Summary
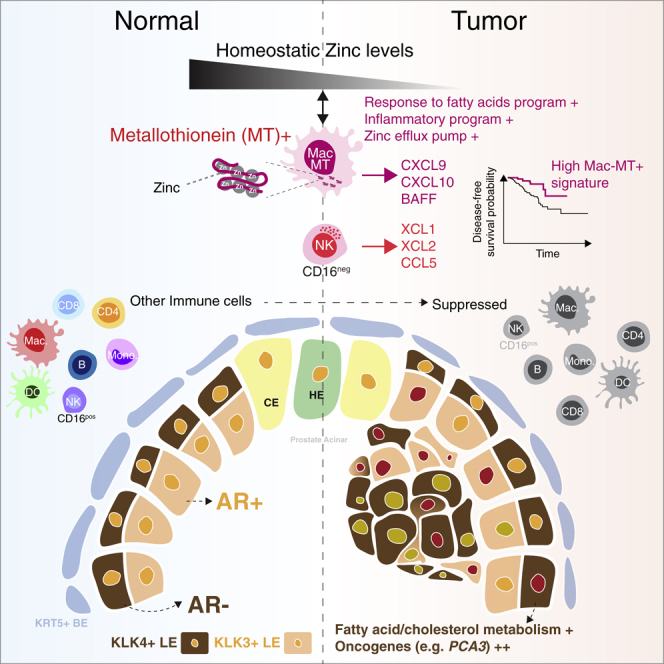
The prostate gland produces prostatic fluid, high in zinc and citrate and essential for the maintenance of spermatozoa. Prostate cancer is a common condition with limited treatment efficacy in castration-resistant metastatic disease, including with immune checkpoint inhibitors. Using single-cell RNA-sequencing to perform an unbiased assessment of the cellular landscape of human prostate, we identify a subset of tumor-enriched androgen receptor-negative luminal epithelial cells with increased expression of cancer-associated genes. We also find a variety of innate and adaptive immune cells in normal prostate that were transcriptionally perturbed in prostate cancer. An exception is a prostate-specific, zinc transporter-expressing macrophage population (MAC-MT) that contributes to tissue zinc accumulation in homeostasis but shows enhanced inflammatory gene expression in tumors, including T cell-recruiting chemokines. Remarkably, enrichment of the MAC-MT signature in cancer biopsies is associated with improved disease-free survival, suggesting beneficial antitumor functions.
Keywords: single-cell RNA sequencing, macrophage, zinc, immune landscape, human prostate, prostate cancer
Graphical abstract
Highlights
-
•
An immune cell atlas of healthy human prostate and prostate cancer
-
•
Low androgen receptor-expressing luminal epithelial cell present in prostate cancer
-
•
Metallothionein-expressing macrophage subset (MAC-MT) regulates prostate zinc
-
•
MAC-MT gene signature in prostate cancer associated with better outcomes
Tuong et al. generated a single-cell transcriptomic map of the human prostate immune landscape in health and show how this is perturbed in cancer. They identify a prostate-specific macrophage population that helps maintain tissue zinc and is associated with better outcomes in cancer.
Introduction
The prostate gland is critical for human reproduction, generating prostatic fluid that is high in zinc and citrate. This forms an essential component of seminal fluid that is required for the maintenance of spermatozoa (Costello and Franklin, 2016). Prostatic acini are comprised of an outer basal cell layerand inner layers of secretory luminal epithelial (LE) cells; as well as neuroendocrine, stromal, immune, endothelial, and nerve cells (Shen and Abate-Shen, 2010, Henry et al., 2018). Prostate cancer is a major cause of cancer-related mortality and morbidity in men (Ferlay et al., 2015) and is characterized by a reduction in zinc and citrate concentration in both glandular tissue and prostatic fluid (Costello and Franklin, 2016). Cancer cells display a luminal phenotype but the cellular origin of prostate cancer is debated with lineage tracing studies in mice indicating that they may arise from both basal and luminal cells (Choi et al., 2012, Wang et al., 2013, Wang et al., 2014).
There is limited information on the nature and composition of the tissue-resident immune cell compartment in the healthy human prostate (Shen and Abate-Shen, 2010, Henry et al., 2018). However, several tumor-associated immune cell subsets have been reported, including T and B lymphocytes, regulatory T cells, monocytes, macrophages, dendritic cells (DCs), and natural killer (NK) cells (Hussein et al., 2009, Solinas et al., 2017), some of which correlate with a worse prognosis—for example, regulatory T cells (Davidsson et al., 2013) and CD163-positive M2 macrophages (Erlandsson et al., 2019).
Prostate cancer is classified according to the ISUP Grade Group system (Epstein et al., 2016) and the Gleason Grading System, based on the extent to which tissue architecture and cellular morphology are disrupted, with higher scores associated with more aggressive tumor growth and worse outcomes. While primary localized disease has a generally good prognosis, men with locally advanced and metastatic disease have much worse 10-year survival rates (Gnanapragasam et al., 2016). Androgen deprivation therapy is the mainstay of treatment for de novo metastatic prostate cancer, but a proportion of patients progress to castration-resistant disease, although the mechanisms underpinning this are unclear (Watson et al., 2015). Immune checkpoint blockade with antibodies against cytotoxic-T-lymphocyte-associated protein 4 (CTLA4) or programmed cell death 1 (PD-1)/PD-1 ligand 1 (PD-L1) has been used in these patients, but results have largely been disappointing (Kwon et al., 2014, Beer et al., 2017), consistent with reports that less than a third of tumors show evidence of PD1 or PDL1 expression (Haffner et al., 2018). Therefore, there is an urgent need to better understand tissue immunity in the healthy prostate and the nature of its perturbation in prostate cancer to inform future therapeutic strategies. Here, we applied single-cell RNA sequencing (scRNaseq) to paired human prostate biopsies collected at the time of cancer diagnosis, to comprehensively profile the cellular landscape of normal human prostate and to determine how this becomes disrupted in cancer. We validated our findings using flow cytometry, immunofluorescence microscopy, spatial transcriptomics; and by cross-species studies of mouse prostate specimens.
Results
Single-cell landscape of healthy prostate and prostate cancer
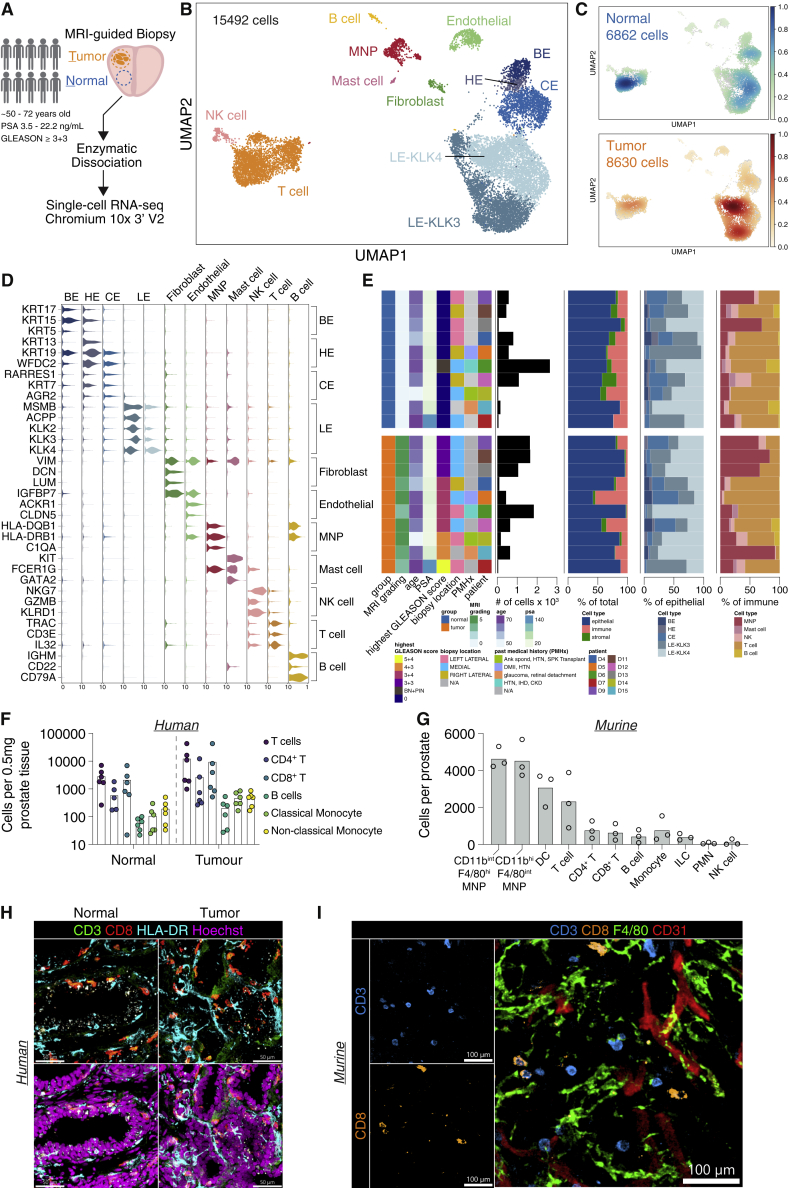
We performed scRNaseq on paired cancer biopsy and adjacent normal prostate tissue in n = 10 subjects aged 50–72 years of age (Figure 1A, Table S1) generating data on 15,492 cells post-QC. We identified 14 cell clusters (Figure 1B), all of which contained cells from both normal and cancer biopsies (Figures 1C and S1A). This included immune cells, endothelial cells, fibroblasts, as well as several epithelial cell subtypes (Figure 1B), that were annotated based on canonical marker expression and comparison with a previously published scRNA seq dataset of young, healthy human prostate tissue (Henry et al., 2018) (Figures 1D and S1B).
Figure 1.
Single-cell RNA sequencing reveals immune and epithelial cell heterogeneity in paired normal-cancer samples
(A) Schematic describing experimental set-up for 10x genomic single-cell RNaseq of matched tumor-normal prostate samples from n = 10 patients.
(B) UMAP of 15,492 cells post-QC from all prostate samples.
(C) UMAP of embedding density of source of samples (normal—green to blue gradient, top; tumor—orange to red gradient, bottom).
(D) Violin plot of canonical marker genes for each cell types found in prostate. Gene expression values per cell are standardized to a range from 0 to 10.
(E) Patient demographics displayed as a color-coded heatmap and stacked bar charts of single-cell cell type proportions.
(F) Quantification of absolute cells counts by flow cytometry per 0.5mg of normal and malignant human prostatic tissue for the indicated immune subsets. Data are a combination of n = 6 donors with each dot representing an individual donor.
(G) Quantification of absolute cells counts by flow cytometry per murine prostate lobe for the indicated immune subsets. Each dot represents an individual mouse (n = 3 biological replicates). Abbreviations: MNP—mononuclear phagocyte; DC—dendritic cell; ILC—innate-like lymphoid cell; PMN—polymorphonuclear; NK natural killer (H) Confocal imaging of CD3, CD4 and CD8 in human normal and tumor prostate. Scale bars, 50 μm. (I) Confocal imaging of CD3, CD8, F4/80 and CD31 in normal murine prostate. Scale bars, 100 μm. See also Figure S1 and Table S1.
Immune cell clusters included mononuclear phagocytes (MNPs), mast cells, NK cells, B cells, and T cells (Figure 1D) that were present in both normal and cancer samples with similar frequency (Figure 1E). We used flow cytometric analysis and confocal imaging to validate the presence of the major immune cell subsets in normal prostate tissue in human, and performed a cross-species comparison in mice, using a CD45 antibody administered intravenously premortem to label intravascular cells and confirm bona fide tissue residency (Figures 1F, 1G, 1H, 1I, S1C, S1D, and S1E). Together, these data show that the healthy prostate has a rich immune landscape dominated by T cells and MNPs, and that these cells persist in prostate cancer.
Distinct subset of luminal epithelial cells enriched in cancer
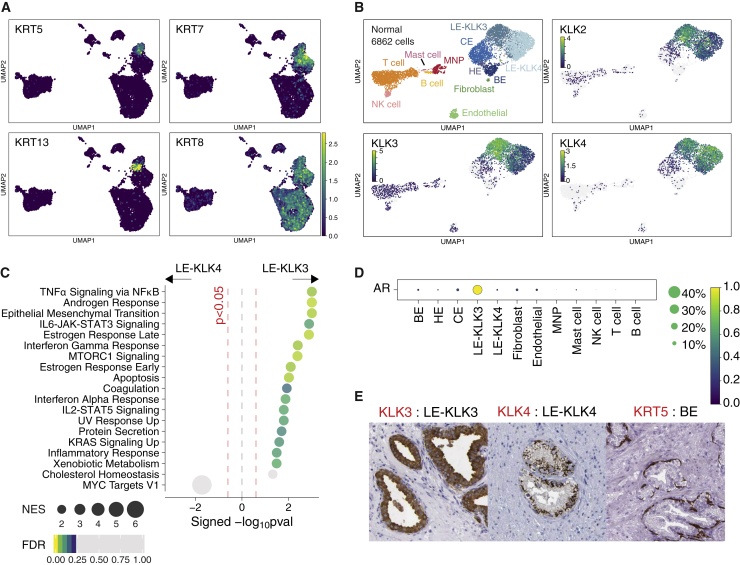
Among nonimmune cell clusters, we identified basal, hillock, and club cell clusters, as noted in previous single cell analyses of human prostate (Henry et al., 2018, Karthaus et al., 2020), but in contrast to published data, we found two distinct clusters of cytokeratin-8+ luminal epithelial (LE) cells, rather than a single LE cell population (Figure 2A). The two LE cell populations were present in normal prostate and prostate cancer samples and were transcriptionally distinct with one cluster expressing high levels of KLK3 (encoding kallikrein related peptidase 3—also known as prostate-specific antigen [PSA]), KLK2, and KLK4 (annotated as LE-KLK3)—and the second cluster expressing high levels of KLK4 and little KLK3 (annotated as LE-KLK4) (Figure 2B). Cells with a high degree of transcriptional similarity to LE-KLK4 were confirmed to be present in the previously published healthy prostate dataset generated from three organs donors aged 18–31 years of age (Henry et al., 2018; Figure S2A), but the limited number of LE-KLK4 cells in this dataset did not enable their identification as a distinct subset in the previously published analysis (Henry et al., 2018). Similarly, examination of previously published prostate cancer single-cell datasets (Karthaus et al., 2020, Chen et al., 2021, Crowley et al., 2020) also identified cells with high transcriptional similarity to LE-KLK3 and LE-KLK4 (Figures S2B–S2D). LE-KLK3 demonstrated enrichment for several immune pathways, including ‘TNFa via NFKB signaling’, ‘IL6-JAK-STAT3 signaling’ ‘interferon gamma response’, as well as ‘androgen and estrogen response’, with LE-KLK4 showing some enrichment for ‘Myc target’ genes suggestive of proliferative activity (Figure 2C). Furthermore, a subset of LE-KLK4 cells enriched for a prostate tumor-associated proliferation ‘Polaris’ signature (Figure S3A) and included cells with a G2/M cell-cycle profile (Figure S1A). In keeping with the high expression of ‘androgen response’ pathway genes in LE-KLK3, the gene encoding the androgen receptor (AR) was also highly expressed in this cluster, with little AR expression in LE-KLK4 (Figure 2D). Protein atlas data demonstrated the presence of KLK4+ cells between KLK5+ basal cells and KLK3+ cells adjacent to the lumen (Figure 2E). Colocalization of LE-KLK3 and LE-KLK4 cells was also confirmed in a previously published spatial transcriptomics dataset (Berglund et al., 2018; Figure S3B). The closer spatial proximity of LE-KLK3 to the lumen compared with LE-KLK4 is consistent with the transcriptional enrichment in immune defense genes observed in LE-KLK3, as these cells present an interface with the external environment and potential pathogen challenge.
