Graphical abstract
Keywords: Intestinal microbiota, Type 2 diabetes mellitus, Predictive model, Coronary heart disease, CORDIOPREV
Highlights
-
•
Type 2 diabetes (T2DM) increases the risk of recurrence in myocardial infarction patients.
-
•
A gut microbiota profile is associated to the further T2DM development.
-
•
Microbiome data improved the prediction of T2DM development when added to clinical parameters.
-
•
A risk score including the most predictive genera was associated with the probability of T2DM.
-
•
A high risk score was associated with a higher hepatic insulin resistance and β-cell dysfunction.
Abstract
Introduction
A distinctive gut microbiome have been linked to type 2 diabetes mellitus (T2DM).
Objectives
We aimed to evaluate whether gut microbiota composition, in addition to clinical biomarkers, could improve the prediction of new incident cases of diabetes in patients with coronary heart disease.
Methods
All the patients from the CORDIOPREV (Clinical Trials.gov.Identifier: NCT00924937) study without T2DM at baseline were included (n = 462). Overall, 107 patients developed it after a median of 60 months. The gut microbiota composition was determined by 16S rRNA gene sequencing and predictive models were created using hold-out method.
Results
A gut microbiota profile associated with T2DM development was determined through a microbiome-based predictive model. The addition of microbiome data to clinical parameters (variables included in FINDRISC risk score and the diabetes risk score of the American Diabetes Association, HDL, triglycerides and HbA1c) improved the prediction increasing the area under the curve from 0.632 to 0.946. Furthermore, a microbiome-based risk score including the ten most discriminant genera, was associated with the probability of develop T2DM.
Conclusion
These results suggest that a microbiota profile is associated to the T2DM development. An integrate predictive model of microbiome and clinical data that can improve the prediction of T2DM is also proposed, if is validated in independent populations to prevent this disease.
Introduction
One of the metabolic disorders with a higher incidence which has a strong public health impact worldwide is Type 2 diabetes mellitus (T2DM). Insulin resistance (IR) along with impaired beta-cell function are critical determinants of this disease [1]. Moreover, obesity has been associated with the chronic activation of inflammatory pathways causally linked to IR [2].
The concurrent presence of coronary heart disease (CHD) and T2DM significantly raises the risk of macrovascular complications and mortality [3]. It is therefore especially important to design strategies to prevent the development of this disease in patients with CHD. The current standard approaches for identifying patients at higher risk of T2DM include risk scores such as that provided by the American Diabetes Association (ADA) [4] or Finnish Diabetes Risk Score (FINDRISC) [5]. However, these tools are not always able to accurately predict the development of the disease, which varies in different ethnic and population groups [5], [6], [7]. There is therefore an urgent need to improve the early detection of T2DM risk in such patients. In this context, we recently reported that postprandial endotoxemia has a considerable potential to asses diabetes risk [6].
Our intestines harbor a vast microbial community, which interacts with the host exerting major metabolic functions. Human studies have shown that the microbiome composition determines the functionality of the gut microbiota associated with obesity and T2DM [8], [9].
A growing number of cross-sectional case-control studies have appeared in past few years showing that the intestinal microbiota of diabetic patients, differs from non-diabetic individuals [8]. However, although most of the studies did not assign a causal role to the gut microbiota, and they might have also been influenced by an anti-diabetic drug, such as metformin, that has been demonstrated to modify gut microbiota composition [10], it has been recently reported that microbiota of T2DM patients is also altered in absence of diabetes treatment [11]. Furthermore, several studies showed a transitory improvement in IR following fecal transplantation from healthy individuals to patients with the metabolic syndrome (MetS) suggesting a causal function of the microbiota in T2DM [9], [12].
Recently, it has been suggested the potential use of the microbiome to evaluate the risk of T2DM [13]. However, the predictive value of a microbiota profile derived from cross-sectional analysis in already diagnosed T2DM patients is limited. Nevertheless, prospective studies might improve the selection of the bacterial taxa associated with T2DM development and the robustness of the predictive models. In fact, to the best of our knowledge, no prospective studies have been conducted so far in order to identify a microbiota associated to T2DM development.
In consideration of the possible implication of the gut microbiota in the disease development, we hypothesized that an alteration of its composition might precede the development of this disease and could therefore be used as a predictive associated factor. Therefore, we aimed to evaluate whether gut microbiota composition, in addition to clinical biomarkers, could improve the prediction of new incident cases of T2DM in patients with CHD, within the CORDIOPREV study.
Patients and methods
Study patients
This study was accomplished in the context of the CORDIOPREV study (Clinicaltrials.gov NTC00924937), a prospective randomized controlled and ongoing clinical trial which included 1002 patients with CHD. Patients were randomized to receive during a period of 7 years, in addition to their pharmacological prescription for CHD, the Mediterranean (MED) or a low-fat (LF). The inclusion criteria, methodology and rationale of the trial have been previously described [14].
From the non-diabetic patients at the beginning of the CORDIOPREV study (N = 462), 107 patients were diagnosed with T2DM after a median of 60 months of follow-up. The diagnosis was assessed according to the ADA criteria [4]. From the patients included in this study during the follow-up, 17 patients died, and 7 patients dropped out without being diagnosed with diabetes. Fecal samples were available at baseline for a total of 273 patients who had not received treatment with antibiotics within 3 months before baseline sample collection; among them, 64 patients developed T2DM during the follow-up (Incident-DIAB group). Suppl. Table S1 and Suppl. Table S2 show the baseline characteristics of the patient groups.
Study design
The methods of the study has been previously detailed elsewhere [14]. Briefly, participants were randomized to consume two dietary patterns: a MED diet or an LF diet. The LF diet consisted of a minimum of 55% carbohydrates, 15% protein, and < 30% total fat, specifically < 10% saturated fat, 6–8% PUFA fat and 12%-14% MUFA fat. The MED diet comprised a minimum 35% of calories as fat, specifically 22% of MUFA fat, and the same percentage for the rest of fat types than the LF diet. The amount of cholesterol was < 300 mg per day for the two diets.
Dietary assessment
At baseline and every year of the study, patients were interviewed individually and face-to-face by a nutritionist to complete a previously validated in Spain with 137-items [15], semi-quantitative food frequency questionnaire, and also a validated MED diet adherence questionnaire [16].
The participants received personalized one-on-one interviews at the start and two-yearly and quarterly educational collectively sessions with nutritionists with the same intensive dietary counseling in both intervention groups.
Oral glucose tolerance test (OGTT)
An OGTT was conducted at baseline and every year thereafter to determine plasma glucose and insulin levels. Blood samples were taken before the test and at regular intervals of 30 min during 2 h after taken the glucose solution (A dilution of 75 g of glucose in 250 ml of water). Insulin sensitivity index (ISI), homeostatic model assessment (HOMA-IR), and disposition index (DI), insulinogenic index (IGI), hepatic insulin resistance index (HIRI) and muscle insulin sensitivity index (MISI) were calculated as previously described. [17].
