all-trans-violoxanthin to the all-trans-neoxanthin [40], providing some clues for the exploration of these transition.
The next pivotal gene in the subsequent stages of ABA biosynthesis was firstly cloned from maize viviparous mutant vp14 as NINE-CIS-EPOXYCAROTENOID DIOXYGENASE (NCED9). The vp14 mutant has a fault in the oxidation of 9-cis-epoxycarotenoid during the last few steps in ABA biosynthesis and exhibits reduced ABA content in the dry seed [41]. In Arabidopsis, NCED2, NCED3, NCED5, NCED6, and NCED9 are known as the homologs of VP14 participating in a rate-limiting step in ABA biosynthesis [24] (Fig. 1). Furthermore, the PvNCED1, LeNCED1, and BdNCED1 identified from the bean, tomato, and Brachypodium distachyon, respectively also show the important roles in ABA biosynthesis and seed development [42], [43]. All the above studies have delivered pieces of evidence that the oxidative cleavage of xanthophylls is the main step during ABA biosynthesis regulation for dormancy and development mediation in seeds [44].
Abscisic aldehyde oxidation is the last step of ABA biosynthesis, where abscisic aldehyde is oxidatively converted into ABA (Fig. 1) [45]. Firstly, identified mutants defective in the oxidation of abscisic aldehyde into ABA were flacca and sitiens in tomato [46]. Later on, abscisic aldehyde oxidase3 (AAO3) was identified in Arabidopsis which functions in the last two steps of ABA biosynthesis in seed and its expression was also observed in embryo vascular tissues during mid and late maturation phases [47], [48]. The ABA synthetic pathway offers an active ABA pool during the whole plant development that is controlled by various homologous genes. The identification of cofactors of the enzymatic reactions in the ABA synthesis pathway would be helpful for the understanding of the complete network of ABA synthesis.
Role of ABA signaling components in different seed developmental stages
ABA works via a complex signaling network and initiates the cell response through activating downstream signaling genes to induce the response according to physiological effects [49], [50]. In seed development and maturation, the role of ABA has been recognized by analyzing the mutants that were insensitive to ABA. The ABA insensitive mutants fail to promote ABA response due to defects in the ABA signaling pathway, which steadily affects seed maturation and several other important traits of the dormant seed [19].
Identification and mechanisms of the core components in ABA signaling pathway
The identification of PYL/RCAR family proteins verified that ABA receptor PYLs are essential ABA signaling components and predominantly function in seed [51]. In Arabidopsis, fourteen members of the PYR/PYL/RCAR protein family were documented that have vital roles in seed, such as pyr1/prl1/prl2/prl4 quadruple and pyl duodecuple mutants show reduced seed dormancy and insensitivity to ABA [29], [52]. Furthermore, an ospyl septuple mutant was identified in rice which is insensitive to ABA during seed germination [51].
In the absence of ABA, PYLs proteins release protein phosphatase type 2C (PP2C), another important component in ABA signaling, and activate their functions of phosphatase [29]. PP2Cs proteins including ABA-INSENSITIVE 1/2 (ABI1/2) and ABA-HYPERSENSITIVE GERMINATION3 (AHG3) suppress the activities of downstream ABA signaling proteins SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2s (SnRK2s) by protein phosphorylation, as a result, blocking the function of the downstream ABA signaling network (Fig. 2) [30]. So, PP2Cs function as negative regulators in the ABA signaling system and were identified through ABA insensitive mutants screening, whereas, their knockout mutants exhibited reduced seed dormancy and hypersensitivity to ABA [53]. Recently, it is demonstrated that ENHANCER OF ABA CO-RECEPTOR1 (EAR1) acts together with PP2C proteins (i.e. ABI1/2, HAB1/2, and AHG1/3), to increase the activities of PP2C [54]. Like EAR1, PR5 receptor-like kinase 2 (PR5K2) inhibits ABA-signaling via phosphorylation enhancement of ABI1/2 [55]. On the other hand, DELAY OF GERMINATION1 (DOG1) binds to heme and interacts with the AHG1 to stop its phosphatase function and enhance seed dormancy [56]. These studies concluded that PP2Cs can be regulated either by PYLs receptors or by other proteins, but the complete phenomenon and relationships between PP2Cs, PYLs, and other regulatory factors (DOG1, PR5K2, and EAR1) is unidentified in seed development.
Fig. 2.
The ABA signaling pathway is involved in seed development. Left, in the absence of ABA: Receptors PYLs release and activate protein phosphatase 2C (PP2C) such as ABI1/2 and AGH1/3. Downstream SNF1-RELATED PROTEIN KINASE subfamily (SnRK2s) genes are inactivated by active PP2C which leads to premature germination and the non-dormant seed through repression of lots of transcription factors such as ABI1/2/3/4/5 and bZIP67. Right, in the presence of ABA: Receptors PYR/PYL/RCAR bind ABA and PP2C together to inhibit the activity of PP2C, which release the activity of SnRK2s and downstream transcription factors such as ABI3 by protein phosphorylation, then regulate downstream genes SGR1/2 function to mediate seed de-greening process. Additionally, the active LAFL (ABI3, FUS3, LEC1, and LEC2) network by ABA along with WRI1 regulates the At2S3 gene; an active bZIP22 function downstream of SnRK2s to promote gene transcription of 27-kD γ-zein for protein reserve accumulation in the seed. Along with seed de-greening and storage product accumulation, SnRK2s function upstream of ABI3/5 and ABFs to regulate LEAs and HSPs that are pivotal for desiccation tolerance. In other branches, DOG1 also plays a role upstream of ABI3/5/ABFs as well as functions as a repressor of PP2Cs (AHG1/3) to involve seed desiccation tolerance acquirement. In combination, all key ABA signaling components (SnRK2s, ABI3, ABI4, ABI5, ZmbZIP22, bZIP67, and ABFs) are involved in storage product accumulation, de-greening, and desiccation tolerance with different function pathways to provide a mature and dormant seed. Letters “P” and “T” in the color circles indicate the two manners of protein phosphorylation and gene transcription regulation, respectively. Activated and repressive effects are shown by arrows and bars, respectively.
In the presence of ABA, PYR/PYL/RCAR protein binds with both the ABA and the PP2C proteins to stop the phosphatase activity of the PP2Cs, which releases and enables the SnRK2s function. It is showed that all members of PYLs protein family from Arabidopsis can interact with PP2C family members and function in ABA mediating response [57]. Totally, three SnRK2s (SnRK2.2, SnRK2.3, and SnRK2.6) were found as positive regulators of the ABA signaling network and involved in various seed developmental processes such as the de-greening process, accumulation of seed storage products, seed maturation, desiccation-tolerant, and germination in Arabidopsis [19]. A recent report identified an ABA Signaling Terminator (ABT), a WD40 protein, which can efficiently shut down the ABA signaling and is vital for seed germination and seedling establishment. In a PYR1/PYL/RCAR-PP2C-dependent manner, ABT is induced by ABA and interacts with the PYR1/PYL/RCAR and PP2C proteins, which disturb the interaction between PYR1/4 and ABI1/2, thus cut off ABA signaling [58], which further enriches and illuminates the ABA signaling network.
In addition, the major targets of SnRK2s are ABSCISIC ACID RESPONSIVE ELEMENT (ABRE) binding factors (ABF). ABFs family consists of nine members ABF1, ABF2/ABA–RESPONSIVE ELEMENT BINDING PROTEIN1 (AREB1), ABF3, ABF4/AREB2, AREB3, ABI5, bZIP15, bZIP67, and EEL from bZIP subfamily, predominantly participates in the regulation of ABA-mediated transcription [59]. The transcription of ABI5 can be activated by SnRK2s through specifically binding with ABRE cis-element in ABI5 promoter, in turn, activate the ABA-mediated transcription activity in late seed maturation phase and imbibed seeds in Arabidopsis [60]. Moreover, another key factor-ABI3 interacts with the ABI5 transcription factor and functions collectively with ABI5 to promote transcription of downstream ABA-responsive genes [61], both of them can be regulated by RELATED TO ABI3/VP1 (RAV1) through binding to their promoters. Interestingly, ABI5 also modulates the ABA response through binding to PYL11 and PYL12 promoters to regulate the transcription directly during germination. ABA hypersensitivity resulting from PYL11 and PYL12 overexpression was totally or partially damaged when ABI5 was mutated. Above all these explore a feedback regulation in ABA signaling mediated by ABI5 [62].
By genetic screening, two LEAFY COTYLEDON (LEC1/2) genes and FUSCA3 (FUS3) were identified as key roles in ABA-mediated seed development. ABI3 along with LEC1, LEC2, and FUS3 transcription factors play roles in seed development by mediating ABA biosynthesis in seed tissues. For the above four genes (i.e. ABI3, FUS3, LEC1, and LEC2), a defect in any one leads to abnormal seed development such as altered seed dormancy, failure to attain desiccation tolerance, and a low level of ABA contents [26]. All these supported that there is a positive correlation between these transcription factors and ABA content and signaling transduction [63]. Moreover, ABI3/FUS3/LEC1/2 combine with LEC1-LIKE (L1L) to form a complex transcription control network known as LAFL that control the embryogenesis process, hormone signaling, metabolic pathways, and function upstream of several genes such as PEI1, BABY BOOM (BBM), APETALA2 (AP2), SEED STORAGE PROTEINS (SSP), FLOWERING LOCUS C (FLC), including 2S ALBUMIN STORAGE CRUCIFERIN C (CRC) and PROTEIN 1 (At2S1) that involve seed development [64], [65], [66]. Many members of LAFL network are regulated by BBM during somatic embryogenesis [67], indicating the feedback regulation between BBM and LAFL components. Additionally, they are also regulated by VIVIPAROUS 8 (VP8), a B3 type transcription factor in maize, that show the pleiotropic roles during seed development [68]. In maize, a defective kernel 33 (dek33) mutant was identified and the causal locus was cloned as a pyrimidine reductase in riboflavin biosynthesis. The genetic and molecular research indicated that DEK33 interacts with RGLG2 and SnRK1, influences the ABA synthesis positively to regulate seed development [69], which shed light on the regulation of ABA synthesis other than before in seed development.
Mechanisms of key genes associated with ABA in de-greening process of seed
In seed maturation, SnRK2s and ABI3 genes were identified as an essential component for the de-greening process (Fig. 2) [70]. The snrk2.2/snrk2.3/snrk2.6 triple mutant exhibits ABA insensitivity during seed development and produces green seeds [52], [59]. Additionally, the targeted gene of ABI3- stay-green (SGR1/2), plays an important task in the process of de-greening of seed, whereas, abi3-6 mutant exhibits pleiotropic effects during seed development including immature embryo growth, failure of embryo de-greening, and insensitivity to ABA as well as non-dormant and desiccation intolerant seeds [70], which indicated an important SnRK2s -ABI3 -SGR1/2 pathway associated with ABA in seed de-greening and other traits determination.
Mechanisms of ABA in accumulation of seed storage products and desiccation tolerance achievement
Along with seed maturation, some reserve materials accumulate in the seed's later stages [71]. Six genes belonging to different transcription factors family (ABI3/VP1, ABI4, ABI5, LEC1, LEC2, and FUS3) have been identified that induces the expression of ABA-responsive and seed-specific factors such as LEA and storage protein genes [72]. ABI5, bZIP67 together with ABI3 and ABI4 control the expression of many genes that are involved in ABA-mediated seed storage processes [73], [74]. ABI4, identified from the ABA-insensitive mutants, encoding an ERF/AP2 type transcription factor, expresses transcriptionally in all seed developmental stages [75]. Many studies reported that various transcription factors regulated ABI4 transcription. Interestingly, ABI4 can also activate ABI4 itself expression during the early stages of seedling growth [76]. The bZIP67 transcription factor together with L1L and NUCLEAR FACTOR-YC2 (NF-YC2) transcription factors form a complex to promote FATTY ACID DESATURASE 3 (FAD3) in the seed which functions in the storage of omega-3 fatty acid during maturation [73]. Moreover, induced expression of maize bZIP22 changes endosperm starch content and composition in maize and rice during seed storage and is required for the transcription of a 27-kD γ-zein [77], [78]. Many studies have reported that ABA insensitive mutants snrk2.2/3/6 triple mutant and pyl duodecuple mutants exhibited less level of seed storage products due to defect in ABA signaling [52]. Moreover, the RNA-seq data analysis exhibited that the expression of 12S globulin storage protein was down-regulated in the snrk2.2/3/6 triple mutant [79]. In addition, the induced expression of SnRK2.6 showed increased seed production, on the contrary, snrk2.6 mutant showed 7–25% reduced oil content of seed [80].
Besides above, the lec1 and fus3 mutant embryos exhibit reduced accumulation of storage proteins and lipids compared to wild-type during maturation [81]. Moreover, the LEC2 protein shows synergistic activity for the abundance of storage proteins with ABI3, FUS3, and LEC1 during maturation [82]. FUS3 and LEC1 control the accumulation of ABI3 protein in seeds and function with each other in many physiological processes including lipids formation, anthocyanins synthesis, accumulation of chlorophyll, and storage proteins [81]. Comprehensive studies indicated that the expressions of several storage protein genes such as Arabidopsis 2S storage protein 3 (At2S3) and 12S storage protein gene rely on FUS3, ABI3, and WRINKLED1 (WRI1) transcription factors through an ABA-mediated manner [81], [83], [84] (Fig. 2). Moreover, LEC1, LEC2, and GmDREBL regulate WRI1 to play roles in sugars and oil content storage in seed, as wri1 mutant is revealed 80% less oil content and a higher level of soluble sugar in seed [85], [86]. So, LAFL network regulates the expression of ABA signaling components including PYR/PYL/RCARs, SnRK2s, and ABFs that are involved to induce the expression of LEAs and HSPs genes at the time of seed maturation (Fig. 2) [52], [59], [87], [88].
