Graphical abstract
Keywords: Candida albicans, Helicobacter pylori, Co-aggregation, Polymicrobial biofilm, Functionally-Enhanced derivative spectroscopy, Surface properties, Analytical bio-spectroscopy
Highlights
-
•
C. albicans and H. pylori co-existence has been described from stomach.
-
•
To advance in the understanding of H. pylori and C. albicans polymicrobial biofilms.
-
•
Multiple surface interaction mechanisms were identified
-
•
Anchoring, co-aggregation and formation of yeast-bacterium polymicrobial biofilm
Abstract
Introduction
Polymicrobial biofilms have a significant impact on pathogenesis of infectious microorganisms. Many human diseases are affected by colonization of multi-species communities affecting negatively the treatments and increase the risks for the health. In particular, in the epithelium of the stomach co-existence between C. albicans and H. pylori has been described, which has been associated to a synergistic effect on ulcer pathogenesis.
Objective
The objective of this work was to advance in the understanding of surface interaction between H. pylori and C. albicans for the formation of polymicrobial biofilms.
Methods
Studies of microbial surfaces both bacterium, yeast and co-cultures of them were carried out by infrared spectroscopy, deconvolution analysis, transmission and scanning electron microscopies, and optic microscopy. Additional methods were used to contrast the results as dynamic light scattering, contact angle, agarose gel electrophoresis and gene amplification.
Results
Several surface interaction mechanisms promote the anchoring of H. pylori on C. albicans, cell co-aggregation, and polymicrobial biofilm formation, main identified interactions were: (i) hydrophobic interactions between non-polar peptide chains and lipid structures, characterized by θw among 84.9 ± 1.6 (γ = 22.78 mJ/m2 with 95.3 of dispersive contribution) and 76.6 ± 3.8 (γ = 17.34 mJ/m2, 40.2 of dispersive contribution) for C. albicans and H. pylori, respectively, (ii) hydrogen bonds between surface components of yeast and bacterium (e.g., —S—H⋅⋅⋅NH2— or —S—H⋅⋅⋅O CO—) and (iii) thiol-mediated surface interactions identified by displacements to lower wavenumbers (Δv = 5 cm−1). Evidence of internalization and electrostatic interactions were not evidenced. All observations were congruent with the biofilm formation, including the identification of small-size biostructures (i.e., 122–459 nm) associated with extracellular proteins, extracellular DNA, or outer membrane vesicles were observed characteristic of biofilm formation.
Conclusion
It is concluded that biofilm is formed by co-aggregation after anchoring of H. pylori on C. albicans. Several surface interactions were associated with the prevalence of H. pylori, the possibility to find C. albicans in the stomach epithelium infected by H. pylori, but also, strength interactions could be interfering in experimental observations associated with bacterial-DNA detection in culture mixtures.
Introduction
Bacteria and fungi often are found in the same microhabitats forming dynamic communities known as polymicrobial biofilms. Physical, chemical and biological interactions between these microorganisms (MOs) play a key role in the functioning of numerous ecosystems and impact the health and diseases of plants and animals [1], [2]. In humans, in the absence of disease, bacteria and fungi are usually found on cutaneous and mucosal surfaces as skin, oral cavity, gastrointestinal tract, and reproductive tract [3], [4], [5]. Several studies have evidenced that biochemical and physical interactions between these MOs are the beginning point for the forming of polymicrobial biofilms [3]. It is widely-accepted that the understanding of nature and correct description of bacterium-yeast interaction mechanism are important tools for the design of new strategies of treatment in the case of diseases produced by these pathogens [6], [7].
Candida albicans (C. albicans) is an opportunistic pathogenic yeast forming part of the human gut flora; however, it becomes a pathogenic MO in immunocompromised individuals under a variety of conditions, but also, from medical and economic approaches, it is a serious public health challenge due to its negative impact as consequence the high mortality rates and significant increase of care and hospitalization costs [8], [9]. Candida pathogenicity is eased by a number of virulence factors, being the adherence to tissues and medical devices the most important factors. In concordance with the above, C. albicans is able to produce physical and chemical changes on the environment by formation and secretion of hydrolytic enzymes, bind amino acids and sugars, and form polymicrobial biofilms [9], [10], [11]. Studies have evidenced that enhanced-bacterial virulence of C. albicans is resulting of interaction with S. aureus and E. coli [3]. But also, interaction of C. albicans with different bacteria, including Pseudomonas spp, Acinetobacter baumannii, Lactobacillus rhamnosus, and Staphylococcus epidermidis, are well-known [3], [7], [12], [13], [14].
On the other hand, H. pylori is a Gram-negative pathogenic bacteria colonizing the stomach by various efficiently specialized mechanisms, mainly by urease-mediated biochemical processes generating alkaline micro-domains in the affected zones, being able to stay decades interacting with the gastric mucosa without causing harm to the host (asymptomatic carriers) [15], [16], [17], [18]. It has been estimated that around 50% of the human population is affected by this bacterium. The presence of H. pylori is associated as a predisposing factor for the development of gastric pathologies such as gastritis, peptic ulcers, non-cardiac gastric adenocarcinoma and gastric MALT lymphoma [15], [19]. Since 1994 H. pylori is recognized by the International Agency for Research on Cancer and the World Health Organization (WHO) as a category I carcinogen [20], [21].
Though C. albicans is present in some parts of the gastrointestinal tract is not usually found in the stomach because this is a pathway of passage for MO [7], [22]. However, in vitro studies have evidenced the ability of C. albicans to grow in an acid environment (i.e., pH = 2.0) and colonize H. pylori-associated gastric ulcers [7], [22], [23]. Thus, it is evident that some type of efficient mechanism coexistence and adapting of C. albicans to survive at the low pH of stomach should be acting [22] (see Fig. 1). Recently, it has been proposed that C. albicans and H. pylori could interact by internalization of bacterium inside of yeast which acts as vehicle in bacterial transferring. However, the understanding how this interaction occurs is still a matter of debate since the specific information is very scarce, in fact, this internalization mechanism between H. pylori and C. albicans is rejected by some researchers and defended by others [24], [25], [26]. In presence of H. pylori, the growth of C. albicans is expected to be eased due to pH changes promoted by H. pylori, as a result, the success of drugs used for the treatment of bacterial gastric ulcers is negatively impacted [27], [28], [29]. In addition, this coexistence could have implications on the adaptability in exgastric environment, prevalence and pathogenicity of H. pylori [27], [28], [29].
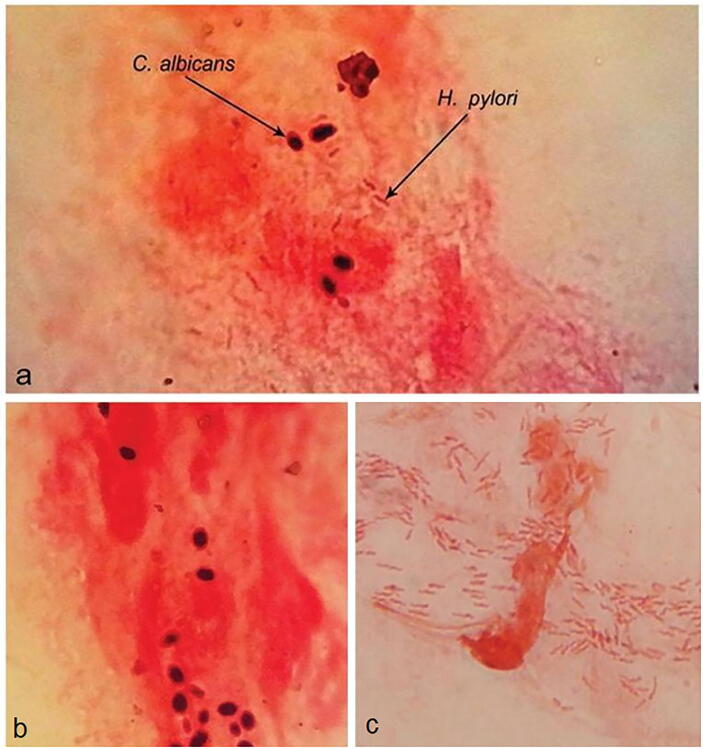
Fig. 1.
Microscopic observation of H. pylori and C. albicans in gastric biopsies from patients with gastritis: (a) simultaneous concurrence from H. pylori and C. albicans, (b) cell of C. albicans and (c) bacillar form H. pylori (Image adapted from S. Massarat, P. Saniee, F. Siavoshi, R. Mokhtari, F. Mansour-Ghanaei, S. Khalili-Samani, Front Microbiol 2016, 7, 801; Copyright © 2016 CCBY).
The objective of this work was to advance in the understanding of surface interaction between H. pylori and C. albicans for the formation of polymicrobial biofilms. At present, studies of dual-species biofilm between C. albicans and H. pylori have not been previously published. In addition, new tools for the biointerfaces by analytical biospectroscopy are introduced.
Experimental section
Microorganisms strain and growing conditions
C. albicans strain: ATCC 90028:C. albicans ATCC 90028 was used as a yeast model. C. albicans strain was grown by inoculating in tryptic soy broth (TSB) (Invitrogen, Carlsbad, CA, USA) and standardized to 0.5 in the Mc-Farland scale by absorbance measurements at 625 nm using turbidity standards prepared from BaCl2 solution (0.048 molL−1) and H2SO4 (0.18 molL−1). Later, sub-samples of yeast were separately inoculated in agar Müller-Hinton and incubated for 24 h at 37 °C. Gram stain was performed when the yeast growth was evidenced.
H. pylori strain: ATCC 43504:H. pylori ATCC 43504 (American Type Culture Collection, USA) was used as bacterial model. H. pylori strain was grown by inoculating in Columbia agar plates (Oxoid, England) supplemented with 7% horse blood and incubated under microaerobic conditions in the presence of 5% O2, 10% CO2 and 85% N2 at 37 °C for 4 days. Bacterial growth was standardized to 0.5 in the Mc-Farland by analogous procedure to that described for C. albicans, and verified by Gram stain, urease test and SEM (Phenom PRO X, ThermoFisher Scientific).
C. albicans and H. pylori co-cultures: Microbial mixtures, (Ca + Hp)BF, were made from suspensions of bacteria and yeasts previously standardized. For the preparation of yeast and bacterial suspensions, colonies of C. albicans and H. pylori were separately dispersed in 50 ml of sterile physiological serum until to obtain a homogeneous suspension. Afterward, the dispersions were standardized to correspond to 1.0x108 CFU/ml by the quantification of scattered-light intensity at 600 nm, using UV–vis spectrophotometry; then, a mixture of suspensions was performed using a 1:1 vol ratio.
After, aliquots of mixed suspension were prepared in Eppendorf tubes (10 µl) and inoculated in BHI broth, incubating at 25, 30 and 37 °C for 48 h under aerobiosis conditions. These tests were performed in triplicate. Microscopic observations of mixtures were performed and captured in digital format for its posterior analysis.
DNA extraction, gene amplification and electrophoresis
DNA extraction was performed using the UltraClean® Microbial DNA Isolation kit (M.O. BIO, USA), according to the manufacturer's instructions. Microbial cells are re-suspended in MicroBead solution (microspheres dispersion), followed by lysis solution. Thus, cell-lysed components are separated by centrifugation. From the lysed cells, the released DNA is bound to a silica Spin Filter. Finally, the filter is washed, and the DNA is recovered in a certified DNA-free Tris-buffer. Later, an amplification of the 16 s rRNA gene was performed using the Sapphire Amp® Fast PCR Master Mix kit (TAKARA BIO INC, Japan). To each sample into working temperatures, 25, 30 and 37 °C, as well as for their respective controls (positive control of H. pylori was H. pylori ATCC 43504 suspension and Lactobacillus sp, and as negative control was used water), 6.25 µl of Master Mix, 0.5 µl of Forward Primer, 0.5 µl of Reverse Primer, 3 µl of each DNA sample and 2.25 µl of PCR grade water were added. Finally, agarose gel electrophoresis was performed (Lonza, USA). Once the gel solidified, 2 µl of 100 bp run marker (MASTROGEN, USA) ranged between 100 bp and 3,000 bp was added to the last well, then 5 µl of the amplified samples was added to each well, and it was run the gel at 90 V for 50 min.
