Abstract
Background: Recently, accumulating evidence confirmed that up-frameshift protein 1 (UPF1) was aberrantly expressed in various cancers. However, the molecular mechanism mediated by UPF1 underlying colorectal carcinogenesis remains unclear. Method: Immunohistochemistry (IHC) and quantitative real-time polymerase chain reaction analysis were used to determine the expression level of UPF1 in colorectal cancer (CRC) tissues. CCK-8, EdU, transwell assay, and flow cytometry were performed to investigate the biological significance of UPF1. Epithelial–mesenchymal transition (EMT) and apoptosis associated markers were detected by western blotting. Results: We found that UPF1 expression was upregulated in CRC tissues and cell lines. Clinical analysis revealed that high UPF1 expression was positively correlated with advanced stage, lymph node metastasis and shorter survival. Knockdown of UPF1 suppressed cell proliferation and cell cycle progression. Functionally, UPF1 promotes tumor metastasis by inducing epithelial to mesenchymal transition. Further investigations revealed that knockdown of UPF1 promoted apoptosis through triggering DNA damage. Conclusions: Taken together, this research revealed that UPF1 plays an oncogenic role in CRC via regulating EMT and apoptosis and may be a potential therapeutic target for CRC.
Keywords: colorectal cancer, up-frameshift protein 1, epithelial–mesenchymal transition, DNA damage, apoptosis
Introduction
Colorectal cancer (CRC) is the third frequent malignancy and is the second leading cause of cancer-related deaths worldwide. 1 Incident rates of CRC rose by 2.2% annually overall. 2 Although screening and treatments of CRC get advanced recently, majority of patients hold a poor prognosis. 2 Therefore, better understanding of potential prognosis biomarkers and therapeutic targets is needed to improve survival of CRC patients.
Up-frameshift protein 1 (UPF1), located at chromosome 19p13.2-p13.11, is a highly conserved and ubiquitously expressed phosphoprotein with RNA/DNA-dependent ATPase and RNA helicase activity. 3 Depending on its ATPase and helicase activities,4,5 UPF1 serves as the core regulator of nonsense-mediated mRNA decay machinery, which is a RNA surveillance pathway protecting cells from damage by premature termination codons. 6 Also, UPF1 is involved in cell proliferation and differentiation.7,8 In addition, UPF1 is required for S phase progression and genome stability, which is essential for DNA damage repair. 9 Previous studies have clarified the exact activities of UPF1 in hepatocellular cancer 10 and gastric cancer, 10 however, UPF1 exact biological activities in human CRC are poorly understood.
DNA damage is the well-known trigger of the intrinsic apoptosis pathway. Signaling to intrinsic apoptosis due to DNA damage stimuli the release of cytochrome C and activate BCL-2 family, promoting the formation of apoptosome.11,12 Then, procaspase 9 is activated, which in turn activates the executioner apoptosis cleaved caspase 3, leading to the death of cells. 13 Double strand breaks (DSB) are wildly recognized to be among the most deadly forms of DNA damage, causing genomic unstability. 14 H2AX is considered to be the hallmark of DNA damage and DSB, its phosphorylation is tightly linked to DNA damage repairs. 15 According to reports, the expression of H2AX is increased in UPF1-depleted cells. 9 These observations arise our interest to explore whether and how UPF1 and H2AX are involved in tumorigenesis of CRC and the underlying mechanism driving the relationships with apoptosis.
In this study, we demonstrated that UPF1 was significantly upregulated in CRC tissues compared with adjacent normal tissues. Subsequently, we investigated the contribution of UPF1 to the proliferation and metastasis of CRC as well as its effect on DNA damage and apoptosis, which provides a new sight for clinical diagnosis and treatment of CRC.
Materials and Methods
Colorectal Clinical Specimens and Cell Lines
Forty-eight pairs of CRC specimens and paired adjacent noncancer tissues were collected from patients of The First Affiliated Hospital of Shantou University Medical College. Tumor staging relies on eighth American Joint Committee on Cancer Tumor-node-metastasis classification (AJCC TNM-8). All the patients who participated in this study had signed the written informed consent voluntarily. The present study was approved by the Medical Ethics Committee of The First Affiliated Hospital of Shantou University Medical College.
The FHC human normal colorectal epithelial cell line and 4 human CRC cell lines (SW480, SW620, RKO, Caco-2) were cultured with DMEM supplemented with 10% fetal bovine serum (Gibco, Australia origin) in a water-saturated environment at 37 °C with 5% CO2. All the cell lines (FHC, SW480, SW620, RKO, Caco-2) were donated by General surgery laboratory of Southern Medical University.