During the last phases of seed development, desiccation tolerance is acquired associated with the accumulation of antioxidants, sugar, and late embryogenesis abundant (LEA) proteins [89]. The genetic analysis of loss and gain of function mutants of LEC1 indicated that LEC1 is a major regulator during seed maturation and accumulation of storage products, desiccation tolerance as well as induction of dormancy [90]. LEC1 functions together with NF-YB, AREB3, bZIP67, and ABI3 to regulate genes required for seed maturation [91]. The FUS3 gene is required for seed maturation and desiccation tolerance during seed development, thus, the fus3 mutant showed premature embryo growth and seeds were desiccation intolerant [63], [92]. The double mutant aba/abi3 shows ABA insensitivity and produces seeds that are desiccation intolerant. ELONGATED HYPOCOTYL 5 (HY5) is an important light signaling component which binds with the promoter of ABI5 to regulate the LEAs gene expression [93]. Further, DOG1 increases the LEA and HSP genes expression through ABI5/ABI3 and speeds up the storage of N-rich compounds in the seed which promotes the dormancy and viability of the seed [94]. In some studies, it is shown that DOG1 expression is controlled by bZIP67 and ETHYLENE RESPONSE FACTOR12 (ERF12) during seed maturation negatively or positively [95], [96]. Moreover, ABA regulates the expression of the DOG1-LIKE 4 (DOGL4) gene and increases the expression of some seed storage proteins including CRUCIFERINs, ALBUMINs, and OLEOSINs during the seed maturation process [97]. From above, some specific factors (e.g., bZIP67, DOG1, NF-YC, AREB3) were identified to function associated with LAFL genes to mediate the seed storage protein accumulation and acquirement of desiccation tolerance related to maturation, which facilitates the elucidation of the regulation network of LAFL genes in the seed different developmental stages.
Mechanisms of ABA in primary seed dormancy induction
Dormancy is an imperative characteristic of wild plant species, prevents the seeds from adverse environmental conditions, and confirms the initiation of a next-generation [98], [99]. Seed dormancy is achieved at the end of the seed maturation when molecular dependence from the mother plant disappears, storage products are synthesized, dehydration occurs, and de novo ABA is stored [24], [100]. After dehydration, the seed enters into a state of dormancy physically and physiologically. The physical structures of the seed such as the testa and endosperm are responsible for the physical dormancy [101], [102], from their ability to enhance seed impermeability or limit water uptake [103]. ABA is the major internal physiological factor inducing seed dormancy through affecting a lot of physiological pathways such as storage proteins and lipids in seed [79], [82].
It is notable that de novo ABA biosynthesis occurs in the embryo and later is utilized during seed maturation and induction of primary dormancy indicating that the dormancy is a characteristic of an embryo and its related tissues [31], [104]. Numerous ABA deficient (aba and nceds) and insensitive (pyls, snrks, and abi3/4/5) mutants show reduced seed dormancy and early germination indicating that ABA exerts a vital role in the induction of dormancy [59]. The AtNCED3 mainly expressed in the seed is perceived as the critical enzyme for ABA synthesis during early seed development, over-expression of which improved ABA contents in seeds and prolonged seed dormancy [44]. Similarly, nced6 and nced9 mutants show decreased ABA level and dormancy in mature dry seed [105]. Furthermore, a recent study speculated that ODR1 (suppressor of RDO5) acted together with bHLH57 and functioned upstream of NCED6 and NCED9 to control the ABA synthesis and seed dormancy in Arabidopsis [106]. Interestingly, the ectopic and over-expression of bean PvNCED1 gene in imbibed seeds of tobacco elevated ABA levels and exhibited delayed seed germination. In tomato, the over-expression of LeNCED1 also enhanced dormancy by enhancing the ABA level in seeds. In addition, MYB96 binds directly with ABA synthesis genes (NCED2, NCED5, NCED6, and NCED9) promoters and inactivates GA biosynthesis genes (GA3ox1 and GA20ox1) to induced primary seed dormancy in Arabidopsis [107]. ABI4 deepens seed dormancy in Arabidopsis through direct interaction with promoter regions of NCED6 to increase ABA biosynthesis and with promoter regions of GA2ox7, a GA-inhibitor gene [108] to inhibit GA biosynthesis [109]. A study reported that peroxiredoxin PER1 improves the primary seed dormancy by inhibiting the ROS which in turn inactivates the ABA catabolism and GA biosynthesis genes in Arabidopsis [110]. In Sorghum bicolor, ABI4 and ABI5 (SbABI4 and SbABI5) enhance the transcription of SbGA2ox3 through directly binding to its promoter and consequently extend seed dormancy [111].
The loss of function mutant lec1 shows premature germination during seed development indicating that LEC1 is required for induction of primary seed dormancy [90]. During germination, the functions of LAFL network can be controlled or repressed by VIVIPAROUS1/ABI3-LIKE1/2/3 (VAL1/2/3) [66]. Consistently, Members of LAFL genes are regulated by VP8 in maize [68]. The mutations in VP8 homolog gene PLASTOCHRON3/GOLIATH (PLA3/GO) in rice and ALTEREDMERISTEM PROGRAM1 (sAMP1) in Arabidopsis show reduced dormancy phenotype [112]. Interestingly, VP8 and its homologs (PLA3/GO and AMP1) contained glutamate carboxypeptidase [68], indicating that this peptide might be important for seed maturation and dormancy, but its detailed biochemical mechanism is almost blank. Two individual studies demonstrated that the RAF-like MAPKKKs, RAF10/11 can phosphorylate SnRK2s and ABFs to influence the dormancy of seed [113], [114]. The key factor DOG1 imposes primary seed dormancy by inhibiting AHG1 action to enhance ABA sensitivity [56]. Moreover, HISTONE DEACETYLASE 19 (HDA19) interacts with SIN3-Like 1 (SNL1) to modulate the ABA signaling pathway to promote seed dormancy [115], which contributes to further understanding between epigenetic modifications and ABA signal in seed development. In wheat, TaABI5 transcripts accumulate in seed embryos. Over-expression of TaABI5 in Arabidopsis displayed high sensitivity to ABA and increased dormancy, indicating that TaABI5 playing a positive role in dormancy maintenance as a functional ortholog to Arabidopsis ABI5 [116].
Mechanisms of ABA in seed germination and seedling establishment
Germination is a critical and initial process in the plant life cycle which starts with the uptake of water by mature seed at imbibition and shifts from maturation stage to germination stage via emerging the radicle [117]. During germination, the high levels of ABA in imbibed seeds of strongly dormant A. thaliana ecotype Cvi reduce clearly indicating that seed dormancy in A. thaliana Cvi accession seeds depends on the endogenous ABA level [118], [119], [120].
In many studies, it is proved that ABA catabolism is a crucial step to alter the dormancy state of seed to germination in Hordeum vulagre, Pseudotsuga menziesii, Cupressus nootkatensis, and yellow-cedar [121]. The ABA is degraded through consecutive hydroxylation and conjugation steps. The CYTOCHROME P450, FAMILY 707, SUBFAMILY A (CYP707As) provided with cytochrome P450 monooxygenase and ABA 8 prime-hydroxylase activity catalyzes the ABA to phaseic acid (PA). PA reductase (PAR) ABH2 and glycosyltransferase (GT) then catalyze PA to dihydrophaseic acid (DPA) and DPA-4-O-β-D-glucoside (DPAG), resulting in the ABA degradation (Fig. 3) [122]. The decreased level of ABA at the time of imbibition leads to a higher amount of PA and DPA accumulation in lettuce, Arabidopsis, and H. vulgare seed [27], [123] suggesting the positive role of ABA 8-hydroxylation in germination [124]. Moreover, it was demonstrated that in non-dormant H. vulgare seeds, ABA catabolic enzyme HvABA8′OH-1 expression was preferentially detected in coleorhiza which is an important tissue from where germination starts [125].
Fig. 3.
The function of ABA in seed germination and seedling establishment. Left, Seed completes germination successfully through degradation of active ABA into PA (phaseic acid) and DPA (dihydrophaseic acid)/DPAG with CYP707As regulated by REF6 and phaseic acid reductase (ABH2 and GT) respectively. During germination and seedling establishment, the core ABA signaling component SnRK2s and downstream ABI3/4/5 are activated or repressed by many factors directly or indirectly to promote seed germination and seedling establishment. For example, RAV1 and BASS2 bind to the ABI4 promoter to inhibit ABI4 expression, while MYB96 promotes ABI4 expression through binding to its promoter. A Casein Kinase 2 promotes ABI4 expression indirectly. Furthermore, ANAC060 transcription is activated directly by ABI4 through binding to its promoter to enhance post-germination. Several BTB-A2 proteins can impair SnRK2.3 stability to act as negative regulators of ABA signaling. NRT1.2 is identified as an ABA transporter to regulate downstream factors ABI1-ABI5, RAB18, etc. positively to mediate germination and seedling development. Further, some negative factors such as SUN24, UGT74E2, FOF2, and VQs regulate seed germination and seedling development through repressing ABI3-, ABI5-mediated ABA pathway. Activated and repressive effects are shown by arrows and bars, respectively.
All four members of CYP707As (CYP707A1-CYP707A4) in Arabidopsis play regulatory functions to control the ABA level. The expression of CYP707A1 is absent during zygotic embryogenesis [123] while is present in the embryo predominantly in the middle of seed maturation to inactivates ABA biosynthesis and decreases at maturity [125]. Whereas, CYP707A2 catabolism the ABA during late maturation, and cyp707a2 mutant accumulates less ABA compared to cyp707a1 mutant after imbibition [110]. The over-expression of CYP707A2 decreased the ABA content in seed at maturity and reduced the storage time required to release the dormancy of the seeds whereas, cyp707as mutants required more storage time to reduce the dormancy compared to that of control [123], [126]. In a recent study, it is demonstrated that Arabidopsis RELATIVE OF EARLY FLOWERING6 (AtREF6) directly binds and regulates the key ABA catabolism genes (CYP707A1 and CYP707A3) to promote the catabolism of ABA and seed development [127].
Besides, the key components of ABA signaling also show indispensable roles in seed germination with various mechanisms. Recently, two members of the VQ family, VQ18 and VQ26, were proved to act as direct and negative interactors of the ABI5 to mediate the ABA signaling level and regulate seed germination and early seedling establishment [128]. Likewise, many studies proved that ABA signaling through the ABI4-mediated cascades such as miRNA 165/166, E3 ubiquitin ligase CER9 (ECERIFERUM 9), transcription factors OsAP2-39 and nuclear C2H2 zinc-finger protein ZFP3, and AtGLR3.5 (glutamate receptor homolog 3.5) [129], [130], [131], [132], [133], [134], [135] play important roles during seed germination and post-germination seedling growth, illustrating that ABI4 is a key factor with regard to ABA-mediated regulation of seed germination and early seedling establishment. Another gene CK2 (Casein Kinase 2), positively enhances ABA signaling during seed germination and seedling establishment by enhancing ABI4 expression partially and indirectly [136]. MYB96 increases ABI4 expression during seed germination, while RAV1 and BILE ACID: SODIUM SYMPORTER FAMILY PROTEIN 2 (BASS2) repress ABI4 expression during seedling development by binding to its promoter [76]. As the key terminator of ABA signaling, over-expression of ABT promotes seed germination and seedling greening in the presence of ABA, and knockout of ABT exhibits the contrary effect [58]. Three BTB-A2 (broad-complex, tramtrack, and bric-a-brac-A2) domain family genes BTB-A2.1, BTB-A2.2, and BTB-A2.3 act as negative regulators of ABA signaling by impacting SnRK2.3 stability and subsequently weakening the expression of ABA-responsive genes, for example, btb-a2.1/2/3 triple mutant showed a decrease in ABA-induced inhibition of seed germination by increasing ABA signaling [137]. In tomato, it is reported that MAPK11 also phosphorylates SnRK2s which affects ABA signaling by suppressing the transcription of ABI5 and ultimately influences seed germination [138]; further, IQ67-Domain (IQD) protein SUN24 regulates seed germination by altering the expression of two key ABA signaling genes Solanum lycopersicum ABA-insensitive 3/5 (SlABI3 and SlABI5) in tomato germinating seeds [139]. UDP-glycosyltransferases (UGTs) transferring glucose to indole-3-butyric acid plays key roles in plant development. Overexpression of OsUGT74E2 down-regulated the expression of OsABI3 and OsABI5, and promoted seed germinating with a lower ABA level, indicating a regulation of seed germination involved by UGT74E2 function upstream of ABA signaling in rice but not Arabidopsis [140]. In Medicago truncatula, a ATP-binding cassette (ABC) transporter, MtABCG20, functions as an ABA exporter in germinating seeds. In seeds of mtabcg20, due to the ABA translocation impairment, it showed more sensitivity to ABA in germination [141]. All these novel progresses in species other than Arabidopsis provide more rich knowledge for the mechanism of ABA in seed germination.