Figure 2.
Androgen-receptor-negative prostate luminal epithelial cell type
(A) UMAP expression plot of keratin genes in prostate cells. Increasing color gradient from purple, blue, green to yellow corresponds to increasing (standardized) expression value.
(B) UMAP of normal prostate sample cells. Expression of kallikrein genes marking luminal epithelial cell types, including luminal cell type, is presented as a heatmap where cells with no expression (0 expression) are colored gray and increasing expression is colored according to increasing gradient from purple, blue, green to yellow. (C) Pre-ranked GSEA of hallmark gene sets between normal KLK3+ versus KLK4+ LE clusters. Pathways with FDR < 0.25 are colored from purple, blue, green to yellow according to decreasing FDR value. Grey circles indicate pathways that attained p < 0.05 and FDR > 0.25. Size of circles indicate the significance (signed -log10(p value)).
(D) Mean expression dot plot of gene encoding androgen receptor (AR). Expression values are scaled from 0 to 1. Size of circles indicate percentage of cells expressing the gene and increasing color gradient from purple, blue, green to yellow corresponds to increasing (standardized) expression value.
(E) Immunohistochemistry images of KLK3, KLK4 and KRT5 in prostate tissue. Images are sourced from the Human protein atlas (https://www.proteinatlas.org). See also Figures S2–S3.
Zinc transporter-expressing prostate-specific macrophage population
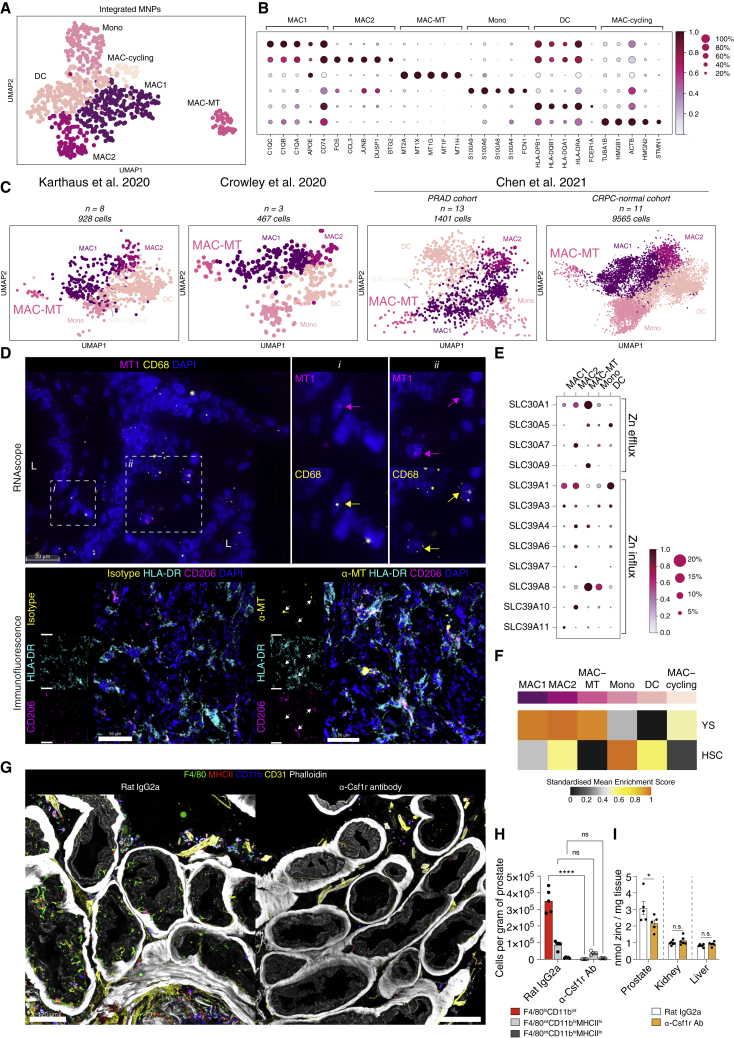
We next considered MNPs in the human prostate in isolation and integrated scRNaseq data from MNPs in a published normal prostate dataset (Henry et al., 2018; Figure S4A). We found six distinct clusters of MNPs that exhibited transcriptional profiles consistent with their identity as monocytes (Mono), conventional DCs (cDCs), proliferating macrophages (MAC-cycling), and macrophages (MAC1, MAC2 and MAC-MT) (Figures 3A, S4B, and S4C). Two major subsets of cDCs are recognized, cDC1 and cDC2. cDC1 express XCR1 and cross-present antigens to CD8 T cells, while cDC2 express CD1c and activate CD4 T cells (Guilliams et al., 2014). In our dataset, we did not find two distinct DC clusters, but both CD1C positive and negative cells were evident within the DC cluster (Figure S4B). Two of the three macrophage populations were transcriptionally similar (MAC1 and MAC2) (Figure 3B), while MAC-MT cells formed a completely distinct cluster (Figure 3A) and expressed high levels of the metallothionein family genes (Figure 3B), encoding cysteine-rich proteins that bind divalent heavy metal ions and are involved in the cellular transport, storage, and metabolism of metal ions (Miles et al., 2000). High expression of metallothioneins has been noted in prostate cancer (Wei et al., 2008, Wang et al., 2018), but the MAC-MT cluster contained cells from both our dataset and that generated from normal, young human prostate tissue (Henry et al., 2018; Figure S4C), confirming that these cells exist in healthy prostate tissue in homeostasis. We also validated the presence of MAC-MT in the three other published scRNaseq datasets of both normal prostate and prostate cancer (Figures 3C and S4D).
Figure 3.
Immune landscape of the prostate includes a prostate-specific macrophage subset enriched in metallothionein transcripts
(A) UMAP of 793 cells in myeloid compartment after integration of myeloid/MNP cells from n = 10 patients with Henry et al. myeloid/MNP cells.
(B) Mean expression dot plot of top five significant marker genes for each myeloid cluster. Marker genes were identified using Wilcoxon rank sum test and p adj < 0.05 was considered statistically significant. Size of circles indicate percentage of cells expressing the gene and increasing color gradient from white to red corresponds to increasing expression value.
(C) UMAP plot of predicted MNP clusters in prostate cancer single cell data from (Karthaus et al., 2020, Chen et al., 2021, Crowley et al., 2020).
(D) (top) Representative RNAscope images of probes targeting MT1 family genes (magenta) and CD68 (yellow). ‘L’ indicates lumen. Arrows point to single cells that are marked by both probes in sub-panels i and ii. Scale bar, 20 μm. (bottom) Representative immunofluorescence microscopy images of a human prostate section labeled for metallothionein (α-MT)/isotype control (yellow), HLA-DR (cyan), CD206 (purple) and DAPI (blue). White arrows point to structure displaying colocalization of α-MT with HLA-DR and/or CD206 labeling. Scale bars, 50 μm.
(E) Mean expression dot plot of Zinc transporter genes for each myeloid cluster. Size of circles indicate percentage of cells expressing the gene and increasing color gradient from white to red corresponds to increasing expression value.
(F) Heatmap of mean AUCell enrichment of F4/80hi/lo gene sets, corresponding to yolk sac (YS) versus hematopoetic stem-cell (HSC) lineage. Row enrichment value is scaled from 0 to 1 and presented as an increasing gradient from black, gray, yellow to orange which corresponds to increasing enrichment score.
(G) Representative immunofluorescence microscopy images of cross sections of mouse prostate labeled for F4/80 (green), MHCII (red), CD11b (blue), CD31 (yellow) and phalloidin. Scale bars, 120 μm.
(H) Cell counts per gram of prostate for rat IgG2a isotype or anti-Csf1r antibody (Ab) treated male mice. N = 5 per group. ∗∗∗∗p < 0.0001; n.s denotes not significant (p > 0.05) (Two-way ANOVA with Tukey’s multiple correction).
(I) Zinc concentration of anterior prostate lobe, liver lobe, and kidney from male mice treated with either rat IgG2a isotype control or anti-Csf1r Ab. N = 6 per group. (shown is representative quantification from one of two independent experiments). ∗p < 0.05; n.s. not significant (Mann-Whitney test). See also Figures S4–S5.
Spatially, MAC-MT cell signatures colocalized with those of LE-KLK3 and LE-KLK4 (Figure S4E), and we confirmed the presence of an MT1+ CD68+ macrophage population in human prostate using both RNA scope and immunofluorescence microscopy, which were localized adjacent to luminal regions (Figure 3D). Zinc accumulation is controlled by two families of zinc transporters, the SLC39 (Zrt- and Irt-like proteins [ZIP]) that increase intracellular zinc, and the SLC30 (ZnT) proteins that lower zinc cellular levels (Liuzzi and Cousins, 2004). Interestingly, among prostate MNPs, the MAC-MT subset expressed the highest level of the zinc transporter genes, SLC39A8 (ZIP-8) and SLC30A1 (ZNT-1) (Figure 3E). MAC-MT cells were unique to the prostate, with no transcriptionally similar cells identified in other human organs (Figure S4F). Together, these data suggest that MAC-MT represent a prostate-specific macrophage subset, adapted to residency within the high zinc environment (Costello and Franklin, 2016).
Tissue macrophages arise prenatally from yolk sac (YS) or fetal liver progenitors, but are variably replaced postnatally by monocyte precursors that adopt tissue-specific transcriptional profiles (Ginhoux and Guilliams, 2016, Mass et al., 2016). All three macrophage clusters in human prostate showed transcriptional similarity to YS-derived macrophages (Schulz et al., 2012), as did the proliferating macrophage cluster, but MAC2 also enriched for the monocyte-derived macrophage signature (Figure 3F). Therefore, MAC1 and MAC-MT may represent prenatally seeded macrophage subsets, while MAC2 may be monocyte-derived, subsequently taking up a tissue-macrophage transcriptional signature. In keeping with this, mouse prostate contained both F4/80highCD11blo and F4/80loCD11bhigh macrophage subsets, (Figure S5A) identified as YS and hematopoetic stem cell (HSC)-derived subsets, respectively, in murine fate-mapping studies (Schulz et al., 2012). An analysis of a murine prostate scRNaseq dataset (Karthaus et al., 2020) also confirmed the presence of macrophage clusters with enrichment for both YS- and monocyte-derived macrophage signatures (Figure S5B). Anatomically, F4/80high macrophages were located among luminal epithelial cells in mouse prostate (Figure 3G). Expression of zinc transporters and metallothionein genes was also evident in mouse prostate macrophages, particularly F4/80hi cells enriching for the YS-derived macrophage signature (Figures S5C and S5D).
We therefore hypothesized that these prostate macrophages may contribute to zinc homeostasis in the organ. To test this, we used an anti-Csf1r antibody which effectively depleted prostate macrophages, particularly the F4/80hi subset (Figure 3G, 3H, S5E, and S5F). This led to a significant reduction in prostate zinc concentration (Figure 3I), showing that prostate macrophages play a role in maintaining tissue zinc levels. The tissue zinc concentration in other organs was unaffected by macrophage depletion (Figures 3I and S5F), indicating this is a prostate specific macrophage function.