Immunohistochemistry
Immunohistochemical staining of 48 pairs of CRC tissues and paired normal tissues was performed on 4 μm thick paraffin sections. All the procedures were referred to manufacturer's protocol using an UltraSensitiveTM S-P kit and DAB (Maixinbio, China). Antibody UPF1(dilution of 1:10000, ab109363, abcam) was applied. Cytoplasm stained with brownish yellow is considered positive. The staining paraffin sections were imaged with microscope (200× magnification). Image J software was used to evaluate the immunohistochemistry (IHC) staining. The percent positively stained areas were scored as “1” (0-25%), “2” (26-50%), and “3” (≥50%). The staining intensity was scored as “0” (no staining), “1” (weak), “2” (moderate), and “3” (strong). Then we calculated the sum of the 2 scores. The total score ≤3 was defined as negative expression, while the total score ≥4 was defined as positive expression. All specimens were evaluated by 2 pathologists who were blinded to the patient identity and clinical outcome.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted from fresh tissue using EasyPure RNA Kit (TransGen) and was reverse-transcribed into cDNA using a PrimeScript RT Master Mix (TaKaRa). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using SYBR Premix Ex Taq II Kit (TaKaRa) on CFX Connect. β-actin was used as an internal control. The relative expression was evaluated using 2−ΔΔct method. The primers used are as follows: β-actin (5’-TGGCACCCAGCACAATGAA-3’; 5’-CTAAGTCATAGTCCGCCTAGAAGCA-3’); UPF1 (5’-AGAGCCTCATGCAGTTCAGCAA-3’; 5’-GGCATCATACATGGCTGTGGTC-3’).
Plasmid Constructions and Transfection
UPF1-shRNAs were designed and synthesized by GeneCopoeia. The sequence of shRNAs used are as follows UPF1-shRNA #1: 5’-GCAGCCACATTGTAAATCACC-3’; UPF1-shRNA #2: 5’-GCGTGGTTTACTGTAATACCA-3’; UPF1-shRNA #3: 5’-CCTATTACACGAAGGACCTCC-3’. CRC cell lines SW620 and RKO were transfected using Lipofectamine 3000 Transfection Reagent (Thermo Fisher). At 48 h after transfection, qRT-PCR and western blotting were used to confirm the expression of UPF1.
Cell Proliferation Assay
CCK-8 assay was used to examine cell proliferation. SW620 and RKO cell lines with UPF1 knockdown were seeded on the 96-well plates at a concentration of 3 × 103 of RKO while 5 × 103 of SW620 per well with 100 μl DMEM containing 10% CCK-8 solution (CCK-8, Dojindo). Then, each well was cultured for 24, 48, 72, and 96 h. Cells with CCK-8 reagent were incubated for 4 h at 37 °C. Finally, the absorbance value (OD) of 450 nm was read on the microplate reader (Bio-Rad).
EdU staining was also used to assess cell proliferation. CRC cells seeded in each well of 24-well plates at a density of 4 × 104 cells/well of RKO while 6 × 104 cells/well of SW620 were stained using Meilun EdU cell proliferation Kit with Alexa Fluor 488 (Meilun, Dalian) referring to manufacturer's protocol. Lastly, the cells were imaged with inverted microscope (100× magnification).
Transwell Assay
The migration and invasion of cells were assessed using transwell chamber (0.8 mm) with Matrigel-coated or not. Transfected RKO cells (4 × 104) and SW620 cells (6 × 104) were separately seeded in upper chamber with 100 μl serum-free medium. The lower chamber was filled with 500 μl medium containing 20% FBS. After 48 h, cells trapped in the chamber membrane surface were fixed with 4% paraformaldehyde for 15 min, stained using 0.1% crystal violet for 15 min and imaged with microscope (400× magnification). Migrated and invaded cells of each sample were counted in 5 random fields for statistical analysis.
Flow Cytometric Analyses
Transfected RKO cells and SW620 cells were collected 48 h after transfection and then stained using cell cycle detection kit (MultiSciences) following the manufacturer's instructions. The percentage of cells in the cell cycle stages was analyzed by a FACSCanto II flow cytometer (BD Biosciences). For the cell apoptosis assays, transfected cells were analyzed using Annexin V-FITC/PI apoptosis detection kit (4A Bio) according to the manufacturer's instructions, cell apoptosis rate was then evaluated by FACSCanto II flow cytometer.