Besides the seed development, ABA synthesis and signaling genes play direct and crucial roles in the establishment of post-germination development but not in the phase conversion from germination to the seedling establishment [142]. In post-germination, ABA plays the role with multiple other participants such as JUMONJI-C domain-containing protein 30 (JMJ30), a histone demethylase activated by ABI3, which inhibits the enrichment of H3K27me3 at the promoter of SnRK2.8, activates the SnRK2.8′s kinase activity and ABI3 function to encourage post-germination growth (Fig. 3) [143]. This study revealed a forward regulatory loop associated with ABI3 in post-germination. F-BOX OF FLOWERING 2 (FOF2), a key factor in flowering, plays an important negative role in ABA-mediated seed germination and early seedling development, partially by repressing the expression of ABI3 and ABI5 [144]. During post-germination, ABI4 enhancec ANAC060 transcription to start post-germination growth by directly interacting with its promoter to reduce ABA sensitivity and glucose-mediated ABA accumulation [145]. Interestingly, another NAC family factor, NAC103 was up-regulated and enhanced in transcription and protein stabilization by ABA treatment respectively. Moreover, NAC103 over-expression plants showed more sensitivity to ABA during seed germination and young seedling growth, which was acquired by regulating some downstream genes such as MYB78, PLP3, and RGL2 in Arabidopsis [146].
Pyrenophoric acid (P-Acid) is one kind of phytotoxic sesquiterpenoids produced by the Pyrenophora semeniperda, an effective mycoherbicide in crop cultivation. An intensive study found that it inhibites seedling establishment through activating the ABA signaling pathway; further, P-Acid B exerts the ABA biosynthesis pathway but not interacts with PYR/PYL receptors to involve the ABA pathway, which also explores the underlying mechanism associated with ABA of parasites in seed [147]. NITRATE TRANSPORTER 1.2 (NRT1.2) is identified as a nitrate transporter and an ABA transporter in Arabidopsis. Some ABA-responsive genes, ABI1-ABI5, RAB18, RD29A, and PHOSPHOLIPASE Dα1 (PLDα1) were up-regulated by over-expression of NRT1.2 as well as exogenous ABA. Consequently, NRT1.2 interacts with PLDα1 at the plasma membrane and positively involves the ABA pathway to mediate germination and seedling development [148]. From above, ABA interacts with different molecules or metabolites to involve seed germination and seedling development antagonistically/synergistically.
Crosstalk between ABA and other phytohormones and signaling molecules in seed germination
As endogenous organic substances, phytohormones play distinct roles in the plant life cycle from seed maturation, seed germination to the floral transition, and abiotic/biotic stress responses [13], [149]. Numerous elegant studies have demonstrated that different phytohormones interact antagonistically and/or synergistically with one another and form complicated networks in seed germination regulation [108], [150], [151], [152]. ABA and gibberellins (GA) are one pair of classic phytohormones, which antagonistically mediate several plant developmental processes and regulate the decision between dormancy and germination [6], [19], [153]. Therefore, the balance between catabolism and synthesis of ABA/GA by regulating signaling pathways stabilizes the balance between germination and dormancy (Fig. 3). Genetic and mutational analyses of the ABA and GA metabolism and signaling genes suggested that some genes are evidenced the importance in the regulation of seed germination and seedling growth [154], [155]. For example, FUS3 plays an important role to maintain ABA:GA balance by inhibiting GA biosynthesis and activating ABA biosynthesis during Arabidopsis seed development [63], [92], and a reduced amount of ABA and increase in GA content in fus3 mutant was detected during seed maturation indicating that FUS3 protein functions as a hub in GA and ABA synthesis in the seed [92], [156]. ABI4 is another central factor mediating the antagonism between ABA and GA by regulating the biosynthesis of both phytohormones, resulting in the precise control of the degree of seed dormancy and post-germination seedling growth [16], [157]. Alteration in Arabidopsis ABI4 accumulates the GA content and reduces ABA content, which retrieves the dormant phenotype of ga1-1 mutant, indicating that ABI4 is also important to maintain the balance between ABA:GA ratio during seed development similar to FUS3 (Fig. 4) [108], [109]. However, the underlying molecular mechanisms between FUS3 and ABI4 in regulating the ABA/GA simultaneously are still a mystery.
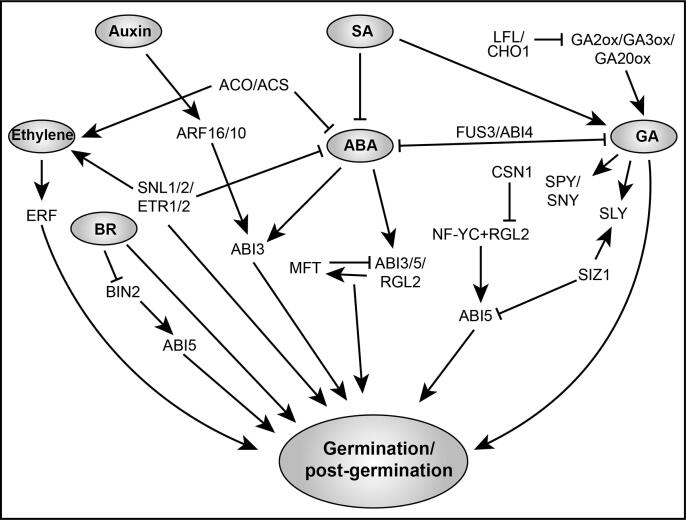
Fig. 4.
The interplay of ABA and other phytohormones (GA, ET, SA, BR, and Auxin) signaling in the regulation of seed germination and post-germination growth. ABA crosstalks with other phytohormones either by affecting their biosynthesis or by interfering with their signaling pathways during germination and post-germination growth. Among these, the interaction between ABA and GA is most studied and important. FUS3 and ABI4 play more vital roles to mediate the antagonism between ABA and GA. SA was found to regulate the content of ABA negatively and GA positively, respectively in seed germination. Both two factors LFL and CHO1 were showed inhibition to GA synthesis to involve seed germination regulation. After that, some downstream factors of GA such as SPY, SNY, SLY, SIZ, RGL2, etc. display different functions in the crosstalk between GA and ABA. Both SIZ and RGL2 can repress and promote ABI5, respectively. Besides, RGL2 can be degraded by CSN1 and regulate ABI5 together with an NF-CY factor. ABA-mediating ABI3, ABI5, and RGL2 regulate MFT by establishing a negative feedback loop to modulate the ABA and GA antagonism, in which, MFT also inhibits ABI5. A study indicated that auxin stimulates ABI3 expression through ARF10 and ARF16 indirectly, which connects the ABA and auxin in seed germination regulation. In some ways, ABA inhibits ACO and ACS to influence ethylene synthesis negatively. Meantime, ETR1/2 and histone deacetylation cofactors SNL1/2 mediate the antagonism between the ABA and ethylene to involve seed germination, in which some ERF factors are involved. BR promotes seed germination as well as inhibits BIN2, which interacts with ABI5 and positively regulates ABA responses during seed germination and post-germination. Activated and repressive effects are shown by arrows and bars, respectively.
Some generic nuclear factors are also involved in the crosstalk between ABA and GA. A study illustrated that GERMINATION DEFECTIVE 1 (GD1) encoding a B3 domain TF suppresses LEC2 and FUS3 like gene (OsLFL1) and modulates GA biosynthesis genes (OsGA3ox, OsGA20ox, and OsGA2ox) expression to regulate germination in rice [158]. Another transcription factor containing AP2 domain CHOTTO1 (CHO1) enhances seed germination by regulating ABA-related genes to suppress GA biosynthesis genes in Arabidopsis [159]. Interestingly, three NUCLEAR FACTOR-Y C (NF-YC) homologs genes in Arabidopsis NF-YC3/4/9 are involved in the regulation of GA-ABA crosstalk during seed which are regulated by GA to suppress ABA signaling [160]. Further research explored that NF-YC9 promotes ABA responses in early seedling growth by binding to ABI5 to increase ABA sensitivity [161] elucidating that the NF-YCs-ABI5 module integrates the antagonistic GA and ABA signaling in seed germination and post-germination stages (Fig. 4). These provide novel information to explore the underlying mechanisms associated with ABI5 of ABA and GA antagonism.
Further, the well-known negative GA signaling components such as DELLA proteins (i.e. GA INSENSITIVE (GAI), REPRESSOR OF GA1-3 (RGA), RGA-LIKE1 (RGL1), RGL2, and RGL3 influence seed dormancy and germination [162], [163] through stimulating the ABA biosynthesis and ABI5 activity, in which NF-YC and RGL2 together promote the expression of ABI5 and enhance the ABA-mediated repression of seed germination [160]. As a result, the rgl2 mutant exhibited a reduced ABA concentration during imbibition, terminated dormancy, and accelerated germination [164]. In addition, in Arabidopsis RGL2 forms a complex with DOF6 transcription factor which positively activates GATA12 transcription to control seed germination [165]. The RGL2 can also be degraded by the COP9 Signalosome 1 (CSN1) which may inhibit ABI5 activity and promote seed germination [166]. However, the mutation in GA signaling gene SLEEPY1 (SLY1), exhibites higher germination and mRNA level of RGL2, indicating that SLY1 functions independently of RGL2 in seed germination [167]. Interestingly, ABA can enhance RGL2 expression, this feedback loop modifies ABA and GA paths in the seed germination process [164]. Epigenetically, it is stated that E3 SUMO ligase SIZ1 sustained ABA: GA level by SUMOylating ABI5 to negatively regulate ABA signaling and SLY1, as well as to positively regulate GA signaling during germination in Arabidopsis [168], [169], [170]. Furthermore, some other genes involved in GA signaling were also identified including SPINDLY (SPY) and SNEEZY (SNE) belonging to F-box proteins involved in seed germination regulation (Fig. 4) [171], [172]. All above these indicate that some factors in the GA pathway participate in the interplay of ABA and GA in seed germination through diverse function patterns.
MOTHER OF FT AND TFL1 (MFT) genes, encoding phosphatidylethanolamine-binding proteins regulate germination in many species by the ABA-mediated pathway. ABI3, ABI5, and RGL2 regulate MFT by establishing a negative feedback loop in the ABA signaling pathway to modulate the ABA and GA signaling and to stimulate embryo growth during germination in Arabidopsis (Fig. 4) [173]. ABI4 reduces MFT gene expression through its effect on ABA, which promotes MFT itself expression, all these indicate the feedback between MFT and ABA signaling [173]. However, later studies revealed that AtMFT inhibits the germination in freshly mature seeds, while reduces the dormancy in after-ripened seeds [174]. In wheat, TaMFT acts as a repressor for seed germination, and a high level of TaMFT expression is correlated with a low germination index. In rice, the OsMFT2 gene plays a function in the regulation of seed germination through interacting with OsbZIP23/66/72 and the ABA-mediated pathway [175]. The above results indicate that MFT may regulate different seed developmental stages with diverse mechanisms and through participating in the antagonism between ABA and GA.
The phytochrome A (PHYA) and PHYB mediated photo-signal are important for seed dormancy and germination regulation, in which PHYTOCHROM-INTERACTING FACTOR1 (PIF1) plays a downstream and vital role. Previous work has shown that ABI3 expression is induced under PHYB and, in turn, ABI3 controls expression of ABA-response related genes including ABI5 [176]. Whereas under light conditions it activated by PHYA, the pattern of expression of ABI4 is opposite to that of ABI3 and ABI5 in both Arabidopsis seed dormancy [177], [178] and Aethionema arabicum light-dependent seed germination [179]. Further study indicated that ABI4 promotes PHYA-dependent germination and inhibits ABA accumulation and MFT gene expression in Arabidopsis [180]. Interestingly, PIF1 is shown to regulate GIBBERELLIN 3-OXIDASE2 (GA3OX2) and GA content. Reciprocally, PIF1 inhibits the transcriptional activity and DNA-binding ability of REVEILLE1 (RVE1), while, RVE1 stimulates the PIF1 DNA binding capability to modify ABI3 expression. As a result, PIF1 and RVE1 coordinately work as a feedback loop to regulate seed germination [181], which is also achieved dependent on the antagonism between ABA and GA. The PHYTOCHROME INTERACTING FACTOR 3-LIKE 5 (PIL5), a basic helix loop helix, exhibits a significant function in germination through phytochrome [182]. In pil5 mutant, the expression of ABA and GA metabolism genes was disturbed meanwhile a defect in GA signaling was also detected. Moreover, the PIL5 acts as an RNA binding protein, activated through phytochrome, to influence the ABA and GA metabolism by directly activating the SOMNUS (SOM) gene transcription [183], [184]. Both bHLH transcription factors, SPATULA (SPT) and PIL5 inhibit GA synthesis genes such as GA3OX1/2 and directly activate the GA catabolism gene (GA2ox2), thereby preventing germination [174], [185], [186]. Furthermore, SPT controls the germination by repressing the expression of ABI4 and RGA and promoting the expression of ABI5 and RGL3 [174]. However, PIL5 deactivates the ABA catabolism gene (CYP707A2) and positively regulates ABA biosynthesis genes (ABA1, NCED6/9) [19] to inhibit the germination, which suggests that PIL5 and SPT functions as a crosslink and fundamental hub during the antagonism of ABA and GA in the context of light condition.