Lymphoid immune landscape of human prostate
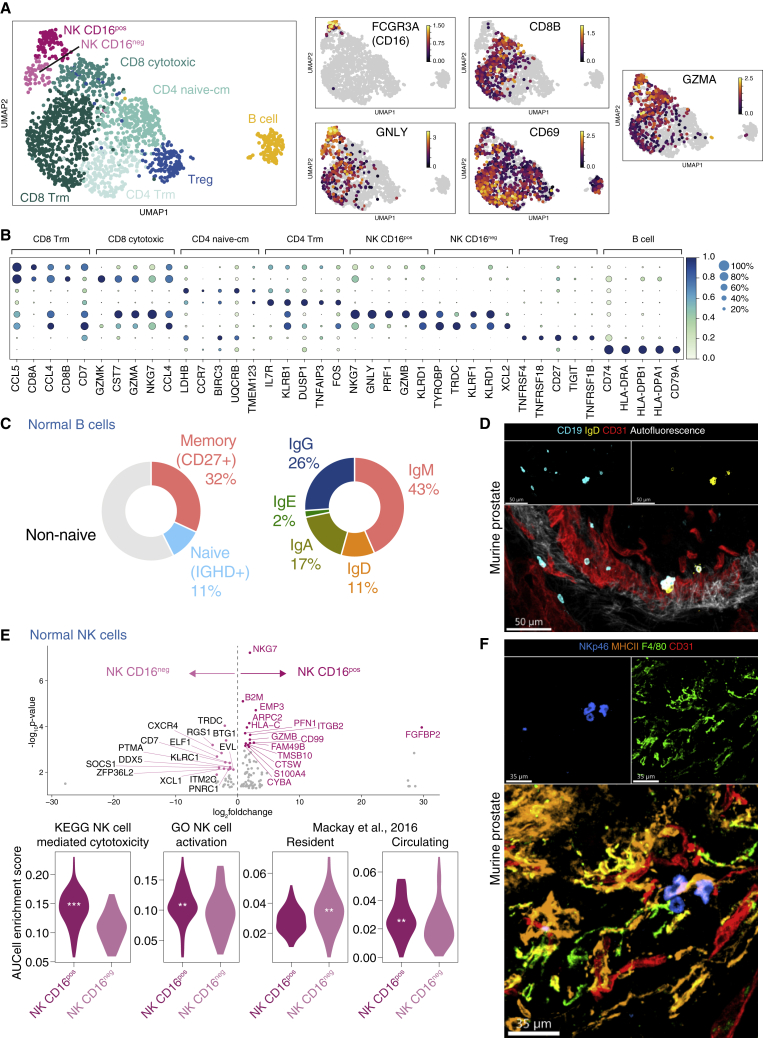
The lymphoid compartment of normal prostate included CD4 and CD8 T cells, two subsets of NK cells (CD16+ and CD16neg) and B cells (Figures 4A and 4B). Further analysis indicated the presence of naive, tissue-resident memory, and regulatory CD4 T cells, as well as cytotoxic and tissue-resident memory CD8 T-cell clusters (Figures 4A, 4B, and S6A) (Kumar et al., 2017) (Mackay et al., 2013) (Szabo et al., 2019). We categorized the B cells according to CD27 (a marker of memory B cells) and IGHD (a marker of naive B cells) expression and observed that ∼30% and 10% of B cells were memory (CD27+IGHD-) and naive (CD27-IGHD+), respectively, with the remaining fraction mature non-naive B cells (Figures 4C and S6B). The majority of IGHM-expressing cells were CD27+ IgM memory cells, and there were also some class-switched B cells, with only a handful of BLIMP1-expressing plasma cells observed (Figure S6B). In the mouse prostate, extravascular naive and non-naive B cells were also evident adjacent to luminal epithelial cells (Figure 4D).
Figure 4.
Lymphoid single-cell landscape of normal prostate and prostate cancer
(A) UMAP of 1694 lymphoid cells from n = 7 patients. Expression of marker genes for NK cells (FCGR3A, GNLY), CD8 T cells (CD8B), tissue residency and activation (CD69) and cytolytic molecule (GZMA) are shown as a heatmap where gray indicates no expression and increasing expression is colored from purple, orange to yellow.
(B) Dot plot of top five significant marker genes for each lymphoid clusters. Marker genes were identified using Wilcoxon rank sum test and p adj < 0.05 was considered statistically significant. Size of circles indicate percentage of cells expressing the gene and increasing color gradient from white to blue corresponds to increasing expression value.
(C) Pie chart showing proportion of cells expressing markers for (left) memory (CD27+IGHD-), naive (IGHD+CD27-), non-naive (remainder) and (right) heavy gene constant gene expression.
(D) Confocal imaging of CD19, IgG and CD31 in normal murine prostate section. Scale bars, 50 μm.
(E) (Top) Volcano plot showing top 15 significant DEGs between NK CD16pos and NK CD16neg (normal only). (Bottom) Violin plots of gene set testing (AUCell) for NK cell gene sets (KEGG and GO) and lymphocyte tissue residency gene sets from (Mackay et al., 2016). Significance is denoted by ∗∗p < 0.01; ∗∗∗p < 0.001 (Mann-Whitney test). Position of asterisks indicate the group with higher expression.
(F) Confocal imaging of NKp46, MHCII, F4/80 and CD31 in normal murine prostate section. Scale bars, 35 μm. See also Figure S6.
The identification of two subsets of NK cells in normal prostate is consistent with previous descriptions of NK cells in blood and other organs; the majority of peripheral blood NK cells are CD56dimCD16+, with a small subset of CD56brightCD16neg NK cells, termed tissue-resident NK cells that are also present in spleen, uterus, and liver (Shi et al., 2011, Cuff et al., 2016). Functionally, these CD16neg tissue-resident NK cells differ from conventional CD16+ NK cells, with reduced or altered cytotoxicity and prominent cytokine and chemokine production (Fauriat et al., 2010). In the prostate, CD16+ NK cells had a higher expression of genes associated with NK cell activation compared to CD16neg NK cells, with significant enrichment of NK cell cytotoxicity and activation gene sets (Narni-Mancinelli et al., 2012; Figure 4E). In addition, CD16neg NK cells showed enrichment for a universal lymphocyte tissue-residency signature (Mackay et al., 2016; Figure 4E), confirming that this subset represents tissue-resident NK cells and extending the list of tissues in which these cells have been identified in homeostasis (Dogra et al., 2020). The presence of CD16+ and CD16- subsets of NK cell was confirmed in an independent normal prostate single-cell dataset (Figure S6C), and NK cells were also evident in normal mouse prostate samples (Figure 4F).
Immune perturbation in prostate cancer
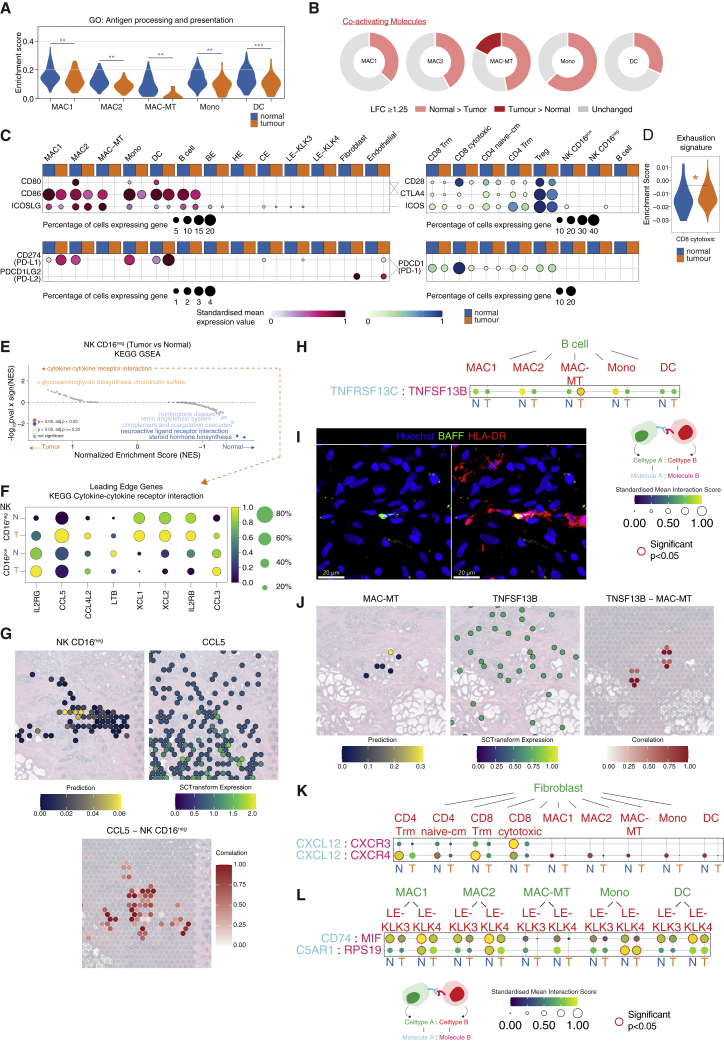
We next sought to determine how prostate tissue immune cells were perturbed in cancer. Although there was no significant difference in immune cell number in prostate cancer samples (Figure 1E), several transcriptional differences in immune cells were evident; Antigen presentation and processing pathway genes were significantly reduced in prostate MNPs in cancer samples compared with normal prostate (Figure 5A), consistent with a widespread attenuation of CD4 T cell activation, a process critical for the generation of antitumor adaptive immune responses. In line with this, the expression of many co-activating receptors was higher in normal prostate MNPs compared with those in prostate cancer, the exception being MAC-MT, which showed higher expression of a subset of co-activating receptors in tumor samples, including CD40 (Figures 5B and S7A). There was increased expression of inhibitory PDL1 in MAC1 and DCs in tumor (Figure 5C and S7A), little PDL1 expression in non-immune –tumor cells, but some PDL2 expression detectable in tumor fibroblasts and endothelial cells (Figures 5C, S7A, and S7B). In naive/Tcm CD4 T cells, there was reduced expression of CD28, ICOS, and OX40 in tumor compared with normal, with little PD1 expression and reduced CTLA4 expression in tumor T cells (Figures 5C and S7A). Overall, in the lymphoid compartment, immune-related GO term genes were downregulated in tumor-associated lymphocyte subsets (Figure S7C), and there was significant enrichment for exhaustion signature genes in cytotoxic CD8 T cells (Figure 5D). Interestingly, in NK cells, we observed a significant enrichment of the ‘cytokine-cytokine receptor interaction’ gene set in the CD16neg NK cell subset in tumor compared to normal (Figure 5E) and the leading-edge genes included several chemokine transcripts related to DC recruitment, including CCL5, XCL1, and XCL2 (Figure 5F). Spatial transcriptomic analysis of prostate cancer confirmed colocalization of the CD16neg NK cell signature and CCL5 transcripts (Figure 5G). This suggests that the CD16neg resident-like NK cells in the prostate may promote the recruitment of tumor antigen cross-presenting cDC1, with potential beneficial anti-tumor effects, as described in melanoma, breast, and colon cancer (Böttcher et al., 2018).
Figure 5.
Perturbed immune cell transcriptomes and cellular interactions in prostate tumor
(A) Violin plot of gene module score of GO term corresponding to antigen processing and presentation in tumor versus normal in myeloid cells. Kruskal-Wallis test was performed between normal and tumor for each cluster and p < 0.05 was considered statistically significant.
(B) Pie chart of co-activating and co-inhibitory DEGs between normal (light colored sections) and tumor (dark colored sections) in myeloid clusters. Darker sections indicate genes that were that were upregulated in tumor versus normal and lighter section indicate upregulation is in normal versus tumor (> = 1.25 log2 fold change). Grey sections indicate < 1.25 log2 fold change.
(C) Mean expression dot plot of costimulatory/coinhibitory molecules. Increasing expression corresponds with increasing gradient from white to red (MNP/B cell/epithelial/stromal) or white to blue (T/NK/B cell) corresponding to increasing expression value. Size of circles indicate the percentage of cells expression the gene.
(D) Violin plot of gene set test (AUCell) results in CD8 cytotoxic T cell cluster for murine CD8 T cell exhaustion gene set curated from (Doering et al., 2012). Significance is denoted by ∗p < 0.05 (Mann-Whitney test).
(E) GSEA of KEGG pathways for NK CD16neg tumor versus normal. Statistically significant pathways are colored and labeled.
(F) Mean expression dot plot of leading edge genes in cytokine-cytokine receptor interaction as in (E). Only genes that are expressed by at least 20% of cells are plotted.
(G) (Top) Expression of CCL5 and prediction/label transfer score of NK CD16neg cells in visium data of normal and tumor prostate sections. (Bottom) Spatial correlation of CCL5 with NK CD16neg cells in prostate cancer visium data. Only positive correlations are plotted; increasing value of correlation is shown as a gradient from white to red.
(H) CellPhoneDB receptor-ligand interaction analysis between B cell and myeloid clusters.
(I) Representative immunofluorescence confocal microscopy of BAFF and HLA-DR in human prostate tumor section. Scale bars, 20 μm.
(J–L) (J) Expression of TNFSF13B and prediction/label transfer score of MAC-MT cells, and correlation of TNFSF13B with MAC-MT cells in prostate cancer visium data. Only positive correlations are plotted; increasing value of correlation is shown as a gradient from white to red. CellPhoneDB receptor-ligand interaction analysis between (K) fibroblasts and T cell clusters, and fibroblast and myeloid clusters and (L) myeloid clusters with LE clusters split by group (N = normal; T = tumor). The order of the receptor-ligand interactions corresponds to the order of the cell-types i.e., cell type A expressing molecule A interacts with cell type B expressing molecule B. Size of circles and color gradient corresponds to the receptor-ligand interaction score, which purple, blue, green to yellow for increasing values. Significant interactions (p < 0.05) are highlighted in red. See also Figure S7.
Analysis of predicted MAC-MT interactions with other immune cells based on receptor-ligand expression using CellPhoneDB (Vento-Tormo et al., 2018) demonstrated that MAC-MT within tumor samples had increased expression of the gene encoding BAFF (TNFSF13B) with the potential to support BAFF-R-expressing B cells (Figures 5H and S7D). BAFF protein expression was evident in prostate adenocarcinoma samples (Figure S7E) and BAFF staining colocalized with MHCII staining, marking MNPs (Figure 5I-J). In contrast, analysis of immune-fibroblast interactions highlighted CXCL12 expression by fibroblasts, with the potential to recruit CXCR3/4-expressing CD8 T cells and MNPs that was reduced in tumor (Figure 5K). LE-KLK3 and LE-KLK4 both expressed macrophage inhibitory factor (MIF), a macrophage survival, activation and recruiting factor (Gregory et al., 2006), with its receptor CD74 expressed by all prostate MNPs, and this interaction was attenuated in cancer samples (Figure 5L). LE-KLK4 also expressed RPS19, with monocyte-recruiting activity (Yamamoto, 2007), and this too was reduced in tumor samples (Figures 5L and S7F).
In summary, these data indicate widespread immune transcriptional perturbation in prostate cancer, with reduced antigen presentation gene expression in MNP subsets, increased expression of exhaustion-associated genes in CD8 T cells, and reduced expression of immune-recruiting and activating chemokines and cytokines by fibroblast and epithelial cells in prostate cancer. However, in contrast, CD16neg NK cells in tumor had increased expression of cDC1-recruiting chemokines and MAC-MT higher expression of the B cell survival factor BAFF, both with potential beneficial antitumor effects.