Western Blotting
Total proteins in cells were extracted by radio-immunoprecipitation assay buffer (RIPA) and the proteins concentration was measured using the bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific). The targeted proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (5% stacking and 12% separating gels) and transferred onto PVDF membranes which were blocked with 5% bovine serum albumin (BSA) solution for 1 h. Then the polyvinylidene fluoride (PVDF) membranes with targeted proteins were incubated with primary antibody including UPF1 (dilution of 1:10000, ab109363, abcam), β-actin (dilution of 1:1000, 4ab000001, 4A Biotech), E-cadherin (dilution of 1:1000, #3195, cell signaling), Vimentin (dilution of 1:1000, #5741, cell signaling), H2AX (dilution of 1:1000, #7631, cell signaling), Bax(dilution of 1:1000, #5023, cell signaling), Bcl-2 (dilution of 1:1000, Ab32124, abcam), Caspase3 (dilution of 1:1000, #14220, cell signaling), Cleaved Caspase3 (dilution of 1:1000, #9661, cell signaling), and paired secondary antibody goat anti-rabbit/mouse IgG H&L (HRP) (dilution of 1:2000, SA00001-1/ SA00001-2, Proteintech). Finally, the proteins bands were detected using ECL chemiluminescence system.
Statistical Analysis
All experiments were repeated at least 3 times and the data were expressed as the means ± standard deviation. The data were evaluated using Student's t-test and X2-test. And P < .05 was considered to be significant. Statistical graphs were performed using GraphPad Prism software.
Results
UPF1 Expression is Significantly Upregulated in CRC and Correlates with Patients’ Prognosis
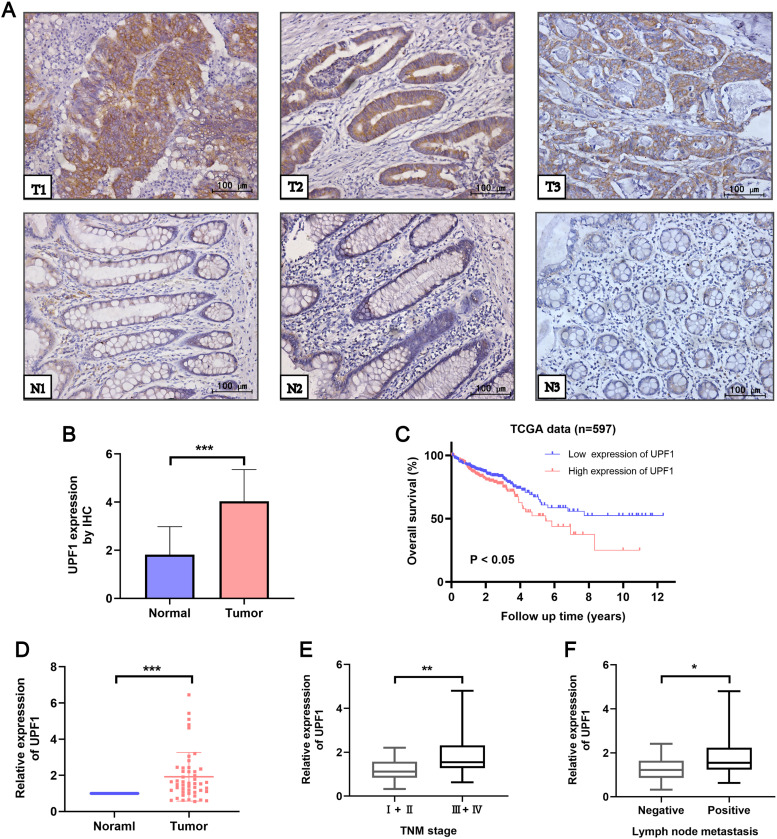
To evaluate the expression and clinical significances of UPF1 in CRC, we first measured its expression level in 48 CRC patients using IHC staining. The results showed that UPF1 was significantly upregulated in CRC tissues compared to adjacent normal tissues (Figure 1A and B). qRT-PCR was further performed to examine the expression of UPF1 in 48 CRC patients. As shown in Figure 1D, UPF1 expression was higher in CRC tissues than in adjacent normal mucosa. Next, we separated these patients into UPF1 high (n = 23) and low (n = 25) group according to median value. Clinical data showed that aberrant expression of UPF1 was positively associated with the positive lymph node metastasis and TMN stage (Figure 1E and F, Table 1). Then, we downloaded the data containing 597 patients from the TCGA-COAD dataset. Kaplan-Meier and log-rank test analyses showed that patients with higher expression of UPF1 had significantly shorter overall survival (OS) (Figure 1C).