Additionally, DOG1 functions downstream of PIL5 and increased expression of it inhibit the GA biosynthesis and activate ABI3 and ABI5 to control seed dormancy and germination [187]. A study demonstrated that a Dof-type transcription factor DOF AFFECTING GERMINATION 1 (DAG1) functions downstream of PIL5 to inhibit the synthesis of GA by binding with the promoter of GA3OX1, meanwhile, the dag1 mutant exhibited the up-regulated expression of ABA catabolism gene and down-regulated expression of ABA biosynthesis genes [186]. The above results indicate that the antagonistic role of ABA and GA during seed germination is also regulated by some factors activated by light signals such as PIF1 and PIL5 exemplifying that seed germination is also regulated partially by light-mediated pathway.
Besides GA, ABA also interacts with ET by the regulation of important ET biosynthesis and signaling genes such as 1-AMINOCYCLOPROPANE-1-CARBOXYLIC OXIDASE (ACO) and 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACIDSYNTHASE (ACS), and ETHYLENE RESPONSE FACTOR 11 (ERF11) to regulate the ABA-ET mediated seed ripening [132], [188], [189], [190], [191]. Mutation in ERA3 (enhanced response to ABA3) belonging to ETHYLENE INSENSITIVE2 locus showed increased sensitivity to ABA, which illustrated that ET is a negative regulator of ABA [192], [193]. In tomato, the ethylene response factor (ERF) Pti4 is involved in the regulation of seed germination by mediating ABA synthesis and signaling positively [194]. In addition, ETR1/2 and SNL1/2 regulate the ABA-ET crosstalk between dormancy and germination [115], [131]. Thus, the crosstalk between ABA and ET is also important in maintaining the hormonal level of each other for finalizing decisions on dormancy and germination (Fig. 4) [195], [196].
Glucose-6-phosphate dehydrogenase (G6PDH) plays a key role in reactive oxygen species (ROS) scavenging as the supply of NADPH. A study found that a null mutant g6pd5 is more sensitive to ABA during seed germination, whereas over-expression of G6PD5 showed hyposensitive to ABA compared to WT. Furthermore, it is found that G6PD5 restrain the expression of ABI5 to repress the ABA signaling in seed germination [197]. GLUTATHIONE S-TRANSFERASE (GST) plays pivotal roles in redox associated processes, metabolism, and detoxification in plants. AtGSTU7, a member of GST, whose null mutant (atgstu7) showed hyposensitivity to ABA in germinating seeds dependent on ABI3 [140]. These indicate a potential correlation between ROS and ABA in seed germination regulation. Moreover, a study reported that phytohormone salicylic acid (SA) together with hydrogen peroxide (H2O2) up-regulated transcription of both the GA biosynthesis gene ZmGA20ox1 and the ABA catabolism gene ZmCYP707A2, while down-regulated the expression of the GA catabolism gene ZmGA2ox1 [198], indicating the interplay among SA, ROS, ABA and GA.
Furthermore, ABA works antagonistically with auxin to regulate developmental processes and to contribute to the survival of seeds [199], [200]. For instance, the core ABA signaling gene ABI3 is an auxin-regulated, ABRE-based transcription factor that plays important role in seed dormancy [152]. An intensive study showed that through recruiting auxin responsive factors 10 (ARF10) and ARF16 with ABI3, auxin regulates the seed dormancy in synergy with ABA [201]. Brassinosteroids (BRs) play a critical antagonistic function in the seed germination inhibition of ABA [202], [203]. The advanced study showed that Glycogen Synthase Kinase 3-like kinase BRASSINOSTEROID INSENSITIVE2 (BIN2), a critical repressor of BR signaling, interacts with ABI5, and functions upstream of ABI5 to positively regulate ABA responses during seed germination and post - germinative growth. Accordingly, BRs repress the BIN2-ABI5 cascade to antagonize ABA-inhibited seed germination and seedling establishment (Fig. 4) [204]. All these progresses uncover some mask of the interaction of ABA and other hormones (e.g., SA, auxin, BR) in the seed development.
Concluding remarks and future prospects
ABA is the most important hormone and shows versatile roles in seed development as well as the seed germination. Plants synthesize their ABA through indirect pathways in embryo and endosperm during seed development which accumulates continuously in seed late maturation. For many years, the functions of ABA have been studied comprehensively, in which metabolism and signaling pathways were focused to understand the regulation of different traits in seed development. Some important proteins involved in the different stages of ABA metabolism were identified (e.g., ZEP, ABAs, NCEDs, AAOs, and CYP707As), most of which usually display specific roles in special developmental stages, but how they are regulated differentially and specifically is unknown. Moreover, lots of downstream signaling-related genes were identified, in which some PYR/PYLs, PP2Cs, SnRK2s, and components in LAFL hub show vital roles in multiple developmental stages of seed. Firstly, PYR/PYLs are responsible to accept the ABA signal redundantly. After that, the PP2C activity is inhibited by dephosphorylation to activate downstream SnRK2s proteins and several other transcription factors including ABIs and bZIPs that play crucial roles in many processes during seed development such as accumulation of seed storage products, seed maturation, seed de-greening, desiccation-tolerant acquirement, maintenance and induction of primary dormancy and germination (Table 1). The finding of the new ABA signaling terminator – ABT further enriched the understanding of the ABA signaling pathway. So, both ABA synthesis and signaling all show very complicated networks in plant development. Here, we systematically summarize the updated progresses in ABA synthesis and ABA signaling regulation, as well as their interaction in seed, which still needs much work to explore the detailed regulators and intrinsic mechanisms.
Table 1.
Associated ABA metabolism (synthesis and catabolism) and response genes in this paper.
| Gene Name | Protein/Enzyme | Mutant | Mutant phenotype | Specie | Reference |
|---|---|---|---|---|---|
| ABA Biosynthesis | |||||
| ABA1 | Zeaxanthin epoxidase (ZEP) | aba1, vp2/5/7/9 | Reduced dormancy | Arabidopsis, Maize, Tobacco | [104] |
| ABA2 | Short-chain dehydrogenase reductase (AB-SDR) | aba2 | Reduced dormancy | Arabidopsis | [31], [37] |
| ABA2 | Zeaxanthin epoxidase (ZEP) | aba2 | Reduced dormancy | Tobacco | [37] |
| VP14 | 9-cis Epoxycarotenoid dioxygenase | vp14 | Reduced dormancy | Maize | [35], [205] |
| NCED | 9-cis Epoxycarotenoid dioxygenase | nced1-9 | Reduced dormancy | Arabidopsis, Bean, Brachypodium distachyon | [44] |
| AAO3 | Aldehyde oxidase 3 | aao3-1 | Slightly reduced dormancy | Arabidopsis | [47] |
| ABA Catabolism | |||||
| CYP707A2 | ABA 8′-hydroxylase | cyp707a2-1/2 | Enhanced dormancy | Arabidopsis | [27] |
| CYP707A1 | ABA 8′-hydroxylase | cyp707a1 | Enhanced dormancy | Arabidopsis | [28], [206] |
| ABA signaling components | |||||
| PYR, PYL/RCAR | ABA receptors | Pyls | Reduced dormancy and ABA sensitivity | Arabidopsis , Rice | [29], [30] |
| ABI1 | Protein phosphatase 2C | abi1-1 | Reduced dormancy and ABA sensitivity | Arabidopsis | [207], [208], [209] |
| ABI2 | Protein phosphatase 2C | abi2-1 | Reduced dormancy and ABA sensitivity | Arabidopsis | [209], [210], [211] |
| AHG1 | Protein phosphatase 2C | ahg1-1/2/3/4/5 | Enhanced dormancy and ABA sensitivity | Arabidopsis | [54], [55] |
| AHG3 | Protein phosphatase 2C | Ahg3-1/2 | Enhanced dormancy and ABA sensitivity | Arabidopsis | [212], [213] |
| SnRK2s | Protein kinase | snrk2.2, 2.3, 2.6 triple mutant | Green seed, Reduced dormancy and ABA sensitivity | Arabidopsis | [214], [215] |
| Transcription factors | |||||
| ABI3/VP1 | B3 domain | abi3-1 to 17, vp1 | Green seed, Reduced dormancy and ABA sensitivity | Arabidopsis, Rice, Maize | [60], [216], [217] |
| ABI4 | ERF/APETALA domain | abi4-1 | ABA insensitive | Arabidopsis | [75], [218] |
| ABI5 | ABF, bZIP | abi5-1/7/8 | ABA insensitive | Arabidopsis | [60], [219] |
| FUS3 | B3 domain | fus3-3/8 | Reduced dormancy | Arabidopsis | |
| LEC1 | B3 domain | lec1-1/2 | Reduced dormancy | Arabidopsis | [68], [220] |
| LEC2 | B3 domain | lec2-1/3 | Reduced dormancy | Arabidopsis | [26], [220] |
Although ABA shows versatile roles in plant development, we focus on the biological roles and underlying mechanisms of ABA in seed-related straits. In seed development, dormancy is a decisive factor influencing seed vigor and plant propagation. Alteration in the state of dormancy (dormant to non-dormant) is an active process that involves variations in the expression of genes in after-ripening dry seeds, in this period, ABA-associated ways play crucial roles. The level of dormancy is severely poor in ABA biosynthesis and signaling mutants, indicating the direct and important function of ABA in seed dormancy maintenance and induction. Both ABA catabolism and stability between ABA⁄GA crosstalk both put an impact on the level of seed dormancy. So, the exploration of the detailed interaction between ABA and GA could also facilitate the illumination of mechanisms of ABA in seed development including dormancy. From numerous studies, it is proved that ABA activates some key proteins (e.g., ABI3, LEC1, LEC2, and FUS3) comprising a LAFL hub that plays roles in ABA metabolism, showing the reciprocal effect between ABA signaling and syntheses, but, the complete work model of these proteins is not clear yet. More comprehensive biochemical and genetic analysis for the key genes would be helpful for the elucidation of the detailed interplay of ABA synthesis and signaling in seed development.
The crosstalks between ABA and other phytohormones such as GA, ET, and auxin are also important for finalizing decisions of the seed dormancy, germination, or seedling establishment [133]. Studies about light signal-related factors (e.g., HY5 and PIF1) have provided some clues for interpretation of the interaction between ABA and exogenous signal in seed development. Here, we provide a comprehensive and updated network for crosstalks between ABA and other phytohormones in seed development, which indicates the bona fide case that it is common of pleiotropism and the reciprocal regulation between different factors or signals. But, how the temperature, humidity and other exogenous factors (e.g., oxygen, calcium, and pollutants) influence the ABA/GA balance or the interaction between ABA and other hormones is still in a blank.
During post-germination growth, ABI3 activated by ABA activates JMJ30 and accelerates SnRK2.8 expression through H3K27me3 demethylation, promoting downstream ABI3 expression to enhance post-germination growth as a feedback loop, which provides some hints to reveal the relationship of ABA and epigenetic modification machinery to improve the network of ABA in seed straits as well as shed light for the regulation of ABA pathway in genomic level. Due to the technology limitation of ABA visualization, the regulation of ABA metabolism has not been well studied, especially at cellular and tissue levels. Along with the advance of technology, uncovering the profiles of GA and ABA in different tissues during seed development would provide direct evidence for the antagonism between them. Compared to fresh harvest seeds, the seeds after-ripening or stratification display decreased dormancy and increased seed germination, how activation of GA biosynthesis impels the break of dormancy is still an open question during this period. Expectedly, the low temperature and high humidity play some roles in GA activation, but the underlying factors and mechanisms are still unclear.
Collectively, although great progress of roles of ABA in understanding the regulation of seed development has been done, some open questions remain unanswered. For example, it is still unclear how ABA regulates downstream genes such as components of LAFL, and the function of ABA signaling factors in response to ABA against dormancy. Many genes influenced dormancy, but, it is unknown how these genes interact with each other in detail. In the future, studies should be focused on the questions discussed above to improve the understandings of the mechanisms by which ABA and genetic factors regulate and maintain seed dormancy and germination, which would be conducive to a better presentation of the system of ABA function and continuous agricultural productivity.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Declaration of Competing Interest
All the authors in the manuscript have no conflicts of interest.
Acknowledgments
Acknowledgments
We apologize to colleagues whose work we do not cite here due to space limitations. This work is supported by the the National Natural Science Foundation of China (31621005, 31690093, and 32072022), Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences, Central Public-interest Scientific Institution Basal Research Fund (1610162021008), and Zhengzhou University (32410196).
Author’s contribution
Z.W. and F.L. conceived the manuscript; F.A, G.Q, and Z.W. drafted the manuscript and prepared the figures. All authors revised and approved the final manuscript.
Biographies

Faiza Ali was born and raised in Multan, Punjab, Pakistan. She has completed her Bachelor of Science (B.Sc.) in 2012 and subsequently received her Master of Science (M.Sc.) degree in 2014 in Plant Breeding and Genetics from Bahauddin Zakariya University, Multan, Punjab, Pakistan. She is currently pursuing her Doctorate in Cotton Biochemistry and Molecular Biology at the Institute of Cotton Research (ICR) Anyang, Chinese Academy of Agricultural Sciences, Beijing, China. Here she is working on genes involved in seed germination and vigor in Arabidopsis and cotton. She has four published research articles.