Ethics statement
All procedures followed were in conformity with the Helsinki Declaration of 1975, as revised in 2008. The written informed consent to take part of the study was obtained from all patients. Reina Sofia University Hospital Ethics Committee, regional the responsible committee on human experimentation, approved the trial protocol and all amendments (No. 1496/27/03/2009).
Microbiota analysis
We used the QIAamp DNAStool Mini Kit Handbook (QIAGEN, Hilden, Germany) for the DNA extraction of fecal samples, which were collected at baseline, following the manufacturer’s instructions. The 16S rRNA gene sequencing performed use the V4 hypervariable region that were amplified using the primers F515 (5′- TAT GGT AAT TGT GTG CCA GCM GCC GCG GTA A −3′) and R806 (5′- AGT CAG TCA GCC GGA CTA CHV GGG TWT CTA AT −3′) primers [18] to generate an amplification library. The composite of the above specific primers included Illumina adapters and a unique 8-nucleotide sample index sequence key [18]. The PCR amplification was performed using the Phusion High-Fidelity PCR Master mix (New England Biolabs, Ipswich, MA, USA) with 100 pg template DNA. The amplification program conducted was initial denaturation during 30 s at 98 °C; 30 cycles of amplification (10 s at 98 °C, 30 s at 55 °C and 30 s at 72 °C); and a final elongation of 5 min at 72 °C. PCR products were examined on a Fragment Analyzer (Advanced Analytical Technologies Inc., Ankeny, IA, USA) to estimate DNA concentration. The amplicon libraries were pooled in equimolar amounts and purified utilizing in a first step with the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA, USA) and then Agencourt AMPure magnetic beads (Beckman Coulter, Brea, CA, USA). DNA concentration of the pool was calculated on a fluorometer applying the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA).
The purified bar-coded amplicons obtained were paired-end sequenced using the Illumina MiSeq platform (Illumina Technologies, San Diego, CA, USA) using 2 × 250 cycle paired-end settings. The sequence data was processed and analyzed using Mothur version 1.36.1. The taxonomic identification was conducted employing the Ribosomal Database Proyect (RDP) tool and the SILVA 16S rRNA gene database up to the level of the genera. To exclude any bacterial taxa that were not detected in most of the samples, a cut-off for exclusion was set; only bacterial taxa containing sequence readings in at least 75% of total samples were considered.
Predictive models development
A random forest classifier was performed by hold-out analysis using the script of the caret package in R, in which the model was trained on 70% of the data and the remaining 30% for testing to prevent overfitting. Other machine learning methods such as Support Vector Machine and Neural Networks were also tested, throwing worse results than the random forest classifier. To obtain more precise curves and assess the performance of the models on unseen data, we used repeated 10-fold cross-validation that consists of ten train and test splits, so that each data has been used in the test set once. The cross-validation error curves (average of ten validation sets each) and performance were averaged. The predictive value of each variable in the random forest models was calculated by Mean Decrease in Accuracy. The model's performance was further evaluated through the AUC on the test set. pROC R package was used to calculate the confidence intervals for ROC curves.
Microbiota-based risk score development
We created a risk score based on patients’ microbiome profile from the ten bacterial taxa with the highest importance on a microbiome-based predictive model. The predictive importance was assessed by Mean Decrease Accuracy values. We categorized patients according to the abundance of these bacterial taxa in ascending tertiles, and the detrimental or beneficial role was determined according to a higher mean baseline abundance in Incident-DIAB or Non-DIAB group, respectively. In this way, for a detrimental genus: tertil 1 was scored as −1 (protective), tertil 2 as 0 (neutral effect) and tertil 3 as 1 (risk), and the opposite for a beneficial genus: tertil 1 as 1 (risk), tertil 2 as 0 (neutral effect) and tertil 3 as −1 (protective). The score was calculated by adding the Mean Decrease Accuracy for each bacterial taxa, with a positive or negative value, according to the above risk or protective tertiles and disregarding the tertiles with neutral effect (added as 0).
Statistical analysis
The statistical analysis of the data was carried out with SPSS statistical software (IBM SPSS Statistics version 21.0). In all statical analysis, p-values ≤ 0.05 were statistically significant. Kolmogorov-Smirnov test was used to assess whether variables follow a normal distribution. One-way ANOVA was performed to calculate the statistical differences of the quantitative anthropometric and metabolic variables between groups, while qualitative data were analyzed using the Chi-Square analysis. LEfSe (Linear discriminant analysis Effect Size) [19] was conducted for the identification of the most differently abundant taxa between groups. Furthermore, Cox proportional hazard regression analysis was run incorporating as covariables: age, gender, BMI, diet, HDL, triglycerides and the intensity of statin therapy defined by the American Heart Association (AHA). In addition, a nomogram was performed with R based on the results of a Weibull survival model fit with psm function employing survival package [20]. The relationship between the microbiome-based T2DM risk score and baseline abundance of the top ten most discriminant bacterial taxa with OGTT-derived indexes was analyzed by ANOVA for repeated measures.
Results
Baseline characteristics of the participants
Baseline body weight, waist circumference, glycosylated haemoglobin (HbA1c), fasting glucose, body mass index (BMI), insulin, HIRI and HOMA-IR were higher, whereas DI, ISI and IGI were lower in the Incident-DIAB in comparison with the Non-DIAB group (p < 0.05). No differences were found for age or gender between the groups (Suppl. Table S1). Furthermore, the patient population with available fecal samples at baseline and who had not received antibiotic treatment within 3 months before sample collection, was representative of the total population for the T2DM incidence study [6] (Suppl. Table S2).
Differences in gut microbiota composition between study groups
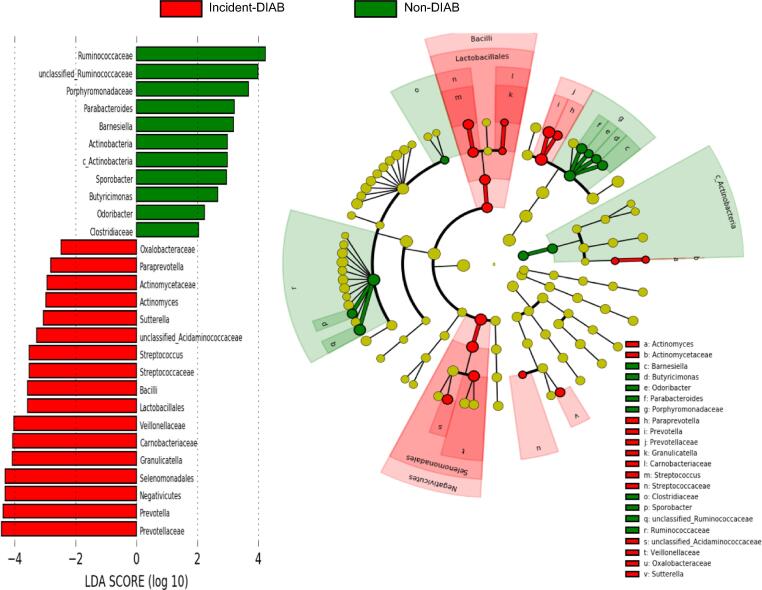
LEfSe was performed to determine the most differently abundant phylotypes and the taxa of the gut microbiota at baseline between the Incident-DIAB and the Non-DIAB group.