Biofilm formation
Microbial biofilms were performed by deposition of individual colonies on ultrafiltration cellulose membranes at 37 °C per 48 h (1 cm2, MWCO: 10 kDa, Millipore, Merck). In order to eliminate residual composition of growth medium on the surface, after forming the biolayer on the support, samples were successively washed with deionized water and dried using a laminar flow oven with temperature control at 40 °C per 12 h. To warrant this stage, spectra of support and blank experiments, i.e., growth medium without bacterium, were compared with the respective spectra of biofilm samples. Similar procedure was used for mixtures of C. albicans and H. pylori. In all cases, four replicates were prepared.
Biofilm study by infrared spectroscopy
Recording of spectra: By the use infrared (IR) spectroscopy, average spectra were obtained for biofilms of strains of H. pylori, C. albicans and (Ca + Hp)BF. IR spectra of four replicates were obtained using a spectrophotometer with Attenuated Total Reflectance (ATR) with ZnSe crystal (IR-Affinity, Shimadzu Co). Each spectrum was recorded by 20 scans from 4000 to 500 cm−1, with a maximum resolution of 0.5 cm−1. In addition, Michelson interferometer (30° incident angle) equipped with Dynamic Alignment System Sealed interferometer with Automatic Dehumidifier and DLATGS detector equipped with temperature control mechanism were used. Data were extracted in file format .txt in order to carry out the analysis using a spreadsheet.
Selecting of region for spectral analysis: A strong overlap of the signals is expected due to the compositional complexity of the biofilms [30], [31], [32]. A first comparison of differentiation capacity of IR spectra was performed by chemometric analysis using Principal Component Analysis (PCA). To study the biofilms, the region between 3100 and 2400 cm−1 was selected, in this region, signals associated with cysteine units have been previously described for H. pylori by FEDS [30]. In particular, H. pylori contains cysteine-rich proteins on its external membrane which are associated to disulfide bonds (—S—S—) which can be monitored from IR spectra in conjunction with deconvolution techniques. Disulfide bonds are important in this research because they have been related with the anchoring of H. pylori to surfaces [30], [33]. In addition, in this spectral range, signals associated with invariant structural units can be easily identified, such as CH, CH2 and CH3, which can be used as internal markers for the analysis of signal displacements since they are weakly affected by neighboring electronegative groups. However, in multicomponent systems with a strong overlap of signals, the vibration associated with —SH is often difficult to identify due to the overlap and the weak signal related with the stretching of S—H. In order to achieve an adequate comparative analysis of the biofilms, deconvolution by Functionally Enhanced Derivative Spectroscopy (FEDS) was carried out.
FEDS deconvolution of IR Spectra: To carry out the deconvolution, FEDS was used. By the use of FEDS is possible to carry out the transformation of the spectrum of the biofilms allowing the construction of “spectral fingerprints” with higher resolution of signals in comparison with IR spectra. Details of this procedure have recently been published previously for H. pylori and C. albicans using artificial biofilms [30], [31]. Here, the analysis is focused in the building spectral fingerprints of biofilms and dual-species biofilms promoted from aqueous environments. Spectral data, both wavenumber (v) and absorbance (a), were smoothed using Average-Based Spectral Filter (ABSF) with a data window of 3 and 20 cycles; later, self-scaling of data of absorbance was carried out and, finally, self-scaled data were used as in-put data for the deconvolution function permitting to obtain the corresponding FEDS spectra. Details of this procedure have been previously published [34], [35], [36], [37]. In order to define one standard criterion to identify the occurrence of shifts, the following aspects were taken in considering: (i) small differences are usual in IR spectra, (ii) spectral resolution is defined by two consecutive wavenumbers, being in our case, Δv = 1.93 cm−1, and (iii) by FEDS a variability around Δv cm−1 is unavoidably produced due to differentiation procedure. In consequence, intrinsic variability of signals in FEDS is ±2Δv cm−1 = ±3.86 cm−1 ≈ ±4.0 cm−1. By the above, Δv lower than ±4.0 cm−1 were assumed to be not significant.
Biofilm study by microscopy: SEM, TEM and optic microscopy
Samples obtained from C. albicans, H. pylori and (Ca + Hp)BF were analyzed by Scanning Electron Microscopy (SEM) (JEOL 6380LV with resolution of 3.0 nm), Transmission Electron Microscopy (TEM) (JEOL-JEM 1200EXII with resolution of 0.15 nm), and optic microscopy (Nikon Eclipse E600 microscope). Samples were pretreated by the following procedure: For SEM, chemical fixation was performed with paraformaldehyde, followed of dehydration with acetone, dried at 40 °C in temperature-controlled oven and coated with an ultrathin layer of gold; whereas for TEM, chemical fixation was performed using paraformaldehyde and osmium tetroxide, followed of dehydration with acetone and embedding in EMbed-812 [30], [31].
Size, cell wall thickness and cell volume of MOs were obtained from images of SEM and TEM using several size descriptors: Equivalent Sphere Radius (ESR) defined to be the radius of sphere with the same projected area than the object in the image (1D descriptor), Equivalent Circle Area (ECA) defined to be the area of circle with the same area than object in the image (2D descriptor), and Equivalent Sphere Volume (ESV) defined as the sphere with the same volume defined by the ECA (3D descriptor) [38].
Optic microscopy videos were recorded using a digital camera DSC-H300 (Sony, 100x of digital zoom and 20.1 megapixels). From video, image sequences were extracted and analyzed. In all cases, image digital analysis was performed using Digital Micrograph 3.7.0 (Gatan Software Team; Gatan Inc., USA).
Study by dynamic light scattering (DLS)
Size and size distribution of particles in suspension for H. pylori, C. albicans and (Ca + Hp)BF were obtained using a 90 plus/BI-MAS equipped with Nano-hpp v330 software and detecting of particles from 1 nm to 6 µm. In all cases, measurements were performed in quadruplicate.
Study of surface hydrophilic-hydrophobic properties
To study the hydrophobic or hydrophilic nature of the biofilm surface, the sessile drop method was used. For this, three testing liquids were deposited, separately, on the surface of the biofilms. The procedure was recorded in video format using a digital camera DSC-H300 (Sony, 100x of digital zoom and 20.1 megapixels). After, the pictures were extracted for the analysis of the contact angle of the liquid–air-biofilm interface. As working liquids, water, ethylene glycol and propane-1,3-diol were used since they have different polarity. In all cases, measurements were performed in quadruplicate. Total surface free energy of biofilm was calculated using the van Oss-Chaudhury-Good theory, contact angles, surface free energy of working liquids and their components [39], [40]. Software Surface-Q was used for calculations (Mindtech s.a.s., Colombia).
Results and discussion
Study by IR spectroscopy and FEDS
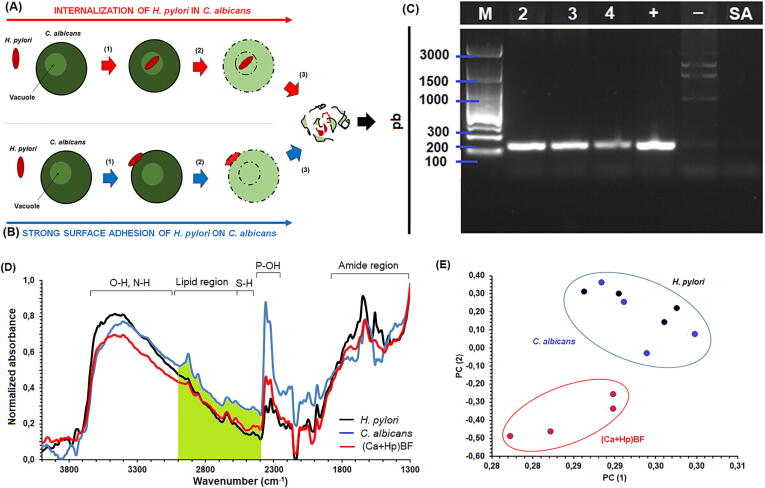
H. pylori DNA mixed with C. albicans DNA have been detected from mixed growth between the bacterium and the yeast. This observation has been interpreted as evidence of internalization of H. pylori in the inner of the yeast (see Fig. 2A and C). However, a mixture of genetic material also is expected if H. pylori and C. albicans experience a strong surface interaction, which could avoid the complete elimination of H. pylori from the growth medium, and since cell lysis for extraction of DNA is not a selective procedure, a partial elimination of the bacterial cells is a potential contamination and experimental error source (see Fig. 2B). By experiments using (Ca + Hp)BF samples, which were analyzed by agarose gel electrophoresis, the obtaining of mixtures of DNA was verified. However, it is possible to establish a second hypothesis, which is related with the release of extracellular DNA from H. pylori during the formation of biofilms.
Fig. 2.
Hypotheses explaining the observation of mixed DNA from mixtures of C. albicans and H. pylori: (A) hypothesis of internalization and (B) hypothesis of surface anchoring (release of extracellular DNA has not illustrated). (D) IR spectra of H. pylori, C. albicans and (Ca + Hp)BF, and (C) comparison of variability of spectra by analysis of principal component.
By IR spectroscopy, IR spectra of H. pylori, C. albicans and (Ca + Hp)BF were obtained (see Fig. 2D). A complete analysis for individual MOs was previously published from artificial biolayers [30], [31]; however, in these experiments, though main signals are the same, a strong overlap and increasing of absorbances were observed below 1300 cm−1. By analysis of PCA was determined that 95% of variability of H. pylori, C. albicans and (Ca + Hp)BF spectra can be described by two components. Results evidence that H. pylori and C. albicans spectra cannot be differenced whereas (Ca + Hp)BF spectrum showed a great variability in comparison with the spectra of individual MOs (see Fig. 2E). The above suggests that derivative deconvolution techniques are an alternative to increase the spectral differentiation without loss information from IR spectra [34].
Analysis of IR spectra were performed by three stages: (i) assignation of signals easily identified in the IR spectra from previously published information, (ii) comparison of FEDS spectra of H. pylori and C. albicans to identify characteristic signals, which can be: signals appearing in each IR and FEDS spectra (c, h and m for C. albicans, H. pylori and (Ca + Hp)BF, respectively), signals observed at the same wavenumbers from FEDS spectra of individual MOs (k), and signals observed in only one from FEDS spectra (d). In particular, any shift of signals can be used to identify possible types of interactions associated with functional groups appearing in the spectrum.