Ghulam Qanmber was born in 1991 and raised in Multan, Punjab, Pakistan. He obtained his B.Sc. (Hons) from Bahauddin Zakariya University (BZU) Multan and later M.Sc. (Hons) in Plant Breeding and Genetics from Bahauddin Zakariya University (BZU) Multan. He completed his Ph.D. from the Chinese Academy of Agricultural Sciences (CAAS), China in Plant Breeding and Genetics in 2019. During his Ph.D. he was awarded Outstanding Student of the Year. He has more than 20 peer-reviewed research articles and review papers in well-reputed journals. Nowadays, his current interest is focused on seed development, germination, dormancy, and plant hormones and signaling pathways regulating seed development and growth.

Dr. Zhi Wang, professor and doctoral supervisor in Institute of cotton research, Chinese agricultural academy of sciences. He gained his doctorate of nature science at the Institute of Botany, Chinese academy of sciences in 2010, he experienced two years of postdoctoral study at the Beijing Academy of Agriculture and Forestry Sciences. From 2009 to 2017, Zhi Wang worked at the Institute of Botany, Chinese academy of sciences, and obtained the associate professor position in 2017; from 2017 to now, Dr. Zhi Wang was employed in the Institute of cotton research, Chinese agricultural academy of sciences as a distinguished fellow. Since 2009, Zhi Wang has focused on seed development and trichome development studies for more than ten years. Dr. Wang has identified the molecular mechanisms and correlation of epigenetic modification (e.g., histone deacetylation) and phytohormones (e.g., ABA, ethylene, auxin) in seed dormancy and seed germination after post-maturation. Moreover, he is focusing on the interaction and molecular mechanism between seed development and seed coat trichome development in cotton. He has been supported by some funders such as the National Natural Science Foundation of China (No.3090106 and No.32072022).

Dr. Fuguang Li, Professor in Institute of Cotton Research, Chinese Academy of Agricultural Sciences. He is a plant geneticist by training, received his BSc degree of Science (1989) from the China Agricultural University, China, and his Ph.D. in Molecular Biology (2003) from the Graduate School of Chinese Academy of Agricultural Sciences, China. Dr. Li has established a system of a large scale of transformation on cotton. Using the purified lines from CCRI 24, a system of a large scale of transformation on cotton has been established, this makes cotton become a crop which could be genetically engineered by a large scale of candidate genes. 156 candidate genes came from 24 labs in China have been validated in Dr. Li's lab. As the leading scientist of the innovative team of molecular genetic improvement of cotton, he also researched the mechanism of cotton fiber development, seed development, drought tolerance and provided many transgenic materials to the breeder in recent 5 years. He has been funded by many funders such as the National Science Fund for Distinguished Young Scholars (Grant no. 31125020) and the Major Program of Joint Funds (Sinkiang) of the National Natural Science Foundation of China (Grant No.U1303282) so on.
Footnotes
Peer review under responsibility of Cairo University.
Contributor Information
Fuguang Li, Email: aylifug@caas.cn.
Zhi Wang, Email: wangzhi01@caas.cn.
References
- 1.Graeber K., Nakabayashi K., Miatton E., Leubner-Metzger G., Soppe W.J. Molecular mechanisms of seed dormancy. Plant, Cell Environ. 2012;35(10):1769–1786. doi: 10.1111/j.1365-3040.2012.02542.x. [DOI] [PubMed] [Google Scholar]
- 2.Qanmber G., Lu L., Liu Z., Yu D., Zhou K., Huo P., et al. Genome-wide identification of GhAAI genes reveals that GhAAI66 triggers a phase transition to induce early flowering. J Exp Bot. 2019;70(18):4721–4736. doi: 10.1093/jxb/erz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faiza A., Qanmber G., Yonghui L., Shuya M., Lili L., Zuoren Y., et al. Genome-wide identification of Gossypium INDETERMINATE DOMAIN genes and their expression profiles in ovule development and abiotic stress responses. J Cotton Res. 2019 [Google Scholar]
- 4.Koornneef M., Bentsink L., Hilhorst H. Seed dormancy and germination. Curr Opin Plant Biol. 2002;5(1):33–36. doi: 10.1016/s1369-5266(01)00219-9. [DOI] [PubMed] [Google Scholar]
- 5.Righetti K., Vu J.L., Pelletier S., Vu B.L., Glaab E., Lalanne D., et al. Inference of longevity-related genes from a robust coexpression network of seed maturation identifies regulators linking seed storability to biotic defense-related pathways. Plant Cell. 2015;27(10):2692–2708. doi: 10.1105/tpc.15.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finch-Savage W.E., Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171(3):501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- 7.Bewley J.D. Seed germination and dormancy. Plant Cell. 1997;9(7):1055. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donohue K., Rubio de Casas R., Burghardt L., Kovach K., Willis C.G. Germination, postgermination adaptation, and species ecological ranges. Annu Rev Ecol Evol Syst. 2010;41:293–319. [Google Scholar]
- 9.Baskin C.C., Seeds Baskin JM. Ecology, Biogeography, and Evolution of Dormancy and Germination. 1998 6/10/1998. [Google Scholar]
- 10.Hilhorst H.W. A critical update on seed dormancy. I. Primary dormancy. Seed Sci Res. 1995;5(2):61–73. [Google Scholar]
- 11.Kucera B., Cohn M.A., Leubner-Metzger G. Plant hormone interactions during seed dormancy release and germination. Seed Sci Res. 2005;15(4):281–307. [Google Scholar]
- 12.Holdsworth M.J., Finch-Savage W.E., Grappin P., Job D. Post-genomics dissection of seed dormancy and germination. Trends Plant Sci. 2008;13(1):7–13. doi: 10.1016/j.tplants.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Yang C., Li L. Hormonal regulation in shade avoidance. Front Plant Sci. 2017;8:1527. doi: 10.3389/fpls.2017.01527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kendall S.L., Hellwege A., Marriot P., Whalley C., Graham I.A., Penfield S. Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell. 2011;23(7):2568–2580. doi: 10.1105/tpc.111.087643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shu K., Luo X., Meng Y., Yang W. Toward a molecular understanding of abscisic acid actions in floral transition. Plant Cell Physiol. 2018;59(2):215–221. doi: 10.1093/pcp/pcy007. [DOI] [PubMed] [Google Scholar]
- 16.Shu K., Liu X.D., Xie Q., He Z.H. Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant. 2016;9(1):34–45. doi: 10.1016/j.molp.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Wang H., Zhang Y., Xiao N., Zhang G., Wang F., Chen X., et al. Rice GERMIN-LIKE PROTEIN 2–1 functions in seed dormancy under the control of abscisic acid and gibberellic acid signaling pathways. Plant Physiol. 2020;183(3):1157–1170. doi: 10.1104/pp.20.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson R.L., Kim H., Bakshi A., Binder B.M. The Ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of arabidopsis during salt stress. Plant Physiol. 2014;165(3):1353–1366. doi: 10.1104/pp.114.241695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finkelstein R., Reeves W., Ariizumi T., Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 20.Vishal B., Kumar P.P. Regulation of seed germination and abiotic stresses by gibberellins and abscisic acid. Front Plant Sci. 2018;9(838) doi: 10.3389/fpls.2018.00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kermode A.R. Role of abscisic acid in seed dormancy. J Plant Growth Regul. 2005;24(4):319–344. [Google Scholar]
- 22.Nonogaki H. Seed biology updates – highlights and new discoveries in seed dormancy and germination research. Front Plant Sci. 2017;8(524) doi: 10.3389/fpls.2017.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodríguez-Gacio Md.C., Matilla-Vázquez M.A., Matilla A.J. Seed dormancy and ABA signaling: the breakthrough goes on. Plant Signaling Behav. 2009;4(11):1035–1048. doi: 10.4161/psb.4.11.9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nambara E., Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 25.Vishwakarma K., Upadhyay N., Kumar N., Yadav G., Singh J., Mishra R.K., et al. Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front Plant Sci. 2017;8(161) doi: 10.3389/fpls.2017.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holdsworth M.J., Bentsink L., Soppe W.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008;179(1):33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- 27.Kushiro T., Okamoto M., Nakabayashi K., Yamagishi K., Kitamura S., Asami T., et al. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. The EMBO journal. 2004;23(7):1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamoto M., Kuwahara A., Seo M., Kushiro T., Asami T., Hirai N., et al. CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006;141(1):97–107. doi: 10.1104/pp.106.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324(5930):1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 30.Park S.-Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frey A., Godin B., Bonnet M., Sotta B., Marion-Poll A. Maternal synthesis of abscisic acid controls seed development and yield in Nicotiana plumbaginifolia. Planta. 2004;218(6):958–964. doi: 10.1007/s00425-003-1180-7. [DOI] [PubMed] [Google Scholar]
- 32.Kermode AR. Regulatory mechanisms in the transition from seed development to germination: interactions between the embryo and the seed environment. Seed development and germination: Routledge; 2017. p. 273-332.
- 33.Walton D.C. In: Plant Hormones and their Role in Plant Growth and Development. Davies P.J., editor. Springer, Netherlands; Dordrecht: 1987. Abscisic Acid Biosynthesis and Metabolism; pp. 113–131. [Google Scholar]
- 34.Marion-Poll A., Leung J. Abscisic acid synthesis, metabolism and signal transduction. Plant Hormone Signalling. Ann Plant Rev. 2006;24:1–35. [Google Scholar]
- 35.Schwartz S.H., Tan B.C., Gage D.A., Zeevaart J.A.D., McCarty D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;276(5320):1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- 36.Audran C., Liotenberg S., Gonneau M., North H., Frey A., Tap-Waksman K., et al. Localisation and expression of zeaxanthin epoxidase mRNA in Arabidopsis in response to drought stress and during seed development. Funct Plant Biol. 2001;28(12):1161–1173. [Google Scholar]
- 37.Marin E., Nussaume L., Quesada A., Gonneau M., Sotta B., Hugueney P., et al. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. Embo j. 1996;15(10):2331–2342. [PMC free article] [PubMed] [Google Scholar]
- 38.Agrawal G.K., Yamazaki M., Kobayashi M., Hirochika R., Miyao A., Hirochika H. Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel ostatc gene. Plant Physiol. 2001;125(3):1248–1257. doi: 10.1104/pp.125.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reid J.B. Phytohormone mutants in plant research. J Plant Growth Regul. 1990;9(1):97. [Google Scholar]
- 40.North H.M., De Almeida A., Boutin J.P., Frey A., To A., Botran L., et al. The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. The Plant J Cell Mol Biol. 2007;50(5):810–824. doi: 10.1111/j.1365-313X.2007.03094.x. [DOI] [PubMed] [Google Scholar]
- 41.Tan B.C., Schwartz S.H., Zeevaart J.A., McCarty D.R. Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci. 1997;94(22):12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin X., Zeevaart J.A. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 2002;128(2):544–551. doi: 10.1104/pp.010663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrero J.M., Jacobsen J.V., Talbot M.J., White R.G., Swain S.M., Garvin D.F., et al. Grain dormancy and light quality effects on germination in the model grass Brachypodium distachyon. New Phytol. 2012;193(2):376–386. doi: 10.1111/j.1469-8137.2011.03938.x. [DOI] [PubMed] [Google Scholar]
- 44.Tan B.C., Joseph L.M., Deng W.T., Liu L., Li Q.B., Cline K., et al. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J Cell Molecular Biol. 2003;35(1):44–56. doi: 10.1046/j.1365-313x.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 45.Seo M., Koshiba T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002;7(1):41–48. doi: 10.1016/s1360-1385(01)02187-2. [DOI] [PubMed] [Google Scholar]
- 46.Taylor I.B., Linforth R.S.T., Al-Naieb R.J., Bowman W.R., Marples B.A. The wilty tomato mutants flacca and sitiens are impaired in the oxidation of ABA-aldehyde to ABA. Plant, Cell Environ. 1988;11(8):739–745. [Google Scholar]