Baseline gut microbiota in the Incident-DIAB group was characterized by the preponderance of the Negativicutes and Bacilli classes, as well as the Selenomonadales and Lactobacillales orders. Regarding family, it was characterized by a higher abundance of Prevotellaceae, Carnobacteriaceae, Veillonellaceae, Streptococcaceae, Actinomycetaceae and Oxalobacteraceae, as well as by Prevotella, Granulicatella, Streptococcus, Acidaminococcaceae unclassified genus, Sutterella, Actinomyces and Paraprevotella. In contrast, baseline gut microbiota in the Non-DIAB group was enriched in Actinobacteria at phylum and class levels, as well as characterized by a predominance of Clostridiaceae, Porphyromonadaceae and Ruminococcaceae families; in terms of genus, Odoribacter, Butyricimonas, Sporobacter and Barnesiella, Parabacteroides, as well as an unknown genus of the Ruminococcaceae family were predominant (Fig. 1). However, the bacterial richness and diversity assessed by the main α diversity indexes were similar between the groups (Mean ± standard deviation (SD) in the Incident-DIAB group and the Non-DIAB group of Chao1: 164.6 ± 23.26 and 162.6 ± 23.86 respectively, p = 0.94; Simpson: 0.926 ± 0.01 and 0.928 ± 0.01, respectively, p = 0.63; Shannon: 3.12 ± 0.22 and 3.14 ± 0.20, respectively, p = 0.71).
Fig. 1.
Differently abundant taxa identified using LEfSe analysis. The most differently abundant taxa between the groups of study are represented in a bar graph according to the LDA score (log 10), an estimation of the effect size and in a taxonomic cladogram. Only taxa meeting a p < 0.05 and LDA score significant threshold |>2| are shown. The colors represent the group in which the indicated taxa is more abundant compared to the other group. In a taxonomic cladogram, each successive circle represents a different phylogenetic level. The order from the center to the outside is phylum, class, family and genus levels. Differing taxa are listed on the right side of the cladogram. c_Actinobacteria: Actinobacteria class.
Random forest predictive model for T2DM development
Several random forest classifier models were created based on the following data: (1) clinical variables (encompassed in the FINDRISC and ADA scores: BMI, waist circumference, use of antihypertensive medication, age, dietary consumption of fruit and vegetables, physical activity, family history of diabetes, history of gestational diabetes, history of high blood glucose (i.e. whether participant has ever been determined to have high blood glucose in a health test), High Density Lipoprotein (HDL), triglycerides and HbA1c), (2) The indexes calculated from the Oral Glucose Tolerance Test (OGTT) (i.e. IGI, HOMA-IR, ISI, HIRI, DI and MISI), (3) the microbiome (bacterial composition at genus level expressed as abundance), (4) clinical variables along with microbiome, and (5) OGTT-derived insulin sensitivity indexes combined with the microbiome.
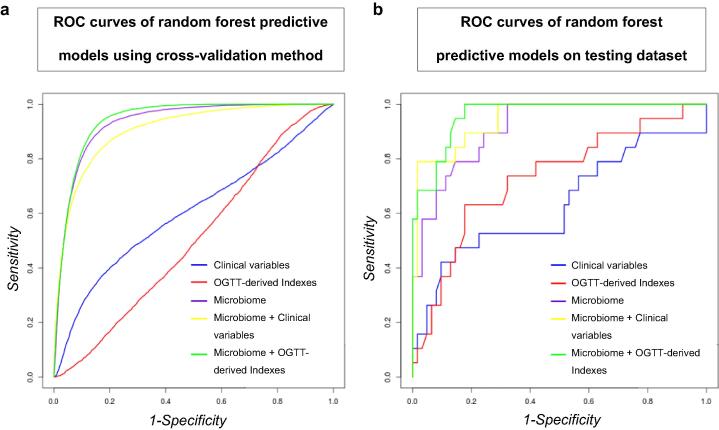
Firstly, we ran all the models in a training dataset that accounted for 70% of the patients. The models were performed using a 10-fold cross-validation method to evaluate the model's predictive accuracy and generalization performance. Diet and intensity of statin treatment were included in the analyses (Table 1a and Fig. 2a). Validation of the models was then performed to confirm the results and rule out the possibility of over-fitting models. This validation was performed in the 30% of the patients not included in the training set (N = 82; 19 incident cases) (Table 1b and Fig. 2b).
Table 1.
ROC analysis of the Random Forest classification models. (a) ROC performed using cross-validation method. (b) ROC performed on testing dataset.
| a | |||||
|---|---|---|---|---|---|
| AUC | Sensitivity | Specificity | Accuracy | Kappa coefficient | |
| Clinical variables | 0.614 (0.139) | 0.92 (0.07) | 0.24 (0.19) | 0.758 (0.068) | 0.178 (0.225) |
| OGTT-derived indexes | 0.511 (0.151) | 0.67 (0.13) | 0.30 (0.22) | 0.586 (0.114) | −0.021 (0.224) |
| Microbiome | 0.952 (0.048) | 0.94 (0.07) | 0.77 (0.18) | 0.896 (0.064) | 0.707 (0.178) |
| Microbiome + Clinical variables | 0.925 (0.075) | 0.92 (0.07) | 0.72 (0.24) | 0.874 (0.078) | 0.637 (0.238) |
| Microbiome + OGTT-derived indexes | 0.958 (0.038) | 0.91 (0.07) | 0.89 (0.15) | 0.905 (0.060) | 0.753 (0.154) |
| b | ||||||
| AUC | 95% CI | Sensitivity | Specificity | Accuracy | Kappa coefficient | |
| Clinical variables | 0.632 | 0.467–0.797 | 0.95 | 0.26 | 0.790 | 0.269 |
| OGTT-derived indexes | 0.729 | 0.593–0.866 | 0.81 | 0.63 | 0.765 | 0.401 |
| Microbiome | 0.913 | 0.850–0.976 | 0.92 | 0.58 | 0.840 | 0.527 |
| Microbiome + Clinical variables | 0.946 | 0.895–0.997 | 0.95 | 0.79 | 0.914 | 0.755 |
| Microbiome + OGTT-derived indexes | 0.961 | 0.924–0.997 | 0.92 | 0.79 | 0.889 | 0.696 |
Data are mean (SD). The models were adjusted by diet and intensity of statin treatment. Clinical variables (variables included in the FINDRISC and ADA scores: body mass index, waist circumference, dietary consumption of fruit and vegetables, age, use of antihypertensive medication, family history of diabetes, history of high blood glucose, physical activity in addition to gestational diabetes, high density lipoprotein, triglycerides and HbA1c); OGTT-derived indexes: HOMA-IR, Homeostasis model assessment-insulin resistance; HIRI, Hepatic insulin resistance index; ISI, Insulin sensitivity index; MISI, Muscle insulin sensitivity index; IGI, Insulinogenic index; DI, Disposition index; AUC: area under the curve in the ROC analysis; CI: confidence interval.