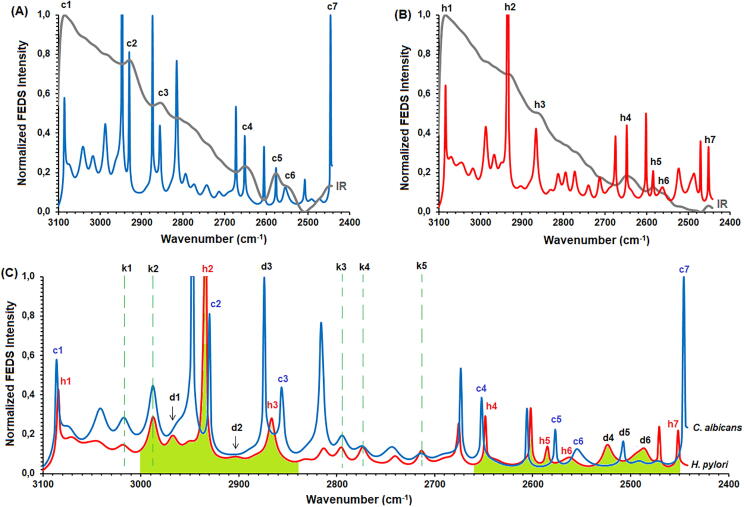
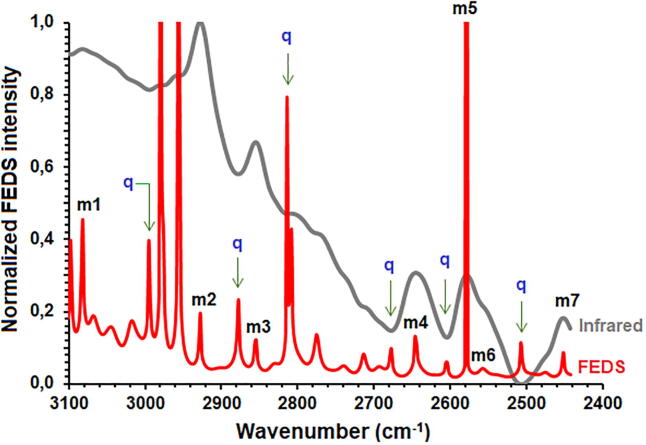
Normalized IR and FEDS spectra, between 3100 and 2400 cm−1, obtained from biofilms of H. pylori and C. albicans are shown in Fig. 3A and 3B. As it is expected, a strong overlap of signals is evidenced in the IR spectra. However, seven signals can be recognized and easily correlated with their respective FEDS spectra. For C. albicans spectrum, the observed signals were c1 (3086 cm−1), c2 (2930 cm−1), c3 (2858 cm−1), c4 (2652 cm−1), c5 (2577 cm−1), c6 (2555 cm−1), and c7 (2445 cm−1); whereas for H. pylori spectrum the observed signals were h1 (3084 cm−1), h2 (2934 cm−1), h3 (2868 cm−1), h4 (2649 cm−1), h5 (2585 cm−1), h6 (2563 cm−1), and h7 (2450 cm−1). In general, with exception of c1 and h1 significant differences in the wavenumbers were observed. Spectral assignation of signals is summarized in Table 1.
Fig. 3.
Normalized FEDS spectra of C. albicans and H. pylori biofilms between 3100 and 2450 cm−1 overlapping with their respective normalized IR spectra, A and B, respectively. Comparison of FEDS spectra is shown in C. Signals c, h, k and d denote Candida, Helicobacter, ‘no-shift signal’ and ‘characteristic signal’, respectively.
Table 1.
Summarize of signals identified in infrared and FEDS spectra of C. albicans and H. pylori.
|
C. albicans |
H. pylori |
Assignation | ||
|---|---|---|---|---|
| Signal | v (cm−1) | Signal | v (cm−1) | |
| c1 | 3086 | h1 | 3084 | Amide B (Fermi resonance between the first overtone of Amide II and N-H stretching) |
| c2 | 2930 | h2 | 2934 | Asymmetric stretching of C-H in CH2 and CH3 |
| c3 | 2858 | h3 | 2868 | Symmetric stretching of C-H in CH2 and CH3 |
| c4 | 2652 | h4 | 2649 | NH stretching vibrations of amino acids |
| c5 | 2577 | h5 | 2585 | SH stretching vibrations |
| c6 | 2555 | h6 | 2563 | SH stretching vibrations forming hydrogen bonds (S-H⋅⋅⋅NH2) |
| c7 | 2445 | h7 | 2450 | It could be associated to P-OH bonds as result of stress or some metabolic process |
Signals c1 and h1 were associated to Amide B vibration which is the result of Fermi resonance between the first overtone of Amide II and the N-H stretching vibration; this signal is expected to be identified in all MOs since all contain some type of peptide on their surface [41], [42], [43]. Signals c2 and h2 are associated with asymmetric stretches, whereas c3 and h3 are associated to symmetric stretches from C-H in CH2 and CH3 on the side chains of amino acids such as alanine, aspartic acid, or glycine [44], [45], [46], fatty acids [47] and metabolites like to N-Acetyl-D-glucosamine (GlcNAc) which is a structural fragment constituting glycosylated proteins and chitin of C. albicans, but also, lipopolysaccharides in form of phosphorylated glucosamine of H. pylori [48]. In consequence, in term of nature of structural units, c2, c3, h2 and h3 do not provide important information for the differentiation of H. pylori and C. albicans biofilms formed from individually cultures nor from mixture of them, however, it is small shifts of signals could be used for the recognition of MOs on the surface.
On the other hand, by Density Functional Theory, or DFT, have been predicted for cysteine five signals: three signals associated with SH stretching vibrations (2543, 2545 and 2552 cm−1) and two signals associated to NH stretching vibrations at 2639 cm−1 and 2960 cm−1 [49], [50]. Nevertheless, signal at 2960 cm−1 was not identified by the strong overlap; c4 and h4 were associated to NH stretching, and c5, c6, h5 and h6 were associated to SH stretching. Due to formation of hydrogen bonds these signals can appear between 2600 and 2550 cm−1 [51]. Finally, c7 and h7 are not completely explained. However, it is suggested that could be associated with P-H bonds appearing in the range 2410–2442 cm−1 [52]. Some authors have described vibrations of P-OH around 2400 cm−1 [53]. The above can be explained by the surface structures like polyphosphates, phospholipomannan of C. albicans, Lipid A 1-Phosphatase of H. pylori among others [54], [55], [56], [57].
On the other hand, several type of signals can be identified by comparison of FEDS spectra of C. albicans and H. pylori with their corresponding IR spectra. The first set of signals is inherited from IR spectra; these are easily identified overlapping FEDS and IR spectra. An advantage of the FEDS analysis is that, although the FEDS spectra are more complex in terms of the number of signals compared with IR spectra, the main signals of the IR spectra are contained in their FEDS spectra, and therefore, their meaning and interpretation are maintained (see Fig. 3C).
A second group of signals, named as k1-k5, are situated at exactly the same wavelengths in both spectra. By these signals was possible to corroborate that there is no systematic error in the transformation of the spectra and, consequently, the differences in the wavelengths between equivalent signals allowed to identify the occurrence of shifts of C. albicans and H. pylori respect to the spectrum of (Ca + Hp)BF. According to their origin, two types of signals are obtained by FEDS deconvolution: (i) signals associated with relative maxima, or primary signals, which are signals obtained from the IR spectrum and can be directly correlated with the different vibrational modes of the sample components, and (ii) signals associated with relative minima, or overlap signals, which do not have a direct relationship with vibrational modes of bonds or structural groups, nevertheless, they do have a direct relationship with the position of two adjacent primary signals and with the degree of overlap between them; therefore, the combination of the primary signals and the overlapping signals allows to define a more specific signal pattern and easier to compare due to the sharp shape of them. It is important to note that overlap signals are not always visible, since for two strongly-overlap adjacent signals, the signal is hidden below a critical separation distance; in consequence, for correct separation of signals is initially necessary the identification of primary signals from respective IR spectra [36]. An important observation from FEDS spectra of C. albicans and H. pylori is the enormous similarity in the spectral range analyzed. Nevertheless, the greatest differentiation of them is observed in two ranges of data: 3000 cm−1 and 2840 cm−1, and 2660 cm−1 and 2450 cm−1. Signals associated with these differences have been named as d1 (2968 cm−1 in H. pylori), d2 (2903 cm−1 in H. pylori), d3 (2875 cm−1 in C. albicans), d4 (2524 cm−1 in H. pylori), d5 (2509 cm−1 in C. albicans), and d6 (2487 cm−1 in H. pylori).
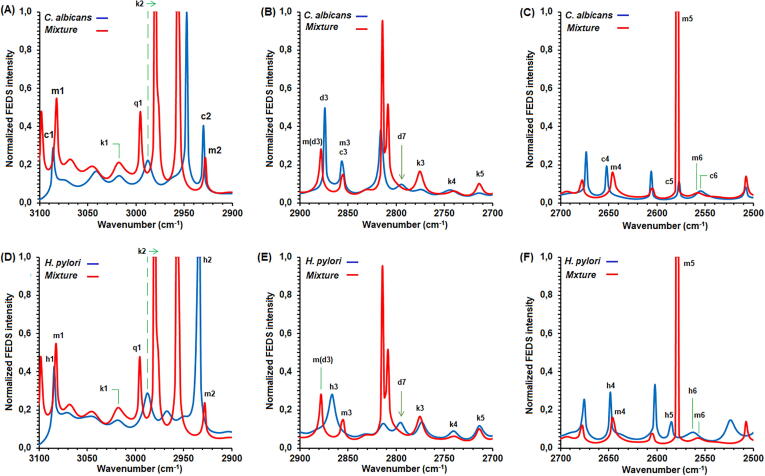
On the other hand, IR and FEDS spectra of (Ca + Hp)BF are shown in Fig. 4. It can be seen that the same primary signals identified for C. albicans and H. pylori can be assigned. Besides, IR and FEDS spectra of (Ca + Hp)BF showed the same signal patterns observed in IR and FEDS spectra for the target MOs. Comparison of the FEDS spectra of individual MOs and (Ca + Hp)BF is shown in Fig. 5. A detailed comparison shows that the (Ca + Hp)BF spectrum is different from those obtained for C. albicans (see Fig. 5A, 5B and 5C) and H. pylori (see Fig. 5D, 5E and 5F).
Fig. 4.
Normalized infrared and FEDS spectra of (Ca + Hp)BF. Signals FEDS associated with relative-minimum absorbance values showing the same patter observed for C. albicans and H. pylori spectra (q).
Fig. 5.
Normalized FEDS spectra of C. albicans and (Ca + Hp)BF (A, B and C) and H. pylori and (Ca + Hp)BF (D, E, F).
From Fig. 5A is observed that Δv between c1 and m1, and between c2 and m2 is 2 cm−1, therefore, it is concluded that there is no significant difference between signals. The same was observed between h1 and m1 but not between h2 and m2 (Δv = 7 cm−1) (Fig. 5D). Since c1 and c2 are associated with C—H bonds of —CH2— and —CH3, which do not form hydrogen bonds due to the relatively low electronegativity of the carbon atom compared with N or O atoms, it is expected that any perturbation of these signals is related with hydrophobic interactions. However, while k1 does not change, k2 is shifted to lower wavenumbers, from 2988 cm−1 to 2980 cm−1 (Δv = 8 cm−1), resulting in strongly overlapping adjacent signals (q1). By this line of analysis, the presence of k2 in the spectra of C. albicans and H. pylori suggests that q1 is important to understand the molecular mechanisms promoting the co-aggregation stage for the formation of the (Ca + Hp)BF. One drawback is that this signal is not normally described in IR studies of complex systems, being the usual practice the grouping of a set of signals, all associated to C—H stretches, in a wavenumber range. However, it is proposed that the C—H bonds associated with k2 are not part of aliphatic fatty acid chains characteristic of biological membrane systems but they should be associated with polar structures from proteins or peptides. Therefore, bonds associated with O—C—H and N—C—H are candidates for this hypothesis because, in addition to amino acids forming proteins, glycosylphosphatidylinositol proteins (Gpi-P) of C. albicans contains units of terminal ethanolamine phosphate (EtAm-P) linked to sugars [58], [59]. The above is consistent with the composition of cell wall of C. albicans, that is constituted by threonine (18.1%), serine (13.3%), glutamic acid (10.4%) and proline (10.1%), i.e., 51.9% of polar amino acids [60]. Nevertheless, it is important to note that approximately 33% of the cell wall of C. albicans is made up of non-polar amino acids: leucine (4.2%), proline (10.1%), isoleucine (6.1%), glycine (6.7%), valine (5.7%), and alanine (~0.5%) [60], but also, it is consistent with the affinity of C. albicans to effectively adsorb non-polar aliphatic amino acids (alanine, leucine, and proline) [61]. In consequence, the k2 shift suggests that a potential route for the co-aggregation of C. albicans and H. pylori for the formation of dual-species biofilm is through hydrophobic interactions between non-polar peptide chains. Similar results are observed in the comparison between the H. pylori biofilm and (Ca + Hp)BF (see Fig. 5D). Furthermore, the signal associated with C-H vibrations denoted by h2 is shifted from 2934 cm−1 to 2927 cm−1, reaffirming the hypothesis of co-aggregation mediated by hydrophobic interactions. However, it is suggested that these interactions should be through peptide segments of outer non-polar proteins since hydrophilic surface is expected for H. pylori from structural analysis of lipopolysaccharides (LPSs). It is well-known that LPSs are the main component of the bacterial cell wall of gram-negative bacteria, but also, the presence of multiple outer membrane proteins has been described for H. pylori [62]. Evidence experimental demonstrating an intimate contact at level surface between C. albicans and H. pylori is obtained from contact angle data, TEM and SEM. These results are shown and discussed in later sections.