- 47.Seo M., Aoki H., Koiwai H., Kamiya Y., Nambara E., Koshiba T. Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol. 2004;45(11):1694–1703. doi: 10.1093/pcp/pch198. [DOI] [PubMed] [Google Scholar]
- 48.Seo M., Hanada A., Kuwahara A., Endo A., Okamoto M., Yamauchi Y., et al. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J Cell Molecular Biol. 2006;48(3):354–366. doi: 10.1111/j.1365-313X.2006.02881.x. [DOI] [PubMed] [Google Scholar]
- 49.Himmelbach A., Yang Y., Grill E. Relay and control of abscisic acid signaling. Curr Opin Plant Biol. 2003;6(5):470–479. doi: 10.1016/s1369-5266(03)00090-6. [DOI] [PubMed] [Google Scholar]
- 50.Shinozaki K., Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58(2):221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 51.Miao C., Xiao L., Hua K., Zou C., Zhao Y., Bressan R.A., et al. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc Natl Acad Sci. 2018;115(23):6058–6063. doi: 10.1073/pnas.1804774115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y., Zhang Z., Gao J., Wang P., Hu T., Wang Z., et al. Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-independent SnRK2 activity. Cell Rep. 2018;23(11) doi: 10.1016/j.celrep.2018.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida T., Nishimura N., Kitahata N., Kuromori T., Ito T., Asami T., et al. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006;140(1):115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang K., He J., Zhao Y., Wu T., Zhou X., Ding Y., et al. EAR1 negatively regulates ABA signaling by enhancing 2C protein phosphatase activity. Plant Cell. 2018;30(4):815–834. doi: 10.1105/tpc.17.00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baek D., Kim M.C., Kumar D., Park B., Cheong M.S., Choi W., et al. AtPR5K2, a PR5-like receptor kinase, modulates plant responses to drought stress by phosphorylating protein phosphatase 2Cs. Front Plant Sci. 2019;10:1146. doi: 10.3389/fpls.2019.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishimura N., Tsuchiya W., Moresco J.J., Hayashi Y., Satoh K., Kaiwa N., et al. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat Commun. 2018;9(1):2132. doi: 10.1038/s41467-018-04437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Y., Chan Z., Xing L., Liu X., Hou Y.-J., Chinnusamy V., et al. The unique mode of action of a divergent member of the ABA-receptor protein family in ABA and stress signaling. Cell Res. 2013;23(12):1380–1395. doi: 10.1038/cr.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z., Ren Z., Cheng C., Wang T., Ji H., Zhao Y., et al. Counteraction of ABA-mediated inhibition of seed germination and seedling establishment by ABA signaling terminator in arabidopsis. Mol Plant. 2020;13(9):1284–1297. doi: 10.1016/j.molp.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 59.Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., et al. Three arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50(7):1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- 60.Lopez-Molina L, Mongrand S, Chua N-H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in <em>Arabidopsis</em>. Proceedings of the National Academy of Sciences. 2001;98(8):4782. [DOI] [PMC free article] [PubMed]
- 61.Nakamura S., Lynch T.J., Finkelstein R.R. Physical interactions between ABA response loci of Arabidopsis. Plant J. 2001;26(6):627–635. doi: 10.1046/j.1365-313x.2001.01069.x. [DOI] [PubMed] [Google Scholar]
- 62.Zhao H., Nie K., Zhou H., Yan X., Zhan Q., Zheng Y., et al. ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol. 2020;228(2):596–608. doi: 10.1111/nph.16713. [DOI] [PubMed] [Google Scholar]
- 63.Gazzarrini S., Tsuchiya Y., Lumba S., Okamoto M., McCourt P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell. 2004;7(3):373–385. doi: 10.1016/j.devcel.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 64.Kwong R.W., Bui A.Q., Lee H., Kwong L.W., Fischer R.L., Goldberg R.B., et al. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell. 2003;15(1):5–18. doi: 10.1105/tpc.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jia H., McCarty D.R., Suzuki M. Distinct roles of LAFL network genes in promoting the embryonic seedling fate in the absence of VAL repression. Plant Physiol. 2013;163(3):1293–1305. doi: 10.1104/pp.113.220988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jia H., Suzuki M., McCarty D.R. Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks. Wiley interdisciplinary reviews Develop Biol. 2014;3(1):135–145. doi: 10.1002/wdev.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Horstman A., Li M., Heidmann I., Weemen M., Chen B., Muino J.M., et al. The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 2017;175(2):848–857. doi: 10.1104/pp.17.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki M., Latshaw S., Sato Y., Settles A.M., Koch K.E., Hannah L.C., et al. The maize Viviparous8 locus, encoding a putative ALTERED MERISTEM PROGRAM1-like peptidase, regulates abscisic acid accumulation and coordinates embryo and endosperm development. Plant Physiol. 2008;146(3):1193–1206. doi: 10.1104/pp.107.114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dai D., Tong H., Cheng L., Peng F., Zhang T., Qi W., et al. Maize Dek33 encodes a pyrimidine reductase in riboflavin biosynthesis that is essential for oil-body formation and ABA biosynthesis during seed development. J Exp Bot. 2019;70(19):5173–5187. doi: 10.1093/jxb/erz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Delmas F., Sankaranarayanan S., Deb S., Widdup E., Bournonville C., Bollier N., et al. ABI3 controls embryo degreening through Mendel's I locus. Proc Natl Acad Sci USA. 2013;110(40):E3888–E3894. doi: 10.1073/pnas.1308114110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M.M., Seki M., et al. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17(12):3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Finkelstein R.R., Gampala S.S., Rock C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14(Suppl 1):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mendes A., Kelly A.A., van Erp H., Shaw E., Powers S.J., Kurup S., et al. bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase3. Plant Cell. 2013;25(8):3104–3116. doi: 10.1105/tpc.113.116343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zinsmeister J., Lalanne D., Terrasson E., Chatelain E., Vandecasteele C., Vu B.L., et al. ABI5 Is a Regulator of Seed Maturation and Longevity in Legumes. Plant Cell. 2016;28(11):2735–2754. doi: 10.1105/tpc.16.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Söderman E.M., Brocard I.M., Lynch T.J., Finkelstein R.R. Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 2000;124(4):1752–1765. doi: 10.1104/pp.124.4.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao Y., Ai X., Wang M., Xiao L., Xia G. A putative pyruvate transporter TaBASS2 positively regulates salinity tolerance in wheat via modulation of ABI4 expression. BMC Plant Biol. 2016;16(1):109. doi: 10.1186/s12870-016-0795-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li C., Yue Y., Chen H., Qi W., Song R. The ZmbZIP22 transcription factor regulates 27-kD γ-Zein gene transcription during maize endosperm development. Plant Cell. 2018;30(10):2402–2424. doi: 10.1105/tpc.18.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dong Q., Xu Q., Kong J., Peng X., Zhou W., Chen L., et al. Overexpression of ZmbZIP22 gene alters endosperm starch content and composition in maize and rice. Plant Sci. 2019;283:407–415. doi: 10.1016/j.plantsci.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 79.Bies-Etheve N., da Silva Conceicao A, Giraudat J., Ome Koornneef M., Léon-Kloosterziel K., et al. Importance of the B2 domain of the Arabidopsis ABI3 protein for Em and 2S albumin gene regulation. Plant Mol Biol. 1999;40(6):1045–1054. doi: 10.1023/a:1006252512202. [DOI] [PubMed] [Google Scholar]
- 80.Zheng Z., Xu X., Crosley R.A., Greenwalt S.A., Sun Y., Blakeslee B., et al. The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiol. 2010;153(1):99–113. doi: 10.1104/pp.109.150789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parcy F., Valon C., Kohara A., Miséra S., Giraudat J. The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell. 1997;9(8):1265–1277. doi: 10.1105/tpc.9.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Santos Mendoza M., Dubreucq B., Miquel M., Caboche M., Lepiniec L. LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 2005;579(21):4666–4670. doi: 10.1016/j.febslet.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 83.Kagaya Y., Okuda R., Ban A., Toyoshima R., Tsutsumida K., Usui H., et al. Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant Cell Physiol. 2005;46(2):300–311. doi: 10.1093/pcp/pci031. [DOI] [PubMed] [Google Scholar]
- 84.Mu J., Tan H., Zheng Q., Fu F., Liang Y., Zhang J., et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008;148(2):1042–1054. doi: 10.1104/pp.108.126342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.To A., Joubès J., Barthole G., Lécureuil A., Scagnelli A., Jasinski S., et al. WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell. 2012;24(12):5007–5023. doi: 10.1105/tpc.112.106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y.Q., Lu X., Zhao F.Y., Li Q.T., Niu S.L., Wei W., et al. Soybean GmDREBL increases lipid content in seeds of transgenic Arabidopsis. Sci Rep. 2016;6:34307. doi: 10.1038/srep34307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wehmeyer N., Vierling E. The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol. 2000;122(4):1099–1108. doi: 10.1104/pp.122.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maia J., Dekkers B.J.W., Dolle M.J., Ligterink W., Hilhorst H.W.M. Abscisic acid (ABA) sensitivity regulates desiccation tolerance in germinated Arabidopsis seeds. New Phytol. 2014;203(1):81–93. doi: 10.1111/nph.12785. [DOI] [PubMed] [Google Scholar]
- 89.Bewley JD, Bradford KJ, Hilhorst HWM, Nonogaki H. Synthesis of Storage Reserves. Seeds: Physiology of Development, Germination and Dormancy, 3rd ed. New York, NY: Springer New York; 2013. p. 85-131.
- 90.West M., Yee K.M., Danao J., Zimmerman J.L., Fischer R.L., Goldberg R.B., et al. LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in arabidopsis. Plant Cell. 1994;6(12):1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jo L., Pelletier J.M., Hsu S.-W., Baden R., Goldberg R.B., Harada J.J. Combinatorial interactions of the LEC1 transcription factor specify diverse developmental programs during soybean seed development. Proc Natl Acad Sci. 2020;117(2):1223–1232. doi: 10.1073/pnas.1918441117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Curaba J., Moritz T., Blervaque R., Parcy F., Raz V., Herzog M., et al. AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 2004;136(3):3660–3669. doi: 10.1104/pp.104.047266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen H., Zhang J., Neff M.M., Hong S.W., Zhang H., Deng X.W., et al. Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci USA. 2008;105(11):4495–4500. doi: 10.1073/pnas.0710778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dekkers B.J.W., He H., Hanson J., Willems L.A.J., Jamar D.C.L., Cueff G., et al. The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J. 2016;85(4):451–465. doi: 10.1111/tpj.13118. [DOI] [PubMed] [Google Scholar]
- 95.Li X., Chen T., Li Y., Wang Z., Cao H., Chen F., et al. ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-TPL complex on DELAY OF GERMINATION1 expression. Plant Cell. 2019;31(4):832–847. doi: 10.1105/tpc.18.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bryant F.M., Hughes D., Hassani-Pak K., Eastmond P.J. Basic LEUCINE ZIPPER TRANSCRIPTION FACTOR67 transactivates DELAY OF GERMINATION1 to establish primary seed dormancy in Arabidopsis. Plant Cell. 2019;31(6):1276–1288. doi: 10.1105/tpc.18.00892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sall K., Dekkers B.J.W., Nonogaki M., Katsuragawa Y., Koyari R., Hendrix D., et al. DELAY OF GERMINATION 1-LIKE 4 acts as an inducer of seed reserve accumulation. Plant J. 2019;100(1):7–19. doi: 10.1111/tpj.14485. [DOI] [PubMed] [Google Scholar]
- 98.Dickie J. The ecology of seeds.: Fenner M, Thompson K. 2005. Cambridge: Cambridge University Press. £26 (softback) £55 (hardback) 260 pp. Annals of botany. 2006;97(1):151-2.