MAC-MT in tumor associated with improved outcomes
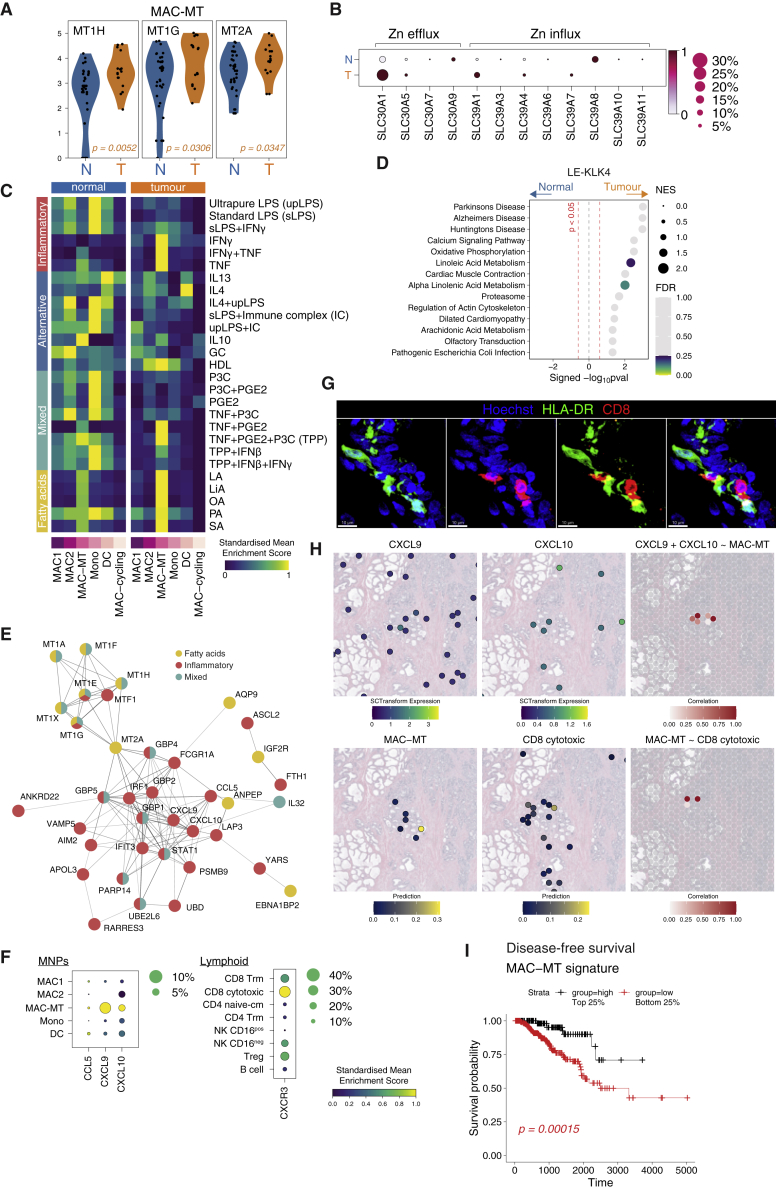
To further probe the effect of the tumor microenvironment on MNPs, we compared GO term enrichment in normal and tumor MNPs. This analysis showed attenuated expression of all pathways enriched in homeostasis in every MNP subset except for MAC-MT (Figure S8A). Indeed, metallothionein genes were increased in tumor-associated MAC-MT compared with those in normal prostate (Figures 6A and S8B). SLC30A1 expression was increased and SLC39A8 decreased in MAC-MT cells in cancer compared to non-tumor samples (Figure 6B), the overall effect of which may be to increase zinc efflux, potentially counteracting the known decreased zinc concentration associated with prostate cancer.
Figure 6.
MT1-expressing macrophages in tumor have increased metallothionein and pro-inflammatory gene expression and are associated with improved tumor event-free survival
(A) Violin plots show genes than achieved a p adj < 0.05 after statistical analyses with Wilcoxon Rank Sum Tests. Color of adjusted p value indicates the group where expression is higher (orange = tumor).
(B) Mean expression dot plot of metal ion transport genes in MAC-MT cluster separated by normal (N) or tumor (T) in rows. Size of circle indicates the percentage of cells expressing the genes and color indicates which group (normal or tumor) expresses higher (dark red) levels of the genes.
(C) Heatmap of mean AUCell enrichment of 27 macrophage-stimulation signatures split by normal or tumor. Row expression value is scaled from 0 to 1 and presented as a gradient from purple, blue, green to yellow.
(D) GSEA of KEGG pathways in tumor versus normal for LE-KLK4. Pathways were considered statistically significant if p value < 0.05 (marked by vertical dashed red line). Size of circles indicate normalized enrichment score (NES) and colors indicate if pathways achieved FDR < 0.25 starting from purple, blue, green to yellow as significance values decreases.
(E) String-DB analysis of leading edge genes from selected pathways enriched in tumor MAC-MT.
(F) Mean expression dot plot of CCL5, CXCL9, CXCL10 in MNP clusters and CXCR3 in lymphoid clusters. Size of circle indicates the percentage of cells expressing the genes and increasing expression (scaled from 0 to 1) corresponds to increasing color gradient from purple, blue, green to yellow.
(G) Representative immunofluorescence confocal microscopy of CD8 and HLA-DR in human prostate tumor section. Scale bars, 10 μm.
(H) Expression of CXCL9 and CXCL10 and prediction/label transfer scores of MAC-MT and CD8 cytotoxic cells in visium data of tumor prostate sections. (Bottom) Spatial correlation of CXCL9 and CXCL10 with MAC-MT or MAC-MT with CD8 cytotoxic cells in prostate cancer visium data. Only positive correlations are plotted; increasing value of correlation is shown as a gradient from white to red.
(I) Kaplan-Meier survival curve for TCGA-PRAD disease free index with deconvolved MAC-MT score. Samples were categorised into high (black, top 25%) and low (red, bottom 25%) of deconvolved score. Statistical analysis was performed with log rank test and p < 0.05 was considered statistically significant. See also Figures S8–S9 and Table S2.
Transcriptional alignment of healthy prostate MNPs with human macrophages activated with a variety of stimuli (Xue et al., 2014) demonstrated that MAC2 enriched for inflammatory LPS- and IFNγ-stimulated M1-like macrophage signatures, as well as IL4 and glucocorticoid-stimulated macrophage signatures (Figure 6C). MAC1 showed little enrichment for any signatures except for the glucocorticoid-stimulated macrophage signature, while MAC-MT were completely distinct, enriching for TNF-, IL10-, and fatty acid–stimulated macrophage signatures (Figure 6C). In contrast to normal prostate, MNPs in tumor tissue demonstrated a global reduction in most macrophage activation signatures, consistent with a broad immune-suppressive effect of the tumor environment (Figure 6C). However, the remarkable exception to this tumor-associated suppression was the MAC-MT subset, which showed a significant increase in the expression in IFNγ-, TNF-, and fatty acid–stimulated macrophage gene signatures (Figure 6C). We observed an increase in cholesterol homeostasis pathway genes (Hallmarks, Figure S8C) and linoleic acid (a fatty acid) metabolism pathway genes (KEGG, Figure 6D) in prostate cancer–associated LE-KLK4 cells, raising the possibility that the fatty acid response gene enrichment in MAC-MT cells may arise due to fatty acid generation by local LE-KLK4 cells. Of note, increased fatty acid production from de novo lipogenesis has been described in prostate cancer (Rossi et al., 2003) and inhibition of lipogenesis reduced cancer growth in vitro (Zadra et al., 2014).
STRING analysis of leading-edge genes (Figure S8D) in these macrophage activation signatures, showed that fatty acid stimulation genes upregulated in MAC-MT in tumor were dominated by metallothionine genes, while inflammatory stimulation genes included several chemokines (CXCL9, CXCL10; Figures 6E and 6F). CD8 cytotoxic T cells were the principle immune cell subset expressing CXCR3, the receptor for these chemokines (Figure 6F), suggesting that MAC-MT activation may promote CD8 T cell recruitment to tumors (Figure S8E). Consistent with this, some/many CD8 T cells were located adjacent to MNPs (Figure 6G) and spatial transcriptomic analysis of prostate cancer confirmed colocalization of the MAC-MT signature with CXCL9/10 and with CD8 T cells (Figure 6H).
Given the pro-inflammatory transcriptional profile of MAC-MT cells, we hypothesized that they may have an antitumor effect, promoting anti-cancer immune responses and counteracting cancer-associated perturbations in zinc concentration. In keeping with this, cellular deconvolution (Figure S9A) of The Cancer Genome Atlas (TCGA) data (Figure S9B) indicated that prostate cancer biopsies with higher MAC-MT enrichment had a lower Gleason score (Figure S9C) and improved disease-free survival (Figure 6I; Table S2). No other individual immune cell subset signature had prognostic significance in this dataset, although the limited number of samples with a high CD16neg NK cell signature precluded a robust analysis of the prognostic association of these cells (Figure S9C).
Discussion
Our single-cell analysis of the human prostate delivered several remarkable findings; our sample processing protocol enriched for immune cells enabling us to deliver the most comprehensive overview of the immune landscape of normal human prostate to date. Our prostate immune cell atlas delineates a range of innate and adaptive immune cells, with several CD4 and CD8 T cell subsets, two subsets of NK cells, and a prostate-specific macrophage subset, which we designated MAC-MT. The latter subset was transcriptionally similar to YS-derived murine macrophages, consistent with the conclusion that they may be prenatally seeded, although transcriptional profile does not definitively prove ontogeny. They also expressed high levels of metallothionein and zinc transporter genes, suggesting that they could contribute to zinc homeostasis in the prostate. To test this hypothesis, we depleted prostate macrophages in mice, and confirmed a reduction in tissue zinc levels. In prostatic epithelial cells, the high zinc concentration acts to inhibit mitochondrial aconitase, truncating the Krebs cycle to generate citrate which is secreted into prostatic fluid maintaining spermatozoa (Costello and Franklin, 2016). The bioenergetic consequence of this is a reduction in ATP generation; therefore, aerobic glycolysis is increased in prostatic epithelium (Costello and Franklin, 2016). MAC-MT also enriched for glycolysis genes and may therefore directly contribute to prostate zinc and citrate via a similar mechanism, although we did not explore this in the current study. Of note, this homeostatic role of MAC-MT in maintaining prostate zinc adds to other examples of prenatally seeded tissue macrophages that contribute to organ physiology and function; for example, in the heart, a subset of macrophages act to buffer calcium ions within the conducting system (Hulsmans et al., 2017), and in the intestine, muscularis macrophages regulate the steady-state peristaltic activity of the colon (Muller et al., 2014).
In prostate cancer, the concentration of both citrate and zinc are markedly decreased (Franklin et al., 2005). We found that in prostate cancer, MAC-MT markedly increased the expression of the zinc efflux transporter SLC30A1, which may represent a mechanism to counteract the disrupted zinc transport present in tumor luminal epithelial cells. MAC-MT were also largely resistant to the immunosuppressive effect of the tumor environment. In fact, they become transcriptionally more inflammatory, expressing the B cell survival factor BAFF and lymphocyte-recruiting chemokines. Our analyses suggest that this response may be driven by the metabolic changes observed in KLK4 cells in tumor, namely the production of fatty acids. This increase in fatty acid metabolism has been previously described in prostate cancer (Rossi et al., 2003). We found that MAC-MT in tumor showed a transcriptional profile similar to that observed in macrophages stimulated with fatty acids. Remarkably, enrichment of the MAC-MT signature in prostate cancer biopsies was associated with improved disease-free survival, supporting the conclusion that their pro-inflammatory effects could be beneficial in the context of prostate cancer. The effects of MAC-MT enrichment are in contrast to previous studies investigating MNPs in prostate cancer which have associated the presence of myeloid-derived suppressor cells (Brusa et al., 2013) and CD163-positive M2 macrophages (Erlandsson et al., 2019) with worse survival, and emphasize the value of scRNA seq in delineating distinct cell subsets with important transcriptional and functional differences (Montoro et al., 2018). Our data have translational relevance as the antitumor effects of MAC-MT could potentially be harnessed as an immunotherapeutic strategy in advanced progressive prostate cancer.
We also identified a tissue-resident CD16neg NK cell subset in normal prostate. These cells showed increased expression of CCL5, XCL1, and XCL2 in prostate cancer, with the potential to recruit cDC1. This is reminiscent of a recent study in mice showing that NK cells in implanted melanomas, breast, and colon cancers played a critical role in recruiting cDC1 via the production of CCL5 and XCL1, with important antitumor effects (Böttcher et al., 2018). cDC1 express XCR1 (Dorner et al., 2009), as well as CCR1 and CCR5, both of which bind CCL5 (McColl, 2002) and have antitumor functions; they can attract and activate tumor-specific CD8 T cells (Spranger et al., 2017) (Broz et al., 2014), and internalize and transport tumor antigens to lymph nodes, where they may cross-prime CD8 T cells (Roberts et al., 2016). The potential importance of NK cells in antitumor responses in prostate cancer has been suggested by previous studies associating lower numbers of peripheral blood NK cells, particularly CD16neg NK cells (Koo et al., 2013), or reduced activation capacity of circulating NK cells, with a higher risk of prostate cancer on biopsy (Vidal et al., 2019). NK cells within prostate cancer tissue have not been well studied, but enumeration via immunohistochemical staining indicated an association with a lower risk of disease progression (Gannon et al., 2009), and there is one previous flow cytometric assessment of prostate tumor NK cells, demonstrating an enrichment of the CD56bright subset, relative to blood (Pasero et al., 2016). Our study confirms the presence of a tissue-resident CD56bright CD16neg NK cell subset in the prostate in health and in cancer, and provides a transcriptional assessment of these cells, revealing a potentially important antitumor function in cDC1 recruitment.