Figure 1.
UPF1 expression is upregulated in CRC tissues compared to adjacent normal tissues. (A,B) The protein expression level of UPF1 was detected in 48 pairs of CRC and adjacent normal tissues via IHC staining (200×). (C) Kaplan-Meier curves showed that CRC patients with higher expression of UPF1 had worse overall survival (P < .05, log-rank test). (D) The mRNA expression level of UPF1 was measured in 48 pairs of CRC and adjacent normal tissues via RT-qPCR. (E) Correlation between UPF1 mRNA expression level in CRC patients and clinical TNM stage. (F) Correlation between UPF1 mRNA expression level in CRC patients and lymph node metastasis. *P < .05, **P < .01, ***P < .001.
Table 1.
Relation Between Clinicopathological Features and UPF1 Expression in CRC Patients (n = 48).
| Parameters | Cases | UPF1 expression | P value | |
|---|---|---|---|---|
| Low (n = 25) | High (n = 23) | |||
| Gender | ||||
| Female | 16 | 6 | 10 | .153 |
| Male | 32 | 19 | 13 | |
| Age (years) | ||||
| <60 | 17 | 7 | 10 | .263 |
| ≥60 | 31 | 18 | 13 | |
| Stage | ||||
| I + II | 20 | 15 | 5 | .007* |
| III + IV | 28 | 10 | 18 | |
| Tumor size | ||||
| <5 cm | 25 | 14 | 11 | .571 |
| ≥5 cm | 23 | 11 | 12 | |
| Lymphatic status | ||||
| positive | 26 | 9 | 17 | .008* |
| negative | 22 | 16 | 6 | |
*P<.05
Knockdown of UPF1 Inhibits CRC Cell Proliferation and Cell Cycle Progression
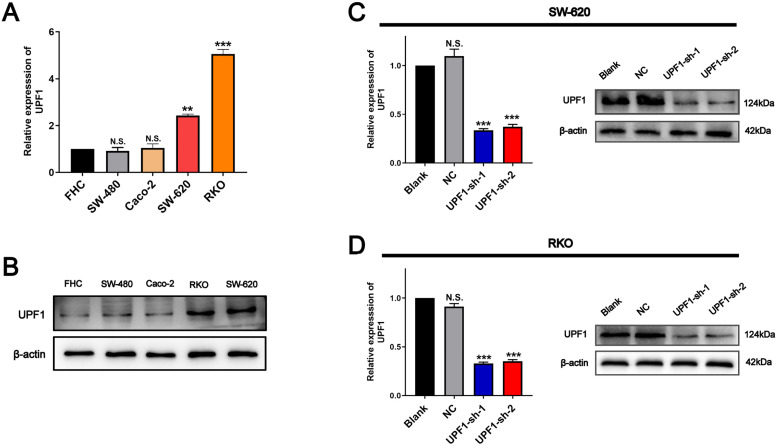
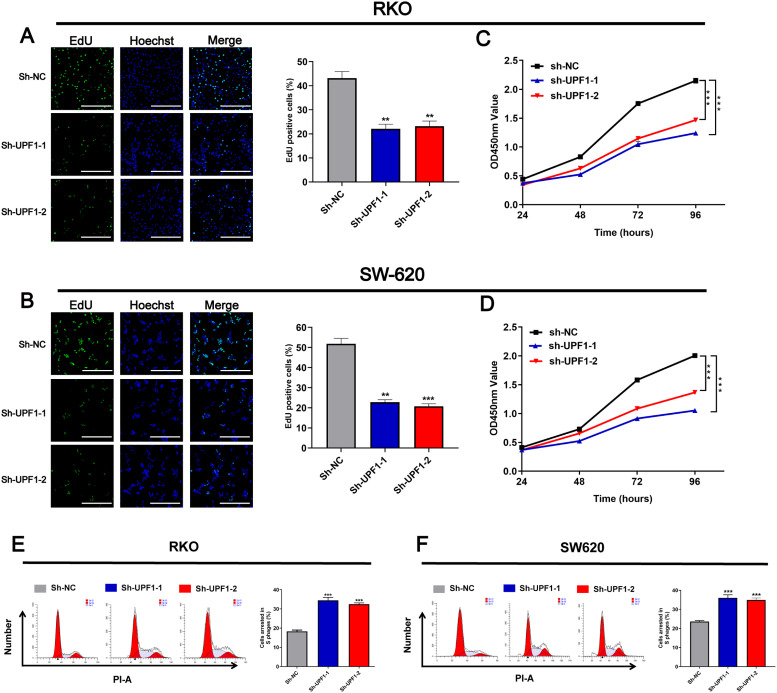
Similarly, qRT-PCR showed that UPF1 expression was higher in a group of CRC cell lines compared with FHC cell line, which is an immortalized normal human colorectal cell line (Figure 2A). Western blotting further proved the higher expression of UPF1 in CRC cell lines (Figure 2B), especially in SW620 and RKO. Therefore, these 2 cell lines were employed to confirm the oncogene role of UPF1 in vitro. To further investigate the biological function of UPF1, we knocked down UPF1 in SW620 and RKO cells by transfecting specific shRNA against UPF1. The results showed that the mRNA and protein levels were significantly reduced in 2 CRC cells transfected with sh-UPF1-1 and sh-UPF1-2 (Figure 2C and D). CCK8 and EdU proliferation assays revealed that silencing UPF1 inhibited cell proliferation relative to the negative control (NC) groups (Figure 3A-D). In addition, the cell cycle assays showed that silencing of UPF1 increased the proportion of cells in S phase (Figure 3E and F).