- 99.Vaughan D.A., Balázs E., Heslop-Harrison J.S. From crop domestication to super-domestication. Ann Bot. 2007;100(5):893–901. doi: 10.1093/aob/mcm224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gutierrez L., Van Wuytswinkel O., Castelain M., Bellini C. Combined networks regulating seed maturation. Trends Plant Sci. 2007;12(7):294–300. doi: 10.1016/j.tplants.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 101.Vu D.T., Velusamy V., Park E. Structure and chemical composition of wild soybean seed coat related to its permeability. Pak J Bot. 2014;46:1847–1857. [Google Scholar]
- 102.Chandra S., Yadav R., Poonia S., Pal Y., Rathod D., Gupta A., et al. Seed coat permeability studies in wild and cultivated species of soybean. Int J Curr Microbiol Appl Sci. 2017;6:2358–2363. [Google Scholar]
- 103.Kebede H., Smith J.R., Ray J.D. Identification of a single gene for seed coat impermeability in soybean PI 594619. Theor Appl Genet. 2014;127(9):1991–2003. doi: 10.1007/s00122-014-2355-2. [DOI] [PubMed] [Google Scholar]
- 104.Karssen C., Brinkhorst-Van der Swan D., Breekland A., Koornneef M. Induction of dormancy during seed development by endogenous abscisic acid: studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta. 1983;157(2):158–165. doi: 10.1007/BF00393650. [DOI] [PubMed] [Google Scholar]
- 105.Lefebvre V., North H., Frey A., Sotta B., Seo M., Okamoto M., et al. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006;45(3):309–319. doi: 10.1111/j.1365-313X.2005.02622.x. [DOI] [PubMed] [Google Scholar]
- 106.Liu F., Zhang H., Ding L., Soppe W., Xiang Y. REVERSAL OF RDO5 1, a homolog of rice seed dormancy 4, interacts with bHLH57 and controls ABA biosynthesis and seed dormancy in Arabidopsis. Plant Cell. 2020 doi: 10.1105/tpc.20.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee H.G., Lee K., Seo P.J. The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol Biol. 2015;87(4–5):371–381. doi: 10.1007/s11103-015-0283-4. [DOI] [PubMed] [Google Scholar]
- 108.Shu K., Chen Q., Wu Y., Liu R., Zhang H., Wang P., et al. ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 2016;85(3):348–361. doi: 10.1111/tpj.13109. [DOI] [PubMed] [Google Scholar]
- 109.Shu K., Zhang H., Wang S., Chen M., Wu Y., Tang S., et al. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in arabidopsis. PLoS Genet. 2013;9(6):e1003577. doi: 10.1371/journal.pgen.1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen H., Ruan J., Chu P., Fu W., Liang Z., Li Y., et al. AtPER1 enhances primary seed dormancy and reduces seed germination by suppressing the ABA catabolism and GA biosynthesis in Arabidopsis seeds. Plant J Cell Molecular Biol. 2020;101(2):310–323. doi: 10.1111/tpj.14542. [DOI] [PubMed] [Google Scholar]
- 111.Cantoro R., Crocco C.D., Benech-Arnold R.L., Rodríguez M.V. In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: possible role of this interaction in the expression of seed dormancy. J Exp Bot. 2013;64(18):5721–5735. doi: 10.1093/jxb/ert347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Griffiths J., Barrero J.M., Taylor J., Helliwell C.A., Gubler F. ALTERED MERISTEM PROGRAM 1 is involved in development of seed dormancy in Arabidopsis. PLoS ONE. 2011;6(5) doi: 10.1371/journal.pone.0020408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee S-j, Lee M.H., Kim J.-I., Kim S.Y. Arabidopsis putative MAP kinase kinase kinases Raf10 and Raf11 are positive regulators of seed dormancy and ABA response. Plant Cell Physiol. 2014;56(1):84–97. doi: 10.1093/pcp/pcu148. [DOI] [PubMed] [Google Scholar]
- 114.Nguyen Q.T.C., Lee S.J., Choi S.W., Na Y.J., Song M.R., Hoang Q.T.N., et al. Arabidopsis Raf-like kinase Raf10 is a regulatory component of core ABA aignaling. Mol Cells. 2019;42(9):646–660. doi: 10.14348/molcells.2019.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang Z., Cao H., Sun Y., Li X., Chen F., Carles A., et al. Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell. 2013;25(1):149–166. doi: 10.1105/tpc.112.108191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Utsugi S., Ashikawa I., Nakamura S., Shibasaka M. TaABI5, a wheat homolog of Arabidopsis thaliana ABA insensitive 5, controls seed germination. J Plant Res. 2020;133(2):245–256. doi: 10.1007/s10265-020-01166-3. [DOI] [PubMed] [Google Scholar]
- 117.Nonogaki H., Bassel G.W., Bewley J.D. Germination—still a mystery. Plant Sci. 2010;179(6):574–581. [Google Scholar]
- 118.Cadman C.S., Toorop P.E., Hilhorst H.W., Finch-Savage W.E. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J Cell Molecular Biol. 2006;46(5):805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- 119.Ali-Rachedi S., Bouinot D., Wagner M.-H., Bonnet M., Sotta B., Grappin P., et al. Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta. 2004;219(3):479–488. doi: 10.1007/s00425-004-1251-4. [DOI] [PubMed] [Google Scholar]
- 120.Alonso-Blanco C., Bentsink L., Hanhart C.J., Blankestijn-de Vries H., Koornneef M. Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics. 2003;164(2):711–729. doi: 10.1093/genetics/164.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schmitz N., Abrams S.R., Kermode A.R. Changes in ABA turnover and sensitivity that accompany dormancy termination of yellow-cedar (Chamaecyparis nootkatensis) seeds. J Exp Bot. 2002;53(366):89–101. [PubMed] [Google Scholar]
- 122.Weng J.-K., Ye M., Li B., Noel J.P. Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity. Cell. 2016;166(4):881–893. doi: 10.1016/j.cell.2016.06.027. [DOI] [PubMed] [Google Scholar]
- 123.Saito S., Hirai N., Matsumoto C., Ohigashi H., Ohta D., Sakata K., et al. Arabidopsis CYP707As encode (+)-abscisic acid 8'-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004;134(4):1439–1449. doi: 10.1104/pp.103.037614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gonai T., Kawahara S., Tougou M., Satoh S., Hashiba T., Hirai N., et al. Abscisic acid in the thermoinhibition of lettuce seed germination and enhancement of its catabolism by gibberellin. J Exp Bot. 2004;55(394):111–118. doi: 10.1093/jxb/erh023. [DOI] [PubMed] [Google Scholar]
- 125.Jacobsen J.V., Pearce D.W., Poole A.T., Pharis R.P., Mander L.N. Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol Plant. 2002;115(3):428–441. doi: 10.1034/j.1399-3054.2002.1150313.x. [DOI] [PubMed] [Google Scholar]
- 126.Millar A.A., Jacobsen J.V., Ross J.J., Helliwell C.A., Poole A.T., Scofield G., et al. Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. Plant J. 2006;45(6):942–954. doi: 10.1111/j.1365-313X.2006.02659.x. [DOI] [PubMed] [Google Scholar]
- 127.Chen H., Tong J., Fu W., Liang Z., Ruan J., Yu Y. The H3K27me3 demethylase RELATIVE OF EARLY FLOWERING6 suppresses seed dormancy by inducing abscisic. Acid Catabolism. 2020;184(4):1969–1978. doi: 10.1104/pp.20.01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pan J., Wang H., Hu Y., Yu D. Arabidopsis VQ18 and VQ26 proteins interact with ABI5 transcription factor to negatively modulate ABA response during seed germination. Plant J Cell Molecular Biol. 2018;95(3):529–544. doi: 10.1111/tpj.13969. [DOI] [PubMed] [Google Scholar]
- 129.Yaish M.W., El-Kereamy A., Zhu T., Beatty P.H., Good A.G., Bi Y.M., et al. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010;6(9):e1001098. doi: 10.1371/journal.pgen.1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Feng C.Z., Chen Y., Wang C., Kong Y.H., Wu W.H., Chen Y.F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J Cell Molecular Biol. 2014;80(4):654–668. doi: 10.1111/tpj.12670. [DOI] [PubMed] [Google Scholar]
- 131.Joseph M.P., Papdi C., Kozma-Bognár L., Nagy I., López-Carbonell M., Rigó G., et al. The Arabidopsis ZINC FINGER PROTEIN3 interferes with abscisic acid and light signaling in seed germination and plant development. Plant Physiol. 2014;165(3):1203–1220. doi: 10.1104/pp.113.234294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhao H., Zhang H., Cui P., Ding F., Wang G., Li R., et al. The putative E3 ubiquitin ligase ECERIFERUM9 regulates abscisic acid biosynthesis and response during seed germination and postgermination growth in arabidopsis. Plant Physiol. 2014;165(3):1255–1268. doi: 10.1104/pp.114.239699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kong D., Ju C., Parihar A., Kim S., Cho D., Kwak J.M. Arabidopsis glutamate receptor homolog3.5 modulates cytosolic Ca2+ level to counteract effect of abscisic acid in seed germination. Plant Physiol. 2015;167(4):1630–1642. doi: 10.1104/pp.114.251298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee K, Lee HG, Yoon S. The Arabidopsis MYB96 Transcription Factor Is a Positive Regulator of ABSCISIC ACID-INSENSITIVE4 in the Control of Seed Germination. 2015;168(2):677-89. [DOI] [PMC free article] [PubMed]
- 135.Yan J, Zhao C. The miR165/166 Mediated Regulatory Module Plays Critical Roles in ABA Homeostasis and Response in Arabidopsis thaliana. 2016;12(11):e1006416. [DOI] [PMC free article] [PubMed]
- 136.Wang Y., Chang H., Hu S., Lu X., Yuan C., Zhang C., et al. Plastid casein kinase 2 knockout reduces abscisic acid (ABA) sensitivity, thermotolerance, and expression of ABA- and heat-stress-responsive nuclear genes. J Exp Bot. 2014;65(15):4159–4175. doi: 10.1093/jxb/eru190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cai G, Wang Y, Tu G, Chen P, Luan S, Lan W. Type A2 BTB Members Decrease the ABA Response during Seed Germination by Affecting the Stability of SnRK2.3 in Arabidopsis. Int J Molecular Sci. 2020;21(9):3153. [DOI] [PMC free article] [PubMed]
- 138.Song J., Shang L., Wang X., Xing Y., Xu W., Zhang Y., et al. MAPK11 regulates seed germination and ABA signaling in tomato by phosphorylating SnRKs. J Exp Bot. 2020 doi: 10.1093/jxb/eraa564. [DOI] [PubMed] [Google Scholar]
- 139.Bi L, Weng L, Jiang Z, Xiao H. The tomato IQD gene SUN24 regulates seed germination through ABA signaling pathway. 2018;248(4):919-31. [DOI] [PubMed]
- 140.Wu J., Zhang N., Liu Z., Liu S., Liu C., Lin J., et al. The AtGSTU7 gene influences glutathione-dependent seed germination under ABA and osmotic stress in Arabidopsis. Biochem Biophys Res Commun. 2020;528(3):538–544. doi: 10.1016/j.bbrc.2020.05.153. [DOI] [PubMed] [Google Scholar]
- 141.Pawela A., Banasiak J., Biała W., Martinoia E., Jasiński M. MtABCG20 is an ABA exporter influencing root morphology and seed germination of Medicago truncatula. Plant J Cell Molecular Biol. 2019;98(3):511–523. doi: 10.1111/tpj.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Barrero J.M., Piqueras P., González-Guzmán M., Serrano R., Rodríguez P.L., Ponce M.R., et al. A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. J Exp Bot. 2005;56(418):2071–2083. doi: 10.1093/jxb/eri206. [DOI] [PubMed] [Google Scholar]
- 143.Wu J., Ichihashi Y., Suzuki T., Shibata A., Shirasu K., Yamaguchi N., et al. Abscisic acid-dependent histone demethylation during postgermination growth arrest in Arabidopsis. Plant, Cell Environ. 2019;42(7):2198–2214. doi: 10.1111/pce.13547. [DOI] [PubMed] [Google Scholar]
- 144.Qu L., Sun M., Li X., He R., Zhong M., Luo D., et al. The Arabidopsis F-box protein FOF2 regulates ABA-mediated seed germination and drought tolerance. Plant Sci. 2020;301:110643. doi: 10.1016/j.plantsci.2020.110643. [DOI] [PubMed] [Google Scholar]
- 145.Li P., Zhou H., Shi X., Yu B., Zhou Y., Chen S., et al. The ABI4-induced Arabidopsis ANAC060 transcription factor attenuates ABA signaling and renders seedlings sugar insensitive when present in the nucleus. PLoS Genet. 2014;10(3):e1004213. doi: 10.1371/journal.pgen.1004213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sun L., Liu L.-P., Wang Y.-Z., Yang L., Wang M.-J., Liu J.-X. NAC103, a NAC family transcription factor, regulates ABA response during seed germination and seedling growth in Arabidopsis. Planta. 2020;252(6):95. doi: 10.1007/s00425-020-03502-2. [DOI] [PubMed] [Google Scholar]
- 147.Lozano-Juste J., Masi M., Cimmino A., Clement S., Fernández M.A., Antoni R., et al. The fungal sesquiterpenoid pyrenophoric acid B uses the plant ABA biosynthetic pathway to inhibit seed germination. J Exp Bot. 2019;70(19):5487–5494. doi: 10.1093/jxb/erz306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li J., Zhao C., Hu S., Song X., Lv M., Yao D., et al. Arabidopsis NRT1.2 interacts with the PHOSPHOLIPASE Dα1 (PLDα1) to positively regulate seed germination and seedling development in response to ABA treatment. Biochem Biophys Res Commun. 2020;533(1):104–109. doi: 10.1016/j.bbrc.2020.08.025. [DOI] [PubMed] [Google Scholar]
- 149.Shu K., Zhou W., Chen F., Luo X., Yang W. Abscisic acid and gibberellins antagonistically mediate plant development and abiotic stress responses. Front Plant Sci. 2018;9:416. doi: 10.3389/fpls.2018.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chitnis V.R., Gao F., Yao Z., Jordan M.C., Park S., Ayele B.T. After-ripening induced transcriptional changes of hormonal genes in wheat seeds: the cases of brassinosteroids, ethylene, cytokinin and salicylic acid. PLoS ONE. 2014;9(1):e87543. doi: 10.1371/journal.pone.0087543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shan X., Yan J., Xie D. Comparison of phytohormone signaling mechanisms. Curr Opin Plant Biol. 2012;15(1):84–91. doi: 10.1016/j.pbi.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 152.Cui D., Zhao J., Jing Y., Fan M., Liu J., Wang Z., et al. The arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PLoS Genet. 2013;9(9):e1003759. doi: 10.1371/journal.pgen.1003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yazaki J., Kikuchi S. The genomic view of genes responsive to the antagonistic phytohormones, abscisic acid, and gibberellin. Vitam Horm. 2005;72:1–30. doi: 10.1016/S0083-6729(05)72001-X. [DOI] [PubMed] [Google Scholar]
- 154.Nambara E., Okamoto M., Tatematsu K., Yano R., Seo M., Kamiya Y. Abscisic acid and the control of seed dormany and germination. Seed Sci Res - SEED SCI RES. 2010;20 [Google Scholar]
- 155.Matilla A.J., Carrillo-Barral N., Rodríguez-Gacio Md.C. An update on the role of NCED and CYP707A ABA metabolism genes in seed dormancy induction and the response to after-ripening and nitrate. J Plant Growth Regul. 2015;34(2):274–293. [Google Scholar]
- 156.Nambara E., Hayama R., Tsuchiya Y., Nishimura M., Kawaide H., Kamiya Y., et al. The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev Biol. 2000;220(2):412–423. doi: 10.1006/dbio.2000.9632. [DOI] [PubMed] [Google Scholar]
- 157.Shu K., Zhou W., Yang W. APETALA 2-domain-containing transcription factors: focusing on abscisic acid and gibberellins antagonism. New Phytol. 2018;217(3):977–983. doi: 10.1111/nph.14880. [DOI] [PubMed] [Google Scholar]