To date, metastatic prostate cancer has shown variable responsiveness to checkpoint blockade (Kwon et al., 2014, Beer et al., 2017). Evaluation of PD-L1 expression in prostate cancer previously identified its presence in less than 10% of primary cancer and up to a third of metastatic cancers (Haffner et al., 2018). Our analysis enabled simultaneous assessment of all PD1/2 ligands and receptors across cell types. PDL1 was expression was upregulated in tumor MAC2 and DCs, and we identified de novo PDL2 expression in tumor samples, suggesting that this may be a more relevant therapeutic target in prostate cancer than PDL1. This is consistent with a recent analysis of bulk RNA seq data from prostate cancer biopsies that found an increase in PDL2 transcripts compared with normal tissue, and that higher PDL2 expression was associated with worse outcomes (Zhao et al., 2019). Our analysis enabled the specific identification of tumor fibroblasts and endothelial cells as key expressors of this immunoinhibitory molecule.
Aside from tissue-resident immune cells, we also identified a subset of luminal epithelial cells that lack expression of AR that may interact metabolically with the prostate-specific MAC-MT. Our dataset delineates several cell-specific markers that should be investigated for their utility as biomarkers for the early identification of this cell subset to assess their contributions to development of castration-resistant disease.
In summary, we define the immune cell landscape in normal human prostate and describe its perturbation in cancer. Our study revealed the presence of a prostate-specific macrophage subset, marked by high expression of metallothionein genes that, in contrast to other MNP subsets, increases inflammatory gene expression in cancer with potential beneficial prognostic effects.
Limitations of the study
While our study used a tissue processing strategy that enriched for immune cells on n = 10 paired normal and prostate cancer samples and successfully created a single-cell prostate immune atlas, there are several limitations; the patients recruited to our study had predominantly moderate prostate cancer and did not receive androgen deprivation therapy, in contrast to the other prostate cancer single-cell datasets which primarily consist of samples from more advanced disease. Furthermore, we discovered a metallothionein- expressing, zinc-regulating macrophage (MAC-MT) population, the MAC-MT gene signature was associated with improved outcomes, and experimental depletion in mice showed that prostate macrophages contribute to zinc homeostasis in normal prostate. However, further work is needed to investigate the role of these macrophages in the context of prostate cancer.
STAR★Methods
Key resources table
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| anti-human CD14 (FITC) | Invitrogen | Cat#11-0419-42; clone 61D3; RRID: AB_10597597 |
| anti-human CD45 (APC-eFluor780) | eBioscience | Cat#47-0459-42; clone HI30; RRID: AB_1944368 |
| anti-human CD19 (eFluor450) | eBioscience | Cat#48-0199-42; clone HIB19; RRID: AB_1272053 |
| anti-human CD3 (BV785) | BioLegend | Cat#317330; clone OKT3; RRID: AB_2563507 |
| anti-human CD8 (PE) | BioLegend | Cat#300908; clone HIT8a; RRID: AB_314112 |
| anti-human CD16 (PE-Cyanine7) | Invitrogen | Cat#25-0168-42; clone CB16; RRID: AB_10714839 |
| anti-mouse Ly-6G/Ly-6C (Gr1) (FITC) | BioLegend | Cat#108406; clone RB6-8C5; RRID: AB_313371 |
| anti-mouse CD11b (PerCP-Cy5.5) | eBioscience | Cat#45-0112-82; clone M1/70; RRID: AB_953558 |
| anti-mouse CD3e (APC) | eBioscience | Cat#17-0031-82; clone 145-2C11; RRID: AB_469315 |
| anti-mouse CD19 (APC) | BioLegend | Cat#152410; clone 1D3; RRID: AB_2629839 |
| anti-mouse CD45 (APC-eFluor780) | Invitrogen | Cat#47-0451-82; clone 30-F11; RRID: AB_1548781 |
| anti-mouse I-A/I-E (Pacific Blue) | BioLegend | Cat#107620; clone M5/114.15.2; RRID: AB_493527 |
| anti-mouse CD11c (PE) | BioLegend | Cat#117308; clone N418; RRID: AB_313777 |
| anti-mouse F4/80 (PE-Cyanine7) | Invitrogen | Cat#25-4801-82; clone BM8; AB_469653 |
| anti-mouse CD4 (FITC) | eBioscience | Cat#11-0041-82; clone GK1.5; RRID: AB_464892 |
| anti-mouse NK1.1 (Pacific Blue) | BioLegend | Cat#108722; clone PK136; RRID: AB_2132712 |
| anti-mouse CD8a (BV785) | BioLegend | Cat#100750; clone 53-6.7; RRID: AB_2562610 |
| anti-mouse NKp46 (PE/Dazzle) | BioLegend | Cat#137630; clone 29A1.4; RRID: AB_2616666 |
| anti-mouse CD3 (PE-Cyanine7) | BD PharMingen | Cat#560591; clone 17A2; RRID: AB_1727462 |
| anti-mouse Ly-6G/Ly-6C (Gr-1) (APC) | BioLegend | Cat#108412; clone RB6-8C5; RRID: AB_313377 |
| anti-mouse CSF1R | BioXCell | Cat#BE0213; clone AFS98; RRID: AB_2687699 |
| Rat IgG2a isotype control | BioXCell | Cat#BE0089; clone 2A3; RRID: AB_1107769 |
| anti-human HLA-DR (AF647) | Abcam | Cat#ab20181; clone TAL 1B5; RRID: AB_445401 |
| anti-human CD206 (PE/Dazzle) | BioLegend | Cat#321130; clone 15-2; RRID: AB_2616867 |
| anti-human MT1 | Abcam | Cat#ab12228; clone UC1MT; RRID: AB_298949 |
| anti-human CD3 (AF488) | BioLegend | Cat#300415; clone UCHT1; RRID: AB_389310 |
| anti-human BAFF (polyclonal rabbit) | Bioss | Cat#bs-2431R; RRID: AB_10855666 |
| Mouse IgG1 kappa monoclonal isotype control | Abcam | Cat#ab170190; clone 15-6E10A7; RRID: AB_2736870 |
| Goat anti-mouse IgG secondary (FITC, polyclonal) | Invitrogen | Cat#31569; RRID: AB_228306 |
| anti-mouse CD31 (AF594) | BioLegend | Cat#102520; clone MEC13.3; RRID: AB_2563319 |
| anti-mouse F4/80 (AF647) | Abcam | Cat#ab204467; clone F4/80; RRID: AB_2810932 |
| anti-mouse CD19 (AF594) | BioLegend | Cat#115552; clone 6D5; RRID: AB_2563459 |
| anti-mouse IgD (AF488) | BioLegend | Cat#405718; clone 11-26c.2a; RRID: AB_10730619 |
| anti-mouse NKp46 (PE) | eBioscience | Cat#12-3351-82; clone 29A1.4; RRID: AB_1210743 |
| anti-mouse CD8 (FITC) | eBioscience | Cat#11-0081-82; clone 53-6.7; RRID: AB_464915 |
| anti-mouse CD3 (Pacific Blue) | BioLegend | Cat#100214; clone 17A2; RRID: AB_493645 |
| Flash Phalloidin 488 | BioLegend | Cat#424201 |
| Hoechst 33258 | Biotium | Cat#40044 |
| DAPI (in mounting medium) | Invitrogen | Cat#00-4959-52 |
| LIVE/DEAD Aqua | Invitrogen | Cat#L34957 |
| Biological samples | ||
| Human normal and cancer prostate tissues | Cambridge University Hospitals NHS Foundation Trust | N/A |
| Critical commercial assays | ||
| Zinc Assay Kit | Abcam | Cat#ab102507 |
| Chromium Single Cell 3′ Library & Gel Bead Kit v2 | 10X Genomics | Cat#PN-120237 |
| Chromium Single Cell A Chip Kit, 16 rxns | 10X Genomics | Cat#PN-1000009 |
| Chromium i7 Multiplex Kit, 96 rxns | 10X Genomics | Cat#PN-120262 |
| RNAscope® 2.5 LS Multiplex Reagent Kit | Advanced Cell Diagnostics | Cat#322800 |
| RNAscope® LS 4-Plex Ancillary Kit Multiplex Reagent Kit | Advanced Cell Diagnostics | Cat#322830 |
| RNAscope® 2.5 LS Probe- Hs-CD68-C2 | Advanced Cell Diagnostics | Cat#560598-C2 |
| RNAscope® 2.5 LS Probe- Hs-MT1-C3 (custom probe) | Advanced Cell Diagnostics | Cat#831088 |
| Deposited data | ||
| Matched normal and tumor prostate scRNaseq data | This paper, European Genome–Phenome Archive | EGAS00001005787, prostatecellatlas.org |
| Spatial Gene Expression Dataset by Space Ranger 1.3.0, Human Prostate Cancer, Adenocarcinoma with Invasive Carcinoma (FFPE) | 10X Genomics | N/A |
| Prostate Cancer Spatial Transcriptomics data (L1.2) (Berglund et al., 2018) | European Genome–Phenome Archive | EGAS0000100300 |
| Normal human prostate scRNaseq data (Henry et al., 2018) | GEO | GSE120716 |
| Human prostate cancer scRNaseq data (Chen et al., 2021) | GEO | GSE141445 |
| Human prostate cancer scRNaseq data (Crowley et al., 2020) | GEO | GSE150692 |
| Human prostate cancer scRNaseq data (Karthaus et al., 2020) | https://singlecell.broadinstitute.org/ | SCP864 |
| Mouse prostate scRNaseq data (Karthaus et al., 2020) | GEO, https://singlecell.broadinstitute.org/ | GSE146811, SCP859 |
| Human prostate cancer Bulk RNaseq | TCGAbiolinks (R/Bioconductor) | TCGA-PRAD |
| Human spleen and lung scRNaseq data (Madissoon et al., 2019) | Human Cell Atlas Data Portal, https://www.tissuestabilitycellatlas.org/ | N/A |
| Human kidney scRNaseq data (Stewart et al., 2019) | Human Cell Atlas Data Portal, https://www.kidneycellatlas.org/ | N/A |
| Human liver scRNaseq data (MacParland et al., 2018) | GEO | GSE115469 |
| Human heart scRNaseq data (Wang et al., 2020) | GEO | GSE109816 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 (B6) | Jackson Laboratories | Stock No: 000664 |
| Software and algorithms | ||
| seurat | CRAN | V3.2.3 |
| glmnet | CRAN | V4.1-2 |
| survival | CRAN | V2.41-3 |
| survminer | CRAN | V0.4.6 |
| pheatmap | CRAN | V1.0.12 |
| AUCell | Bioconductor | V1.14.0 |
| fgsea | Bioconductor | V1.18.0 |
| clusterProfiler | Bioconductor | V4.0.5 |
| biomaRt | Bioconductor | V2.48.3 |
| TCGAbiolinks | Bioconductor | V2.15.3 |
| String-DB | https://string-db.org | V11 |
| scanpy | https://github.com/theislab/scanpy | V1.4.5.post2 and V.1.7.2 |
| soupx | https://github.com/constantamateur/soupx | V1.2.1 |
| scrublet | https://github.com/swolock/scrublet | V0.2.1 |
| umap | https://github.com/lmcinnes/umap | V3.10.0 |
| gseapy | https://github.com/zqfang/gseapy | V0.10.5 |
| stLearn | https://github.com/BiomedicalMachineLearning/stLearn | V0.3.2 |
| CellPhoneDB | https://github.com/Teichlab/cellphonedb | V2.0.5 |
| MuSiC | https://github.com/xuranw/MuSiC | V0.1.1 |
| Cellranger | 10X Genomics | V2.1.0 |
| CASAVA | Illumina | V1.8.2 |
| FlowJo | BD | V10 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Menna R. Clatworthy (mrc38@cam.ac.uk).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Participants
Fifteen men (50 – 74 years old) undergoing image guided prostate biopsies for suspicion of prostate cancer were enrolled in the DIAMOND study (NHS National Research Ethics Service reference 03/018) (CI:Gnanapragasam). All participants had previously undergone multi-parametric magnetic resonance imaging (mpMRI) of the prostate on a 3T magnet (Discovery MR750, GE Healthcare), using a 32-channel phased array coil. T2-weighted, contrast-enhanced, and diffusion-weighted imaging was acquired using the LIKERT scoring system according to Prostate Imaging–Reporting and Data System (PI-RADS) guidelines (Barrett et al., 2019). Only men with a positive MRI were approached and recruited for this study, defined as a PI-RADS score 3 or greater.
Each participant underwent transperineal biopsy under general anesthesia using the Biopsee fusion platform (Medcom, Darmstadt, Germany) according to the Ginsburg protocol, with a variable number of biopsies cores taken in order to obtain an appropriate tissue diagnosis for that individual (Kuru et al., 2013). All targets were defined by radiologists pre-procedure using T2-weighted imaging as the primary source images, using Biopsee fusion software. Patient/sample characteristics are summarized in Table S1. Samples from 5 men were used for sample preparation optimization and remaining 10 were used for sequencing experiments.
Mice
All murine research was conducted under the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012 following ethical review by the University of Cambridge Animal Welfare and Ethical Review Body (AWERB). Mice were housed at Cambridge Biomedical Services under specific-pathogen-free conditions. Wild-type C57BL/6 male mice aged 8 – 12 weeks were obtained from Jackson Laboratories (Margate, UK),
Method details
Sample collection
Each participant underwent prostate biopsy with a variable number of biopsies (20-30) cores taken in order to obtain an appropriate tissue diagnosis. Samples were taken from systematic and targeted biopsies as standard of care. Men were consented to have additional cores taken from the “Target” (area where cancer was suspected on the MRI) and from an “off-target” area to provide a normal prostate tissue comparator. The number of biopsies taken from both areas ranged from 1 to 6 additional cores. Biopsies were placed into phosphate buffered saline and placed on ice immediately.
Tissue disaggregation of human tissue
Prostate tissue was received in ice cold PBS, minced into approximately 5 mm3 pieces and digested for 20 min at 37°C with agitation in a digestion solution containing 32.5 μg/mL Liberase TM and 50 μg/mL DNase in RPMI. Following incubation samples were passed through a 100 μm cell strainer using a 1 mL syringe plunger and washed by centrifugation with PBS. Live cells were enriched using a Dead Cell Removal kit (Miltenyi Biotec) as per manufactures instructions. This was followed by a 44% Percoll density-gradient for 30 min at room temperature. Enriched live cells were washed and counted using a haemocytometer with trypan blue. Cells were then blocked with human FcR block (Miltenyi Biotech) prior to surface staining for flow cytometry. Cell counts per gram were calculated with the addition of 123count eBeads.