Fig. 2.
Multivariate ROC models built based on the Random Forest Algorithm. (a) ROC curves of the models built using a cross-validation method in a training dataset that accounted for 70% of the total patients. (b) ROC curves of the models obtained in the validation performed on a testing dataset composed of patients not used to build the models (30% remaining patients not included in the training dataset). Clinical: clinical variables included in the FINDRISC and ADA scores: age, BMI, waist circumference, physical activity, dietary consumption of fruit and vegetables, use of antihypertensive medication, history of high blood glucose, family history of diabetes in addition to gestational diabetes, HDL, triglycerides and HbA1c; Indexes: OGTT-derived indexes (HOMA-IR, Homeostasis model assessment- insulin resistance; ISI, Insulin sensitivity index; IGI, Insulinogenic index; HIRI, Hepatic insulin resistance index; MISI, Muscle insulin sensitivity index; DI, Disposition index). The models were adjusted by diet and intensity of the statin treatment including these variables in all the models.
Clinical variables produced an area under the curve (AUC) of 0.632, i.e. lower than the AUC obtained for OGTT-derived insulin sensitivity indexes (AUC = 0.729), and the microbiome (AUC = 0.913). In addition, the combination of clinical variables with the microbiome yielded an AUC of 0.946. The AUC reached 0.961 when the OGTT-derived insulin sensitivity indexes were combined with the microbiome. The increase in the AUC observed by the addition of the microbiome to the clinical variables was evaluated using Venkatrama's permutation test and Delong's test with a significant AUC difference observed (p < 0.001). We also observed that the model combining clinical variables and microbiome obtained a 95% sensitivity and a 79% specificity on the test dataset, whereas the model exclusively based on clinical variables a 95% sensitivity and a 26% specificity.
When models included the patients with available fecal samples without removing those who had any antibiotic prescription in the three months before the baseline sampling, the results were consistent with those obtained previously (Suppl. Table S3).
Microbiome-based risk assessment of T2DM development
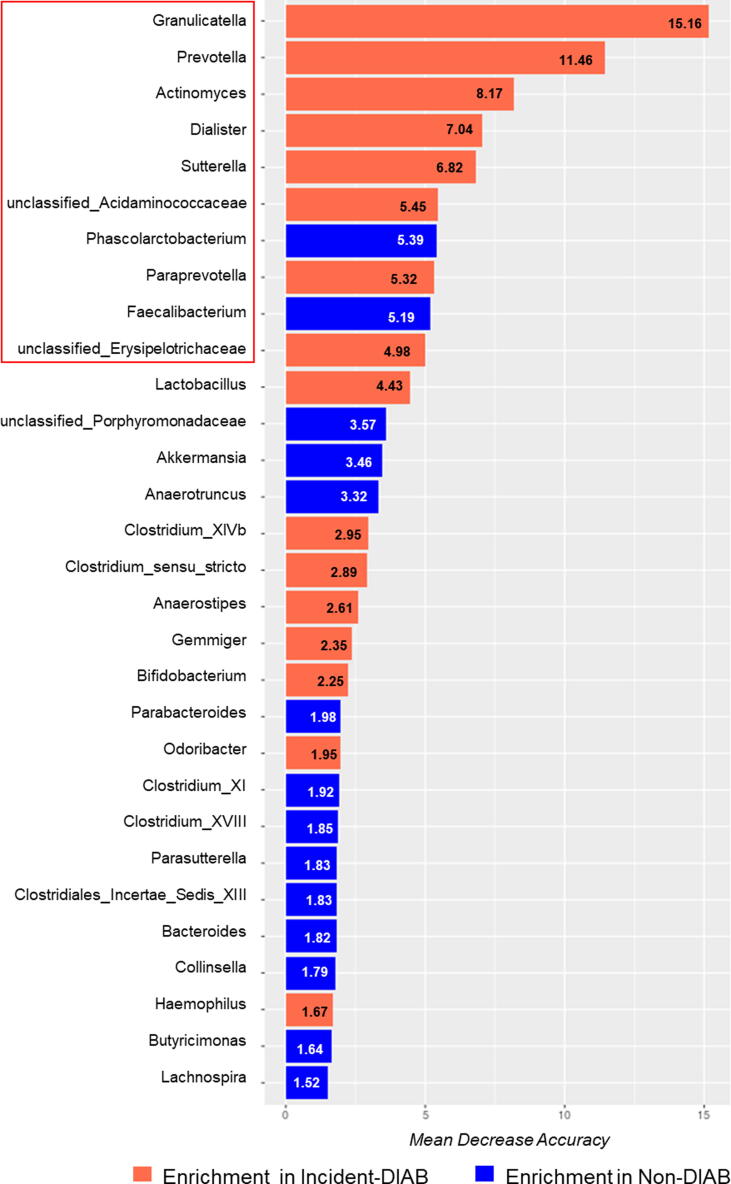
We also analyzed the potential usefulness of the microbiome to evaluate the risk of T2DM development by COX regression analysis. The ten most discriminant bacterial taxa by the Mean Decrease Accuracy were selected (Fig. 3). Furthermore, we classified patients by ascending tertiles according to the abundance for each genus: T1, low abundance; T2, intermediate abundance; T3, high abundance. When we calculated the Cox proportional hazards regression analysis for each of the ten genera, we observed that higher abundance of Granulicatella and Prevotella was linked with a greater risk of diabetes development [Hazard Ratio (HR) unadjusted T1 vs. T3: 2.310 and 1.844, 95% Confidence Interval (CI): 1.278–4.175 and 1.001–3.395, respectively; HR adjusted T1 vs. T3: 1.963 and 2.147, 95% CI: 1.073–3.593 and 1.134–4.064, respectively] (Suppl. Fig. S1).
Fig. 3.