Figure 2.
UPF1 is upregulated in CRC cell lines. (A, B) UPF1 expression level was upregulated in CRC cell lines, especially in RKO and SW620. (C, D) Knockdown efficiency of UPF1 was confirmed via RT-qPCR. N.S., not significant. **P < .01, ***P < .001.
Figure 3.
Knockdown UPF1 inhabited cell proliferation and cell cycle of CRC cell lines. (A, B) EdU staining showed that UPF1 depletion inhabited cell growth in RKO and SW620 cells (100×). Scale bar = 200 μm. (C, D) CCK8 assay showed that UPF1 depletion inhabited cell growth in RKO and SW620 cells. (E, F) Flow cytometry analysis showing the proportion of CRC cells in the G1, S, and G2/M phases when UPF1 was silenced in RKO and SW620 cells. **P < .01, ***P < .001.
Knockdown of UPF1 Inhibits Cell Migration, Invasion, and EMT in CRC Cells
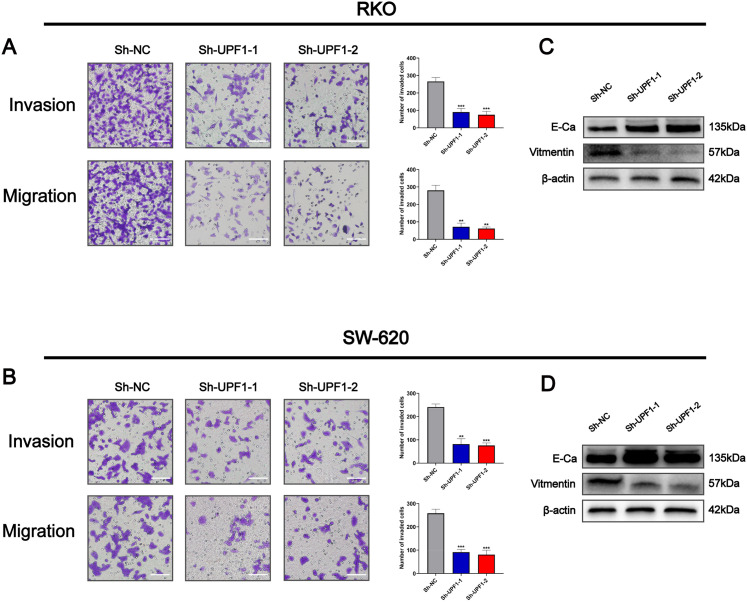
To further examine the effects of UPF1 on tumor metastasis, we evaluated the biological activity of UPF1 in regulating migration and invasion through transwell assays. As shown in Figure 4, silencing of UPF1 repressed the migration and invasion abilities of SW620 and RKO cells (Figure 4A and B). As EMT is well known for tumor metastasis, the protein level of EMT markers was evaluated by western blotting. Silencing of UPF1 increased the expression of epithelial marker E-cadherin, while repressed the expression of mesenchymal marker vimentin (Figure 4C and D). Taken together, our results revealed that knockdown of UPF1 inhibits cell migration, invasion, and EMT in CRC.
Figure 4.
Knockdown UPF1 inhabited cell migration, invasion, and EMT in CRC cell lines. (A, B) The ability of cell migration and invasion was inhibited when UPF1 was silenced in RKO and SW620 cells (400×). Scale bar = 200 μm. (C, D) UPF1 depletion upregulated E-cadherin expression while downregulated the expression of N-cadherin and vimentin. **P < .01, ***P < .001.