The previous analysis agrees with observations from Fig. 5B and 5C. Since the C-H bonds show symmetric and antisymmetric vibrations, if the previous approach is correct, it is expected that the c3 signal in the C. albicans spectrum does not show a significant difference with the m3 signal of the (Ca + Hp)BF, and at the same time, the h3 signal should differ significantly of m3. From the spectra FEDS, a signal denoted d3 is identified to the left of c3, which for (Ca + Hp)BF was denoted as m(d3) to indicate that it is the same signal d3 but identified in the spectrum obtained from microorganism mixture. This signal corresponds to a relative minimum in the IR spectra, though their identification is easy from the IR spectra of yeast and (Ca + Hp)BF, from spectrum of H. pylori is not evident. The small displacement of d3 to m(d3), Δv = 4.3 cm−1, suggests that some type of disturbance occurred and produced the modification of IR spectrum, or if preferred, the function of line that describes the spectrum was modified for the perturbation of surface C. albicans. This disturbance can be explained by the presence of the bacteria, since the signal associated with the respective vibration, h3, is observed at 2867 cm−1 (Δv = 9 cm−1). Thus, the superposition of the contributions in the spectrum of (Ca + Hp)BF produces a small displacement of the signal due to the widening of m3 in its respective IR spectrum. A result of great importance is the disappearance of h3, which reaffirms the hypothesis non-polar peptide-mediated hydrophobic interactions. Likewise, signals ranged between 2820 cm−1 and 2795 cm−1 are associated with N-CH2 in aliphatic secondary and tertiary amines which are typical of peptide and proteins on the surface of MOs [31]. In particular, for C. albicans spectra, units of EtAm-P should be shown this signal, however, the fact that signal is present in H. pylori spectra suggests that it is related strongly with amino acids like Arginine, Lysine, Asparginine or Glutamine. In consequence, the disappearing of signal at 2795 cm−1 (d7), both for C. albicans and H. pylori spectra, is interpreted to be an evidence of a second type of interaction mediated by polar domains associated with C—N—H bonds, besides multiples hydrogen bonds resulting of polar domains on surface of microorganisms.
In the Fig. 5C and 5F are shown the FEDS spectra of C. albicans, H. pylori and (Ca + Hp)BF between 2700 cm−1 and 2500 cm−1. This spectral region is important because permits explore the possibility of covalent interactions between C. albicans and H. pylori via formation of disulfide bonds. It can be seen that signal c4 which is associated with N—H vibrations is displaced to lower wavenumbers (Δv = 5 cm−1). In addition, Δv = 2 cm−1 when h4 is compared with m4. This suggests the possibility of peptide-mediated interaction between C. albicans and H. pylori. Though this signal can be assumed to be a direct contribution of H. pylori, it should be noted that whereas spectra of C. albicans and (Ca + Hp)BF show the same signals for wavenumber lower than 2610 cm−1, spectra of H. pylori and (Ca + Hp)BF are very different with multiple displacements. In consequence, a third interaction is possible by the hydrogen bonds (e.g., —S—H⋅⋅⋅NH2— or —S—H⋅⋅⋅O CO—) and disulfide bonds, which can be formed spontaneously under alkaline conditions (—SH + —SH → —S—S—). The previous interaction mechanisms are possible due to the presence of cysteine-rich outer membrane proteins and extracellular urease on the surface of H. pylori [22], [63] and the presence of cysteine on the surface of C. albicans (2.7%) [62].
From IR analysis is shown that at least three possible interactions routes are possible between H. pylori and C. albicans to form dual-species biofilm: (i) hydrophobic interaction between peptide components from cell wall of C. albicans and H. pylori (mechanism 1); (ii) interaction by hydrogen bonds between polar amino acids from cell wall of C. albicans and outer-membrane proteins of H. pylori, and between polar domains associated with Gpi-P and LPSs (mechanism 2), and (iii) covalent interaction between thiol groups on the surface of MOs (mechanism 3).
In relation with the mechanism 1, it has been reported that the union of alkyl groups to polar groups, e.g., carbonyl, is affected in their vibratory movements as a function of the length of the alkyl chain [64]. In this way, for shorter chain length is greater the effect of polar groups, and conversely, as the chain length increases, the vibratory movements become independent of the influence of polar groups. Therefore, vibration wavelengths will be different for C-H bonds from CH2 or CH3 groups depending if they are part of amino acids or EtAm-P; for fatty acids, the effect is lower, and consequently, through the spectroscopic analysis carried out, evidence of disturbance at the level of the lipid bilayer was not evidenced. Likewise, for amino acids, shifts of wavenumbers would be expected since they have non-polar short chains. Similarly, it has been reported that the displacements of the signals associated with CH bonds are related to molecular packaging [64], [65]; thus, a decrease in packing will be associated with greater freedom of movement and consequently greater vibrational energy (displacement of signals at higher wavenumbers) [65]. The above is important because a weakening of the yeast cell envelope of C. albicans, due to the presence of H. pylori, could manifest itself in a shift to larger wavenumbers; likewise, a restriction of freedom of movement would be associated with shifts to lower wavenumbers. However, an effect of this nature between MOs would only be appreciable if it occurs in a significant proportion of the sample, because the IR technique obtains information and builds the spectrum from the population of MOs and not from a single cell. Since the technique allows the monitoring of MOs without practically any disturbance, beyond the drying of the sample, these can have secreted substances for the conditioning of their immediate environment and, consequently, metabolic processes could mask any changes in terms of cell integrity when the concentration of extracellularly-secreted substance is significant. The biofilm matrix of C. albicans has been described to be composed of carbohydrates, such as β-glucans and mannan, as well as proteins, lipids, and extracellular DNA, and hexosamines, uronic acid, and phosphorus [66]. But also, protein–protein interaction between C. albicans surface and foreign proteins have been described to occur though outer GPI proteins [58], in consequence, small displacements of IR signals, associated with CH2 or CH3 units on biofilm surface, allow to establish that exists a contribution of non-polar amino acids in the interaction between C. albicans and H. pylori.
In relation with mechanism 2 previously indicated, due to numerous polar groups on the surface associated with LPSs, outer membrane proteins and Gpi-P, formation of hydrogen bonds is the most evident interaction from a molecular point of view. In particular, LPSs of H. pylori are composed of three domains: a hydrophobic domain termed lipid A (or endotoxin), non-repeating core oligosaccharide formed by short chain of sugars, which connects the lipid A anchor to O-antigen and can be divided into inner and outer core regions, and a variable outermost polysaccharide (or O-antigen) constituting hydrophilic domain of the LPSs [48]. Composition of monosaccharides is very variate, however, different H. pylori strains contain xylose (Xyl), mannose (Man), galactose (Gal), glucose (Glc), heptose (Hep), inositol, and N-Acetylglucosamine (GlcNAc) [48]. All component contains hydrogen atoms linked to highly electronegative oxygen, and therefore, multiples hydrogen bonds can be formed. In addition, it has been described that outer membrane of H. pylori is composed of LPSs contributing with the colonization and persistence in the stomach, in addition, it is recognized that LPSs mediate several interactions between the bacterium and its surrounding environment [48], [67]. On the other hand, the cell wall of C. albicans is a bilayer structure with an external protein coat consisting mainly of Gpi-Ps; which are often strongly mannosylated and phosphorylated, and are covalently linked to highly-branched β-1,6-glucan molecules which are linked to internal network of β-1,3-glucan molecules [58], [59], [66]. In addition, polypeptide content of the wall is estimated at 2.4–3.3% [58].
In relation with mechanism 3, covalent interaction between thiol groups, the formation of disulfide links and hydrogen bonds mediated by thiols (-SH) requires the presence of thiolated amino acid chains. It has been suggested that abundant cell wall proteins are interconnected by disulfide bridges, e.g., protein Pga59 and Pga62 which are GPI cell wall protein with cysteine residues, in addition, Pga59 and Pga62 are expressed to much higher levels in biofilms than in planktonic cultures [59] and they are required for cell wall integrity [58]. The above is consistent with the observations about effect of N-acethylcysteine on C. albicans biofilms [68], [69]. In general, anti-biofilm activity of N-acethylcysteine is based on the breaking of disulfide [68], [69]. But in term of amino acid composition, it has been determined that the content of thiolated amino acids, i.e., cysteine and tyrosine, forming the wall cell are 2.7 and 3.4%, respectively [60]. In consequence, formation of specific disulfide bonds is low probably though possible. However, results suggest that surface interactions by the interpenetration of outer cell structures implies interactions of hydrogen on S-H with a rich surface in polar atoms. Finally, evidence of electrostatic interaction was not observed because analyzed range not offers information about charged groups.
On the other hand, IR and FEDS analysis do not allow to establish with total certainty the occurrence of these interactions. One of the main limitations of FEDS is its relatively recent development and application to the analysis of complex biological systems, and therefore, like other deconvolution techniques, additional information is required in order to achieve an adequate interpretation, however, usually the information is not available or has been established through different conditions, being this a second current limitation. In addition, systems studied in this research are characterized to be dynamic, with high-variability in function of growth stage or adaptation; in consequence, differences between planktonic cells and biofilms are expected. For example, whereas for C. albicans in planktonic state is characterized by Gpi-Ps and phosphodiester bridges in N-linked carbohydrate side-chains of cell-wall proteins, the biofilm matrix of C. albicans is composed of carbohydrates, such as β-glucans and mannan, as well as proteins, lipids, and extracellular DNA, and hexosamines, uronic acid, and phosphorus [66].
The fact that the FEDS analysis is carried out with practically zero disturbance of the sample, it suggests that is a promissory alternative for the in situ study of MO and biofilms. Therefore, by IR and FEDS can be concluded that the samples described as (Ca + Hp)BF correspond to the dual-species biofilm formed by C. albicans and H. pylori. But also, it is evidenced that in the formation of dual-species biofilm between C. albicans and H. pylori, the yeast cells mostly constitute the biofilm because biofilm of C. albicans has a higher similarity with (Ca-Hp)BF, besides, in concordantly with microscopy observations, C. albicans acts as surface of anchoring of H. pylori. In order to contrast the results obtained in this first part, different experiments are analyzed and discussed in the next sections.