Variable Importance values of microbiome model. Variable Importance is represented by the mean decrease in accuracy of the models when these taxa are removed. The higher the mean decrease in accuracy or bar length, the greater the importance of the variable. The ten most discriminant genera were highlighted.
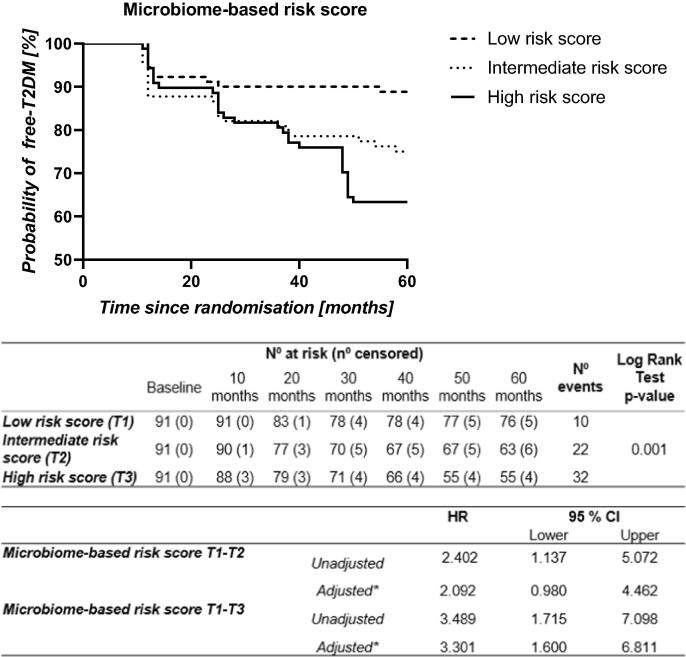
A risk score was then created based on the ten most discriminant bacterial taxa (Fig. 3). We then evaluated T2DM development risk according to the score generated by COX regressions, categorizing patients by ascending tertiles of the score: T1, low risk score; T2, intermediate risk score; T3, high risk score. We found an unadjusted HR of 2.402 (95% CI 1.137–5.072) between the tertiles of T1 vs. T2, and 3.489 (95% CI 1.715–7.098) between the tertiles of T1 vs. T3. Moreover, we obtained an adjusted HR of 2.092 (95% CI 0.980–4.462) between the tertiles of T1 vs. T2, and 3.301 (95% CI 1.600–6.811) between the tertiles of T1 vs. T3 (Fig. 4). Moreover, we also built a nomogram based on the microbiome-based risk score and the clinical variables included in the adjusted COX model. The nomogram showed the contribution of the value of each variable to a total T2DM risk points that assess probability of T2DM development at 30 months and 60 months (Suppl. Fig. S2).
Fig. 4.
Probability of T2DM development by COX regression analysis according to the microbiome-based risk score. The microbiome-based risk score was built with the top ten most discriminant bacterial taxa. The data represent risk score values by ascending terciles: T1, low risk score; T2, intermediate risk score; T3, high risk score. N° at risk: number of patients remaining non-diabetic. N° censored: cumulative number of censored patients because not completing the follow-up period (dropout or death). N° events: number of patients who were diagnosed as diabetic during the follow-up. CI: confidence interval. HR: Hazard ratio. *This model was adjusted by age, gender, diet, BMI, HDL, triglycerides and intensity of statin treatment.
The relationship between microbiome-based T2DM risk score, insulin sensitivity and beta-cell function indexes
We also evaluated whether a microbiota profile, based on the highly discriminant bacterial taxa identified by the predictive model, may affect the pathophysiological mechanisms underlying T2DM development. For this purpose, we examined the relationship between the risk score, created from the ten most discriminant bacterial taxa, and the OGTT-derived indexes. Patients with a high score risk for T2DM development had a significantly lower DI (p = 0.048) and a significantly higher HIRI than patients with a low-risk score (p = 0.046). Furthermore, we found that a lower ISI was related to a high richness of Suterella (p = 0.038), and a low abundance of Phascolarctobacterium (p = 0.040) and Faecalibacterium (p = 0.007). In addition, a lower DI was linked to a high abundance of Dialister and Erysipelotrichaceae unclassified genus (p = 0.032 and p = 0.021, respectively), whereas a higher HIRI was associated with a high abundance of Prevotella (p = 0.029) and low abundances of Phascolarctobacterium and Faecalibacterium (p = 0.010 and p = 0.019) (Suppl. Fig. S3).
Discussion
The present study conducted on CHD patients showed a different baseline gut microbiota profile between groups. These results therefore suggest that a preceding gut microbiota profile is associated with T2DM development several years before its clinical diagnosis. Moreover, the addition of the baseline microbiome data to the traditional clinical risk parameters included in the ADA and the FINDRISC risk scores, significantly improved the prediction of diabetes development. A microbiome-based score, including the ten most relevant bacterial taxa in the microbiome model, showed that patients with a certain “harmful” gut microbiota profile had a significantly increased risk of incident T2DM.
The clinical biomarkers used to identify patients at T2DM risk, such as those contained in the FINDRISC score, are not able to accurately predict disease development in every population [5], [6], [7]. Given the need to identify new biomarkers for the early detection diabetes risk, we previously showed that the curves of probability of disease-free status based on the plasma lipopolysaccharides (LPS) postprandial fold change, enhanced diabetes risk assessment compared with the previously cited FINDRISC score [6]. Current evidence links the development of metabolic diseases, including T2DM, to modifications in the composition of the gut microbiota [8], [9]. In the same context, the gut microbiota have potential to distinguish T2DM patients from non-diabetic individuals supported by the results of several studies [8]. However, these studies cannot conclusively prove the causality of the gut microbiota in T2DM development, since they were performed in already diagnosed T2DM patients whose gut microbiota could also be altered by several confounding factors, including the disease duration and antidiabetic drug treatment [10]. In contrast, our study has the added value of have being performed years before diabetes clinical diagnosis, supporting a stronger evidence of an association between a microbiota profile and T2DM development.
We developed a predictive model combining the baseline microbiome data and the traditional clinical risk parameters of ADA and FINRDRIS scores for T2DM. This model was able to predict accurately with a 95% sensitivity and a 79% specificity T2DM development on the test dataset.
In fact, our study applied the traditional clinical risk parameters of ADA and FINRDRISC scores for T2DM to patients with CHD and high T2DM risk. The analysis results indicated that the addition of baseline microbiome to the traditional clinical risk parameters significantly improve the prediction in CHD patients.
The performance obtained with the model of this study improve the recently reported models based on microbiome features combined with traditional risk factors to identify type 2 diabetes patients [11], [13]. It is noteworthy that, all the above reported models were obtained from cross-sectional studies. A prospective clinical investigation of gut microbiota as predictive factor of T2DM development had not been previously conducted. Therefore, in view of these findings, prospective studies might be more appropriate for the selection of the bacterial taxa associated, the robustness of predictive models and microbiome risk score construction.