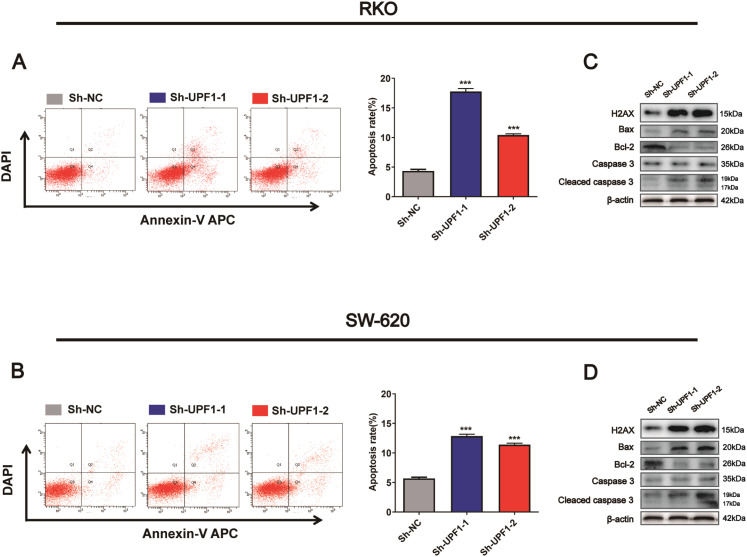
Knockdown of UPF1 Promotes Apoptosis by Inducing DNA Damage in CRC
According to the study by Azzalin and his colleagues, UPF1 was essential to DNA stability, and DNA damage might be triggered by silencing of UPF1. 16 Previous studies had demonstrated that UPF1 was involved in apoptosis regulation,10,11 however, whether UPF1 could regulate apoptosis through triggering DNA damage promotes us to explore the underlying mechanism in CRC cells. As shown, the expression of DNA damage marker H2AX was significantly upregulated when UPF1 was silenced in SW620 and RKO cells (Figure 5C and D). Then, apoptosis flow cytometry analysis (FACS) showed that UPF1-knockdown induced apoptosis in the 2 cells (Figure 5A and B). We also evaluated the expression of apoptosis markers in cells transfected with UPF1-shRNA. The results showed that knockdown of UPF1 increased the pro-apoptosis marker BAX protein expression while decreased the anti-apoptosis marker Bcl-2 protein expression. In addition, cleaved caspase 3 were upregulated in UPF1-knockdown CRC cells compared to control cells, while the protein levels of procaspase 3 were not significantly altered (Figure 5C and D).
Figure 5.
Knockdown UPF1 promoted cell apoptosis by triggering DNA damage. (A, B) Knockdown UPF1 induced apoptosis in RKO and SW320. (C, D) Western blotting analysis of H2AX protein expression and apoptosis marker proteins following transfection of UPF1-shRNA. ***P < .001.
In addition, DNA mismatch repair (MMR) is also implicated in apoptosis after DNA damage. 17 In order to explore the relationship between UPF1 and MMR proteins, we performed analysis using GEPIA database. As shown in Supplemental Figure S1, expression of UPF1 was positively correlated with MMR proteins including MLH1, MSH2, MSH6, and PMS2 (Supplemental Fig. S1 A-D). Therefore, we concluded that knockdown of UPF1 caused damage to DNA, and consequently promoted apoptosis in CRC cells.
Discussion
Our study showed that UPF1 was upregulated in CRC tissues and cell line. Moreover, high expression promoted cell growth and metastasis of malignant CRC cells, which agreed with the clinicopathological parameter. The expression of UPF1 was associated closely with TMN stages and lymphatic penetration in CRC. Therefore, our data demonstrated that UPF1 has a potential to act as a prognostic biomarker for CRC. However, more samples should be included to determine the clinical value of UPF1 in CRC.
EMT is recognized to be a crucial mechanism that plays an important role in cancer invasion and metastasis. 16 In this process, cells which lose epithelial characteristics gain migratory and invasive properties. 18 Our study elucidated that knockdown of UPF1 increased expression E-cadherin with reduced expression of Vimentin. These data suggested that UPF1 could activate process of EMT in CRC, leading to the enhanced metastasis. Tumor budding is a highly aggressive subpopulation of tumor cells with migratory and invasive capacities. 19 According to researches of Gurzu et al., EMT was associated with the tumor-budding degree in CRC. Budding degree could be used to evaluate EMT in CRC.20,21 However, whether UPF1 was correlated with tumor budding and the mechanism behind EMT are unclear, which should be further investigated in the following research.