Study by microscopy: SEM, TEM and optic microscopy
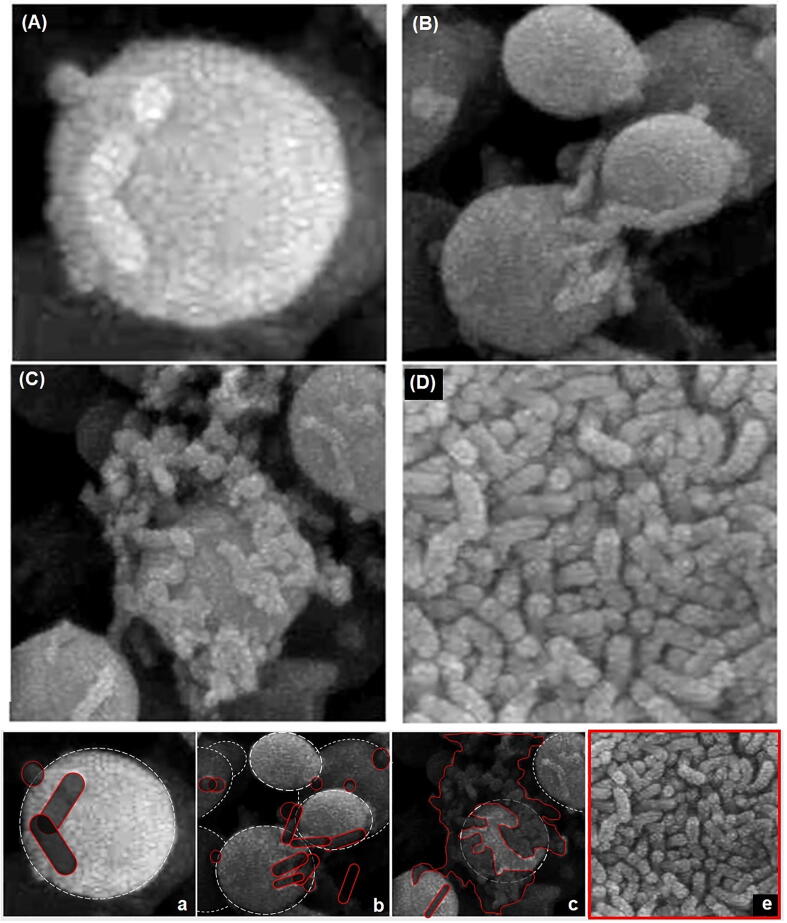
Three different microscopy studies were performed: SEM, TEM and optic microscopy. SEM images of C. albicans, H. pylori and (Ca + Hp)BF are shown in Fig. 6. In images 6A, 6B and 6C are shown the C. albicans cells with a quasi-spherical shape and a smooth surface, whereas in the images 6D, 6E and 6F are shown the H. pylori cells with bacillar morphology. From images different size descriptors were calculated (see Table 2). An interesting observation is shown in Fig. 7 corresponding to a detailed analysis of image shown in Fig. 6G. For the formation of H. pylori biofilm in the mixture, different stages can be identified: (i) anchoring of H. pylori on the surface of C. albicans, (ii) co-aggregation of cells forming clusters, (iii) growth of H. pylori biofilm, and (iv) total colonization of surface, situation which is achieved since H. pylori cells are the single identified. By size analysis in Fig. 7a, it is obtained that σ2 is 3.6, 12.8 and 43.2 for ESR, ECA and ESV, respectively. Two important aspects are concluded by SEM: (i) it is verified the formation of biofilms of H. pylori in unfavorable conditions and in presence of C. albicans, and (ii) it was possible to identify the anchoring of bacterium cell on the surface of the yeast.
Fig. 6.
SEM images of C. albicans (A, B and C), H. pylori (D, E and F) and (Ca + Hp)BF (G, H and I).
Table 2.
Size descriptors for C. albicans and H. pylori from individual cultures and mixtures.
| Culture | Size descriptor | ESR (µm) | ECA (µm2) | ESV (µm3) |
|---|---|---|---|---|
| C. albicans | Mean cell size (SCa) | 0.31 ± 0.03 | 0.30 ± 0.05 | 0.13 ± 0.03 |
| H. pylori | Mean cell size (SCa) | 0.50 ± 0.03 | 0.79 ± 0.09 | 0.53 ± 0.09 |
| σ1 = SCa/SHp | 0.62 | 0.38 | 0.24 | |
| (Ca + Hp)BF | Mean cell size (SCa) | 1.59 ± 0.39 | 8.38 ± 3.69 | 19.39 ± 11.82 |
| Mean cell size (SHp) | 0.60 ± 0.10 | 1.16 ± 0.38 | 0.97 ± 0.47 | |
| σ2 = SCa/SHp | 2.7 | 7.2 | 20.0 | |
Fig. 7.
SEM images from Fig. 5G illustrating different stage of H. pylori biofilm formation: (A) anchoring on the surface of C. albicans, (B) co-aggregation of cells forming clusters, (C) growth of H. pylori biofilm and (E) total colonization of surface. Figures a-e highlights the particles in the image to ease the comparison.
It can be seen that in its bacillar morphology the bacterium is a very large entity to achieve an adequate internalization without causing a significant damage to the yeast wall. However, these results should be taken with caution and at least three aspects must be considered: (i) the size obtained by SEM is a geometric projection in two dimensions of a three-dimensional body; (ii) hypothesis of H. pylori internalization in C. albicans, without a comprehensive explaining about internalization mechanism for this fact could be associated with a different morphology, i.e., coccoid [7], [24], but also, extracellular DNA or outer membrane vesicle could be masking the results of Fluorescence Microscopy. Extracellular DNA has been identified as part of biofilm matrix of H. pylori and outer membrane vesicle with 20–500 nm in diameter containing phospholipids, proteins, LPSs, and DNA; these outer membrane vesicles are produced from both biofilm and planktonic systems [29]; and (iii) the bacteria and yeast can be found in different growth stages with which the difference in size becomes more significant compared with size data obtained from individual cultures (see Fig. 6H). In Table 2 it can be seen that, for the system (Ca + Hp)BF, the size difference varies significantly with respect to the first estimate. These differences are explained due to the differences in the growth conditions since, for the mixture of MOs, the incubation conditions are favorable for the yeast and unfavorable for the bacteria. It can be observed that, consistently with the SEM images, it is possible, in term of size, the internalization of bacteria; however, information or clues about how such a process would occur were not identified.
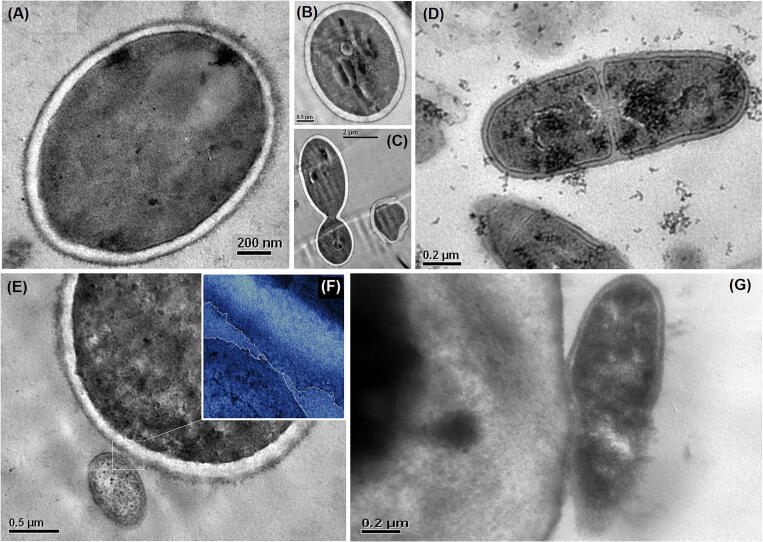
TEM images are shown in Fig. 8, from individual cultures, of C. albicans (Fig. 8A, B and C) and H. pylori (Fig. 8D); TEM images of (Ca + Hp)BF are shown in Fig. 8E, F and G. By TEM, morphological aspects were calculated in order to try elucidating the possible mechanisms of interaction between H. pylori and C. albicans. In particular, as it was explored the viability of internalization of H. pylori in the yeast, which is a hypothesis proposed from experiments of fluorescence microscopy where bacterium-like objects have been described in the inner of yeast vacuoles [7], [24]. Previously by SEM was evidenced that is physically possible the internalization in term of size (see value of σ2 in Table 2 and Fig. 7). However, it is well- known that the cell wall is the first physical barrier that bacteria must cross to achieve the inner yeast. Cell wall of C. albicans represents a challenge for the bacteria due to its enormous relative size constituting 16% of the yeast cell volume and which was calculated from TEM images to be 16.04 ± 9.27 µm3 (see Fig. 8). From TEM analysis was determined that thickness of cell wall of C. albicans is 0.20 ± 0.02 µm (~200 nm); whereas the size H. pylori, in term of ESV, is 0.97 ± 0.47 µm3 with a cell wall thickness of 34.1 ± 4.4 nm, in consequence, the cell wall of C. albicans is ~7 times larger than cell wall of H. pylori (see Fig. 8E and G). In addition, no significant change in the thickness of the wall cell of C. albicans was evidenced during cellular division (see Fig. 8C and Table 3).
Fig. 8.
TEM images of C. albicans (A, B and C), H. pylori (D) from individual cultures and from mixtures (E, F and G).
Table 3.
Cell dimensions for C. albicans and H. pylori determined by TEM (MCWT = Mean cell wall thickness).
| Microorganism | Parameter evaluated | Size descriptor |
||
|---|---|---|---|---|
| ESR (µm) | ECA (µm2) | ESV (µm3) | ||
| C. albicans | Mean cell size (SCa) | 1.55 ± 0.23 | 7.61 ± 2.21 | 16.04 ± 6.88 |
| MCWT | 0.20 ± 0.02 | – | – | |
| MCWT during cell division | 0.25 ± 0.06 | – | – | |
| Mean inner volume | – | – | 13.54 ± 6.88 | |
| Mean cell wall volume | – | – | 2.5 ± 2.39 | |
| H. pylori | Mean cell size (SCa) | 0.42 ± 0.38 | 0.55 ± 0.45 | 0.31 ± 6.88 |
| MCWT | 0.03 ± 0.01 | – | – | |
| Mean inner volume | – | – | 0.25 ± 0.13 | |
| Mean cell wall volume | – | – | 0.06 ± 0.01 | |
On the other hand, from the TEM images, two important aspects related to the formation of dual-species biofilm between C. albicans and H. pylori were verified: (i) The anchoring of H. pylori on the surface of C. albicans takes place by direct contact interfacial suggesting a control of this first stage at surface level (see Fig. 8F), inconsequence, the multiple interactions could promote this stage; and (ii) after surface contact, H. pylori adheres to the surface (see Fig. 8G). These two observations imply that some anchoring mechanism should be occurring in conjunction with a high affinity between the surfaces in contact; however, the surface affinity could be a result of the molecular nature of the cell interface or it may be due to surface conditioning subsequent to the attachment of H. pylori. Nevertheless, both SEM and TEM are fix observation of system latter to pre-treatment of samples, consequently, information about how the system reaches the observed state must be obtained through other types of techniques.