Several of the bacterial taxa showing a high predictive power according to our model have previously been associated with metabolic diseases. For example, an abundance of Granulicatella has been positively related to MetS [21], body fat percentage and fasting glucose levels [22]. Prevotella has recently been related to IR [23]. The Incident-DIAB group was characterized by the preponderance of these two genera, and a high abundance of these was associated with a greater risk of diabetes development. The Sutterella genus has also been positively associated with MetS [24] and prediabetes [25]. Furthermore, the abundance of Prevotella and Paraprevotella, both highly discriminant in our model and more abundant in the Incident-DIAB group, has been reported to decrease after duodenal-endoluminal sleeve surgery in parallel with an improvement in glucose homeostasis [26]. The abundance of the Faecalibacterium [9] and Phascolarctobacterium [27], previously associated with beneficial effects in terms of Short-Chain Fatty Acid (SCFA) production, were also identified as significant in our model; the Incident-DIAB group had a significantly lower concentration of these bacteria. This is supported by the fact that Faecalibacterium is a butyrate-producing genus reduced in diabetic patients [8]. Of note, butyrate exerts anti-inflammatory properties [28]. Phascolarctobacterium is an acetate and propionate producer [29]; acetate is essential for butyrate synthesis via the butyryl CoA:acetate CoA transferase pathway, used by the Faecalibacterium genus [30].
Even though some of the highly predictive bacteria in our model differ from previous studies comparing microbiome from diagnosed diabetic patients with controls [8]. However, these differences can be attributed to the fact that, unlike the previous studies, the predictive models used in this study involved microbiome composition data that preceding the clinical diagnosis of T2DM. Overall, the microbiota profile related to T2DM incidence in our study is in agreement with the functionality previously proposed for these bacterial taxa.
In this context, Akkermansia has been reported by several studies to act beneficially through its involvement in intestinal barrier integrity and immune response [31]. However, in another study, diabetic patients exhibited a high abundance of Akkermansia, probably due to the effects of metformin which can increase these bacteria [8]. Notably, a major strength of our study is that microbiome was analyzed years before the clinical diagnosis, thus avoiding any effect produced by antidiabetic drug treatment. Despite the potential role of this bacterial genus in gut barrier integrity, Akkermansia appeared in the fifteenth position in our predictive model.
Moreover, we created a microbiome-based risk score with the ten most important bacterial taxa of the microbiome-based model. A high risk score, implying a harmful effect of these bacteria, was related with a greater risk of becoming diabetic, highlighting the potentiality of certain gut microbiota to identify high risk patients in a CHD population. To explore the underlying pathophysiological mechanisms that might support these results, we analyzed the relationship between the microbiome-based risk score and the OGTT-derived indexes. Our results suggested that patients with a high risk microbiome-based score had increased beta-cell dysfunction and IR throughout the study, as evidenced by a lower DI and a higher HIRI, respectively. However, despite these observations and the elevated incidence of T2DM in our CHD population, higher than the observed in the whole population [32], we observed a higher AUC in the ROC analysis of the models including microbiome dataset than those including OGTT-derived indexes, suggesting gut microbiome as a major associated factor of T2DM development.
We also found that a lower DI was related to a high baseline abundance of Dialister. Dialister invisus is a non-SCFA producer species, whose growth is enhanced by succinate [33]. It is plausible that a higher abundance of Dialister genus, as observed at baseline in Incident-DIAB group, may represent a higher abundance of this succinate-consuming species, a precursor of propionate and butyrate production [34]. Consequently, the production of these SCFAs may be decreased, leading to a depletion of glucagon-like peptide 1 (GLP-1) and insulin production, since these SCFAs can lead to the secretion of GLP-1 by binding to G-protein coupled receptors (GPCRs) from L-cells; GLP-1 promotes insulin secretion and beta-cell proliferation [9]. This suggestion is supported by the reduction of D. invisus abundance in post-bariatric surgery diabetic patients, together with an improvement in their metabolic status [35]. Furthermore, the abundance of Dialister was positively correlated with a high HbA1c in prediabetic patients [36]. Besides an increased abundance of this genus was also observed in diabetic patients without metformin treatment compared with controls [36].
Intestinal microbiota may regulate beta-cell functionality through bile acid metabolism. Certain secondary bile acids synthesized by the gut microbiota have been recognized as ligands for the membrane-bound GPCR TGR5 in the lumen, whose activation leads to GLP-1 production [37]. We also observed a lower DI in patients with a high baseline abundance of an unidentified genus belonging to the Erysipelotrichaceae family, that has been linked to host lipid and cholesterol metabolism [38]. Actually, the abundance of this bacterial family is negatively related to fecal cholesterol excretion [39]. Given this association, a higher abundance of the Erysipelotrichaceae unclassified genus may reflect reduced bile acid fecal excretion, therefore, bile acid levels in the colon and, consequently, lower secondary bile acid production. This, in turn, may downregulate TGR5 and decrease the secretion of GLP-1 and glucose-stimulated insulin. This suggested pathway has been supported by the fact that bile acid sequestrants (enhancing fecal excretion of bile acids) produces a glucose-lowering effect and increase GLP-1 levels in diabetes [40].
Apart from other potential mechanisms by which a specific microbiota pattern may trigger T2DM development, metabolic endotoxemia may play a crucial role. In this context, gut microbiota can influence intestinal permeability, hence, affect the absorption of pro-inflammatory bacterial components, such as LPS, which have been shown to induce hepatic IR [41]. We have previously shown in the same population that higher postprandial endotoxemia precedes the development of T2DM [6]. The increased LPS levels may be partially responsible for the higher HIRI observed in this study in patients with a high risk score, because the liver is known to be the first target of LPS-induced IR [41]. Furthermore, the baseline abundance of Phascolarctobacterium and Faecalibacterium had a positive effect on HIRI progression, presumably through the butyrate-induced reduction in inflammation and the improved intestinal barrier integrity [28], [42]. Another potential mechanism linking the gut microbiota profile to hepatic IR may involve the modulation of serum Branched-Chain Amino Acids (BCAA) levels. Our study showed that HIRI could also be negatively affected by the baseline abundance of Prevotella, which has been reported as a potential biosynthetic genus for BCAAs, driving the positive association between elevated BCAAs levels and IR [23].
This study has limitations. Really, it was a secondary study conducted in the non-diabetic subgroup at baseline, as T2DM prevention was not the main objective of the CORDIOPREV study. Therefore, it limits our findings to individuals with this comorbidity. Nevertheless, T2DM prediction is essential, since patients with concurrent CHD and diabetes have a considerably greater risk of having a new cardiovascular event than those without diabetes [3]. It would be ideal to validate it in a cohort without CHD in order to unequivocally confirm and extend our findings to the general population.