Single-cell sequencing
10X Chromium Chip Single-cell library generation and preparation were performed on the single-cell suspension according to 10X Chromium 3′ solution (V2 kit) as per manufacturer’s instructions with an aim to capture 5000-10000 cells/channel. Sequencing was performed at the Cancer Research UK Cambridge Institute on the Illumina HiSeq4000 platform.
Following sequencing BCL files were demutiplexed to Fastq files using CASAVA. Subsequently splitting to single cells and mapping and quantification of genes was carried out using Cellranger software package (10X genomics). This generated count tables of unique molecular identifiers (UMI) for each gene per droplet.
Tissue disaggregation of murine tissue
The left and right anterior prostate lobes were harvested from mice and minced into approximately 15 mm3 pieces. Samples were digested for 20 min at room temperature in a digestion solution containing 0.1 M HEPES, 32.5 μg/mL Liberase TM and 50 μg/mL DNase in RPMI. Following incubation samples were passed through a 100 μm cell strainer using a 1 mL syringe plunger, washed by centrifugation with PBS and blocked with 50:50 mix of normal mouse and rat serum prior to staining. Cell counts per organ / gram of tissue were calculated with the addition of 123count eBeads (Invitrogen).
Flow cytometry
After blocking cells were incubated with live/dead cell staining (Live/Dead Aqua 405, Invitrogen) for 15 minutes on ice. Cell surface staining occurred on ice for 30 minutes. All samples were acquired on an LSR 4/5 laser Fortessa (BD) and data analyzed using FlowJo v10. Human antibody: anti-CD14 FITC (61D3, Invitrogen), anti-CD45 APC-eFluor780 (HI30, eBioscience), anti-CD19 eFluor450 (HIB19, eBioscience), anti-CD3 BV785 (OKT3, BioLegend), anti-CD8 PE (HIT8a, BioLegend), anti-CD16 PE-Cyanine7 (CB16, Invitrogen). Murine antibody (myeloid panel): anti-Gr1 FITC (RB6-8C5, BioLegend), anti-CD11b PerCP-Cy5.5 (M1/70, eBioscience), anti-CD3e APC (145-2C11, eBioscience), anti-CD19 APC (1D3, BioLegend), anti-CD45 APC-eFluor780 (30-F11, Invitrogen), anti-I-A/I-E Pacific Blue (M5/114.15.2, Biolegend), anti-CD11c PE (N418, BioLegend), anti-F4/80 PE-Cyanine7 (BM8, Invitrogen). Murine antibody (lymphoid panel): anti-CD4 FITC (GK1.5, eBioscience), anti-CD19 APC (1D3, BioLegend), anti-CD45 APC-eFluor780 (30-F11, Invitrogen), anti-NK1.1- Pacific Blue (PK136, BioLegend), anti-CD8a BV785 (53-6.7, BioLegend), anti-NKp46 PE/Dazzle (29A1.4, BioLegend), anti-CD3 PE-Cyanine7 (17A2, BD PharMingen).
Single-cell data analysis and preprocessing
The single-cell data (10X cellranger output) processed with EmptyDrops (Lun et al., 2019) and then corrected for ambient RNA expression using SoupX (v1.2.1) (Young and Behjati, 2020). Contamination fractions were estimated using the following genes: haemoglobin genes: HBA1, HBA2 and HBB; immunoglobulin genes: IGKC, IGLC1, IGLC2, IGLC3, IGLC4, IGLC5, IGLC6 and IGLC7; sperm genes: STMN1; prostate specific antigen gene: KLK3. SoupX was run with clustering information derived from a generic processing workflow in Seurat (Stuart et al., 2019). After SoupX, doublet detection was performed using scrublet (v0.2.1) (Wolock et al., 2019) with adaptations outlined in (Popescu et al., 2019) – Briefly, after scrublet was performed, the data was iteratively sub-clustered using standard Seurat-inspired scanpy (v.1.4.5.post2) workflow (Wolf et al., 2018, Stuart et al., 2019) and a median scrublet score for each sub-cluster was computed. Median absolute deviation (MAD) scores were computed from the cluster scrublet scores and a one tailed t test was performed with Benjamini-Hochberg (BH) correction (Benjamini and Hochberg, 1995) applied and cells with significantly outlying cluster scrublet scores (BH pval < 0.1) were flagged as potential doublets. The data was then processed using scanpy with standard quality control steps; cells were filtered if number of genes > 2500 or < 200. Percentage mitochondrial content cut-off was set at < 30%. Genes were retained if they are expressed by at least 3 cells. Genes counts for each cell were normalized to contain a total count equal to the median of total counts in cells before normalization. This led to a working dataset of 17,108 cells. We filtered out 1,616 cells that we could annotate as sperm cells from seminal fluid contamination from a single normal sample before resulting in the final set of 15,492 cells. Highly variable genes were selected based on the following parameters: minimum and maximum mean expression are > = 0.0125 and ≤ 3 respectively; minimum dispersion of genes = 0.5. The number of principal components used for neighborhood graph construction and dimensional reduction was set at 50. Batch correction was performed using bbknn with patients as the batch term with all other parameters as per default settings (Polański et al., 2020) (for all cells and for lymphoid analysis). Clustering was performed using Leiden algorithm (Traag et al., 2019) with resolution set at 1.0 (for all cells) or 0.5 (for myeloid and lymphoid). Henry et al. (Henry et al., 2018) dataset was identically processed for comparison except that cell type identities were used as published and sub-clustering of leukocytes was performed to extract MNPs after marker gene identification with Wilcoxon Rank Sum tests.
In all cases where Uniform Manifold Approximation and Projection (UMAP; v3.10.0) (McInnes et al., 2018) was used for dimensional reduction and visualization, the minimum distance was set at 0.3 and all other parameters as per default settings in scanpy.
For integration of MNPs, standard SCTransform workflow implemented in Seurat (v3.2.3) was used (Hafemeister and Satija, 2019). Calculation of PCA, UMAP and neighborhood graphs post integration was performed in scanpy using the SCTransform normalized data. Neighborhood graph graphs were constructed with 10 neighbors. For analysis of lymphoid cells, cells from three patients were excluded due to low numbers of cells (< 10 cells; D7 and D14) or low-quality information from cells (D6; insufficient cell-cell heterogeneity). Further sub-clustering was also performed on NK cells, CD8 T cells and non-CD8 T cells (annotated as CD4 T cells) separately to obtain the final lymphoid clusters via specifying the restrict_to option with resolution set at 0.3 in scanpy.
Differential gene testing
Differential gene testing was performed using the Wilcoxon test rank sum test implemented in scanpy’s rank_genes_groups module.
Cell type similarity assessment
We used a logistic regression approach to test for cell type similarity. This is done with L2-regularised logistic regression (ridge regression) multinomial models with the glmnet R package (Friedman et al., 2010) (i.e., alpha parameter = 0). Models were trained on normalized gene expression data with 10-fold cross-validation to obtain the appropriate lamda coefficient (lambda.1se; within 1 standard error from best model) for prediction. Gene expression values were standardized in both the training and test sets. The average of 50 iterations was used for the final score. To determine if predictions were significant, a median prediction probability score for each cluster was calculated and MAD-outliers were identified using a one-tailed t test. Cells were considered to be significantly similar if the BH p value was < 0.05 and the probability was > 50%. Re-embedding of each relevant dataset into UMAP or tSNE space were performed where possible with standard scanpy workflow and cluster identities were used as published. Label transfer for prostate cancer datasets (Chen et al., 2021, Crowley et al., 2020, Karthaus et al., 2020) was performed with default ingest protocol in implemented in scanpy. Pre-processing of these additional datasets were performed as close as possible to the dataset in this manuscript except for the exclusion of ambient RNA correction due to inavailability of raw data.
Gene set enrichment and pathway analyses
Gene module scores of gene sets used were obtained using scanpy’s score_genes module or AUCell (Aibar et al., 2017). Kruskal-Wallis test or Mann Whitney U tests were performed to test for significance of enrichment where appropriate using Prism software (v8). P value < 0.05 were considered as statistically significant. Typically, gene sets were retrieved and used as published in the original articles; in cases where murine gene sets were used, murine genes were converted to human orthologs using biomaRt (Durinck et al., 2009).
Pre-ranked gene set analysis (prGSEA) on hallmark genesets (Liberzon et al., 2015) and macrophage stimulation genesets (Murray et al., 2014) were performed with gseapy (https://github.com/zqfang/GSEApy/).
Gene ontology and KEGG pathway analyses were performed using fgsea (Korotkevich et al., 2019) or over-representation analysis implemented in clusterProfiler (Yu et al., 2012) R package. Genes were pre-ranked according to signed -log10Pvalues for all prGSEA procedures.
String-DB (v11) analysis was performed using the web browser tool (https://string-db.org/).
Spatial transcriptomics data analysis
We compared our scRNA-seq analysis with spatial gene expression data generated by spatial transcriptomics protocol (Ståhl et al., 2016) for prostate cancer tissues (Berglund et al., 2018). The tissues were from radical prostatectomy for a patient with adenocarcinoma. We used the tissue section L1.2, which was pathologically annotated as at cancer stage GLEASON score 3+3. For accurately mapping spot expression data to the tissue H&E image, we used spot count matrices with adjusted spot coordinates. The coordinate adjustment was based on the alignment of the H&E image with the corresponding spot-fluorescent image, where each spot was detected by Cy3 fluorescence signal. The alignment accounted for manufacturing variation that caused the differences between expected coordinates and the actual coordinates of spots printed onto the spatial gene-expression slide.
To classify cell-types in each spot, we used the anchor-based data integration method and calculated probabilistic transfer scores of discrete cell-type labels from cell types information in our reference scRNA-seq data to spot data (Stuart et al., 2019). For each spot, the class probability of the spot belonging to each of the 12 cell types was calculated. For estimating the abundance of two cell types in each spot as in the pie chart, we calculate the probability of the spot to be of the cell types of interest and scaled each value to 1. The script for anchor-based label transferring and for plotting cell type proportion to tissue is available at https://github.com/BiomedicalMachineLearning/stLearn and described in stLearn package (Pham et al., 2020).
For label transfer of the FFPE visium datasets available from 10X resource page, SCTransform was used to normalize both the reference (prostate single-cell dataset) and spatial data prior to integration as per instructions for Seurat v3.2.3. Expression values plotted are SCTransformed normalized values. For calculation of spatial correlation, k = 5 nearest neighborhoods were extracted from a kNN graph computed from the spatial location of each voxel. Peason’s correlation was then performed on each neighborhood using the gene expression value and cell type prediction value, followed by averaging across neighborhoods. Correlation values will not be returned if expression value was not detected in all neighborhoods or expression value was uniform across all voxels. For the CXCL9 + CXCL10 comparison, the value from Seurat’s AddModuleScore of the two genes was used for computing the correlation.
CellPhoneDB analysis
Normalized expression values from cell types found in this dataset were subjected to CellPhoneDB analysis (v2.0.0) (Efremova et al., 2020). The minimum threshold was set at 30% and results were considered statistically significant if p < 0.05.
Survival analysis
The Cancer Genome Atlas (TCGA) expression and clinical data for Prostate Adenocarcinoma (PRAD) were downloaded with TCGAbiolinks (v2.15.3) (Colaprico et al., 2016). Single-cell deconvolution was performed using Multi-subject Single Cell deconvolution R package (MuSiC, v0.1.1) (Wang et al., 2019) with raw counts as instructed in the package. Disease free survival indices were extracted from days_to_new_tumor_event contained in the TCGA clinical data; samples without days_to_new_tumor_event entries were removed. Events were considered if days_to_new_tumor_event_dx was annotated as ‘YES’ and all other samples were censored. Outcome for events were generally from biochemical evidence of disease but also included distant metastasis, locoregional recurrence, primary tumor and ‘not available’. All events were considered regardless of treatment received (radiological or pharmaceutical). Kaplan-Meier survival analyses were performed using the survival (v2.41-3) and survminer (v0.4.6) R packages where the deconvolved scores were categorised into a ‘high’ or ‘low’ group, which corresponds to top and bottom 25% MuSiC deconvolved scores. The results/info are tabulated and summarized in Table S2.
RNA in situ hybridization
Simultaneous detection of human CD68 and MT1 family genes were performed on FFPE sections using Advanced Cell Diagnostics (ACD) RNAscope® 2.5 LS Multiplex Reagent Kit (Cat No. 322800), RNAscope® LS 4-Plex Ancillary Kit Multiplex Reagent Kit (Cat No. 322830), RNAscope® 2.5 LS Probes (ACD, Hayward, CA, USA) at the histopathology/in situ hybridization core facility at Cancer Research UK – Cambridge Institute. Because it was not possible to design a specific RNAscope probe for Hs-MT1H due to high homology to other genes in the MT1 family, we used a probe that recognizes multiple MT1 genes. Briefly, sections were cut at 3 μm thick, baked for 1 h at 60°C before loading onto a Bond RX instrument (Leica Biosystems). Slides were deparaffinised and rehydrated on board prior to pre-treatments using Epitope Retrieval Solution 2 (Cat No. AR9640, Leica Biosystems) at 95°C for 15 min, and ACD Enzyme from the Multiplex Reagent kit at 40°C for 15 min. Probes were visualized using Opal fluorophores (Opal 570 and Opal 650 Akoya Biosciences Cat No. FP1488001KT and FP1496001KT respectively) diluted to 1:1000 using RNAscope LS Multiplex TSA Buffer. Probe hybridization, signal amplification and detection was performed on the Bond Rx according to the ACD protocol. Slides were then removed from the Bond Rx and mounted using Prolong Diamond (ThermoFisher Cat No P36965). The slides were imaged on the AxioScan (Zeiss) to create whole slide images. Images were captured at 40x magnification, with a resolution of 0.25 μm per pixel. ISH validation was carried out on one slide from the Cambridge 109 Prostate TMA (slide-1), consisting of paired sets of benign and tumor tissue cores that were collected under PrompT ethics (MREC/01/4/061).