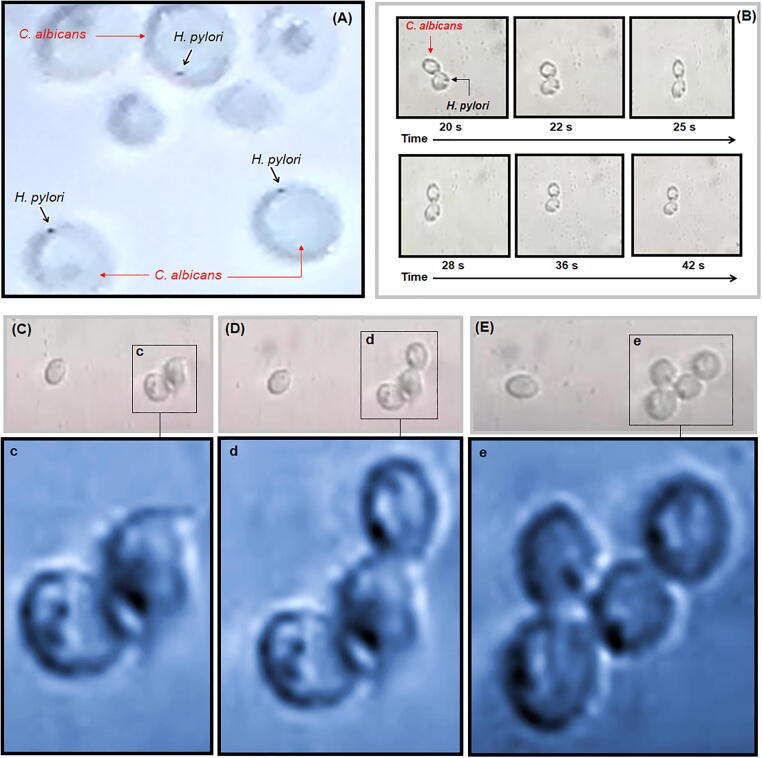
By optic microscopy, cultures corresponding to (Ca + Hp)BF were directly observed. Water flux was applied in order to evaluate the strong of anchoring of H. pylori on C. albicans. However, dispersion of cell was not possible. In Fig. 9A can be seen the cell-like objects associated with H. pylori anchored on C. albicans cells. In Fig. 9B, it is shown the co-aggregation over time of C. albicans and H. pylori. Changes of spatial orientation evidences the fix positions of H. pylori and its anchoring on the surface. However, recording in video format for longer times permitted to obtain evidence of simultaneous co-aggregation of cells (Fig. 9C-9E). Thus, it is concluded that anchoring of H. pylori occurs in aqueous water previous to formation of biofilms by C. albicans, in consequence, co-aggregation of C. albicans can be defined as a mixed process of cellular aggregation that would lead to the formation of dual-species biofilm completely integrated by the two MOs instead of a process direct to the formation of yeast biofilm acting as a surface or substrate for the formation of a second biofilm.
Fig. 9.
Image of optic microscopy of (Ca + Hp)BF (A), time-sequence of anchoring of C. albicans and H. pylori (B), time-sequence of co-aggregation of C. albicans and H. pylori (C, D, and E). Magnification of images is shown in c, d and e, respectively.
Study by microscopy DLS
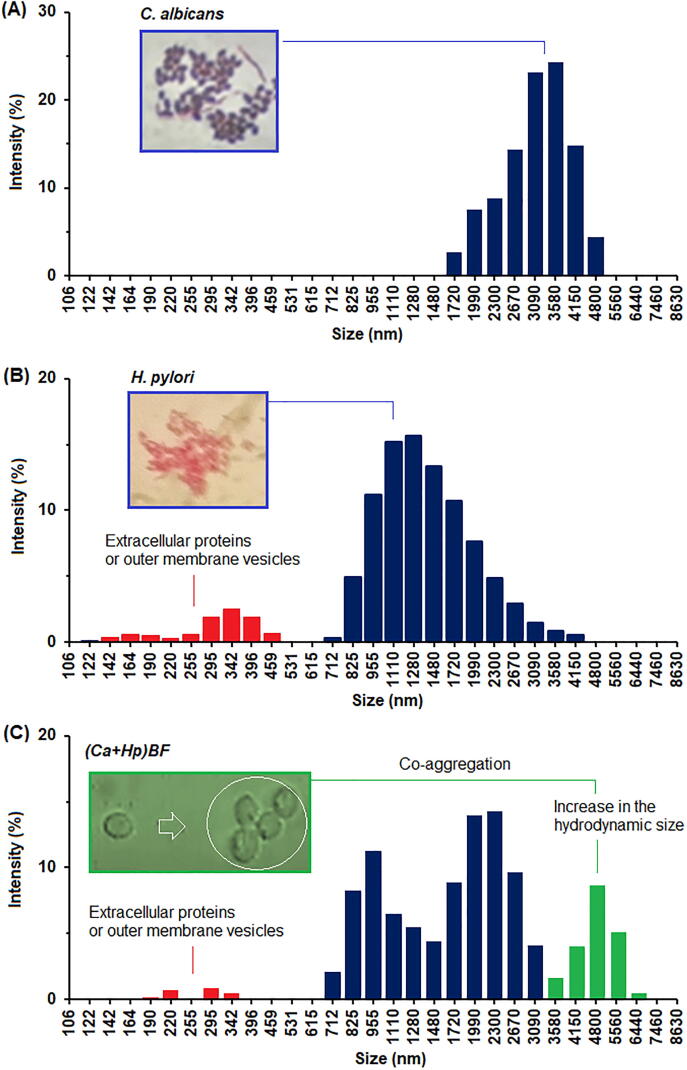
Size and size distribution by DLS are shown in Fig. 10. It can be seen that alike to results obtained by SEM and TEM, the mean size of C. albicans is higher than mean size of H. pylori. By DLS, size changes from 1.7 to 4.8 µm for C. albicans and from 0.71 to 4.1 µm for H. pylori were obtained. For H. pylori, substances with lower size are observed. These showed a size from 122 nm to 459 nm and were associated with extracellular proteins, extracellular DNA, and outer membrane vesicles [29].
Fig. 10.
Size and size distribution by DLS of C. albicans (A), H. pylori (B) and (Ca + Hp)BF (C).
For (Ca + Hp)BF the size distribution was characterized to be multimodal. Several aspects must be highlighted: (i) displacement of distribution to higher size is observed, which can be explained by aggregation of cells, (ii) planktonic cells of C. albicans (from 0.71 to 1.48 µm) and H. pylori (from 1.48 to 3.1 µm) apparently are presents and (iii) amount of extracellular lipoproteins, extracellular DNA and outer membrane vesicles were strongly decreased. Two situations could explain the above observation, the first is that concentration of extracellular substances decreased in the mixture due to reduction of some metabolic processes, and the second, that decrease of extracellular substances is associated with the interaction between microorganisms, e.g., absorption by C. albicans of extracellular substances. It has been demonstrated that ATCC43504 biofilm contain proteomannans consisting of β-1,3- and β-1,6-glucan structures linked to proteoglycans [29], since these are analogous substances to that forming the inner layer of the C. albicans cell envelope is suggested that high degree of affinity between these substances and C. albicans cells is expected [29], [70].
Study of hydrophilic-lipophilic properties of the surface
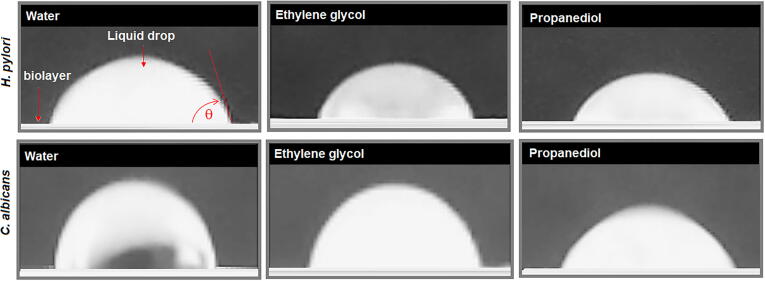
Measurements of contact angle for C. albicans and H. pylori biofilms are shown in Table 4. Illustration of sessile drop method for the determination of contact angle for each working liquid is shown in the Fig. 11. It can be seen from water contact angle that biofilm surface can be described in both cases to be with low hydrophilic surfaces (45 < θ < 90) [71]. However, definition of hydrophobic or hydrophilic surface is not a physical definition, therefore, a better interpretation of contact angle is obtained by comparison with known surfaces (see Table 5). A comparison with non-polar polymeric surfaces permits conclude that in term of wettability with water both C. albicans and H. pylori surfaces are alike to poly(ethylene terephthalate) (PET) and poly(styrene) (PS), which are surface characterized to show a very low wettability. In consequence, results suggest that C. albicans and H. pylori surfaces have a high hydrophobic character. The above is even an aspect that differentiates H. pylori from other gram-negative bacteria such as E. coli, and C. albicans from other yeasts of the same genus such as C. tropicalis. But at the same time, the surface affinity between C. albicans and H. pylori towards hydrophobic surfaces, and between them, is higher. These results are consistent with description of lipid profiles of H. pylori, from which has been evidenced a high relative content of lysophospholipids which have been associated with a better adherence to epithelial cells in vitro [72], but also, with the fact that H. pylori strains produce several characteristic lipids, e.g., steryl glycosides and cholesteryl glucosides [73]. For the case of C. albicans, its surface hydrophobicity is well-known and has been associated to the promotion of adhesion process on surfaces; however, for C. albicans, the presence of negative charges on its surface also have been documented [61], [74]. In addition, these results are consistent with those obtained from IR spectroscopy and FEDS. It can be observed that, while in E. coli, C. tropicalis and H. pylori the contribution of the dispersive and polar components is very similar, in C. albicans the dispersive contribution is much more significant. In comparison with polymeric surface, dispersive component is similar to observed for polypropylene (PP), PS and poly(vinyl alcohol), which are associated with London dispersion and hydrogen bonds.
Table 4.
Measurements of contact angle (θ) of biolayer of C. albicans and H. pylori using three solvents with different polarities. Coefficient of variation appears in parenthesis followed of measured of contact angle, ε is dielectric constant of solvent at 25 °C [76], [77].
| Water | Ethylene glycol | 1,3-Propanediol | |
|---|---|---|---|
| Microorganism | (ε = 78.3) | (ε = 41.2) | (ε = 30.2) |
| C. albicans | 84.9 ± 1.6 (1.9%) | 77.5 ± 3.6 (4.6%) | 74.0 ± 4.3 (5.8%) |
| H. pylori | 76.6 ± 3.8 (4.9%) | 79.7 ± 3.9 (4.9%) | 77.2 ± 2.7 (3.5%) |
Fig. 11.
Contact angle measurements for H. pylori and C. albicans using water, ethylene glycol and 1,3-propanediol.
Table 5.