Previous studies have demonstrated that UPF1 was essential in various biological processes including embryonic and brain development. 22 In Drosophila melanogaster, UPF1 loss-of-function suppressed cell growth and triggered apoptosis. 23 However, dysfunction of UPF1 also contributed in tumorigenesis through regulating apoptosis. Actually, how UPF1 inducing apoptosis still remains unclear. As Azzalin and his colleagues mentioned, UPF1 was necessary to genome stability and UPF1-depletion might trigger DNA damage.9,24 In our study, we found that UPF1 silencing induced DNA damage response and increased the proportion of cells in S phase. Intriguingly, apoptotic rate in UPF1-depleted cell was significantly increased. It is known that intrinsic apoptosis pathway signal could arise from DNA damage. 12 Taken together, we concluded that UPF1-depletion regulated apoptosis through inducing DNA damage. However, more downstream functional experiments still determine to be clarified in the following research.
Actually, the function of UPF1 is required for genome stability and is dual in tumorigenesis. Chang et al. clarified that UPF1 could suppress HCC invasion, migration, and proliferation through activating the TGF-β/Smad pathway by eliminating LncRNA SNHG6. 25 Whereas, Shao and his colleagues demonstrated that UPF1 increased cell viability and promoted metastasis via enhancing the stability of Linc-00313 in glioma. 26 Our results showed that knockdown of UPF1 suppressed the malignant biological behaviors of CRC cells via triggering DNA damage. It seems that UPF1 phosphorylated by different kinases lead to different DNA or RNA metabolism processes. On the one hand, phosphorylated by SMG-1, UPF1 recognizes targeted mRNA and then works with UPF2/UPF3 to trigger the decay of aberrant RNA.27,28 On the other, modified by ATR phosphorylation, UPF1 is essential in DNA stability. 24 Herein, we elucidated regulatory roles of UPF1 in CRC. However, whether DNA damage caused by UPF1 knockdown was induced by ATR phosphorylation in CRC cells should be further investigated in the future.
Conclusion
Our study suggested that the upregulation of UPF1 in tumor tissues was significantly correlated with poor prognosis. Furthermore, UPF1 depletion was involved in CRC progression through activated apoptosis and EMT progression. All the results indicated that UPF1 played an important role in CRC carcinogenesis and may serve as a potential target for CRC diagnosis and therapy.
Supplemental Material
Supplemental material, sj-docx-1-tct-10.1177_15330338211064438 for Up-frameshift Protein 1 Promotes Tumor Progression by Regulating Apoptosis and Epithelial–Mesenchymal Transition of Colorectal Cancer by Binlie Chen, Huaiming Wang, Danfeng Li, Xiaosheng Lin, Zhiyan Ma and Yongming Zeng in Technology in Cancer Research & Treatment
Acknowledgments
We are grateful to all the patients who participated in this study.
Abbreviations
- ATR
ataxia telangiectasia mutated and RAD3-related
- BAX
bcl-2-associated X protein
- BCL-2
B-cell lymphoma-2
- CRC
colorectal cancer
- DSB
double strand breaks
- EMT
epithelial–mesenchymal transition
- FACS
flow cytometry analysis
- HCC
hepatocellular cancer
- LncRNA
long noncoding RNA
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- NMD
nonsense-mediated mRNA decay
- OS
overall survival
- PTC
premature termination codons
- SMG-1
suppressor with morphogenetic effect on genitalia-1
- SNHG6
small nucleolar RNAs host gene 6
- TGF-β
transforming growth factor beta
- UPF2/UPF3
up-frameshift protein 2/3
- ZFX
zinc finger protein X
- 53BP1
p53-binding protein 1.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received funding through the Shantou Science and Technology Bureau (No. 200624155260702).
Ethics approval and informed consent: The present study was authorized by the Research Ethics Review Board of The First Affiliated Hospital of Shantou University Medical College (No. 2020-102). All the patients who enrolled in this research had signed the written informed consent voluntarily.