- 158.Guo X., Hou X., Fang J., Wei P., Xu B., Chen M., et al. The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism. Plant J Cell molecular Biol. 2013;75(3):403–416. doi: 10.1111/tpj.12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yano R., Kanno Y., Jikumaru Y., Nakabayashi K., Kamiya Y., Nambara E. CHOTTO1, a putative double APETALA2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis. Plant Physiol. 2009;151(2):641–654. doi: 10.1104/pp.109.142018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu X., Hu P., Huang M., Tang Y., Li Y., Li L., et al. The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat Commun. 2016;7:12768. doi: 10.1038/ncomms12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bi C., Ma Y., Wang X.-F., Zhang D.-P. Overexpression of the transcription factor NF-YC9 confers abscisic acid hypersensitivity in Arabidopsis. Plant Mol Biol. 2017;95(4):425–439. doi: 10.1007/s11103-017-0661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ariizumi T., Hauvermale A.L., Nelson S.K., Hanada A., Yamaguchi S., Steber C.M. Lifting della repression of Arabidopsis seed germination by nonproteolytic gibberellin signaling. Plant Physiol. 2013;162(4):2125–2139. doi: 10.1104/pp.113.219451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tyler L., Thomas S.G., Hu J., Dill A., Alonso J.M., Ecker J.R., et al. DELLA proteins and gibberellin-regulated seed germination and floral development in arabidopsis. Plant Physiol. 2004;135(2):1008–1019. doi: 10.1104/pp.104.039578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lee K.P., Piskurewicz U., Turecková V., Strnad M., Lopez-Molina L. A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc Natl Acad Sci USA. 2010;107(44):19108–19113. doi: 10.1073/pnas.1012896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ravindran P., Verma V., Stamm P., Kumar P.P. A Novel RGL2-DOF6 complex contributes to primary seed dormancy in arabidopsis thaliana by regulating a GATA transcription factor. Mol Plant. 2017;10(10):1307–1320. doi: 10.1016/j.molp.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 166.Jin D, Wu M, Li B, Bücker B, Keil P, Zhang S, et al. The COP9 Signalosome regulates seed germination by facilitating protein degradation of RGL2 and ABI5. 2018;14(2):e1007237. [DOI] [PMC free article] [PubMed]
- 167.Ariizumi T., Steber C.M. Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell. 2007;19(3):791–804. doi: 10.1105/tpc.106.048009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Miura K., Lee J., Jin J.B., Yoo C.Y., Miura T., Hasegawa P.M. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. PNAS. 2009;106(13):5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liu X., Hou X. Antagonistic regulation of ABA and GA in metabolism and signaling pathways. Front Plant Sci. 2018;9:251. doi: 10.3389/fpls.2018.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kim S.I., Park B.S., Kim D.Y., Yeu S.Y., Song S.I., Song J.T., et al. E3 SUMO ligase AtSIZ1 positively regulates SLY1-mediated GA signalling and plant development. Biochem J. 2015;469(2):299–314. doi: 10.1042/BJ20141302. [DOI] [PubMed] [Google Scholar]
- 171.Silverstone A.L., Tseng T.S., Swain S.M., Dill A., Jeong S.Y., Olszewski N.E., et al. Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 2007;143(2):987–1000. doi: 10.1104/pp.106.091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ariizumi T., Lawrence P.K., Steber C.M. The role of two f-box proteins, SLEEPY1 and SNEEZY, Arabidopsis gibberellin signalling. Plant Physiol. 2011;155(2):765–775. doi: 10.1104/pp.110.166272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xi W., Liu C., Hou X., Yu H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell. 2010;22(6):1733–1748. doi: 10.1105/tpc.109.073072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vaistij F.E., Gan Y., Penfield S., Gilday A.D., Dave A., He Z., et al. Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc Natl Acad Sci USA. 2013;110(26):10866–10871. doi: 10.1073/pnas.1301647110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Song S., Wang G., Wu H., Fan X., Liang L., Zhao H., et al. OsMFT2 is involved in the regulation of ABA signaling-mediated seed germination through interacting with OsbZIP23/66/72 in rice. Plant J Cell Molecular Biol. 2020;103(2):532–546. doi: 10.1111/tpj.14748. [DOI] [PubMed] [Google Scholar]
- 176.Piskurewicz U., Turecková V., Lacombe E., Lopez-Molina L. Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. Embo J. 2009;28(15):2259–2271. doi: 10.1038/emboj.2009.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Footitt S., Clay H.A., Dent K., Finch-Savage W.E. Environment sensing in spring-dispersed seeds of a winter annual Arabidopsis influences the regulation of dormancy to align germination potential with seasonal changes. New Phyto. 2014;202(3):929–939. doi: 10.1111/nph.12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Footitt S., Douterelo-Soler I., Clay H., Finch-Savage W.E. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc Natl Acad Sci. 2011;108(50):20236–20241. doi: 10.1073/pnas.1116325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mérai Z., Graeber K., Wilhelmsson P., Ullrich K.K., Arshad W., Grosche C., et al. Aethionema arabicum: a novel model plant to study the light control of seed germination. J Exp Bot. 2019;70(12):3313–3328. doi: 10.1093/jxb/erz146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Barros-Galvão T., Dave A., Gilday A.D., Harvey D., Vaistij F.E., Graham I.A. ABA INSENSITIVE4 promotes rather than represses PHYA-dependent seed germination in Arabidopsis thaliana. New Phytol. 2020;226(4):953–956. doi: 10.1111/nph.16363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yang L., Jiang Z., Jing Y., Lin R. PIF1 and RVE1 form a transcriptional feedback loop to control light-mediated seed germination in Arabidopsis. J Integr Plant Biol. 2020;62(9):1372–1384. doi: 10.1111/jipb.12938. [DOI] [PubMed] [Google Scholar]
- 182.Oh E., Kim J., Park E., Kim J.I., Kang C., Choi G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell. 2004;16(11):3045–3058. doi: 10.1105/tpc.104.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kim D.H., Yamaguchi S., Lim S., Oh E., Park J., Hanada A., et al. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell. 2008;20(5):1260–1277. doi: 10.1105/tpc.108.058859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., et al. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell. 2007;19(4):1192–1208. doi: 10.1105/tpc.107.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Penfield S., Josse E.-M., Kannangara R., Gilday A.D., Halliday K.J., Graham I.A. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol. 2005;15(22):1998–2006. doi: 10.1016/j.cub.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 186.Gabriele S., Rizza A., Martone J., Circelli P., Costantino P., Vittorioso P. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 2010;61(2):312–323. doi: 10.1111/j.1365-313X.2009.04055.x. [DOI] [PubMed] [Google Scholar]
- 187.Bentsink L., Jowett J., Hanhart C.J., Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA. 2006;103(45):17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tanaka Y., Sano T., Tamaoki M., Nakajima N., Kondo N., Hasezawa S. Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 2005;138(4):2337–2343. doi: 10.1104/pp.105.063503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Jiang Y., Joyce D.C. ABA effects on ethylene production, PAL activity, anthocyanin and phenolic contents of strawberry fruit. Plant Growth Regul. 2003;39(2):171–174. [Google Scholar]
- 190.Zhang M., Yuan B., Leng P. The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J Exp Bot. 2009;60(6):1579–1588. doi: 10.1093/jxb/erp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li Z., Zhang L., Yu Y., Quan R., Zhang Z., Zhang H., et al. The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J Cell Molecular Biol. 2011;68(1):88–99. doi: 10.1111/j.1365-313X.2011.04670.x. [DOI] [PubMed] [Google Scholar]
- 192.Ghassemian M., Nambara E., Cutler S., Kawaide H., Kamiya Y., McCourt P. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell. 2000;12(7):1117–1126. doi: 10.1105/tpc.12.7.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Harrison M.A. In: Phytohormones and Abiotic Stress Tolerance in Plants. Khan N.A., Nazar R., Iqbal N., Anjum N.A., editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2012. Cross-Talk Between Phytohormone Signaling Pathways Under Both Optimal and Stressful Environmental Conditions; pp. 49–76. [Google Scholar]
- 194.Sun Y., Liang B., Wang J., Kai W., Chen P., Jiang L., et al. SlPti4 affects regulation of fruit ripening, seed germination and stress responses by modulating ABA signaling in tomato. Plant Cell Physiol. 2018;59(10):1956–1965. doi: 10.1093/pcp/pcy111. [DOI] [PubMed] [Google Scholar]
- 195.Arc E., Sechet J., Corbineau F., Rajjou L., Marion-Poll A. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front Plant Sci. 2013;4:63. doi: 10.3389/fpls.2013.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Dolferus R. To grow or not to grow: a stressful decision for plants. Plant Sci. 2014;229:247–261. doi: 10.1016/j.plantsci.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 197.Yang L., Wang S., Sun L., Ruan M., Li S., He R., et al. Involvement of G6PD5 in ABA response during seed germination and root growth in Arabidopsis. BMC Plant Biol. 2019;19(1):44. doi: 10.1186/s12870-019-1647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li Z., Xu J., Gao Y., Wang C., Guo G., Luo Y., et al. The synergistic priming effect of exogenous salicylic acid and H(2)O(2) on chilling tolerance enhancement during maize (Zea mays L.) seed germination. Front Plant Sci. 2017;8:1153. doi: 10.3389/fpls.2017.01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Daminato M., Guzzo F., Casadoro G. A SHATTERPROOF-like gene controls ripening in non-climacteric strawberries, and auxin and abscisic acid antagonistically affect its expression. J Exp Bot. 2013;64(12):3775–3786. doi: 10.1093/jxb/ert214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sun J., Li C. In: Abscisic Acid: Metabolism, Transport and Signaling. Zhang D.-.P., editor. Springer, Netherlands; Dordrecht: 2014. Cross Talk of Signaling Pathways Between ABA and Other Phytohormones; pp. 243–253. [Google Scholar]
- 201.Liu X., Zhang H., Zhao Y., Feng Z., Li Q., Yang H.Q., et al. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. PNAS. 2013;110(38):15485–15490. doi: 10.1073/pnas.1304651110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Clouse S.D., Langford M., McMorris T.C. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111(3):671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhang S., Cai Z., Wang X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc Natl Acad Sci USA. 2009;106(11):4543–4548. doi: 10.1073/pnas.0900349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hu Y., Yu D. BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell. 2014;26(11):4394–4408. doi: 10.1105/tpc.114.130849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.White C.N., Proebsting W.M., Hedden P., Rivin C.J. Gibberellins and seed development in maize. I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiol. 2000;122(4):1081–1088. doi: 10.1104/pp.122.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Koornneef M., Reuling G., Karssen C. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant. 1984;61(3):377–383. [Google Scholar]
- 207.Giraudat J., Parcy F., Bertauche N., Gosti F., Leung J., Morris P.-C., et al. Current advances in abscisic acid action and signalling. Plant Mol Biol. 1994;26(5):1557–1577. doi: 10.1007/BF00016490. [DOI] [PubMed] [Google Scholar]
- 208.Leung J., Giraudat J. Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- 209.Steber C.M., Cooney S.E., McCourt P. Isolation of the GA-Response Mutant sly1 as a Suppressor of ABI1-1 in Arabidopsis thaliana. Genetics. 1998;149(2):509–521. doi: 10.1093/genetics/149.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Nambara E., Suzuki M., Abrams S., McCarty D.R., Kamiya Y., McCourt P. A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics. 2002;161(3):1247–1255. doi: 10.1093/genetics/161.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Beaudoin N., Serizet C., Gosti F., Giraudat J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell. 2000;12:1103–1115. doi: 10.1105/tpc.12.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Nishimura N, Tsuchiya W, Moresco JJ, Hayashi Y, Satoh K, Kaiwa N, et al. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. 2018;9(1):2132. [DOI] [PMC free article] [PubMed]
- 213.Wang X., Guo C., Peng J., Li C., Wan F., Zhang S., et al. ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 2019;221(1):341–355. doi: 10.1111/nph.15345. [DOI] [PubMed] [Google Scholar]
- 214.Koornneef M., Jorna M.L., Brinkhorst-van der Swan D.L.C., Karssen C.M. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) heynh. Theor Appl Genet. 1982;61(4):385–393. doi: 10.1007/BF00272861. [DOI] [PubMed] [Google Scholar]
- 215.Léon-Kloosterziel K.M., Gil M.A., Ruijs G.J., Jacobsen S.E., Olszewski N.E., Schwartz S.H., et al. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J Cell Molecular Biol. 1996;10(4):655–661. doi: 10.1046/j.1365-313x.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- 216.Holdsworth M., Lenton J., Flintham J., Gale M., Kurup S., McKibbin R., et al. Genetic control mechanisms regulating the initiation of germination. J Plant Physiol. 2001;158:439–445. [Google Scholar]
- 217.McCarty D.R. Genetic control and integration of maturation and germination pathways in seed development. Ann Rev Plant Physiol Plant Molecular Biol. 1995;46(1):71–93. [Google Scholar]
- 218.Ohta M., Ohme-Takagi M., Shinshi H. Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J Cell Molecular Biol. 2000;22:29–38. doi: 10.1046/j.1365-313x.2000.00709.x. [DOI] [PubMed] [Google Scholar]
- 219.Finkelstein R.R., Li Wang M., Lynch T.J., Rao S., Goodman H.M. The Arabidopsis abscisic acid response locus the arabidopsis abscisic acid response locus (ABI4) encodes an APETALA2 domain protein. Plant Cell. 1998;10(6):1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Raz V., Bergervoet J., Koornneef M. Sequential steps for developmental arrest in Arabidopsis seeds. Development. 2001;128(2):243–252. doi: 10.1242/dev.128.2.243. [DOI] [PubMed] [Google Scholar]