Total surface free energy (γtotal) and their components according to van Oss-Chaudhury-Good theory: dispersive component (γLW), acid-base component (γAB), acidic component (γs−), basic component (γs+). All values are in mJ/m2. Percentages of contribution of γLW and γAB to γtotal are shown in parenthesis. Contact angle with water (θw).
| θw | ||||||
|---|---|---|---|---|---|---|
| C. albicans | 84.9 ± 1.6 | 22.78 | 21.72 (95.3%) | 1.06 (4.7%) | 13.62 | 0.02 |
| H. pylori | 76.6 ± 3.8 | 17.34 | 6.97 (40.2%) | 10.37 (59.8%) | 30.25 | 0.89 |
| PVA | 51 | 42 | 42.0 (100%) | 0.0 (0.0%) | – | – |
| PVAc | 60.6 | 37.7 ± 2.9 | 24.8 ± 1.6 (65.7%) | 13.0 ± 1.5 (34.3%) | – | – |
| PEG | 63 | 43.9 ± 1.9 | 37.7 ± 8.1 (86.0%) | 6.9 ± 6.4 (14.0%) | – | – |
| PET | 69.8 ± 14 | 43.2 ± 2.5 | 37.6 ± 5.8 (86.8%) | 5.0 ± 4.2 (13.2%) | – | – |
| PS | 89.0 ± 10 | 41.8 ± 1.2 | 37.6 ± 3.8 (90.0%) | 4.2 ± 3.3 (10.0%) | – | – |
| PP | 101.5 ± 9 | 31.0 ± 3.9 | 28.9 ± 3.7 (93.2%) | 2.1 ± 2.5 (6.8%) | – | – |
| S. aureus | – | 43.9 ± 0.5 | 39.6 ± 0.4 (90.2%) | 4.3 ± 0.5 (9.8%) | 73.5 | 0.07 |
| P. aeruginosa | – | 39.3 ± 0.8 | 34.8 ± 0.5 (88.5%) | 4.4 ± 0.8 (11.5%) | 69.1 | 0.09 |
| E. coli | 19.1 ± 0.9 | 47.9 | 25.7 (53.7%) | 22.2 (46.3%) | 123.2 | 0.0 |
| C. tropicalis | 48.4 ± 0.7 | 40.4 | 20.5 ± 0.2 (50.8%) | 19.8 (49.2%) | 39.5 | 2.5 |
These results evidence that co-aggregation surface between H. pylori and C. albicans in aqueous environments is mainly promoted by dispersive forces, though a small electrostatic contribution could be present, which is expected to be small compared with S. aureus, P. aeruginosa, E. coli and C. tropicalis (see Table 5). The above is consistent with electrostatic adsorption of arginine by C. albicans [61], and the small amount of arginine and lysine in H. pylori biofilm (3.5% and 7.9%, respectively) [75].
Conclusions
Results evidence that hydrophobic or van der Waals interactions between non-polar peptide chains of cell wall of C. albicans and lipid substance or non-polar peptide chains from membrane of H. pylori influence the co-aggregation for the formation of dual-species biofilm between these MOs. It is suggested that this interaction occurs simultaneously with the formation of hydrogen bonds, and therefore, a strong contact among cell surfaces should be observed. After to co-aggregation stage, stronger interactions could take place as a result of the interaction of cysteine residues present in the outer proteins of both microorganisms. In consequence, anchoring of H. pylori is promoted previous to the formation of growth and consolidation of biofilm. Our results show that biofilm formed corresponds to dual-species biofilm, which though it is similar to biofilm of individual microorganisms by IR spectroscopy and FEDS was demonstrated that (Ca + Hp)BF corresponds to different system. It is concluded that the formation of C. albicans and H. pylori dual-species biofilm occurs through the combination of different mechanisms at the surface level: hydrophobic interactions between non-polar amino acids chains and lipid structures, formation of hydrogen bonds and covalent anchoring through the formation of disulfide bonds. No significant effect of electrostatic interaction or internalization were evidenced from our experiments. In addition, results suggest that the strength interaction could be interfering in some experimental observations associated with DNA detection.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Sixta Palencia thanks to Conicyt-Chile, Universidad de Concepción (Chile) and Universidad del Valle (Colombia) by the funds for the performing of the project and doctoral scholarship. Authors thanks to Mindtech s.a.s. by the support into acquisition of spectral marker.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Brogden K.A., Guthmiller J.M., Taylor C.E. Human polymicrobial infections. Lancet. 2005;365:253–255. doi: 10.1016/S0140-6736(05)70155-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deveau A., Bonito G., Uehling J., Paoletti M., Becker M., et al. Bacterial-fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018;42:335–352. doi: 10.1093/femsre/fuy008. [DOI] [PubMed] [Google Scholar]
- 3.Peleg A.Y., Hogan D.A., Mylonakis E. Medically important bacterial-fungal interactions. Nature Rev. Microbial. 2010;8:340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 4.Peters B.M., Jabra-Rizk M.A., O’May G.A., Costerton J.W., Shirtliff M.E. Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev. 2012;25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krüger W., Vielreicher S., Kapitan M., Jacobsen I.D., Niemiec M.J. Fungal-bacterial interactions in health and disease. Pathogens. 2019;8:1–45. doi: 10.3390/pathogens8020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arvanitis A., Mylonakis E. Fungal–bacterial interactions and their relevance in health. Cellular Microbiol. 2015;17:1442–1446. doi: 10.1111/cmi.12493. [DOI] [PubMed] [Google Scholar]
- 7.Allison D., Willems H., Jayatilake J., Bruno V., Peters B.M., Shirtliff M. Candida-bacteria interactions: their impact on human disease. Microbiol. Spectr. 2016;4 doi: 10.1128/microbiolspec.VMBF-0030-2016. [DOI] [PubMed] [Google Scholar]
- 8.Romeo O., Crisen G. Molecular epidemiology of candida albicans and its closely related yeasts candida dubliniensis and Candida africana. J. Clin. Microbiol. 2009;47:212–214. doi: 10.1128/JCM.01540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sardi J.C.O., Scorzoni L., Bernardi T., Fusco-Almeida A.M., Mendes M.J.S. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J. Med. Microbiol. 2013;62:10–24. doi: 10.1099/jmm.0.045054-0. [DOI] [PubMed] [Google Scholar]
- 10.Pfaller M.A., Diekema D.J. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai C.C., Wang C.Y., Liu W.L., Huang Y.T., Hsueh P.R. Time to positivity of blood cultures of different Candida species causing fungaemia. J. Med. Microbiol. 2012;61:701–704. doi: 10.1099/jmm.0.038166-0. [DOI] [PubMed] [Google Scholar]
- 12.Adam B., Baillie G.S., Douglas L.J. Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J. Med. Microbiol. 2002;51 doi: 10.1099/0022-1317-51-4-344. [DOI] [PubMed] [Google Scholar]
- 13.Gaddy J.A., Tomaras A.P., Actis L.A. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 2009;77:3150–3160. doi: 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribeiro F.C., de Barros P.P., Rossoni R.D., Junqueira J.C., Jorge A.O.C. Lactobacillus rhamnosus inhibits Candida albicans virulence factors in vitro and modulates immune system in Galleria mellonella. J. Appl. Microbiol. 2017;122:201–211. doi: 10.1111/jam.13324. [DOI] [PubMed] [Google Scholar]
- 15.Mahnaz A.M., Chamanrokh P., Whitehouse C.A., Huq A. Methods for detecting the environmental coccoid form of Helicobacter pylori. Front. Publ. Health. 2015;3:147. doi: 10.3389/fpubh.2015.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cover T.L., Blaser M.J. Helicobacter pylori in health and disease. Gastroenterol. 2009;136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham D., Miftahussurur M. Helicobacter pylori urease for diagnosis of Helicobacter pylori infection: a mini review. J. Adv. Res. 2018;13:51–57. doi: 10.1016/j.jare.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nardone G., Compare D. The human gastric microbiota: is it time to rethink the pathogenesis of stomach diseases? United Eur. Gastroenterol. J. 2015;3:255–260. doi: 10.1177/2050640614566846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atherton J.C. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Ann. Rev. Pathol: Mechanis. Disease. 2006;1:63–96. doi: 10.1146/annurev.pathol.1.110304.100125. [DOI] [PubMed] [Google Scholar]
- 20.Hsu P.I., Lai K.H., Hsu P.N., Lo G.H., Yu H.C., Chen W.C., et al. Helicobacter pylori infection and the risk of gastric malignancy. Am. J. Gastroenterol. 2007;102:725–730. doi: 10.1111/j.1572-0241.2006.01109.x. [DOI] [PubMed] [Google Scholar]
- 21.K. Stave, M.E. Berrio, S.L. Palencia, Methods of diagnosis of Helicobacter pylori in children: current status and prospects 2 (2017) 24–43. 10.34294/j.jsta.17.2.12. [DOI]
- 22.Krajewska B., Ureases I. Functional, catalytic and kinetic properties: a review. J. Mol. Catal. B: Enzym. 2009;59:9–21. doi: 10.1016/j.molcatb.2009.01.003. [DOI] [Google Scholar]
- 23.Massarat S., Saniee P., Siavoshi F., Mokhtari R., Mansour-Ghanaei F., Khalili-Samani S. The effect of helicobacter pylori infection, aging, and consumption of proton pump inhibitor on fungal colonization in the stomach of dyspeptic patients. Front. Microbiol. 2016;7:801. doi: 10.3389/fmicb.2016.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siavoshi F., Saniee P. Vacuoles of Candida yeast as a specialized niche for Helicobacter pylori. World J. Gastroenterol. 2014;20:5263–5273. doi: 10.3748/wjg.v20.i18.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palencia S.L., Lagos G., Garcia A. Fungi-bacterium interactions: Helycobacter pyroli – Candida albicans. J. Sci. Technol. Appl. 2016;1:53–69. doi: 10.34294/j.jsta.16.1.5. [DOI] [Google Scholar]
- 26.Siavoshi F., Parastoo S. Candida accommodates non-culturable Helicobacter pylori in its vacuole-Koch’s postulates aren’t applicable. World J. Gastrienterol. 2018;24:310–314. doi: 10.3748/wjg.v24.i2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia A., Salas-Jara M.J., Herrera C., González C. Biofilm and Helicobacter pylori: from environment to human host. World J. Gastroenterol. 2014;20:5632–5638. doi: 10.3748/wjg.v20.i19.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yonezawa H., Osaki T., Kamiya S. Biofilm formation by Helicobacter pylori and its involvement for antibiotic resistance. Biomed. Res. Int. 2015:914791. doi: 10.1155/2015/914791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hathroubi S., Servetas S.L., Windham I., Merrell D.S., Ottemann N. Helicobacter pylori biofilm formation and its potential role in pathogenesis. Microbiol. Mol. Biol. Rev. 2018;82 doi: 10.1128/MMBR.00001-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palencia S.L., García A., Palencia M. Vibrational spectrum characterization of outer surface of Helicobacter pylori biofilms by Functionally-Enhanced Derivative Spectroscopy (FEDS) J. Chilean Chem. Soc. 2020;5015–5022 [Google Scholar]
- 31.Palencia S.L., García A., Palencia M. Mid-infrared vibrational spectrum characterization of outer surface of Candida albicans by Functionally-Enhanced Derivative Spectroscopy (FEDS) J. Appl. Spectr. 2021 (Acepted) [Google Scholar]