In-vivo macrophage depletion
5-6 C57BL/6 male mice aged 16 weeks received 0.5 mg of depleting anti-CSF1R mAb (BioXCell, clone AFS98) or isotype control (BioXCell, Rat IgG2a) intraperitoneally on day −7, −4 and −2 prior to euthanasia per experiment (n = 2 independent replicates). Following terminal procedure, kidneys, liver and prostate from each mouse were harvested and divided for either flow cytometry, microscopy or zinc quantification. Tissues for flow cytometry was minced finely and digested in RPMI containing 0.1 mg/mL DNase I, 32.5 mg/mL Liberase TM and 10 mM HEPES for 25 min at room temperature. Organs were then mechanically dissociated through a 70 μm cell strainer, washed in PBS and red blood cell lysis performed. Single cell suspensions were blocked for 30 min with 50 μL normal mouse serum in PBS 2% FBS on ice then stained with live/dead fixable aqua (Invitrogen), anti-CD45 APC-eFluor780 (30-F11, eBioscience), anti-I-A/I-E Pacific Blue (M5/114.15.2, Biolegend), anti-CD11b PerCP-Cy5.5 (M1/70, Invitrogen), anti-F4/80 PE/Cyanine7 (BM8, Invitrogen), anti-CD3e APC (145-2C11, Invitrogen), anti-CD19 APC (1D3, eBioscience) and anti-Ly-6G/Ly-6C APC (Rb6-8C5, Biolegend).
Tissue zinc quantification
A single anterior prostate lobe, one kidney and one lobe of the liver per mouse was weighed and homogenized in 300 μL of T-PER lysis buffer using the Precellys homogenizer system. Samples were then centrifuged at 1500 x g for 10 min to remove contaminating material and supernatants used for zinc analysis. Zinc levels were analyzed using the Zinc Quantification Kit (ab102507, abcam) as per the manufacturer’s instructions. Zinc concentration expressed as nmol per gram of tissue.
Immunofluorescence microscopy
Samples were fixed in AntigenFix for 30 min at 4°C, rinsed in PBS for 5 min then transferred into 30% sucrose in PBS for 24 h and embedded in optimal cutting temperature compound for cutting. 30 μm sections were permeabilised and blocked in blocking buffer containing 0.1 M TRIS, 0.1% Triton, 1% normal mouse serum, 1% normal rat serum, 1% BSA for 1 h at room temperature. Staining was performed in blocking buffer for 2 h at room temperature prior to washing in PBS and mounting in Fluoromount-G or Fluoromount-G with DAPI. When required, a secondary staining was performed in blocking buffer for 2 h at room temperature prior to washing and mounting. Images were acquired using a TCS SP8 confocal microscope and raw images were processed using Imaris. Sections for the human sample were obtained from a 15 mm x 1 mm core needle biopsy as per sample collection described above. Sections for mouse sample were obtained from cross section of the prostate across the lateral/ventral and dorsal prostate region. Antibodies used include – mouse: MHC II-Pacific Blue (clone M5/114.15.2, 1/50 dilution, BioLegend), CD11b-PE (clone M1/70, 1/50 dilution, Invitrogen), CD31-AF594 (clone MEC13.3, 1/100 dilution, BioLegend), F4/80-AF647 (clone F4/80, 1/50 dilution, Abcam), CD19-AF594 (clone 6D5, 1/100 dilution, BioLegend), IgD-AF488 (clone 11-26c.2a, 1/50 dilution, Biolegend), NKp46-PE (clone 29A1.4, 1/50 dilution, eBioscience), CD8-FITC (clone 53-6.7, 1/50 dilution, eBioscience), CD3-Pacific Blue (clone 17A2, 1/50 dilution, BioLegend); human: HLA-DR-AF647 (clone TAL 1B5, 1/50 dilution, Abcam), CD206-PE Dazzle (clone 15-2, 1/50 dilution, BioLegend), MT1 (clone UC1MT, 1/50 dilution, Abcam), CD8-PE (clone HIT8a, dilution 1/100, BioLegend), CD3-AF488 (clone UCHT1, dilution 1/100, BioLegend), BAFF (polyclonal rabbit, dilution 1/50, Bioss), Mouse IgG1 kappa monoclonal isotype control (clone 15-6E10A7, 1/50 dilution, abcam), Goat anti-mouse IgG secondary-FITC (polyclonal, 1/200 dilution, Invitrogen). Dyes: Flash Phalloidin 488 (1/300 dilution, BioLegend), Hoechst 33258 (dilution 1/10 000, cat# 40044, Biotum), DAPI (in mounting medium, cat# 00-4959-52, Invitrogen).
Other data visualization
Results were generated using R packages or python modules and organized as figures using Adobe Illustrator. Combined scatter-,box-, violin-plots were generated in R using code based on (Allen et al., 2018) and violin plots and dot plots were generated using plotting modules implemented in scanpy or with R ggplot2-based functions. Heatmaps were generated using pheatmap (v1.0.12) R package or matplotlib (v3.0.3) modules in python. Bar plots were generated using Prism (v8).
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism software, R, or python and have been described in the relevant methods sections and figure legends accordingly. In general, unless otherwise specified, non-parametric tests were used and p value after false discovery correction procedures < 0.05 were considered statistically significant. Sample sizes for mice experiments can be found in figure legends.
Additional resources
An interactive version and h5ad files of the single-cell RNaseq data is available at prostatecellatlas.org.
Acknowledgments
The Clatworthy Lab is based in the University of Cambridge Molecular Immunity Unit in the MRC Laboratory of Molecular Biology and is grateful for the use of the core facilities. Z.K.T. and M.R.C. are supported by a Medical Research Council Human Cell Atlas Research Grant (MR/S035842/1), K.W.L is supported by a Kidney Research UK Clinical Training Fellowship (TF_013_20171124), N.R. was supported by a Wellcome Fellowship (106809/Z/15/Z), M.R.C. is supported by a Versus Arthritis Cure Challenge Research Grant (21777), and an NIHR Research Professorship (RP-2017-08-ST2-002). AYW is supported by the Cancer Research UK Cambridge Centre (C9685/A25177) and NIHR Cambridge Biomedical Research Centre (BRC-1215-20014). Z.K.T. holds an honorary research fellow appointment with The University of Queensland Diamantina Institute. We are grateful for infrastructure support from the Cambridge NIHR Biomedical Campus and Cancer Research UK Cambridge Centre. The NIHR Cambridge Biomedical Research Centre (BRC) is a partnership between Cambridge University Hospitals NHS Foundation Trust and the University of Cambridge, funded by the National Institute for Health Research (NIHR).
Author contributions
Formal analysis: Z.K.T., X.T., Q.N.; Investigation: K.W.L., B.B., J.J., N.R., K.K., S.F., P.C., R.G., A.G.L., A.Y.W.; Resources and Data curation: K.W.L., B.B., S.H., A.G., A.B., T.B., V.G., C.M.; Writing—original draft preparation: Z.K.T., M.R.C.; Writing—review and editing: Z.K.T., K.W.L., A.G.L., V.G., C.M., M.R.C.; Project administration: C.M., M.R.C.; Funding acquisition: C.M., M.R.C.; Conceptualization: V.G., C.M., M.R.C.; Supervision: C.M., M.R.C.
Declaration of interests
The authors declare no competing interests.
Published: December 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110132.
Contributor Information
Charlie Massie, Email: cem45@hutchison-mrc.cam.ac.uk.
Menna R. Clatworthy, Email: mrc38@cam.ac.uk.
Supplemental information
Data and code availability
-
•
The raw single-cell RNA-sequencing data reported in this paper is deposited at the European Genome-Phenome Archive under the accession id EGA: EGAS00001005787 with restricted data access control and will be made available by the data access committee, including the lead contact, upon reasonable request. The count data and single-cell objects are available at www.prostatecellatlas.org.
-
•
All code used for the study are available at https://github.com/clatworthylab/prostateimmuneatlas.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.C., Geurts P., Aerts J., et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M., Poggiali D., Whitaker K., Marshall T.R., Kievit R. Raincloud plots: a multi-platform tool for robust data visualization. PeerJ Preprints. 2018;6:e27137v1. doi: 10.12688/wellcomeopenres.15191.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Rajesh A., Rosenkrantz A.B., Choyke P.L., Turkbey B. PI-RADS version 2.1: one small step for prostate MRI. Clin. Radiol. 2019;74:841–852. doi: 10.1016/j.crad.2019.05.019. [DOI] [PubMed] [Google Scholar]
- Beer T.M., Kwon E.D., Drake C.G., Fizazi K., Logothetis C., Gravis G., Ganju V., Polikoff J., Saad F., Humanski P., et al. Randomized, Double-Blind, Phase III Trial of Ipilimumab Versus Placebo in Asymptomatic or Minimally Symptomatic Patients With Metastatic Chemotherapy-Naive Castration-Resistant Prostate Cancer. J. Clin. Oncol. 2017;35:40–47. doi: 10.1200/JCO.2016.69.1584. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- Berglund E., Maaskola J., Schultz N., Friedrich S., Marklund M., Bergenstråhle J., Tarish F., Tanoglidi A., Vickovic S., Larsson L., et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 2018;9:2419. doi: 10.1038/s41467-018-04724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher J.P., Bonavita E., Chakravarty P., Blees H., Cabeza-Cabrerizo M., Sammicheli S., Rogers N.C., Sahai E., Zelenay S., Reis e Sousa C. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell. 2018;172:1022–1037.e14. doi: 10.1016/j.cell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz M.L., Binnewies M., Boldajipour B., Nelson A.E., Pollack J.L., Erle D.J., Barczak A., Rosenblum M.D., Daud A., Barber D.L., et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell. 2014;26:638–652. doi: 10.1016/j.ccell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusa D., Simone M., Gontero P., Spadi R., Racca P., Micari J., Degiuli M., Carletto S., Tizzani A., Matera L. Circulating immunosuppressive cells of prostate cancer patients before and after radical prostatectomy: profile comparison. Int. J. Urol. 2013;20:971–978. doi: 10.1111/iju.12086. [DOI] [PubMed] [Google Scholar]
- Chen S., Zhu G., Yang Y., Wang F., Xiao Y.-T., Zhang N., Bian X., Zhu Y., Yu Y., Liu F., et al. Single-cell analysis reveals transcriptomic remodellings in distinct cell types that contribute to human prostate cancer progression. Nat. Cell Biol. 2021;23:87–98. doi: 10.1038/s41556-020-00613-6. [DOI] [PubMed] [Google Scholar]
- Choi N., Zhang B., Zhang L., Ittmann M., Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I., et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello L.C., Franklin R.B. A comprehensive review of the role of zinc in normal prostate function and metabolism; and its implications in prostate cancer. Arch. Biochem. Biophys. 2016;611:100–112. doi: 10.1016/j.abb.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley L., Cambuli F., Aparicio L., Shibata M., Robinson B.D., Xuan S., Li W., Hibshoosh H., Loda M., Rabadan R., Shen M.M. A single-cell atlas of the mouse and human prostate reveals heterogeneity and conservation of epithelial progenitors. eLife. 2020;9:e59465. doi: 10.7554/eLife.59465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff A.O., Robertson F.P., Stegmann K.A., Pallett L.J., Maini M.K., Davidson B.R., Male V. Eomeshi NK Cells in Human Liver Are Long-Lived and Do Not Recirculate but Can Be Replenished from the Circulation. J. Immunol. 2016;197:4283–4291. doi: 10.4049/jimmunol.1601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsson S., Ohlson A.L., Andersson S.O., Fall K., Meisner A., Fiorentino M., Andrén O., Rider J.R. CD4 helper T cells, CD8 cytotoxic T cells, and FOXP3(+) regulatory T cells with respect to lethal prostate cancer. Mod. Pathol. 2013;26:448–455. doi: 10.1038/modpathol.2012.164. [DOI] [PubMed] [Google Scholar]
- Doering T.A., Crawford A., Angelosanto J.M., Paley M.A., Ziegler C.G., Wherry E.J. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37:1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra P., Rancan C., Ma W., Toth M., Senda T., Carpenter D.J., Kubota M., Matsumoto R., Thapa P., Szabo P.A., et al. Tissue Determinants of Human NK Cell Development, Function, and Residence. Cell. 2020;180:749–763.e13. doi: 10.1016/j.cell.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner B.G., Dorner M.B., Zhou X., Opitz C., Mora A., Güttler S., Hutloff A., Mages H.W., Ranke K., Schaefer M., et al. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity. 2009;31:823–833. doi: 10.1016/j.immuni.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- Epstein J.I., Zelefsky M.J., Sjoberg D.D., Nelson J.B., Egevad L., Magi-Galluzzi C., Vickers A.J., Parwani A.V., Reuter V.E., Fine S.W., et al. A Contemporary Prostate Cancer Grading System: A Validated Alternative to the Gleason Score. Eur. Urol. 2016;69:428–435. doi: 10.1016/j.eururo.2015.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandsson A., Carlsson J., Lundholm M., Fält A., Andersson S.O., Andrén O., Davidsson S. M2 macrophages and regulatory T cells in lethal prostate cancer. Prostate. 2019;79:363–369. doi: 10.1002/pros.23742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauriat C., Long E.O., Ljunggren H.G., Bryceson Y.T. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115:2167–2176. doi: 10.1182/blood-2009-08-238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Franklin R.B., Feng P., Milon B., Desouki M.M., Singh K.K., Kajdacsy-Balla A., Bagasra O., Costello L.C. hZIP1 zinc uptake transporter down regulation and zinc depletion in prostate cancer. Mol. Cancer. 2005;4:32. doi: 10.1186/1476-4598-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J., Hastie T., Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Gannon P.O., Poisson A.O., Delvoye N., Lapointe R., Mes-Masson A.M., Saad F. Characterization of the intra-prostatic immune cell infiltration in androgen-deprived prostate cancer patients. J. Immunol. Methods. 2009;348:9–17. doi: 10.1016/j.jim.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Guilliams M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity. 2016;44:439–449. doi: 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- Gnanapragasam V.J., Lophatananon A., Wright K.A., Muir K.R., Gavin A., Greenberg D.C. Improving Clinical Risk Stratification at Diagnosis in Primary Prostate Cancer: A Prognostic Modelling Study. PLoS Med. 2016;13:e1002063. doi: 10.1371/journal.pmed.1002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory J.L., Morand E.F., McKeown S.J., Ralph J.A., Hall P., Yang Y.H., McColl S.R., Hickey M.J. Macrophage migration inhibitory factor induces macrophage recruitment via CC chemokine ligand 2. J. Immunol. 2006;177:8072–8079. doi: 10.4049/jimmunol.177.11.8072. [DOI] [PubMed] [Google Scholar]