Conclusion
In conclusion, the current study supports a potential role of the intestinal microbiota as an associated factor of T2DM development. The results showed a microbiota profile associated to the T2DM that improved the prediction of diabetes development when added to traditional clinical parameters. Moreover, we built a risk score based on the ten most important bacteria of the predictive model representative of the likelihood of developing diabetes. In fact, patients with high risk score values had higher the hepatic insulin resistance and beta-cell disruption. Therefore, the use of microbiome data combined with clinical parameters may improve the early identification of patients at risk for T2DM in clinical practice.
Ethical statement
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). Informed consent was obtained from all patients for being included in the study.
CRediT authorship contribution statement
Cristina Vals-Delgado: Investigation, Writing - original draft, Investigation, Data curation, Writing - review & editing, Conceptualization. Juan F. Alcala-Diaz: Investigation, Conceptualization. Helena Molina-Abril: Writing - original draft. Irene Roncero-Ramos: Investigation. Martien P.M. Caspers: Data curation, Formal analysis. Frank H.J. Schuren: Data curation, Formal analysis. Tim J. Van den Broek: Data curation, Formal analysis. Raul Luque: Methodology. Pablo Perez-Martinez: Methodology. Niki Katsiki: Methodology. Javier Delgado-Lista: Methodology. Jose M. Ordovas: Methodology. Ben van Ommen: Writing - review & editing. Antonio Camargo: Investigation, Investigation, Data curation, Methodology, Writing - review & editing. Jose Lopez-Miranda: Investigation, Data curation, Writing - review & editing, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The CIBEROBN is an initiative of the Instituto de Salud Carlos III, Madrid, Spain. We want to acknowledge the Biobank of the Sistema Sanitario Público de Andalucía (Andalusia, Spain) (Córdoba branch), for the human biological samples provided. We also thank the performed the randomization process by the EASP (Escuela Andaluza de Salud Publica), Granada, Spain. The CORDIOPREV study is supported by Ministerio de Economía y Competitividad (PIE14/00005, 14/00031, and AGL2012/39615, to J. L.-M.; AGL2015-67896-P to J.L.-M. and A.C.; FIS PI13/00023 to J.D.-L., PI16/01777 to P.P.-M. FIS PI19/00299 to A.C.; DTS19/00007 to A.C., CP14/00114 to A.C.); Fundación Patrimonio Comunal Olivarero, Diputaciones de Jaén y Córdoba, Junta de Andalucía (Consejería de Salud, Consejeria de Innovación, Ciencia y Empresa Consejería de Agricultura y Pesca,), Ministerio de Medio Ambiente, Medio Rural y Marino, Gobierno de España; Consejeria de Innovación, Ciencia y Empresa, and Centro de Excelencia en Investigación sobre Aceite de Oliva y Salud, , Junta de Andalucía (Proyectos de Investigación de Excelencia CVI-7450 to J.L.-M.); J.M.O. is supported by the US Department of Agriculture, under agreement no. 8050-51000-098-00D. We also thank the European Reginal Development Fond. We also thank Jose Andrés Morales Martínez for his technical support.
Data availability
The sequences obtained for this study are open available in the NCBI Sequence Read Archive (SRA) repository at https://www.ncbi.nlm.nih.gov/sra/PRJNA675744, reference number PRJNA675744.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.05.001.
Contributor Information
Antonio Camargo, Email: antonio.camargo@imibic.org.
Jose Lopez-Miranda, Email: jlopezmir@uco.es.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Halban P.A., Polonsky K.S., Bowden D.W., Hawkins M.A., Ling C., Mather K.J., et al. Β-cell failure in type 2 diabetes: Postulated mechanisms and prospects for prevention and treatment. Diab Care. 2014;37:1751–1758. doi: 10.2337/dc14-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samuel Varman T., Shulman Gerald I. Mechanisms for insulin resistance: Common threads and missing links. Cell. 2012;148:852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeFronzo RA. Insulin resistance, lipotoxicity, type 2 diabetes and atherosclerosis: The missing links. The Claude Bernard Lecture 2009. Diabetologia 2010;53:1270–1287. [DOI] [PMC free article] [PubMed]
- 4.American Diabetes Association. 2. Classification and diagnosis of diabetes: Standards of medical care in diabetes—2019. Diab Care 2019;42 (Suppl 1):S13–S28. [DOI] [PubMed]
- 5.Jølle A., Midthjell K., Holmen J., Carlsen S.M., Tuomilehto J., Bjørngaard J.H., et al. Validity of the FINDRISC as a prediction tool for diabetes in a contemporary Norwegian population: A 10-year follow-up of the HUNT study. BMJ Open Diab Res Care. 2019;7 doi: 10.1136/bmjdrc-2019-000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camargo A., Jimenez-Lucena R., Alcala-Diaz J.F., Rangel-Zuñiga O.A., Garcia-Carpintero S., Lopez-Moreno J., et al. Postprandial endotoxemia may influence the development of type 2 diabetes mellitus: From the CORDIOPREV study. Clin Nutr. 2019;38:529–538. doi: 10.1016/j.clnu.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 7.Bennet L., Groop L., Lindblad U., Agardh C.D., Franks P.W. Ethnicity is an independent risk indicator when estimating diabetes risk with FINDRISC scores: A cross sectional study comparing immigrants from the Middle East and native Swedes. Prim Care Diabetes. 2014;8:231–238. doi: 10.1016/j.pcd.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Tilg H., Moschen A.R. Microbiota and diabetes: An evolving relationship. Gut. 2014;63:1513–1521. doi: 10.1136/gutjnl-2014-306928. [DOI] [PubMed] [Google Scholar]
- 9.Tremaroli V., Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 10.Forslund K., Hildebrand F., Nielsen T., Falony G., Le Chatelier E., Sunagawa S., et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H., Tremaroli V., Schmidt C., Lundqvist A., Olsson L.M., Krämer M., et al. The gut microbiota in prediabetes and diabetes: A population-based cross-sectional study. Cell Metab. 2020;32(379–90) doi: 10.1016/j.cmet.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Vrieze A., Van Nood E., Holleman F., Salojärvi J., Kootte R.S., Bartelsman J.F.W.M., et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(913–6) doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 13.Gou W., Ling C-w, He Y., Jiang Z., Fu Y., Xu F., et al. Interpretable machine learning framework reveals robust gut microbiome features associated with type 2 diabetes. Diabetes Care. 2021;44:358–366. doi: 10.2337/dc20-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delgado-Lista J., Perez-Martinez P., Garcia-Rios A., Alcala-Diaz J.F., Perez-Caballero A.I., Gomez-Delgado F., et al. CORonary Diet Intervention with Olive oil and cardiovascular PREvention study (the CORDIOPREV Study): Rationale, methods, and baseline characteristics: A clinical trial comparing the efficacy of a Mediterranean diet rich in olive oil versus a low-fat diet on cardiovascular disease in coronary patients. Am Heart J. 2016;177:42–50. doi: 10.1016/j.ahj.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernández-Ballart J.D., Piñol J.L., Zazpe I., Corella D., Carrasco P., Toledo E., et al. Relative validity of a semi-quantitative food-frequency questionnaire in an elderly Mediterranean population of Spain. Br J Nutr. 2010;103:1808–1816. doi: 10.1017/S0007114509993837. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Gonzalez M.A., Fernandez-Jarne E., Serrano-Martinez M., Wright M., Gomez-Gracia E. Development of a short dietary intake questionnaire for the quantitative estimation of adherence to a cardioprotective Mediterranean diet. Eur J Clin Nutr. 2004;58:1550–1552. doi: 10.1038/sj.ejcn.1602004. [DOI] [PubMed] [Google Scholar]