- 32.Moonens K., Gideonsson P., Subedi S., et al. Structural insights into polymorphic ABO glycan binding by Helicobacter pylori. Cell Host Microbe. 2017;19:55–66. doi: 10.1016/j.chom.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamway Y., Taxauer K., Neumeyer V., Fischer W., et al. Cysteine residues in helicobacter pylori Adhesin HopQ are required for CEACAM-HopQ interaction and subsequent CagA translocation. Microorganisms. 2020;8:465. doi: 10.3390/microorganisms8040465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palencia M. Functional transformation of Fourier-transform mid-infrared spectrum for improving spectral specificity by simple algorithm based on wavelet-like functions. J. Adv. Res. 2018;14:53–62. doi: 10.1016/j.jare.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Quintero A., Combatt E., Palencia M. Structural study of humin and its interaction with humic acids by Fourier-transform mid-infrared spectroscopy. J. Sci. Technol. Appl. 2018;4 doi: 10.34294/j.jsta.18.4.28. [DOI] [Google Scholar]
- 36.Otalora A., Palencia M. Application of functionally-enhanced derivative spectroscopy (FEDS) to the problem of the overlap of spectral signals in binary mixtures: Triethylamine-acetone. J. Sci. Technol. Appl. 2019;6 doi: 10.34294/j.jsta.19.6.44. [DOI] [Google Scholar]
- 37.Anaya L.A., Libreros K., Palencia V.J., Atencio V.J., Palencia M. Mid-infrared spectral characterization of fish scales: “Bocachico” (Prochilodus magdalenae) by functionally-enhanced derivative spectroscopy (FEDS) – a methodological approach. J. Sci. Technol. Appl. 2019;6 doi: 10.34294/j.jsta.19.6.39. [DOI] [Google Scholar]
- 38.Mustard A.T., Anderson T. Use of spherical and spheroidal models to calculate zooplankton biovolume from particle equivalent spherical diameter as measured by an optical plankton counter. Limnol. Oceanogr.: Met. 2005;3:183–189. [Google Scholar]
- 39.Palencia M. Surface free energy of solids by contact angle measurements. J. Sci. Technol. Appl. 2017;2:84–93. doi: 10.34294/j.jsta.17.2.17. [DOI] [Google Scholar]
- 40.Cavitt T.B., Carlisle J.G., Dodds A.R., Faulkner R.A., Garfiel T.C., et al. Thermodynamic surface analyses to inform biofilm resistance. IScience. 2020;23 doi: 10.1016/j.isci.2020.101702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arrondo J.L., Goñi F.M. Structure and dynamics of membrane proteins as studied by infrared spectroscopy. Prog. Biophys. Mol. Biol. 1992;72:367–405. doi: 10.1016/s0079-6107(99)00007-3. [DOI] [PubMed] [Google Scholar]
- 42.Arrondo J.L., Muga A., Castresana J., Goñi F.M. Quantitative studies of the structure of proteins in solution by Fourier-transform infrared spectroscopy. Prog. Biophys. Mol. Biol. 1993;59:23–56. doi: 10.1016/0079-6107(93)90006-6. [DOI] [PubMed] [Google Scholar]
- 43.Kong J., Yu S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sinica. 2007;39:549–559. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 44.Barth A. The infrared absorption of amino acid side chains. Prog. Biophys. Mol. Biol. 2000;74:141–173. doi: 10.1016/s0079-6107(00)00021-3. [DOI] [PubMed] [Google Scholar]
- 45.Kumar S., Rai A.K., Singh V.B., Rai S.B. Vibrational spectrum of glycine molecule. Spectroch. Acta A. 2005;61:2741–2746. doi: 10.1016/j.saa.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 46.Mohamed M.E., Mohammed A. Experimental and computational vibration study of amino acids. Int. Lett. Chem., Phys. Astron. 2013;10:1–17. doi: 10.18052/www.scipress.com/ILCPA.15.1. [DOI] [Google Scholar]
- 47.Adt I., Toubas D., Pinon J.M., Manfait M., Sockalingum G.D. FTIR spectroscopy as a potential tool to analyse structural modifications during morphogenesis of Candida albicans. Arch. Microbiol. 2006;185 doi: 10.1007/s00203-006-0094-8. [DOI] [PubMed] [Google Scholar]
- 48.Leker K.Y., Lozano-Pope I., Bandyopadhyay K., Choudhury B.P., Obonyo M. Comparison of lipopolysaccharides composition of two different strains of Helicobacter pylori. BMC Microbiol. 2017;17:226. doi: 10.1186/s12866-017-1135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker S.F. Assignment of the vibrational spectrum of L-cysteine. Chem. Phys. 2013;424:75–79. doi: 10.1016/j.chemphys.2013.04.020. [DOI] [Google Scholar]
- 50.Wohlmeister D., Barreto D.R., Helfer V.E., Calil L.N., Buffon A., Fuentefria A.M., et al. Differentiation of Candida albicans, Candida glabrata, and Candida krusei by FT-IR and chemometrics by CHROMagar™ Candida. J. Microbiol. Met. 2017;141:121–125. doi: 10.1016/j.mimet.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 51.Nyquist R.A. Chapter 4 – Thiols, sulfides and disulfides, alkanethiols, and alkanedithiols (S-H stretching) Int. Infrared Raman Nucl. Mag. Resonance Spectra Interpret. Infrared Raman Nucl. Mag. Resonance Spectra. 2001:65–83. doi: 10.1016/B978-012523475-7/50184-4. [DOI] [Google Scholar]
- 52.Nyquist R.A. Chapter 8 – Phosphorus compounds. Int. Infrared Raman Nucl. Mag. Resonance Spectra Interpret. Infrared Raman Nucl. Mag. Resonance Spectra. 2001:231–350. doi: 10.1016/B978-012523475-7/50188-1. [DOI] [Google Scholar]
- 53.Rungrodnimitchai S. Rapid preparation of biosorbents with high ion exchange capacity from rice straw and bagasse for removal of heavy metals. Scient. World J. 2014;634837:1–9. doi: 10.1155/2014/634837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bode G., Mauch F., Ditschuneit H., Malfertheiner P. Identification of structures containing polyphosphate in Helicobacter pylori. J. General Microbiol. 1993;139:3029–3033. doi: 10.1099/00221287-139-12-3029. [DOI] [PubMed] [Google Scholar]
- 55.McGrath J.W., Quinn J.P. Intracellular accumulation of polyphosphate by the yeast Candida humicola G-1 in response to Acid pH. Appl. Environ. Microbiol. 2000;66:4068–4073. doi: 10.1128/AEM.66.9.4068-4073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tran A., Whittimore J.D., Wyrick P.B., McGrath S.C., Cotter R.J., Trent M.S. The lipid A 1-phosphatase of Helicobacter pylori is required for resistance to the antimicrobial peptide polymyxin. J. Bacteriorl. 2006;188:4531–4541. doi: 10.1128/JB.00146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gannedi V., Ali A., Singh P.P., Vishwakarma R. Total synthesis of phospholipomannan of the Candida albicans. The J. Org. Chem. 2020 doi: 10.1021/acs.joc.0c00402. [DOI] [PubMed] [Google Scholar]
- 58.Moreno-Ruiz E., Ortu G., De Groot P.W., et al. The GPI-modified proteins Pga59 and Pga62 of Candida albicans are required for cell wall integrity. Microbiol. 2009;155:2004–2020. doi: 10.1099/mic.0.028902-0. [DOI] [PubMed] [Google Scholar]
- 59.Klis F.M., Sosinska G.J., De Groot P.W.J., Brul S. Covalentlylinked cellwall proteins of Candida albicans and their role in virulence. FEMS Yeast Res. 2009:1013–1028. doi: 10.1111/j.1567-1364.2009.00541.x. [DOI] [PubMed] [Google Scholar]
- 60.Ruiz-Herrera J., Mormeneo S., Vanaclocha P., Font-de-Mora J., Iranzo M., Puertes I., et al. Structural organization of the components of the cell wall from Candida albicans. Microbiol. 1994;140:1513–1523. doi: 10.1099/13500872-140-7-1513. [DOI] [PubMed] [Google Scholar]
- 61.Hawser S., Islam K. Binding of Candida albicans to immobilized amino acids and bovine serum albumin. Infect. Immun. 1998;66:140–144. doi: 10.1128/IAI.66.1.140-144.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oleastro M., Menard A. The role of helicobacter pylori outer membrane proteins in adherence and pathogenesis. Biology. 2013;2:1110–1134. doi: 10.3390/biology2031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kao C.Y., Sheu B.S., Wu J.J. Helicobacter pylori infection: an overview of bacterial virulence factors and pathogenesis. Biomed. J. 2016;39:14–23. doi: 10.1016/j.bj.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nelson P.N. Chain length and thermal sensitivity of the infrared spectra of a homologous series of anhydrous silver(I) n-alkanoates. Int. J. Spectr. 2016;3068430:1–9. doi: 10.1155/2016/3068430. [DOI] [Google Scholar]
- 65.Rommel V., Da Silva A., Pimentel A. Infrared spectroscopy of anionic, cationic, and zwitterionic surfactants. Adv. Phys. Chem. 2012;903272:1–14. doi: 10.1155/2012/903272. [DOI] [Google Scholar]
- 66.Pierce C.G., Vila T., Romo J.A., Montelongo-Jauregui D., Wall G., Ramasubramanian A., et al. The Candida albicans biofilm matrix: composition, structure and function. J. Fungi. (Basel) 2017;3:14. doi: 10.3390/jof3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H., Liao T., Debowski A., Tang H., Nilsson H.O., Stubbs K., Marshall B.J., Benhezal M. Lipopolysaccharide structure and biosynthesis in helicobacter pylori. Helicobacter. 2016:1–17. doi: 10.1111/hel.12301. [DOI] [PubMed] [Google Scholar]
- 68.El-Baky R.M., Dalia M.M., Gamal F. N-acetylcysteine inhibits and eradicates Candida albicans biofilms. Am. J. Infect. Dis. Microbiol. 2014;2:122–130. doi: 10.12691/AJIDM-2-5-5. [DOI] [Google Scholar]
- 69.Santos T.S., Matheus L., Vega-Chacon Y., Garcia de Oliveira W. Fungistatic action of N-acetylcysteine on candida albicans biofilms and its interaction with antifungal agents. Microorganisms. 2020;8 doi: 10.3390/microorganisms8070980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raa J. Immune modulation by non-digestible and non-absorbable beta-1,3/1,6-glucan. Microbial Ecol. Health Dis. 2015;26:27824. doi: 10.3402/mehd.v26.27824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Law K.Y. Definitions for hydrophilicity, hydrophobicity, and superhydrophobicity: getting the basics right. J. Phys. Chem. Lett. 2014;5:686–688. doi: 10.1021/jz402762h. [DOI] [PubMed] [Google Scholar]
- 72.Tannaes T., Bukholm I.K., Bukholm G. High relative content of lysophospholipids of Helicobacter pylori mediates increased risk for ulcer disease. FEMS Immunol. Med. Microbiol. 2006;44:17–23. doi: 10.1016/j.femsim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 73.Huang Z., Zhang X.S., Blaser M., London E. Helicobacter pylori lipids can form ordered membrane domains (rafts) Biochim. Biophys. Acta Biomembr. 2019;1861:183050. doi: 10.1016/j.bbamem.2019.183050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hobden C., Teevan C., Jones L., O’shea P. Hydrophobic properties of the cell surface of Candida albicans: a role in aggregation. Microbiol. 2005;141:1875–1881. doi: 10.1099/13500872-141-8-1875. [DOI] [PubMed] [Google Scholar]
- 75.Stark R.M., Gerwig G.J., Pitman R.S., Potts L.F., Williams N.A., Greenman J., et al. Biofilm formation by Helicobater pylori. Lett. Appl. Microbiol. 1999;28:121–126. doi: 10.1046/j.1365-2672.1999.00481.x. [DOI] [PubMed] [Google Scholar]
- 76.Sengwa R.J. A comparative dielectric study of ethylene glycol and propylene glycol at different temperatures. J. Mol. Liq. 2003;108:47–60. doi: 10.1016/S0167-7322(03)00173-9. [DOI] [Google Scholar]
- 77.Jazi B., Amiri H., Mohsen-Nia M. Dielectric constants of water, methanol, ethanol, butanol and acetone: measurement and computational study. J. Sol. Chem. 2010;39:701–708. doi: 10.1007/s10953-010-9538-5. [DOI] [Google Scholar]
- 78.ADT. Base data: Accu Dyne Test, 2020. Available from: http://www.accudynetest.com/)
- 79.Moreira J., Simoes M., Melo L.F., Mergulhao F. Escherichia coli adhesion to surfaces–a thermodynamic assessment. Colloid Polym. Sci. 2015;293:177–185. doi: 10.1007/s00396-014-3390-x. [DOI] [Google Scholar]
- 80.Asri M., Elabed A., Ghachtouli N.E., Koraichi S.I., Bahafid W., Elabed S. Theoretical and experimental adhesion of yeast strainswith high chromium removal potential. Environ. Eng. Sci. 2017;34:693–702. doi: 10.1089/ees.2016.0515. [DOI] [Google Scholar]