- Guilliams M., Ginhoux F., Jakubzick C., Naik S.H., Onai N., Schraml B.U., Segura E., Tussiwand R., Yona S. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol. 2014;14:571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner M.C., Guner G., Taheri D., Netto G.J., Palsgrove D.N., Zheng Q., Guedes L.B., Kim K., Tsai H., Esopi D.M., et al. Comprehensive Evaluation of Programmed Death-Ligand 1 Expression in Primary and Metastatic Prostate Cancer. Am. J. Pathol. 2018;188:1478–1485. doi: 10.1016/j.ajpath.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry G.H., Malewska A., Joseph D.B., Malladi V.S., Lee J., Torrealba J., Mauck R.J., Gahan J.C., Raj G.V., Roehrborn C.G., et al. A Cellular Anatomy of the Normal Adult Human Prostate and Prostatic Urethra. Cell Rep. 2018;25:3530–3542.e5. doi: 10.1016/j.celrep.2018.11.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsmans M., Clauss S., Xiao L., Aguirre A.D., King K.R., Hanley A., Hucker W.J., Wülfers E.M., Seemann G., Courties G., et al. Macrophages Facilitate Electrical Conduction in the Heart. Cell. 2017;169:510–522.e20. doi: 10.1016/j.cell.2017.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein M.R., Al-Assiri M., Musalam A.O. Phenotypic characterization of the infiltrating immune cells in normal prostate, benign nodular prostatic hyperplasia and prostatic adenocarcinoma. Exp. Mol. Pathol. 2009;86:108–113. doi: 10.1016/j.yexmp.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Karthaus W.R., Hofree M., Choi D., Linton E.L., Turkekul M., Bejnood A., Carver B., Gopalan A., Abida W., Laudone V., et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science. 2020;368:497–505. doi: 10.1126/science.aay0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo K.C., Shim D.H., Yang C.M., Lee S.B., Kim S.M., Shin T.Y., Kim K.H., Yoon H.G., Rha K.H., Lee J.M., Hong S.J. Reduction of the CD16(-)CD56bright NK cell subset precedes NK cell dysfunction in prostate cancer. PLoS ONE. 2013;8:e78049. doi: 10.1371/journal.pone.0078049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G., Sukhov V., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2019:060012. [Google Scholar]
- Kumar B.V., Ma W., Miron M., Granot T., Guyer R.S., Carpenter D.J., Senda T., Sun X., Ho S.H., Lerner H., et al. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep. 2017;20:2921–2934. doi: 10.1016/j.celrep.2017.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru T.H., Wadhwa K., Chang R.T., Echeverria L.M., Roethke M., Polson A., Rottenberg G., Koo B., Lawrence E.M., Seidenader J., et al. Definitions of terms, processes and a minimum dataset for transperineal prostate biopsies: a standardization approach of the Ginsburg Study Group for Enhanced Prostate Diagnostics. BJU Int. 2013;112:568–577. doi: 10.1111/bju.12132. [DOI] [PubMed] [Google Scholar]
- Kwon E.D., Drake C.G., Scher H.I., Fizazi K., Bossi A., van den Eertwegh A.J., Krainer M., Houede N., Santos R., Mahammedi H., et al. CA184-043 Investigators Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 2014;15:700–712. doi: 10.1016/S1470-2045(14)70189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liuzzi J.P., Cousins R.J. Mammalian zinc transporters. Annu. Rev. Nutr. 2004;24:151–172. doi: 10.1146/annurev.nutr.24.012003.132402. [DOI] [PubMed] [Google Scholar]
- Lun A.T.L., Riesenfeld S., Andrews T., Dao T.P., Gomes T., Marioni J.C., participants in the 1st Human Cell Atlas Jamboree EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 2019;20:63. doi: 10.1186/s13059-019-1662-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay L.K., Rahimpour A., Ma J.Z., Collins N., Stock A.T., Hafon M.L., Vega-Ramos J., Lauzurica P., Mueller S.N., Stefanovic T., et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 2013;14:1294–1301. doi: 10.1038/ni.2744. [DOI] [PubMed] [Google Scholar]
- Mackay L.K., Minnich M., Kragten N.A., Liao Y., Nota B., Seillet C., Zaid A., Man K., Preston S., Freestone D., et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 2016;352:459–463. doi: 10.1126/science.aad2035. [DOI] [PubMed] [Google Scholar]
- MacParland S.A., Liu J.C., Ma X.Z., Innes B.T., Bartczak A.M., Gage B.K., Manuel J., Khuu N., Echeverri J., Linares I., et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018;9:4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madissoon E., Wilbrey-Clark A., Miragaia R.J., Saeb-Parsy K., Mahbubani K.T., Georgakopoulos N., Harding P., Polanski K., Huang N., Nowicki-Osuch K., et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol. 2019;21:1. doi: 10.1186/s13059-019-1906-x. Published online December 31, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass E., Ballesteros I., Farlik M., Halbritter F., Günther P., Crozet L., Jacome-Galarza C.E., Händler K., Klughammer J., Kobayashi Y., et al. Specification of tissue-resident macrophages during organogenesis. Science. 2016;353 doi: 10.1126/science.aaf4238. Published online September 9, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl S.R. Chemokines and dendritic cells: a crucial alliance. Immunol. Cell Biol. 2002;80:489–496. doi: 10.1046/j.1440-1711.2002.01113.x. [DOI] [PubMed] [Google Scholar]
- McInnes L., Healy J., Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv. 2018 https://arxiv.org/abs/1802.03426v3 [Google Scholar]
- Miles A.T., Hawksworth G.M., Beattie J.H., Rodilla V. Induction, regulation, degradation, and biological significance of mammalian metallothioneins. Crit. Rev. Biochem. Mol. Biol. 2000;35:35–70. doi: 10.1080/10409230091169168. [DOI] [PubMed] [Google Scholar]
- Montoro D.T., Haber A.L., Biton M., Vinarsky V., Lin B., Birket S.E., Yuan F., Chen S., Leung H.M., Villoria J., et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P.A., Koscsó B., Rajani G.M., Stevanovic K., Berres M.L., Hashimoto D., Mortha A., Leboeuf M., Li X.M., Mucida D., et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 2014;158:300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T., et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narni-Mancinelli E., Jaeger B.N., Bernat C., Fenis A., Kung S., De Gassart A., Mahmood S., Gut M., Heath S.C., Estellé J., et al. Tuning of natural killer cell reactivity by NKp46 and Helios calibrates T cell responses. Science. 2012;335:344–348. doi: 10.1126/science.1215621. [DOI] [PubMed] [Google Scholar]
- Pasero C., Gravis G., Guerin M., Granjeaud S., Thomassin-Piana J., Rocchi P., Paciencia-Gros M., Poizat F., Bentobji M., Azario-Cheillan F., et al. Inherent and Tumor-Driven Immune Tolerance in the Prostate Microenvironment Impairs Natural Killer Cell Antitumor Activity. Cancer Res. 2016;76:2153–2165. doi: 10.1158/0008-5472.CAN-15-1965. [DOI] [PubMed] [Google Scholar]
- Pham D., Tan X., Xu J., Grice L.F., Lam P.Y., Raghubar A., Vukovic J., Ruitenberg M.J., Nguyen Q. stLearn: integrating spatial location, tissue morphology and gene expression to find cell types, cell-cell interactions and spatial trajectories within undissociated tissues. bioRxiv. 2020 2020.05.31.125658. [Google Scholar]
- Polański K., Young M.D., Miao Z., Meyer K.B., Teichmann S.A., Park J.E. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics. 2020;36:964–965. doi: 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu D.M., Botting R.A., Stephenson E., Green K., Webb S., Jardine L., Calderbank E.F., Polanski K., Goh I., Efremova M., et al. Decoding human fetal liver haematopoiesis. Nature. 2019;574:365–371. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E.W., Broz M.L., Binnewies M., Headley M.B., Nelson A.E., Wolf D.M., Kaisho T., Bogunovic D., Bhardwaj N., Krummel M.F. Critical Role for CD103(+)/CD141(+) Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell. 2016;30:324–336. doi: 10.1016/j.ccell.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S., Graner E., Febbo P., Weinstein L., Bhattacharya N., Onody T., Bubley G., Balk S., Loda M. Fatty acid synthase expression defines distinct molecular signatures in prostate cancer. Mol. Cancer Res. 2003;1:707–715. [PubMed] [Google Scholar]
- Schulz C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S.E., Pollard J.W., et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- Shen M.M., Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F.D., Ljunggren H.G., La Cava A., Van Kaer L. Organ-specific features of natural killer cells. Nat. Rev. Immunol. 2011;11:658–671. doi: 10.1038/nri3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas C., Chanzá N.M., Awada A., Scartozzi M. The immune infiltrate in prostate, bladder and testicular tumors: An old friend for new challenges. Cancer Treat. Rev. 2017;53:138–145. doi: 10.1016/j.ctrv.2016.12.004. [DOI] [PubMed] [Google Scholar]
- Spranger S., Dai D., Horton B., Gajewski T.F. Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer Cell. 2017;31:711–723.e4. doi: 10.1016/j.ccell.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl P.L., Salmén F., Vickovic S., Lundmark A., Navarro J.F., Magnusson J., Giacomello S., Asp M., Westholm J.O., Huss M., et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- Stewart B.J., Ferdinand J.R., Young M.D., Mitchell T.J., Loudon K.W., Riding A.M., Richoz N., Frazer G.L., Staniforth J.U.L., Vieira Braga F.A., et al. Spatiotemporal immune zonation of the human kidney. Science. 2019;365:1461–1466. doi: 10.1126/science.aat5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P.A., Levitin H.M., Miron M., Snyder M.E., Senda T., Yuan J., Cheng Y.L., Bush E.C., Dogra P., Thapa P., et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. 2019;10:4706. doi: 10.1038/s41467-019-12464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag V.A., Waltman L., van Eck N.J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 2019;9:5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.E., Stephenson E., Polański K., Goncalves A., et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal A.C., Howard L.E., Wiggins E., De Hoedt A.M., Shiao S.L., Knott S., Taioli E., Fowke J.H., Freedland S.J. Natural killer cell activity and prostate cancer risk in veteran men undergoing prostate biopsy. Cancer Epidemiol. 2019;62:101578. doi: 10.1016/j.canep.2019.101578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.A., Mitrofanova A., Bergren S.K., Abate-Shen C., Cardiff R.D., Califano A., Shen M.M. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 2013;15:274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.A., Toivanen R., Bergren S.K., Chambon P., Shen M.M. Luminal cells are favored as the cell of origin for prostate cancer. Cell Rep. 2014;8:1339–1346. doi: 10.1016/j.celrep.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Xin F., Lin N., Wang Y., Liu X., Liu J. Metallothioneins may be a potential prognostic biomarker for tumors: A Prisma-compliant meta-analysis. Medicine (Baltimore) 2018;97:e13786. doi: 10.1097/MD.0000000000013786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Park J., Susztak K., Zhang N.R., Li M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun. 2019;10:380. doi: 10.1038/s41467-018-08023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yu P., Zhou B., Song J., Li Z., Zhang M., Guo G., Wang Y., Chen X., Han L., Hu S. Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat. Cell Biol. 2020;22:108–119. doi: 10.1038/s41556-019-0446-7. [DOI] [PubMed] [Google Scholar]
- Watson P.A., Arora V.K., Sawyers C.L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Desouki M.M., Lin S., Xiao D., Franklin R.B., Feng P. Differential expression of metallothioneins (MTs) 1, 2, and 3 in response to zinc treatment in human prostate normal and malignant cells and tissues. Mol. Cancer. 2008;7:7. doi: 10.1186/1476-4598-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock S.L., Lopez R., Klein A.M. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 2019;8:281–291.e9. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J., Schmidt S.V., Sander J., Draffehn A., Krebs W., Quester I., De Nardo D., Gohel T.D., Emde M., Schmidleithner L., et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T. Roles of the ribosomal protein S19 dimer and the C5a receptor in pathophysiological functions of phagocytic leukocytes. Pathol. Int. 2007;57:1–11. doi: 10.1111/j.1440-1827.2007.02049.x. [DOI] [PubMed] [Google Scholar]
- Young M.D., Behjati S. SoupX removes ambient RNA contamination from droplet based single-cell RNA sequencing data. bioRxiv. 2020:303727. doi: 10.1093/gigascience/giaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadra G., Photopoulos C., Tyekucheva S., Heidari P., Weng Q.P., Fedele G., Liu H., Scaglia N., Priolo C., Sicinska E., et al. A novel direct activator of AMPK inhibits prostate cancer growth by blocking lipogenesis. EMBO Mol. Med. 2014;6:519–538. doi: 10.1002/emmm.201302734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S.G., Lehrer J., Chang S.L., Das R., Erho N., Liu Y., Sjöström M., Den R.B., Freedland S.J., Klein E.A., et al. The Immune Landscape of Prostate Cancer and Nomination of PD-L2 as a Potential Therapeutic Target. J. Natl. Cancer Inst. 2019;111:301–310. doi: 10.1093/jnci/djy141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw single-cell RNA-sequencing data reported in this paper is deposited at the European Genome-Phenome Archive under the accession id EGA: EGAS00001005787 with restricted data access control and will be made available by the data access committee, including the lead contact, upon reasonable request. The count data and single-cell objects are available at www.prostatecellatlas.org.
-
•
All code used for the study are available at https://github.com/clatworthylab/prostateimmuneatlas.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.