ORCID iD: Yongming Zeng https://orcid.org/0000-0001-6704-1403
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145-164. [DOI] [PubMed] [Google Scholar]
- 3.Applequist SE, Selg M, Raman C, Jäck HM. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mrna-reducing UPF1 protein. Nucleic Acids Res. 1997;25(4):814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brogna S, mcleod T, Petric M. The meaning of NMD: translate or perish. Trends Genet. 2016;32(7):395-407. [DOI] [PubMed] [Google Scholar]
- 5.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36(8):801-808. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci. 2010;67(5):677-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Q, Jagannathan S, Bradley RK. The RNA surveillance factor UPF1 represses myogenesis via its E3 ubiquitin ligase activity. Mol Cell. 2017;67(2):239-251.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10(2):99-105. [DOI] [PubMed] [Google Scholar]
- 9.Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol. 2006;16(4):433-439. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Geng Y, Feng R, et al. The human RNA surveillance factor UPF1 modulates gastric cancer progression by targeting long non-coding RNA MALAT1. Cell Physiol Biochem. 2017;42(6):2194-2206. [DOI] [PubMed] [Google Scholar]
- 11.Zou H, Li Y, Liu X, Wang X. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274(17):11549-11556. [DOI] [PubMed] [Google Scholar]
- 12.Fennell DA. Caspase regulation in Non–small cell lung cancer and its potential for therapeutic exploitation. Clin Cancer Res. 2005;11(6):2097-2105. [DOI] [PubMed] [Google Scholar]
- 13.Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8(5):405-413. [DOI] [PubMed] [Google Scholar]
- 14.Karagiannis TC, El-Osta A. Double-strand breaks: signaling pathways and repair mechanisms. Cell Mol Life Sci. 2004;61(17):2137-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10(15):886-895. [DOI] [PubMed] [Google Scholar]
- 16.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871-890. [DOI] [PubMed] [Google Scholar]
- 17.Tennen RI, Haye JE, Wijayatilake HD, Arlow T, Ponzio D, Gammie AE. Cell-cycle and DNA damage regulation of the DNA mismatch repair protein Msh2 occurs at the transcriptional and post-transcriptional level. DNA Repair (Amst). 2013;12(2):97-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niwa Y, Yamada S, Koike M, et al. Epithelial to mesenchymal transition correlates with tumor budding and predicts prognosis in esophageal squamous cell carcinoma. J Surg Oncol. 2014;110(6):764-769. [DOI] [PubMed] [Google Scholar]
- 19.Zlobec I, Lugli A. Epithelial mesenchymal transition and tumor budding in aggressive colorectal cancer: tumor budding as oncotarget. Oncotarget. 2010;1(7):651-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banias L, Jung I, Bara T, et al. Immunohistochemical-based molecular subtyping of colorectal carcinoma using maspin and markers of epithelial-mesenchymal transition. Oncol Lett. 2020;19(2):1487-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banias L, Gurzu S, Kovacs Z, Bara T, Bara T, Jr., Jung I. Nuclear maspin expression: a biomarker for budding assessment in colorectal cancer specimens. Pathol Res Pract. 2017;213(9):1227-1230. [DOI] [PubMed] [Google Scholar]
- 22.Karousis ED, Nasif S, Muhlemann O. Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdiscip Rev RNA. 2016;7(5):661-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avery P, Vicente-Crespo M, Francis D, Nashchekina O, Alonso CR, Palacios IM. Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner. RNA. 2011;17(4):624-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azzalin CM, Lingner J. The double life of UPF1 in RNA and DNA stability pathways. Cell Cycle. 2006;5(14):1496-1498. [DOI] [PubMed] [Google Scholar]
- 25.Chang L, Yuan Y, Li C, et al. Upregulation of SNHG6 regulates ZEB1 expression by competitively binding mir-101-3p and interacting with UPF1 in hepatocellular carcinoma. Cancer Lett. 2016;383(2):183-194. [DOI] [PubMed] [Google Scholar]
- 26.Shao L, He Q, Liu Y, et al. UPF1 Regulates the malignant biological behaviors of glioblastoma cells via enhancing the stability of linc-00313. Cell Death Dis. 2019;10(9):629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denning G, Jamieson L, Maquat LE, Thompson EA, Fields AP. Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J Biol Chem. 2001;276(25):22709-22714. [DOI] [PubMed] [Google Scholar]
- 28.Kashima I, Yamashita A, Izumi N, et al. Binding of a novel SMG-1-Upf1-erf1-erf3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20(3):355-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tct-10.1177_15330338211064438 for Up-frameshift Protein 1 Promotes Tumor Progression by Regulating Apoptosis and Epithelial–Mesenchymal Transition of Colorectal Cancer by Binlie Chen, Huaiming Wang, Danfeng Li, Xiaosheng Lin, Zhiyan Ma and Yongming Zeng in Technology in Cancer Research & Treatment