- 17.Blanco-Rojo R., Alcala-Diaz J.F., Wopereis S., Perez-Martinez P., Quintana-Navarro G.M., Marin C., et al. The insulin resistance phenotype (muscle or liver) interacts with the type of diet to determine changes in disposition index after 2 years of intervention: The CORDIOPREV-DIAB randomised clinical trial. Diabetologia. 2016;59:67–76. doi: 10.1007/s00125-015-3776-4. [DOI] [PubMed] [Google Scholar]
- 18.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the Miseq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z., Kattan M.W. Drawing nomograms with R: Applications to categorical outcome and survival data. Ann Transl Med. 2017;5:211. doi: 10.21037/atm.2017.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Si J., Lee C., Ko G. Oral microbiota: Microbial biomarkers of metabolic syndrome independent of host genetic factors. Front Cell Infect Microbiol. 2017;7:516. doi: 10.3389/fcimb.2017.00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaakoush N.O., Lecomte V., Maloney C.A., Morris M.J. Cross-talk among metabolic parameters, esophageal microbiota, and host gene expression following chronic exposure to an obesogenic diet. Sci Rep. 2017;7:45753. doi: 10.1038/srep45753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedersen H.K., Gudmundsdottir V., Nielsen H.B., Hyotylainen T., Nielsen T., Jensen B.A.H., et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 24.Lim M.Y., You H.J., Yoon H.S., Kwon B., Lee J.Y., Lee S., et al. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut. 2017;66:1031–1038. doi: 10.1136/gutjnl-2015-311326. [DOI] [PubMed] [Google Scholar]
- 25.Allin K.H., Tremaroli V., Caesar R., Jensen B.A.H., Damgaard M.T.F., Bahl M.I., et al. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia. 2018;61:810–820. doi: 10.1007/s00125-018-4550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim T., Holleman C.L., Ptacek T., Morrow C.D., Habegger K.M. Duodenal endoluminal barrier sleeve alters gut microbiota of ZDF rats. Int J Obes. 2017;41:381–389. doi: 10.1038/ijo.2016.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X., Zhao Y., Xu J., Xue Z., Zhang M., Pang X., et al. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci Rep. 2015;5:14405. doi: 10.1038/srep14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan H., Ajuwon K.M. Butyrate modifies intestinal barrier function in IPEC-J2 cells through a selective upregulation of tight junction proteins and activation of the Akt signaling pathway. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0179586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe Y., Nagai F., Morotomi M. Characterization of Phascolarctobacterium succinatutens sp. nov., an asaccharolytic, succinate-utilizing bacterium isolated from human feces. Appl Environ Microbiol. 2012;78:511–518. doi: 10.1128/AEM.06035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duncan S.H., Holtrop G., Lobley G.E., Calder A.G., Stewart C.S., Flint H.J. Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr. 2007;91:915–923. doi: 10.1079/BJN20041150. [DOI] [PubMed] [Google Scholar]
- 31.Derrien M., Belzer C., de Vos W.M. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog. 2017;106:171–181. doi: 10.1016/j.micpath.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Razquin C., Toledo E., Clish C.B., Ruiz-Canela M., Dennis C., Corella D., et al. Plasma lipidomic profiling and risk of type 2 diabetes in the PREDIMED trial. Diabetes Care. 2018;41:2617–2624. doi: 10.2337/dc18-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jumas-Bilak E., Jean-Pierre H., Carlier J.-P., Teyssier C., Bernard K., Gay B., et al. Dialister micraerophilus sp. nov. and Dialister propionicifaciens sp. nov., isolated from human clinical samples. Int J Syst Evol Microbiol. 2005;55:2471–2478. doi: 10.1099/ijs.0.63715-0. [DOI] [PubMed] [Google Scholar]
- 34.Louis P., Flint H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 35.Graessler J., Qin Y., Zhong H., Zhang J., Licinio J., Wong M.L., et al. Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: Correlation with inflammatory and metabolic parameters. Pharmacogenomics J. 2013;13:514–522. doi: 10.1038/tpj.2012.43. [DOI] [PubMed] [Google Scholar]
- 36.Barengolts E., Green S.J., Eisenberg Y., Akbar A., Reddivari B., Layden B.T., et al. Gut microbiota varies by opioid use, circulating leptin and oxytocin in African American men with diabetes and high burden of chronic disease. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0194171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wahlström A., Sayin Sama I., Marschall H.-U., Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 38.Kaakoush N.O. Insights into the role of Erysipelotrichaceae in the human host. Fronti Cell Infect Microbiol. 2015;5:84. doi: 10.3389/fcimb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martínez I., Perdicaro D.J., Brown A.W., Hammons S., Carden T.J., Carr T.P., et al. Diet-induced alterations of host cholesterol metabolism are likely to affect the gut microbiota composition in hamsters. Appl Environ Microbiol. 2013;79:516–524. doi: 10.1128/AEM.03046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prawitt J., Caron S., Staels B. Glucose-lowering effects of intestinal bile acid sequestration through enhancement of splanchnic glucose utilization. Trends Endocrinol Metab. 2014;25:235–244. doi: 10.1016/j.tem.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Cani P.D., Amar J., Iglesias M.A., Poggi M., Knauf C., Bastelica D., et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 42.Plöger S., Stumpff F., Penner G.B., Schulzke J.-D., Gäbel G., Martens H., et al. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci. 2012;1258:52–59. doi: 10.1111/j.1749-6632.2012.06553.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences obtained for this study are open available in the NCBI Sequence Read Archive (SRA) repository at https://www.ncbi.nlm.nih.gov/sra/PRJNA675744, reference number PRJNA675744.